Significance
Many microtubule (MT) plus-end regulators have been described, but regulation of MT minus-ends remains poorly understood. Recently, the related calmodulin-regulated spectrin-associated proteins (CAMSAPs) CAMSAP2/3 (vertebrate) and Patronin (Drosophila) were shown to bind minus-ends, but their effects on MT dynamics in vitro have not been studied. Here, by using in vitro assays, we show that minus-end binding and stabilization are properties of all CAMSAPs. However, their effects on minus-end growth and decoration are variable among different members of this family. Together, these studies solidify the role of CAMSAPs/Patronin as MT minus-end regulatory proteins and reveal how evolution has tuned their activities to affect minus-ends in distinct ways.
Keywords: tubulin polymerization, cytoskeletal regulation, TIRF microscopy
Abstract
The microtubule (MT) cytoskeleton plays an essential role in mitosis, intracellular transport, cell shape, and cell migration. The assembly and disassembly of MTs, which can occur through the addition or loss of subunits at the plus- or minus-ends of the polymer, is essential for MTs to carry out their biological functions. A variety of proteins act on MT ends to regulate their dynamics, including a recently described family of MT minus-end binding proteins called calmodulin-regulated spectrin-associated protein (CAMSAP)/Patronin/Nezha. Patronin, the single member of this family in Drosophila, was previously shown to stabilize MT minus-ends against depolymerization in vitro and in vivo. Here, we show that all three mammalian CAMSAP family members also bind specifically to MT minus-ends and protect them against kinesin-13–induced depolymerization. However, these proteins differ in their abilities to suppress tubulin addition at minus-ends and to dissociate from MTs. CAMSAP1 does not interfere with polymerization and tracks along growing minus-ends. CAMSAP2 and CAMSAP3 decrease the rate of tubulin incorporation and remain bound, thereby creating stretches of decorated MT minus-ends. By using truncation analysis, we find that somewhat different minimal domains of CAMSAP and Patronin are involved in minus-end localization. However, we find that, in both cases, a highly conserved C-terminal domain and a more variable central domain cooperate to suppress minus-end dynamics in vitro and that both regions are required to stabilize minus-ends in Drosophila S2 cells. These results show that members of the CAMSAP/Patronin family all localize to and protect minus-ends but have evolved distinct effects on MT dynamics.
Microtubules (MTs) are cellular polymers that are important for a variety of functions, including cargo transport and mitotic spindle formation. MTs are composed of dimers of α- and β-tubulin that assemble head-to-tail, creating a polar protofilament. Protofilaments then assemble laterally to form the canonical 13-protofilament MT structure, with β-tubulin exposed at fast-growing plus-ends and α-tubulin exposed at slow-growing minus-ends (1). MTs exhibit an intriguing property termed “dynamic instability,” whereby the polymer can abruptly switch between episodes of net growth and shrinkage (2). The rates of growth and shrinkage as well as the frequency of transitions between these two states are regulated by numerous MT-associated proteins, many of which bind to the ends of the polymer (3, 4).
The dynamics of MT plus-ends are regulated by a well-characterized network of plus-end tracking proteins (+TIPs) (5). End-binding proteins recognize a tubulin conformation unique to the growing ends of MTs and can affect the dynamics of plus-ends by intrinsically altering the structure of the MT end (6–8) as well as recruiting other interacting proteins (9). In contrast, TOG domain-containing proteins, such as XMAP215, promote MT growth and have been suggested to act as MT “polymerases” (10, 11). Conversely, kinesin-13s [e.g., mitotic centromere-associated kinesin (MCAK) from hamster] increase instability of MT ends, leading to increased catastrophe frequency (12, 13). Thus, regulation of these and other +TIPs can dramatically affect the stability and turnover of the MT network (14, 15).
In comparison with the well-characterized +TIPs, much less is known about regulation of MT minus-ends. In many cells, minus-ends in vivo are anchored at the centrosome by the γ-tubulin ring complex (γ-TuRC). However, cells such as epithelial cells and neurons have noncentrosomal MT arrays, and many mitotic spindle MTs are not directly connected to centrosomes. Minus-ends that are not connected to the centrosome appear to be highly stable, in contrast to the behavior of minus-ends composed of pure tubulin. For example, newly created minus-ends formed by breakage or laser severing tend to neither grow nor shrink, whereas newly created plus-ends tend to rapidly depolymerize (16–20). It is unclear how this stability is mediated and whether minus-end stability is regulated to control MT turnover in cells.
Previous work in our laboratory identified the Drosophila protein Patronin (from the Latin patronus, protect) in a whole-genome RNAi screen for mitotic spindle formation (21) (originally named ssp4) and showed that this protein binds to and protects the MT minus-end against depolymerization by kinesin-13 in vitro and in vivo (22). Further work in flies has shown that Patronin antagonizes kinesin-13 during mitosis and that regulation of Patronin activity facilitates spindle elongation in anaphase B (23). In mammals, Takeichi and coworkers (24, 25) have shown that a homologous protein, which they termed Nezha, anchors minus-ends of MTs in cell–cell adherens junctions. The protein family was also identified through a spectrin binding activity and named calmodulin-regulated spectrin-associated protein (CAMSAP) (26, 27). Bioinformatic analyses suggest that this protein family first evolved in metazoans; invertebrates possess a single gene whereas vertebrates possess three CAMSAP genes (CAMSAP1, CAMSAP2, and CAMSAP3/Nezha; Fig. 1A) (24, 26). Recent work has demonstrated that CAMSAP2 and CAMSAP3 bind MT minus-ends and that depletion of these proteins causes a reduction in MT numbers and changes the MT organization in epithelial cells (28, 29). The Caenorhabditis elegans homolog of Patronin/CAMSAP (PTRN-1) stabilizes MTs in neurons and promotes neurite and synapse stability (30, 31).
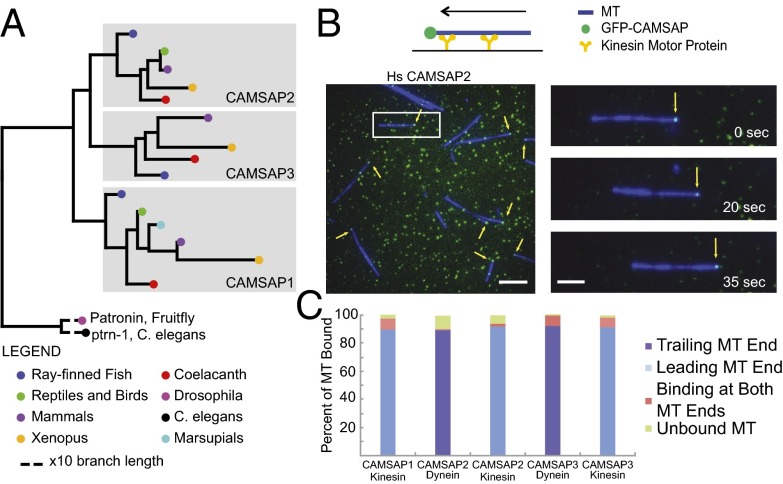
Fig. 1.
CAMSAPs have a conserved function of binding MT minus-ends. (A) Most vertebrate species possess three CAMSAP/Patronin homologs, whereas Drosophila and C. elegans each have only one. (B) Motor gliding assay for determining the end of the MT bound by CAMSAPs; in a kinesin gliding assay, the minus-end of the GMPCPP-stabilized MT is the leading end; polarity is reversed in a dynein gliding assay. Kinesin assay for GFP-CAMSAP2 is shown. Arrows show GFP-CAMSAP2 on MT minus-ends. (Scale bar: 5 μm.) Movement of an MT–CAMSAP2 complex over time (Scale bar: Inset, 2 μm.) (C) Quantification of number of MT with GFP-CAMSAPs on their ends. Results compiled from two independent experiments (>100 MTs scored per assay).
Although these studies demonstrate that CAMSAPs and Patronin stabilize MT minus-ends in vivo, many questions remain as to how these proteins recognize minus-ends and affect their dynamics. Here, we show that all vertebrate CAMSAP family members bind to MT minus-ends in vitro. However, we find differences between the CAMSAPs and Patronin with regard to their effects on minus-end dynamics. These results indicate that minus-end binding and regulation is universal for all CAMSAP members but that this activity is tunable in ways that might be exploited by cells to regulate MT organization. While the present paper was in submission, Jiang et al. (32) reported similar findings for CAMSAP proteins in vitro; we compare our findings in the Discussion.
Results
All Vertebrate CAMSAP Family Members Bind MT Minus-Ends in Vitro.
Previous studies have shown that CAMSAP2 and CAMSAP3 bind to MT minus-ends in vivo (24, 28). To determine whether these proteins alone are sufficient for this activity and whether this property is also true for CAMSAP1 (Fig. 1A), we purified full-length (FL), N-terminal GFP-tagged mammalian CAMSAP1 (human), CAMSAP2 (human), and CAMSAP3 [mouse; chosen because of previous in vivo work performed on this protein (24)]. The GFP-CAMSAPs (10–12 nM) were incubated with fluorescently labeled, guanylyl 5′-α,β methylenediphosphonate (GMPCPP)-stabilized MTs, which were then bound to and moved along a glass surface coated with kinesin or dynein motors. In the kinesin gliding assay, the MT minus-end is leading in the direction of motion, whereas the gliding direction is opposite for dynein. These gliding assays allowed us to distinguish the polarity of end binding and determine whether the proteins were stably bound as the MT translocated across the surface. For all three GFP-tagged CAMSAPs, a bright fluorescent spot was observed at the minus-ends of the majority of gliding MTs, and, in rarer cases (<10%), was observed at both ends (Fig. 1 B and C and Movie S1). Some MTs also had GFP spots along their length, but these spots tended not to be stably bound as the MT moved along the surface and were more sensitive to salt-induced dissociation. The minus-end–bound GFP punctae were approximately two to fourfold brighter than GFP-CAMSAP fluorescent spots on the glass (Fig. S1A), suggesting that one or a few CAMSAP molecules are sufficient to bind at the minus-end of a stabilized MT. These data suggest that that binding of CAMSAPs to minus-ends differs from γ-tubulin, which, as part of a γ-tubulin ring complex, caps each of the 13 protofilaments on the minus-end (33). In summary, we find that all vertebrate CAMSAPs specifically recognize the minus-ends of nondynamic MTs.
CAMSAPs Differentially Regulate Dynamic MT Minus-Ends.
Previous in vitro work with Patronin and CAMSAP3, as well as the work described here earlier, were performed with stabilized MT (22, 24, 26). However, in vivo, MTs are dynamic polymers; therefore, we wanted to determine if CAMSAP proteins affected the incorporation of tubulin into growing MTs in vitro. For this experiment, a GMPCPP-stabilized “seed” MT with incorporated Alexa 647-tubulin was adhered onto the cover glass surface. Then tubulin monomers were added, a subset of which were labeled with Alexa 561-tubulin to discern new subunit addition to the MT ends; GFP-CAMSAP was added into the mixture with the tubulin monomers (Fig. 2A). Triple color images, to visualize MT seeds, new tubulin addition, and CAMSAP, were acquired for 3–4 min by total internal reflection microscopy (TIRF), allowing accurate measurements of plus- and minus-end growth of MTs. In the assay buffer and the concentrations of tubulin (20–25 μM) used in this assay, the majority (>80%) of MTs grew from both ends (Fig. 2 B, D, and E and Movie S2); catastrophes (rapid depolymerization) were occasionally observed, but were rare. Plus-ends could be differentiated from minus-ends by their twofold faster growth rates (Fig. 2 D and E). Interestingly, when GFP-CAMSAP1 was added, fluorescent molecules were observed tracking at the tip of growing MT minus-ends (Fig. 2D), but did not affect the rate of tubulin incorporation at the minus-end (Fig. 2G). In contrast, upon the addition of GFP-CAMSAP2 and GFP-CAMSAP3, most MT minus-ends (∼70% and 80%, respectively) did not display growth at the limit of detection (∼0.5 μm growth) over the ∼4 min time of image acquisition (Fig. 2 C–E). However, the fluorescence intensity of the GFP-CAMSAP at the minus-ends in these dynamic assays with monomeric tubulin (Fig. 2C) was brighter than that observed on minus-ends of GMPCPP-stabilized MTs (Fig. 1B), suggesting that perhaps some minus-end tubulin addition occurred and deposited CAMSAPs on the MT. None of the CAMSAPs affected plus-end growth (Fig. 2G).
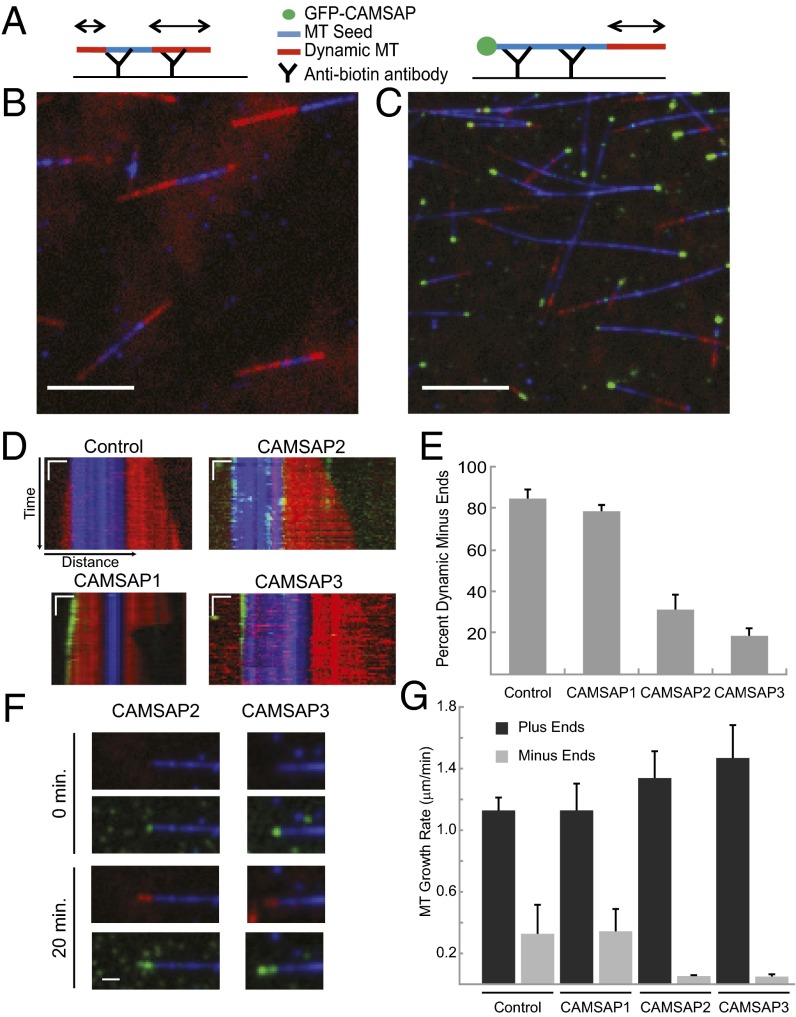
Fig. 2.
CAMSAPs differentially affect tubulin subunit addition at MT minus-ends. (A) GMPCPP seed MTs (with Alexa-647 tubulin and biotin tubulin) were attached to an anti-biotin antibody–coated coverslip (Materials and Methods). Unpolymerized tubulin (23 μM) was added to the chamber with Alexa 561-labeled tubulin (2 μM) and allowed to polymerize at room temperature. (B) Control field of polymerizing MT. GMPCPP seeds shown in blue and newly polymerized tubulin are shown in red. Image was background-corrected in Fiji (SI Materials and Materials). Image shown ∼3–4 min after reaction setup. (Scale bar: 5 μm.) (C) Dynamic assay performed with addition of 10–12 nM GFP-CAMSAP3 (green spot at MT minus-ends) shows little growth after 3–4 min of polymerization. (Scale bar: 5 μm.) (D) Representative kymographs showing polymerization of control MT and MTs bound by GFP-CAMSAPs. Photobleaching of CAMSAP2 was partially corrected by using Fiji plugin (SI Materials and Materials). Minus-ends are oriented to the left. (Scale bars: horizontal, 1 μm; vertical, 20 s.) (E) Quantification of MT minus-ends showing visible growth in <4-min rapid acquisition window (control, n = 4; CAMSAP1, n = 2; CAMSAP2, n = 3; CAMSAP3, n = 4, where n is number of experimental days; n = 19–138 MTs per day). Minus-ends were scored as dynamic if they underwent growth of >0.5 µm over the course of imaging (200 s). Mean and SEM are shown for averages of each experimental day. Only MTs with GFP at the minus-end were scored. (F) Long-term (20 min) imaging of MT seeds (blue), newly polymerized tubulin (red), and GFP-CAMSAP2 and -3 (green). Minus-ends are oriented to the left. (Scale bar: 1 μm.) (G) Quantification of MT plus- and minus-end growth rates; short-term imaging (<4 min) was measured for plus- and minus-end growth rates; long-term imaging was used for CAMSAP2 and CAMSAP3 minus-ends. Number of independent experiments on separate days are as follows. Plus ends: control, n = 6; CAMSAP1, n = 2; CAMSAP2, n = 5; CAMSAP3, n = 5. Minus-ends: control, n = 6; CAMSAP1, n = 2; CAMSAP2, n = 2; CAMSAP3, n = 2, for n = 19–77 MTs per day. Mean and SEM of independent days of measurement are shown.
These experiments showed that CAMSAP2 and CAMSAP3 dramatically suppress MT minus-end growth, whereas CAMSAP1 has little effect despite its presence at the MT minus-end. To determine the extent of growth suppression more accurately for CAMSAP2 and CAMSAP3, we performed longer-term imaging experiments (Fig. 2F). Over the course of 20 min, discernable tubulin incorporation at the minus-end could be observed for the majority of MTs that had GFP-CAMPSAP2 or GFP-CAMPSAP3 at the end. GFP-CAMSAP2 coated the entire stretch of new tubulin incorporation, suggesting that, when bound to the minus-end, it can remain associated with the MT lattice after new tubulins have been added on (Fig. 2F). For GFP-CAMSAP3, the decoration appeared more discontinuous, suggesting that occasionally tubulin could grow past a GFP-CAMSAP3 cap and then become recapped with GFP-CAMSAP3 (Fig. 2F). These longer time-lapse experiments revealed that CAMSAP2 and CAMSAP3 reduce minus-end growth by 86% and 93%, respectively, compared with control MTs (Fig. 2G).
In summary, CAMSAP2 and CAMSAP3 strongly suppress minus-end growth, but do not affect plus-end growth. In contrast, CAMSAP1 tracks along the growing tips of minus-ends without significantly affecting the polymerization rate. These differences in the dynamic assay suggest that the three CAMSAP proteins, despite their shared ability to bind selectively to MT minus-ends, regulate minus-end dynamics in distinct ways.
Cooperation Between CAMSAP Domains Enhances Minus-End Localization and Suppression of MT Minus-End Dynamics.
The three vertebrate CAMSAP proteins and the single Patronin protein from invertebrates all share a similar overall domain structure of an N-terminal calponin-homology (CH) domain [which has actin binding activity in some proteins (34)], a central domain of three coiled coils with a predicted poorly structured region between coils 2 and 3 [herein referred to as the coiled-coil (CC) domain], and a C-terminal conserved globular domain known as the CKK domain (26). Previous studies have shown that Patronin’s CH domain is diffusely localized when expressed in cells (22). The CKK domain has been shown to bind to MT in vivo (22) and in vitro (26), but minus-end localization was not carefully examined in these studies.
To understand the roles of each domain in minus-end binding, we expressed and purified three truncations of CAMSAP3: the CKK domain, the CC domain, and the CC+CKK domains (Fig. 3). We first incubated these GFP-tagged, purified proteins with GMPCPP-stabilized MTs and performed kinesin gliding assays. We found that the CKK domain of CAMSAP3 (CKK3) bound to MTs along their entire lengths, although MTs showed a higher concentration of GFP-CKK3 at their minus-ends (Fig. 3B and Movie S3). However, this association was very sensitive to salt; virtually all GFP-CKK dissociated from the MTs when 60 mM KCl was added to the buffer (Fig. 3B and Movie S3). Similar results were also obtained for GFP-CKK domains from CAMSAP1 and CAMSAP2 (Fig. S1). In contrast, the CC domain from CAMSAP3 did not bind to MTs (Fig. 3B). However, the CC+CKK domain from CAMSAP3 (Fig. 3B) and CAMSAP2 (Fig. S1C) bound to MT minus-ends at 60 mM KCl and few molecules were found along the length of the MT. These data indicate that the CKK domain is responsible for MT localization of mammalian CAMSAP proteins, but that the CC domain enhances the strength and specificity of minus-end binding at higher, more physiological ionic strengths.
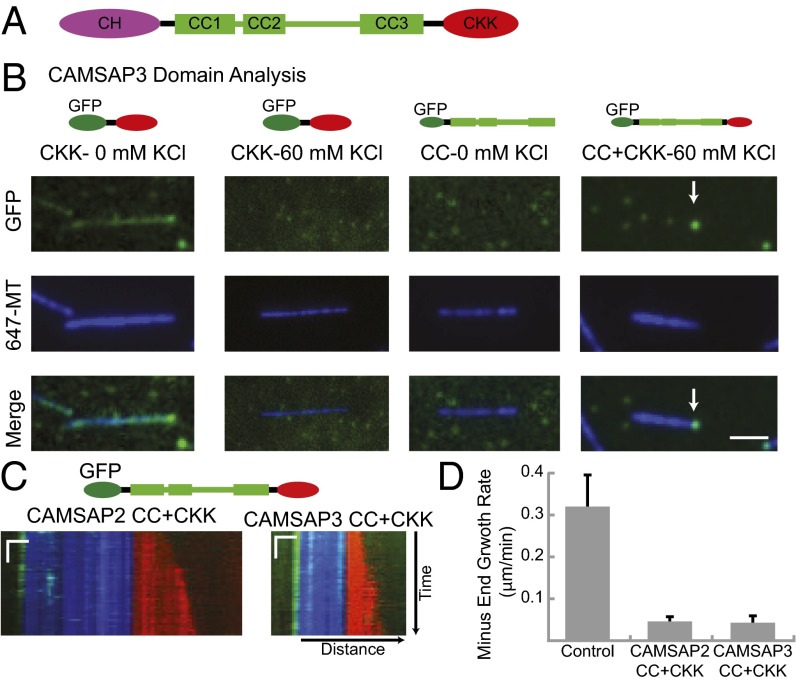
Fig. 3.
CAMSAP domains involved in minus-end MT binding and regulation of dynamics. (A) CAMSAP domain structure; predicted CC regions shown as cylinders. (B) The CKK domain of CAMSAP3 (20 nM) binds along the length of MTs with an accumulation at MT minus-ends, as shown by TIRF microscopy. CKK binding is abolished in the presence of 60 mM KCl. CAMSAP3 CC does not exhibit MT binding, but the CC+CKK domain shows specific MT minus-end binding in the presence of 60 mM KCl. (Scale bar: 2 μm.) (C) Representative kymographs of MTs capped by the CAMSAP2 and CAMSAP3 CC+CKK domains. For both CAMSAPs, the CC+CKK domains recapitulate minus-end capping to the same extent as the FL proteins. (Scale bars: horizontal, 1 μm; vertical, 20 s.) (D) Quantification of minus-end growth rates determined by long-term (20 min) imaging of MT dynamics (control growth rate from Fig. 2, for CAMSAP2 and -3 CC+CKK, n = 2, n = 14–32 MTs per day). Mean and SEM of each experimental day are shown. Only MTs with GFP initially at the minus-end were analyzed.
We next tested the CC+CKK domains from CAMSAP2 and CAMSAP3 on dynamic MTs and found that they both suppressed MT minus-end growth (Fig. 3 C and D). Thus, the CH domain is not needed for minus-end localization or suppression of dynamics.
CAMSAP Proteins Protect Stabilized MT from Depolymerization by Kinesin-13.
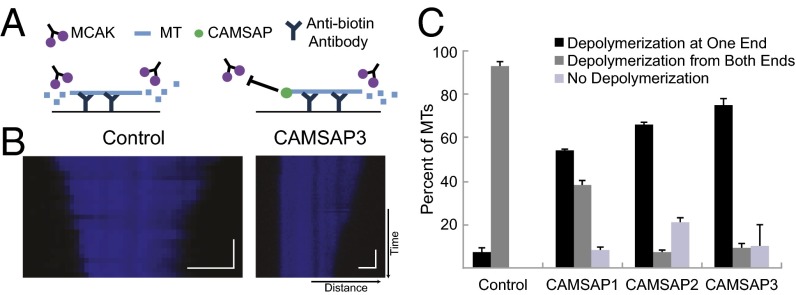
Previous work has shown that Patronin protects MT minus-ends from depolymerization by MCAK, a kinesin-13 protein that causes subunit loss and destabilizes MT ends (22). We next investigated whether the three mammalian CAMSAPs are able to protect MT minus-ends against kinesin-13–induced depolymerization. GMPCPP-stabilized, fluorescent MTs with incorporated biotinylated tubulin were adhered to coverslips; kinesin-13 (MCAK; 6 nM) was added, and depolymerization of MT ends was followed by TIRF microscopy (Fig. 4A). For control MTs, 93% of the MTs were depolymerized from both ends and 7% did not depolymerize at either end; depolymerization from just one end was never observed (Fig. 4 B and C). In contrast, the majority of MT depolymerized from just one end in the presence of CAMSAP3 (74%), CAMSAP2 (63%), and CAMSAP1 (54%; Fig. 4 B and C and Movie S4). These results suggest that the function of protecting MTs from kinesin-13–mediated depolymerization is conserved throughout all members of the family.
Fig. 4.
CAMSAPs protect MT minus-ends from depolymerization by MCAK. (A) Schematic of assay. (B) Representative kymographs of control MT depolymerizing from both ends (Left), and MT protected in the presence of CAMSAP3 (Right). In the low-salt conditions needed for MCAK depolymerization, CAMSAPs tend to bind along MT length; hence, only the MT imaging channel is shown. (Scale bars: horizontal, 1 μm; vertical, 60 s.) (C) Quantification of MTs exhibiting kinesin-13-induced end-wise depolymerization behaviors. Number of independent experiments on separate days are as follows: control, n = 3; CAMSAP1, n = 2; CAMSAP2, n = 2; CAMSAP3, n = 2, for n = 17–142 MTs per day. Mean and SEM are shown for the different experimental days.
Suppression of MT Minus-End Dynamics Is Essential for the Cell Biological Activity of Drosophila Patronin.
Drosophila has a single CAMSAP homolog, Patronin, which is most closely aligned with CAMSAP2 and CAMSAP3 (Fig. 1A). The presence of a single gene, coupled with a high efficiency of RNAi-mediated protein knockdown, makes Drosophila S2 cells a good system for studying the roles of these minus-end regulatory proteins. Previous RNAi knockdown of Patronin was shown to decrease the number of interphase MTs and cause the appearance of short, “treadmilling” MT fragments (22). The latter phenotype is easily scored by time-lapse microscopy and provides a clear in vivo assay for Patronin function.
We first conducted a functional analysis of purified Patronin domains (Fig. 5A). As was true for CAMSAP3, we found that the CC+CKK domain bound tightly and specifically to MT minus-ends (Fig. 5B). In contrast to what we found for CAMSAP3, the Patronin GFP-CKK domain (10 nM) did not exhibit observable binding to MT in a TIRF assay, even in the absence of KCl (Fig. S1C). Also in contrast to the mammalian CAMSAPs, the GFP-CC domain alone bound highly specifically to MT minus-ends (Fig. 5B). Through truncation analysis, we identified an ∼70-kDa fragment in the C-terminal half of the CC domain (amino acids 868–1457) that is necessary and sufficient for binding selectively to MT minus-ends (Fig. S2 A and B, construct 4). This region eliminates the first two predicted coiled coils and contains a predicted unstructured region plus the C-terminal coiled coil; the C-terminal coiled coil (amino acids 1288–1457) alone, however, was insufficient to confer MT minus-end binding (Fig. S2B).
Fig. 5.
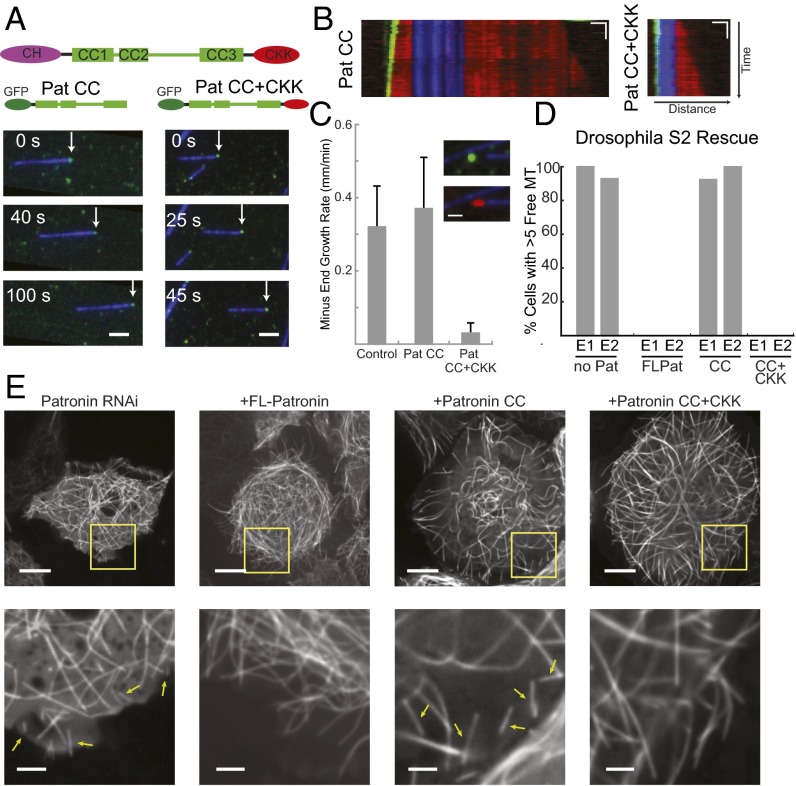
Domains required for Drosophila Patronin minus-end binding, suppression of growth in vitro, and MT stability in S2 cells. (A) Schematic of Patronin domain structure. The Patronin CC domain and CC+CKK domains bind to MT minus-ends (white arrows) in a kinesin gliding assay. (Scale bar: 2 μm.) (B) Representative kymographs of dynamic MTs capped by Patronin GFP-CC and GFP-CC+CKK. (Scale bars: horizontal, 1 μm; vertical, 20 s.) (C) Quantification of minus-end growth rates in the presence of Patronin truncations. Short-term images were scored for Pat CC; long-term images were scored for Patronin CC+CKK. For control, n = 6; Pat CC, n = 1; Pat CC+CKK, n = 2; n = 10–39 MTs per day; mean and SD shown for each experimental day. (Inset) MT growth (red) from stabilized seeds (blue) after 15 min in presence of Pat CC+CKK (green). (Scale bar: 1 μm.) (D) Quantification of Drosophila rescue experiments. Experiment was performed in duplicate (n = 10–12 cells scored on each day); results from each day are shown as separate bars. (E) Representative cells from rescue experiments. (Insets) Zoom of cell edge; yellow arrows mark representative MT fragments used for scoring. (Scale bars: full cell, 10 μm; Inset, 2.5 μm.)
To assess how these domains affect MT dynamics, we performed MT growth assays, as previously described for the CAMSAP proteins. Surprisingly, we found that the Patronin CC domain tracked growing MT minus-ends in vitro, but did not change the rate of minus-end growth (Fig. 5C and Movie S5), similar to the behavior observed for CAMSAP1. In contrast, the Patronin CC+CKK domain bound to minus-ends and suppressed tubulin incorporation by ∼90% compared with control MT minus-ends (Fig. 5C). The GFP-CC+CKK domain tracked along the tip of these very slow-growing minus-ends, but did not coat the region of new growth like CAMSAP2 (Fig. 5C). These results show that the Patronin CC domain can bind to MT minus-ends, but must work in coordination with the CKK domain to suppress minus-end dynamics.
The Patronin domain analysis uncovered a construct that binds MT minus-ends but does not suppress growth (Pat CC) and a construct that binds and suppresses minus-end dynamics (Pat CC+CKK). We performed rescue experiments with these constructs, as well as FL Patronin, to determine which domains are necessary to rescue the interphase Patronin RNAi phenotype. Endogenous Patronin was depleted by using dsRNAs against the 3′ and 5′ UTRs, so that rescue experiments could be performed with Patronin constructs lacking these UTR sequences. Similar to our earlier results (22), Patronin knock-down with UTR sequences caused the appearance of small, treadmilling MT fragments in the cytoplasm (Fig. 5 D and E and Movie S6). Transfection of the FL and the CC+CKK constructions led to full rescue (no treadmilling, short MTs; Fig. 5 D and E and Movie S6). In contrast, the CC domain, which localizes to minus-ends but does not suppress minus-end dynamics, did not rescue the knock-down phenotype (Fig. 5 D and E). These results demonstrate that suppression of MT minus-end dynamics is essential for Patronin's function in stabilizing MT minus-ends in vivo.
Discussion
This work, in combination with several other recent studies, provides support for the model that metazoan CAMSAPs/Patronins act as stabilizing proteins for MT minus-ends. Previous work has shown that Drosophila Patronin stabilizes minus-ends against depolymerization by kinesin-13 (22). Here, we show that this kinesin-13 protection activity extends to the mammalian CAMSAPs and that Patronin and CAMSAP2 and CAMSAP3 also strongly suppress minus-end growth. While this paper was in submission, Jiang et al. (32) showed that CAMSAP2 and CAMSAP3 decrease minus-end polymerization in vitro and protect minus-ends from depolymerization after a laser-induced cut of MTs in vitro and in vivo. They also showed that a stretch of MT coated with CAMSAP2 is protected against depolymerization traveling toward it from the plus end following a catastrophe.
In addition to these in vitro studies, the MT minus-end protection activity of CAMSAPs/Patronin has been shown to be important for maintaining noncentrosomal MTs (22, 28, 32), and the loss of this activity impacts cell migration (32), neurite morphology (30, 31), and mitosis (21, 23). Interestingly, although Patronin plays an important role in mitosis in flies, Jiang et al. did not find mitotic defects of CAMSAP2 knockdown (32). Although this result may reveal a difference in the biological function of Drosophila vs. mammalian proteins, it is possible that the knockdown of all three CAMSAPs may be needed to uncover a mitotic phenotype. CAMSAPs also have been reported to interact biochemically with spectrins (an activity from which the CAMSAPs derive their name), and overexpression studies have suggested that this interaction may be important in vivo (27). However, evidence supporting a physiological connection of this protein family with spectrin is less clear than their role in regulating MT minus-ends. Thus, it may be warranted to develop a new unifying nomenclature for the invertebrate and vertebrate proteins reflecting their MT function.
Despite their common activity in binding MT minus-ends, the mammalian CAMSAPs exhibit different properties with respect to their effectiveness as a “cap” for growing MT minus-ends. We and Jiang et al. (32) show that CAMSAP1 tracks at the tips of growing minus-ends, whereas CAMSAP2 and CAMSAP3 suppress minus-end growth, with CAMSAP3 being more robust in its activity. Thus, all the CAMSAPs can remain bound to the MT end and still allow the addition of new tubulins to varying extents. The different CAMSAPs/Patronin also exhibit different off-rates from the MT lattice [present work and Jiang et al. (28)]. CAMSAP1 and Patronin bind at the very tip of the minus-end, and thus, in a similar manner to +TIP tracking proteins, must dissociate after allowing tubulin incorporation. CAMSAP2 and CAMSAP3, however, appear to remain tightly bound to the MT lattice after tubulin incorporation, creating an elongated stretch of bound molecules.
The mechanism of minus-end recognition (where α-tubulin is exposed) by CAMSAP/Patronin is unclear, although several results suggest a complex binding mechanism. The CAMSAPs and Patronin bind to the ends of nondynamic, GMPCPP-stabilized MT in substoichiometric (n < 13) numbers, suggesting that they can recognize some feature that is unique to the exposed α-tubulin surface at the minus-end. In a dynamic MT assay, however, tubulin addition results in increasing numbers of CAMSAPs being deposited near the minus end. Because not all these CAMSAPs can bind to the limited number of exposed α-tubulins at the very minus-end, they must be retained by other means. The lattice affinity of individual CAMSAPs on stabilized MTs appears to be relatively weak (Fig. 1). However, when they have been deposited at the exposed minus-end by tubulin subunit addition, adjacent CAMSAPs might interact cooperatively with one another to be retained on the MT lattice.
Truncation experiments to identify a minimal domain involved in minus-end binding led to somewhat different results for the mammalian CAMSAPs and invertebrate Patronin. For CAMSAP, the CKK domain binds to MTs weakly and shows preferential minus-end localization [present results and those of Jiang et al. (32)], but the Patronin CKK domain does not bind MTs under the same conditions. Conversely, the Patronin CC domain shows clear minus-end binding, whereas the CAMSAP3 CC domain shows no detectable MT binding activity in the microscopy assay. However, for both CAMSAPs and Patronin, a combination of the CC+CKK produces robust minus-end recognition and suppression of minus-end dynamics in vitro [present results and those of Jiang et al. (32)] and in vivo (Fig. 5). It seems plausible that mammalian CAMSAPs and invertebrate Patronin have two domains that facilitate minus-end binding; the evolution of different binding strengths and cooperativity of these domains may account for the distinct minus-end behavior of invertebrate Patronin and mammalian CAMSAPs, and between the mammalian CAMSAPs. Consistent with this idea, a region in the CC domain that is important for CAMSAP function and conserved among vertebrate CAMSAPs is not conserved in Drosophila Patronin (32). Interestingly, the N-terminal CH domain, which is conserved among all invertebrate and vertebrate CAMSAPS, does not appear to be necessary for MT minus-end regulation and perhaps is involved in a yet unidentified function of these proteins.
Our work, as well as the recent studies by Jiang et al. (32) and Tanaka et al. (28), suggest that MT minus-ends may be subject to more regulation than previously thought. The vertebrate and invertebrate CAMSAPs all appear to strongly protect minus-ends against depolymerization caused by MT depolymerases or breaks. However, CAMSAPs and Patronin permit tubulin subunit addition at the minus-end at very different rates. These differences could lead to a tunable system for regulation of minus-end dynamics in cells. We also find somewhat different efficiencies of protection against kinesin-13–mediated depolymerization by the CAMSAPs. The different off-rates of CAMSAPs from MTs also may have a physiological role, as elegantly demonstrated by Jiang et al. (32), who showed that long, decorated stretches of CAMSAP2 at minus-ends can create stable seeds that can undergo repeated rounds of growth after plus-end catastrophes. The interplay of CAMSAPs within a single cell may play an important role in regulating the MT network. For example, Tanaka et al. have documented cooperation between CAMSAPs in epithelial cells and observed that MT minus-ends are decorated with similar sized, short stretches of CAMSAP2 and CAMSAP3, but that the length of minus-end decoration by CAMSAP2 increased considerably after CAMSAP3 depletion by RNAi (28). This result suggests that the size of these minus-end caps may be governed by the ratios of the CAMSAPs in cells. Future studies will likely reveal how different cell types use and regulate the different activities of CAMSAP proteins to control noncentrosomal MTs during cell division, migration, and differentiation.
Materials and Methods
Plasmid Construction, Protein Expression. and Protein Preparation.
Information on plasmid construction, protein expression. and protein preparation is provided in SI Materials and Materials.
In Vitro Assays.
Single-molecule assays were imaged by using a Nikon Ti microscope (1.49 N.A., 100× objective) by TIRF; images were acquired with an Andor EM-CCD camera and MicroManager software (35). Flow chambers (∼8 μL) were created by using double-stick tape and silanized coverslips by using a procedure modified from Gell et al. (36). Briefly, coverslips were washed by sonication in 1 M NaOH, rinsed, dried and plasma cleaned, silanized for 1 h using trichloroethylene plus 0.1% dimethyldichlorosilane, washed in three consecutive baths with methanol and sonication, and then dried.
All assays were completed in assay buffer [80 mM piperazine-N,N'-bis(ethanesulfonic acid) (PIPES), 2 mM MgCl2, 1 mM EGTA (BRB80), with 60 mM KCl] as specified. For motor-driven gliding assays, flow chambers were incubated sequentially (5 min each) with 0.2 mg/mL of anti-His antibody and 1 mg/mL κ-casein, followed by ∼100–200 pM of a truncated, dimeric human kinesin (SI Materials and Materials). For dynein gliding assays, the motor domain of dynein (SI Materials and Materials) was incubated in a flow cell with acid-base washed coverslips, followed by 1 mg/mL κ-casein and addition of motility mix. Motility mix contains assay buffer plus CAMSAP proteins (10–12 nM), MTs, 4 mM ATP, and 2 mM protocatechuic acid (PCA)/protocatechuate-3,4-dioxygenase (PCD)/Trolox [to reduce photobleaching, (37)]; images were acquired at 2, 5 or 10 s intervals for 20–50 frames.
For in vitro assays, we used a modification of the assay developed by Gell et al. (36). Flow chambers were incubated (5 min each) sequentially with 0.2 mg/mL anti-biotin antibody, 1 mg/mL κ-casein, and biotinylated MT seeds. For dynamic assays, tubulin dynamics were measured in assay buffer plus 25 μM unlabeled tubulin, 2 μM Cy5 or Alexa 561-labeled tubulin, 1 mg/mL κ-casein, 0.1% methyl cellulose, 2–4 mM GTP, and 2 mM PCA/PCD/Trolox. CAMSAP and Patronin GFP-tagged proteins were added at a final concentration of 10–20 nM. Imaging was performed at room temperature. Images were acquired at 2-s intervals for 100–200 s. For long-term imaging, flow chambers were sealed using valap; images were acquired every 1–5 min for 20 min. For kinesin-13 assays, chambers were prepared as described previously. BRB80 plus 6 nM MCAK (38), 2 mM ATP, 1 mg/mL κ-casein, and Trolox/PCA/PCD was flowed into the chamber for imaging. Imaging was done in TIRF at 10-s intervals for 10–20 min.
Image Analysis.
Kymographs of MT growth or depolymerization were made using the Fiji Reslice function (39). Rates of growth for long- and short-term imaging experiments were measured over a period for which the MT could be clearly resolved by making a rectangle in which the corners marked the beginning and end of growth; height of the rectangle corresponded to time elapsed, and width corresponded to the growth distance. For depolymerization assays with kinesin-13, MTs were assessed for depolymerization by kymograph analysis.
Patronin RNAi Experiments in Drosophila S2 Cells.
Drosophila S2 cells stably expressing GFP-tubulin and mCherry–Patronin constructs were generated and cultured as previously described (22). For RNAi experiments, dsRNAs against the 3′ and 5′ UTRs of Patronin were synthesized as described in SI Materials and Materials. Cells were subjected to two rounds of RNAi over the course of 8 d and plated on concanavalin A for imaging. Cells were imaged on a Zeiss spinning-disk confocal microscope with a 100× 1.45 N.A. objective by using an Andor EM CCD camera at 5–10-s intervals.
Supplementary Material
Acknowledgments
We thank M. Sirajuddin, E. Jonsson, and W. Huynh for contributing reagents; N. Stuurman for microscopy assistance; S. Petry and S. Goodwin for helpful advice and discussion; and F. Collman for assistance with data analysis. This work was supported by the Howard Hughes Medical Institute (R.D.V.), the National Institutes of Health Grant 38499 (to R.D.V.), and the National Science Foundation Graduate Research Fellowship Program (to M.C.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404133111/-/DCSupplemental.
References
- 1.Mitchison TJ. Localization of an exchangeable GTP binding site at the plus end of microtubules. Science. 1993;261(5124):1044–1047. doi: 10.1126/science.8102497. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 3.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450(7172):1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 4.Akhmanova A, Steinmetz MO. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9(4):309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 5.Schuyler SC, Pellman D. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 2001;105(4):421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 6.Vitre B, et al. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol. 2008;10(4):415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- 7.Blake-Hodek KA, Cassimeris L, Huffaker TC. Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins. Mol Biol Cell. 2010;21(12):2013–2023. doi: 10.1091/mbc.E10-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149(2):371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138(2):366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 10.Tournebize R, et al. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2(1):13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- 11.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132(1):79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter AW, et al. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11(2):445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moores CA, et al. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5(16):1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- 14.Heald R, Nogales E. Microtubule dynamics. J Cell Sci. 2002;115(pt 1):3–4. doi: 10.1242/jcs.115.1.3. [DOI] [PubMed] [Google Scholar]
- 15.Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422(6933):753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- 16.Keating TJ, Peloquin JG, Rodionov VI, Momcilovic D, Borisy GG. Microtubule release from the centrosome. Proc Natl Acad Sci USA. 1997;94(10):5078–5083. doi: 10.1073/pnas.94.10.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterman-Storer CM, Salmon ED. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139(2):417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yvon AM, Wadsworth P. Non-centrosomal microtubule formation and measurement of minus end microtubule dynamics in A498 cells. J Cell Sci. 1997;110(pt 19):2391–2401. doi: 10.1242/jcs.110.19.2391. [DOI] [PubMed] [Google Scholar]
- 19.Rodionov V, Nadezhdina E, Borisy G. Centrosomal control of microtubule dynamics. Proc Natl Acad Sci USA. 1999;96(1):115–120. doi: 10.1073/pnas.96.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorobjev IA, Rodionov VI, Maly IV, Borisy GG. Contribution of plus and minus end pathways to microtubule turnover. J Cell Sci. 1999;112(pt 14):2277–2289. doi: 10.1242/jcs.112.14.2277. [DOI] [PubMed] [Google Scholar]
- 21.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316(5823):417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143(2):263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM. Patronin mediates a switch from kinesin-13-dependent poleward flux to anaphase B spindle elongation. J Cell Biol. 2013;203(1):35–46. doi: 10.1083/jcb.201306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135(5):948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 25.Shahbazi MN, et al. CLASP2 interacts with p120-catenin and governs microtubule dynamics at adherens junctions. J Cell Biol. 2013;203(6):1043–1061. doi: 10.1083/jcb.201306019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baines AJ, et al. The CKK domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol Biol Evol. 2009;26(9):2005–2014. doi: 10.1093/molbev/msp115. [DOI] [PubMed] [Google Scholar]
- 27.King MD, et al. A conserved sequence in calmodulin regulated spectrin-associated protein 1 links its interaction with spectrin and calmodulin to neurite outgrowth. J Neurochem. 2014;128(3):391–402. doi: 10.1111/jnc.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka N, Meng W, Nagae S, Takeichi M. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc Natl Acad Sci USA. 2012;109(49):20029–20034. doi: 10.1073/pnas.1218017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagae S, Meng W, Takeichi M. Non-centrosomal microtubules regulate F-actin organization through the suppression of GEF-H1 activity. Genes Cells. 2013;18(5):387–396. doi: 10.1111/gtc.12044. [DOI] [PubMed] [Google Scholar]
- 30.Marcette JD, Chen JJ, Nonet ML. The Caenorhabditis elegans microtubule minus-end binding homolog PTRN-1 stabilizes synapses and neurites. Elife. 2014;3:e01637. doi: 10.7554/eLife.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson CE, et al. PTRN-1, a microtubule minus end-binding CAMSAP homolog, promotes microtubule function in Caenorhabditis elegans neurons. Elife. 2014;3:e01498. doi: 10.7554/eLife.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang K, et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev Cell. 2014;28(3):295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466(7308):879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ. Functional plasticity of CH domains. FEBS Lett. 2002;513(1):98–106. doi: 10.1016/s0014-5793(01)03240-9. [DOI] [PubMed] [Google Scholar]
- 35.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol. 2010;chapter 14(14):20. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gell C, et al. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 37.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94(5):1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peris L, et al. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185(7):1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.