This work uses Hsp70 proteins with altered affinities for ATP to demonstrate that Hsp70 chaperones underlie the requirement for ATP in plastid protein import.
Abstract
The 70-kD family of heat shock proteins (Hsp70s) is involved in a number of seemingly disparate cellular functions, including folding of nascent proteins, breakup of misfolded protein aggregates, and translocation of proteins across membranes. They act through the binding and release of substrate proteins, accompanied by hydrolysis of ATP. Chloroplast stromal Hsp70 plays a crucial role in the import of proteins into plastids. Mutations of an ATP binding domain Thr were previously reported to result in an increase in the Km for ATP and a decrease in the enzyme’s kcat. To ask which chloroplast stromal chaperone, Hsp70 or Hsp93, both of which are ATPases, dominates the energetics of the motor responsible for protein import, we made transgenic moss (Physcomitrella patens) harboring the Km-altering mutation in the essential stromal Hsp70-2 and measured the effect on the amount of ATP required for protein import into chloroplasts. Here, we report that increasing the Km for ATP hydrolysis of Hsp70 translated into an increased Km for ATP usage by chloroplasts for protein import. This thus directly demonstrates that the ATP-derived energy long known to be required for chloroplast protein import is delivered via the Hsp70 chaperones and that the chaperone’s ATPase activity dominates the energetics of the reaction.
INTRODUCTION
Chloroplasts are organelles in plants responsible for numerous metabolic activities in addition to photosynthesis. Because they are of endosymbiotic origin, chloroplasts possess their own genome and are capable of protein biosynthesis. Despite this, over 95% of chloroplast proteins are encoded in the nucleus, synthesized in the cytoplasm as precursors, and posttranslationally translocated into the organelle via the translocation machineries in the outer (Toc) and inner (Tic) envelope membranes (reviewed in Jarvis, 2008; Li and Chiu, 2010; Shi and Theg, 2013a). Precursor import into chloroplasts has been proposed to be similar to the transport of proteins into mitochondria and the endoplasmic reticulum in terms of its coupling to ATP hydrolysis by a motor protein poised at the exit of the translocation channel. In mitochondria and the endoplasmic reticulum, this motor is thought to be a member of the highly conserved family of 70-kD heat shock proteins (Hsp70s) (Kang et al., 1990; Berthold et al., 1995; Bagola et al., 2011; Becker et al., 2012). Although a chloroplast stromal Hsp70 was identified more than 20 years ago (Marshall and Keegstra, 1992), evidence for its role in chloroplast protein import was lacking until relatively recently (Shi and Theg, 2010; Su and Li, 2010). Instead, a member of the Hsp100 family of chaperones called ClpC or Hsp93 was regularly isolated with other components of the translocation machinery (Akita et al., 1997; Nielsen et al., 1997), and the postulate was put forward that this protein served as the trans-side motor powering the protein import process (Flores-Pérez and Jarvis, 2013). Recently, this issue has been reexamined in two laboratories in both moss (Physcomitrella patens) and Arabidopsis thaliana (Shi and Theg, 2010; Su and Li, 2010). Using a combination of biochemical and reverse genetics techniques, compelling evidence has been obtained indicating that stromal Hsp70 does indeed participate in the protein import process.
Recognition that the stromal Hsp70 plays a significant role in chloroplast protein import raises the question of the nature of that role and the distribution of the motor function between Hsp70 and Hsp93, both of which are ATPases. In particular, it is not clear whether these two chaperones act in parallel or series, whether their actions are synergistic, and whether one or the other dominates in the delivery of ATP-derived energy to the import reaction. To address this latter question, we looked to a study of the bovine (Bos taurus) Hsc70 in which mutation of a Thr in the nucleotide binding pocket was discovered to increase the Km for ATP hydrolysis (O’Brien and McKay, 1993). We reasoned that if a similar Km-raising mutation in the stromal Hsp70 resulted in an increase in the ATP requirement for chloroplast protein import, we could conclude that Hsp70 is the chaperone largely responsible for the ATP dependence of the import reaction. Furthermore, it would provide direct confirmation of the long-held view that the energy required for chloroplast protein import is delivered through ATP hydrolysis by a stromal chaperone.
In this article, we describe the consequences of a targeted replacement of wild-type Hsp70 in the stroma of chloroplasts with two such high-Km mutants in the moss P. patens. We report that the mutant moss grow somewhat more slowly than the wild-type plants. Also, in vitro experiments with isolated chaperones indicated that the Thr mutants display a lower Vmax and increased Km compared with wild-type Hsp70. Of most significance to this study, we found that chloroplasts isolated from the Km mutant plants required more ATP to import proteins than did those from wild-type plants. This indicates that the energetics of chloroplast protein import is dominated by the properties of the stromal Hsp70.
RESULTS
Rescue of the Lethal Hsp70 Knockout by Wild-Type and Thr Mutant Hsp70 cDNA
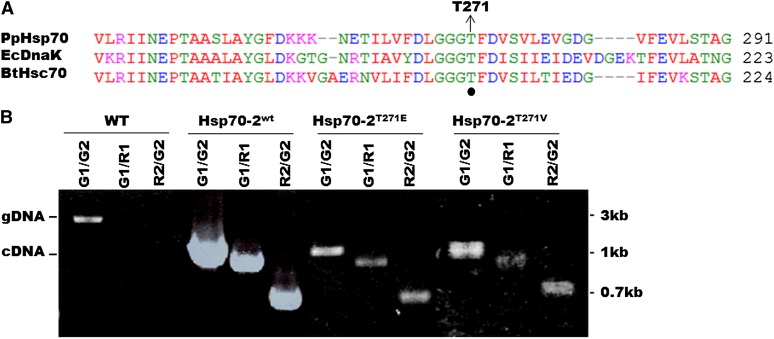
In an earlier study (Shi and Theg, 2010), we demonstrated that one of the three Hsp70 chaperones present in the stroma of P. patens chloroplasts, termed Hsp70-2, is required for protein import into chloroplasts. From sequence alignments, we determined that the moss Hsp70-2 protein sequence shares 50 and 45% identity with Escherichia coli DnaK and bovine Hsc70, respectively. Residue Thr-271 in Pp-Hsp70-2 corresponds to Thr-199 in Ec-DnaK and Thr-204 in Bt-Hsc70, both of which have been implicated in ATP binding and hydrolysis (Figure 1A) (McCarty and Walker, 1991; O’Brien and McKay, 1993). Since Thr-to-Val (T204V) and Thr-to-Glu (T204E) mutations in Bt-Hsc70 caused the Km for ATP hydrolysis to increase from 1 to 90 μM, we expected similar mutations at this position in Pp-Hsp70-2 to cause a similar ATP affinity decrease without destroying the function of the enzyme.
Figure 1.
Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T271V cDNA Rescues the Hsp70-2 KO in P. patens Plants.
(A) ClustalW alignment of a portion of the nucleotide binding sites of Pp-Hsp70-2 (P. patens), Ec-DnaK (E. coli), and Bt-Hsc70 (bovine). The position of T271 in the moss gene is identified.
(B) Genotyping of moss strains generated from Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T271V cDNA-rescued KO plants. Primer pairs are indicated above the gel; G1, G2, R1, and R2 are forward and reverse primers in the genomic DNA and the resistance gene used for the KO, respectively. gDNA, PCR product amplified from the genomic locus (lane 1); cDNA, PCR product amplified from the cDNA construct rescuing the plant from the lethal Hsp70-2wt KO (lanes 4, 7, and 10).
In moss, Hsp70-2 is an essential protein (Shi and Theg, 2010). This allows us to functionally replace the chaperone by cotransforming the moss with a knockout (KO) construct to inactivate the chromosomal gene and simultaneously rescue the plant with a cDNA copy of the gene containing desired modifications. In this manner, we replaced the chromosomal Hsp70-2 with either wild-type cDNA as a control or with cDNA versions harboring T271V and T271E mutations. The transgenic plants were genotyped (Figure 1B) to confirm the KO of the native chromosomal gene. Since this gene is essential, these transgenic plants survive on the basis of the critical Hsp70 activity residing in the cDNA-derived proteins. Accordingly, we can conclude that both of the Thr mutants can complement inactivated wild-type Hsp70-2. These transgenic plants are hereafter referred to as Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T271V, according to their inserted cDNA (Figure 1B).
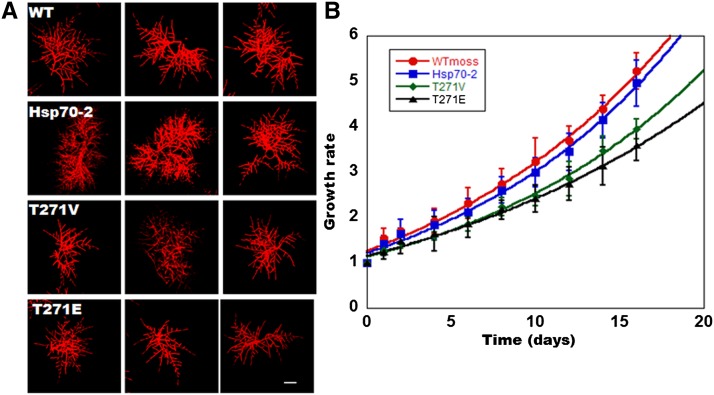
To determine the growth phenotype of Hsp70-2wt–, Hsp70-2T271E–, and Hsp70-2T271V–rescued transgenic plants, we used a quantitative analysis of plant morphology and area to measure the growth rate. In order to standardize the starting material, colonies were started on plates from individual protoplasts. Figure 2A shows that each of the transgenic plants recovered normally from protoplasts. We then followed the growth of the colonies for 14 d by microscopy imaging and extracting chlorophyll autofluorescence via the red channel using ImageJ software. The Thr mutants of Hsp70 complemented the lethal Hsp70-2 KO plants quite well, displaying normal morphology and with the only apparent phenotype in the living plants being a slightly reduced growth rate (Figure 2B).
Figure 2.
Growth Rate Measurement for Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T27V cDNA-Rescued KO Transgenic Plants.
(A) Three representative micrographs showing chlorophyll fluorescence (red) of 1-week-old plants. Hsp70-2, Hsp70-2wt; T271E, Hsp70-2T271E; T271V, Hsp70-2T271V. Bar = 100 μm.
(B) Growth rate of the indicated plants on BCD (phototrophic) media. Colony area was measured every 2 d for 16 d and normalized to the initial area on day 0. Data represent the means and standard errors from three colonies for each moss strain.
Thr Mutants of Hsp70 Display Lower Vmax and Increased Km Compared with the Wild Type
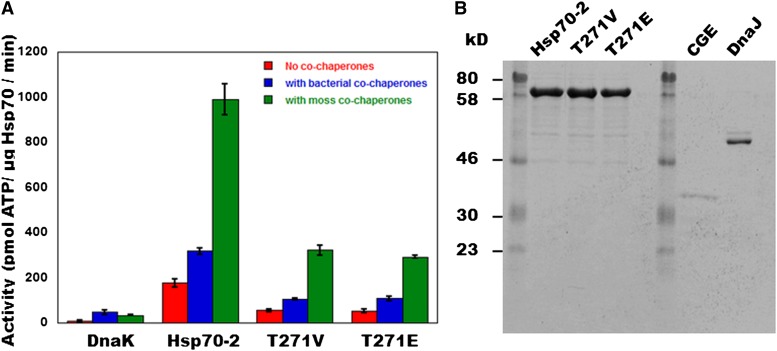
The functions of Hsp70 chaperones are assisted by their cognate cochaperones: DnaJ and GrpE in E. coli (Liberek et al., 1991) and DnaJ and CGE in plants (Schroda et al., 2001). In order to test the ATPase activity of the Thr mutants, we cloned Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T271V and the corresponding cochaperones from moss into expression vectors and produced them in bacteria (Figure 3B). The overexpressed and purified proteins were then used in ATPase activity assays supplemented with the model substrate NRLLLTG (O’Brien and McKay, 1993). Commercial DnaK, DnaJ, and GrpE from E. coli also were used as positive controls for the measurement. As expected, DnaK, Hsp70-2, and the Hsp70-2 Thr variants exhibited some ATPase activity on their own, which was stimulated ∼2- to 6-fold by the additions of the cochaperones (Figure 3A). Interestingly, the cochaperones cloned from moss were considerably more effective in stimulating ATP hydrolysis by the moss chaperones than were bacterial DnaJ and GrpE. Conversely, the moss cochaperones were slightly less effective at stimulating bacterial DnaK as were DnaJ and GrpE. The basis of this cochaperone selectivity is currently unknown, but we note that the divergence between Ec-GrpE and Pp-CGE (26%) and Pp-DnaJ and Ec-DnaJ (46%) is significantly greater than that between moss Hsp70-2 and bacterial DnaK (63%). Figure 3A also shows that both Thr mutants, Hsp70-2T271E and Hsp70-2T271V, displayed cochaperone-mediated ATPase activity, although neither was as active as the wild-type protein. This is consistent with the experiment of Figure 2 showing that these mutants complement the wild-type Hsp70-2 KO in vivo, supporting a somewhat reduced growth rate.
Figure 3.
Effect of Cochaperones on the in Vitro ATPase Activity of Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T27V.
(A) Comparison of ATPase activities of bacterially produced moss Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T271V and E. coli DnaK with cochaperones from E. coli and moss. ATPase activity of these proteins was assayed colorimetrically with malachite green in a medium containing HEPES-KOH (40 mM, pH 7.0), KCl (75 mM), Mg(CH3COO)2, denatured NR substrate peptide (NRLLLTG) (200 nM), ATP (1 mM), and when added, DnaK or Hsp70 (0.3 μM), E. coli or P. patens DnaJ (1 μM), and GrpE or CGE (1 μM). Shown are means and standard errors from three replicate experiments.
(B) Recombinant moss proteins used in the ATPase assays. Proteins were purified after overexpression in bacteria; the figure shows 1 μg of each on a Coomassie blue–stained 12.5% SDS-PAGE gel.
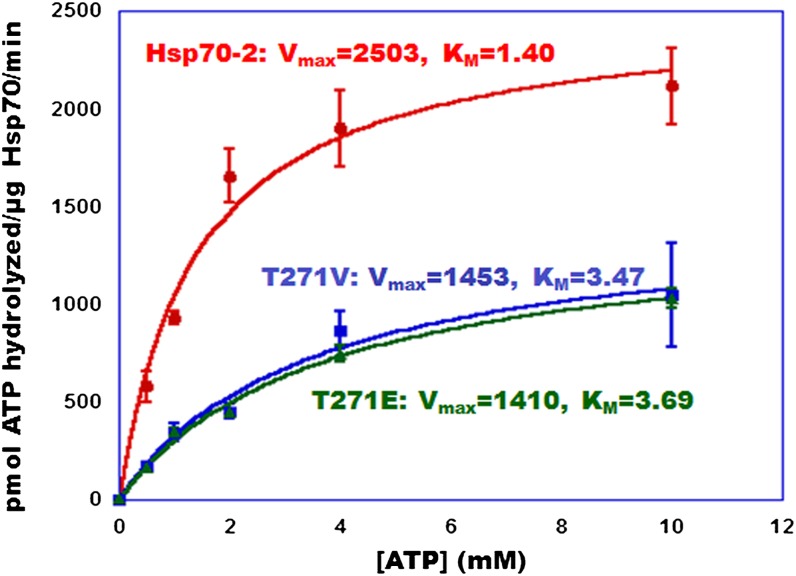
We then measured the effect of the Thr mutations on the kinetic parameters that describe the chaperones’ ATPase activity. To this end, we measured the initial velocity of the ATPase reaction as a function of ATP concentration using both the natural import substrate prSSU (Figure 4) and the substrate analog peptide NR (Gragerov et al., 1994) (Supplemental Figure 1; compared with Methods); the results using the two substrates were indistinguishable. Extraction of the kinetic parameters from best fits to the Michaelis-Menten equation for the data obtained with the two substrates revealed that the Km for ATP hydrolysis increased from an average of 1.40 mM with the wild-type protein to an average of 3.58 mM in the two Thr mutants. At the same time, the reaction Vmax decreased from an average of 2.50 nmol ATP hydrolyzed/μg protein⋅min by the Hsp70-2wt proteins to an average of 1.43 nmol ATP hydrolyzed/μg protein⋅min by the mutants. These results are consistent with the increase in Km and decline in Vmax observed in the bovine Hsc70 when analogous Thr mutations were introduced into that protein.
Figure 4.
Thr Hsp70 Mutants Display Lower Vmax and Increased Km for ATP Hydrolysis Compared with the Wild Type.
Moss proteins were purified after overexpression in bacteria. ATPase activity was assayed with moss cochaperones as in Figure 3 using prSSU as the substrate. Data were fitted to the Michaelis-Menten equation with the indicated parameters using KaleidaGraph. Circles, squares, and triangles correspond to Hsp70-2wt, Hsp70-2T271V, and Hsp70-2T271E, respectively. The data represent means and standard errors from three replicates.
Chloroplasts Isolated from Thr Mutants Require More ATP for Protein Import Than Do Those from Wild-Type Plants or Transgenic Hsp70 KO Plants Rescued with Hsp70-2wt cDNA
Since the in vitro ATPase assays showed that the Km of the Thr mutants increased compared with Hsp70-2wt, we wanted to test the effect these mutations had on the ATP requirement for protein import into isolated chloroplasts. To this end, chloroplasts from Thr mutant– and Hsp70-2wt–rescued KO plants were isolated, and an import assay was performed using a chimeric precursor consisting of the transit peptide of the small subunit of Rubisco placed in front of GFP (tp22GFP; Shi and Theg, 2013b). As it was important to control the amount of ATP in the assay, we removed the ATP present in the wheat germ translation mixture by passing the precursor through a desalting column (Theg et al., 1989). We also excluded light during the import reactions and supplied the required ATP exogenously. Figure 5 compares the import of tp22GFP into isolated chloroplasts in the light and the dark and confirms that, as in chloroplasts from flowering plants, the reaction can be driven efficiently in moss chloroplasts by exogenous ATP (Shi and Theg, 2013b).
Figure 5.
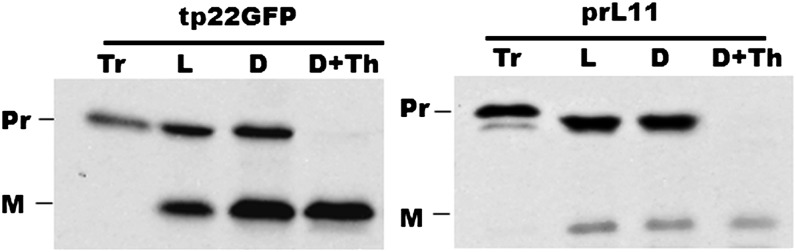
ATP-Dependent Import of tp22GFP and prL11 into 7-d-Old Wild-Type Moss Chloroplasts.
L, import assay in the light; D, import assay in the dark with 3 mM added ATP; D+Th, import assay in the dark followed by thermolysin treatments to digest the unimported proteins; Pr, precursor; M, mature protein. ATP was removed from the precursors by gel filtration.
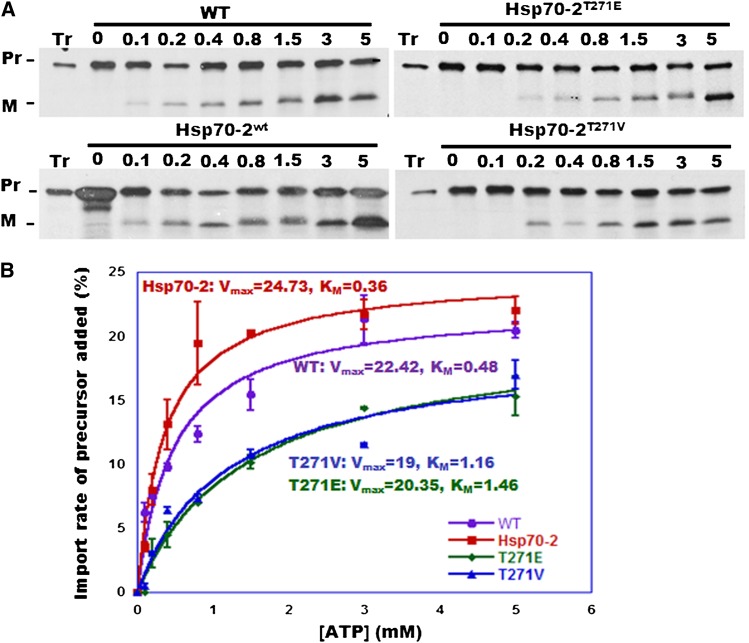
We then performed assays using tp22GFP to measure the initial velocity of the import reaction in the dark as a function of ATP concentration using chloroplasts from wild-type, transgenic wild-type, and Thr mutant plants. The ATP dependence of the import reaction was similar for wild-type and Hsp70-2wt–rescued KO chloroplasts, with visible import at 0.1 mM ATP and reaching a maximum at ∼3 mM added ATP (Figure 6A). By contrast, chloroplasts from both Thr mutants displayed visible import only at ≥0.2 mM and the reactions reached their maxima at ≥5 mM. Quantification of the reactions revealed Km values of 0.36 and 0.48 mM ATP for wild-type and Hsp70-2wt KO chloroplasts, respectively, and 1.16 and 1.46 mM ATP for chloroplasts carrying the Hsp70-2T271V and Hsp70-2T271E chaperones, respectively (Figure 6B). Unlike the in vitro ATPase assays, the Vmax for the protein import reactions was essentially the same in the wild-type and Hsp70-2 mutant chloroplasts (19 to 24.73% of the added precursor imported in 5 min).
Figure 6.
Import of tp22GFP into Wild-Type and Transgenic Moss Chloroplasts as a Function of Added ATP.
(A) ATP dependence of tp22GFP import in the dark into chloroplasts isolated from 7-d-old wild-type plants and from transgenic KO moss rescued with Hsp70-2wt, Hsp70-2T271E, or Hsp70-2T271V cDNA. Pr, precursor; M, mature protein.
(B) Quantification of (A). Data from three replicates (±se) were fitted to the Michaelis-Menten equation using KaleidaGraph; resulting values for Km and Vmax are indicated.
The ATP Requirement for Protein Import Follows the Km of Hsp70 for Nonphotosynthetic Proteins
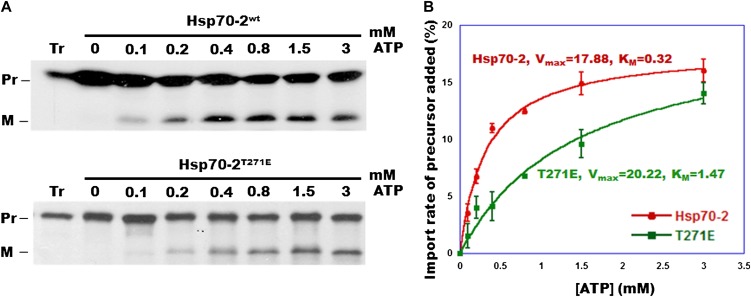
It has been observed in some experiments that the import characteristics of photosynthetic proteins can be different from those of nonphotosynthetic or housekeeping proteins (Kubis et al., 2003; Jarvis, 2008; Li and Chiu, 2010). Since the precursor we used for the experiment in Figure 6 utilizes the transit peptide from the prSSU, tp22GFP can be considered to be a photosynthetic protein. In order to ask whether stromal Hsp70 dominates the energy requirements for import of a housekeeping protein as well, we repeated these ATP titration experiments using the precursor to L11, a subunit of the 50S chloroplast ribosome (Pesaresi et al., 2001). As with tp22GFP, the Km for ATP utilization for the import of prL11 was more than 3-fold higher in the Thr mutant chloroplasts compared with those from the wild type (data not shown) or Hsp70-wt–rescued KO plants, while the reaction Vmax was essentially unchanged (Figure 7). These data indicate that the measurable ATP requirement for the import of both photosynthetic and housekeeping proteins into isolated chloroplasts is determined by the affinity of Hsp70 for ATP. This in turn suggests that this chaperone carries the major responsibility for supplying energy to the chloroplast import reaction.
Figure 7.
Import of prL11 into Transgenic Moss Chloroplasts as a Function of Added ATP.
(A) Import of prL11 to chloroplasts isolated from 7-d-old transgenic KO moss rescued with Hsp70-2wt and Hsp70-2T271E cDNA. Pr, precursor; M, mature protein.
(B) Quantification of (A). Data from three replicates (±se) were fitted to the Michaelis-Menten equation using KaleidaGraph; resulting values for Km and Vmax are indicated.
Overexpression of Hsp70 Has No Effect on the Amount of Hsp93 in the Transgenic Moss
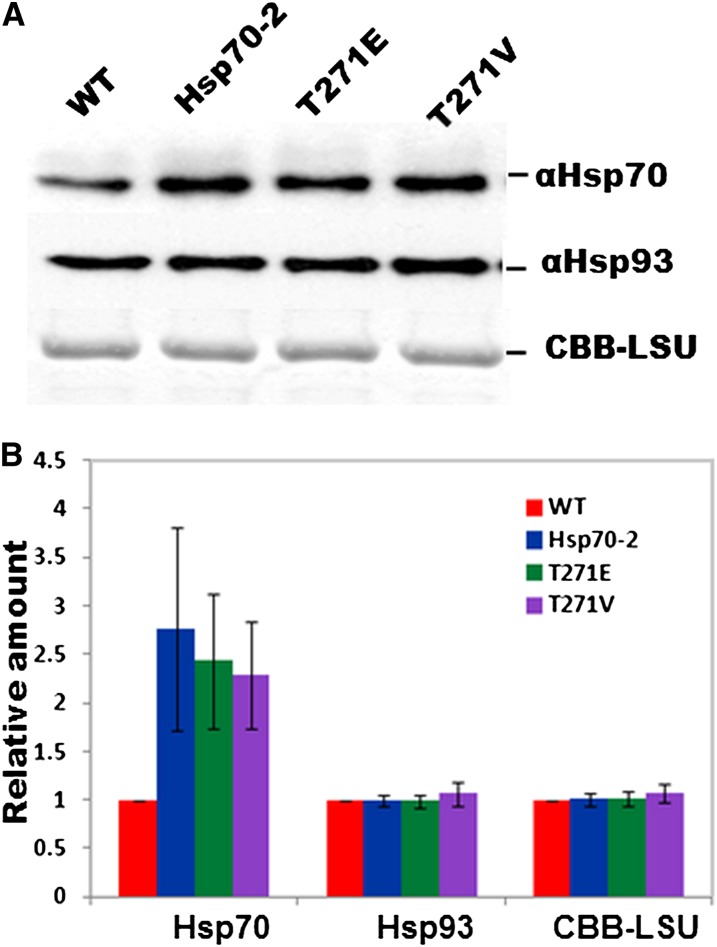
Each of the Hsp70-2 KO plants used in this study was rescued with cDNA versions of the gene, either wild-type or those containing the Thr mutations, driven by the constitutive cauliflower mosaic virus 35S promoter (Odell et al., 1985). This is known to be a strong promoter in flowering plants (Benfey and Chua, 1990) but is relatively less active in moss (Horstmann et al., 2004). In order to monitor the amount of stromal Hsp70-2 produced in transgenic plants, we probed immunoblots with Hsp70-2–specific antibodies (Figure 8A). Quantitation of the signal on the immunoblots indicated that compared with wild-type plants the level of Hsp70-2 was more than doubled in each of the transgenic plants (Figure 8B). We also asked whether the level of Hsp93 changed in response to changes in the activity of the Hsp70-2 mutants. To this end, we probed the immunoblots with antibodies specific to Hsp93. As seen in Figure 8, none of the transgenic plants, which have higher levels of Hsp70-2 as well as lower chaperone-specific activities, accumulated Hsp93 at levels different than that seen in the control wild-type plants.
Figure 8.
Immunoblot Showing Relative Protein Abundance of Hsp70 and Hsp93 in Chloroplasts.
(A) Immunoblot using antibodies against Hsp70-2 and Hsp93. Chloroplasts were isolated from wild-type plants and KO plants rescued by Hsp70-2wt, Hsp70-2T271E, and Hsp70-2T271V cDNA, respectively; CBB-LSU, loading control showing the large subunit of Rubisco stained with Coomassie blue.
(B) Quantification of (A) showing means and standard errors from three independent experiments and normalized to the wild-type value.
DISCUSSION
Until relatively recently the motor function for protein import into chloroplasts was proposed to reside with Hsp93, a member of the Hsp100 family of molecular chaperones. This view changed in 2009 with the publications of two articles demonstrating that Hsp70s are part of the import motor in both P. patens and Arabidopsis (Shi and Theg, 2010; Su and Li, 2010). This study was initiated with the aim of clarifying the roles of these two chaperones in this process. The moss genome encodes three Hsp70s targeted to the chloroplast stroma. Only one of these, which we have termed Hsp70-2, is an essential protein and is implicated in protein import. This, combined with the ability to create targeted KOs in moss, allows us to investigate Hsp70s with altered properties in the import reaction. It has been demonstrated that mutation of a Thr in the ATP binding domain of bovine (O’Brien and McKay, 1993) and E. coli (McCarty and Walker, 1991) Hsp70s results in lowered ATPase activity and, in the case of the bovine protein, a decreased affinity of the chaperone for ATP. We made equivalent mutations in the moss Hsp70-2 and showed that they also resulted in an increase in the Km and a lowered Vmax for ATP in the hydrolysis reaction.
The Thr-mutated Hsp70s complemented the chaperone KO mutant well, as the transgenic plants harboring these mutations displayed only a slightly reduced growth rate phenotype. Since the stromal Hsp70s are expected to perform other functions in addition to their role in protein import, it was not possible to determine whether the slower growth in the mutants was due to a limitation in protein import not observed in vitro or to some other altered chaperone-dependent process.
As has been amply demonstrated by previous researchers, we found that Hsp70-2wt has little intrinsic ATPase activity (Mayer, 2010). This activity is stimulated by addition of the cochaperones CgE and DnaJ. The moss chaperone displays the signs of promiscuity that the yeast and bacterial homologs exhibit (Krzewska et al., 2001). That is, the ATPase activity of the moss Hsp70-2wt is stimulated by both its native cochaperones and those from bacteria. However, the moss cochaperones are considerably more efficacious with the moss Hsp70. By contrast, the moss cochaperones stimulate the activity of DnaK about as much as the bacterial cochaperones do. There are several instances known in which a more basal enzyme has been observed to have a higher promiscuity for substrates, and we see this specificity for cochaperones as a similar case (Khersonsky and Tawfik, 2010; Voordeckers et al., 2012). In this view, we postulate that Hsp70-2 may have evolved for more specific purposes than its bacterial counterpart DnaK and thus may prefer the moss cochaperones. This specialization of eukaryotic chaperones is consistent with our observation that a KO of Hsp70-2 is lethal, whereas KOs of the other three moss stromal Hsp70s, either individually or together, lead to minimal phenotypes (Shi and Theg, 2010).
Compared with its activity in angiosperms, the 35S promoter is not considered a strong promoter in moss (Horstmann et al., 2004). Nonetheless, all of our transgenic lines that produced Hsp70-2wt or its mutants using the 35S promoter exhibited elevated levels of the chaperone in their chloroplasts compared with wild-type plants. Even so, the protein import activity of the chloroplasts from Hsp70-2wt transgenic plants was similar to that from wild-type plants, from which we can conclude that the amount of exogenous Hsp70-2 in wild-type chloroplasts is not limiting for import. That is, neither the rate of import nor the amount of ATP required was altered by the increased quantity of Hsp70, suggesting that a limited number of chaperone binding sites are present in the translocon and that these are generally saturated by the wild-type levels of Hsp70. This is also evidenced by the observation that the ATP-saturated import rates were relatively unchanged in chloroplasts containing wild type or Thr-mutated Hsp70s, despite the relative decrease in the mutants’ Vmax for ATP hydrolysis with the isolated proteins. Interestingly, the level of Hsp93 remained unchanged between the wild-type and transgenic lines, thereby showing no tendency to adjust in reciprocal manner to changes in either Hsp70 levels or activities.
Our observation that the ATP requirement for protein import follows the Km of the stromal Hsp70 could in principle be due to pleiotropic effects of the mutations on folding of chloroplast proteins that coincidentally resulted in an increase in the amount of ATP utilized by the translocation machinery. However, it seems a far simpler and more likely explanation that we have made an alteration in the import motor itself. With this interpretation, our results are noteworthy for a number of reasons. First, because molecular chaperones possess ATPase activity, it has long been assumed that the well-known ATP requirement for protein import into chloroplasts is fulfilled by the stromal chaperones (Flügge and Hinz, 1986; Pain and Blobel, 1987; Schindler et al., 1987; Theg et al., 1988; Leheny and Theg, 1994; Scott and Theg, 1996; Shi and Theg, 2010). This view is supported by experiments showing that the chaperones Hsp93, Hsp70, and, more recently, Hsp90 are required for protein import. The experiments presented here correlating the Km for ATP hydrolysis with the ATP requirement for import provide direct proof that the delivery of ATP-derived energy to the reaction is made through Hsp70. Whether this is the principal or only protein that can deliver this energy remains to be elucidated. Second, our data showed that the Km-induced change in the import ATP requirement applied to both photosynthetic and housekeeping proteins. This rules out the possibility that the motor functions of Hsp70 and Hsp93 are divided between these two classes of proteins, as in the situation that has been postulated for the receptors made up of combinations of Toc 159/132/120 and Toc 33/34 (Kubis et al., 2003, 2004). Finally, our data appear to be incompatible with the idea that Hsp70 and Hsp93 operate in parallel, at least in a model in which each precursor could follow one pathway or the other depending on its availability. This model was suggested by Su and Li (2010) based on their findings that mutants in both chaperones displayed a more severe phenotype than did mutants in either gene independently. However, in the parallel chaperone model, an increase in the Km of one chaperone would be expected to force the precursor onto the other chaperone’s pathway at limiting ATP concentrations, and no change in the ATP requirement for import would be expected. We observed just the opposite behavior, and it appears that the precursors examined were required to pass through an Hsp70-dependent step during the import reaction. Accordingly, we believe that our data are consistent only with models in which the two chaperones act sequentially in series or together at the same step to provide the driving force for protein import.
In summary, we have shown that the ATP requirement for protein import into isolated chloroplasts is determined by the Km for ATP hydrolysis of the Hsp70 operating in the import motor. This provides a direct demonstration that the ATP requirement for protein import into chloroplasts is fulfilled by this trans-side chaperone. Experiments are currently underway to determine whether Hsp93 and Hsp70 bring about protein import by operating sequentially or at the same time in the same step.
METHODS
Sequence Analysis
Protein sequences were compared with ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The sequences compared were Pp-Hsp70 (Hsp70-2) from Physcomitrella patens, Ec-DnaK from Escherichia coli, and Bt-Hsc70 from bovine (Bos taurus).
Gene Construction
For bacterial overexpression, moss Hsp70-2 without its transit peptide (encoded by the first 222 bp) was amplified and cloned to the SpeI-BamHI sites of the vector pTYB12 (New England Biolabs), which was named pTYB12-Hsp70. Km mutants T271V and T271E were generated from this construct using the QuikChange kit (Stratagene), which were named pTYB12-TV and pTYB12-TE. Primers used in PCR are shown in Supplemental Table 1: pTYB12-HSP70F and pTYB12-HSP70R (for pTYB12-Hsp70), QCTVF and QCTVR (for pTYB12-TV), and QCTEF and QCTER (for pTYB12-TE).
For moss transformation, KO and rescue plasmids from Shi and Theg (2010) were used, which were named pBJIR4 and pART7-Hsp70, respectively. Rescuing plasmids for Km mutants T271V and T271E were generated from pART7-Hsp70 using the QuikChange kit (Stratagene), named pBJIR4-TV and pBJIR4-TE. Primers used for mutations are shown in Supplemental Table 1: QCTVF and QCTVR (for pBJIR4-TV) and QCTEF and QCTER (for pTYB12-TE).
The moss cochaperones DnaJ and CGE used in the in vitro ATPase assay were separately inserted into the pTYB12 vectors. DnaJ cDNA without its transit peptide coding region (the first 144 bp) was PCR amplified with primers PpDnaJ EcoRI_F and PpDnaJ XhoI_R. After subcloning into PCR TOPO II BLUNT, the PCR product was cloned into the EcoRI-XhoI sites of pTYB12. Similarly, CGE cDNA without its transit peptide-encoding region (the first 156 bp) was PCR amplified with primers PpGrpE-1 NdeI_F and PpGrpE-1 SalI_R and cloned to the NdeI-SalI sites of pTYB12. Hsp70-2 was coexpressed with HEP2 in a pET9 vector (Sichting et al., 2005; Willmund et al., 2008).
Moss Growth and Transformation
P. patens (subspecies patens, Gransden) was grown at 23°C under continuous light (60 to 80 μmol photons m−2 s−1) and maintained as previously described (Hofmann and Theg, 2003). Polyethylene glycol–mediated transformation of protoplasts was performed to generate the rescued transgenic plants as described by Shi and Theg (2010). Protoplasts were isolated from 7-d-old subcultured protonemal tissues and adjusted to 1.6 × 106 cells/mL. Then, 300 μL of protoplasts was mixed with 300 μL of polyethylene glycol solution, 10 μg of linear KO DNA, and 10 μg of rescuing plasmids. The mixture was heat shocked for 5 min at 45°C and then cooled to room temperature. At 10-min intervals 1, 2, and 7 mL of 8% mannitol were added to the cooled mixture and gently mixed to recover the protoplasts. After collecting and resuspending the protoplasts in 0.5 mL of 8% mannitol, 10 mL of top agar warmed to 42°C was added. The mixture was divided into three Petri dishes containing regeneration medium. These Petri dishes were incubated at 23°C under continuous light for 1 week before transfer to selective medium containing 30 mg/L of G418 for 14 d. Colonies were then transferred to antibiotic-free medium for 1 week and then to selective medium containing 30 mg/L of G418 for 14 d. The surviving transformants were harvested for genotyping with primers G1, G2, R1, and R2 following the method previously described (Shi and Theg, 2010). The colonies were then grown on nonselective medium.
ATPase Assay
Plasmids pTYB12-Hsp70, pTYB12-TV, and pTYB12-TE were transformed into BL21 (DE3) competent cells (Promega). Proteins Hsp70, T271V, and T271E were induced by 1mM isopropyl β-d-1-thiogalactopyranoside and then purified using chitin beads following the instructions (New England Biolabs). Purified proteins were concentrated with Amicon Ultra-4 centrifugal filter devices (Millipore), and then the concentration was measured by Bradford assay using BSA as the standard. ATPase activity was measured by a modification of the malachite green assay as described (Chang et al., 2008). Briefly, reactions were performed in a mixture containing ATP (varied from 0 to 10 mM), DnaK or Hsp70 (0.3 μM), DnaJ (1 μM), GrpE (1 μM), prSSU (100 nM), or NR substrate (NRLLLTG, 200 nM; synthesized by Biomatic Technologies) into a total volume of 25 μL in the reaction buffer. The reaction buffer contained 40 mM HEPES-KOH, pH 7.0, 75 mM KCl, and 4.5 mM Mg(CH3COO)2. After 1 h of incubation at 37°C, 80 μL of malachite green reagent, which was a mixture of malachite green (0.081% [w/v]), polyvinyl alcohol (2.3% [w/v]), ammonium heptamolybdate tetrahydrate (5.7% [w/v] in 6 M HCl), and water in the ratio of 2:1:1:2, was added into the ATPase reaction tube. Then, 10 μL 34% sodium citrate was added to halt the nonenzymatic hydrolysis of ATP. The samples were mixed thoroughly and incubated at 37°C for 15 min before measuring OD620.
Moss Import Assay
Plasmid pet23a-tp22GFP was linearized with restriction enzyme XhoI and transcribed (Promega) in vitro under the control of SP6 promoter. Plasmid prL11-BSK was a gift from P. Jarvis’s lab (Aronsson and Jarvis, 2002). The prL11 fragment was obtained by PCR using primers T7 and T3 and then transcribed under the control of T7 promoter. The radiolabeled precursor proteins tp22GFP and prL11 were translated in wheat germ in the presence of 3H-Leu and other nonradioactive amino acids (Promega). ATP was removed from the in vitro–translated precursors by Zeba spin desalting columns (Thermo Fisher Scientific). Intact chloroplasts were isolated from 7-d-old moss protonemal tissues as described previously (Hofmann and Theg, 2003). Radiolabeled precursors were incubated with moss chloroplasts for 30 min in the dark in the presence of the indicated concentrations of ATP. Then, 600 μL of import buffer (330 mM sorbitol, 50 mM Tricine, and 3 mM MgCl2, pH 8.0) was added to stop the reaction, and a pellet was then collected by centrifuging at 3,000g for 5 min. The pellets were resuspended in sample buffer (50 mM Tris-HCl at pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue) and boiled for 10 min, followed by SDS-PAGE and fluorography. Thermolysin treatments were performed as described previously (Alder and Theg, 2003). Quantification of gel bands was performed using ImageJ. The data were plotted in KaleidaGraph.
Measurement of Moss Growth Rate
Protoplasts from 7-d-old tissues were made and regenerated following a modified protocol from (Schaefer et al., 1991). The concentration of protoplasts was adjusted to 1.6 × 10−6/mL. After plating to Petri dishes, colonies were imaged every 2 d using a SteREO Discovery-V12 microscope (Carl Zeiss) equipped with an X-Cite 120 fluorescence illuminator. Moss tissue area was measured by image analysis with ImageJ as described (Vidali et al., 2009).
Immunoblotting
Immunoblotting was performed according to Shi and Theg (2010). PAGE was performed with 12.5% SDS gels using a mini-slab gel system (Mini Protean III; Bio-Rad). Following electrophoresis, chloroplasts were transferred onto Poly-Screen PVDF membrane by a Bio-Rad Transblot cell with a transfer buffer containing 25 mM Tris-HCl, pH 8.3, 192 mM Gly, and 20% methanol. Blots were probed with antibodies against Hsp93 and Hsp70. Hsp93 antibody was raised against pea (Pisum sativum) ClpC (Shanklin et al., 1995), and Hsp70 antibody was raised against moss Hsp70-2 (Shi and Theg, 2010). Goat-anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad) diluted to 1:15,000 was used for immunodecorating both proteins. Blots were developed by enhanced chemiluminescence (ECL; Amersham), and the band intensities quantitated with ImageJ software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Pp-Hsp70 (Hsp70-2), ADB23406; Ec-DnaK, BAA01595; Bt-Hsc70, NP_776770; CGE, GU223151; and DnaJ, Pp_vt0409_25007_3036.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Thr Hsp70 Mutants Display Lower Vmax and Increased Km for ATP Hydrolysis Compared with the Wild Type Using the Artificial NR Substrate.
Supplemental Table 1. Primers Used in the Study.
Supplemental Table 2. Constructs.
Supplementary Material
Acknowledgments
We thank Magdalena Bezanilla for providing ImageJ macros for quantitation of moss colony growth and Terry Bricker for many helpful discussions. Expert technical assistance was provided by Tatiana Wilson and Dung Nguyen. This work was supported by National Science Foundation Grant MCB-0956484 to S.M.T.
AUTHOR CONTRIBUTIONS
L.L., L.-X.S., and S.M.T. designed the research. L.L. and R.T.M. performed the research. L.L. and S.M.T. wrote the article. L.-X.S. and R.T.M edited the article.
Glossary
- KO
knockout
Footnotes
Online version contains Web-only data.
References
- Akita M., Nielsen E., Keegstra K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder N.N., Theg S.M. (2003). Protein transport via the cpTat pathway displays cooperativity and is stimulated by transport-incompetent substrate. FEBS Lett. 540: 96–100. [DOI] [PubMed] [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220. [DOI] [PubMed] [Google Scholar]
- Bagola K., Mehnert M., Jarosch E., Sommer T. (2011). Protein dislocation from the ER. Biochim. Biophys. Acta 1808: 925–936. [DOI] [PubMed] [Google Scholar]
- Becker T., Böttinger L., Pfanner N. (2012). Mitochondrial protein import: From transport pathways to an integrated network. Trends Biochem. Sci. 37: 85–91. [DOI] [PubMed] [Google Scholar]
- Benfey P.N., Chua N.H. (1990). The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250: 959–966. [DOI] [PubMed] [Google Scholar]
- Berthold J., Bauer M.F., Schneider H.C., Klaus C., Dietmeier K., Neupert W., Brunner M. (1995). The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell 81: 1085–1093. [DOI] [PubMed] [Google Scholar]
- Chang L., Bertelsen E.B., Wisén S., Larsen E.M., Zuiderweg E.R., Gestwicki J.E. (2008). High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal. Biochem. 372: 167–176. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú., Jarvis P. (2013). Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833: 332–340. [DOI] [PubMed] [Google Scholar]
- Flügge U.I., Hinz G. (1986). Energy dependence of protein translocation into chloroplasts. Eur. J. Biochem. 160: 563–570. [DOI] [PubMed] [Google Scholar]
- Gragerov A., Zeng L., Zhao X., Burkholder W., Gottesman M.E. (1994). Specificity of DnaK-peptide binding. J. Mol. Biol. 235: 848–854. [DOI] [PubMed] [Google Scholar]
- Hofmann N.R., Theg S.M. (2003). Physcomitrella patens as a model for the study of chloroplast protein transport: Conserved machineries between vascular and non-vascular plants. Plant Mol. Biol. 53: 621–632. [DOI] [PubMed] [Google Scholar]
- Horstmann V., Huether C.M., Jost W., Reski R., Decker E.L. (2004). Quantitative promoter analysis in Physcomitrella patens: A set of plant vectors activating gene expression within three orders of magnitude. BMC Biotechnol. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179: 257–285. [DOI] [PubMed] [Google Scholar]
- Kang P.J., Ostermann J., Shilling J., Neupert W., Craig E.A., Pfanner N. (1990). Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348: 137–143. [DOI] [PubMed] [Google Scholar]
- Krzewska J., Langer T., Liberek K. (2001). Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 489: 92–96. [DOI] [PubMed] [Google Scholar]
- Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., Kurth J., Leister D., Jarvis P. (2003). The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15: 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., Biehl A., Leister D., Ríos G., Koncz C., Jarvis P. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leheny E.A., Theg S.M. (1994). Apparent inhibition of chloroplast protein import by cold temperatures is due to energetic considerations, not by membrane fluidity. Plant Cell 6: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Chiu C.C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61: 157–180. [DOI] [PubMed] [Google Scholar]
- Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991). Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA 88: 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.S., Keegstra K. (1992). Isolation and characterization of a cDNA clone encoding the major hsp70 of the pea chloroplastic stroma. Plant Physiol. 100: 1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P. (2010). Gymnastics of molecular chaperones. Mol. Cell 39: 321–331. [DOI] [PubMed] [Google Scholar]
- McCarty J.S., Walker G.C. (1991). DnaK as a thermometer: Threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc. Natl. Acad. Sci. USA 88: 9513–9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16: 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M.C., McKay D.B. (1993). Threonine 204 of the chaperone protein Hsc70 influences the structure of the active site, but is not essential for ATP hydrolysis. J. Biol. Chem. 268: 24323–24329. [PubMed] [Google Scholar]
- Odell J.T., Nagy F., Chua N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812. [DOI] [PubMed] [Google Scholar]
- Pain D., Blobel G. (1987). Protein import into chloroplasts requires a chloroplast ATPase. Proc. Natl. Acad. Sci. USA 84: 3288–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Varotto C., Meurer J., Jahns P., Salamini F., Leister D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: Effects on mRNA translation and photosynthesis. Plant J. 27: 179–189. [DOI] [PubMed] [Google Scholar]
- Schaefer D., Zryd J.-P., Knight C.D., Cove D.J. (1991). Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 226: 418–424. [DOI] [PubMed] [Google Scholar]
- Schindler C., Hracky R., Soll J. (1987). Protein transport in chloroplasts: ATP is prerequisit. Z. Naturforsch. 42: 103–108. [Google Scholar]
- Schroda M., Vallon O., Whitelegge J.P., Beck C.F., Wollman F.A. (2001). The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell 13: 2823–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Theg S.M. (1996). A new chloroplast protein import intermediate reveals distinct translocation machineries in the two envelope membranes: Energetics and mechanistic implications. J. Cell Biol. 132: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J., DeWitt N.D., Flanagan J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2013a). The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2013b). Energetic cost of protein import across the envelope membranes of chloroplasts. Proc. Natl. Acad. Sci. USA 110: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichting M., Mokranjac D., Azem A., Neupert W., Hell K. (2005). Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 24: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O., Tawfik D.S. (2010). Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79: 471–505. [DOI] [PubMed] [Google Scholar]
- Theg S.M., Bauerle C., Olsen L.J., Selman B.R., Keegstra K. (1989). Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264: 6730–6736. [PubMed] [Google Scholar]
- Theg S.M., Chiang G., Dilley R.A. (1988). Protons in the thylakoid membrane-sequestered domains can directly pass through the coupling factor during ATP synthesis in flashing light. J. Biol. Chem. 263: 673–681. [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Fay S.N., Franco P., Pattavina K.A., Bezanilla M. (2009). Rapid screening for temperature-sensitive alleles in plants. Plant Physiol. 151: 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K., Brown C.A., Vanneste K., van der Zande E., Voet A., Maere S., Verstrepen K.J. (2012). Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 10: e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F., Hinnenberger M., Nick S., Schulz-Raffelt M., Mühlhaus T., Schroda M. (2008). Assistance for a chaperone: Chlamydomonas HEP2 activates plastidic HSP70B for cochaperone binding. J. Biol. Chem. 283: 16363–16373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.