Abstract
Understanding stem cell (SC) population dynamics is essential for developing models that can be used in basic science and medicine, to aid in predicting cells fate. These models can be used as tools e.g. in studying patho-physiological events at the cellular and tissue level, predicting (mal)functions along the developmental course, and personalized regenerative medicine. Using time-lapsed imaging and statistical tools, we show that the dynamics of SC populations involve a heterogeneous structure consisting of multiple sub-population behaviors. Using non-Gaussian statistical approaches, we identify the co-existence of fast and slow dividing subpopulations, and quiescent cells, in stem cells from three species. The mathematical analysis also shows that, instead of developing independently, SCs exhibit a time-dependent fractal behavior as they interact with each other through molecular and tactile signals. These findings suggest that more sophisticated models of SC dynamics should view SC populations as a collective and avoid the simplifying homogeneity assumption by accounting for the presence of more than one dividing sub-population, and their multi-fractal characteristics.
Stem cells are classically defined as unspecialized cells that can self-renew and give rise to differentiated cell types during embryogenesis, and in the adult, during tissue homeostasis or injury repair. These functions make them highly attractive to study for the purposes of understanding ontogeny and development, or for their potential use in regenerative medicine and tissue engineering.
After more than 25 years of extensive research of numerous stem cell types, the field still struggles with how to define stem cells based on a molecular or chemical signature. Defining stem cells using molecular surface markers is a challenge. The lack of consistency in marker expression may be due the changing expression of markers during stem cell manipulation, or maturation, or to population heterogeneity. Technical differences between laboratories' methods and reagents can also contribute to challenges in defining stem cells based on markers. This study takes a system-level view on stem cells and particularly focuses on heterogeneity and population dynamics which are poorly understood and contribute to ambiguity in the identification of cells responsible for specific functions.
The notion of a stem cell population which is comprised of a network of cells with interacting functions is rarely considered ex vivo. In vivo, it is well established that stem cells reside within a niche or microenvironment consisting of different cell types that provide physical and chemical supportive factors. However, the in vitro study of stem cells often does not consider a niche environment. Rather, attempts to study stem cells have predominantly focused on the isolation of purified subsets of cells with specific markers or functions1,2,3,4,5,6,7,8,9,10. Yet, several reports suggest that a population level exists for various stem cell types including hematopoietic stem cells (HSCs), mesenchymal stem cells (MSCs)11,12,13and muscle stem cells14,15,16,17,18,19,20. In support of this, several groups have shown that an individual cell from a stem cell population can re-establish the heterogeneous parent population21,22,23,24,25.
The basic science challenges with population heterogeneity subsequently lead to issues related to their use in regenerative medicine, e.g., in ensuring cell potency or predicting ex vivo expansion or growth rates. Producing therapeutic doses of stem cells by ex vivo expansion requires what the FDA terms ‘more-than-minimal manipulation’26,27which carries with it the risks of stem cells becoming contaminated, genetically transformed, or functionally changed. Bio-manufacturing methods must predict the time required to obtain potent dose(s) of stem cells, yet minimize the amount of time that cells are manipulated ex vivo. Indeed, models which can accurately predict the growth rate of a heterogeneous population will be valuable tools in the development of a manufacturing process that minimizes cell culture time and reduces exposure to foreign materials. Until now, very few approaches examine nonlinear behavior of stem cell growth28,29,30. Rather, the basic exponential model which is used extensively in cell biology assumes a constant division time, and that all cells are dividing. As such, the proliferative heterogeneity of stem cell populations has only been addressed superficially by segregating populations into dividing and nondividing cells in compartment models30,31,32,33,34,35,36,37,38,39,40,41, structure population models5,32,42,43,44,45,46,47,48,49, and agent-based models50,51. Few have addressed the potential existence of distinct dividing subpopulations within the heterogeneous stem cell population. For example, Glauche et al.52 developed a nonlinear, adaptive model which accounts for two functional states–quiescence and proliferative–to explain HSC Bromodeoxy-Uridine (BrdU)-label dilution data. Wilson et al.53 and Foudi et al.54 show that similar label dilution data can be modeled by a two subpopulations with distinct division rates.
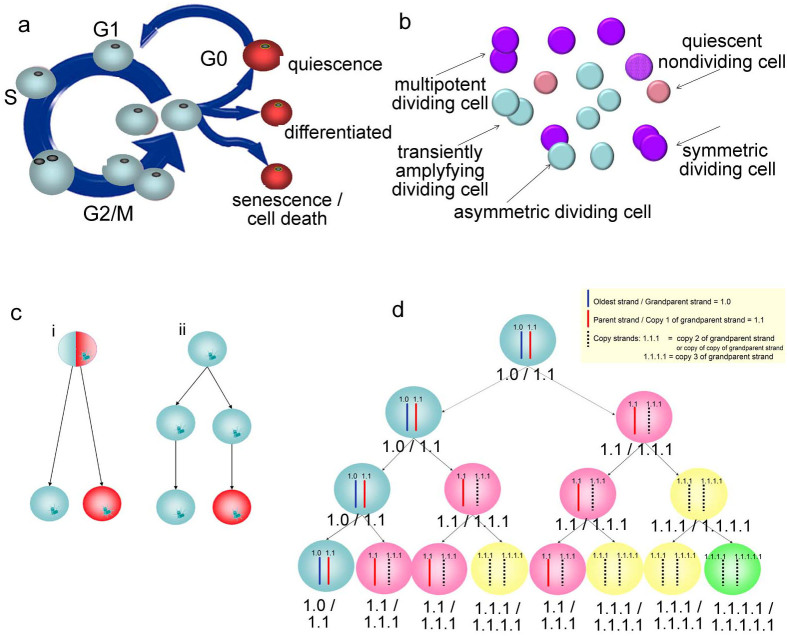
Mathematical models must consider population heterogeneity that may manifest in individual cell proliferative behavior, in molecular activity and metabolism, and/or in cell morphology. In terms of proliferative heterogeneity, stem cells may progress through the cell cycle at different rates, or may exit the cell cycle to various fates (Fig. 1a). Stem cell populations consist of mitotically active (dividing) and mitotically inactive (nondividing) cells. Included in the non-dividing fraction are 1) quiescent cells, which have the theoretical potential to re-generate the entire population, 2) differentiated cells, and 3) senescent cells. All these subpopulations are present in a typical asynchronously growing population of cells (Fig. 1b). The dividing fraction can be further distinguished into cells that give rise to symmetric and asymmetric divisions/fates3,4,5,6,7,8,9,10,11 (Fig. 1b&c). Asymmetric divisions/fates lead to population heterogeneity, which can be characterized using different parameters including molecular, morphological or proliferative features. Asymmetry may result from a number of mechanisms including the immortal strand hypothesis55 (Fig. 1d). Proliferative heterogeneity, which we examine in this paper, results from the presence of all these subpopulations and asymmetric cell fates30.
Figure 1. Heterogeneity in Stem Cell Populations.
(a) Stem cell populations are comprised of mitotically active (dividing) and mitotically inactive (nondividing: quiescent cells, terminally differentiated cells, senescent cells, and dying cells). (b) Proliferative heterogeneity results from asynchrony in life stage which leads, in part, to the presence of these subpopulations. (c) More specifically, asymmetric divisions and fates or symmetric divisions1,2,3,4,5,6,7,8,9 lead to different subpopulations. Intrinsic or extrinsic factors play a role in whether the daughter cells are different due to internal cues or environmental cues. (d) An example of development of heterogeneity from intrinsic cell differences. Asymmetry in DNA strand segregation would allow for stem cell self-renewal. The (blue) stem cell would retain the oldest DNA strands. This cartoon shows segregation of 1 chromosome: the oldest/grandparent strand is blue and designated 1.0, and the parent strand (a copy of the grandparent strand) is red and designated 1.1. All other copies are dashed lines and designated copy numbers are 1.1.1, 1.1.1.1, 1.1.1.1.1 etc. If non-random strand segregation occurs among all chromatids in the cell, the result is asymmetric divisions and self-renewal of the stem cell. We show here how heterogeneity could result in the expanding population. If distinct phenotypes occur based on the DNA strand copy numbers, then these phenotypes can be categorized based on the template and copy number. E.g., after the 3 divisions shown in the lineage tree above, there would be 1 stem cell (1.0 and 1.1 strands, p0 phenotype), 3 pI cells would have (1.1/1.1.1 stands), 3 pII cells (1.1.1/1.1.1.1) and 1 pIII cell (1.1.1.1/1.1.1.1.1).
In this paper, we demonstrate that the combination of statistical tools and time-lapsed imaging of individual cells' dynamics allows us to study stem cells as a population or network of cells. This combination of imaging-based approaches as previously used by Bahnson et al.56 or, more recently, Scherf et al.57 and Rappaport et al.58, with rigorous multivariate statistics, and data-driven modeling has been recognized recently as being of fundamental importance for the future of cell biology59. As such, we hypothesize that higher level cell-cell interactions are at work to coordinate cell fate decisions leading to re-establishing heterogeneity. Indeed, we show here that multiscale phenomena do exist for individual cells in the population for certain cell characteristics, namely cell division time (DT); this process is power-law in nature and provides evidence for interactions between subpopulations. Consequently, in this study, we show that the dynamics of stem cell populations displays a heterogeneous structure consisting of multiple sub-population behaviors and suggest that rather than defining stem cells by exclusive cell markers, efforts should be made to understand the stem cell population, its subpopulations, and the interactions among them.
Results
Stem cell dynamics display a heterogeneous structure
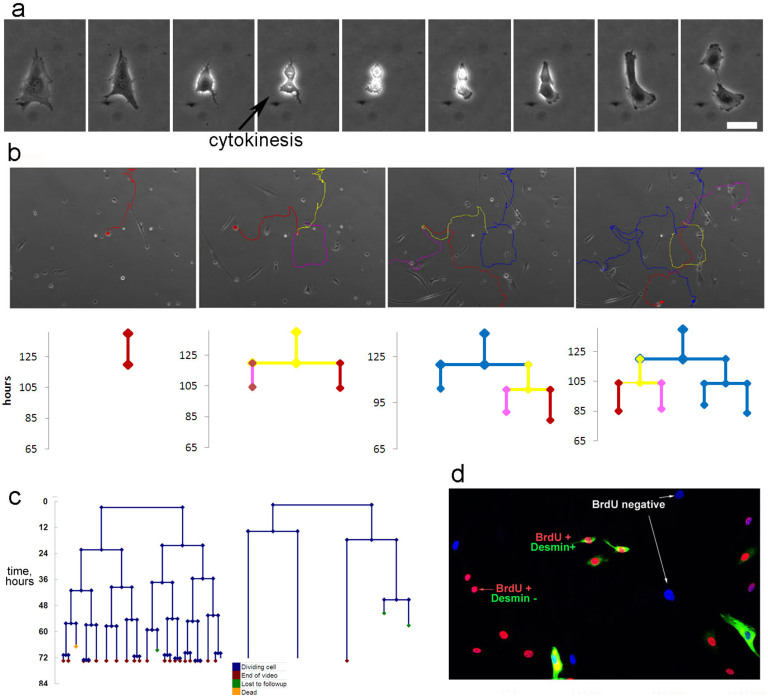
Using time-lapsed imaging60,61, we recorded the cell growth and examined the structure and cell division dynamics for both muscle stem cells62,63 from mouse and rat, and human mesenchymal stem cells64,65. Time-lapsed images were acquired at 10-minute intervals over the course of 5 days. From these images, we obtained direct measurements of cell cycle times (division times) as time elapsed between cytokinetic events (Fig. 2a). Based on these data, we were able to construct cell lineage trees of divisional history (Fig. 2b).
Figure 2. Time-lapsed image analysis of stem cell populations show heterogeneity in proliferative activity.
(a) Cell division occurring in a human muscle stem cell population. Total time lapsed is 4.5 hours, scale bar represents 50 microns. (b) Individual cell tracking (Cytotracker,) through 4 generations. The paths for parent and daughter cells are shown as an overlay on phase contrast images. Division times are used to construct cell lineage trees. The vertical distance on the corresponding lineage trees represents the cell division time or the amount of time the cell was on screen. lpar; c) Heterogeneity in cell division times can be detected by cell lineage analysis (human muscle stem cells are shown here). Individual cells were examined using time-lapsed imaging over 3 days. Lineage history trees show the presence of both dividing and nondividing cells in the population. Variation in cell division times is also detected. Most current growth models assume homogeneity with a constant division time, and exponential growth. (d) Identification of dividing and nondividing cells in time-lapsed images after addition of BrdU to media for 48 hours and immunostaining for BrdU. Muscle stem cell populations contain both dividing (BrdU + cells, and nondividing (BrdU-) cells. Desmin staining illustrates that molecular heterogeneity for the myogenic cytoplasmic protein does not correspond with proliferative heterogeneity.
Heterogeneity was detected in cell lineage analysis of all stem cells (Fig. 2c). Lineage history trees show the presence of both dividing and nondividing cells in the population. Variation in DTs is also detected. Proliferative heterogeneity was also identified by BrdU labeling as we observed both BrdU+ and BrdU– cells; this did not correlate well with molecular heterogeneity in expression of the cytoplasmic myogenic protein desmin (Fig. 2d), which illustrates the complexity of understanding heterogeneous characteristics.
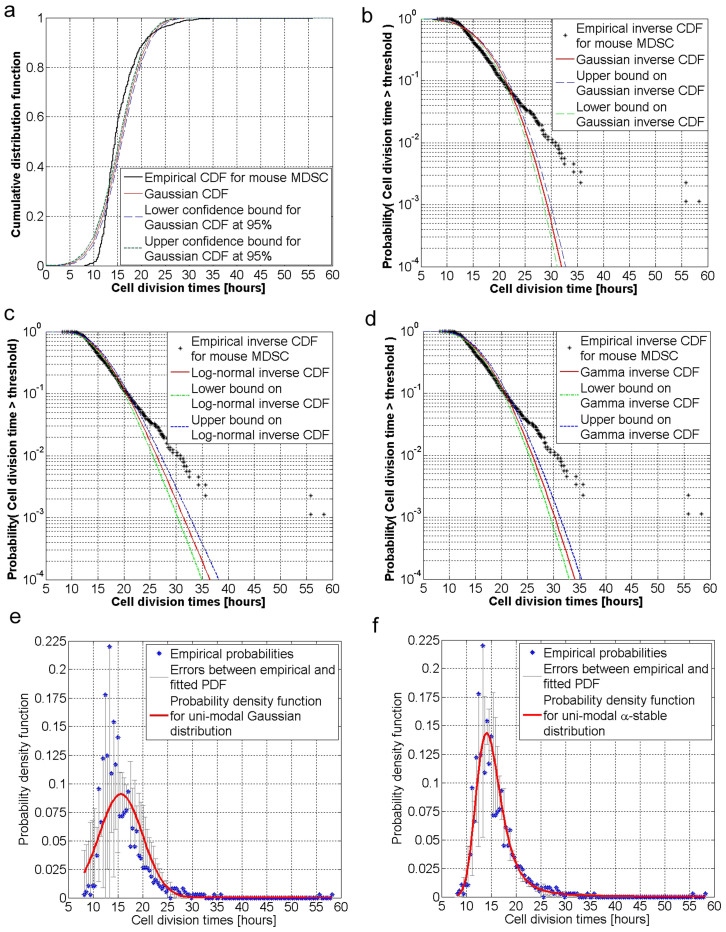
For the stem cell DTs from all populations examined, we first estimate the empirical cumulative distribution function (see Figure 3.a and Supplementary Material Figures 1.a, 2.a, and 3.a), the empirical probability of observing a cell DT larger than a given threshold (also called exceedance probability), then perform a maximum likelihood estimation (MLE) of well-known distributions and investigate their goodness-of-fit with respect to the actual data (see Supplementary Material Figures 1, 2, 3 and 4 and Notes 1, 2 and 3). The main reason for analyzing the exceedance probabilities is to quantify whether the stem cell growth exhibits an exponential or a power law type of behavior. By analyzing the exceedance probability of cell DTs for all three species, we find that the cell DTs are not well approximated by exponential type of distributions (Supplementary Material Figures. 1b, 2b, 3b) as one would assume. This is also shown by the deviations of the empirical exceedance probabilities from the Gaussian (Figures 3.a–b), log-normal (Figure 3.c) and gamma (Figure 3.d) distributions. Instead, the graphical analysis in Figures 3.e–f, and the Supplementary Figures 1, 2 and 3 and the statistical results in Supplementary Tables 1, 2 and 3 show that the exceedance probabilities are better fitted by long-tail distributions such as α-stable, Student-t and generalized extreme value (Gev) distributions. However, to better assess the validity of modeling stem cell DTs via uni-modal distributions, we estimate the statistical goodness-of-fit for each MLE fit which implies analyzing the fitting errors between the empirical and postulated probability density function. These results suggest that although the stem cell DTs exhibit a long-tail type of behavior, this cannot be well fitted by uni-modal distributions alone (see Supplementary Notes 1, 2 and 3, and Tables 1, 2, and 3).
Figure 3. Statistical investigation and interpretation of tracked stem cell division times.
(a) Comparison between the cumulative distribution function (CDF) and the 95% confidence intervals for the Gaussian distribution fitted via maximum likelihood estimation method and the empirical CDF of the mouse stem cell division times. (b) Comparison between the exceedance probability (i.e., probability of observing a stem cell division time greater than a specific threshold) and the 95% confidence interval for the fitted Gaussian distribution and the empirical inverse CDF of the mouse stem cell division times. (c) Comparison between the exceedance probability and its 95% confidence intervals for the fitted Log-normal distribution and the empirical inverse CDF of the mouse stem cell division times. (d) Comparison between the exceedance probability and its 95% confidence intervals for the fitted Gamma distribution and the empirical inverse CDF of the mouse stem cell division times. (e) Maximum likelihood fitting and the error bars for the uni-modal Gaussian probability density function (PDF). The empirical PDF is shown by the blue dots. (f) Maximum likelihood fitting and the error bars for the α-stable PDF. Rejection of uni-modal mathematical modeling approaches for stem cell growth is motivated not only by the inconsistencies observed via graphical inspection of the empirical PDF against postulated PDF, but also by the very small p-value probabilities of the Kolmogorov-Smirnov (K-S) test (see the Supplementary Material).
A similar conclusion can be reached by analyzing the graphical fittings in Figures 1.c, 2.c and 3.c in Supplementary Material of several well known unimodal distributions (i.e., Gaussian, Gamma, Log-normal, Generalized extreme value, Student-t, Weibull, asymmetric α-stable distribution) for the mouse and human cell DTs. In addition to graphical evidence, the Kolmogorov-Smirnov test, the Akaike Information Criterion and Hartigan's Dip test66 (see Notes 4, 5, and 6 in the Supplementary Material) suggest that stem cell DTs cannot be modeled by an uni-modal distribution. This suggests that there actually may exist two dividing subpopulations of stem cells, each characterized by distinct dynamics of stem cells division rate; this is reflected by different probability density functions (PDFs) of the DTs.
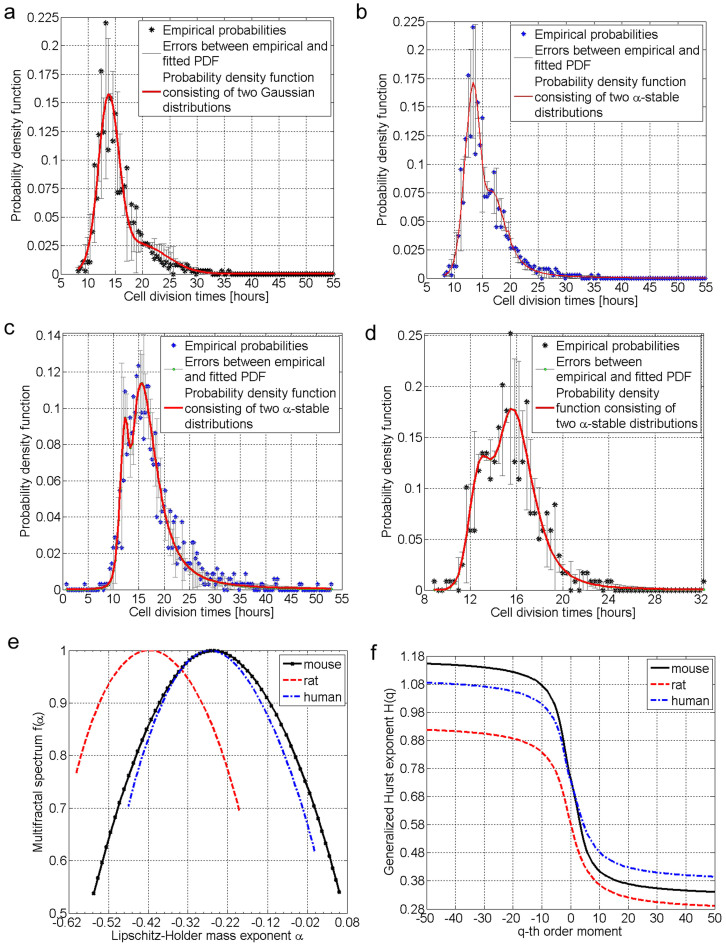
To overcome the poor fitting of uni-modal distributions and explain the observed heterogeneity in the PDFs of stem cell DTs, we consider the bi-modal statistical investigation and two PDF candidates: i) a bi-modal Gaussian distribution (Figure 4.a) and ii) a bi-modal asymmetric α-stable distributions (Figures 4.b–d). By employing the MLE strategy for a mixture of two asymmetric α-stable distributions and selecting the best fit according to the best Kolmogorov-Smirnov (K-S) test result, we observe that the empirical PDFs of mouse, rat, and human stem cell DTs are well fitted by the bi-modal α-stable distribution. Indeed, both graphically and statistically by looking at the error bars, we can see that the fitting involving the α-stable distribution captures much better the overall trend than the Gaussian one (Figure 4.a).
Figure 4. Statistical analysis of bi-modal asymmetric α-stable distribution fitting and multi-fractal analysis of stem cell division times.
(a) Comparison between the empirical PDF (black dots) of mouse stem cell division times (DTs) and the bi-modal Gaussian distribution (red line). (b) Comparison between the empirical PDF of mouse stem cell DTs and the bi-modal α-stable distribution for all 889 samples. The high p-value of 0.78 for the K-S test shows that the postulated bi-modal α-stable distribution cannot be rejected as a model. (c) Comparison between the empirical PDF of rat stem cell DTs and the bi-modal α-stable distribution for all 1055 samples. The p-value of 0.11 for the K-S test shows that the postulated bi-modal α-stable distribution cannot be rejected as a model. (d) Comparison between the empirical PDF of human MSC DTs and the bi-modal α-stable distribution for all 350 samples. The p-value of 0.81 for the K-S test shows that the postulated bi-modal α-stable distribution cannot be rejected as a model. (e) Multi-fractal spectrum as a function of Lipschitz-Holder mass exponent for mouse, rat and human muscle derived stem cell division times. (f) Generalized Hurst exponent as a function of q-th order moment for mouse, rat and human muscle derived stem cell division times.
In sum, the magnitude of p-values of the K-S tests imply not only that we cannot reject the bi-modal α-stable distribution as a viable model for stem cell DTs, but also that the stem cells population exhibits a heterogeneous structure consisting of at least two sub-populations, namely a sub-population of stem cells that divide faster (with an average DT of approximately 12 hours and corresponding to first peak in the PDF plot of Figure 4.b), and a second sub-population of stem cells with a slower dynamics (with an average DT of approximately 18 hours corresponding to the second peak in the PDF plot of Figure 4.b) which exhibits a long-tail type of behavior.
Stem cell division times exhibit non-stationary behavior
Besides the heterogeneous structure of stem cells population, we also observe that the empirical PDF estimated from stem cell DTs exhibits a pronounced time dependent behavior (Supplementary Material Figure 5). This time dependent behavior is illustrated not only by the changes in the shape of the empirical PDF at different time windows, but also by the estimated values of the bi-modal PDF. Indeed, the PDF investigation shows that stem cell growth is not a stationary process, but rather a highly dynamic one characterized by time-dependent PDF variation.
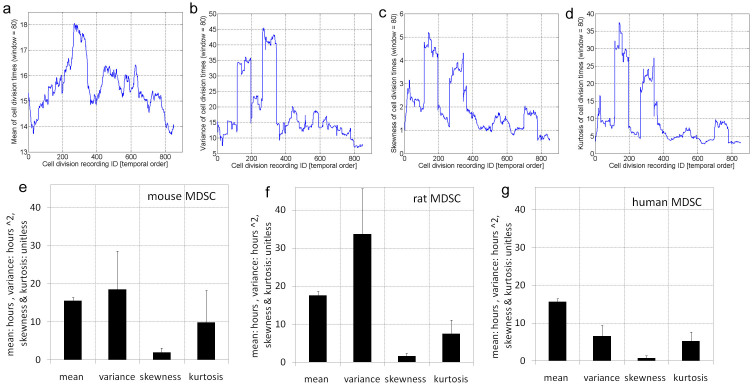
To further investigate the existence of a non-stationary behavior, we measure the mean (Figure 5.a), variance (Figure 5.b), skewness (Figure 5.c), and kurtosis (Figure 5.d) over a sliding window of 80 cell DT recordings (from the time-ordered mouse stem cell DT data). Additionally, Figures 5.e–g show the error bars in the time variation of mean, variance, skewness, and kurtosis for mouse, rat, and human cells. We observe that these higher order moments vary with time which suggests that the cell division is a non-stationary process (see Supplementary Materials Figures 6 and 7). For instance, the mean of the rat stem cell DTs exhibit a wide range variation from 15 hours in the beginning to almost 19 hours at later times. Similarly, the variance, skewness, and kurtosis exhibit a spiky behavior which cannot be modeled if one assumes a stationary behavior. In addition, the non-zero values for skewness and kurtosis show that the rat stem cell DTs do not display a Gaussian behavior; this supports our predictions concerning the existence of a non-Gaussian dynamics of stem cell growth shown in Fig. 3. Further, the non-zero skewness shows that the stem cell DTs are not symmetrically distributed around the mean and may exhibit different tails at both ends of the PDF curve. Similarly, the non-zero kurtosis implies that the stem cell DTs can exhibit large values and so a better modeling approach than the Gaussian-based framework needs to rely on long-tail distributions.
Figure 5. Higher order moment analysis of mouse, rat, and human stem cells division times.
(a) Moving average analysis of mean of mouse stem cell division times over a time window of 80 samples. (b) Moving average analysis of variance of mouse stem cell division times over a time window of 80 samples. (c) Moving average analysis of skewness of mouse stem cell division times over a time window of 80 samples. (d) Moving average analysis of kurtosis of mouse stem cell division times over a time window of 80 samples. Moving average analysis of higher order moments of stem cell division times shows that cell growth exhibits a pronounced time dependent behavior. (e) Average values and errors for the mean, variance, skewness and kurtosis of the mouse stem cell division times. (f) Average values and errors for the mean, variance, skewness and kurtosis of the rat stem cell division times. (g) Average values and errors for the mean, variance, skewness and kurtosis of the human MSC division times.
Moreover, the observed variability and spiky dynamics of these higher moments imply that the stem cell DTs exhibit a heteroscedastic dynamics (i.e., some sub-populations exhibit different moment variability compared to others). An alternative proof of existence of heteroscedastic dynamics is shown in the Supplementary Material Figure 8 where shuffled stem cell DT series lose their correlation structure when compared to the initial data sets. In general terms, the heteroscedastic dynamics means that the statistical moments of the stem cell DTs and, implicitly, those associated with stem cell growth exhibit a local variation which is very different from the global one or asymptotic limits. In addition, the very existence of this heteroscedastic behavior may not only confirm the heterogeneity of stem cell population, but also be indicative of an aging phenomenon that is characteristic to all multi-cellular organisms. As we later show in the paper, this aspect can play a crucial role not only in constructing an accurate mathematical model of stem cells growth, but also for predicting the structure and size of the entire population over time with high confidence while accounting through non-stationary methods and tools for the age of a patient.
Stem cell growth rates possesses multi-fractal characteristics
For a comprehensive investigation of the heteroscedastic dynamics of stem cell growth, we investigate the relationship between the higher order moments of stem cells dynamics and their order; we also estimate both the multi-fractal spectrum and generalized Hurst exponent function [A fractal is a geometrical object or stochastic process that displays self-similarity, on all scales and is characterized by a single fractal dimension. The fractal dimension is a mathematical concept measuring the unique features such as geometrical shape of an object or irregularity of a stochastic process. The object need not exhibit exactly the same structure at all scales, but the same “type” of structures must appear on all scales. Similarly, a stochastic process is regarded as fractal if its probability distribution function measured at two scales is self-similar and related to each other via a power law relationship. In contrast, a multi-fractal object or stochastic process is characterized by a series of fractal dimensions.]. The rationale for estimating the generalized Hurst function in Figure 4.f is two-fold: (1) It allows us to detect the existence of multi-fractality. For instance, if the generalized exponent is independent of the order q of the moment (see Figure 4.f), then the stochastic process is mono-fractal. In contrast, if the generalized Hurst exponent is dependent of the order q of the moment, then the process is called multi-fractal (see Figure 4.f). (2) It allows us to compute the Hurst parameter and discriminate between short-range and long-range memory effects. For instance, if the Hurst exponent equals 0.5 then we can state that the dynamics of stem cell populations is governed by short-range memory or Markovian models. In contrast, if the Hurst exponent ranges between 0.5 and 1, then the dynamics of stem cell populations is said to possess long-range memory characteristics and non-Markovian dynamical models are needed. While mono-fractality implies that a single Lipschitz-Holder exponent67 (i.e., a function f(t) is said to have Lipschitz-Holder exponent a around point t if f(t + x) − f(t) ~ xa) characterizes the entire cell dynamics, the multi-fractal behavior shows that the existing temporal heterogeneity in stem cell population leads to multiple Lipschitz-Holder exponents. The multi-fractal spectrum encapsulates in its distribution of Lipschitz-Holder exponents information about multiple point correlations existing both in space and time. Implicitly, it is also a measure of the memory characterizing the stem cell dynamics.
By measuring the dependency of the higher order statistical moments on their order value and using the Legendre transformation, we estimate the multi-fractal spectrum for the mouse, rat, and human stem cell DTs (Figure 4.e and Supplementary Material Figure 9). One can clearly observe from Figure 4.e not only that the stem cell DTs exhibit a wide multi-fractal spectrum, but also the Lipschitz-Holder exponents are mostly negative. The fact that the stem cell DTs display a significant range of negative Lipschitz-Holder exponents implies that the stem cell growth process exhibits an oscillating behavior consisting of active periods of growth (when the majority of the cells are actively dividing) followed by periods when fewer cells are recruited into the cell cycle. Alternatively, the results in Figure 4.f (and Supplementary Material Figure 10) display a complex nonlinear dependency between the generalized Hurst exponent and the q-th order moment, which again supports the existence of multi-fractal behavior in stem cell growth.
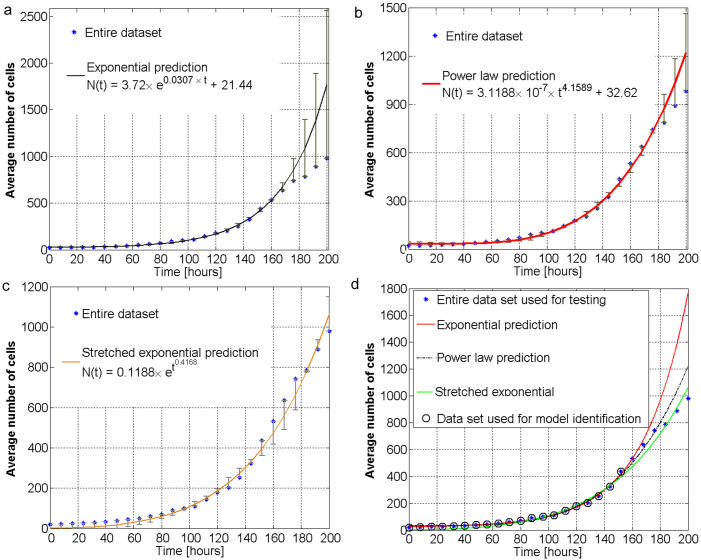
Based on these findings, our subsequent question is whether the current models relying on the assumption of either having constant or exponential growth in stem cell population size are suitable for predicting the population size; answering this question is critical for designing cell therapies or tissue engineered constructs for future personalized medicine. Therefore, we next considered the time lapsed traces of the number of stem cells in the population over time and investigate the goodness-of-fit for three mathematical modeling approaches: i) exponential growth associated with a non-fractal dynamics (Figure 6.a); ii) power law growth corresponding to a mono-fractal dynamics (Figure 6.b); and iii) a stretched exponential hinting towards a multi-fractal approach (Figure 6.c). We observe that the stretched exponential and power law type of models offer better prediction capabilities than the state-of-the art exponential models (see Figure 6.d). By ignoring the multiscale biological dynamics and spatio-temporal correlations, the exponential models can be misleading. Indeed, the exponential growth assumption can lead to very optimistic predictions concerning the population size of stem cells and this may have detrimental effects on the therapeutic strategies. Our results suggest that the multi-fractal behavior of stem cell dynamics can be modeled as a superposition of several stretched exponential functions.
Figure 6. Implications of multi-fractal behavior on population size and their contrasting differences when considering simplified models.
(a) Comparison between the empirical average number of rat stem cells as a function of time and the exponential growth model. (b) Comparison between the empirical average number of rat stem cells as a function of time and the power-law growth model. (c) Comparison between the empirical average number of rat stem cells as a function of time and the stretched exponential growth model. (d) Empirical average number of rat stem cells as a function of time and three mathematical interpolations, i.e. exponential (red line), power law (black dotted line), and stretched exponential (green line). Their best parameters are shown in the legends in plots a–c. The first 75% of the data samples (~150 hours) also shown in black circles are used to identify the parameters of each model. The remaining 25% of the data samples (~50 hours) are used to test the prediction capabilities of each model. One can notice that the stretched exponential and power law models have better prediction capabilities than the simple exponential model. This is in agreement with our multi-fractal analysis which predicts that the intrinsic growth of stem cell population is characterized by non-exponential features.
Discussion
Here, we show for the first time that distinct subpopulations of dividing cells exist within several heterogeneous stem cell populations. We report that the populations show time-dependent and fractal behavior. Both newborn derived mesenchymal stem cells and adult derived muscle stem cells demonstrate these population characteristics. By employing non-Gaussian statistics, statistical hypothesis testing and statistical inference, we identify the presence of two unique dividing subpopulations of stem cells, each with a distinct cell division rate and characterized by a different probability density function of DTs. Previous reports treated stem cells as homogeneous populations and assumed either a constant division time for the whole population or Gaussian distributed DTs. In terms of improving how stem cells are defined, our study strongly supports the notion of defining cells from a population level perspective in order to account for cell-cell interactions that affect the population structure (composition of different subpopulations) and dynamics (interactions of and changes in the subpopulations).
Bacteria and yeast cell division and growth rates have been examined extensively for more than 50 years; the distributions appear to be rather homogeneous68,69,70,71 and, to the best of our knowledge, there are no reports of bi– or multi–modal growth rates. For instance, Tsuru et al.72 recognize the existence of fluctuations in E.coli growth rates and investigated how the changes in cell volume of individual bacteria are correlated with fluctuations in protein concentration. Complementing such a phenotypic diversity, a report by Niven et al. describes skewed cell division distributions in E. coli in the presence of environmental factors73. The authors examined several distributions to fit their data (also obtained via time-lapsed cell imaging) and no definitive model distribution was identified, possibly due to the small sample size73.
More recent studies of differentiated cells, particularly those that use similar technology to permit the collection of a dataset of individual cell measurements, also did not report bi-modality56,59,74,75. For instance, Tyson et al.76 emphasized the importance of investigating the statistics of intermitotic times and modeled these times via an exponentially modified Gaussian distribution. Based on their analysis, Tyson et al. concluded that their proposed exponentially modified Gaussian models are not definitive and require further improvement to accurately accommodate age structure and dynamics of stem cell populations. Along the same lines, Chang et al.77 demonstrate the existence of cell-to-cell variability in clonal populations of mouse hematopoietic progenitor cells and showed a link to heterogeneity of gene expression. Tan et al.78 used single-cell gene expression to show functional heterogeneity in epidermal cells, while Koschmieder et al.79 and Cantor et al.80 investigated it in the hematopoietic lineage. We see that such mechanism might be an underlying cause of the heterogeneity in our populations of stem cells. Furthermore, in our own previous reports, we used both standard Gaussian and non-parametric statistics to examine the distributions of cell cycle times in human and mouse stem cells. However, lack of sophistication in our previous statistical analyses and the small size29,30, did not allow us to identify bi-modality (let alone multi-modality) in stem cells dynamics which we report here.
Stem cell population heterogeneity may develop through the process of spontaneous progression of stem cell differentiation. It is worth mentioning that the interaction of the cells with the culture substrate and the rigidity of the substrate are important factors in modulating differentiation and cell phenotype heterogeneity as has been shown by Gilbert et al.81. In most scenarios, non-dividing quiescent cells are presumed to become activated to slowly dividing multipotent stem cells which then progress to transiently amplifying (TA) committed progenitors and finally non-dividing differentiated cells. Here, we detected, in all populations the presence of a non-dividing quiescent subpopulation that could re-enter the cell cycle. The slowly dividing subpopulation for MSCs (16.8 hrs), rat stem cells (21.2 hrs), or mouse stem cells (21.9 hrs) was at least 4 hours slower than the fast diving subpopulation (human MSC 12.8 hrs, rat stem cells 11.7 hrs, mouse stem cells 13.2 hrs). It is possible that the slow dividing fraction represents multipotent cells, while the faster dividing cells represent committed precursors or transiently–amplifying cells. However, at this point, there are no clear methods to physically separate these subpopulations, and BrdU pulsing experiments (Fig. 2) did not show a correlation between the myogenic precursor marker Desmin and BrdU rate of incorporation in stem cell populations. Nevertheless, the statistical analysis demonstrates the presence of two distinct populations which grow at different rates. This was detected in different species, and in different stem cell types, which suggests that it may be indeed a universal characteristic.
The existence of these different phenotypes may allow for both short-term and long-term functions of the stem cell populations in response to tissue injury. Indeed, fast dividing TA cells can provide immediate restoration of damaged/lost differentiated cells, while the dynamic of TA cell depletion for acute repair may illicit a faster transition rate for cells in the multi-potent state to move to the TA state to re-establish some equilibrium of subpopulations. In this scenario, because dividing cells expand according to a stretched exponential, the quiescent stem cell activation could be conservative and occur only in the case of severe tissue loss or damage. In the case of normal tissue homeostasis, we could expect to observe oscillating dynamics (Figure 5.a or Supplementary Material Figures 6.a–c and 7.a–c) related to cell division behavior in the population, as normal cell death occurs, and lost cells are replaced.
Further, our statistical analysis demonstrates that the population subsets display a non-stationary (i.e., time-dependent statistics). Non-stationary statistical analysis shows that not only do individual cells change over time, intrinsically, but the stem cell population also changes over time. In part, the non-stationary phenomenon finding shows that stem cells exhibit an aging process which is characteristic to all living organisms. Consequently, the rates of transition between subpopulation states will be important for understanding their evolution over time.
The fractal behavior we observe in stem cell DTs identifies a stochastic process that displays a probability density function that is self-similar in nature82,83, i.e., the distribution of cell DTs at one scale can be retrieved from that of another scale by using the fractal dimension and a scaling coefficient. Simply speaking, the existence of fractal statistics in stem cell DTs implies that the birth of a new cell is not a random (i.e., uncorrelated) event from the development of previous ones. In other words, there is some form of correlation between cell divisions and this leads to a complex behavior which cannot be quantified by current mathematical models of stem cell growth. Nevertheless, due to numerous sources of variation (e.g., intrinsic differences, environmental interactions, nutrient availability, cell density), stem cell DTs cannot be characterized by a single fractal dimension, but by a set of scaling exponents which also demonstrates the existence of heterogeneity in stem cell population structure. Taking into account such characteristics allows us to construct an accurate dynamical system approach as described by Furusawa and Kaneko84,85 which may explain and, more importantly, predict the stem cell dynamics. Furthermore, the newly proposed model can be extended to include cell cycle and cell volume, along the lines of Halter et al.86 and Anderson et al.87; these parameters may be used to show the multi-fractal behavior as the cells interact with each other.
The value for teasing out the subsets of cells, their interactions, time-dependent and fractal behavior has implications for applied cell therapeutics. First, it is needed to identify/define the stem cell product. Second, it is necessary to accurately predict ex vivo growth rates of the stem cell populations. In regards to developing an FDA-approved cell therapeutic, purity and potency criteria need to be established. Recognizing that subpopulations, and hence multiple targets, exist in the stem cell population is essential to developing tools to control the fate of stem cells. Although, we and others have shown that the quiescent population exists and requires nonlinear growth models28,29,30,88, our study is the first to show quantitatively and qualitatively that more sophisticated growth models are warranted due to the presence of multiple phenotypes. Models which account for population dynamics can better predict whether cytokines or growth factors that increase proliferation rates (e.g., for the purposes of obtaining certain cell doses for transplantations) cause perturbations in the dynamics of subpopulations, and how this affects the behavior of the entire cells population.
This statistical approach may also be exploited for personalized medicine strategies. Using high throughput live cell imaging and our computational analyses, inter-patient variability and intrinsic intra-patient variation in stem cells dynamics can be studied. In the era of individualized medicine, the computational modeling may also include genomics and proteomics information and the dynamics of an individual biopsy-derived stem cell growth can be assessed before developing the cell therapy approach. Personalized medicine could also exploit our statistical investigation for estimating the richness of fractal behavior at the cellular level and make predictions at higher scales. By early detection of losing fractal richness at the level of stem cell evolution, one could predict the signs of genetic instability and onset of a disease well in advance of actual symptoms in the patient, when it may be hard, or impossible, to intervene.
Further studies are needed to examine the molecular phenotype and transition rates of the two dividing subsets identified here. Epigenetic modifications may account for differences in phenotype or division rates, or the cell-cell communications may modulate division rates. The oscillating time-dependent behavior together with the finding that these subpopulations interact with each other (i.e., cells do not grow in isolation, but instead interact at various scales). In addition to that, the cells indeed interact with their microenvironment57 including the extracellular matrix89, growth factors, chemokines90, and the substrate they are adhering to81. These interactions have been shown to affect the differentiation and fate decisions of stem cells. All of these factors call for developing new mathematical models describing their interaction and overall population growth. A challenge to these studies will be the isolation of these subsets and maintenance without re-establishing the heterogeneous parent population.
Methods
We used time-lapsed imaging60,61 to directly examine the cell growth of mouse and rat stem cells isolated from skeletal muscle tissue and human mesenchymal stem cells. (Additional detail regarding imaging experimental set-up is included in60,61 and stem cell isolation62,63 is included in the Supplementary Material Note).Time-lapsed images were acquired at 10-minute intervals over the course of 5 days on collagen-coated surfaces. To determine the divisional status of the cells, and the cell cycle length of dividing cells, we used a custom developed program, i.e., ImageView (Karios Instruments, Harmar, PA). Cell cycle length was recorded as the time elapsed between cytokinetic events, as previously reported29,30,91. Dividing cells were classified as cells with a visible DT. Non-dividing cells were classified as cells that were visible on screen for over twice the average DT.
After determining DTs, the cells were tracked in order to perform lineage analysis and determine differences between dividing and non-dividing cells while also identifying the quiescent cells. A custom built image analysis program, CytoTracker, (Karios Instruments, Harmar,PA), was used to perform lineage analysis. A cell was selected and followed for the entire time it was visible on screen. When a cell divides, the division event is recorded, and each daughter cell was subsequently tracked. The entire cell lineage is tracked until either the end of the video or the cells are lost to follow up. Each cell is assigned a name in order to keep track of the lineage.
The initial parent cell is 1.0. The two daughter cells are named 1.1 and 1.2. Their daughter cells are named 1.11, 1.12 and 1.21, 1.22 respectively. The tracks/locations (x and y location) for the entire family of cells are recorded along with the cell name for each jpeg image.This allows a cells lineage to be determined. The cell histories can then be converted to a graphical display, or lineage tree which displays all the cells in the family, and their DTs, or their time on screen if the cell was lost to follow up (Figure 2). In the graphical displays, the vertical lines represent a cell, and the length of the line corresponds to how long the cell was on screen. Lineage trees allow the divisional status of a cell to be determined. A tree with many cells represents a cell that is actively dividing and giving rise to progeny that continue to divide and contribute to renewal of the population. The lineage trees also allow DTs to be compared, or to identify cells that are non-dividing and visible on screen for over twice the division time. Also, asymmetrical divisions can easily be identified.
Author Contributions
P.B., B.D., B.G. and R.M. contributed to the discussion, design the experiments and writing of the manuscript. B.D., T.R. and B.G. contributed to performing the biological experiments, saving and extracting the biological datasets. P.B. and R.M. contributed to developing the statistical analysis of the biological experimental data. P.B., B.D., B.G. and R.M. prepared the figures, wrote their captions and organized the information in both main text and supplementary material.
Supplementary Material
Supplementary Material
Acknowledgments
Authors acknowledge the help of Dr. Johnny Huard, director of the Stem Cell Research Center for allowing access to some of his lab resources.
References
- Qian X., Goderie S. K., Shen Q., Stern J. H. & Temple S. Intrinsic programs of patterned cell lineages in isolated vertebrate CNS ventricular zone cells. Development 125, 3143–3152 (1998). [DOI] [PubMed] [Google Scholar]
- Zhong W., Jiang M. M., Weinmaster G., Jan L. Y. & Jan Y. N. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development 124, 1887–1897 (1997). [DOI] [PubMed] [Google Scholar]
- Jan Y. N. & Jan L. Y. Asymmetric cell division. Nature 392, 775–778 (1998). [DOI] [PubMed] [Google Scholar]
- Watt F. M. & Hogan B. L. Out of Eden: stem cells and their niches. Science 287, 1427–1430 (2000). [DOI] [PubMed] [Google Scholar]
- Hackney J. A. et al. A molecular profile of a hematopoietic stem cell niche. Proc Natl Acad Sci U S A 99, 13061–13066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzel M. et al. The myeloid-lymphoid initiating cell (ML-IC) assay assesses the fate of multipotent human progenitors in vitro. Blood 93, 3750–3756 (1999). [PubMed] [Google Scholar]
- Kiger A. A., White-Cooper H. & Fuller M. T. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature 407, 750–754 (2000). [DOI] [PubMed] [Google Scholar]
- Spradling A., Drummond-Barbosa D. & Kai T. Stem cells find their niche. Nature 414, 98–104 (2001). [DOI] [PubMed] [Google Scholar]
- Lee H. S. et al. Clonal expansion of adult rat hepatic stem cell lines by suppression of asymmetric cell kinetics (SACK). Biotechnol Bioeng 83, 760–771 (2003). [DOI] [PubMed] [Google Scholar]
- Hope K. & Bhatia M. Clonal interrogation of stem cells. Nat Methods 8, S36–40, 10.1038/nmeth.1590 (2011). [DOI] [PubMed] [Google Scholar]
- Phinney D. G. Functional heterogeneity of mesenchymal stem cells: implications for cell therapy. J Cell Biochem 113, 2806–2812, 10.1002/jcb.24166 (2012). [DOI] [PubMed] [Google Scholar]
- Benz C. et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell stem cell 10, 273–283, 10.1016/j.stem.2012.02.007 (2012). [DOI] [PubMed] [Google Scholar]
- Qian H. et al. Molecular characterization of prospectively isolated multipotent mesenchymal progenitors provides new insight into the cellular identity of mesenchymal stem cells in mouse bone marrow. Mol Cell Biol. 33, 661–677, 10.1128/MCB.01287-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. A. et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122, 289–301, 10.1016/j.cell.2005.05.010 (2005). [DOI] [PubMed] [Google Scholar]
- Mitchell P. O. et al. Sca-1 negatively regulates proliferation and differentiation of muscle cells. Dev Biol 283, 240–252 (2005). [DOI] [PubMed] [Google Scholar]
- Molnar G., Ho M. L. & Schroedl N. A. Evidence for multiple satellite cell populations and a non-myogenic cell type that is regulated differently in regenerating and growing skeletal muscle. Tissue Cell 28, 547–556 (1996). [DOI] [PubMed] [Google Scholar]
- Schultz E. Satellite cell proliferative compartments in growing skeletal muscles. Dev Biol 175, 84–94 (1996). [DOI] [PubMed] [Google Scholar]
- Deasy B. M., Li Y. & Huard J. Tissue engineering with muscle-derived stem cells. Curr Opin Biotechnol 15, 419–423 (2004). [DOI] [PubMed] [Google Scholar]
- Zammit P. & Beauchamp J. The skeletal muscle satellite cell: stem cell or son of stem cell? Differentiation 68, 193–204 (2001). [DOI] [PubMed] [Google Scholar]
- Wagers A. J. & Conboy I. M. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell 122, 659–667 (2005). [DOI] [PubMed] [Google Scholar]
- Ezine S., Jerabek L. & Weissman I. The phenotype of thymocytes derived from a single clonogenic precursor. J Immunol 139, 2195–2199 (1987). [PubMed] [Google Scholar]
- Jankowski R. J., Deasy B. M., Cao B., Gates C. & Huard J. The role of CD34 expression and cellular fusion in the regeneration capacity of myogenic progenitor cells. J Cell Sci 115, 4361–4374 (2002). [DOI] [PubMed] [Google Scholar]
- Baroffio A. et al. Identification of self-renewing myoblasts in the progeny of single human muscle satellite cells. Differentiation 60, 47–57, 10.1046/j.1432-0436.1996.6010047.x (1996). [DOI] [PubMed] [Google Scholar]
- Wagers A. J., Sherwood R. I., Christensen J. L. & Weissman I. L. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science 297, 2256–2259 (2002). [DOI] [PubMed] [Google Scholar]
- Sherwood R. I. et al. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119, 543–554, 10.1016/j.cell.2004.10.021 (2004). [DOI] [PubMed] [Google Scholar]
- Committee on Ranking FDA Product Categories Based on Health Consequences, Phase II; National Research Council. . A risk-characterization framework for decision-making at the Food and Drug Administration. (National Academies Press, Washington, D.C., 2011). [PubMed]
- US Food and Drug Administration. . US Food and Drug Administration. Guidance for Industry. Current Good Tissue Practice (CGTP) and Additional Requirements for Manufacturers of Human Cells, Tissues, and Cellular and Tissue-Based Products (HCT/Ps) (US FDA, Washington, D.C., 2011). Accessed on December 2011: http://www.fda.gov/downloads/biologicsbloodvaccines/guidancecomplianceregulatoryinformation/guidances/tissue/ucm285223.pdf.
- Sherley J. L., Stadler P. B. & Stadler J. S. A quantitative method for the analysis of mammalian cell proliferation in culture in terms of dividing and non-dividing cells. Cell Prolif 28, 137–144 (1995). [DOI] [PubMed] [Google Scholar]
- Deasy B. M., Qu-Peterson Z., Greenberger J. S. & Huard J. Mechanisms of muscle stem cell expansion with cytokines. Stem Cells 20, 50–60 (2002). [DOI] [PubMed] [Google Scholar]
- Deasy B. M. et al. Modeling stem cell population growth: incorporating terms for proliferative heterogeneity. Stem Cells 21, 536–545 (2003). [DOI] [PubMed] [Google Scholar]
- MacArthur B. D., Please C. P., Taylor M. & Oreffo R. O. Mathematical modelling of skeletal repair. Biochem Biophys Res Commun 313, 825–833 (2004). [DOI] [PubMed] [Google Scholar]
- Mancuso L. et al. Experimental analysis and modelling of in vitro proliferation of mesenchymal stem cells. Cell Prolif 42, 602–616, 10.1111/j.1365-2184.2009.00626.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M. & Kawanabe M. Machine Learning in Non-Stationary Environments: Introduction to Covariate Shift Adaptation. (MIT Press, 2012). [Google Scholar]
- Gyllenberg M. & Webb G. F. Quiescence as an explanation of Gompertzian tumor growth. Growth Dev Aging 53, 25–33 (1989). [PubMed] [Google Scholar]
- Gyllenberg M. & Webb G. F. A nonlinear structured population model of tumor growth with quiescence. J Math Biol 28, 671–694 (1990). [DOI] [PubMed] [Google Scholar]
- Izquierdo J. M. & Perez C. A stochastic approach for the interpretation of single pulse experiments in morphological multicompartments of renewing and exponentially growing cell populations. J Theor Biol 101, 39–75 (1983). [DOI] [PubMed] [Google Scholar]
- Morris V. B. & Cowan R. A growth curve of cell numbers in the neural retina of embryonic chicks. Cell Tissue Kinet 17, 199–208 (1984). [DOI] [PubMed] [Google Scholar]
- Cowan R. & Morris V. B. Determination of proliferative parameters from growth curves. Cell Tissue Kinet 20, 153–159 (1987). [DOI] [PubMed] [Google Scholar]
- Hirsch H. R. Influence of the existence of a resting state on the decay of synchronization in cell culture. J Theor Biol 111, 61–79 (1984). [DOI] [PubMed] [Google Scholar]
- Jones R. B. & Smith J. R. A stochastic model of cellular senescence. II. Concordance with experimental data. J Theor Biol 96, 443–460 (1982). [DOI] [PubMed] [Google Scholar]
- Voit E. O. Cell cycles and growth laws: the CCC model. J Theor Biol 114, 589–599 (1985). [DOI] [PubMed] [Google Scholar]
- Abu-Absi N. R. & Srienc F. Instantaneous evaluation of mammalian cell culture growth rates through analysis of the mitotic index. J Biotechnol 95, 63–84 (2002). [DOI] [PubMed] [Google Scholar]
- Setty Y., Dalfo D., Korta D. Z., Hubbard E. J. & Kugler H. A model of stem cell population dynamics: in silico analysis and in vivo validation. Development 139, 47–56, 10.1242/dev.067512 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busini V., Arosio P. & Masi M. Mechanistic modelling of avascular tumor growth and pharmacokinetics influence-Part I. Chem. Eng. Sci 62, 1877–1886 (2007). [Google Scholar]
- Sidoli F., Asprey S. & Mantalaris A. A coupled single cell-population-balance model for mammalian cell cultures. Ind. Eng. Chem. Res. 45, 5801–5811 (2006). [Google Scholar]
- Lemon G., Waters S. L., Rose F. R. & King J. R. Mathematical modelling of human mesenchymal stem cell proliferation and differentiation inside artificial porous scaffolds. J Theor Biol 249, 543–553, 10.1016/j.jtbi.2007.08.015 (2007). [DOI] [PubMed] [Google Scholar]
- Liu Y. H., Bi J. X., Zeng A. P. & Yuan J. Q. A population balance model describing the cell cycle dynamics of myeloma cell cultivation. Biotechnol Prog 23, 1198–1209, 10.1021/bp070152z (2007). [DOI] [PubMed] [Google Scholar]
- Basse B., Baguley B. C., Marshall E. S., Wake G. C. & Wall D. J. Modelling cell population growth with applications to cancer therapy in human tumour cell lines. Prog Biophys Mol Biol 85, 353–368, 10.1016/j.pbiomolbio.2004.01.017 (2004). [DOI] [PubMed] [Google Scholar]
- Cipollina C., Vai M., Porro D. & Hatzis C. Towards understanding of the complex structure of growing yeast populations. J Biotechnol 128, 393–402, 10.1016/j.jbiotec.2006.10.012 (2007). [DOI] [PubMed] [Google Scholar]
- Galle J., Hoffmann M. & Krinner A. Mesenchymal Stem Cell Heterogeneity and Ageing In Vitro: A Model Approach. In Computational Modeling in Tissue Engineering Vol. 10 183–205 (Springer-Verlag 2013).
- Cao Y. et al. Modeling spatial population dynamics of stem cell lineage in tissue growth. Conf Proc IEEE Eng Med Biol Soc 2012, 5502–5505, 10.1109/EMBC.2012.6347240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauche I. et al. Stem cell proliferation and quiescence--two sides of the same coin. PLoS Comp Biol 5, e1000447, 10.1371/journal.pcbi.1000447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129, 10.1016/j.cell.2008.10.048 (2008). [DOI] [PubMed] [Google Scholar]
- Foudi A. et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol 27, 84–90, 10.1038/nbt.1517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh Y. H., Cohen J. & Sherley J. L. Higher 5-hydroxymethylcytosine identifies immortal DNA strand chromosomes in asymmetrically self-renewing distributed stem cells. Proc Natl Acad Sci U S A 110, 16862–16867, 10.1073/pnas.1310323110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahnson A. et al. Automated measurement of cell motility and proliferation. BMC Cell Biol 6, 19, 10.1186/1471-2121-6-19 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf N. et al. On the symmetry of siblings: automated single-cell tracking to quantify the behavior of hematopoietic stem cells in a biomimetic setup. Exp Hematol 40, 119–130 e119, 10.1016/j.exphem.2011.10.009 (2012). [DOI] [PubMed] [Google Scholar]
- Rapoport D. H., Becker T., Madany Mamlouk A., Schicktanz S. & Kruse C. A novel validation algorithm allows for automated cell tracking and the extraction of biologically meaningful parameters. PloS one 6, e27315, 10.1371/journal.pone.0027315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot F. D. et al. A time-series method for automated measurement of changes in mitotic and interphase duration from time-lapse movies. PloS one 6, e25511, 10.1371/journal.pone.0025511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirieleison S. M. et al. Automated live cell imaging systems reveal dynamic cell behavior. Biotechnol Prog 27, 913–924, 10.1002/btpr.629 (2011). [DOI] [PubMed] [Google Scholar]
- Deasy B. M. et al. Long-term self-renewal of postnatal muscle-derived stem cells. Mol Biol Cell 16, 3323–3333 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharaibeh B. et al. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nature protocols 3, 1501–1509, 10.1038/nprot.2008.142 (2008). [DOI] [PubMed] [Google Scholar]
- Qu-Petersen Z. et al. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. J Cell Biol 157, 851–864 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. I. Mesenchymal stem cells. J Orthop Res 9, 641–650, 10.1002/jor.1100090504 (1991). [DOI] [PubMed] [Google Scholar]
- Pittenger M. F. et al. Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 (1999). [DOI] [PubMed] [Google Scholar]
- Hartigan J. A. & Hartigan P. M. The dip test of unimodality. Ann. Statist. 13, 70–84 (1985). [Google Scholar]
- Feder J. Fractals. (Plenum Press, 1988). [Google Scholar]
- Brock T. D. Microbial growth rates in nature. Bacteriol Rev 35, 39–58 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. J., Marr A. G. & Painter P. R. Kinetics of growth of individual cells of Escherichia coli and Azotobacter agilis. J Bacteriol 93, 605–617 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueba F. J., Neijssel O. M. & Woldringh C. L. Generality of the growth kinetics of the average individual cell in different bacterial populations. J Bacteriol 150, 1048–1055 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D., Gottlieb P. & Rosenberg E. Division cycle of Myxococcus xanthus. 3. Kinetics of cell growth and protein synthesis. J Bacteriol 105, 811–819 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuru S. et al. Noisy cell growth rate leads to fluctuating protein concentration in bacteria. Phys Biol 6, 036015, 10.1088/1478-3975/6/3/036015 (2009). [DOI] [PubMed] [Google Scholar]
- Niven G. W., Morton J. S., Fuks T. & Mackey B. M. Influence of environmental stress on distributions of times to first division in Escherichia coli populations, as determined by digital-image analysis of individual cells. Appl Environ Microbiol 74, 3757–3763, 10.1128/aem.02551-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau G. et al. Dynamical modelling of phenotypes in a genome-wide RNAi live-cell imaging assay. BMC Bioinformatics 14, 308, 10.1186/1471-2105-14-308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder N. et al. Automatic analysis of dividing cells in live cell movies to detect mitotic delays and correlate phenotypes in time. Genome Res 19, 2113–2124, 10.1101/gr.092494.109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson D. R., Garbett S. P., Frick P. L. & Quaranta V. Fractional proliferation: a method to deconvolve cell population dynamics from single-cell data. Nat Methods 9, 923–928, 10.1038/nmeth.2138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. H., Hemberg M., Barahona M., Ingber D. E. & Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547, 10.1038/nature06965 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D. W. et al. Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development 140, 1433–1444, 10.1242/dev.087551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmieder S., Rosenbauer F., Steidl U., Owens B. M. & Tenen D. G. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int J hematol 81, 368–377 (2005). [DOI] [PubMed] [Google Scholar]
- Cantor A. B., Katz S. G. & Orkin S. H. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol 22, 4268–4279 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P. M. et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081, 10.1126/science.1191035 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbrot B. How long is the coast of britain? Statistical self-similarity and fractional dimension. Science 156, 636–638 (1967). [DOI] [PubMed] [Google Scholar]
- Mandelbrot B. B. The fractal geometry of nature. (W.H. Freeman and Company, 1982). [Google Scholar]
- Furusawa C. & Kaneko K. Theory of robustness of irreversible differentiation in a stem cell system: chaos hypothesis. J Theor Biol 209, 395–416, 10.1006/jtbi.2001.2264 (2001). [DOI] [PubMed] [Google Scholar]
- Furusawa C. & Kaneko K. Epigenetic feedback regulation accelerates adaptation and evolution. PloS one 8, e61251, 10.1371/journal.pone.0061251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter M., Elliott J. T., Hubbard J. B., Tona A. & Plant A. L. Cell volume distributions reveal cell growth rates and division times. J Theor Biol 257, 124–130, 10.1016/j.jtbi.2008.10.031 (2009). [DOI] [PubMed] [Google Scholar]
- Anderson E. C. & Petersen D. F. Cell growth and division. II. Experimental studies of cell volume distributions in mammalian suspension cultures. Biophys J 7, 353–364, 10.1016/S0006-3495(67)86593-7 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. et al. Cell population tracking and lineage construction with spatiotemporal context. Med Image Anal 12, 546–566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prewitz M. C. et al. Tightly anchored tissue-mimetic matrices as instructive stem cell microenvironments. Nat Methods 10, 788–794, 10.1038/nmeth.2523 (2013). [DOI] [PubMed] [Google Scholar]
- Kurth I., Franke K., Pompe T., Bornhauser M. & Werner C. Hematopoietic stem and progenitor cells in adhesive microcavities. Integr Biol (Camb) 1, 427–434, 10.1039/b903711j (2009). [DOI] [PubMed] [Google Scholar]
- Deasy B., Chirieleison S., Witt A., Peyton M. & Bissell T. Tracking stem cell function with computers via live cell imaging: Identifying donor variability in human stem cells. OperTech Orthop 20, 127–135 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material