SUMMARY
Inflammasomes elicit host defense inside cells by activating caspase-1 for cytokine maturation and cell death. AIM2 and NLRP3 are representative sensor proteins in two major families of inflammasomes. The adaptor protein ASC bridges the sensor proteins and caspase-1 to form ternary inflammasome complexes, achieved through pyrindomain (PYD) interactions between sensors and ASC, and caspase activation and recruitment domain (CARD) interactions between ASC and caspase-1. We found that PYD and CARD both form filaments. Activated AIM2 and NLRP3 nucleate PYD filaments of ASC, which in turn cluster the CARD of ASC. ASC thus nucleates CARD filaments of caspase-1 leading to proximity-induced activation. Endogenous NLRP3 inflammasome is also filamentous. The cryo-EM structure of ASCPYD filament at near-atomic resolution provides a template for homo- and hetero-PYD/PYD associations, as confirmed by structure-guided mutagenesis. We propose that ASC-dependent inflammasomes in both families share a unified assembly mechanism that involves two successive steps of nucleation-induced polymerization.
INTRODUCTION
The immune system provides protection from the environment and is critically important for multiple aspects of mammalian biology. It consists of an adaptive component that generates specific antibodies and cells through clonal selection, and an innate component that utilizes preformed receptors. Innate immunity offers the first line of defense against infections and hazards by directly recognizing conserved pathogen- and danger-associated molecular patterns (PAMPs and DAMPs) to alert the immune system (Medzhitov and Janeway, 2000). Inflammasomes are key components of innate immunity inside the cell. They are formed in response to PAMPs and DAMPs, and activate inflammatory caspases such as caspase-1 and -11 (Franchi et al., 2012; Lamkanfi and Dixit, 2012; Rathinam et al., 2012). Caspase activation can lead to proteolytic maturation of cytokines IL-1β and IL-18, and elicit the inflammatory form of cell death pyroptosis, as ways to control exogenous and endogenous invasions.
Inflammasomes are supramolecular assemblies composed of at least a sensor protein and a caspase, and often the adapter protein ASC. Based on the domain architecture of the sensor protein, inflammasomes maybe divided into two families. The first family is known as ALR [Absent in Melanoma 2 (AIM2)-like receptor], named after the first identified member (Figure 1A). ALRs are composed of an N-terminal PYD and one or two HIN domains (Rathinam et al., 2012). AIM2 directly senses the cytosolic PAMPs dsDNAs, such as those associated with viruses, using its HIN domain (Jin et al., 2012; Rathinam et al., 2012). The second class of inflammasomes contains receptors in the NLR [nucleotide-binding domain (NBD) and leucine rich repeat (LRR) – containing receptors] family (Figure 1A). NBD belongs to the AAA+ superfamily of ATPase domains. Most NLRs contain an N-terminal PYD and are known as NLRPs. The best-studied NLRP3 inflammasome is activated following a wide range of pathogen and danger signals including extra cellular ATP and uric acid crystals (Franchi et al., 2012; Lamkanfi and Dixit, 2012; Rathinam et al., 2012). Upon activation, both AIM2 and NLRP3 recruit the PYD- and CARD-containing bipartite adapter ASC (apoptosis-associated speck-like protein containing a CARD) through PYD/PYD interactions (Masumoto et al., 1999).ASC in turn recruits caspase-1 through CARD/CARD interactions (Figure 1A). PYD and CARD both belong to the death domain (DD) fold superfamily (Ferrao and Wu, 2012), for which structures of two defined DD helical assemblies are known (Lin et al., 2010; Park et al., 2007).
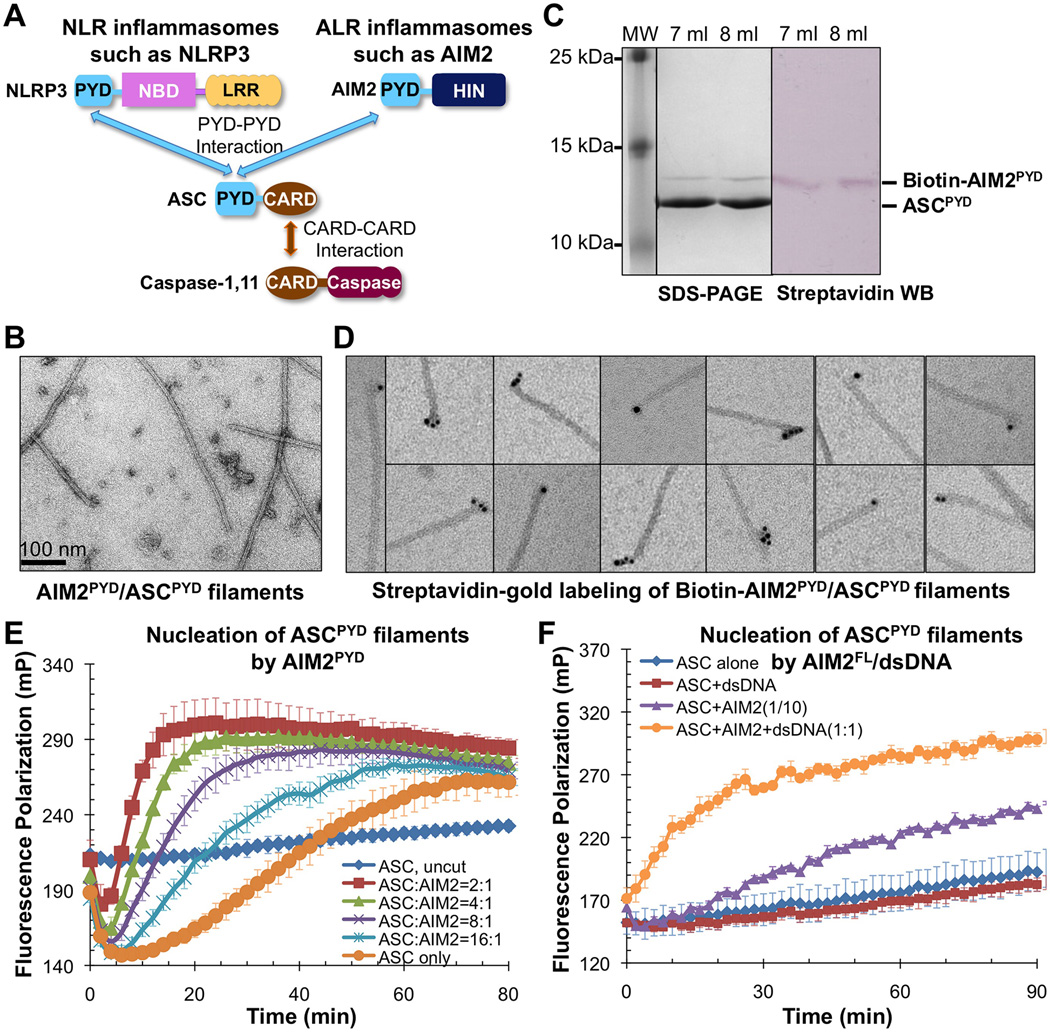
Figure 1. AIM2 Promotes Formation of ASCPYD Filaments.
A. Domain composition and interaction hierarchy of NLRP3 and AIM2 inflammasomes.
B. An electron micrograph of the AIM2PYD/ASCPYD binary complex.
C. Gel filtration fractions of biotinylated AIM2PYD/ASCPYD complex as visualized by Coomassie Blue-stained SDS-PAGE (left) and streptavidin-alkaline phosphatase Western blot (right). D. Labeling of biotinylated AIM2PYD/ASCPYD binary complex by streptavidin-gold conjugate (6nm).
E. Fluorescence polarization (FP) assay of AIM2PYD-nucleated ASCPYD filament formation. mP: unit for FP. Data are represented as mean±SD (N=3).
F. Effect of dsDNA on AIM2FL-nucleated ASCPYD filament formation. 2 µM of AIM2FL monomer from gel filtration was incubated with or without equimolar300-bp dsDNA (assuming a 10-bp footprint of AIM2 for molar calculation) for 30 minutes before diluting to a working concentration 0.1 µM (ASCPYD:AIM2FL=10:1) for the FP assay. Data are represented as mean±SD (N=3).
See also Figure S1.
Malfunctioning of inflammasomes is associated with serious human diseases (Strowig et al., 2012). Mutations in inflammasome proteins NLRP3, NLRP12 and MEFV (also known as Pyrin) are linked to auto inflammatory and fever syndromes (Rathinam et al., 2012). Aberrations in NLR inflammasome activation have been connected to psoriasis, type II diabetes, inflammatory bowel diseases and Alzheimer’s disease (Franchi et al., 2012; Lamkanfi and Dixit, 2012; Rathinam et al., 2012; Strowig et al., 2012).The PYD-less ALR member, mouse p202, interacts with the HIN domain of AIM2 to inhibit inflammasome and potentiatelupus (Yin et al., 2013).
Immunofluorescence microscopy showed that transfected full-length ASC (ASCFL) and endogenous ASC upon stimulation both form speck-like aggregates (Masumoto et al., 1999). Because transfected PYD and CARD-only ASC fragments are filamentous (Masumoto et al., 2001), the specks are most likely dense, cross linked composites of PYD and CARD filaments. Because of the strong tendency of ASC to aggregate, the structures of ASC PYD (ASCPYD) and ASCFL were solved in monomeric states using nuclear magnetic resonance (NMR) at acidic conditions (de Alba, 2009; Liepinsh et al., 2003). Although additional monomeric PYD structures have been reported, including those of NLRP3 (Bae and Park, 2011) and AIM2 (Jin et al., 2013), the mode of homo- and hetero-PYD associations is entirely unknown.
Here, we used in vitro reconstitution, electron microscopy (EM) and polymerization assays to address assembly mechanisms for ASC-dependent AIM2 and NLRP3 inflammasomes. In contrast to the presumption that the different domain structures of AIM2 and NLRP3may lead to considerable differences in the inflammasome architectures, we showed that both AIM2 upon dsDNA interaction and NLRP3 oligomerized through its NBD nucleate ASCPYD filaments. This is particularly surprising for NLRP3; due to the domain similarity of NLRs to Apaf-1-like molecules that form ring-like platforms (Yuan and Akey, 2013), the overarching paradigm had presumed that NLR inflammasomes are also ring-like structures. The flexibly linked ASC CARD (ASCCARD) then clusters along the ASCPYD filament to act as the platform for caspase-1CARD filament formation, leading to proximity-induced caspase dimerization and activation. The ternary inflammasome complex showed star-shaped branched filamentous morphology, and exhibited unequal stoichiometries among the component proteins. We determined the cryo-EM structure of the ASCPYD filament at near atomic resolution through helical reconstruction. The structure revealed molecular details of ASCPYD/ASCPYD interactions and allowed modeling of AIM2PYD/ASCPYD and NLRP3PYD/ASCPYD interactions. Structure-based mutagenesis confirmed the importance of ASCPYD/ASCPYD, AIM2PYD/ASCPYD and NLRP3PYD/ASCPYD interactions in vitro and in cells. EM of immuno precipitated endogenous NLRP3 inflammasome showed similar filamentous morphology as in vitro reconstituted inflammasomes and quantitative Western blotting confirmed the over-stoichiometry of caspase-1 to ASC. Our studies collectively revealed a universal assembly process for ASC-dependent inflammasomes in both ALR and NLR families that involves nucleation-induced polymerization.
RESULTS
The AIM2PYD/ASCPYD Complex is Filamentous with End Location of AIM2PYD
To elucidate the assembly mechanisms for ASC-containing inflammasomes, we first reconstituted the interaction between AIM2 and ASC. Co-expression of AIM2PYD with ASCPYD showed that the complex eluted at the void position of a Superdex 200 gel filtration column (Figure S1A), suggesting formation of large “aggregates”. We used EM to visualize the negatively stained AIM2PYD/ASCPYD complex, which revealed filaments with uniform diameters of ~9 nm (Figure 1B).
AIM2PYD exists in sub-stoichiometric molar ratio in the AIM2PYD/ASCPYD complex (Figure S1A, 1C). To understand this observation, we generated a construct of AIM2PYD capable of enzymatic biotinylation during expression. Co-expression of the AIM2PYD construct with ASCPYD generated a complex with specific biotinylation of AIM2PYD, shown by streptavidin Western blotting (Figure 1C). Labeling biotinylatedAIM2PYD by 6 nm streptavidin-gold particles showed that AIM2PYD is localized at one end of the filaments (Figure 1D). The number of bound gold particles varies between one and several, consistent with the ability of AIM2PYD to form filaments when expressed alone (Figure S1B).In the presence of ASCPYD, AIM2PYD preferentially associated with ASCPYD to generate short heterogeneous AIM2PYD filaments in complex with much longer ASCPYD filaments. In contrast, Ni-NTA-gold labeling of His-tagged ASCPYD in the biotinylated AIM2PYD/ASCPYD complex showed uniform distribution along the filaments (Figure S1C), confirming that ASCPYD forms the main filament body.
AIM2PYD and the Full-length AIM2/dsDNA Complex Nucleate ASCPYD Filaments
End labeling of AIM2PYD suggested its role as the nucleator for directional polymerization of ASCPYD.To quantitatively assess ASCPYD filament formation, we set up a fluorescence polarization (FP) assay in vitro using a His-MBP-ASCPYD fusion construct with an added C-terminal Cys for conjugating with Alexa488 fluorophore. The large fusion tag MBP inhibited ASCPYD polymerization to enable His-MBP-ASCPYD to be expressed in the monomeric form (Figure S1D). Polymerization of Alexa488-labeled monomeric ASCPYD was initiated by addition of the TEV protease to remove His-MBP from the fusion protein. The increase in FP, which indicates ASCPYD polymerization, was monitored as a function of time (Figure 1E). Although ASCPYD did polymerize on its own upon His-MBP removal, the rates of polymerization were dramatically enhanced in the presence of increasing amounts of sub-stoichiometric AIM2PYD (1/16 – 1/2 molar ratios)(Figure 1E). The initial drop in FP corresponded with His-MBP removal by TEV and the decrease in size of ASCPYD. At 5 min after TEV addition, about 75% His-MBP was removed from the fusion protein (Figure S1E). The AIM2PYD/ASCPYD filaments generated from the polymerization assay (Figure S1F) showed similar morphology to the co-expressed complex (Figure 1B).
Full-length AIM2 (AIM2FL) is a cytosolic dsDNA sensor in which the interaction of its C-terminal HIN domain with dsDNA induces ASC recruitment and inflammasome formation. The PYD in AIM2 has been shown to interact with its HIN domain to provide auto inhibition in the absence of dsDNA binding (Jin et al., 2013). To reconstitute AIM2 inflammasome activation in vitro, we expressed AIM2FL as a His-MBP fusion. Purified His-MBP-AIM2FL was first incubated with equimolar 300-bp dsDNA (molar ratio calculated based on 10-bp footprint of AIM2 on dsDNA), followed by mixing withAlexa488-labeled His-MBP-ASCPYD.TEV was added to remove His-MBP to initiate ASCPYD polymerization as monitored by FP (Figure 1F).A dramatic increase in FP was observed upon activation of AIM2FL by dsDNA, recapitulating the cellular event of inflammasome activation. These data suggest that overcome of auto inhibition and oligomerization of AIM2 by dsDNA sare crucial for inducing ASCPYD polymerization.
NLRP3PYD-NBD Nucleates and End-labels ASCPYD Filaments
NLRs share similar domain architecture sand are recognized to be auto inhibited in the absence of suitable ligands. In NLRC4, an NLR with a CARD, its LRR domain plays an important role in inhibiting NLR oligomerization (Hu et al., 2013). Due to their domain similarity to Apaf-1-like molecules that form ring-like platforms through the NBDs to induce caspase activation and apoptosis (Yuan and Akey, 2013),the overarching paradigm appears to presume that NLRP inflammasomes are also ring-like structures organized by the NBD. Formation of filamentous structures in the AIM2PYD/ASCPYD interaction prompted us to examine ASC-dependent NLRP inflammasomes using the prototypical member NLRP3.
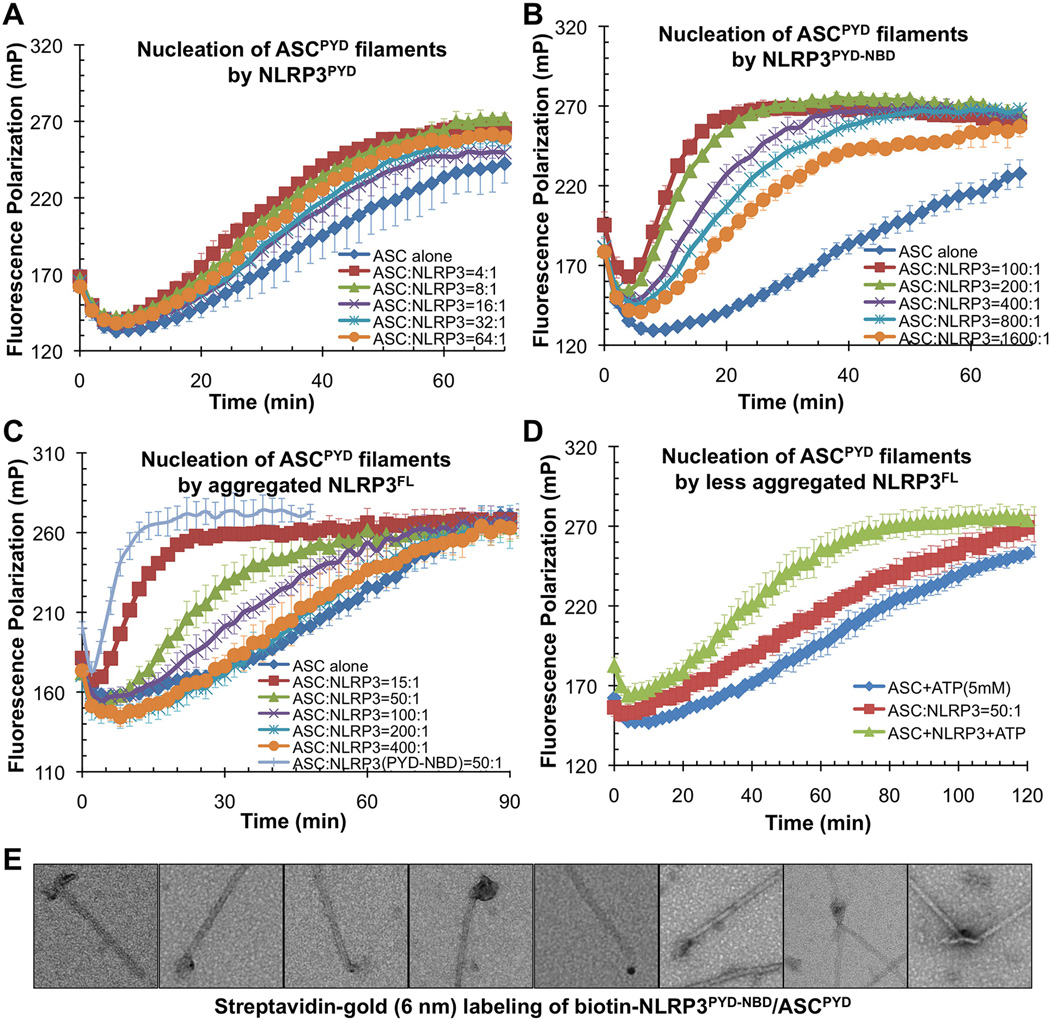
We expressed and purified NLRP3PYD, NLRP3PYD-NBD and NLRP3FL. While AIM2PYD exists as filamentous oligomers and was sufficient in promoting ASCPYD polymerization, NLRP3PYD is a monomer and did not cause significant enhancement in ASCPYD polymerization (Figure 2A). Both insect cell and E. coli expressed NLRP3PYD-NBD with inclusion of the NB Deluted from the void position of a Superdex 200 gel filtration column, and induced greatly increased ASCPYD polymerization (Figure 2B). In comparison, NLRP3PYD-NBD is a much stronger promoter of ASCPYD polymerization than AIM2PYD; it caused significant enhancement of ASCPYD polymerization at a low 1:1,600 molar ratio (Figure 2B).Notably, under the physiological intracellular condition of 140 mM KCl and 10 mM NaCl at pH 7.4 and a lower ASC concentration, ASCPYD did not significantly polymerize unless increasing amounts of NLRP3PYD-NBD were added (Figure S2A), suggesting that ASC does not polymerize under steady physiological state without stimulation. The PYD of NLRP3 is required for ASCPYD polymerization because NLRP3PYD-NBD proteins with mutations on PYD are compromised in this function (see below), suggesting that NBD-oligomerized NLRP3PYD forms the platform for ASCPYD polymerization.
Figure 2. NLPR3FL and NLRP3PYD-NBD but not NLRP3PYD Promote ASCPYD Filament Formation.
A, B, C.Nucleation of ASCPYD filaments by titrating increasing amounts of NLRP3PYD (A), NLRP3PYD-NBD (B), NLRP3FL (C) as monitored by fluorescence polarization. Data are represented as mean±SD (N=3).
D. A less aggregated gel filtration fraction of NLRP3 was subjected to ASCPYD polymerization assay with or without 5mM ATP. Data are represented as mean±SD (N=3).
E. Streptavidin-gold (6nm) labeling of biotinylated NLRP3PYD-NBD/ASCPYD binary complex.
See also Figure S2.
Insect cell expressed NLRP3FL showed a wide distribution on a Superdex 200 gel filtration column (Figure S2B). In keeping with auto inhibition in NLRP3FL as in NLRC4 (Hu et al., 2013),we found that even the highly aggregated NLRP3FL showed less activity than NLRP3PYD-NBD in promoting ASCPYD polymerization because more molar quantities of NLRP3FL were required to achieve similar degrees of FP enhancement (Figure 2B, 2C). Despite being activated by an extensive list of stimuli, it is uncertain what constitutes the direct activator of NLRP3 (Rathinam et al., 2012). We found that addition of ATP enhanced the less aggregated fractions of recombinant NLRP3FL in activating ASCPYD polymerization (Figure 2D); however this activation is minimal in comparison. It is likely that ATP binding by the NBD is associated with, but not sufficient for, NLRP3 activation.
Induction of ASCPYD polymerization by NLRP3 suggests that NLRP3 may reside at the end of ASCPYD filaments. We generated a His-MBP-NLRP3PYD-NBD construct capable of enzymatic biotinylation during expression. We mixed purified His-MBP-NLRP3PYD-NBD-Biotin with His-MBP-ASCPYD and added the TEV protease to cleave off His-MBP to allow ASCPYD polymerization. The purified NLRP3-Biotin/ASC complex was subjected to negative stain EM and 6 nm streptavidin-gold labeling, which confirmed localization of NLRP3 to the end of ASCPYD filaments (Figure 2E).Using negative stain EM, we showed that purified NLRP3PYD-NBD is heterogeneous with a mixture of disk-like structures and filaments (Figure S2C). The latter may represent the spiral, lock washer-like mode of oligomerization of the NBD.
Cryo-Electron Microscopy Structure of ASCPYD at Near Atomic Resolution
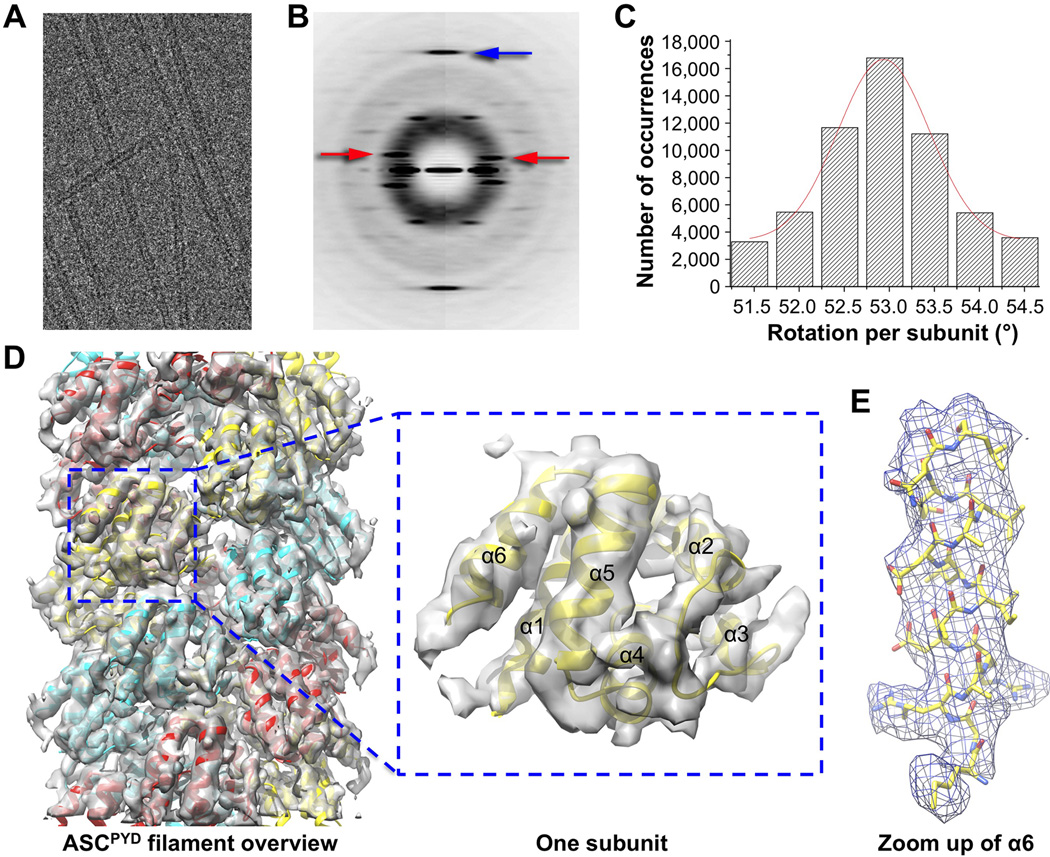
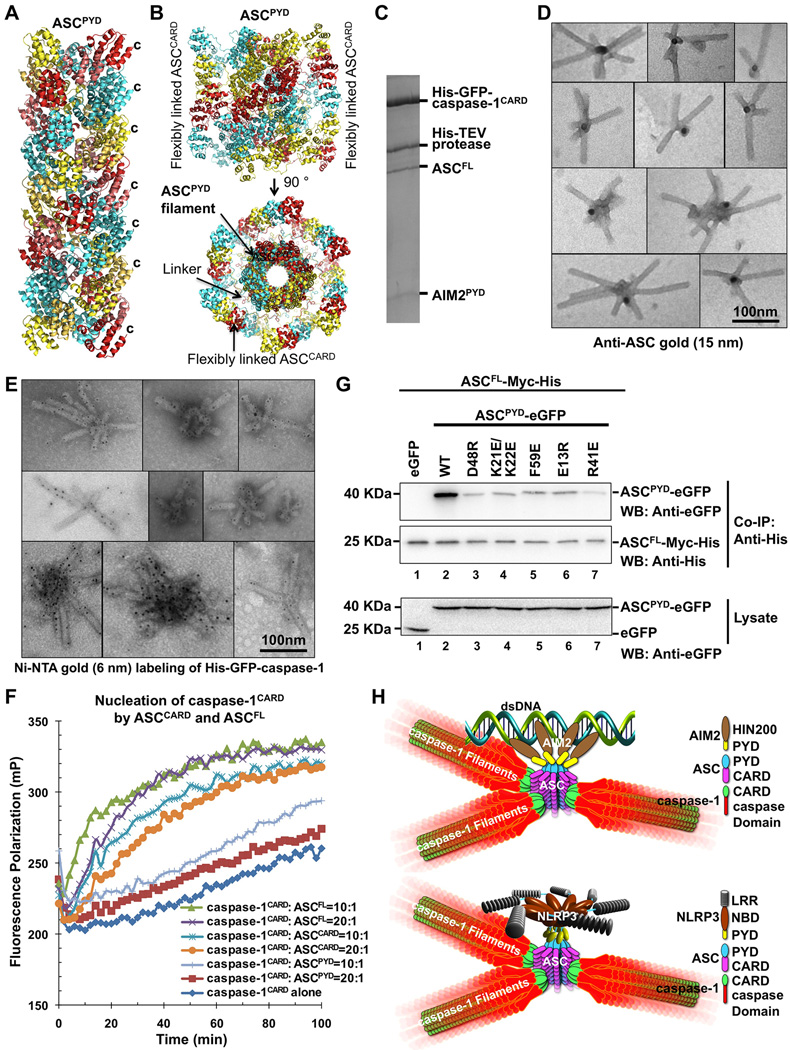
To generate a homogeneous population of ASCPYD filaments without the AIM2 or NLRP3 nucleators, we used in vitro ASCPYD polymerization starting from purified monomeric His-MBP-ASCPYD (Figure S1D). Upon TEV treatment to cleave off His-MBP, ASCPYD filaments spontaneously formed as shown by cryo-EM (Figure 3A). Cryo-EM images were collected using automated data acquisition on a Titan Krios with a back thinned CMOS direct electron detector. Averaged power spectra of segments from cryo-EM images of the helical filaments showed a strong meridional reflection at 1/13.9 Å−1, which corresponds to the reciprocal of the axial rise, but exhibited variable twist (Figure 3B, 3C), like many other helical polymers (Egelman et al., 1982). The magnitude of this variation can be seen in movie S1.
Figure 3. Cryo-EM Structure of the ASCPYD Filament at Near Atomic Resolution.
A. A cryo-EM image of ASCPYD filaments.
B. Average power spectra of ASCPYD filaments in two twist bins (left and right halves) showing constant axial rise per subunit (blue arrow) and variable long-range twist features (red arrows).
C. Filament segments can be divided into separate twist bins according to azimuth angle, or rotation per subunit.
D. Cryo-EM reconstruction of the ASCPYD filament, superimposed with the final atomic model shown in three colors each for one start of the three-start helical assembly.
E. A zoom up view of helix α6 shown in stick model and superimposed with the EM density.
Images were processed using the Iterative Helical Real Space Reconstruction (IHRSR) method with a solid cylinder as the initial reference (Egelman, 2000). The helical heterogeneity was dealt with by sorting images by twist to generate a subset with similar helical parameters, resulting in a map at ~6Å resolution. Correction of out of plane tilt was applied to further improve the map to a conservatively estimated resolution of ~3.8 Å as determined by both Fourier shell correlation (FSC) (Rosenthal and Henderson, 2003) (Figure S3A), and comparison with the final atomic model (Figure S3B, S3C).
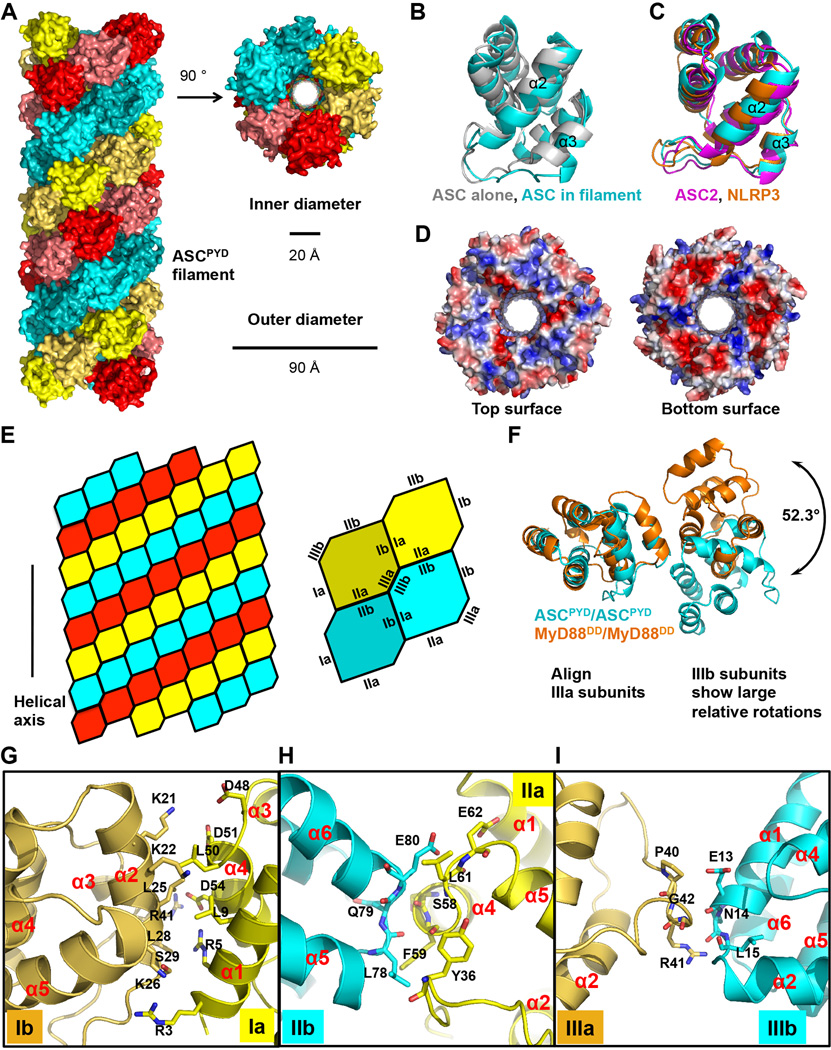
Rigid body fitting of the NMR structure of ASCPYD (Liepinsh et al., 2003) into the cryo- EM map generated a pseudo atomic model of the ASCPYD filament. The fit of the NMR structure resolved the enantiomorphic ambiguity in EM reconstructions, but even without the NMR structure the hand of the α-helices in the reconstruction was clear eliminating such ambiguities. The rigid-body fit was followed by real-space refinement (Schröder et al., 2007) to generate a final atomic model with clearly defined side chain densities (Figure 3D, 3E, S3D). The ASCPYD filament is hollow with inner and outer diameters of ~20 Å and ~90 Å, respectively (Figure 4A). The polar filament has a C3 point group symmetry with 53° right-handed rotation and 14.0 Å axial rise per subunit, after correcting for a mean out-of-plane tilt of ~6°.
Figure 4. Detailed Cryo-EM Model of the ASCPYD Filament.
A. The ASCPYD filament is a three-start helical assembly with C3 symmetry as shown in a surface representation. The three-start helical strands are denoted by red, cyan, and yellow, respectively, with alternating darker and lighter shades to show subunit boundaries.
B. Comparison of the initial ASCPYD subunit model (gray, PDB: 1UCP) and the subunit structure after refinement against the cryo-EM density (cyan).
C.Structures of ASC2PYD (magenta) and NLRP3PYD (orange) are similar to the ASCPYD subunit structure in the filament (cyan).
D.Electrostatic surface representations of approximate cross sections of the filament.
E. A schematic diagram of the ASCPYD filament and the three types of asymmetric interactions, defined in accordance with the previously observed DD/DD interactions.
F. Comparison of the Type III interactions in the ASCPYD filament (cyan) and in the MyD88/IRAK4/IRAK2 DD complex (orange).
G, H, I. Detailed interactions in Type I, II, and III interfaces, respectively. Side chains of interfacial residues are shown as stick models and labeled.
See also Figure S4.
The structure of ASCPYD in the filament exhibits conformational differences with that of ASCPYD alone (Figure 4B). This is apparent in the highly variable α2-α3 loop and the short α3 helix, with clear cryo-EM density (Figure S4A). PYDs share a unique feature: the α3 helix is shortened or missing, and follows the long and flexible α2-α3 loop (Figure S4B). The conformational changes are likely due to participation of this region in all three types of interactions in the filament (see below and Figure S3D). Although the ASCPYD alone structure was determined at a pH below 4.0 (Liepinsh et al., 2003), lack of significant conformational differences else where and absence of acidic residues in α3 helix support the structural changes as due to filament formation. Among known PYD structures, NLRP3PYD and ASC2PYD possess a conformation similar to the filament conformation of ASCPYD (Figure 4C), suggesting that NLRP3PYD and ASC2PYD may be better interactors with ASCPYD.The former similarity may account for the high efficiency of NLRP3PYD-NBD in promoting ASCPYD polymerization (Figure 2B).ASC2 is a PYD only protein that is highly homologous to ASCPYD and has been shown to associate with ASC to modulate caspase-1 activation (Stehlik et al., 2003).If ASC2 can be incorporated into ASCPYD filaments but lacks the effect or CARD, it could inhibit caspase-1 recruitment and activation. One face of a cross section of the filament is largely negatively charged while the opposite face is largely positively charged (Figure 4D), suggesting the role of charge complementarity in filament assembly.
Detailed Interactions in the ASCPYD Filament
There are three major asymmetric interfaces (types I, II, III) in the filament, one within each of the three-start helical strands (type I), and two between the strands (type II and III) (Figure 4E). Remarkably, despite being within the DD superfamily, the PYD/PYD interactions show remarkable differences to the DD complex structures (Figure 4F and S4C–E, Table S1). If one of the subunits is aligned, the corresponding partner subunit would need to rotate by 15–26°, 21–35° and 18–52° for the type I, II and III interfaces respectively, relative to the corresponding interfaces in the MyD88/IRAK4/IRAK2 DD complex and the PIDD/RAIDD DD complex (Ferrao and Wu, 2012; Lin et al., 2010; Park et al., 2007) (Figure S4C–E, Table S1). Relatively, the structural superposition indicates that the type I interaction is the most conserved, which is also the most dominant, burying about 880 Å2 of surface area. Type II and III interactions are highly variable and bury 540 Å2 and 360 Å2 of surface area, respectively. Structural differences between PYD and other members of the DD fold superfamily, and formation of a substantial central cavity may have shifted the relative orientations of the subunits in the type I, II and III interactions.
In the previously observed DD/DD interactions, type I is mediated by residues at α1 and α4 (type Ia) and residues at α2 and α3 (type Ib) (Ferrao and Wu, 2012; Lin et al., 2010; Park et al., 2007). Despite being the most conserved, the relative shift in orientation and the structural differences between DDs and PYDs minimized the involvement of α3 in the intra-strand type I PYD/PYD interaction (Figure 4G, S3D). In the inter-strand type II PYD/PYD interaction, residues at the α4-α5corner of one ASCPYD (type IIa) and residues at the α5-α6 corner of the second ASCPYD (type IIb) mediate this interaction (Figure 4H, Fig. S3D). In the inter-strand type III PYD/PYD interaction, α2-α3 corner of one ASCPYD (type IIIa) interacts with the α1-α2 corner (type IIIb) of the other subunit (Figure 4I,S3D). Prominently, the PYD-unique and highly variable α2-α3 loop participates in all three types of PYD/PYD interactions (Figure S3D), which may explain the conformational changes in this region upon filament formation (Figure 4B). Overall the interactions contain charged, hydrophilic and hydrophobic components, with charge interactions playing an important role (Figure S4F–H). Consistently, ASCPYD polymerization exhibits salt dependence (Figure S4I).
Structure-based Mutagenesis in Vitro and in Cells
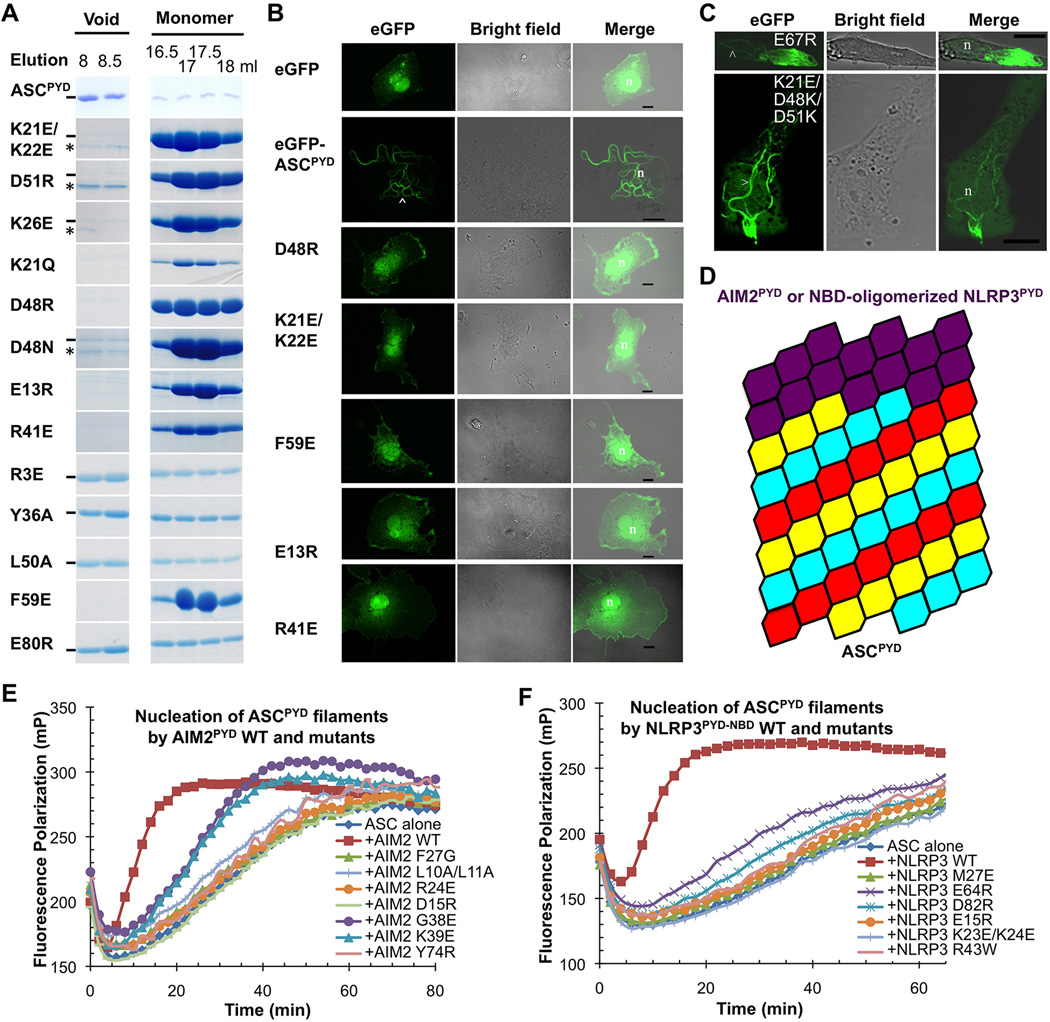
Structure-guided mutagenesis in vitro confirmed the role of type I, II and III interactions in ASCPYD filament formation, as shown by elution in more monomeric fractions (Figure 5A). In particular, K21Q, K21E/K22E, K26E, R41E, D48R, D48N and D51R of the type I interface, F59E of the type II interface, and E13R and R41E of the type III interface, abolished filament formation (Figure 5A, Table S2). Additional mutations, R3E and L50A of the type I interface, and Y36A and E80R of the type II interface, weakened filament formation as shown by increased presence in the monomeric fractions in comparison with the wild type (WT) (Figure 5A, Table S2). Mutations that disrupted filament formation in vitro also abrogated the ability of eGFP-ASCPYD to form filaments in cells as shown by confocal and fluorescence microscopy (Figure 5B, S5A–B) and by EM of immuno purified samples (Figure S5C–D). A previous extensive mutagenesis study on surface exposed charged residues confirmed the importance of additional observed interfacial residues in ASCPYD filament formation in cells (Moriya et al., 2005) (Table S2).
Figure 5. Structure-based Mutations Disrupts ASCPYD Filament Formation, AIM2PYD/ASCPYD Interaction and NLRP3PYD/ASCPYD Interaction in Vitro and in Cells.
A. Size-exclusion chromatography of WT and mutant ASCPYD showing both filamentous (void) and monomeric fractions from a Superdex 200 column. Hyphen denotes ASCPYD and asterisk denotes a contaminant.
B. Morphology of transfected WT and mutant eGFP-tagged ASCPYD constructs visualized by confocal laser scanning microscopy. The arrowhead depicts filaments. n: nucleus; scale bars = 10µm.
C.Morphology of transfected eGFP-tagged ASCPYD visualized by confocal laser scanning microscopy. Top: ASCPYD with charge reversal mutation on a residue outside the filament interface. Bottom: ASCPYD with triple charge reversal mutation that rescued the defectiveness of the single mutants. Arrow heads depict filaments.
D.A schematic model of AIM2PYD/ASCPYD or NLRP3PYD/ASCPYD filaments composed of a top AIM2PYD or NLRP3PYD layer extended by ASCPYD filament body.
E,F. Mutations of conserved interfacial residues on AIM2PYD (E) and NLRP3PYD-NBD (F) reduced or abolished their ability to nucleate ASCPYD filaments.
See also Figure S5.
In contrast to disruptive phenotypes of mutations on interfacial residues, the charge reversal mutation E67Routside the interface did not affect eGFP-ASCPYD filament formation (Figure 5C). At the type I interface, K21, D48 and D51 interact with each other (Figure 4G) and mutations on each of the residues disrupt filament formation (Figure 5A, 5B). Remarkably, the triple charge reversal mutant K21E/D48K/D51K rescued eGFP-ASCPYD filament formation in cells (Figure 5C), supporting the validity of the structurally observed interactions.
Modeled AIM2PYD/ASCPYD and NLRP3PYD-NBD/ASCPYD Interactions
The ASCPYD filament structure provides a template for modeling the AIM2PYD/ASCPYD and NLRP3PYD/ASCPYD hetero-PYD/PYD interactions using the published crystal structures of AIM2PYD (Jin et al., 2013) and NLRP3PYD (Bae and Park, 2011). End locations of AIM2PYD and NLRP3PYD in their complexes with ASCPYD filaments suggest that the PYDs in AIM2 and NLRP3 continue the helical arrangement seen in the ASCPYD filament using a combination of the same type I, II and III interactions (Figure 5D). Given the observed conformational changes at the α2-α3 corner, which points down in the helical diagram (Figure 4D), we reasoned that AIM2 and NLRP3 PYDs should reside above the ASCPYD filament (Figure 5D).
We selected residues in AIM2PYD and NLRP3PYD structures corresponding to those in ASCPYD that caused impairment in filament formation when mutated (Figure 5A, 5B). Assaying the ability of AIM2PYD and NLRP3PYD-NBD mutants in promoting ASCPYD polymerization showed that mutations on each of the predicted interfaces in AIM2PYD, including L10A/L11A, R24E and F27G of the type I interface, Y74R of the type II interface, and G38E, K39E and D15R of the type III interface, either abolished or showed greatly reduced promotion of ASCPYD polymerization (Figure 5E, Table S3). Additionally, in a recently published study on AIM2PYD, the D19A/E20A/E21A/D23A mutation, which harbors mostly type I interface residues, abolished the interaction with ASCPYD (Jin et al., 2013). Similarly, mutations at each of the predicted type I, II and III interfacial residues on NLRP3PYD, including K23E/K24E and M27E of the type I interface, E64R and D82R of the type II interface, and R43W and E15R of the type III interface, caused almost complete impairment in promoting ASCPYD polymerization by the NLRP3PYD-NBD construct (Figure 5F, Table S3). It should be noted because AIM2PYD also aggregates into similar filaments (Figure S1B), the same mutations would likely affect both AIM2/ASC interaction and AIM2 self-association. For NLRP3, the PYD does not self-associate while the NBD mediates self-association; therefore the PYD mutations in NLRP3 would directly affect ASC interaction. Collectively, these data support that the interactions in the ASCPYD filament also define the mode of hetero-oligomerization in the AIM2PYD/ASCPYD and NLRP3PYD/ASCPYD interaction pairs.
Reconstitution of the TernaryAIM2 Inflammasome
The C-termini of ASCPYD subunits extend prominently outward from the filament (Figure6A), providing a connection to the CARD in ASC after a 23-residue linker. Superposition of the NMR structure of ASCFL (de Alba, 2009) with ASCPYD in the filament displayed the flexibly linked, peripheral ASCCARD (Figure 6B). To reveal the structural architecture of full ternary inflammasomes, we expressed and purified His-GFP-caspase-1CARD, His-MBP-ASCFL and His-MBP-AIM2PYD. We mixed the three proteins with the TEV protease to allow His-MBP removal and formation of a ternary complex. His-tag pull down showed that His-GFP-caspase-1CARD interacted with ASCFL and AIM2PYD (Figure 6C). The stoichiometry between ASCFL and AIM2PYD in the ternary complex is consistent with that in the AIM2PYD/ASCPYD binary complex with AIM2PYD under-stoichiometric (Figure 1C). ASC in turn appeared under-stoichiometric tocaspase-1. EM showed that the ternary complex is star-shaped (Figure 6D). Anti-ASC immuno gold-labeling (15 nm) showed that ASC resides in the center of the stars (Figure 6D). In contrast, Ni-NTA conjugated with 6 nm gold particles labeled His-GFP-caspase-1CARD along the arms of the stars (Figure 6E). These data suggest that AIM2PYD nucleates short filaments of ASCFL through PYD/PYD interactions and ASCCARD further initiates caspase-1CARD filaments to promote caspase-1 activation. Because the linker between ASC PYD and CARD is flexible, the outer CARDs should be able to cluster together and act as the platform for caspase-1 polymerization (Figure 6B).
Figure 6. Reconstitution of the Full Ternary AIM2 Inflammasome.
A. The ASCPYD filament structure in a ribbon representation. The protruding C-termini for connecting to ASCCARD are labeled for the subunits at the right.
B. ASCFL NMR structure (PDB: 2KN6) is superimposed on the ASCPYD model to show the outward located ASCCARD.
C. Pull-down of the core AIM2 inflammasome in vitro as visualized on Coomassie-Blue stained SDS-PAGE.
D, E.Electron micrographs of His-GFP-caspase-1CARD/ASCFL/AIM2PYD ternary complex labeled with anti-ASC gold (D) and Ni-NTA gold (E).
F. Promotion ofHis-MBP-caspase-1CARD-Sumo (3 µM) polymerization by ASCFL or ASCCARD at sub stoichiometric ratios of 1:20 and 1:10 upon removal of His-MBP by TEV. ASCPYD did not enhance caspase-1CARD polymerization.
G. Mutations in ASCPYD reduced its binding to ASCFL. ASCFL-Myc-His was co-transfected with WT and mutant ASCPYD-eGFP. Immuno precipitation and Western blotting was carried out using anti-His and anti-eGFP antibodies respectively.
H. Model of inflammasome assembly. Up stream sensing proteins such as AIM2 and NLRP3 oligomerize upon activation to form a platform of PYDs that induces ASC filament assembly through PYD/PYD interactions. Multiple ASCCARD molecules cluster to promotecaspase-1 filament formation through CARD/CARD interactions. Proximity induced dimerization of the caspase domain activates the enzyme followed by auto-cleavage.
See also Figure S6.
We tested the role of ASCCARD in inflammasome assembly using a caspase-1CARD polymerization assay. We used “sandwich”-tagged His-MBP-caspase-1CARD-Sumo construct because N-terminally tagged His-MBP-caspase-1CARD construct still formed filaments. Asortase motif was added for fluorophore labeling (Theile et al., 2013). Polymerization of caspase-1CARD was initiated by addition of TEV to cleave off His-MBP and monitored by fluorescence polarization. In the presence ASCFL or ASCCARD at sub-stoichiometric ratios, caspase-1CARD polymerization was greatly enhanced (Figure 6F), consistent with nucleation of caspase-1 polymerization by ASC.
It is intriguing that the ASCFL component in the ternary complex does not display as long filaments as in the binary AIM2PYD/ASCPYD or NLRP3PYD-NBD/ASCPYD complexes. We reasoned that since His-MBP-ASCFL forms bundled clusters minutes after removal of the His-MBP tag (Figure S6A) and precipitates, likely due to the CARD and its potential to crosslink filaments, ASCFL might only exist as short filaments such that almost all ASCCARD molecules are in complex with caspase-1CARD. To determine if the same ASCPYD interactions in the observed filament govern those in the context of ASCFL, we co-transfected Myc-His-tagged ASCFL with WT and mutant ASCPYD-eGFP in 293T cells. Immuno precipitated with anti-His antibody followed by anti-eGFP Western showed that Myc-His-ASCFL pulled down WT ASCPYD-eGFP, but was severely impaired in interacting with ASCPYD-eGFP that harbors mutations on residues important for the ASCPYD filament formation (Figure 6G, S6B). We further tested the effects of PYD mutations in ASCFL using the in vitro inflammasome reconstitution assay. His-GFP-caspase-1CARD pulled down AIM2PYD in the presence of WT ASCFL, but not mutant ASCFL defective in formation of PYD filaments (Figure S6C), demonstrating that the same interactions in the PYD filaments govern the interaction in the ternary inflammasome complex.
Morphology and Stoichiometry of Endogenous NLRP3 Inflammasome
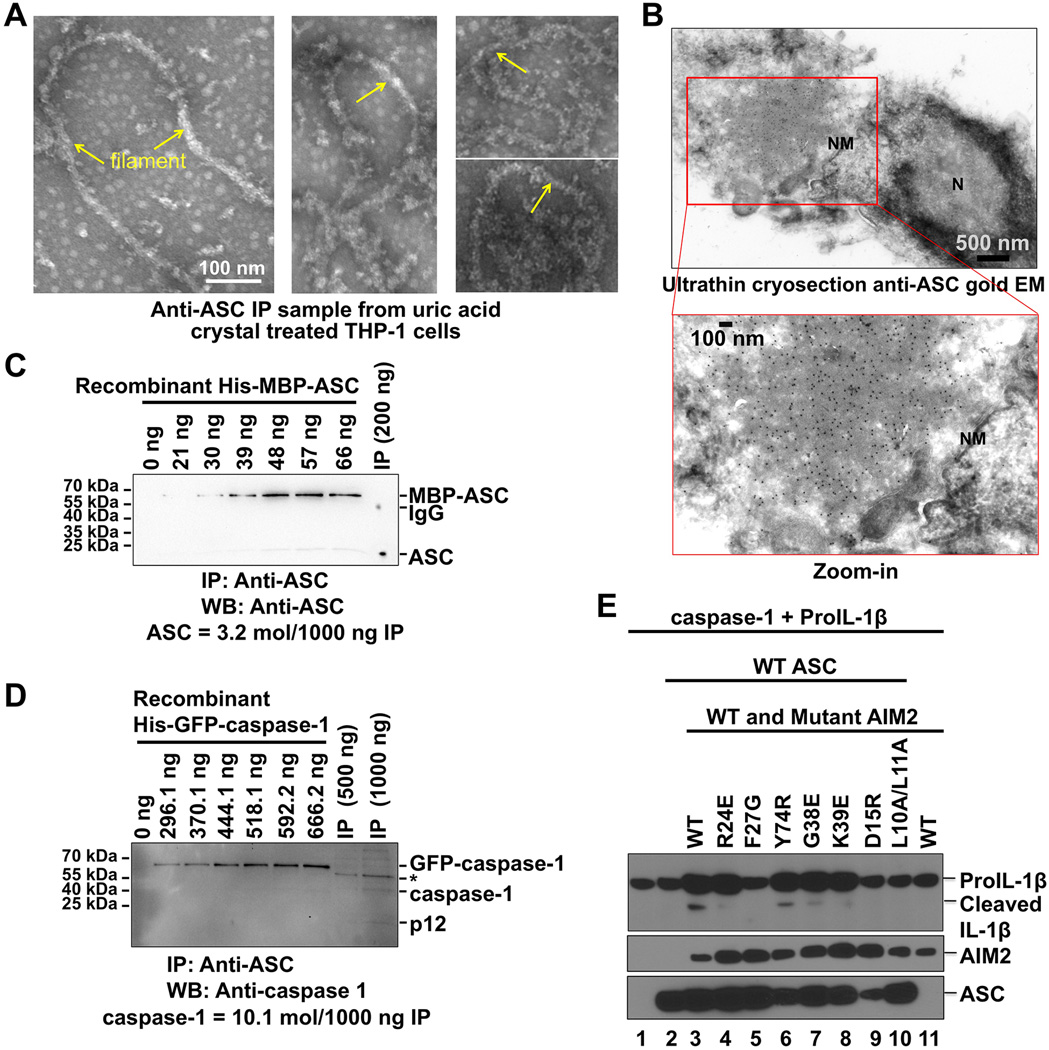
Our data suggest a unified model of inflammasome assembly in which AIM2 upon dsDNA interaction or NLRP3 upon activation nucleates ASC helical clusters through PYD/PYD interactions (Figure 6H). The oligomerized ASC CARDs then form the platform for caspase-1CARD to nucleate into filaments, which in turn bring caspase domains into proximity for dimerization, transauto-cleavage and activation (Figure 6H). To elucidate the morphology of endogenous inflammasomes, we stimulated THP-1 cells with uric acid crystals, immuno precipitated the activated NLRP3 inflammasome using anti-ASC antibody, and subjected the immuno precipitated sample to negative stain EM. The images contained both single filaments (Figure 7A) and intertwined filaments (FS7A); the former resemble sub complexes of in vitro reconstituted inflammasomes and the latter resemble clustered, ball of yarn-like reconstituted inflammasomes that form upon overnight incubation (FS7B).
Figure 7. Morphology, Stoichiometry and ProIL-1β Processing in Inflammasomes.
A. Morphology of anti-ASC immuno precipitated NLRP3 inflammasomes from uric acid crystal activated THP-1 cells analyzed by negative stain EM. Arrows denote filaments.
B. Immuno gold EM on ultra thin cryo sections from ASCFL-eGFP transfected COS-1 cells. The ASC-containing compact structure is densely decorated by gold particles (10 nm). N, nucleus; NM, nuclear membrane.
C, D. Quantification of immuno precipitated ASC-containing complex (IP) from uric acid crystal activated THP-1 cells using quantitative anti-ASC (C)and anti-caspase-1 p12 (D) Western blotting. Known amounts of recombinant His-MBP-ASC and His-GFP-caspase-1 were Western blotted to generate standard curves. The full-length caspase-1 and the cleaved p12 bands were both included in the quantification.
E. AIM2 inflammasome reconstitution in HEK293T cells to define the functional consequence of structure-based mutations in AIM2. Cells were co-transfected with plasmids encoding proIL-1β and caspase-1 (lane 1), plus ASC alone (lane 2), or WT AIM2 alone (lane 11), or ASC together with WT or indicated AIM2 mutants (lanes 3 to 10). Maturation of proIL-1βinto biologically active IL-1β was detected by Western blotting using anti-IL-1β antibody (top panel).The expression levels of HA-ASC and Flag-AIM2 were detected by Western blotting using anti-HA and anti-Flag antibodies (lower panels).
See also Figure S7.
It has been shown previously that upon stimulation, each cell forms one gigantic NLRP3 punctum adjacent to the nucleus (Fernandes-Alnemri et al., 2007). To visualize the structure of such a punctum in situ, we expressed ASC-eGFP in COS-1 cells and performed immuno gold EM on ultra thin cryo sections that preserve native structures. Control cells transfected with eGFP alone showed neither punctum nor anti-ASC gold labeling, and ASC-eGFP transfected cells did not exhibit gold labeling in the absence of the anti-ASC primary antibody (Figure S7C).Specific gold labeling was shown in ASC-eGFP transfected cells in the presence of anti-ASC primary antibody and protein A-gold (10 nm) treatment (Figure 7B). The labeling revealed a densely packed perinuclear punctum of about 2 µm in size, in contrast to the hollow structure implicated previously (Masumoto et al., 1999). The dense structures are consistent with the ball of yarn-like architectures of in vitro reconstituted and in cell immuno precipitated inflammasomes.
In vitro reconstitution of the ternary inflammasome suggests an over-stoichiometry of caspase-1 to ASC (Figure 6C). To determine if endogenous ASC-dependent inflammasomes from cells also possess the similar property, we stimulated THP-1 cells with uric acid crystals, immuno precipitated the activated NLRP3 inflammasome using anti-ASC antibody, and performed quantitative Western blotting using recombinant caspase-1 and ASC as standards. These experiments showed that caspase-1 is over-stoichiometric to ASC, by ~3.5-fold in the current experiment (Figure 7C, 7D, S7D, S7E). It should be noted that an anti-ASC antibody would have precipitated both ASC alone and its complex with caspase-1 and therefore the measured 3.5- fold over-stoichiometry should be an under estimation.
Structure-guided PYD Mutations Compromise IL-1β Processing
One main biological consequence of inflammasome activation is the processing of prolL-1β by caspase-1 to IL-1β. To address the consequence of structure-based mutations in biological function, we used a co-transfection strategy to assay IL-1β processing (Jin et al., 2012). Co-transfect ion of caspase-1, proIL-1β and ASC did not cause significant cleavage of proIL-1β into mature IL-1β (Figure 7E). While addition of WT AIM2 activated the inflammasome and led to IL-1β production, co-transfection of PYD-interaction defective AIM2 mutants compromised IL-1β conversion (Figure 7E), demonstrating the functional consequence of observed PYD/PYD interactions.
DISCUSSION
A Near Atomic Resolution Structure by Cryo-EM
Our reconstruction of a small, structurally variable biological sample represents a significant advance in high-resolution structure determination by cryo-EM made possible by automated microscopy (Potter et al., 1999), a state-of-the-art electron microscope (the Titan Krios), and a new generation of direct electron detectors (Bai et al., 2013; Bammes et al., 2012; Li et al., 2013; Liao et al., 2013), combined with existing computational approaches for variable twist polymers (Egelman, 2000; Egelman et al., 1982). Most of the structures that have been currently solved by cryo-EM to near-atomic resolution are icosahedral viruses that are highly ordered and with a high degree of internal symmetry (Zhou, 2011). In the absence of any mechanism to maintain long-range order all biological polymers will display cumulative disorder (Egelman and DeRosier, 1982). We think that the new hardware and software advances in cryo-EM will have an enormous impact in allowing many biological polymers, including those whose helical symmetry could not even be determined with confidence, to now be reconstructed at near-atomic resolution.
PYD/PYD Interactions
The ASCPYD filament structure presented here provides insights into molecular mechanisms of homo- and hetero-PYD associations in inflammasomes. Among the PYDs with known structures, NLRP3,NLRP 12, AIM2 and ASC2 have been shown to interact with ASC (Rathinam et al., 2012; Stehlik et al., 2003). Consistently, they exhibit the highest sequence conservation at the ASC-interaction surfaces with 61%, 54%, 50%, and 89% homology, respectively (Table S4). The ASCPYD structure may also provide a template for other PYDs with no structures such as the IFI16PYD filament cooperatively assembled on dsDNA (Morrone et al., 2013).
A number of mutations in NLRP3, NLRP12 and MEFV have been shown to associate with hereditary periodic fever syndromes. For NLRP3, all mutations are dominant and likely cause activation by overcoming auto inhibition (Touitou et al., 2004). For NLRP12, a nonsense mutation and a splicing defect generate truncated proteins at residues 284 and 646, respectively, and cause spontaneous inflammation (Jeru et al., 2008), suggesting that the PYD and part of the NBD are sufficient for inflammasome formation and activation. For MEFV, hundreds of variants, most of which are associated with Familial Mediterranean Fever (FMF), have been identified (Touitou et al., 2004). Gene insertion “knockin” (KI) mouse models with three frequent FMF-associated mutations (M680I, M694V, and V726A) showed that they caused severe spontaneous inflammatory phenotypes (Chae et al., 2011). Most relevant to the PYD interactions, six mutants, T12I, Y19C, K25R, R39G, E84K and A89T, have been mapped to the PYD of MEFV (Touitou et al., 2004). None of these residues directly map to the PYD/PYD interaction surfaces (Figure S3D), and may therefore act by overcoming auto inhibition.
A Unified Assembly Mechanism for Inflammasomes
Our data here present a mechanism for the assembly of ASC-dependent inflammasomes, in which AIM2 and NLRP3 both nucleate helical ASC clusters through PYD/PYD interactions, and ASC in turn nucleates caspase-1 filaments through CARD/CARD interactions (Figure 6H). These minimal structures coalesce to form the micron-sized, dense structures we observed in situ.We propose that CARD-containing NLRs (NLRCs), which are independent of ASC, may also form filamentous structures by directly promoting caspase-1 polymerization through CARD/CARD interactions. Therefore, the mechanism of nucleation-induced filament formation may extend beyond ASC-dependent inflammasomes. It has been shown that uncleavedcaspase-1 catalytic domain forms dimers in crystals (Elliott et al., 2009); the dimerization may occur within caspase-1 filaments in inflammasomes, resulting in intradimer cleavage, stabilization of dimerization, and enhancement of enzymatic activity.
Recent studies have revealed that in many innate immune pathways, multiple intracellular signaling proteins assemble into higher-order signaling machines for transmission of receptor activation information to cellular responses, with implicated new molecular mechanisms for threshold behavior, time delay of activation, and temporal and spatial control of signal transduction (Wu, 2013). Here we show that inflammasomes also assemble into higher-order signal osomes that likely impart similar properties to its activation and kinetics. In this scenario, upon reaching the NLRP3 or AIM2 activation threshold, caspase-1 may polymerize until its concentrations falls below the dissociation constant. Given that caspase-1is over-stoichiometric to ASC by just a few fold, the average lengths of individual ASC nucleated caspase-1 filaments in cells may be shorter than those reconstituted in vitro, leading to punctate, rather than filamentous morphology of intact inflammasomes. Once formed, inflammasomes may require active processes such as autophagy for their degradation (Saitoh et al., 2008).This scenario is reminiscent of the case for the filamentous CARMA1/Bcl10/MALT1 signal osome (Qiao et al., 2013), and may represent a general mechanism of disassembly of higher-order signalosomes in innate immunity to terminate signaling.
EXPERIMENTAL PROCEDURES
Recombinant Protein Expression and Purification
Various His-, His-MBP- and His-MBP-Sumo “sandwich”-tag fusion constructs or co-expression constructs of AIM2, ASC, caspase-1 and NLRP3 were expressed in E. coli and insect cells. Biotinylation was performed in E. coil by co-expression with biotin ligase. All mutations in this construct were introduced using the QuikChange mutagenesis protocol.
Polymerization Assays
His-MBP-ASCPYD and His-MBP-caspase-1CARD-Sumo were labeled with Alexa488 and TAMRA fluorophores, respectively. Filament formation was monitored using fluorescence polarization upon addition of TEV protease to remove His-MBP, in the presence and absence of various nucleators of polymerization.
Nanogold and Immunogold EM
Standard protocols were used for streptavidin-gold labeling of biotinylated proteins, Ni-NTA-nanogold labeling of His-tagged proteins, and immunogold labeling by appropriate antibodies using negative stain EM. ASC-transfected COS-1 cells were pelleted, fixed, flash frozen and ultra thin sectioned for immuno gold EM and contrasting with uranylacetete.
Cryo-EM, Image Processing and Refinement
Grids containing ASCPYD were imaged using an FEI Titan Krios electron microscope operating at 300 keV, and recorded using a 4k × 4k Falcon II direct electron detector with a back thinned CMOS chip. The images were processed with SPIDER (Frank et al., 1996) and the IHRSR algorithm (Egelman, 2000) was used for helical reconstruction. The ASCPYD NMR structure (PDB ID 1UCP) (Liepinsh et al., 2003) was chosen as the starting model and the refinement was carried out using DireX (Schröder et al., 2007). We estimate the resolution of the reconstruction at ~3.8 Åas determined by both Fourier shell correlation and comparison with the final atomic model.
AIM2 Inflammasome Reconstitution
HEK293T cells were transfected with pEFBOS-C-term-Guassia luciferase/Flag pro-IL-1β (~54 kDa), pro-caspase-1, HA-ASC, and the full-length wild type or mutant Flag-AIM2 expression constructs using Gene Juice (Novagen). Cell lysates were probed with mouse anti-IL1β monoclonal antibody. Expression of ASC and AIM2 was detected using anti-Flag and anti-HA antibodies, respectively.
Supplementary Material
AIM2 and NLRP3 inflammasomes are filamentous assemblies in vitro and in cells
3.8 Å Cryo-EM structure of ASCPYD depicts the underlying oligomerization mechanism
Fluorescence polarization assays suggest nucleation-induced filament formation
ASC-dependent inflammasomes use a polymerization mechanism for caspase-1 activation
ACKNOWLEDGEMENTS
We thank NatachaOpalka, Seth Darst, Leona Cohen-Gould, Maria Ericsson, William Rice and MarienaSilvestry Ramos for help with EM imaging, Harry Leung for help with confocal microscopy, Gabriel Nuñez for providing cDNAs, and the National Institutes of Health for funding supports (EB001567 to E.H.E and AI083713 to K.A.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The cryo-EMmap of ASCPYD filament was deposited to EMDataBank under accession code EMD-5830. The corresponding refined structure of ASCPYD in the filament was deposited in the Protein Data Bank with ID code 3J63.
REFERENCES
- Bae JY, Park HH. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem. 2011;286:39528–39536. doi: 10.1074/jbc.M111.278812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J Struct Biol. 2012;177:589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae JJ, Cho YH, Lee GS, Cheng J, Liu PP, Feigenbaum L, Katz SI, Kastner DL. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1beta activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH, DeRosier DJ. The Fourier transform of actin and other helical systems with cumulative random angular disorder. Acta Cryst. 1982;A38:796–799. [Google Scholar]
- Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Elliott JM, Rouge L, Wiesmann C, Scheer JM. Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation. J Biol Chem. 2009;284:6546–6553. doi: 10.1074/jbc.M806121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- Jeru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, Grimprel E, Landman-Parker J, Hentgen V, Marlin S, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A. 2008;105:1614–1619. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Smith PT, Jiang J, Xiao TS. Structure of the AIM2 pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013 doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–1163. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T, Sagara J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- Masumoto J, Taniguchi S, Sagara J. Pyrin N-terminal homology domain-and caspase recruitment domain-dependent oligomerization of ASC. Biochem Biophys Res Commun. 2001;280:652–655. doi: 10.1006/bbrc.2000.4190. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C., Jr Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- Moriya M, Taniguchi S, Wu P, Liepinsh E, Otting G, Sagara J. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 2005;44:575–583. doi: 10.1021/bi048374i. [DOI] [PubMed] [Google Scholar]
- Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, Bracken C, Rosen M, Melnick A, Egelman EH, et al. Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003;373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- Theile CS, Witte MD, Blom AE, Kundrat L, Ploegh HL, Guimaraes CP. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8:1800–1807. doi: 10.1038/nprot.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou I, Lesage S, McDermott M, Cuisset L, Hoffman H, Dode C, Shoham N, Aganna E, Hugot JP, Wise C, et al. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum Mutat. 2004;24:194–198. doi: 10.1002/humu.20080. [DOI] [PubMed] [Google Scholar]
- Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, Sagulenko V, Thygesen SJ, Choubey D, Hornung V, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure. 2013;21:501–515. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Atomic resolution cryo electron microscopy of macromolecular complexes. Adv Protein Chem Struct Biol. 2011;82:1–35. doi: 10.1016/B978-0-12-386507-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.