Abstract
The squid Euprymna scolopes has evolved independent sets of tissues capable of light detection, including a complex eye and a photophore or ‘light organ’, which houses the luminous bacterial symbiont Vibrio fischeri. As the eye and light organ originate from different embryonic tissues, we examined whether the eye-specification genes, pax6, eya, six, and dac, are shared by these two organs, and if so, whether they are regulated in the light organ by symbiosis. We obtained sequences of the four genes with PCR, confirmed orthology with phylogenetic analysis, and determined that each was expressed in the eye and light organ. With in situ hybridization (ISH), we localized the gene transcripts in developing embryos, comparing the patterns of expression in the two organs. The four transcripts localized to similar tissues, including those associated with the visual system ~1/4 into embryogenesis (Naef stage 18) and the light organ ~3/4 into embryogenesis (Naef stage 26). We used ISH and quantitative real-time PCR to examine transcript expression and differential regulation in postembryonic light organs in response to the following colonization conditions: wild-type, luminescent V. fischeri; a mutant strain defective in light production; and as a control, no symbiont. In ISH experiments light organs showed down regulation of the pax6, eya, and six transcripts in response to wild-type V. fischeri. Mutant strains also induced down regulation of the pax6 and eya transcripts, but not of the six transcript. Thus, luminescence was required for down regulation of the six transcript. We discuss these results in the context of symbiont-induced light-organ development. Our study indicates that the eye-specification genes are expressed in light-interacting tissues independent of their embryonic origin and are capable of responding to bacterial cues. These results offer evidence for evolutionary tinkering or the recruitment of eye development genes for use in a light-sensing photophore.
Keywords: immunity, microbe, photoreceptor, rhabdomere, retina, vision
1. Introduction
Research on visual systems has revealed remarkable conservation of eye-associated genes throughout much of the animal kingdom. Among such genes are pax6 (paired box gene 6), eya (eyes absent), six (sine oculis), and dac (dachshund); transcription factors that interact in a regulatory network (Fig. 1A) and are critical for eye morphogenesis (Donner and Maas, 2004). Among other locations, these genes are expressed in both simple and complex eyes of diverse taxa, and are often referred to as “eye-specification genes” (e.g. Kumar and Moses, 2001). For example, the well-studied gene pax6, is found in animals with simple pigment-cup eyes (e.g., Platynereis dumerilii, Arendt et al., 2002), invertebrate compound eyes (e.g., Drosophila sp., Halder et al., 1995; Gehring and Ikeo, 1999), and vertebrate camera eyes (e.g., Mus musculus, Donner and Maas, 2004). Although eyes have been studied at the level of gene expression and in an evolutionary context (e.g., see Spady et al., 2005; Harzsch et al., 2006; Porter et al., 2012), few such studies have focused on photophores, light-emitting organs that have ocular attributes (but see Tong et al., 2009; Schnitzler et al., 2012).
Figure 1.
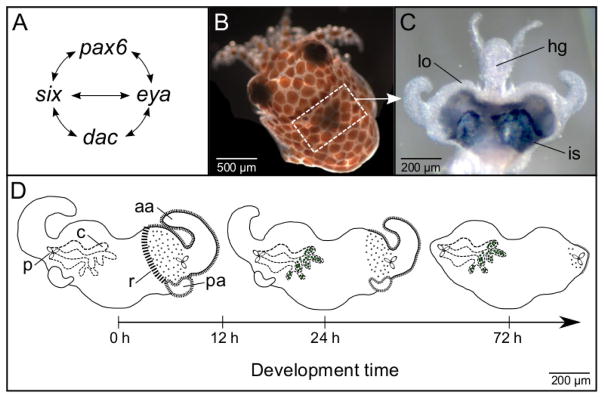
Study system. (A) Regulatory network of the eye-specification genes (adapted from Donner and Maas, 2004). (B) Juvenile E. scolopes. The light organ is located within the white-dash boxed region and is associated with the ink sac, which is visible as a slightly darkened area. (C) Juvenile light organ (lo) surrounded by the ink sac (is) and attached to the hindgut (hg). (D) Early postembryonic development of the juvenile light organ. The juvenile light organ has three pores (p) that enable bacterial symbiont V. fischeri to enter the internal crypt spaces (c) (left side of light organ). V. fischeri are shown as green dots in the crypt spaces at 24 and 72 h of development. The surface tissues of the juvenile light organ include the anterior (aa) and posterior appendages (pa), and the ciliated ridges (r) (right side of light organ), all of which regress during the first several days of development. The developmental time of 12 h post hatching marks the point at which regression of the appendages is no longer reversible and can proceed in the absence of wild-type V. fischeri.
The Hawaiian bobtail squid, Euprymna scolopes (Fig. 1B), is a model invertebrate species with complex eyes and a photophore or ‘light organ’ (Fig. 1C) that houses the luminous bacterial symbiont Vibrio fischeri. The light emitted by V. fischeri matches down-welling moonlight and starlight, and camouflages the squid while active at night in an anti-predatory phenomenon called counter-illumination (McFall-Ngai and Ruby, 1991; Jones and Nishiguchi, 2004; see also Johnsen et al., 2004). The bacterial symbionts, which are harvested anew each generation, enter through pores on either side of the light organ, and ultimately reside along the apical surfaces of polarized epithelia in the crypt spaces (Fig. 1D). Striking anatomical, biochemical, molecular, and physiological similarities exist between the eye and light organ. These similarities include a lens with crystallin proteins (Montgomery and McFall-Ngai, 1992), an analog of the tapetum with ‘reflectin’ proteins (Crookes et al., 2004), genes and proteins involved in phototransduction (Tong et al., 2009), and the physiological ability to respond to light (Tong et al., 2009). In addition, the ink sac, which surrounds a portion of the light organ, functions as both an iris and a choroid (McFall-Ngai and Montgomery, 1990). Such features in the light organ are thought to enable E. scolopes to detect and, in turn, control the light emitted by V. fischeri.
Although the eyes and light organ have notable similarities, they also have key differences. The eyes and light organ are not homologous, developing from ectoderm and mesendoderm, respectively (Montgomery and McFall-Ngai, 1993). The organs also develop at slightly different stages during embryogenesis (Table 1). In addition, light stimuli for the eyes and light organ come from different sources, environmental light and the luminous bacterial symbiont, respectively. Light stimulus is important in the development of both sets of tissues. Environmental light contributes to maturation of the vertebrate eye (e.g., see Grün, 1979; Tian 2004), and therefore might also influence squid eye development. Endogenous light serves as a critical cue for morphogenesis of the E. scolopes light organ (Visick et al., 2000; Koropatnick et al., 2007).
Table 1.
Stages of E. scolopes during embryogenesis and corresponding developmental events occurring in eye and light organ (Montgomery and McFall-Ngai, 1993; Arnold et al., 1972; Lee et al., 2009).
| Stage | Event in Eye | Event in light organ |
|---|---|---|
| 17 | Eye primordia appear as two bilaterally positioned oval ectodermal thickenings | |
| 19 | Eye ectodermal placodes internalize as the annular folds thicken around periphery, forming the optic vesicle | Paired lateral mesoderm of hindgut-ink sac complex begins to proliferate |
| 21 | Retinas are pigmented; paired optic lobe primordia are evident | |
| 22 | Lens primordia are visible | Anterior epithelial appendage and first pair of crypts begins to form |
| 23 | Folds of iris have formed | |
| 24 | Pigmentation of retina continues to increase | Anterior and posterior appendages are evident and partially ciliated; epithelial cells of crypts form extensive brush borders; cells toward site of future ducts form cilia; reflector cells begin to differentiate |
| 26 | Second pair of crypts begins formation; epithelial cells forming crypts appear fully differentiated; many reflector cells differentiated; ink cells are functional | |
| 27 | Eyes can move freely in orbits | |
| 28 | Primary lid covers the eye; patches of iridophores present on the eye | Third pair of crypts begins formation |
| 30 | Three crypts are present on each side; entire lateral surface of each side of light organ is ciliated |
In addition to luminescence, postembryonic development of the light organ depends on other morphogenic signals presented by V. fischeri, which are also molecules associated with animal immune responses. For example, the microbe-associated molecular patterns (MAMPs), specifically the cell-envelope molecules lipopolysaccharide (LPS) and tracheal cytotoxin (TCT), induce morphological changes throughout the first several days of development. These changes include apoptosis within and regression of the juvenile-specific light-organ appendages and ciliated ridges (Fig. 1D), which are surface tissues that facilitate symbiont colonization of the host (Foster et al., 2000; Koropatnick et al., 2004). In other host-microbe associations, immune-related responses contribute to development of host tissues with which the microbes interact (Stappenbeck et al., 2002; Bouskra et al., 2008). Thus, the E. scolopes-V. fischeri system offers an opportunity to examine the effects of microbes on the development of ocular-like tissues in the light organ.
We examined whether the eye-specification genes, pax6, eya, six, and dac, are expressed in the E. scolopes light organ during embryogenesis and whether they are regulated by symbiosis during early postembryonic development. After identifying the four genes in the eye and light organ, we quantified gene expression and differential regulation in the light organ in response to bacterial cues, including luminescence. Our study suggests that the eye-specification genes are present during development of light-interacting tissues, independent of their embryonic origin, and that these genes respond to bacterial cues, including light, that mediate morphogenesis of the light organ.
2. Experimental Procedures
2.1. Sample preparation and generation of full-length cDNA sequences
We used embryos and juveniles produced from mature, wild-caught E. scolopes, which were captured and maintained as previously described (Montgomery and McFall-Ngai, 1993). We stored excised tissues in RNAlater (Life Technologies) at −80°C until use in experiments. Unless otherwise noted, we used a TRIzol Reagent protocol (Life Technologies) for RNA extractions. We determined RNA concentration with a NanoDrop ND-1000 spectrophotometer and tested the quality of the RNA by agarose gel electrophoresis. We removed any contaminating DNA by DNase treatment with Ambion TURBO DNase Kit (Life Technologies).
Although annotation of a previously constructed EST database provided evidence that the pax6, eya, six, and dac transcripts are expressed in light-organ tissues (Chun et al., 2006), we sought to amplify full-length cDNA of all four genes from both eye and light-organ tissues. The full-length pax6 cDNA sequence in E. scolopes had been determined previously from mRNA expressed in the eye, but not from light-organ mRNA (Hartmann et al., 2003). Therefore, we used standard PCR with primers designed to the eye sequence (Table S1) to determine if the same pax6 gene was also present in the light organ. We used RACE-PCR (Rapid Amplification of cDNA Ends – PCR; Invitrogen GeneRacer Kit, Life Technologies) with gene-specific primers designed to the candidate light-organ sequences to generate the eya, six, and dac cDNA sequences (Table S1). To determine whether the eya, six, and dac genes were present in the eye, we used standard PCR with primers designed to sequences known from light organs (Table S1). RNA extracted from ~50 juvenile light organs and 25 eyes was sufficient for the production of RACE-ready cDNA. For additional details see Supplementary Material section S1.1.
2.2. Sequence analysis and phylogenetic reconstructions
Analysis of the derived Pax6, Eya, Six, and Dac protein sequences allowed us to determine whether functional domains are conserved in E. scolopes. We aligned each E. scolopes protein sequence with those of other well-studied exemplar species from the Deuterostomia, Ecdysozoa, and Lophotrochozoa superphyla. The alignments were generated using CLC Sequence Viewer software and analyzed for domain structure using the Pfam website (http://pfam.sanger.ac.uk/). We next conducted phylogenetic analyses to test hypotheses of orthology between E. scolopes proteins and previously published eye-specification proteins. See Supplementary Material section S1.2 for detailed methods.
2.3. Whole-mount in situ hybridization
We examined expression patterns of the pax6, eya, six, and dac gene transcripts in E. scolopes during embryogenesis and in the light organ during early postembryonic development. For both sets of experiments, we used whole-mount colorimetric in situ hybridization (ISH). Details involving specific ISH steps are provided in the Supplementary Material section S1.3 and more briefly here as follows:
Embryonic development
We examined whether the four gene transcripts could be detected in both the eye and light organ of developing E. scolopes embryos. The embryonic stages of development spanned eye and light organ morphogenesis (Naef stages 18, 22, 26, 29, Table 1; Arnold et al., 1972; Lee et al., 2009). To minimize variation in expression patterns due to genetic effects, we tested each gene using samples from a single clutch in this and all subsequent ISH experiments. We performed two replicate experiments each using 6 to 10 embryos per condition per gene (N=14 to 19 total embryos per condition per gene). Reverse transcription-PCR (RT-PCR) confirmed the presence of each transcript in the various tissues at each of the four embryonic stages.
Postembryonic development
We also examined whether expression of the four gene transcripts in the postembryonic light organ were affected by V. fischeri cues. We distributed juvenile squid among four groups: newly hatched, and aposymbiotic (uncolonized) and symbiotic [colonized with either wild-type V. fischeri or with a V. fischeri mutant defective in light production, Δlux (Bose et al., 2008)] after 24 or 72 h of development (Fig. 1D). By convention, we use the term aposymbiotic and symbiotic to refer to animals exposed to environmental bacteria in the absence and presence of V. fischeri, respectively. To determine the effect of bacterial light on gene expression, we compared light organs colonized by wild-type versus Δlux V. fischeri. To determine the effect of bacterial cues other than light production on gene expression (e.g., MAMPs), we compared light organs colonized with wild-type and Δlux V. fischeri versus those left uncolonized. The bacterial colonization protocol was performed as described in a previous study (Ruby and Asato, 1993) using 5,000 colony-forming units of V. fischeri per mL of artificial seawater. We performed two replicate experiments (N=14 to 19 total juvenile light organs per condition per gene).
From preserved juvenile squid we used excised light organs for whole-mount ISH. Following probe-signal development, for each gene, we simultaneously terminated the reactions to determine if transcript expression differed statistically among the four experimental colonization conditions. Specifically, we examined transcript expression in the surface tissues of the light organ, including the appendages and ciliated ridges of the superficial ciliated epithelium, and the pores (see Fig. 1D). We scored labeling of each transcript in each light-organ tissue as presence or absence of signal. Because gene expression by ISH can be difficult to quantify, we did not attempt to score levels of signal other than presence or absence.
2.4. Sectioned light-organ in situ hybridization
Within the light organ, the crypt epithelia are the direct recipient tissues of V. fischeri MAMPs and luminescence. We were interested in whether the four gene transcripts localized to these tissues and whether they might be affected by such cues. By whole-mount ISH, the crypts are difficult to view. Therefore, we performed ISH with sectioned animals to test for transcript signal in the crypts. Preserved juvenile squid were embedded in paraffin wax, sectioned (5 μm), and mounted on slides at the University of Wisconsin, School of Veterinary Medicine Histology Laboratory. We performed two replicate experiments (N=12 to 18 total light organs per condition per gene). See Supplementary Material section S1.3 for remaining details.
2.5. Quantitative Real-Time PCR
We examined the pax6, eya, six, and dac transcripts by quantitative real-time PCR (qRT-PCR) to determine if differential regulation throughout the whole light organ was affected by symbiosis with V. fischeri. The qRT-PCR steps were performed following MIQE guidelines (Bustin et al., 2010). We collected juveniles from several clutches and divided them equally among four groups: newly hatched, 24-h aposymbiotic, and 24-h symbiotic colonized by either wild-type or Δlux V. fischeri. To obtain five independent biological replicates, we performed these collections on five independent days. Symbiotic animals were colonized as noted previously in section 2.3 with wild-type or Δlux V. fischeri. The qRT-PCR reactions of interest contained cDNA from the four light-organ colonization conditions: hatchlings, aposymbiotic, symbiotic with wild-type V. fischeri, and symbiotic with Δlux V. fischeri, each with five biological replicates and three technical replicates. We designed primers (see Table S1) such that amplicon sizes ranged from 81 to 188 bp and efficiencies ranged from 97 to 102%. The PCR reactions were run with a BioRAD CFX connect as follows: (Step 1) 95°C for 3 min; (Step 2) 95°C for 10 s, 60°C for 10 s, 68°C for 15 s (‘Step 2’ for 40 cycles); and (Step 3) 95°C for 10 s followed by a temperature gradient from 65°C to 95°C at 5°C intervals, 5 s per interval. In addition to the four eye-specification genes, we performed similar qRT-PCR reactions for two housekeeping genes, ribosomal 40s and serine HMT (Table S1). We normalized the expression of each eye-specification gene to the geometric mean of the expression of the two housekeeping genes. We used the comparative method (ΔΔCq method) (Pfaffl, 2001) to analyze the data. For additional methods see Supplementary Material section S1.4.
2.6. Analysis of gene transcript expression and regulation data in the light organ
We used the statistical package R (Core Development Team, 2008) to test whether gene transcript expression (whole-mount ISH) and differential regulation (qRT-PCR) in the light organ differed among experimental colonization conditions. We used a Fisher Exact Test to test whether the number of light organs indicating presence versus absence of transcript signal at 24 h post hatching depended on colonization condition (see ISH data). This analysis provided a statistical means for quantifying the effect of bacterial cues on gene expression in light-organ surface tissues (i.e., anterior appendages, posterior appendages, ciliated ridges, pores). The model for gene expression in a given tissue type, Et, as the dependent variable was:
where colonization condition (xc) was the independent variable. We used an analysis of variance (ANOVA) to test whether differential regulation of each eye-specification gene depended on colonization condition and replicate (see qRT-PCR data). The model for normalized expression, R, as the dependent variable was:
where independent variables were colonization condition (xc) and replicate (xr). For each gene, xr represented the five independent biological replicates, each consisting of the three pooled, technical replicates. For both tests, maximum likelihood parameter estimates were β0, β1, and β2. All variables were treated as fixed effects. We performed Tukey pairwise comparisons for Et and R between colonization conditions following the Fisher Exact Test and ANOVA, respectively.
3. Results
3.1. Conservation of eye-specification genes in the eye and light organ
Our experiments yielded evidence for full-length transcripts of the four eye-specification genes, pax6, eya, six, and dac, to be expressed in both the eye and light organ of E. scolopes. The predicted proteins contained conserved domains and functional amino acid residues characteristic of the eye-specification proteins (Fig. S1). In addition, phylogenetic analyses indicated orthology with known eye-specification genes (Figs. S2–S6). See also Supplementary Material sections S2.1 and S2.2.
3.2. Localization of gene transcripts throughout development
We observed transcript expression throughout embryonic and postembryonic development. During embryogenesis the eye develops at an earlier stage than the light organ (Table1). Thus, we predicted that the eye-specification gene transcripts would localize to the region of the head and eye early in development and to the light organ later in development.
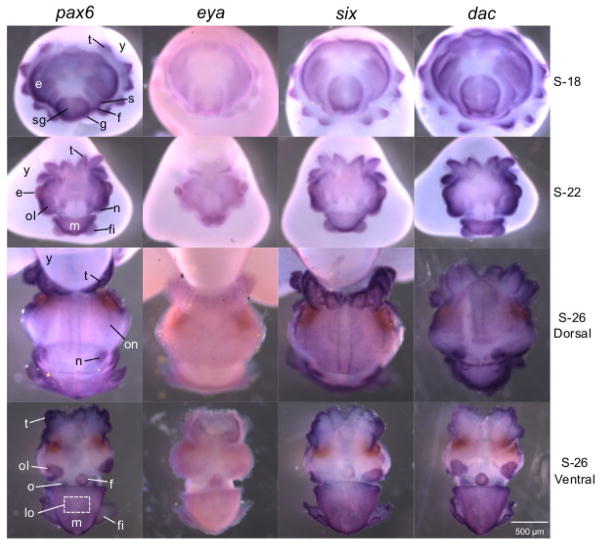
Under the conditions of these experiments, the eye-specification genes generally showed patterns of expression similar to one another in developing embryos, although there was variation in the timing of labeling. By stage 18 (~1/4 of embryogenesis) the eye-specification gene transcripts localized to the eye primordia, and across the skin, including the tentacles and the shell gland, which gives rise to the mantle (Fig. 2, stage 18). We also observed transcript expression in the statocysts, funnel folds, and gill primordia (Fig. 2, stage 18). At stage 22 (~1/2 of embryogenesis) all four transcripts localized to the eye, optic lobe, nuchal organ, mantle, and except for eya, in the tentacles and fins (Fig. 2, stage 22). At stage 26 (~3/4 of embryogenesis) labeling was present in the optic nerve for the pax6 and six transcripts, and in the olfactory organ for the pax6, six, and dac transcripts (Fig. 2, stage 26 dorsal and ventral views). In addition, all transcripts were detectable in the nuchal organ, mantle, distal tentacles, optic lobe, funnel, and fins (Fig. 2, stage 26 dorsal and ventral views). The sense controls showed very low or no detectable labeling (Fig. S7).
Figure 2.
Expression of the pax6, eya, six, and dac gene transcripts in developing embryos by whole-mount in situ hybridization. Embryos were sampled at progressive stages of development, S-18, S-22, and S-26. Tissues showing transcript signal include the eye (e), optic lobe (ol), tentacles (t), statocysts (s), shell gland (sg), mantle (m), funnel fold (f), gill (g), fin (f), nuchal organ (n), optic nerve (on), and olfactory organ (o). The developing light organ is located within the white-dash boxed region, but is covered by the mantle (m). Before hatching the embryo acquires its nutrition from the yolk (y), to which it is attached. See Supplementary Material for sense images (Fig. S7).
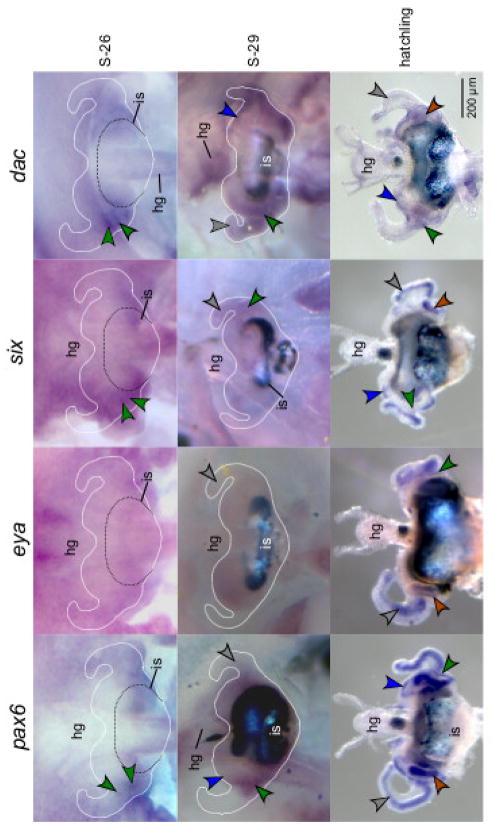
The four eye-specification gene transcripts also localized to the light organ (Fig. 3). Although development of the light organ begins at about stage 22 (Table 1), expression of the pax6, six, and dac transcripts was first discernable by ISH at stage 26 in the pores (Fig. 3, stage 26). At stage 26 labeling of the eya transcript was diffuse across the light organ and the different tissues were difficult to distinguish (Fig. 3, stage 26). By stage 29 all four transcripts localized to the anterior appendages (Fig. 3, stage 29). The pax6, six, and dac transcripts remained visible in the pores (Fig. 3, stage 29). In addition, the pax6 and dac transcripts were expressed in the ciliated ridges (Fig. 3, stage 29). Upon hatching, the four transcripts were detectable in the anterior and posterior appendages, around the pores, and except for eya, in the ciliated ridges (Fig. 3, Hatchlings). The sense controls showed undetectable labeling of these tissues (Fig. S8).
Figure 3.
Light organs showing pax6, eya, six, and dac gene transcript expression by whole mount in situ hybridization at different stages of development (S-26, S-29, hatching). The colored arrows indicate transcript signal in the anterior appendages (grey arrows), posterior appendages (brown arrows), ciliated ridges (blue arrows), and pores (green arrows). The hindgut (hg) develops alongside the light organ and the ink sac (is), visible by stage 29, surrounds a portion of the light organ. For clarity, an outline of the light organ, still attached to the embryos, is indicated at S-26 and S-29. See Supplementary Material for sense images (Fig. S8).
3.3. Effect of symbiosis and symbiont light production on eye-specification genes
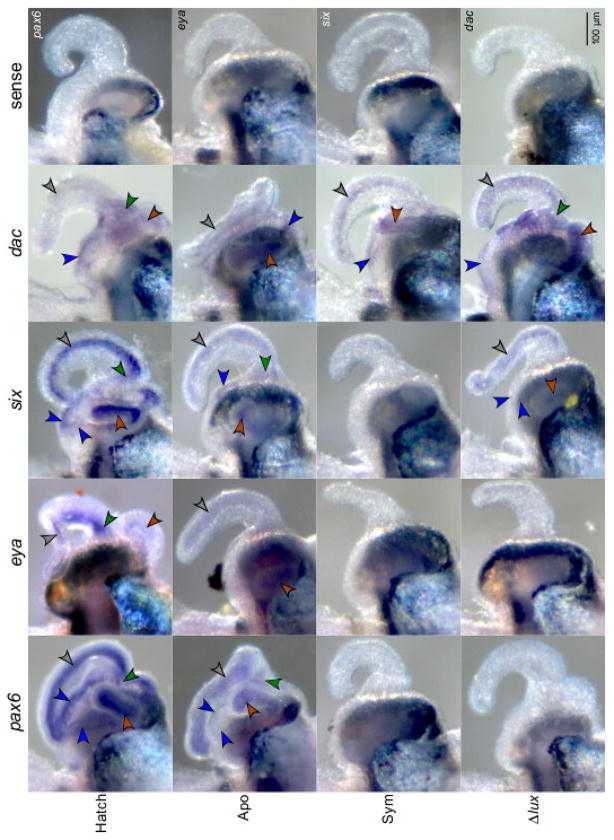
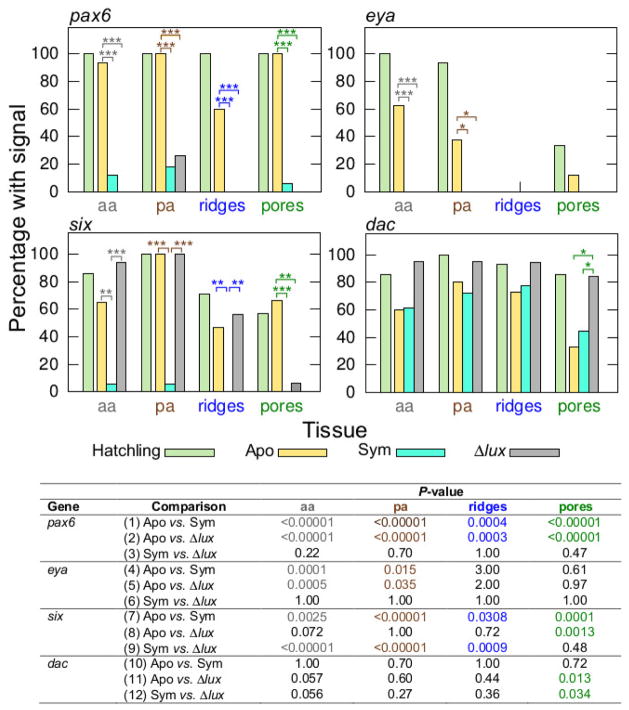
In response to symbiosis, the eye-specification genes showed loss of expression in surface tissues of the light organ, although the region of attenuation varied among the genes. In addition the genes differed in their response to V. fischeri cues. Scoring for the presence/absence of detectable labeling, we observed significant attenuation of labeling (i.e., no detection above background) of the pax6 transcript in symbiotic light organs, both wild type and Δlux V. fischeri, relative to those aposymbiotic (Fisher Exact Test: P<0.00001 all tissue types) (Figs. 4, 5, comparisons 1, 2). Wild-type and Δlux V. fischeri colonized animals did not differ significantly from one another in pax6 expression (Fig. 5, comparison 3). These data indicate an effect of the bacterium (e.g., MAMPs) on pax6 expression. Similar to the pax6 transcript, the eya transcript showed attenuation of labeling in symbiotic light organs with both wild-type and Δlux V. fischeri relative to those aposymbiotic, but with statistical significance only for the anterior (Fisher Exact Test: P<0.00001) and posterior appendages (Fisher Exact Test: P=0.00082) (Figs. 4, 5, comparisons 4, 5). Wild-type and Δlux V. fischeri colonized animals did not differ significantly from one another in eya expression (Fig. 5, comparison 6).
Figure 4.
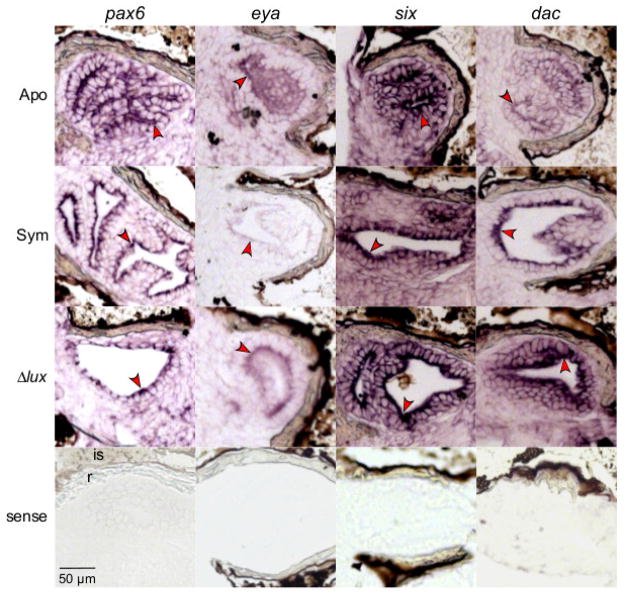
Expression of the pax6, eya, six, and dac gene transcripts in surface tissues of light organs by whole-mount in situ hybridization. Light-organ tissues showing signal development include anterior appendages (grey arrows), posterior appendages (brown arrows), ciliated ridges (blue arrows), and pores (green arrows). Conditions consisted of newly hatched juveniles (‘Hatch’), and 24 h juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with wild-type V. fischeri (‘Sym’), and symbiotic with Δlux V. fischeri (‘Δlux’). Representative light organs exposed to the sense probes are included for each gene transcript.
Figure 5.
Percentage of light organs that indicated pax6, eya, six, and dac expression by whole-mount in situ hybridization in various surface tissues (see Fig. 4). Light-organ tissues of interest include anterior appendages (‘aa’, grey text), posterior appendages (‘pa’, brown text), ciliated ridges (blue text) and pores (green text). The colored text corresponds with the colored arrows in Fig. 4. Conditions consisted of newly hatched juveniles (‘Hatchling’), and 24 h juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with wild-type V. fischeri (‘Sym’), and symbiotic with Δlux V. fischeri (‘Δlux’). Post hoc pairwise comparisons are for 24 h conditions only (***, P<0.001; **, 0.001<P<0.01; *, 0.01<P<0.05). Data consist of two replicates pooled. Sample sizes of light organs for the different conditions (Hatchling, Apo, Sym, Δlux) consisted of the following, respectively, for each transcript: pax6, N=19, 15, 17, 19; eya, N=15, 16, 19, 15; six, N=14, 15, 17, 16; and dac, N=14, 15, 18, 19.
In contrast, we observed significant attenuation of labeling of the six transcript in light organs symbiotic with wild-type, but not Δlux V. fischeri, relative to those aposymbiotic (Fisher Exact Test: Anterior appendages, P<0.00001; posterior appendages, P<0.00001; ciliated ridges, P=0.0003) (Figs. 4, 5, comparisons 7, 9). This trend occurred in all tissues except for the pores. Aposymbiotic light organs and those colonized with Δlux V. fischeri did not differ significantly from one another in six expression in the appendages and ciliated ridges (Fig. 5, comparison 8). Overall, these results indicate an effect of the light production by V. fischeri on six expression.
Finally, the dac transcript showed a trend toward attenuated labeling in aposymbiotic light organs and those symbiotic with wild-type, but not Δlux V. fischeri. However, this trend was only statistically significant at the pores (Fisher Exact Test: P=0.0056) (Figs. 4, 5, comparisons 11, 12). Aposymbiotic light organs and those symbiotic with wild-type V. fischeri did not differ significantly at the pores among colonization conditions (Fig. 5, comparison 10).
In addition to whole-mount ISH, we performed ISH on sectioned animals to determine if the eye-specification gene transcripts were expressed in the light-organ crypts that directly interact with V. fischeri. Using ISH on sectioned animals, all eye-specification gene transcripts localized to the apical surfaces of the crypt epithelia at 24 h post hatching (Fig. 6). However, the presence of transcript signal was not noticeably altered by symbiosis. The four gene transcripts continued to be expressed in the crypt epithelia at 72 h post hatching (Fig. 7), suggesting that these genes serve a role in the tissues where phototransduction develops for at least several days post hatching.
Figure 6.
In situ hybridization of sectioned light organs showing expression of the pax6, eya, six, and dac gene transcripts at 24 h post hatching. Red arrows indicate labeling of the crypt epithelia (antisense). Experimental conditions include light organs of juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with wild-type V. fischeri (‘Sym’), and symbiotic with Δlux V. fischeri (‘Δlux’) at 24 h post hatching. Newly hatched juveniles (‘Hatch’) are provided for comparison. The light organ reflector (r) and ink sac (is) are indicated in the representative pax6 sense image.
Figure 7.
In situ hybridization of sectioned light organs showing expression of the pax6, eya, six, and dac gene transcripts at 72 h post hatching. Red arrows indicate labeling of the crypt epithelia (antisense). Experimental conditions include light organs of juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with wild-type V. fischeri (‘Sym’), and symbiotic with Δlux V. fischeri (‘Δlux’) at 72 h post hatching. The light organ reflector (r) and ink sac (is) are indicated in the representative pax6 sense image.
Transcriptomic studies of the juvenile light organ indicate that certain genes regulated by symbiosis can be detected when whole light organs are sampled (Chun et al., 2008). Thus, we sought to use qRT-PCR to determine whether changes in the eye-specification gene transcripts could be detected by this additional method. However, differential regulation of the pax6, eya, and six transcripts did not differ significantly among the different colonization conditions (ANOVA: pax6, F=0.20, d.f.=3, P=0.90; eya, F=2.6, d.f.=3, P=0.099; six, F=2.2, d.f.=3, P=0.14) (Fig. 8). In contrast, differential regulation of the dac transcript differed significantly among colonization conditions over the whole light organ (ANOVA: F=5.6, d.f.=3, P=0.012) (Fig. 8). The dac transcript was significantly up regulated in light organs colonized by Δlux V. fischeri compared to light organs of newly hatched juveniles (Tukey: P=0.0075). Thus, the dac gene might be involved in differential regulation of other host responses to bacterial light production (Chun et al., 2008).
Figure 8.
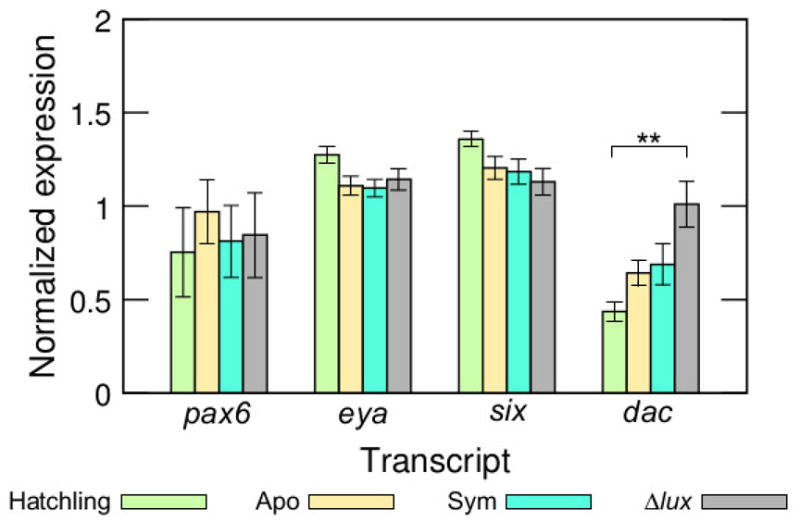
Differential regulation of the pax6, eya, six, and dac transcripts among differently colonized light organs (**, 0.001<P<0.01). Colonization conditions were newly hatched juveniles (‘Hatchling’), and 24 h juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with wild-type V. fischeri (‘Sym’), and symbiotic with Δlux V. fischeri (‘Δlux’). Data consist of five biological replicates and three technical replicates combined per transcript.
4. Discussion
The results of this study indicated that four genes critical for eye development, pax6, eya, six, and dac, are expressed in both the eye and light organ of E. scolopes. The sharing of these genes between the two non-homologous sets of tissues offers evidence for evolutionary tinkering or co-option of a regulatory network at the developmental level (Jacob, 1977; Jacob, 2001; Monteiro, 2011). Within the light organ, gene expression patterns depended on different cues produced by the bacteria. Such differential responses suggest that the eye-specification genes are pivotal members of a greater network of genes regulated by V. fischeri cues, including its luminescence, that affect development of the light organ. Although all four genes have been found in other mollusks, none have been isolated in a photophore. In addition, only pax6 has been described in other cephalopods, including Doryteuthis opalescens, Sepia officinalis, and E. scolopes (Table 2). Thus, this study is the first to identify all four genes in a photophore and to describe the eya, six, and dac genes in a cephalopod.
Table 2.
Tissues in which the pax6, eya, six, and dac genes are expressed in other organisms.
| Gene | Species | Tissues | References |
|---|---|---|---|
| pax6 | Euprymna scolopes | Eye, Optic lobe, Statocysts, Olfactory organ, Tentacles, Mantle, Gills*, Light organ* | Hartmann et al., 2003; Present study* |
| Doryteuthis opalescens | Eye, Optic lobe, Olfactory organ, Tentacles, Mantle | Tomarev et al., 1997 | |
| Sepia officinalis | Eye, Optic lobe, Ganglia, Tentacles, Gills, White body | Navet et al., 2009 | |
| Platynereis dumerilii | Eye | Arendt et al., 2002 | |
| Mus musculus | Eye, Brain, Olfactory epithelium | Duan et al., 2013; Davis and Reed, 1996; Chung et al., 2010 | |
| Drosophila | Eye, Brain, Nerve cord | Quiring et al., 1994 | |
| Danio rerio | Eye, Brain | Amirthalingam et al., 1995; | |
| Schmidtea polychroa | Eye | Martin-Duran et al., 2012 | |
| Dugesia japonica | Eye | Dong et al., 2012 | |
| eya | Euprymna scolopes | Eye, Optic lobe, Statocysts, Olfactory organ, Tentacles, Mantle, Gills, Light organ | Present study |
| Mus musculus | Eye, Ear, Nasal placode, Metanephric mesenchyme | Abdelhak et al., 1997; Xu et al., 1997, 2003; | |
| Drosophila sp. | Muscle | Heanue et al., 1999 | |
| Schmidtea polychroa | Eye | Martin-Durán et al., 2012 | |
| Dugesia japonica | Eye | Dong et al., 2012 | |
| six/so | Euprymna scolopes | Eye, Optic lobe, Statocysts, Olfactory organ, Tentacles, Mantle, Gills, Light organ | Present study |
| Platynereis dumerilii | Eye | Arendt et al., 2002 | |
| Mus musculus | Neural plate, Metanephric mesenchyme | Oliver et al., 1995; Xu et al., 2003 | |
| Drosophila sp. | Muscle | Heanue et al., 1999 | |
| Cladonema radiatum | Eye, Manubrium, Tentacles, Gonads, Umbrella | Stierwald et al., 2004; Graziussi et al., 2012 | |
| Schmidtea polychroa | Eye | Martin-Durán et al., 2012 | |
| Dugesia japonica | Eye | Dong et al., 2012 | |
| dac | Euprymna scolopes | Eye, Optic lobe, Statocysts, Olfactory organ, Tentacles, Mantle, Gills, Light organ | Present study |
| Mus musculus | Eye, Optic cup, Neural crest, Brain, Limb, Otic vesicle, Genitalia | Hammond et al., 1998; Davis et al., 1999; Caubit et al., 1999 | |
| Drosophila sp. | Eye, Limb, Muscle | Mardon et al., 1994; Heanue et al., 1999 | |
| Oryzias latipes | Eye, Central nervous system, Pancreas, Finbuds | Loosli et al., 2002 | |
| Schmidtea polychroa | Eye | Martin-Durán et al., 2012 |
4.1. Eye-specification genes through embryonic development
In general, the eye-specification gene transcripts were expressed in E. scolopes tissues and organs with shared homology to other organisms, especially other cephalopod mollusks (Fig. 2, Table 2). For example, in addition to being expressed in eye-associated tissues (e.g., eye, optic lobe; Fig. 2, Table 2), the four transcripts were expressed within other sensory-related tissues of E. scolopes similar to those of other organisms (e.g., olfactory organ versus nasal placode, statocysts versus inner ear; Fig. 2, Table 2). In the skin of E. scolopes (e.g., over the mantle), the four transcripts might be involved in development of the chromatophores, a group of light-interacting structures that are visible by late embryogenesis (~stages 26 to 30), or ‘photosensitive vesicles’, which occur in the mantle tissue of cephalopods (Young, 1991). Alternatively, there could be other features of the mantle that are regulated by these genes.
Our study also revealed that the eye-specification genes are present in the light-sensing photophore, or light organ, of E. scolopes (Fig. 3). Whereas many animal phyla have autogenic photophores, in which the animal produces the substrates for light production, only cephalopods, fishes, and a tunicate species have bacterial photophores (Herring, 1978). While both types of photophores also feature ocular structures similar to those in the eyes (e.g., anatomical, biochemical, molecular, physiological; Montgomery and McFall-Ngai, 1992; Dove et al., 1993; Nowell et al., 1998; Herring et al., 2002; Cavallaro et al., 2004; Crookes et al., 2004; Tong et al., 2009; Claes et al., 2011a; Claes et al., 2011b), to our knowledge, our study provides the first evidence that the eye-specification genes are present in a photophore. Shared expression between the eye and light organ in E. scolopes suggests that these genes are important for development of light-interacting tissues regardless of their embryonic origin, as the eye and light organ develop from ectodermal and mesendodermal tissues, respectively.
Because the eye-specification genes are associated with development of eyes of distantly related animals, and because the light organ is of much more recent origin (Lindgren et al., 2012), we can infer recent co-option of these genes for use in development of photophores. In addition to the eye-specification genes, phototransduction genes and proteins are identical in the eye and light organ even though the two structures serve different organismal functions (Tong et al., 2009). Whereas in the eyes these proteins respond to environmental light, in the light organ the same proteins likely respond to symbiont light production. The co-option of the eye-specification genes might drive the expression in the light organ of the phototransduction genes, which leads to physiological light responses (Tong et al, 2009).
Our ISH results on sectioned samples showed expression of the eye-specification gene transcripts within the crypt epithelia that are internal to the light organ and directly exposed to V. fischeri cues (Figs. 6, 7). Such expression might indicate that these genes are poised for use in the development of phototransduction for perceiving the light emitted by V. fischeri (Tong et al., 2009). Later in development, the light organ lens forms, which is used for focusing light (Montgomery and McFall-Ngai, 1992; Weis et al., 1993). As the pax6 and six genes are essential in the development of the lens in other organisms (Liu et al., 2006; Huang and Xie, 2010), these genes might also be employed for the same purpose in the E. scolopes light organ.
4.2. Expression of the eye-specification genes in response to bacterial cues
As the juvenile E. scolopes develops postembryonically, discrete surface regions of the light organ undergo distinct morphological changes for which the eye-specification genes could serve a role. Such morphogenesis is accelerated by the presence of V. fischeri (Montgomery and McFall-Ngai, 1994; Doino Lemus and McFall-Ngai, 2000). For example, cells of the anterior and posterior appendages experience apoptosis. This apoptosis leads to regression of the appendages and receding of the ciliated ridges, a process that in wild-type colonized juveniles is visible within 24 h and largely completed within several days post hatching (Fig. 1D). In addition, an initial set of three pores per side of the juvenile light organ eventually coalesces into one pore per side in the adult (McFall-Ngai and Montgomery, 1990). The adult pores and the entire musculature are used in a venting behavior to control the resident population of V. fischeri in the bi-lobed light organ. In the absence of wild-type V. fischeri, the normal morphological changes in these localized surface tissues either fail to occur, such as in aposymbiotic light organs, or proceed at a delayed rate, as in light organs colonized by Δlux V. fischeri (McFall-Ngai et al., 2012).
Our whole-mount ISH results suggested that expression of the pax6 and eya transcripts in surface tissues is affected by V. fischeri cues other than light production (Figs. 4, 5; aposymbiotic versus symbiotic, and aposymbiotic versus Δlux). Such cues likely include microbe-associated molecular patterns (MAMPs), which induce light-organ morphogenesis (Foster et al., 2000; Koropatnick et al., 2004). In addition, MAMPs are potent inducers of immune responses that are conserved across the animal kingdom. Although studies involving the eye-specification genes in host-microbe associations are rare, our findings are in concordance with immune-related literature. For example, proliferation activity and expression of the Pax6 protein is reduced in the cortical progenitors of mouse embryos (Mus musculus, stage E17) following a maternal Poly I:C immunostimulant injection, impacting Pax6-related neurological development in utero (Soumiya et al., 2011). In the Eya protein, threonine-phosphatase activity at the N-terminus regulates immune responses to undigested DNA from apoptotic cells (Okabe et al., 2009; Sander and Blander, 2009). Determining whether analogous residues in the E. scolopes Eya protein are activated during programmed cell death, DNA fragmentation, and regression of the light-organ appendages would be intriguing, although beyond the scope of this study.
Although the pax6 and eya transcripts could be down regulated in direct response to bacterial cues, such an effect could also be triggered by regulators of pax6 and eya. The pax6 gene is known to auto-regulate and to affect the expression of other genes (e.g., eya, six, dac; Fig. 1A). In addition, the notch gene is a known regulator of pax6 expression (Kumar and Moses, 2001; Baker, 2001; Onuma et al., 2002; Mu and Klein 2004), and Notch signaling can control apoptosis (e.g., Drosophila; Koto et al. 2011; Koto and Miura, 2011) and play a key role in inflammatory responses (Kim et al., 2008; Cao et al., 2011). Of particular interest, cultured cells exposed to the MAMP lipopolysaccharide (LPS) exhibit suppressed Notch signaling, and in turn, induce macrophage activity (Kim et al., 2008). Inhibition of Notch signaling also correlates with the production of inflammatory cytokines (e.g., microglial cells; Cao et al., 2011). Previous studies of E. scolopes show LPS to be an essential MAMP that initiates postembryonic development of the light organ in which the appendages undergo regression though cell apoptosis (Foster et al., 2000; Koropatnick et al., 2004). During this process, components of the inflammatory response, including peroxidase, nitric oxide, and NFκB occur in the light organ (Weis 1996; Davidson 2004; Goodson et al., 2005). For example, the presence of V. fischeri MAMPs are required for a symbiosis-induced attenuation of nitric oxide in light-organ tissues (Altura et al., 2011). In E. scolopes, Notch signaling might also be inhibited in response to V. fischeri LPS, enabling inflammatory reactions and apoptosis to occur in the appendages, accompanied by down regulation of the pax6 and eya genes in these tissues. Although testing this prediction would require further investigation, we have identified a candidate notch-delta sequence in the annotated EST database of the E. scolopes light organs.
In contrast to the pax6 and eya transcripts, we found down regulation of the six transcript in response to wild-type, but not Δlux V. fischeri (Figs. 4, 5; aposymbiotic versus wild-type V. fischeri, and Δlux versus wild-type V. fischeri), suggesting that the six transcript responded to bacterial luminescence. As mentioned previously in this section, in the absence of wild-type V. fischeri the light organ fails to develop at a normal rate. Although regression of the appendages occurs in light organs colonized with Δlux V. fischeri, the process proceeds more slowly. Failure of the six gene to be down regulated in the appendages of light organs colonized with Δlux V. fischeri (Figs. 4, 5) might contribute to such delayed regression. As the four eye-specification genes affect one another in a regulatory network (Fig. 1A), down regulation of pax6 and eya expression in the appendages might, with time, induce down regulation of six expression, allowing for the eventual regression of the appendages of light organs colonized with Δlux V. fischeri. Interestingly, in the presence of Δlux V. fischeri, a symbiotic association is initiated with E. scolopes, but does not persist (McFall-Ngai et al., 2012; Heath-Heckmann et al., 2013), suggesting that luminescence, and possibly proper regulation of the eye-specification genes, are critical for the maintenance of the partnership.
Our ISH and qRT-PCR results were not in concordance. This discrepancy might have resulted from opposing developmental or gene-regulatory processes occurring on the surface versus internal crypt epithelia of the light organ. Whole-mount ISH detected the regulation of eye-specification genes in discrete regions of the surface epithelia of postembryonic light organs (Figs. 4, 5), which represent ~10% of the tissue of the whole organ. Discrete regions of the light organ are difficult to subsample for qRT-PCR, because of the sizes and relationships among the light organ tissues. As such, our qRT-PCR results represent amplified message from whole light organs (Fig. 8), including all tissue interacting with bacteria as well as the ink sac, ink gland, and hindgut. A recent transcriptomic study of the light organ, although analyzed at developmental times different from those used in this present study, demonstrated that the interaction of the bacteria with a few epithelial cells causes global changes in transcription throughout the organ (Kremer et al., 2013). Thus, other portions of the organ may experience differential regulation of the eye-specification genes that would obscure the localized changes occurring in the surface epithelia.
4.3. Future studies
Our results suggest several avenues for future study of the convergence of organs that interact with light. For example, the system offers an ideal subject for comparisons of the coordination of the phototransduction and eye-specification genes in the eye and light organ. In addition, the bacterial MAMPs, such as LPS, induce responses in both the mammalian eye (see Pollreisz et al., 2011; Bordone et al., 2012) and squid light organ (for review see McFall-Ngai et al., 2010). Further analyses of the interface between the MAMPs and light-interacting tissues might provide insights into how the immune system tunes the eye in health and disease. From a technical viewpoint, laser capture microdissection of discrete regions of the light organ would likely resolve discrepancies between the ISH and qRT-PCR results.
4.4. Conclusions
Results of this study indicate that four genes essential for eye development, pax6, eya, six, and dac, are expressed in the photophore or light organ of the squid Euprymna scolopes. Such similarities between the eye and light organ provide evidence for evolutionary tinkering at the developmental level and the co-option of genes between sets of non-homologous tissues that differ in organismal function. Whether such conservation of the eye-specification genes between eyes and photophores is unique to E. scolopes or is prevalent throughout the animal kingdom is currently unknown, although the ctenophore Mnemiopsis ledyi also expresses phototransduction genes (Schniztler et al, 2012), which might also be regulated by eye-specification transcription factors. Study systems that involve microbe-driven photophores offer an unusual opportunity to examine the effects of bacteria on the development of ocular structures. We discovered that the eye-specification genes respond to bacterial cues that affect light-organ morphogenesis in E. scolopes, including programmed cell death. Although apoptosis is part of the normal developmental program of the light organ, our results raise questions as to whether microbes and the associated host immune responses affect the development and functioning of light-interacting structures, including highly vascularized tissues such as the eye, in health and disease.
Supplementary Material
Highlights.
The pax6, eya, six, and dac genes are expressed in the eye and photophore of a squid.
Eye and photophore similarities provide an example of evolutionary tinkering.
The eye-specification genes respond to bacterial cues, including luminescence.
Acknowledgments
This project was funded by grants from the National Science Foundation (IOS-0817232) and National Institute of Health (AI-50661) to M. J. M-N. We are especially grateful for laboratory assistance or manuscript comments from M. Altura, R. Augustin, N. Bekiares, E. Heath-Heckmann, B. Krasity, E. Koch, N. Kremer, M. Mandel, A. Pollack-Berti, B. Rader, and two anonymous reviewers. Paraffin sections for in situ hybridization experiments were prepared by S. Mayer and B. Reese at the University of Wisconsin, School of Veterinary Medicine Histology Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ. Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. Cell Microbiol. 2011;13:527–537. doi: 10.1111/j.1462-5822.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirthalingam K, Lorens JB, Sætre BO, Salaneck E, Fjose A. Embryonic expression and DNA-binding properties of zebrafish PAX-6. Biochem Biophys Res Comm. 1995;215:122–128. doi: 10.1006/bbrc.1995.2441. [DOI] [PubMed] [Google Scholar]
- Arendt D, Tessmar K, de Campos-Baptista MI, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development. 2002;129:1143–1154. doi: 10.1242/dev.129.5.1143. [DOI] [PubMed] [Google Scholar]
- Arnold JM, Singley CT, Williams-Arnold LD. Embryonic development and post-hatching survival of the sepiolid squid Euprymna scolopes under laboratory conditions. The Veliger. 1972;14:361–365. [Google Scholar]
- Baker NE. Master regulatory genes; telling them what to do. BioEssays. 2001;23:763–766. doi: 10.1002/bies.1110. [DOI] [PubMed] [Google Scholar]
- Bordone MP, Lanzani MF, López-Costa JJ, Chianelli MS, Franco P, Sáenz DA, Rosenstein RE. Bacterial lipopolysaccharide protects the retina from light-induced damage. J Neurochem. 2012;122:392–403. doi: 10.1111/j.1471-4159.2012.07767.x. [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Bose JL, Rosenberg CS, Stabb EV. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol. 2008;190:169–83. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Kaur C, Wu CY, Lu J, Ling EA. Nuclear factor-kappa β regulates Notch signaling in production of proinflammatory cytokines and nitric oxide in murine BV-2 microglial cells. Neurosci. 2011;192:140–154. doi: 10.1016/j.neuroscience.2011.06.060. [DOI] [PubMed] [Google Scholar]
- Caubit X, Thangarajah R, Theil T, Wirth JU, Nothwang HG, Rüther U, Kraussi S. Mouse dac, a novel nuclear factor with homology to Drosophila dachshund shows a dynamic expression in the neural crest, the eye, the neocortex, and the limb bud. Dev Dyn. 1999;214:66–80. doi: 10.1002/(SICI)1097-0177(199901)214:1<66::AID-DVDY7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Cavallaro M, Mammola CL, Verdiglione R. Structural and ultrastructural comparison of photophores of two species of deep-sea fishes: Argyropelecus hemigymnus and Maurolicus muelleri. J Fish Biol. 2004;64:1552–1567. [Google Scholar]
- Chun CK, Scheetz TE, de Fatima Bonaldo M, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, Janssens B, Kimbell JL, Koropatnick TA, Kucaba T, Smith C, Stewart JJ, Tong D, Troll JV, Webster S, Winhall-Rice J, Yap C, Casavant TL, McFall-Ngai MJ, Soares MB. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo M, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ. Effects of colonization, luminescence and autoinducer on global host transcription in the developing squid-vibrio symbiosis. Proc Natl Acad Sci USA. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Kim CT, Jung YH, Lee NS, Jeong YG. Early cerebellar granule cell migration in the mouse embryonic development. Anat Cell Biol. 2010;43:86–95. doi: 10.5115/acb.2010.43.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes JM, Sato K, Mallefet J. Morphology and control of photogenic structures in a rare dwarf pelagic lantern shark (Etmopterus splendidus) J Exp Mar Biol Ecol. 2011a;406:1–5. [Google Scholar]
- Claes JM, Kronstrom J, Holmgren S, Mallefet J. GABA inhibition of luminescence from lantern shark (Etmopterus spinax) photophores. Comp Biochem Physiol C-Toxicol Pharmacol. 2011b;153:231–236. doi: 10.1016/j.cbpc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Crookes WJ, Ding LL, Huang QL, Kimbell JR, Horwitz J, McFall-Ngai MJ. Reflectins: the unusual proteins of squid reflective tissues. Science. 2004;303:235–238. doi: 10.1126/science.1091288. [DOI] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Shen W, Heanue TA, Mardon G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev Genes Evol. 1999;209:526–536. doi: 10.1007/s004270050285. [DOI] [PubMed] [Google Scholar]
- Doino Lemus J, McFall-Ngai MJ. Alterations in the proteome of the Euprymna scolopes light organ in response to symbiotic Vibrio fischeri. Appl Environ Microbiol. 2000;66:4091–4097. doi: 10.1128/aem.66.9.4091-4097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Maas RL. Conservation and non-conservation of genetic pathways in eye specification. Intl J Dev Biol. 2004;48:743–53. doi: 10.1387/ijdb.041877ad. [DOI] [PubMed] [Google Scholar]
- Dong Z, Yuwen Y, Wang Q, Chen G, Liu D. Eight genes expression patterns during visual system regeneration in Dugesia japonica. Gene Exp Patterns. 2012;12:1–6. doi: 10.1016/j.gep.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Dove S, Horwitz J, McFall-Ngai M. A biochemical characterization of the photophore lenses of the midshipman fish, Porichthys notatus Girard. J Comp Physiol A. 1993;172:565–572. doi: 10.1007/BF00213679. [DOI] [PubMed] [Google Scholar]
- Duan D, Fu Y, Paxinos G, Watson C. Spatiotemporal expression patterns of Pax6 in the brain of embryonic, newborn, and adult mice. Brain Struct Funct. 2013;218:353–372. doi: 10.1007/s00429-012-0397-2. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Goodson MS, Kojadinovic M, Troll JV, Scheetz TE, Casavant TL, Soares MB, McFall-Ngai MJ. Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziussi DF, Suga H, Schmid V, Gehring WJ. The “eyes absent” (eya) gene in the eye-bearing hydrozoan jellyfish Cladonema radiatum: conservation of the retinal determination network. J Exp Zool B Mol Dev Evol. 2012;318:257–267. doi: 10.1002/jez.b.22442. [DOI] [PubMed] [Google Scholar]
- Grün G. Light-induced acceleration of retina development in a mouth-brooding teleost. J Exp Zool. 1979;208:291–302. doi: 10.1002/jez.1402080305. [DOI] [PubMed] [Google Scholar]
- Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. New perspectives on eye evolution. Curr Opin Genet Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in the eye and limb. Mech Dev. 1998;74:121–131. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- Hartmann B, Lee PN, Kang YY, Tomarev S, de Couet HG, Callaerts P. Pax6 in the sepiolid squid Euprymna scolopes: evidence for a role in eye, sensory organ and brain development. Mech Dev. 2003;120:177–83. doi: 10.1016/s0925-4773(02)00456-2. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Vilpoux K, Blackburn DC, Platchetzki D, Brown NL, Melzer R, Kempler KE, Battelle BA. Evolution of arthropod visual systems: development of the eyes and central visual pathways in the horseshoe crab Limulus polyphemus Linnaeus, 1758 (Chelicerata, Xiphosura) Dev Dyn. 2006;235:2641–2655. doi: 10.1002/dvdy.20866. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio. 2013;4:e00167–13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring PJ. Bioluminescence in action. Academic Press; London: 1978. p. 570. [Google Scholar]
- Herring PJ, Dilly PN, Cope C. The photophores of the squid family Cranchiidae (Cephalopoda : Oegopsida) J Zool. 2002;258:73–90. [Google Scholar]
- Huang Y, Xie L. Expression of transcription factors and crystallin proteins during rat lens regeneration. Mol Vis. 2010;16:341–52. [PMC free article] [PubMed] [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Jacob F. Complexity and tinkering. Ann N Y Acad Sci. 2001;929:71–73. doi: 10.1111/j.1749-6632.2001.tb05708.x. [DOI] [PubMed] [Google Scholar]
- Johnsen S, Widder EA, Mobley CD. Propagation and perception of bioluminescence: factors affecting counterillumination as a cryptic strategy. Biol Bull. 2004;207:1–16. doi: 10.2307/1543624. [DOI] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Kim MY, Park JH, Mo JS, Ann EJ, Han SO, Baek SH, Kim KJ, Im SY, Park JW, Choi EJ, Park HS. Downregulation by lipopolysaccharide of Notch signaling, via nitric oxide. J Cell Sci. 2008;121:1466–1476. doi: 10.1242/jcs.019018. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- Koto A, Kuranaga E, Miura M. Apoptosis ensures spacing pattern formation of Drosophila sensory organs. Curr Biol. 2011;21:278–287. doi: 10.1016/j.cub.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Koto A, Miura M. Who lives and who dies. Role of apoptosis in quashing developmental errors. Comm Integ Biol. 2011;4:495–497. doi: 10.4161/cib.4.4.15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EA, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Lee PN, McFall-Ngai MJ, Callaerts P, de Couet HG. Embryonic development of the Hawaiian bobtail squid (Euprymna scolopes) Cold Spring Harb Protoc. 2009;4:1426–1435. doi: 10.1101/pdb.ip77. [DOI] [PubMed] [Google Scholar]
- Lindgren AR, Pankey MS, Hochberg FG, Oakley TH. A multi-gene phylogeny of Cephalopoda supports convergent morphological evolution in association with multiple habitat shifts in the marine environment. BMC Evol Biol. 2012;12:129. doi: 10.1186/1471-2148-12-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Mardon G, Wittbrodt J. Cloning and expression of medaka Dachshund. Mech Dev. 2002;112:203–206. doi: 10.1016/s0925-4773(01)00649-9. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Martín-Durán JM, Monjo F, Romero R. Morphological and molecular development of the eyes during embryogenesis of the freshwater planarian Schmidtea polychroa. Dev Genes Evol. 2012;222:45–54. doi: 10.1007/s00427-012-0389-5. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Montgomery MK. The anatomy and morphology of the adult bacterial light organ of Euprymna scolopes Berry (Cephalopoda: Sepiolidae) Biol Bull. 1990;179:332–339. doi: 10.2307/1542325. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A. Gene regulatory networks reused to build novel traits. Bioessays. 2011;34:181–186. doi: 10.1002/bies.201100160. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. The muscle-derived lens of a squid bioluminescent organ is biochemically convergent with the ocular lens. Evidence for recruitment of aldehyde dehydrogenase as a predominant structural protein. J Biol Chem. 1992;267:20999–21003. [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Sem Cell Dev Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Navet S, Andouche A, Baratte S, Bonnaud L. Shh and Pax6 have unconventional expression patterns in embryonic morphogenesis in Sepia officinalis (Cephalopoda) Gene Expr Patterns. 2009;9:461–467. doi: 10.1016/j.gep.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Nowell MS, Shelton PMJ, Herring PJ. Cuticular photophores of two decapod crustaceans, Oplophorus spinosus and Systellaspis debilis. Biol Bull. 1998;195:290–307. doi: 10.2307/1543141. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature Letters. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- Oliver G, Wehr R, Jenkins NA, Copeland NG, Cheyette BN, Hartenstein V, Zipursky SL, Gruss P. Homeobox genes and connective tissue patterning. Development. 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc Natl Acad Sci. 2002;99:2020–2025. doi: 10.1073/pnas.022626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollreisz A, Rafferty B, Kozarov E, Lalla E. Klebsiella pneumoniae induces an inflammatory response in human retinal-pigmented epithelial cells. Biochem Biophys Res Commun. 2011;418:33–37. doi: 10.1016/j.bbrc.2011.12.102. [DOI] [PubMed] [Google Scholar]
- Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW, Robinson PR. Shedding new light on opsin evolution. Proc Biol Sci. 2012;279:3–14. doi: 10.1098/rspb.2011.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Sander LE, Blander JM. Innate Immune Cells Cast an Eye on DNA. J Mol Cell Biol. 2009;1:77–79. doi: 10.1093/jmcb/mjp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler CE, Pang K, Powers ML, Reitzel AM, Ryan JF, Simmons D, Tada T, Park M, Gupta J, Brooks SY, Blakesley RW, Yokoyama S, Haddock SH, Martindale MQ, Baxevanis AD. Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biol. 2012;21:10, 107. doi: 10.1186/1741-7007-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumiya H, Fukumitsu H, Furukawa S. Prenatal immune challenge compromises the normal course of neurogenesis during development of the mouse cerebral cortex. J Neurosci Res. 2011;89:1575–1585. doi: 10.1002/jnr.22704. [DOI] [PubMed] [Google Scholar]
- Spady TC, Seehausen O, Loew ER, Jordan RC, Kocher TD, Carleton KL. Adaptive molecular evolution in the opsin genes of rapidly speciating cichlid species. Mol Biol Evol. 2005;22:1412–1422. doi: 10.1093/molbev/msi137. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierwald M, Yanze N, Bamert RP, Kammermeier L, Schmid V. The Sine oculis/Six class family of homeobox genes in jellyfish with and without eyes: development and eye regeneration. Dev Biol. 2004;274:70–81. doi: 10.1016/j.ydbio.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Tian N. Visual experience and maturation of retinal pathways. Vis Res. 2004;44:3307–3316. doi: 10.1016/j.visres.2004.07.041. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J. Squid Pax-6 and eye development. Proc Natl Acad Sci USA. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong D, Rozas NS, Oakely TH, Mitchell J, Colley NJ, McFall-Ngai MJ. Evidence for Light Perception in a Bioluminescent Organ. Proc Natl Acad Sci USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bact. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis VM, Montgomery MK, McFall-Ngai MJ. Enhanced production of ALDH-like protein in the bacterial light organ of the sepiolid squid Euprymna scolopes. Biol Bull. 1993;184:309–321. doi: 10.2307/1542449. [DOI] [PubMed] [Google Scholar]
- Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Woo I, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JZ. Light has many meanings for cephalopods. Vis Neurosci. 1991;7:1–12. doi: 10.1017/s0952523800010907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.