Abstract
The interplay between molecular chaperones, ubiquitin/deubiquitinating enzymes, and proteasomes is a critical element in protein homeostasis. Among these factors, the conserved deubiquitinase, Ubp3, has the interesting ability, when overproduced, to suppress the requirement for the major cytosolic Hsp70 chaperones. Here, we show that Ubp3 overproduction counteracts deficiency of Hsp70s by the removal of damaged proteins deposited in inclusion bodies (JUNQ) during both aging and heat stress. Consistent with this, Ubp3 destabilized, deubiquitinated, and diminished the toxicity of the JUNQ-associated misfolded protein Ubc9ts in a proteasome-dependent manner. In contrast, another misfolded model protein, ssCPY*, was stabilized by Ubp3-dependent deubiquitination demonstrating a dual role for Ubp3, saving or destroying aberrant protein species depending on the stage at which the damaged protein is committed for destruction. We present genetic evidence for the former of these activities being key to Ubp3-dependent suppression of heat sensitivity in Hsp70-deficient cells, whereas protein destruction suppresses accelerated aging. We discuss the data in view of how heat stress and aging might elicit differential damage and challenges on the protein homeostasis network.
Keywords: aging, heat stress, protein aggregates, SSA1 SSA2, Ubp3
Introduction
The small protein ubiquitin (Ub) is ubiquitously present in almost all tissues of eukaryotic organisms and is covalently ligated to target proteins by a multienzymatic system consisting of ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes. The “tagging” of a target protein by ubiquitin can direct it to specific cellular locations, trafficking routes, or to the 26S proteasome for destruction. Deubiquitinating enzymes (DUBs), on the other hand, regulate ubiquitin-dependent processes by cleaving ubiquitin-protein bonds. DUBs are members of a large family of cysteine proteases (D'Andrea & Pellman, 1998), divided into two classes: the Ubp family (ubiquitin-specific proteases) and the Uch family (ubiquitin carboxy terminal hydrolases). While Uch enzymes hydrolyze ubiquitin from peptides and small adducts, Ubps cleave ubiquitin from a large range of substrates. This cleavage is important in several processes, in activating the ubiquitin proproteins, recycling of ubiquitin, reversing ubiquitination of target proteins, and regenerating monoubiquitin from unanchored polyubiquitin (Amerik & Hochstrasser, 2004). With respect to proteasome-dependent degradation, the removal of ubiquitin by Ubps can, in principle, have either positive or inhibitory effects on proteolysis. Positive regulation of proteolysis can be achieved through freeing ubiquitin groups that become available for attachment to new substrates targeted for degradation (Leggett et al, 2002) or by facilitating the entry of a specific substrate to the proteasomal cavity by the removal of the ubiquitin chain (Verma et al, 2002; Yao & Cohen, 2002). A negative effect on proteolysis could occur if the reversal of ubiquitination diverts the substrates away from proteasomal degradation.
Yeast is predicted from sequence analysis to harbor 17 Ubp enzymes, displaying both unique and overlapping functions (Baker et al, 1992; Amerik et al, 2000). Among these Ubps, UBP3 was initially isolated in a screen for yeast genes that, when co-expressed with Ub-β-galactosidase in Escherichia coli, resulted in the removal of the ubiquitin moiety (Baker et al, 1992), and later work demonstrated that Ubp3 deficiency resulted in the accumulation of ubiquitin-protein conjugates (Baxter & Craig, 1998). Interestingly, Ubp3 together with its partner Bre5 partakes in, and regulates, a number of seemingly unrelated processes: (i) Cdc48/Rsp5-dependent control of ubiquitin-mediated degradation of the COPII component Sec23 (Cohen et al, 2003; Ossareh-Nazari et al, 2010), (ii) Sir4-dependent transcriptional silencing (Moazed & Johnson, 1996), (iii) stabilization of the microtubule-associated protein Stu1 (Brew & Huffaker, 2002), (iv) selective autophagy targeting ribosomes (Kraft et al, 2008), (v) DNA repair (Mao & Smerdon, 2010), and (vi) regulation of transcription initiation and elongation (Kvint et al, 2008; Chew et al, 2010).
Apart from participating in the above-mentioned processes, UBP3 was identified in a high-copy suppressor screen as a gene able to suppress the temperature sensitivity of yeast cells lacking their major cytosolic Hsp70 chaperons, Ssa1 and Ssa2 (Baxter & Craig, 1998). Ubp3 overproduction was suggested to desensitize ssa1 ssa2 mutants toward heat stress by reversing the ubiquitination of temporarily misfolded proteins, allowing them an opportunity to fold correctly and achieve some residual activity rather than being destroyed by the proteasome. This model highlights that an overzealous protein quality control may have detrimental effects on fitness and that ubiquitin proteases have an important role in balancing such control. An alternative, and not mutually exclusive, model is that ubiquitin proteases facilitate the destruction of proteins, which upon misfolding become cytotoxic. During stress, misfolded proteins can accumulate in distinct aggregates or inclusions such as IPOD (insoluble protein deposit) or JUNQ (juxta-nuclear quality control) in a process that is partly controlled by the ubiquitin-proteasome system (UPS; Kaganovich et al, 2008; Specht et al, 2011; Malinovska et al, 2012). Some forms of such inclusions have been shown to trigger proteotoxicity (Weisberg et al, 2012).
In this work, we have approached the question of how Ubp3 overproduction suppresses Hsp70 deficiency. We demonstrate that Ubp3 is required for clearing misfolded proteins deposited into JUNQ inclusions upon both heat stress and aging and that Ubp3-targeted proteins can be either stabilized or destabilized depending on the protein species; that is, Ubp3-dependent deubiquitination can either “save” a protein from destruction or facilitate its destruction by the 26S proteasome. Furthermore, the data demonstrate that these two diametrically opposed outcomes depend on the genetic context and conditions; during heat stress, Ubp3 overproduction suppresses growth defects by diverting proteins away from destruction, whereas the suppression of accelerated aging instead is linked to Ubp3-dependent protein degradation.
Results
UBP3 is required for counteracting accumulation of JUNQ inclusions upon heat stress and aging
Since overproduction of the ubiquitin protease Ubp3 suppresses the need for cytosolic Hsp70 chaperones during heat stress (Baxter & Craig, 1998) and protein ubiquitination is a key process in the deposition of misfolded proteins into different inclusion bodies (IBs, such as IPOD and JUNQ; Kaganovich et al, 2008), we hypothesized that Ubp3 might be required for efficient management of misfolded proteins accumulating in IBs. To approach this possibility, we used a functional Ssa2p-GFP (Supplementary Fig S1A) as a reporter for protein aggregates/IBs and found that cells lacking Ubp3 displayed a delayed clearance of IBs after a 42°C heat shock (Fig 1A and B). Moreover, analysis of aggregate clearance in cells harboring the catalytically inactive allele of UBP3, ubp3C469A, demonstrated that ubiquitin protease activity is required for efficient Ubp3-dependent aggregate clearance of heat-denatured substrates (Fig 1C). Ubp3 forms a complex with Bre5 (Cohen et al, 2003) and a number of reports have shown that bre5 and ubp3 mutants display identical phenotypes (Cohen et al, 2003; Kvint et al, 2008). Indeed, a bre5Δ mutant phenocopied cells lacking UBP3 also with respect to IB clearance (Supplementary Fig S1B).
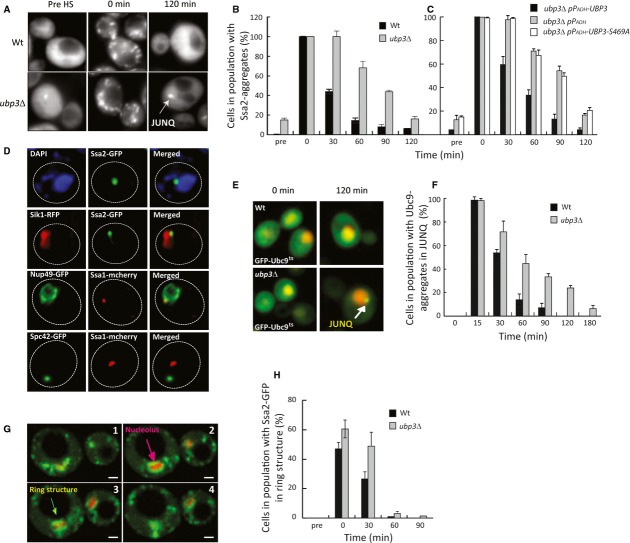
Figure 1. Ubp3 counteracts JUNQ accumulation.
- Exponentially growing wild-type and ubp3 cells with a C-terminal GFP tag of the chromosomal SSA2 were subjected to heat stress (42°C for 30 min), allowed to recover at 30°C, and imaged by wide-field fluorescence microscopy. A putative JUNQ aggregate is denoted by an arrow.
- Quantification of (A) as percentage of cells containing Ssa2-GFP aggregates over time. Data are represented as the mean of at least five experiments ± standard deviation (s.d.).
- The indicated plasmids were inserted in a ubp3 strain with a chromosomal copy of Ssa2-GFP. Cells were monitored and quantified as in (A) and (B). Data are represented as mean ± s.d.
- Co-localization of Hsp70 (Ssa1p or Ssa2p) with DAPI (DNA), Sik1-RFP (nucleolus), Nup49-GFP (nuclear pore complex), or Spc42-GFP (spindle pole body). Representative images are shown for ubp3Δ after 30 min of heat stress at 42°C followed by 60-min recovery at 30°C.
- GFP-Ubc9ts aggregation over time at 37°C in wild-type and ubp3Δ. The nucleus is visualized by a co-expressed NLS-tdTomato.
- Quantification of cells from (E) with respect to percentage of the whole population containing a JUNQ inclusion. Data are represented as the mean ± s.d. of three independent experiments.
- Confocal Z-stack image series (1-μm image spacing) of a representative wild-type cell directly after heat stress (42°C for 30 min) with Ssa2-GFP and the nucleolar marker Sik1-RFP expressed from a low-copy plasmid. Scale bar in the lower right corner represents 1 μm.
- Quantification of cells with Ssa2-GFP surrounding the nucleolus in ring-like structures over time. Data are represented as the mean ± s.d. of three experiments.
One particular inclusion, in close proximity to the nucleus, remained unresolved markedly longer in cells lacking UBP3 (Fig 1D). In addition, a marker for the nucleolus, Sik1-RFP, demonstrated that about 70% of the Hsp70 inclusion was in close proximity to the nucleolus (Fig 1D). Moreover, a marker for the nuclear pore complex, Nup49-GFP, showed that the Ssa2p foci localized on the cytoplasmic side of the nuclear pore complex (Fig 1D) and that these inclusions remained unresolved for approximately 60 min longer when UBP3 was deleted (Fig 1B). In contrast, the spindle pole body marker, Spc42-GFP, did not co-localize with the Hsp70 inclusion (Fig 1D), suggesting that this inclusion is distinct from the aggresome (Johnston et al, 1998; Wang et al, 2009). Therefore, we conclude that cells lacking UBP3 accumulate Hsp70 substrates in juxta-nuclear/nucleolar inclusions that correspond to the JUNQ deposition site described in previous reports (Kaganovich et al, 2008). This notion is supported by the fact that GFP-Ubc9ts, which has been shown previously to translocate from the nucleus to both JUNQ inclusions and peripheral IPOD aggregates at elevated (37°C) temperatures (Kaganovich et al, 2008), remained stable in its juxta-nuclear position in cells lacking Ubp3 (Fig 1E and F). Prior to the formation of inclusions, Ssa2 often accumulated in ring-like structures in the proximity of the nucleolus (Fig 1G). These structures were more abundant in ubp3Δ mutants (Fig 1H). When examining the clearance of GFP-Ubc9ts aggregates in other ubpΔ mutants, we found that ubp3 mutants were worse-off than any other ubpΔ deletion strains, although ubp4Δ, ubp5Δ, and ubp6Δ mutants displayed a modest but significant reduction in their ability to clear GFP-Ubc9ts aggregates (Supplementary Fig S1C).
Recently, it was reported that cAMP levels are elevated in ubp3 mutants leading to hyper-activation of the PKA signaling pathway and that overexpression of PDE2, a phosphodiesterase that hydrolyzes cAMP, counteracts some phenotypes typical of cells lacking UBP3 (Li & Wang, 2013). Therefore, we tested whether overexpression of PDE2 could suppress the prolonged deposition of Ssa2-GFP in JUNQ seen in ubp3 mutants. This was not the case (Supplementary Fig S1D). We conclude that the increased cAMP levels and PKA activity in ubp3 mutants are not the cause of stabilized JUNQ inclusions.
Next we tested whether Ubp3 is also required for counteracting aggregate accumulation during replicative aging by tagging the chromosomal SSA2 allele with GFP in the mother enrichment strain developed by Lindstrom and Gottschling (2009). Wild-type cells accumulated Ssa2-GFP in aggregates to a rather low extent until they reached an age of 16-20 generations whereupon roughly 50% of the cells had at least one Hsp70 containing inclusion (Fig 2A and B). At this stage, the inclusions observed were localized at JUNQ sites (Fig 2A). Later in life, the aged mother cells displayed multiple peripheral aggregates in addition to the juxta-nuclear inclusions (Fig 2C). ubp3 cells, in contrast, accumulated Ssa2-GFP-containing aggregates very early in their replicative life span; JUNQ aggregate was formed already during the first 5 generations, which was followed by the formation of additional peripheral aggregates as aging progressed (Fig 2A–C). The number of cells with multiple aggregates per generation was also higher in ubp3 mutants compared to wild-type cells in all age-matched groups (Fig 2C). Thus, replicative aging is accompanied by the formation of an Hsp70-containing JUNQ inclusion followed by the subsequent accumulation of Hsp70 also in multiple peripheral aggregates. This sequence of events is drastically accelerated in cells lacking UBP3, and accordingly, we found that ubp3 cells display a shortened life span (Fig 2D).
Figure 2. UBP3 counteracts age-induced protein inclusion formation.
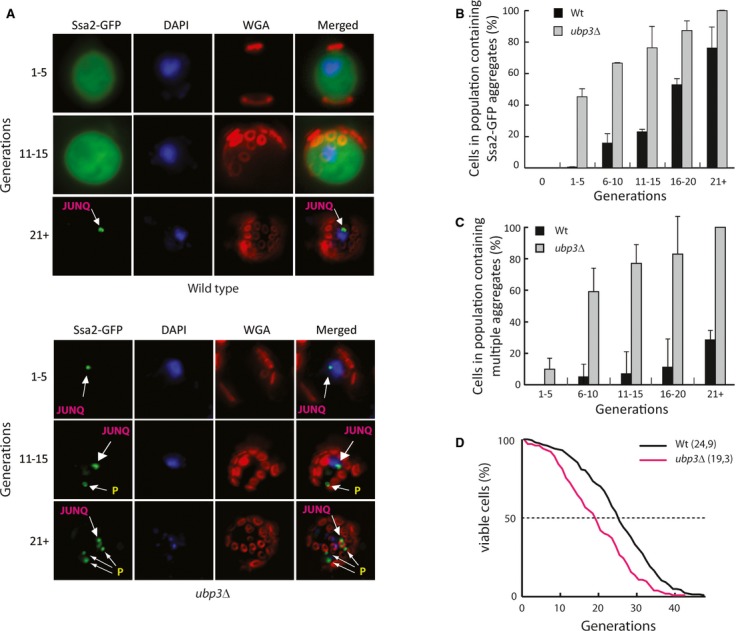
- Images of the mother enrichment strain with a C-terminal GFP tag of the chromosomal SSA2 in the indicated strains. Cells were harvested approximately every five generations and co-stained with DAPI and Alexa Fluor 555 wheat germ agglutinin (WGA) for the visualization of DNA and bud scars, respectively. Images shown are deconvolved maximum intensity projections of Z-stack images taken at the indicated replicative age.
- Quantification of cells from (A) showing the percentage of cells with Ssa2-GFP-containing aggregates in replicatively aged cells. Reported data represent the mean ± s.d. from three independent experiments.
- Quantification of cells in the population containing multiple aggregates (>1) in each replicative age group. Reported data represent the mean ± s.d. from three independent experiments.
- Replicative life span assay (RLS) of wild-type (black line, mean RLS = 24.9, n = 204) and ubp3Δ (red line, mean RLS = 19.3, n = 102). Statistical analysis and P-values can be found in Supplementary Table S4.
Ubp3 overproduction suppresses heat sensitivity, Ubc9ts proteotoxicity, accelerated aging, and the arrested clearance of JUNQ inclusions in cells lacking the major Hsp70s
To test whether UBP3 overproduction, like Ubp3 deficiency, affects management of protein aggregates/inclusions, we constructed a series of strains in which various strong constitutive promoters replaced the native UBP3 promoter of the chromosomal UPB3. Western blotting confirmed Ubp3 overproduction in these strains (Fig 3A). Using these recombinants, Ubp3-dependent suppression of heat sensitivity in cells lacking SSA1 and SSA2 was confirmed in a dosage-dependent manner; the highest level of Ubp3 achieved, using the GPD promoter, allowed some growth of the ssa1 ssa2 double mutant even at 37°C (Fig 3B). Aging, similar to heat stress, causes an increased burden on protein quality control (Erjavec et al, 2007) and we wondered whether elevating Ubp3 levels also counteracted accelerated replicative aging of ssa1 ssa2 cells. Indeed, the replicative life span of ssa1 ssa2 cells was markedly extended by Ubp3 overproduction and almost reached that of wild-type cells (Fig 3C). However, Ubp3 overproduction caused a modest shortening of replicative life span when overproduced in wild-type cells (Supplementary Fig S2A).
Figure 3. UBP3 overproduction suppresses heat sensitivity, Ubc9ts proteotoxicity, accelerated aging, and the arrested clearance of JUNQ inclusions in cells lacking the major Hsp70s.
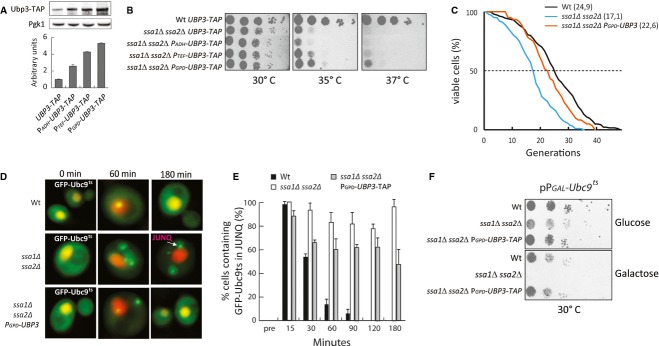
- Western blots and quantification. Ubp3-TAP levels are relative to Pgk1 levels. Wild-type (UBP3-TAP expressed from its native promoter) is set to 1. Data represent the mean ± s.d. from three independent experiments.
- Indicated strains were serially diluted, spotted on YPD plates, and incubated for 2 days at 30°, 35°, and 37°C, respectively.
- RLS of wild-type (black line, mean RLS = 24.9, n = 204), ssa1Δ ssa2Δ (blue line, mean RLS = 17.1, n = 100) ssa1Δ ssa2Δ PGPD-UBP3 (orange line, mean RLS = 22.6, n = 60). Statistical analysis and P-values can be found in Supplementary Table S4. Wild-type RLS data are re-used from Fig 2D as reference.
- Wide-field fluorescence images of indicated strains expressing GFP-Ubc9ts and NLS-tdTomato over time at 37°C.
- Quantification of (D) showing the percentage of cells in each population containing a JUNQ aggregate. Reported data represent the mean ± s.d. from at least three independent experiments.
- The indicated strains were serially diluted and spotted on selective media containing glucose or galactose for GFP-Ubc9ts induction. Plates were incubated for 4–5 days before imaging.
Next, to approach the question of whether Ubp3 overproduction affects JUNQ accumulation, we introduced GFP-Ubc9ts as a reporter (Betting & Seufert, 1996; Kaganovich et al, 2008) in the strains used. We found that the removal of heat-induced Ubc9ts aggregates was severely delayed in the ssa1 ssa2 mutant and that this was counteracted by Ubp3 overproduction (Fig 3D and E). As mentioned above, aggregated Ubc9ts has been shown to become deposited into both peripheral (IPOD) and juxta-nuclear (JUNQ) inclusion bodies (Kaganovich et al, 2008) and we found that it was the latter inclusion that was cleared out faster when Ubp3 was overproduced (Fig 3E), consistent with the fact that Ubp3 deficiency resulted in an increased accumulation of Ubc9ts in JUNQ (Fig 1E). Interestingly, the Ubc9ts protein displayed severe proteotoxicity in cells lacking SSA1 and SSA2 and this toxicity was seen already at low temperatures (30°C). Remarkably, Ubp3 overproduction almost completely suppressed this proteotoxicity (Fig 3F).
The disaggregase Hsp104 fails to associate and resolve protein aggregates in cells lacking SSA1 and SSA2 (Glover & Lindquist, 1998; Winkler et al, 2012). Therefore, we tested whether Ubp3 overexpression suppressed heat sensitivity in these Hsp70-deficient cells by restoring recruitment of Hsp104 to aggregates. This was not the case as Hsp104 failed to associate with aggregates also in the presence of elevated Ubp3 levels (Supplementary Fig S2B).
Ubp3-dependent removal of JUNQ inclusions in Hsp70-deficient cells requires proteasomal activity
Since Ubp3 interacts physically with the 26S proteasome to facilitate degradation of certain substrates (Mao & Smerdon, 2010) and JUNQ inclusions are associated with 26S proteasomes (Kaganovich et al, 2008), we wondered whether Ubp3-dependent suppression of Hsp70 deficiency, including clearance of JUNQ inclusions, is affected by altering the proteasome capacity in the cell. First, we quantified (and confirmed) the levels of the 19S proteasomal subunit Rpt1p in the rpn4Δ mutant reported to exhibit reduced proteasome levels and activity (Wang et al, 2008, Fig 4A). When RPN4 was deleted together with SSA1 and SSA2, overproduction of Ubp3 was no longer effective in enhancing Ubc9ts/JUNQ clearance [Fig 4B (compare Fig 3E)]. The data indicate that aggregate clearance by Ubp3 is intimately linked to proteasome capacity, and we therefore tested, using flow cytometry, whether the stability of Ubc9ts was affected by altering Ubp3 levels (Kaganovich et al, 2008). Indeed, ssa1 ssa2 mutant cells failed to degrade Ubc9ts but overproducing Ubp3 in these cells accelerated degradation to nearly wild-type levels in a proteasome-dependent (Rpn4) manner (Fig 4C). Also, ubp3Δ mutants were slower in degrading Ubc9ts than wild-type cells (Supplementary Fig S2C). Taken together, the results demonstrate that the clearance of Ubc9ts-JUNQ inclusions by the Ubp3 ubiquitin protease is linked to Ubc9ts destruction and requires 26S proteasome activity. Consistent with this, proteasome activity was required for Ubp3 to detoxify Ubc9ts, which was more clearly seen at 37°C (Fig 4D).
Figure 4. UBP3-dependent removal of JUNQ inclusions requires proteasomal activity.
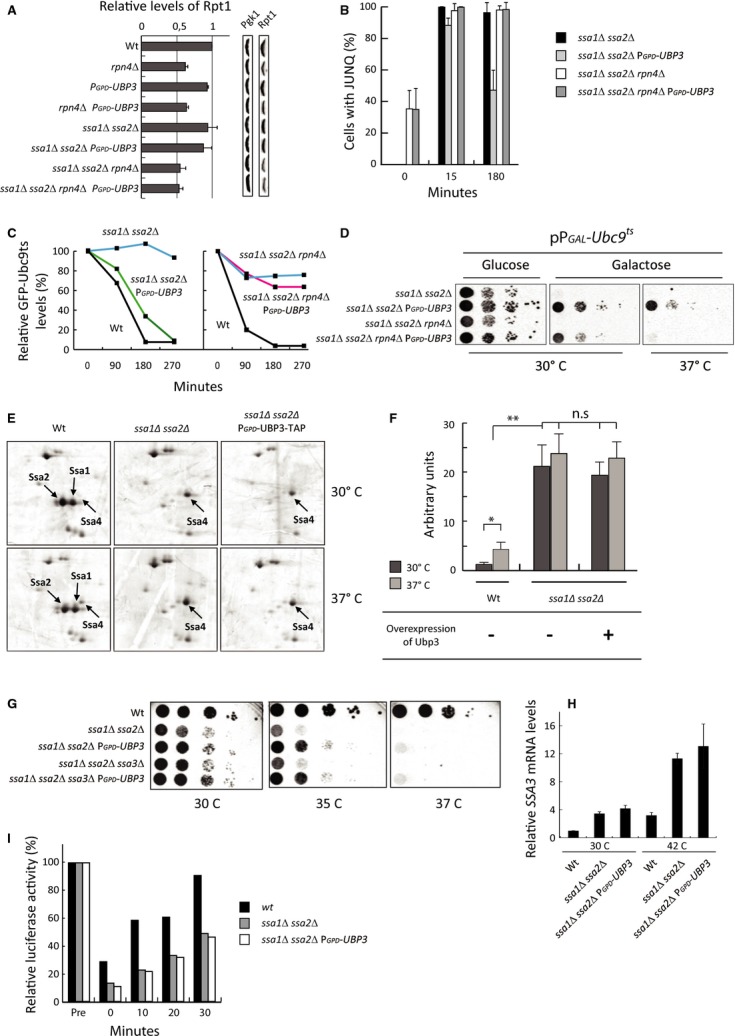
- Quantification of Western blots using antibodies against the 19S subunit protein Rpt1p. Rpt1 values are related to Pgk1. Wild-type levels are set to 1. Reported data represent the mean ± s.d. from at least three independent experiments.
- Quantification of the indicated strains expressing GFP-Ubc9ts 15 and 180 min after shift to 37°C with respect to the percentage of cells displaying a JUNQ aggregate. Reported data represent the mean ± s.d. from at least three independent experiments.
- FACS analysis showing the relative GFP-Ubc9ts signal (levels) over time at 37°C. Representative data from one experiment are shown.
- The indicated strains harboring GFP-Ubc9ts were grown in glucose, serially diluted, and spotted on selective media containing glucose or galactose as indicated. Plates were incubated for 4–5 days at the indicated temperatures prior to imaging.
- 2-D PAGE of the indicated strains and temperatures. Proteins were visualized by silver staining. Ssa1p, Ssa2p, and Ssa4p are indicated by arrows.
- Quantification of silver-stained 2-D gels from (E). Ssa4p levels in the different strains are related to Act1p levels on the same gel and to wild-type Ssa4p at 30°C. Data represent the mean ± s.d. from two to three independent experiments. P-values were obtained by a two-sided t-test with 95% confidence intervals, *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; n.s = P > 0.05. Exact P-values can be found in Supplementary Table S5.
- Indicated strains were serially diluted, spotted on YPD plates, and incubated for 2 days at 30°, 35°, and 37°C, respectively.
- RT-PCR showing relative SSA3 mRNA levels in the indicated strains and temperatures. SSA3 mRNA was normalized to ACT1 mRNA. Wild-type is set to 1. Data represent the mean ± s.d. from three independent experiments.
- Bars represent relative luciferase activity over time. Indicated strains were transformed with the vector pCA837 carrying the gene FLUC-EGFP encoding Firefly luciferase (Gowda et al, 2013). Data shown are from one representative experiment.
It has been reported that SSA4, encoding another cytosolic Hsp70, is induced when SSA1 and SSA2 are absent and that ssa4 is synthetically lethal with ssa1 ssa2 (Werner-Washburne et al, 1987). To determine whether the suppression achieved by Ubp3 overproduction is due to Ubp3 further boosting Ssa4 levels, we quantified the levels of Ssa4 in the strains used. This analysis confirmed that Ssa4 levels are elevated (∽20-fold) in the absence of SSA1 and SSA2 at 30°C (Fig 4E and F). However, Ubp3 overproduction did not significantly increase Ssa4 levels further in the ssa1 ssa2 mutant (Fig 4E and F and Supplementary Table S5). Thus, we conclude that suppression of heat sensitivity by Ubp3 overproduction cannot be explained by elevated levels of Ssa4. Furthermore, overproduction of Ubp3 suppressed reduced viability at elevated temperatures of an ssa1 ssa2 strain also lacking SSA3 and expression of SSA3 was not elevated by Ubp3 overproduction in the ssa1 ssa2 strain background (Fig 4G and H). Finally, we tested whether Ubp3 overproduction boosted general folding activity in an ssa1 ssa2 mutant, using heat-denatured luciferase as a reporter (Gupta et al, 2011), and found that this was not the case (Fig 4I), suggesting that elevated deubiquitination can suppress Ssa1/Ssa2 deficiency in the absence of a compensatory boost in other Hsp70 chaperones and folding activities.
Ubp3 both stabilizes and destabilizes ubiquitin/proteasome system (UPS) substrates in Hsp70-deficient cells
The Ubp3-dependent destruction of Ubc9ts from JUNQ inclusions explains how Ubp3 overproduction overcomes Ubc9ts toxicity in ssa1 ssa2 mutant cells. However, the original model for how Ubp3 suppresses the need for Ssa1/Ssa2 hypothesizes that Ubp3 is doing so by diverting misfolded proteins away from immediate destruction and thereby allowing some folding and sustained residual activity (Baxter & Craig, 1998). To approach this model, we tested whether Ubp3 might actually stabilize other misfolded reporter proteins by fusing them to Leu2 such that growth without leucine (in LEU2 auxotrophs) would be enhanced by stabilizing the protein-Leu2 fusion protein. We used the ssCPY*-Leu2-myc (ΔssCL*) protein, which contains a mutated form of the carboxypeptidase Y (CPY; Stolz & Wolf, 2012) that misfolds in the cytoplasm and is degraded by the UPS due to ubiquitin tagging by the E3 ubiquitin ligase Ubr1 (Eisele & Wolf, 2008). As expected, we found that cells devoid of UBR1 grew better than wild-type cells in the absence of leucine (Fig 5A) and that the ΔssCL* construct was more abundant in ubr1Δ cells (Fig 5B). Next, we tested whether a ubp3Δ deletion affected growth and found that the absence of Ubp3 rendered ΔssCL* cells unable to grow without leucine (Fig 5A). Furthermore, levels of ΔssCL* were drastically reduced in the ubp3Δ deletion mutant (Fig 5B). Overproducing Ubp3 had the opposite effect; cells grew better than wild-type cells (Fig 5A) and ΔssCL* levels were increased (Fig 5B). These data suggest that Ubp3 can deubiquitinate/rescue ΔssCL* and save it from immediate degradation. Next, we tested whether Ubp3 overproduction stabilized ΔssCL* in an ssa1 ssa2 mutant. Ssa1/Ssa2-deficient cells carrying the ΔssCL* fusion displayed similar growth as wild-type cells without leucine (Fig 5C), which is consistent with previously published reports showing that Ssa1 is required for the degradation of different ΔssCPY* species and that their levels remain high in Hsp70-deficient cells (Park et al, 2007). Nevertheless, Ubp3 overproduction in ssa1 ssa2 cells improved growth even further (Fig 5C) and this was accompanied by a retarded degradation rate of the ΔssCPY* fused to GFP (Medicherla et al, 2004; ΔssCG*; Fig 5D). Thus, we conclude that Ubp3 has a dual role in determining protein stability—either stabilizing (i.e., ssCPY* versions) or destabilizing (Ubc9ts) depending on the species of the misfolded protein. We also tested whether other Ubp proteins had an effect on ΔssCL* using the growth assay and found that Ubp2, 6, 8, 14, and 15, like Ubp3, affected the ΔssCL*-dependent growth (Supplementary Fig S2D).
Figure 5. Ubp3 both stabilizes and destabilizes UPS substrates in Hsp70-deficient cells.
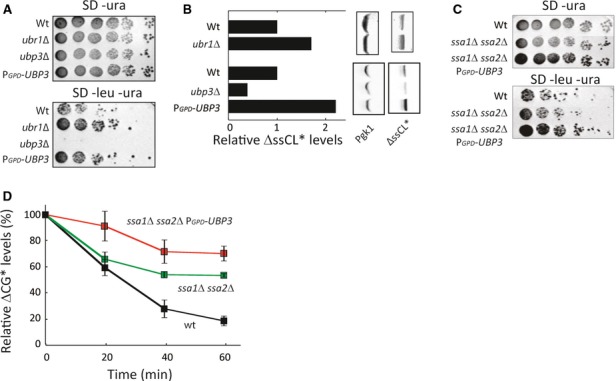
- Cells expressing ΔssCL* under the PRC1 promoter on a centromeric plasmid were spotted in a fivefold serial dilution on indicated media and incubated for 4 days at 30°C.
- Western blots showing ΔssCL* levels related to Pgk1 of logarithmically growing cells of the indicated strains.
- The indicated strains expressing ΔssCL* were spotted in a fivefold serial dilution on indicated media and incubated for 4 days at 30°C.
- FACS analysis showing the relative ΔssCG* signal (levels) over time at 30°C after stopping expression with cycloheximide. Data are represented as the mean ± s.d. of at least three experiments.
Ubp3-dependent deubiquitination results in differential effects depending on the substrate
In view of previous reports (Kvint et al, 2008; Mao & Smerdon, 2010) and the data presented herein about ΔssCPY* species and Ubc9ts, we hypothesized that Ubp3-dependent deubiquitination of substrates may act at different stages toward destruction by the proteasome. If deubiquitination occurs prior to a stage of final “commitment” to destruction, the protein is saved from proteolysis (“rescue pathway”), whereas deubiquitination at the “committed” stage promotes destruction, for example, through facilitating entry into the proteasomal cavity (“destruction pathway”; Verma et al, 2002; Yao & Cohen, 2002; Fig 6A). The predictions of this model are that in the rescue pathway, substrates (like ΔssCPY* species) would display lower ubiquitin levels in cells (e.g., ssa1 ssa2 cells) overproducing Ubp3, while being rapidly committed and degraded in cells lacking Ubp3. In the destruction pathway however, substrates (like Ubc9ts) would become more ubiquitinated in ubp3Δ cells and thus saved from proteolysis, while being rapidly degraded in cells overexpressing Ubp3 and becoming undetectable.
Figure 6. Ubp3-dependent deubiquitination results in differential effects depending on the substrate.
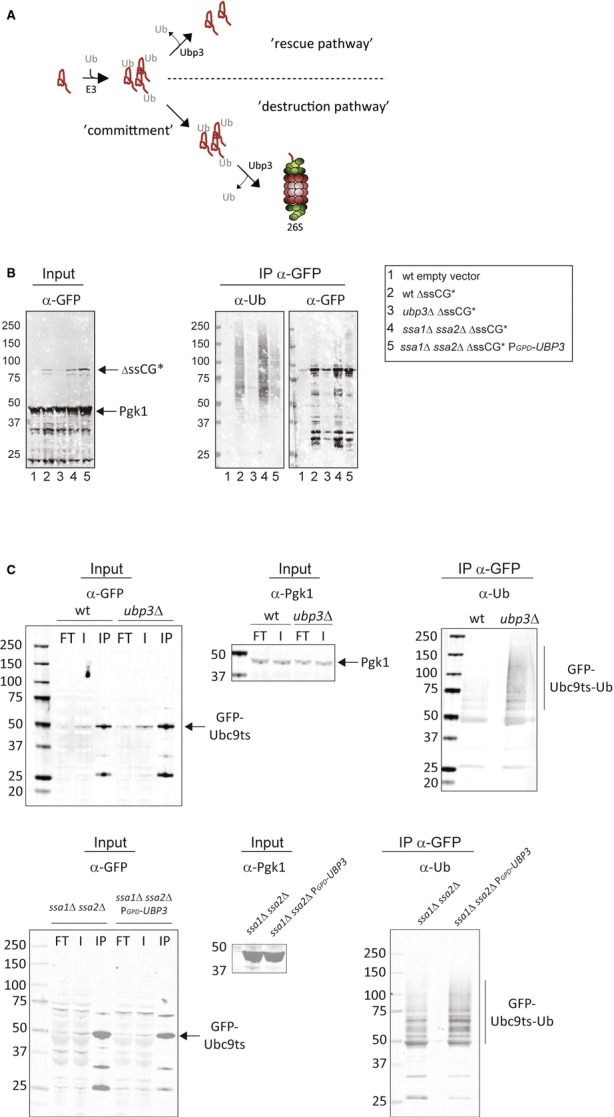
- Illustration of two possible outcomes of deubiquitination by Ubp3p. In one scenario (upper part), Ubp3 rescues a protein from destruction, whereas in another scenario (lower part), the substrate is committed for proteasome-mediated destruction facilitated by Ubp3.
- ΔssCG* was immunoprecipitated from wild-type, ubp3Δ, ssa1Δ ssa2Δ, and ssa1Δ ssa2Δ PGPD-UBP3 strains. A wild-type strain harboring an empty vector served as a control. The immunoblots were analyzed with the indicated antibodies.
- Western blots of immunoprecipitated GFP-Ubc9ts. The same strains as in (B) were analyzed using the indicated antibodies.
Source data are available online for this figure.
To test these predictions, we purified ΔssCG* and Ubc9ts by immunoprecipitation and analyzed their ubiquitin status in the different genetic backgrounds used. This analysis demonstrated that ΔssCG* displayed a reduced ubiquitin status in the Ubp3-overproducing strain, which is consistent with markedly higher levels of the protein in this strain background. In the ubp3 mutant, there was very little ΔssCG* compared to the wild-type strain, most likely due to the fact the ubiquitinated ΔssCG* protein was rapidly destroyed (Figs 5B and 6B). In contrast, the ubiquitin status of Ubc9ts was markedly elevated in a ubp3 mutant (which counteracted degradation), while no significant difference could be observed in the Ubp3-overproducing strain, which facilitated degradation (Fig 6C). These data are consistent with Ubp3 being involved in the deubiquitination of both ΔssCG* and Ubc9ts but that the outcome of this is dependent on the substrate and its stage toward destruction.
Suppression of the heat sensitivity of Hsp70-deficient cells by Ubp3 is due to protein stabilization
Having established that Ubp3-dependent deubiquitination can both stabilize proteins (ssCPY* species) and facilitate their destruction (Ubc9ts), we next asked which of these activities is most important in suppressing the need for Hsp70 during heat stress. The ideas for how this is accomplished posit that Ubp3 is either (i) saving partially aberrant proteins from degradation (allowing residual activity; activity model) or (ii) facilitating degradation of misfolded (and perchance toxic) proteins (toxicity model). A prediction of the activity model is that reducing proteasomal capacity would alleviate the burden of Hsp70 deficiency, while boosting proteasomal activity would further exacerbate Hsp70 deficiency. The toxicity model would predict the exact opposite. To approach these predictions, we genetically altered the proteasomal capacity in wild-type cells and cells deficient for Hsp70 activity. Interestingly, reducing proteasomal levels (by introducing the rpn4Δ deletion) partly suppressed the heat sensitivity of ssa1Δ ssa2Δ cells (Fig 7A). Moreover, based on genetic crossing experiments, we found that boosting proteasomal activity by deleting UBR2 (Wang et al, 2004) was lethal in cells lacking SSA1 and SSA2 (Fig 7B). Remarkably, this lethality was offset by Ubp3 overproduction (Fig 7B). This dataset provides strong genetic support for the activity model proposed by Baxter and Craig (1998) and demonstrates that a key function of Ubp3 in suppressing the need for Hsp70s during a heat shock is to divert protein substrates away from proteasomal destruction.
Figure 7. Elevated proteasome levels are detrimental for viability in strains devoid of SSA1 and SSA2.
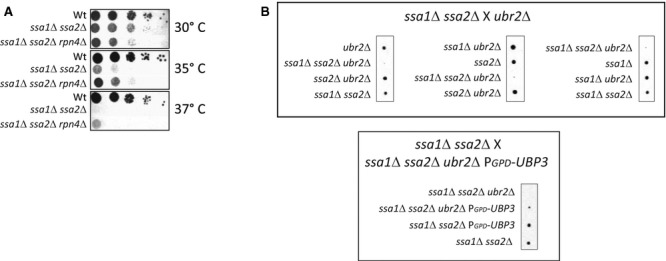
- Spot test of the indicated strains grown in YPD, serially diluted, spotted on YPD plates, and incubated for 2 days at 30°, 35°, and 37°C.
- A representative tetrad dissection of spores generated by mating ssa1 ssa2 with ubr2 (top row) and ssa1Δ ssa2Δ mated with ssa1Δ ssa2Δ ubr2Δ PGPD-UBP3 (bottom row).
Suppression of accelerated aging of Hsp70-deficient cells by Ubp3 requires protein degradation
Since Ubp3 overproduction suppresses accelerated aging of Hsp70-deficient cells, we next tested whether Ubp3 is doing so by the same route as in suppressing heat sensitivity, that is, by saving proteins from proteasomal degradation. In contrast to the effect on heat sensitivity, reducing proteasome activity was detrimental for the longevity of ssa1Δ ssa2Δ cells; that is, aging was further accelerated by removing RPN4 (Fig 8). In addition, overproducing Ubp3 was ineffective in retarding the aging of Hsp70-deficient cells when proteasome activity was limited by deleting RPN4 (Fig 8). These results suggest that Hsp70 deficiency leads to different problems during heat stress and aging, the former of which is exacerbated by proteasomal activity, whereas the latter is counteracted by proteasome activity. Consistent with this, a previous report demonstrated that boosting proteasomal activity (i.e., deletion of UBR2) extends replicative life span (Kruegel et al, 2011).
Figure 8. Suppression of accelerated aging of Hsp70-deficient cells by Ubp3 is proteasome-dependent.
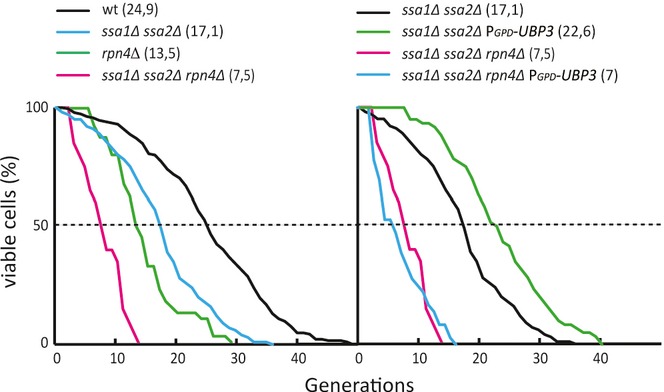
Left graph: RLS data of wild-type (black line, mean RLS = 24.9, n = 204), ssa1 ssa2 (blue line, mean RLS = 17.1, n = 100), rpn4 (green line, mean RLS = 13.5, n = 156), ssa1Δ ssa2Δ rpn4Δ (red line, mean RLS = 7.1, n = 119). Right graph: ssa1Δ ssa2Δ (black line, mean RLS = 17.1, n = 100), ssa1Δ ssa2Δ PGPD-UBP3 (green line, mean RLS = 22.6, n = 60), ssa1Δ ssa2Δ rpn4Δ (red line, mean RLS = 7.1, n = 119), ssa1Δ ssa2Δ rpn4Δ PGPD-UBP3 (blue line, mean RLS = 7.5, n = 60). Wild-type RLS data are re-used from Fig 2D as reference as well as ssa1Δ ssa2Δ and ssa1Δ ssa2Δ PGPD-UBP3 from Fig 3C.
Discussion
In this paper, we have elucidated how elevated Ubp3 levels suppress the need for the major Hsp70 chaperones Ssa1 and Ssa2 (Baxter & Craig, 1998) during both heat stress and aging. In Saccharomyces cerevisiae, there are four cytosolic Ssa chaperones (Ssa1-4). Functional studies on these Hsp70 chaperones suggest that Ssa1 and Ssa2 are more or less functional homologues and that the function of Ssa4 partially overlaps with those of Ssa1 and Ssa2 (Werner-Washburne et al, 1987), while the function of Ssa3 is somewhat obscure. In ssa1 ssa2 cells, SSA4 is strongly upregulated and an ssa4Δ is synthetically lethal with ssa1 ssa2 (Werner-Washburne et al, 1987). A trivial explanation for how Ubp3 overproduction might suppress Ssa1/Ssa2 deficiency, then, could be that Ubp3 further increases the production of Ssa4. However, overproduction of Ubp3 did not induce elevated expression of SSA4 in ssa1 ssa2 cells. In addition, expression of SSA3 was not elevated by Ubp3 overproduction and such overproduction suppressed heat sensitivity also in an ssa1 ssa2 ssa3 triple deletion mutant. Moreover, Ubp3 overproduction in the ssa1 ssa2 mutant did not boost its folding activity, demonstrating that Ubp3 suppresses Ssa1/Ssa2 deficiency by a route other than elevating the levels/activity of residual chaperones. Another heat shock protein central for protein quality control, particularly aggregate clearance, is Hsp104 (Glover & Lindquist, 1998; Schaupp et al, 2007; Wendler et al, 2007) and this disaggregase fails to associate with protein aggregates in cells lacking SSA1 and SSA2 (Winkler et al, 2012). This inability of Hsp104 to associate with aggregates remained in Hsp70-deficient cells when Ubp3 was overproduced (Supplementary Fig S2B), demonstrating that overproducing Ubp3 does not bypass Hsp104's need for SSA1 and SSA2. Instead, the data presented here are in line with a model previously proposed by Baxter and Craig (Baxter & Craig, 1998). The authors proposed that Ubp3 overproduction rescues misfolded, but partly active, proteins required for growth from proteasomal degradation by removing ubiquitin tags. The model highlights that a reduced folding capacity causes a loss of essential protein functions (at least during heat stress) due to an overzealous protein quality control system. We found several lines of experimental support for this model. Firstly, we confirmed that overproduction of Ubp3 suppresses death of ssa1Δ ssa2Δ mutants at elevated temperatures and that Ubp3 does so in a dose-dependent manner. Secondly, the misfolded model protein, ΔssCG*, is deubiquitinated and stabilized by Ubp3 overproduction in an ssa1Δ ssa2Δ mutant. Thirdly, reducing 26S proteasome levels partly suppresses the heat sensitivity of ssa1Δ ssa2Δ cells, whereas boosting proteasomal activity (UBR2 deletion) is lethal in ssa1Δ ssa2Δ cells. Lastly, the synthetic lethality of a ubr2Δ deletion in ssa1Δ ssa2Δ cells is offset by Ubp3 overproduction.
While these data demonstrate that a key function of Ubp3 in suppressing the need for Hsp70s during heat shock is to divert protein substrates away from proteasomal destruction, we found that Ubp3 is also intimately involved in protein destruction and clearance of protein aggregates (Fig 9). This latter activity appears more important in the Ubp3-dependent suppression of accelerated aging. This may be the result of heat and aging generating different problems with respect to protein homeostasis. As ssa1 ssa2 cells subjected to a sudden and transient heat shock do not appear to require protein destruction by the proteasome to recover from such a shock, it appears that no, or very little, toxic protein conformers are formed upon heat stress and/or that such conformers can be easily dealt with by residual chaperone activity. Thus, a central problem in Hsp70-deficient heat-stressed cells might be to maintain protein activity as previously suggested (Baxter & Craig, 1998; Weibezahn et al, 2004). Replicative aging, on the other hand, is characterized by a more progressive decline in protein quality control and the accumulation of protein aggregates (Erjavec et al, 2007) may be caused by a different path than a heat shock-like denaturation of proteins. For example, aging mother cells experience gradually increasing levels of reactive oxygen species (Heeren et al, 2004; Erjavec et al, 2007) that may cause misfolding and aggregation by chemical cross-linking reactions, a modification than can generate toxic conformers that cannot be fixed by chaperones alone. It is also possible that protein aggregation in aging cells is a result of a decline in translational proofreading, which would generate terminally corrupt and aberrant proteins of another kind than heat-denatured ones. In addition, aging causes a progressive decline in 26S proteasome activity (Andersson et al, 2013), which is predicted not only to cause elevated aggregation but also to affect the spatial deposition of misfolded proteins into different compartments for inclusion bodies (Kaganovich et al, 2008): genetically or chemically reducing proteasome activity in heat-stressed cells increases the build-up of terminally damaged proteins in peripheral IPOD aggregates at the expense of 26S proteasome-enriched JUNQ inclusions (Kaganovich et al, 2008). Our data demonstrate that a similar redistribution of misfolded proteins occurs during aging of cells, consistent with the age-related drop in the activity of the ubiquitin-proteasome-dependent system (Andersson et al, 2013). In ubp3Δ cells, JUNQ inclusions appeared much earlier than in wild-type cells during aging and the subsequent formation of peripheral aggregates was drastically accelerated (Fig 2). We suggest that this phenotype is due to the reduced capacity of ubp3Δ cells to facilitate entry of misfolded substrates into the 26S proteasome, which would, at first, cause an expansion of JUNQ inclusions and later a redistribution to peripheral sites of aggregation deposit sites. As the build-up of peripheral aggregates coincides with accelerated aging in ubp3 cells, it is tempting to speculate that these specific aggregates act as aging factors but experimental evidence for or against this notion remains to be obtained. That suppression of accelerated aging by Ubp3 overexpression in ssa1 ssa2 cells required proteasomal activity (Fig 7), is in line with previous results advocating the ubiquitin-proteasome-dependent system as being a bottleneck in life span control (Kruegel et al, 2011). However, it should be noted that direct evidence for damaged/aggregated proteins being bona fide aging factors in yeast is still pending as life span extension by boosting 26S proteasome activity could be due to the destruction of proteins (e.g., regulators of cell cycle progression) other than damaged ones.
Figure 9. A model describing balancing, degradation, and salvation of misfolded proteins by Ubp3.
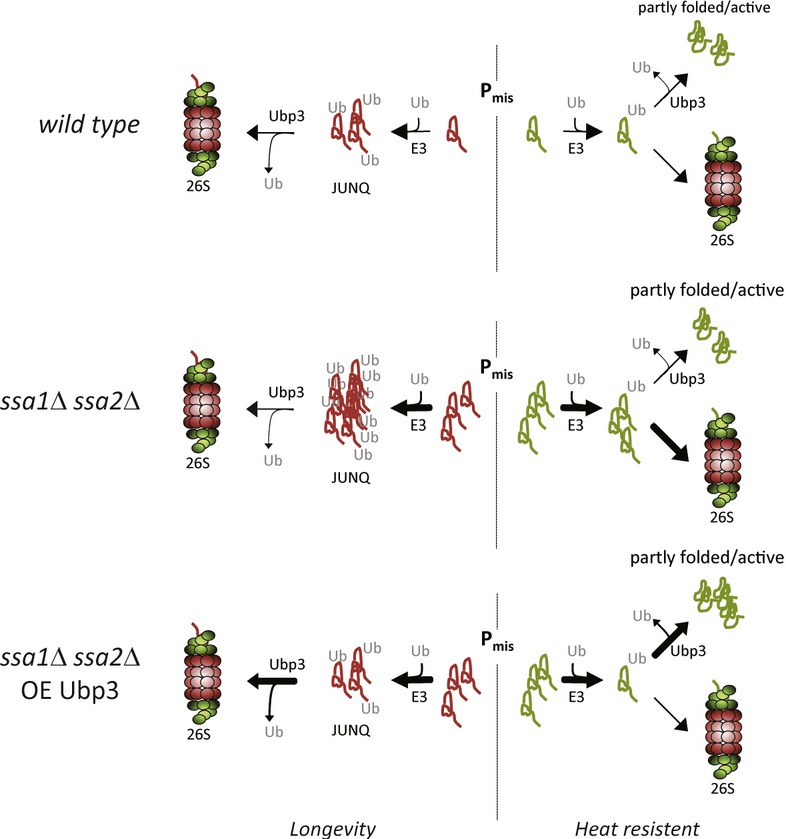
During proteotoxic stress, aberrant proteins are either stabilized or destabilized depending on the protein species. After being ubiquitinated, some misfolded proteins accumulate in JUNQ inclusions whereupon they are resolved/degraded by the 26S proteasome, whereas other misfolded and ubiquitinated proteins may be (transiently) “saved” from destruction by efficient DUB activities. In cells lacking the major Hsp70 chaperones (Ssa1/Ssa2), a larger portion of the proteome becomes misfolded and a concomitant increase in JUNQ inclusions occurs. Ubp3 is key in both facilitating destruction of proteins captured in JUNQ inclusions and saving some misfolded protein species from 26S proteasomal degradation.
The data obtained previously for Rad4 (Mao & Smerdon, 2010) and Rpb1 (Kvint et al, 2008) together with our results concerning ΔssCPY* and Ubpc9ts (this paper) raise the question of how a ubiquitin protease can mechanistically either save or destroy a substrate by the removal of ubiquitin tags. This apparent conundrum may be explained by deubiquitination occurring at different stages toward protein destruction (Fig 6A). The fact that Ubp3 overproduction resulted in elevated levels of deubiquitinated and stabilized ΔssCPY* species is consistent with this deubiquitination occurring at a stage prior to “commitment” for proteasomal destruction. On the other hand, the ubiquitin status of a substrate requiring Ubp3-dependent deubiquitination for entry into the proteasome would “escape” analysis upon Ubp3 overproduction since the deubiquitinated substrate would be rapidly degraded (Fig 6). However, enhanced ubiquitination of such a substrate should be seen in a ubp3Δ cell because the ubiquitinated substrate becomes stabilized (Fig 6A)—this is exactly what we observed for Ubc9ts (Fig 6C). Thus, the data obtained for both ΔssCPY* and Ubc9ts are consistent with Ubp3-dependent deubiquitination effecting different fates of the substrate due to deubiquitination occurring at different stages toward commitment for proteasomal degradation (Fig 6A–C). Follow-up questions are what determines whether Ubp3-dependent deubiquitination occurs at the committed or non-committed stage and do other factors, including other ubiquitin proteases, determine the final outcome of Ubp3-dependent deubiquitination/commitment?
Materials and Methods
Yeast media, plasmids, and strains
Saccharomyces cerevisiae strains and genotypes used in this study are listed in Supplementary Table S1. Deletion mutants were constructed either by PCR-mediated knockout or sporulation. Transformants and dissected spores were verified by PCR. Primers used in this study are listed in Supplementary Table S2. Overexpression strains were constructed by amplifying the ADH, TEF, or GPD promoters from plasmids pYM-N7, pYM-N19, and pYM-N15, respectively, acquired from Euroscarf-PCR toolkit (Janke et al, 2004). The promoter constructs were then inserted replacing the native promoter of the target gene using primers to target insertion directly upstream of the target ORF. The fluorescent proteins used in this study were mainly from Open Biosystems or amplified from GFP-containing plasmids pYM-27 or pYM-28 (Janke et al, 2004). The mCherry C-terminally fused to Ssa1p in this study was amplified from plasmid pBS35 (YRC, NIH). All plasmids used in this study are listed in Supplementary Table S3. For growth in rich media, strains were grown in YPD with 2% glucose. Cells grown in synthetic defined media were grown with yeast nitrogen base + ammonium sulfate and 2% glucose or galactose (w/v). For all experiments using galactose-inducible vectors, expression was shut off by supplementation of glucose to the growth medium.
Fluorescence microscopy and image analysis
All images (unless otherwise stated) were imaged with a Carl Zeiss axiovert 200M wide-field fluorescence microscope with a 100× (NA = 1.4), oil, plan apochromatic correction Zeiss objective. Images were processed with ImageJ (NIH). For deconvolution, the plugin: Iterative deconvolve 3-D was used.
Hsp70 aggregate clearance assay
Cells were grown in YPD (or the appropriate minimal media) to saturation, diluted approximately 50-fold, and then grown to exponential phase. Cultures were then transferred to 42°C for 30 min after which they were transferred back to 30°C. Samples were taken every 30 min as indicated. Samples were centrifuged at 2,700 × g for 1 min, washed and resuspended in PBS prior to imaging.
Confocal imaging
The confocal imaging was performed using a Radiance 2000 confocal system (Bio-Rad), equipped with a Nikon TE300 inverted microscope and a 60× (NA = 1.4), oil, plan apochromatic correction Nikon objective. Images were processed using ImageJ (NIH). Z-stacks were acquired by merging single plane images with 0.5-μm spacing.
Ubc9ts aggregation visualizations
GFP-Ubc9ts aggregation was assayed as described previously (Kaganovich et al, 2008). Briefly, cells were grown in galactose to induce GFP-Ubc9ts (and NLS-tdTomato), shifted to 37°C at which time expression was shut off by the addition of glucose. Cells were harvested roughly every 30 min and resuspended in PBS prior to imaging.
Purification of aged cells
Replicatively aged mother cells were purified as described previously (Lindstrom & Gottschling, 2009). Biotinylated (Pierce) cells were harvested and fixed (formaldehyde final concentration 3.7%) every five generations (every ∽7.5 h) using streptavidin-conjugated paramagnetic beads (Magnabind, Pierce) and a magnetic sorter (Invitrogen). Purified mother cells were stained with DAPI (final concentration 2.5 μg/ml) and Alexa Fluor 555 wheat germ agglutinin (WGA, final concentration 1 μg/ml). Cells were then imaged with a Carl Zeiss axiovert 200M. Z-stack images were processed using maximum intensity projection, and finally, images were deconvoluted appropriately using ImageJ (NIH) plugin Iterative deconvolve 3-D.
Replicative life span assay
A micromanipulator (Singer instruments) was used to remove daughters from mother cells (Kaeberlein et al, 1999). Briefly, cells were grown overnight in YPD, diluted, plated, and allowed to recover on YPD plates before being assayed for RLS which was performed independently at least twice for each strain.
Immunoblotting
Proteins were extracted from pelleted cells washed in ice-cold water. Subsequently, cells were resuspended in 50 μl NaOH (+7% beta-mercaptoethanol) and incubated for exactly 2 min at room temperature (∽25°C). Proteins were then precipitated by the addition of 50 μl TCA (50%) and washed in 250 μl 10 mM Tris–HCl buffer pH 7.5. The extract was then resuspended in 50 μl XT-sample buffer (Bio-Rad) and incubated at 95°C for 5 min. Samples were loaded on SDS–PAGE Criterion XT polyacrylamide precast gels (Bio-Rad) and transferred to a PVDF membrane (Millipore). The blots were saturated in Odyssey blocking buffer (LI-COR) for 1 h at room temperature prior to the addition of primary antibodies. Membranes were washed and incubated with the appropriate secondary antibody (LI-COR, IRdye secondary antibodies). Membranes were scanned on a LI-COR Odyssey scanner, and Western blots were quantified using ImageJ (NIH).
Heat sensitivity assay
Cultures were grown in YPD and diluted in fresh YPD to an optical density (OD) of A600 = 0.1. Cells were then grown to mid-log-phase and serially diluted (tenfold) to OD A600 = 10−1 to 10−5 and spotted on YPD plates that were incubated at the indicated temperatures 2–3 days prior to imaging.
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)
Cell harvesting, protein extraction, and 2D-PAGE were performed as described previously (Maillet et al, 1996; Blomberg, 2002). For the first dimension, strips ranging from pH 3–5.6 were used (GE Healthcare). The second-dimension gels contained 7% polyacrylamide. Total protein was visualized using silver staining. Staining was performed accordingly: fixation in 50% ethanol (v/v) plus 10% acetic acid for 2 h followed by washing in MilliQ water 3 × 20 min. Gels were sensitized in 500 ml DTT solution (5.2 mg/l) for 30 min. Subsequently, gels were incubated for 30 min in 500 ml silver nitrate solution (2 g/l). Gels were washed in 500 ml MilliQ water for 30 s, water poured off and 200 ml sodium bicarbonate (34.7 g/l) with formaldehyde (0.5 g/l) was added until a color development was observed. The solution was discarded and 500 ml fresh sodium bicarbonate with formaldehyde solution was added. Development of gels was stopped after approximately 5–6 min. Silver-stained gels were scanned, aligned, and quantified with ImageJ (NIH). Ssa 1–4 proteins were identified by visual matching with existing yeast 2-D maps.
Quantitative real-time PCR analysis
Total RNA from logarithmically growing cultures was extracted using a hot acid phenol extraction protocol as previously described (Sherman, 1991). The extracted RNA was then isolated a second time using RNeasy mini kits (Qiagen, Valencia, CA) followed by DNase treatment with the Turbo DNA-free kit (Ambion, Austin, TX). 1.5-μg amount of DNase I-treated RNA was subjected to cDNA synthesis, using 0.1 μM strand-specific primers. Primer sequences are available upon request. Next, the DNA product was amplified by standard real-time PCRs using appropriate primers, SYBR green, and the Bio-Rad Q5 system.
Luciferase activity
One OD600 of exponentially growing cells harboring a luciferase construct (Gupta et al, 2011) were harvested and treated according to the manufacturer's instructions (Gold biotechnology D-luciferin, LUCK-100). Briefly, cells were centrifuged for 1 min at 17,900 × g and resuspended in 50 μl yeast suspension buffer (Gold biotechnology, GB-178) supplemented with 0.5 μl beta-mercaptoethanol. Spheroplasts were created using zymolase for 60 min at 30°C. The cell suspension (now spheroplasts) was then collected by centrifugation at 3,500 × g for 5 min. Lysis proceeded by adding 50 μl yeast lysis buffer (Gold biotechnology, GB-178) followed by 3–5 quick inversions and subsequent incubation on ice for 30 min. Finally, lysate was collected by centrifugation for 15 min at 20,800 × g. Luciferase activity was assayed at 25°C using a Synergy 2 plate reader. Assaying was performed according to Gold biotechnology's D-luciferin in vitro protocol.
Immunoprecipitation (IP)
Exponentially growing cultures were harvested (500 or 1000 OD600, ΔssCG*/ Ubc9ts, respectively), lysed with glass beads using a FastPrep (MP Biomedicals) in dilution buffer (ChromoTek) followed by a 20,800 × g centrifugation in order to clear the lysate from debris. For IP, a GFP-Trap A kit (ChromoTek) was used according to the manufacturer's instructions with minor modifications. The IP of GFP-Ubc9ts was conducted for 2–4 h followed by 3 × 20 min wash steps with 1 ml wash buffer of varying NaCl concentrations (100 and 500 mM). For ΔssCG*, the NaCl concentrations were 150, 400, and 150 mM (in that order). Elution of Ubc9ts was performed with 30 μl glycine (200 mM, pH 2.5) according to the manufacturer's instructions. Samples were boiled in loading buffer and prior to Western blotting.
Ubc9ts toxicity
Logarithmically growing cells were serially diluted (tenfold) and spotted on YNB Ura-dropout media with glucose or galactose and incubated for 4–5 days prior to imaging.
Ubc9ts and ΔssCG* stability measurements
ΔssCG* and GFP-Ubc9ts degradation was assayed by FACS (FACSAria, BD Biosciences). For each time point, the percentage of the population emitting a detectable GFP signal above the threshold value (as measured by unstained wild-type cells) was measured. Ubc9ts expression was shut down using glucose, whereas ΔssCG* expression was shut down by the addition of cycloheximide (1.77 mM final). For Ubc9, the NLS-tdTomato signal was monitored to correct for possible dilution of GFP signal via cell proliferation. The NLS-tdTomato signal was stable throughout the full time period. For each time point, 10,000 cells were measured.
Growth test for cells expressing ssCL*
Overnight pre-cultured cells were diluted to an OD of A600 = 0.5 and spotted in a fivefold dilution series on YNB Ura-, or YNB Ura-Leu-media. Plates were incubated at 30°C for 3–4 days.
Acknowledgments
We thank D. Gottschling, C. Dargemont, W. Huh, D. Kaganovich, K. Tatchell, D.H. Wolf, and JR. Glover for providing plasmids and strains used in this study. We thank J. Uhler for suggestions and critical reading of the manuscript. This work was supported by grants from Carl Tryggers Foundation and Wilhelm and Martina Lundgrens Research Foundation to KK and by grants from the Swedish Research Council (VR), the Knut and Alice Wallenberg Foundation (Wallenberg Scholar), and the ERC (Advanced Grant) to TN.
Author contributions
DÖ, FE and KK performed the experiments. DÖ, FE, TN, and KK analyzed the results and participated in the experimental design. DÖ, TN, and KK wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Andersson V, Hanzen S, Liu B, Molin M, Nystrom T. Enhancing protein disaggregation restores proteasome activity in aged cells. Aging. 2013;5:802–812. doi: 10.18632/aging.100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- Baxter BK, Craig EA. Isolation of UBP3, encoding a de-ubiquitinating enzyme, as a multicopy suppressor of a heat-shock mutant strain of S. cerevisiae. Curr Genet. 1998;33:412–419. doi: 10.1007/s002940050354. [DOI] [PubMed] [Google Scholar]
- Betting J, Seufert W. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J Biol Chem. 1996;271:25790–25796. doi: 10.1074/jbc.271.42.25790. [DOI] [PubMed] [Google Scholar]
- Blomberg A. Use of two-dimensional gels in yeast proteomics. Methods Enzymol. 2002;350:559–584. doi: 10.1016/s0076-6879(02)50985-x. [DOI] [PubMed] [Google Scholar]
- Brew CT, Huffaker TC. The yeast ubiquitin protease, Ubp3p, promotes protein stability. Genetics. 2002;162:1079–1089. doi: 10.1093/genetics/162.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew BS, Siew WL, Xiao B, Lehming N. Transcriptional activation requires protection of the TATA-binding protein Tbp1 by the ubiquitin-specific protease Ubp3. Biochem J. 2010;431:391–399. doi: 10.1042/BJ20101152. [DOI] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- D'Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Gowda NK, Kandasamy G, Froehlich MS, Dohmen RJ, Andreasson C. Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc Natl Acad Sci USA. 2013;110:5975–5980. doi: 10.1073/pnas.1216778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Kasturi P, Bracher A, Loew C, Zheng M, Villella A, Garza D, Hartl FU, Raychaudhuri S. Firefly luciferase mutants as sensors of proteome stress. Nat Methods. 2011;8:879–884. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]
- Heeren G, Jarolim S, Laun P, Rinnerthaler M, Stolze K, Perrone GG, Kohlwein SD, Nohl H, Dawes IW, Breitenbach M. The role of respiration, reactive oxygen species and oxidative stress in mother cell-specific ageing of yeast strains defective in the RAS signalling pathway. FEMS Yeast Res. 2004;5:157–167. doi: 10.1016/j.femsyr.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy BK, Schmidt M. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol Cell. 2008;30:498–506. doi: 10.1016/j.molcel.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Y. Ras protein/cAMP-dependent protein kinase signaling is negatively regulated by a deubiquitinating enzyme, Ubp3, in yeast. J Biol Chem. 2013;288:11358–11365. doi: 10.1074/jbc.M112.449751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Gottschling DE. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. 411SI-413SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet I, Lagniel G, Perrot M, Boucherie H, Labarre J. Rapid identification of yeast proteins on two-dimensional gels. J Biol Chem. 1996;271:10263–10270. doi: 10.1074/jbc.271.17.10263. [DOI] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Smerdon MJ. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J Biol Chem. 2010;285:37542–37550. doi: 10.1074/jbc.M110.170175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Cohen M, Dargemont C. The Rsp5 ubiquitin ligase and the AAA-ATPase Cdc48 control the ubiquitin-mediated degradation of the COPII component Sec23. Exp Cell Res. 2010;316:3351–3357. doi: 10.1016/j.yexcr.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaupp A, Marcinowski M, Grimminger V, Bosl B, Walter S. Processing of proteins by the molecular chaperone Hsp104. J Mol Biol. 2007;370:674–686. doi: 10.1016/j.jmb.2007.04.070. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Wolf DH. Use of CPY and its derivatives to study protein quality control in various cell compartments. Methods Mol Biol. 2012;832:489–504. doi: 10.1007/978-1-61779-474-2_35. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, III, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Wang H, Strandin T, Hepojoki J, Lankinen H, Vaheri A. Degradation and aggresome formation of the Gn tail of the apathogenic Tula hantavirus. J Gen Virol. 2009;90:2995–3001. doi: 10.1099/vir.0.012179-0. [DOI] [PubMed] [Google Scholar]
- Wang L, Mao X, Ju D, Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu H, Ju D, Xie Y. Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics. 2008;180:1945–1953. doi: 10.1534/genetics.108.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci USA. 2012;109:15811–15816. doi: 10.1073/pnas.1205829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.