This work shows that the Arabidopsis SORTING NEXIN family proteins are required to recycle the metal transporter IRT1 from the endosomal compartments back to the plasma membrane of root epidermis cells, ensuring efficient iron acquisition.
Abstract
Dicotyledonous plants growing under limited iron availability initiate a response resulting in the solubilization, reduction, and uptake of soil iron. The protein factors responsible for these steps are transmembrane proteins, suggesting that the intracellular trafficking machinery may be involved in iron acquisition. In search for components involved in the regulation of Arabidopsis thaliana iron deficiency responses, we identified the members of the SORTING NEXIN (SNX) protein family. SNX loss-of-function plants display enhanced susceptibility to iron deficiency in comparison to the wild type. The absence of SNX led to reduced iron import efficiency into the root. SNX1 showed partial colocalization with the principal root iron importer IRON-REGULATED TRANSPORTER1 (IRT1). In SNX loss-of-function plants, IRT1 protein levels were decreased compared with the wild type due to enhanced IRT1 degradation. This resulted in diminished amounts of the IRT1 protein at the plasma membrane. snx mutants exhibited enhanced iron deficiency responses compared with the wild type, presumably due to the lower iron uptake through IRT1. Our results reveal a role of SNX1 for the correct trafficking of IRT1 and, thus, for modulating the activity of the iron uptake machinery.
INTRODUCTION
Acquisition of soil nutrients by the root requires tight control mechanisms because of the adverse effects caused by deficiency or overaccumulation. Due to the possibility of changing its oxidation state, Fe is required as a cofactor in a variety of redox enzymes and functions in most of the plant energy management processes, such as photosynthesis and respiration. Dicotyledonous plants, such as the model plant Arabidopsis thaliana, take up iron by a three-step reduction-based mechanism, termed Strategy I. Fe3+ is solubilized from soil particles through the action of plasma membrane (PM) ATPases from the Arabidopsis (H+)-ATPase (AHA) family (Sussman, 1994; Santi and Schmidt, 2009), which extrude protons and lead to rhizosphere acidification. Soluble iron can enter the root apoplast and be reduced to Fe2+ by FERRIC REDUCTASE-OXIDASE2 (FRO2) (Yi and Guerinot, 1996; Robinson et al., 1999) and is imported into the cells by the ZIP family cation transporter IRON-REGULATED TRANSPORTER1 (IRT1) (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). Under iron-deficient conditions, the Strategy I iron uptake system is transcriptionally upregulated. The basic helix-loop-helix (bHLH) family transcription factor FIT was found to be essential but not sufficient for the induction of the genes encoding the Strategy I iron acquisition factors AHA2, FRO2, and IRT1 (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Ivanov et al., 2012a). One or more of the iron deficiency–inducible subgroup Ib bHLH proteins (Heim et al., 2003), bHLH038, bHLH039, bHLH100, and bHLH101, needs to be present and probably forms dimers with FIT for the induction to take place (Wang et al., 2007, 2013; Yuan et al., 2008). This was found to be only a small part of the extensive posttranscriptional regulation of the iron uptake system (Brumbarova and Bauer, 2005; Brumbarova et al., 2008; Donnini et al., 2010; Rellán-Alvarez et al., 2010; Lan et al., 2011). AHA1 and AHA2 activity is regulated by phosphorylation of a single amino acid (Fuglsang et al., 2007; Haruta et al., 2010). FRO2 activity is upregulated only under iron deficiency, even if the gene is artificially expressed under sufficient iron supply (Connolly et al., 2003). The stability of IRT1 is dependent on the ubiquitination of two residues within its large intracellular loop by the ubiquitin ligase IDF1 (Kerkeb et al., 2008; Barberon et al., 2011; Shin et al., 2013). In addition, AHA2, FRO2, and IRT1 are transmembrane proteins that function at the PM. As such, they need to be synthesized within the cell and targeted to their place of action. An HA-tagged FRO2 fusion protein was shown to localize at the PM on the rhizosphere side of epidermal cells (Durrett et al., 2006). Surprisingly, IRT1 was recently found to be predominantly localized in early endosomes. Its PM localization was less strongly pronounced but could be enhanced by abolishing its ubiquitination (Barberon et al., 2011; Shin et al., 2013). These results imply a strong dependence of the iron uptake components on the intracellular trafficking machinery in terms of localization, activity control, and regulation of stability. A key missing point at the moment is knowledge of the protein factors that regulate these processes. Therefore, a very important aspect and long-term aim will be to identify these components. Understanding the molecular basis of these mechanisms will benefit the efforts for crop iron fortification and improving iron bioavailability.
SORTING NEXINs (SNXs) are a family of endosomal regulatory proteins. Unlike their mammalian homologs, plant SNXs form a small family well conserved among land plants. In Arabidopsis, the family contains three members: SNX1, SNX2a, and SNX2b (Vanoosthuyse et al., 2003; Zelazny et al., 2012). SNX1 is a marker for the plant sorting endosome (Jaillais et al., 2008) and was shown to regulate the trafficking of the auxin carrier PIN-FORMED2 (Jaillais et al., 2006; Kleine-Vehn et al., 2008; Ambrose et al., 2013). In the absence of SNX1, the ability of SNX2a or SNX2b to bind the endosomal membrane is abolished; thus, SNX-related defects are observed even in the single snx1 mutant (Pourcher et al., 2010). SNXs usually operate as a part of the multimeric retromer complex, retrieving proteins and routing them back to the Golgi apparatus in a process known as retrograde transport (Cullen and Korswagen, 2012). In plants, the existence of a functional retromer has been suggested by mutant analysis. However, most data support a model wherein SNXs and the core retromer components have developed separate functions (Pourcher et al., 2010).
Our aim was to identify factors involved in regulating iron deficiency responses, with a focus on intracellular trafficking. In a screen among trafficking-related mutants, we found that SNXs play a role in modulating iron deficiency responses in Arabidopsis. We found that a SNX1-GFP (green fluorescent protein) fusion showed partial colocalization with IRT1-mCherry in endosomal compartments. Using an anti-IRT1 antiserum, we could demonstrate that the levels of the IRT1 protein in snx mutants were lower compared with the wild type and that the degradation rate of the protein was enhanced. In support of this, immunohistochemical analysis and membrane fractionation experiments showed reduced IRT1 signal intensity at the PM in the snx1-2 mutant compared with the wild type. Taken together, our data reveal a role of SNX1 in recycling internalized IRT1 and preventing its premature degradation.
RESULTS
Plants Lacking Functional SNX Proteins Exhibit Enhanced Sensitivity to Iron Deficiency
To identify factors responsible for the intracellular translocation of the iron uptake components, we tested the responses of known Arabidopsis trafficking mutants under different iron supply. Among these, SNX loss-of-function plants displayed enhanced sensitivity to iron deficiency in comparison to the wild-type Columbia-0 (Col-0) plants. Unlike many trafficking-related mutants, snx mutants show only mild growth problems and no developmental aberrations, which makes them suitable for physiological analysis. We reasoned that SNX proteins might play a more specific role under iron deficiency stress and are therefore good candidate regulators of the membrane-bound iron uptake–related proteins.
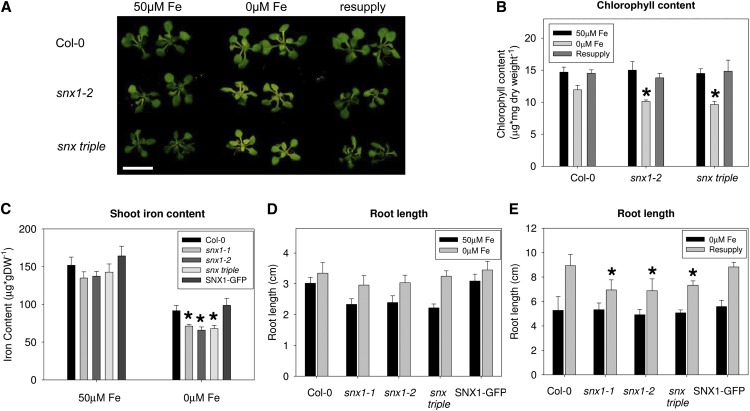
To understand whether the observed low iron sensitivity in the absence of SNX was due to alterations in iron homeostasis, we applied a short-term iron deficiency exposure (Figure 1A). We used seeds from wild type Col-0, snx1-2 mutant, with a disrupted SNX1 gene (Jaillais et al., 2006), and a snx1-2 snx2a-1 snx2b-2 mutant, hereafter named snx triple, where all three Arabidopsis SNX genes are disrupted (Pourcher et al., 2010). Plants were grown in a 2-week growth system (see Methods) that included sufficient iron supply conditions (50 μM Fe), short-term iron deficiency (0 μM Fe), and resupply conditions. Even the short iron deficiency period was enough to provoke differences in the appearance of leaf chlorosis between the wild type and the two snx mutants. We quantified the total amount of chlorophyll (Figure 1B) and found that the two snx mutants displayed a small, 10 to 15%, but statistically significant decrease in the chlorophyll content under iron deficiency compared with the wild type, underlining the potential importance of SNX proteins for iron homeostasis under iron-limiting conditions. No statistically significant differences were observed between the single and the triple mutants or between the snx mutants and Col-0 under iron-sufficient or iron-resupply conditions. We then measured the shoot iron content to see whether the leaf chlorosis was a consequence of compromised iron homeostasis (Figure 1C). For the analysis, two more lines were added, snx1-1, representing an alternative snx1 allele, and the SNX1pro:SNX1-GFP/snx1-1 complementation line (SNX1-GFP; Jaillais et al., 2006). All SNX loss-of-function plants grown under iron-deficient conditions had lower iron content compared with the wild type. In addition, the SNX1-GFP–expressing line showed full complementation of this phenotype, demonstrating that the low iron content phenotype of the snx mutants was a SNX1-dependent effect. These results demonstrate that SNX proteins are needed for maintaining sufficient amounts of iron in the shoots.
Figure 1.
SNX Loss-of-Function Plants Exhibit Iron Deficiency–Related Phenotypes.
(A) Phenotypes of Col-0, snx1-2, and snx triple plants grown in the 2-week growth system. Iron-sufficient (50 μM Fe), iron-deficient (0 μM Fe), and resupply conditions are indicated. Three independent experiments were performed, showing similar results. Bar = 1 cm.
(B) Total chlorophyll content of the plants shown in (A). Error bars represent sd. Asterisks indicate statistical significance (P < 0.05) in comparison to Col-0 under the same condition. Results are based on three biological repetitions.
(C) Shoot iron content of Col-0, snx1-1, snx1-2, snx triple, and SNX1pro:SNX1-GFP/snx1-1 (labeled as SNX1-GFP) plants grown in the 2-week growth system. Error bars represent sd. Asterisks indicate statistical significance (P < 0.05) in comparison to Col-0 under the same condition. Results are based on three biological repetitions. DW, dry weight.
(D) Root lengths of Col-0, snx1-1, snx1-2, snx triple, and SNX1pro:SNX1-GFP/snx1-1 (labeled as SNX1-GFP) plants grown for 5 d under iron-sufficient (50 μM Fe) or iron-deficient (0 μM Fe) conditions. Error bars represent sd. Asterisks indicate statistical significance (P < 0.05) in comparison to Col-0 under the same condition. Results are based on three biological repetitions.
(E) Root lengths of Col-0, snx1-1, snx1-2, snx triple, and SNX1pro:SNX1-GFP/snx1-1 (labeled as SNX1-GFP) plants grown for 7 d under iron-deficient (0 μM Fe) conditions and then resupplied with iron-sufficient (50 μM Fe) medium for 5 d. Error bars represent sd. Asterisks indicate statistical significance (P < 0.05) in comparison to Col-0 under the same condition. Results are based on three biological repetitions.
In addition, we also noted alterations in the primary root growth (Figures 1D and 1E). Under normal nutrient supply, snx mutant roots showed a reduction in length compared with wild type. This is a known phenotype that is complemented by the presence of SNX1-GFP (Jaillais et al., 2006). Roots of iron-deficient plants were longer, and, surprisingly, under these conditions no difference could be observed between mutants and the wild type (Figure 1D). Thus, lack of iron caused compensatory growth that overrides the loss-of-SNX phenotype. Furthermore, we wanted to know whether this growth effect would be permanent or was only manifested under iron limitation. Plants were grown in the absence of iron for 1 week and were then transferred to normal iron supply for another 5 d (Figure 1E). The characteristic snx1 and snx triple mutant growth retardation reappeared after the resupply, suggesting that the effect of growth promotion in the absence of SNX lasts only until plants grow under limited iron supply. In summary, these results show that SNX proteins may be involved in the maintenance of the plant iron homeostasis, since under iron deficiency, the lack of SNX proteins affects the shoot iron content but not the root length response of the plant. This shows that SNX action influences some but not all aspects of plant iron homeostasis and therefore excludes the possibility that the phenotypes are the result of pleiotropic effects. Therefore, our objective was to gain a deeper understanding of the involvement of the SNX proteins in the iron deficiency response of the plant.
SNX1 and SNX2b Expression Patterns Are Consistent with a Role in Iron Uptake
We analyzed the expression patterns of SNX genes in the root with respect to iron deficiency. Gene coexpression networks were generated to investigate whether SNX genes share common regulation with known iron deficiency response genes (Supplemental Figures 1A and 1B). While this was not the case, the analysis revealed coexpression between SNX1 and cell cycle, and DNA damage–related genes. Connections between DNA damage/cell cycle control and iron deficiency have been reported recently (Schuler et al., 2011; Ivanov et al., 2012a). The SNX2b gene was not available for analysis in the ATTED-II tool. The availability of SNXs on tissue level was analyzed using reporter lines in which each of the three SNX genes, including their upstream sequences, was translationally fused to either GFP or β-glucuronidase (GUS) reporters (Jaillais et al., 2006; Pourcher et al., 2010). Expression patterns in 8-d-old plants grown under either iron-sufficient or iron-deficient conditions did not differ significantly (Supplemental Figures 2A to 2R) and correlated well with the previously reported observations for plants grown on Murashige and Skoog plates (Pourcher et al., 2010). SNX1 and SNX2b reporters showed strong accumulation at the root tip and in the vasculature throughout the root (Supplemental Figures 2A to 2C, 2G to 2L, and 2P to 2R), while SNX2a was highly present in the green organs and only the vasculature in the upper part of the root showed visible staining (Supplemental Figures 2D to 2F and 2M to 2O). Importantly, for SNX1 and SNX2b, reporter signal was clearly visible in the epidermal layer and especially in root hair cells in the root differentiation zone (Supplemental Figures 2S to 2X). Expression and localization patterns of key iron uptake regulators and effectors, such as FIT and IRT1 (Vert et al., 2002; Jakoby et al., 2004; Barberon et al., 2011), suggest that these cells are primarily responsible for iron uptake. Therefore, the localization of SNX1 and SNX2b hints at potential involvement in the uptake of iron from the soil.
SNX1 Protein Levels in the Root Are Elevated upon Iron Deficiency
As SNX1 was shown to be the essential factor in the SNX complex assembly (Pourcher et al., 2010), we then analyzed the expression of the SNX1 gene in the root by quantitative RT-PCR. The expression under standard and iron-deficient conditions as well as the dependence of SNX1 on the central iron uptake regulator FIT was tested (Supplemental Figure 3A). We used Col-0, fit-3 (a FIT loss-of-function mutant), and a FITox line, in which the FIT cDNA is overexpressed (Jakoby et al., 2004). Plants were grown continuously at 50 μM Fe or 0 μM Fe for 8 d before the analysis. The expression of SNX1 did not change significantly in any of the samples compared with the Col-0 50 μM Fe condition (Supplemental Figure 3A). We used the SNX1p:SNX1-GFP reporter to estimate the availability of SNX1 protein in the root under the different iron supply conditions by immunoblot (Supplemental Figures 3B and 3C). Since this line complements fully the snx1-1 phenotypes, as well as the iron deficiency–related ones, the GFP fusion can be considered to faithfully represent the fate of the SNX1 protein. Densitometry on exposed films showed that the levels of SNX1-GFP were higher under iron deficiency by ∼25% compared with the sufficient iron condition (Supplemental Figure 3C). This suggests that there might be a higher requirement for SNX1 in the root when iron is limited. Since the SNX1 gene expression is not significantly upregulated under these conditions, the enhanced protein availability might be due to enhanced stability. There is currently no data on the regulation of SNX1 turnover; however, it can be speculated that SNX1, being membrane-attached but also a soluble cytoplasmic protein, may undergo proteasome-dependent degradation. Thus far, immunoblot analysis on SNX1-GFP does not show major variations from the predicted molecular weight of the fusion, which would suggest prominent degradation products or ubiquitinated forms (Supplemental Figure 3).
snx Mutants Display Reduced Iron Uptake Efficiency
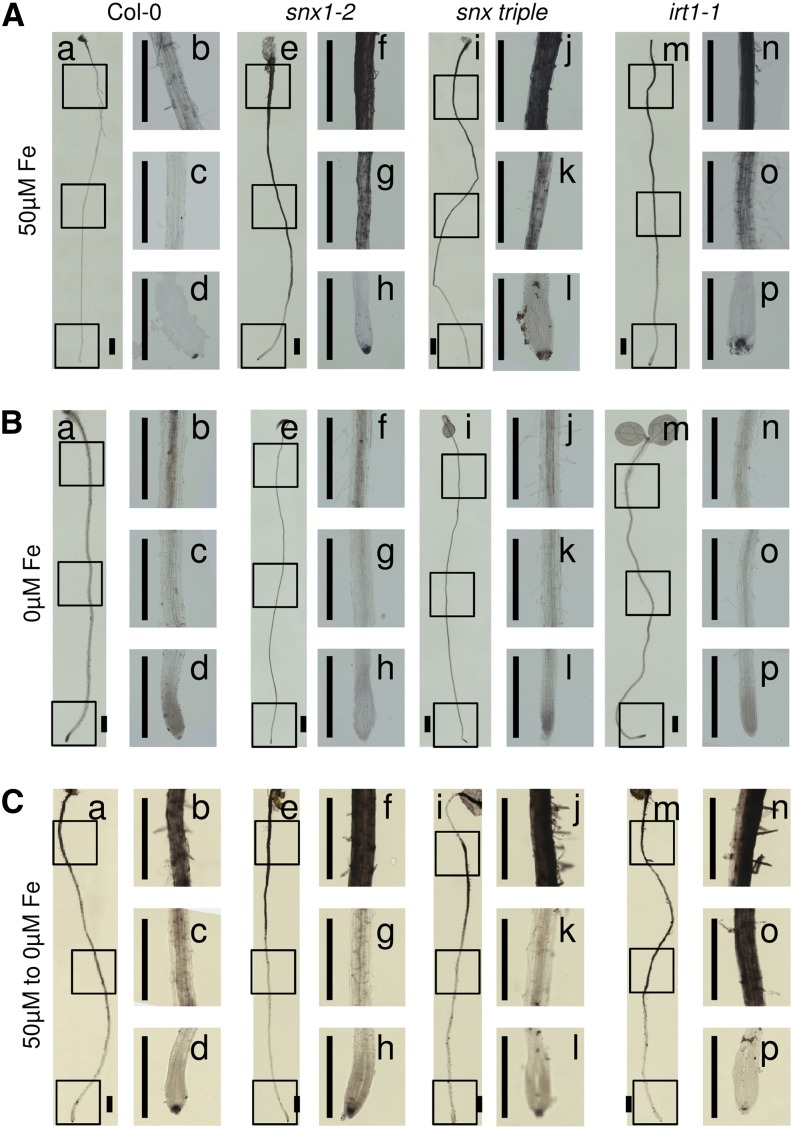
Since two SNX genes are expressed at places of active iron uptake, we tested the uptake efficiency in snx mutant plants. We performed histological staining to visualize the presence of apoplastic iron. Plants from all tested genotypes, grown for 8 d under sufficient iron supply (Figure 2A), displayed very intensive staining of the outer epidermal cell walls, suggesting a strong iron accumulation in the apoplast. The staining was more intense toward the upper parts of the root and became gradually weaker toward the root tip, with the tip itself showing strong signal. This corresponds to the fact that the expression of iron uptake genes, such as IRT1, FIT, BHLH038, BHLH039, BHLH100, and BHLH101, is weak in the basal root zones (Vert et al., 2002; Jakoby et al., 2004; Wang et al., 2007) and suggests that these zones are not primarily used for iron uptake. At the same time, differences could be observed between the wild type (Figure 2A, a to d) and the iron uptake–deficient irt1-1 mutant (Figure 2A, m to p). Roots of irt1-1 were stained much stronger than the wild type, suggesting that Col-0 roots are able to successfully take up the iron from the apoplast, unlike the uptake-incapable mutant. Interestingly, the staining patterns of the snx1-2 and snx triple mutant roots (Figure 2A, e to l) resembled that of irt1-1, showing much stronger staining than that of the wild type throughout the length of the root and suggesting that these plants are not capable of efficiently taking up apoplastic iron. Plants from all tested genotypes, grown continuously under iron-deficient conditions (Figure 2B), showed no staining in the apoplast, even after prolonged development of the color reaction. Clear signal could be observed in the central cylinder of the wild type, snx1-2, and snx triple (Figure 2B, a to l), showing that SNX absence did not cause complete inability to take up iron and that the iron transport within the root tissues was not affected. In comparison, the irt1-1 mutant roots (Figure 2B, m to p) showed only extremely weak signal under these conditions. We wanted to further verify that the snx mutants still possessed an inducible iron uptake system. We grew the plants for 6 d under normal nutrient supply and transferred them to iron-deficient medium for the next 2 d, as such a treatment causes the activation of the iron uptake components (Figure 2C). A strong reduction in the apoplastic signal could be observed in the middle section of the wild type, snx1-2, and snx triple roots (Figure 2C, a to l), whereas staining of the central cylinder could be seen. The overall signal intensity in Col-0 roots was lower compared with the other lines, confirming the previously presented observations. The apoplast of the corresponding central region in the irt1-1 mutant roots (Figure 2C, m to p) remained intensely stained consistent with the known inability of this mutant to take up iron.
Figure 2.
Iron Uptake Is Compromised in snx Mutants.
Visualization of iron in the roots of Col-0, snx1-2, snx triple, and irt1-1 plants grown under iron-sufficient (50 µM Fe; [A]), iron-deficient (0 µM Fe; [B]), or resupply (transferred from 50 µM Fe to 0 µM Fe; [C]) conditions. For each condition and genotype, an overview image of the root (a, Col-0; e, snx1-2; i, snx triple; and m, irt1-1) and three close-ups (b to d, Col-0; f to h, snx1-2; j to l, snx triple; and n to p, irt1-1) are shown. The presence of iron is visible as dark staining along the roots. The experiment was performed on three biological repetitions. In each repetition, a minimum of three plants per genotype and condition was observed. Bars = 2 mm.
This shows that the snx mutants are capable of taking up iron and the system is inducible under iron starvation. Yet, the lack of SNX function leads to overall lower iron intake, consistent with the enhanced sensitivity to iron deficiency.
Expression of Iron Uptake–Related Genes Is Affected by the Absence of SNX Function
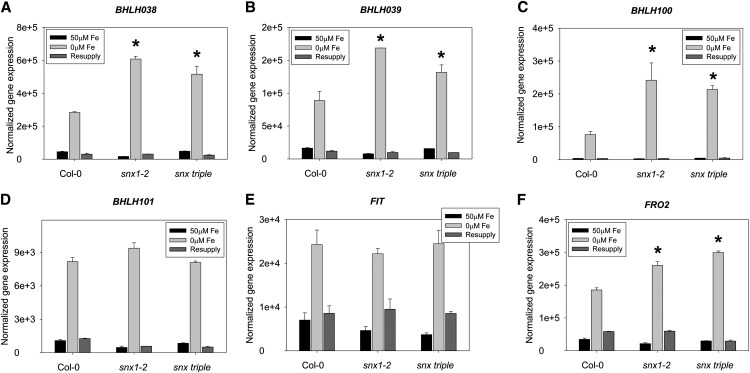
We addressed the question of whether reduced iron uptake in snx mutants resulted in a feedback regulation of the expression of Strategy I genes. We used quantitative RT-PCR to investigate the expression of key iron uptake genes, such as those encoding transcriptional regulators FIT, bHLH038, bHLH039, bHLH100, and bHLH101 and the effectors FRO2 and IRT1 in 2-week-old plants grown under sufficient, deficient, and resupply conditions (Figures 3A to F and 4A). As expected based on previous studies (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Wang et al., 2007), all investigated genes were upregulated under iron deficiency and downregulated when iron was resupplied to iron-deficient plants. Surprisingly, snx mutants showed an expression profile resembling that of wild-type plants, with low gene activity under normal iron supply. The major difference caused by the lack of SNX was the level of induction under iron deficiency. The genes encoding bHLH038, bHLH039, and bHLH100 as well as the targets FRO2 (Figures 3A to 3C and 3F) and IRT1 (Figure 4A) showed significantly elevated transcript levels in both mutant lines compared with the wild type. Interestingly, BHLH101 and FIT (Figures 3D and 3E) transcript abundance under iron limitation was not significantly altered in the mutants, suggesting that multiple signaling inputs might control the expression of the iron uptake regulators and SNX proteins might be involved in modulating some of these signals. The observed enhanced transcript abundance corresponds well to the observed reduced iron uptake capabilities in the absence of SNX. It can be considered as the output of a feedback signaling mechanism aiming to compensate for the impaired iron uptake.
Figure 3.
Enhanced Expression of Iron Uptake–Related Genes in the Absence of SNX Function.
Gene expression analysis by quantitative RT-PCR in the roots of Col-0, snx1-2, and snx triple grown in the 2-week growth system. The genes BHLH038 (A), BHLH039 (B), BHLH100 (C), BHLH101 (D), FIT (E), and FRO2 (F) were investigated. The result of three biological repetitions is presented. Bars represent expression strength under iron-sufficient (50 μM Fe; black), iron-deficient (0 μM Fe; light gray), and resupply (dark gray) conditions. Error bars represent sd. Asterisks indicate statistically significant increase of gene expression in snx1-2 and snx triple, in comparison to Col-0, under iron-deficient conditions (P < 0.005).
Figure 4.
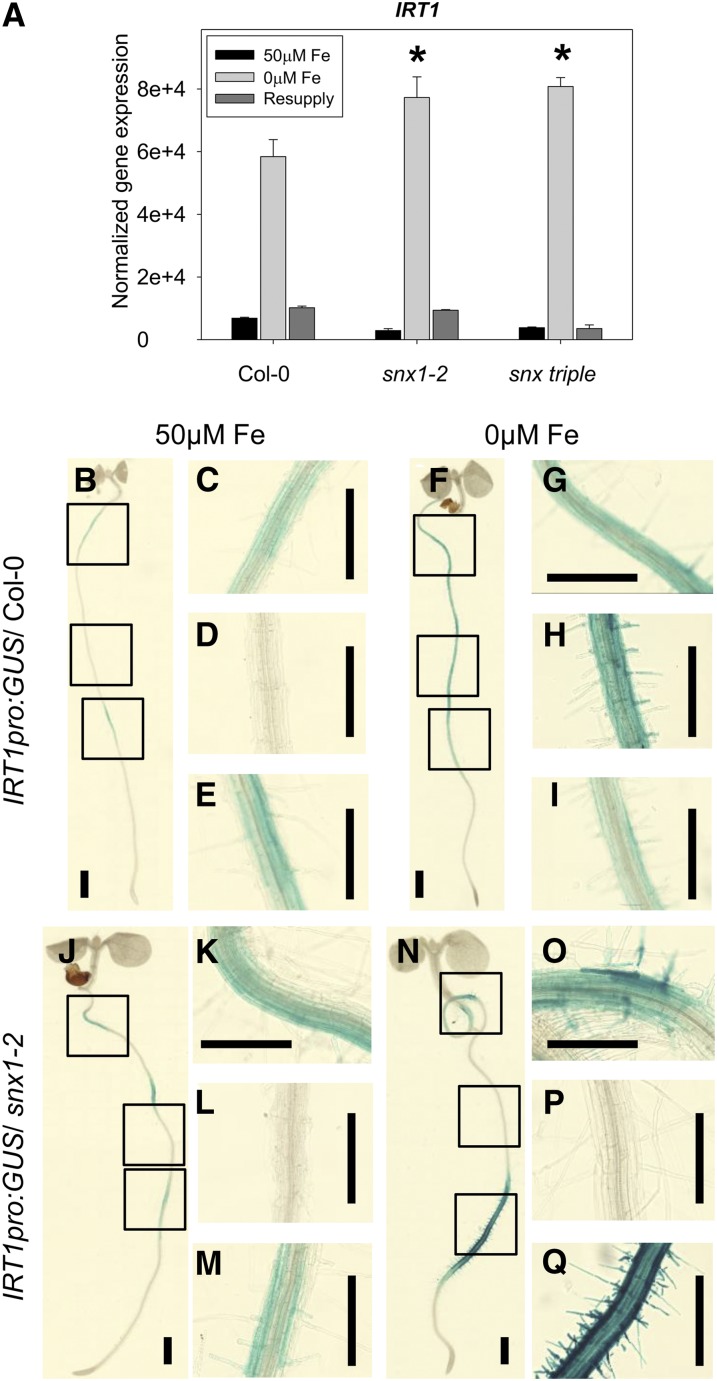
Changes in IRT1 Gene Expression Topology in the Absence of SNX1.
(A) Analysis of IRT1 gene expression by quantitative RT-PCR in the roots of Col-0, snx1-2, and snx triple grown in the 2-week system. The result of three biological repetitions is presented. Bars represent expression strength under iron-sufficient (50 μM Fe; black), iron-deficient (0 μM Fe; light gray), and resupply (dark gray) conditions. Error bars represent sd. Asterisks indicate statistically significant increase of gene expression in snx1-2 and snx triple, in comparison to Col-0, under iron-deficient conditions (P < 0.005).
(B) to (Q) Analysis of reporter gene activity in lines expressing the IRT1pro:GUS construct in Col-0 ([B] to [I]) or snx1-2 mutant background ([J] to [Q]). Plants were grown for 8 d under iron-sufficient (50 μM Fe) or iron-deficient (0 μM Fe) conditions, and reporter gene activity was visualized by bright-field microscopy after a chemical reaction (GUS). Three independent experiments were made, showing similar results. Bars = 2 mm.
Loss of SNX1 Causes Changes in the Topology of IRT1 Gene Expression
In an attempt to elucidate the reason for the observed SNX-related phenotypes, we analyzed in more detail the behavior of the IRT1 gene and its product in snx mutant plants. Similarly to FRO2 (Figure 3F), the expression of IRT1 under iron deficiency was significantly enhanced in the snx1-2 and snx triple mutants compared with the same condition in the wild type (Figure 4A). Considering the fact that IRT1 expression in both snx mutants under normal iron supply was ∼50% of that of the wild type, the net induction of expression was even higher: 45% for snx1-2 and 48% for snx triple. To test whether and how these gene expression changes reflect the domain in the root where IRT1 gene is active, we compared lines expressing GUS reporter gene under the control of the IRT1 promoter in the wild type (Figures 4B to 4I) and snx1-2 background (Figures 4J to 4Q; Supplemental Figures 4A to 4C). In plants with wild-type background grown for 8 d under normal iron supply, IRT1 promoter-driven GUS activity was weak with repeated occurrences and disappearances along the length of the root (Figures 4B to 4E). A dramatic induction of reporter activity could be observed in plants grown in iron-deficient medium. Signals were present starting from the base of the root until just before the end of the root hair zone, while no signals were visible further into the elongation zone and the root tip (Figures 4F to 4I). In snx1-2 background, under normal iron supply the GUS pattern was similar to the control (Figures 4J to 4M). However, under iron deficiency, no activity could be seen in the middle section of the root. Instead, staining was visible in the proximity of the hypocotyl, and especially strong signals could be seen in the last third of the root length finishing just before the end of the root hair zone (Figures 4N to 4Q). Thus, the net increase in IRT1 expression in the snx mutants was due to a strong local induction of IRT1 promoter activity.
IRT1 and SNX1 Partially Colocalize in Cytoplasmic Compartments
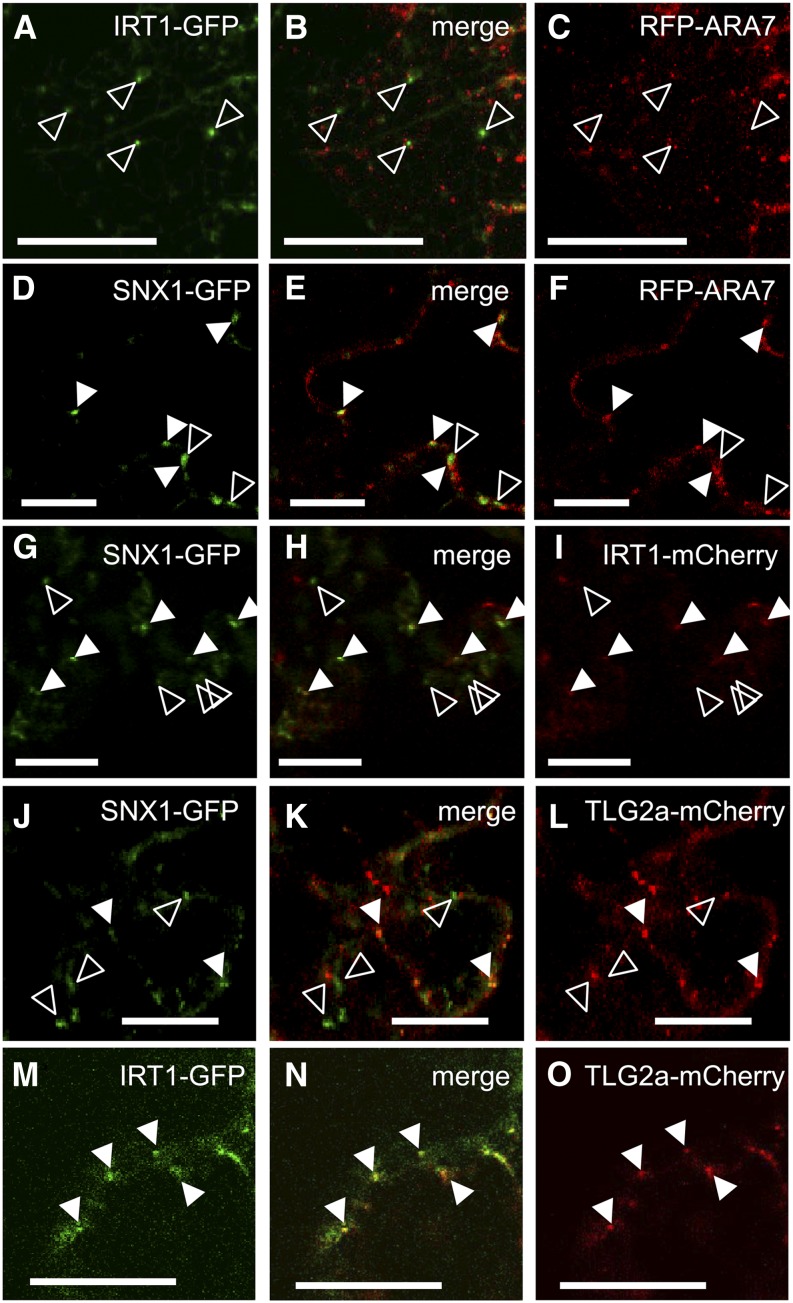
IRT1 was shown to localize in trans-Golgi network (TGN)/early endosomes, while there is some debate concerning SNX1 endosomal localization, which is probably dual, in early and late endosomes (Jaillais et al., 2008; Niemes et al., 2010; Barberon et al., 2011; Stierhof et al., 2013). We tested whether the two proteins IRT1 and SNX1 can be found simultaneously in the same compartments. Translational fusions of IRT1 with GFP and mCherry were used for transformation of tobacco (Nicotiana benthamiana) epidermal cells and colocalization studies. Colocalization of endosomal signals from the GFP and mCherry/mRFP channels was quantified and presented in Supplemental Figure 5K. In preliminary experiments, the two IRT1 fusions were coexpressed to see if they would yield similar localization pattern (Supplemental Figures 5A to 5C). The two fusions showed more than 95% colocalization, thus allowing us to use any of them for further studies. Immunoblot analysis demonstrated that the full-length IRT1-GFP fusion was correctly expressed in the tobacco cells (Supplemental Figure 5J). We tested the late endosome localization of IRT1-GFP using the markers RFP-ARA7 (Figures 5A to 5C) and the ATPase SKD1-mCherry (Supplemental Figures 5D to 5F). No significant overlap in the subcellular distribution in the green and red channels could be observed in both cases (7% or less colocalization). Compartments positive for the marker were negative for IRT1 and vice versa, even though in certain cases they could be seen in close proximity. Next, we tested SNX1-GFP with RFP-ARA7 (Figures 5D to 5F). The two showed a great degree of colocalization (71%), suggesting that unlike IRT1, SNX1 was mainly localized in ARA7-positive compartments. Importantly, ∼29% of the SNX1-positive compartments had no RFP-ARA7 signal showing that part of the SNX1 population is located in different endosomes. We then tested the IRT1-mCherry and SNX1-GFP combination (Figures 5G to 5I). Indeed, 21% of the SNX1-positive and 34% of the IRT1-positive compartments showed signals corresponding to both tested fusions simultaneously. This result demonstrates that IRT1 colocalized with SNX1 within a subpopulation of endosomal compartments. To prove the identity of these structures, we coexpressed each of the two with the TGN marker TLG2a tagged to mCherry. More than 19% of the SNX1-GFP signals colocalized with TLG2a-mCherry, while only 15% of the TGN marker showed SNX1-GFP signals (Figures 5J to 5L). More than 87% of IRT1-GFP–positive endosomes were also TLG2a-mCherry positive, while at the same time, 24% of the TLG2a-mCherry signal could be found in additional compartments, not seen in the GFP channel (Figures 5M to 5O). TLG2a-GFP and ARA7-RFP showed <2% colocalization when coexpressed (Supplemental Figures 5G to 5I). Together, the results show that the majority of IRT1-GFP is located in early endosomes and at least 30% of this population colocalizes with SNX1-GFP, which may represent a prerequisite for an influence of SNX1 on IRT1.
Figure 5.
Colocalization between SNX1 and IRT1.
(A) to (C) Coexpression of IRT1-GFP (A) and RFP-ARA7 ([C]; late endosome marker). Open arrowheads show examples of signals in the green channel that do not correspond to signals in the red channel. Bars = 20 μm.
(D) to (F) Coexpression of SNX1-GFP (D) and RFP-ARA7 (F). Closed arrowheads show examples of colocalization, while open arrowheads show examples of signals in the green channel that do not correspond to signals in the red channel. Bars = 20 μm.
(G) to (I) Coexpression of SNX1-GFP (G) and IRT1-mCherry (I). Closed arrowheads show examples of colocalization, while open arrowheads show examples of signals in the green channel that do not correspond to signals in the red channel. Bars = 20 μm.
(J) to (L) Coexpression of SNX1-GFP (J) and TLG2a-mCherry ([L]; TGN/early endosome marker). Closed arrowheads show examples of colocalization, while open arrowheads show examples of signals in the green channel that do not correspond to signals in the red channel. Bars = 20 μm.
(M) to (O) Coexpression of IRT1-GFP (M) and TLG2a-mCherry (O). Closed arrowheads show examples of colocalization. Bars = 20 μm.
A minimum of 10 images per combination was analyzed.
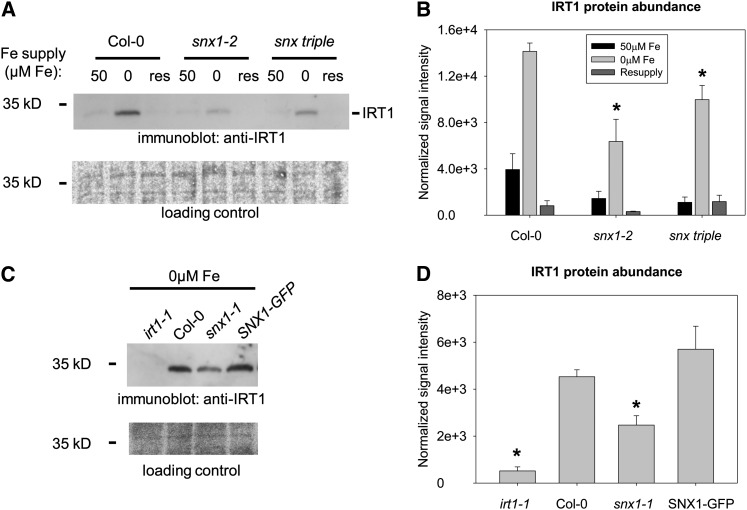
The Abundance of IRT1 Protein Depends on the Availability of SNX Proteins
We aimed to identify a possible effect of SNX proteins on IRT1 that would explain the observed iron uptake defects in snx mutants. For this, we analyzed the abundance of the IRT1 protein by immunoblot in the roots of 2-week-old plants grown under sufficient, iron-limited, and resupply conditions as already described (Figures 6A and 6B). In wild-type plants, IRT1 was barely detectable under sufficient iron supply. A strong increase in IRT1 abundance occurred under limited iron supply, in agreement with earlier reports (Connolly et al., 2002; Séguéla et al., 2008). After resupply, IRT1 abundance was reduced. In snx1-2 mutant plants, this general pattern of IRT1 levels was preserved; however, the overall IRT1 protein abundance was dramatically decreased. IRT1 signal in snx1-2 under iron limitation was ∼45% of the corresponding wild-type levels. The situation was similar in the snx triple mutant roots, though the lack of IRT1 induction under iron deficiency, being ∼70% of the wild type, was not as dramatic as in the single mutant. In addition, we tested the snx1-1 mutant and the SNX1-GFP–expressing complementation line under iron deficiency (Figures 6C and 6D). IRT1 levels were reduced by ∼50% in snx1-1 in comparison with the wild type, and this phenotype was fully complemented in the presence of the GFP-tagged SNX1. irt1-1 mutants served as negative controls for immunoblot analysis using the anti-IRT1 antiserum.
Figure 6.
Reduced IRT1 Protein Amounts in the Absence of SNX Proteins.
(A) Immunoblot analysis of IRT1 protein abundance in roots of Col-0, snx1-2, and snx triple plants grown in the 2-week system. Iron-sufficient (50 μM Fe), iron-deficient (0 μM Fe), and resupply (res) conditions are indicated. Three independent experiments were performed, showing similar results.
(B) Quantification of (A) based on three independent experiments. Bars represent normalized protein abundance under iron-sufficient (50 μM Fe; black), iron-deficient (0 μM Fe; light gray), and resupply (dark gray) conditions. Error bars represent sd. Asterisks indicate statistically significant decrease of IRT1 signal in snx1-2 and snx triple, in comparison to Col-0, under iron-deficient conditions (P < 0.01).
(C) Immunoblot analysis of IRT1 protein abundance in roots of irt1-1, Col-0, snx1-1, and SNX1pro:SNX1-GFP/snx1-1 (labeled as SNX1-GFP) plants grown in the 2-week system and then only under iron deficiency. Three independent experiments were performed, showing similar results.
(D) Quantification of (C) based on three independent experiments. Bars represent normalized protein abundance. Error bars represent sd. Asterisks indicate statistically significant decrease of IRT1 signal in irt1-1 and snx1-1, in comparison to Col-0, under iron-deficient conditions (P < 0.005).
The decreased IRT1 abundance in the absence of SNX1 upon iron deficiency is in good agreement with the reduced iron uptake capacity of the mutants. It suggests that the amount of IRT1 present in the snx mutants is not sufficient for the import of the required quantities of iron under iron deficiency.
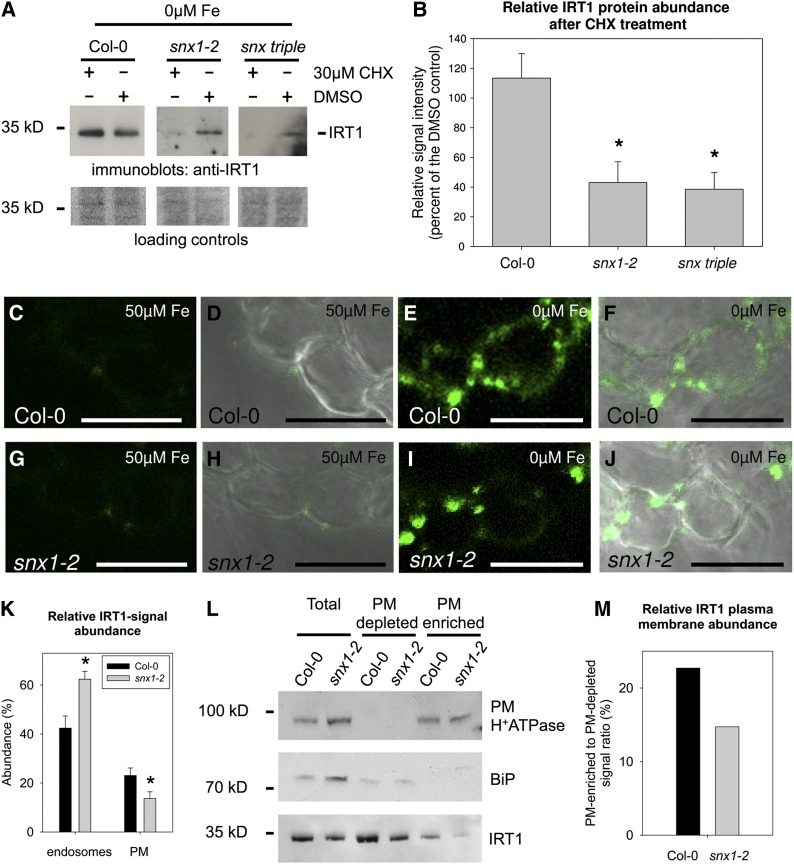
IRT1 Protein Stability Is Decreased in the Absence of SNX Proteins
Furthermore, we wanted to know whether the observed decrease in IRT1 levels could be the result of reduced IRT1 stability. Therefore, IRT1 protein levels were analyzed under iron deficiency when protein synthesis was blocked with the inhibitor cycloheximide (CHX). After a 3-h treatment with CHX, or a mock control treatment with DMSO, samples were analyzed by immunoblot and quantified (Figures 7A and 7B). Surprisingly, IRT1 amounts in CHX-treated Col-0 were slightly elevated in comparison to the corresponding control, suggesting that factors with a high turnover rate might negatively influence IRT1 stability. Similar effects were recently reported for the transcriptional regulation of the IRT1 gene (Sivitz et al., 2012). In the snx1-2 and snx triple mutants, a strong reduction in IRT1 levels was observed after CHX treatment. In the snx1-2 mutant, the residual levels were ∼43% of the control, while in snx triple, they were ∼38%, showing in both cases an enhanced degradation rate of the transporter. In the light of the recent discovery of the role of intracellular trafficking in the regulation of the IRT1 function (Barberon et al., 2011), it can be suggested that in the absence of functional SNX proteins, an active form of IRT1 is misrouted for degradation, instead of being recycled and reused. Such a scenario would mean a decreased abundance of IRT1 at the PM resulting in a reduction in iron uptake.
Figure 7.
Decreased IRT1 Protein Stability and PM Localization in the Absence of SNX Proteins.
(A) Immunoblot analysis of IRT1 protein abundance in roots of Col-0, snx1-2, and snx triple plants grown for 2 weeks under iron-sufficient conditions, transferred for 4 d to iron-deficient conditions, and then treated with protein synthesis inhibitor CHX (+ CHX, − DMSO) or mock treated (− CHX, + DMSO) for 3 h. Three independent experiments were performed, showing similar results.
(B) Quantification of (A) based on three independent experiments. The abundance of IRT1 after CHX treatment is represented as a percentage of the signal in the respective mock treatment. Error bars represent sd. Asterisks indicate statistically significant decrease of IRT1 signal in snx1-2 and snx triple, in comparison to Col-0, after CHX treatment (P < 0.01).
(C) to (J) Immunolocalization of IRT1 in the epidermis of root cross sections from plants grown in the 2-week system. Col-0, 50 µM Fe ([C] and [D]), Col-0, 0 μM Fe ([E] and [F]), snx1-2, 50 μM Fe ([G] and [H]), and snx1-2, 0 μM Fe ([I] and [J]). Bars = 20 μm.
(K) Normalized IRT1 signal intensity in endosomal compartments and the PM in Col-0 and snx1-2 root epidermal cells under iron deficiency, calculated as percentage from the total fluorescence of the cells. Twenty-five cells from each of the two genotypes were used for the quantification. Error bars represent sd. Asterisks indicate statistically significant difference in snx1-2 in comparison to Col-0 (P < 0.01).
(L) Presence of IRT1 in PMs of Col-0 and snx1-2 roots. The input (labeled as “Total”), together with the PM-depleted and PM-enriched fractions are shown. The samples were probed with antibodies against the PM (H+)-ATPase, the endoplasmic reticulum marker BiP, and IRT1. The fractionation experiment was performed twice and yielded identical results.
(M) Quantification of (L). The relative amount of IRT1 at the PM is presented as a percentage of the combined signal in the PM-depleted and PM-enriched fraction for each of the two genotypes.
To test this hypothesis, the subcellular distribution of IRT1 in epidermal cells was investigated by immunolocalization (Figures 7C to 7J). Root cross sections were prepared from the zone showing the strongest IRT1 promoter activity in the snx1-2 mutant. IRT1 signals in roots grown under sufficient iron were weak (Figures 7C, 7D, 7G, and 7H). By contrast, iron-deficient roots showed more intensive staining, due to increased IRT1 abundance (Figures 7E, 7F, 7I, and 7J). In Col-0, the IRT1-signal abundance at the PM relative to the overall cellular signal was ∼23%, while in comparison, in snx1-2, it was only ∼13% (Figure 7K). The relative intensity in the endosomes of snx1-2 was higher than that in Col-0 with 62% versus 42%, respectively.
To verify this observation, we performed PM purification from iron-deficient Col-0 and snx1-2 roots grown in the 2-week system. The resulting fractions were analyzed by immunoblot. Purity of the PM-depleted and the PM-enriched fractions was verified using antibodies against the PM (H+)-ATPase and the endoplasmic reticulum marker BiP, respectively (Figure 7L). Probing against IRT1 showed generally low presence in the PM-enriched fraction in agreement with earlier reports (Barberon et al., 2011; Shin et al., 2013). The amount of IRT1 in the snx1-2 samples was lower, thus corresponding well to our previous observations. We calculated the IRT1 signal intensities in the different fractions to estimate its relative abundance at the PM (Figure 7M). The calculations showed a relative IRT1 PM localization of 22.7% in Col-0 and 14.7% for snx1-2, which is comparable to the immunolocalization results. Together, the data show that in snx1-2 IRT1 accumulates to a higher extent in endosomes, resulting in its diminished amounts at the PM.
Our data thus favor a model in which IRT1 can initially localize to the PM in a SNX-independent manner, but following internalization, its recycling requires the action of SNX proteins in order to avoid premature degradation.
DISCUSSION
Iron Uptake and Intracellular Protein Trafficking
The accumulated data on the posttranscriptional regulation of the iron uptake components, together with the presumed localization of the active AHA2, FRO2, and IRT1 proteins at the PM, already provides strong hints that intracellular trafficking regulators are involved in iron acquisition. Interestingly, only occasionally such regulators show up in large-scale transcriptomic and proteomic analyses as being subject to iron-dependent transcriptional or translational regulation. The reason for this might be that while most of the genes responding to changes in iron supply are a part of a large stress regulon (Ivanov et al., 2012a), the underlying regulatory factors are recruited from the normal plant developmental program, and their transcriptional/translational response to stress may be very weak or nonexistent. In a similar manner, SNX1 gene expression shows no significant iron-dependent change, although the protein is slightly upregulated under low iron supply. Despite that, it has an important function for the regulation of iron uptake through ensuring the stability and PM localization of the principal iron transporter IRT1. This finding adds a layer to the complex picture of posttranscriptional regulation of iron uptake.
A plausible model for the mechanism of SNX-dependent IRT1 stability regulation can be proposed based on the known functions of SNX proteins in mammals, yeast, and Arabidopsis. Similarly to other transmembrane proteins, such as transporters (Geldner et al., 2001; Dhonukshe et al., 2007; Kasai et al., 2011) or receptor kinases (Russinova et al., 2004; Gifford et al., 2005; Ivanov and Gaude, 2009), IRT1 was recently shown to exhibit very complex trafficking behavior. The protein cycles actively between the PM and endosomal compartments where some of the molecules are sent to the vacuole for degradation (Barberon et al., 2011). A lack of SNX function at the endosome would result either in failure of the initial IRT1 PM targeting or in a perturbed IRT1 recycling. We found that snx mutant plants are able to induce their iron uptake capacity under iron limitation but the uptake efficiency is lower than that of the wild type. Based on this, it can be assumed that IRT1 is able to reach the PM in the first place but is not available there in sufficient amounts to meet the root iron uptake requirements. Therefore, the most probable function of SNX proteins in iron uptake regulation is their involvement in recycling of endocytosed IRT1 molecules in the plant-sorting endosome, making them available for another round of activity at the PM. This is well supported by the colocalization between IRT1 and SNX1 in a subset of the cellular endosomal population and the enhanced IRT1 accumulation in endosomes in the absence of SNX1. Barberon et al. (2011) suggested that IRT1 sorting may occur in an endosome different from the early endosome. The fact that IRT1 localizes to at least two different compartments, either containing SNX1 or not, very well supports this idea. Thus, the decision between degradation and recycling will occur in the SNX endosome and the actual exo/endocytosis from/to the early endosome. This additional trafficking step might be related to IRT1 ubiquitination status, since the data from Barberon et al., (2011) suggest that all endocytosed IRT1 is ubiquitinated. However, our localization data support a model of SNX/sorting endosome being, at least in the case of IRT1, different from the late endosome/multivesicular body. In the absence of SNX, IRT1 cannot be further recycled and is first retained in the endosomes and then targeted for degradation. In support of this is the enhanced endosomal localization and reduced stability of IRT1 in snx mutant background. Similar examples of SNX function were shown previously in different organisms. In yeast, the stability and PM localization of the dimeric iron transporter Fet3p-Ftr1p under iron deficiency is maintained by the retromer complex, which includes the SNX1 homolog Vps5p. In the absence of Vps5p, the transporter is targeted to the vacuoles (Strochlic et al., 2007). In Arabidopsis, in the absence of SNX1, an enhanced vacuolar localization and degradation of the auxin efflux carrier PIN2 was observed after gravitropic stimulation (Kleine-Vehn et al., 2008; Ambrose et al., 2013). It is important to note that these PIN2 data suggest that SNX1 might also act as a part of the retromer judging from the similar PIN2 localization phenotypes in snx1 and core retromer mutant lines. In the light of the fact that SNX1 has functions outside of this complex in mammals (Zelazny et al., 2012) and that Arabidopsis core retromer can work independently of SNX (Pourcher et al., 2010), it remains an open question whether the retromer participates in the regulation of iron uptake.
Participation of the SNX Family Members in the Regulation of Iron Uptake
Another point of interest is which members of the SNX family are involved in the regulation of iron uptake. The three Arabidopsis proteins are able to strongly interact with each other and, as already mentioned, the absence of SNX1 abolishes the localization of SNX2a and SNX2b to the endosomal membrane (Pourcher et al., 2010). Thus, even the single mutant displays a complete loss of SNX function. It is at present not clear whether SNX proteins act alone or in the form of homo/heterodimers. Within the retromer, there is a requirement for two SNX proteins (Cullen and Korswagen, 2012), which in Arabidopsis would mean a SNX1-SNX1, SNX1-SNX2a, or SNX1-SNX2b couple at the endosomal membrane. It can be considered that if SNX proteins act independently of the retromer, complexes containing the same combinations would be formed. Importantly, the expression analysis showed that SNX2a presence in the root is limited to the central cylinder in proximity to the hypocotyls. As iron uptake occurs predominantly in the epidermis and generally in the lower portions of the root, it can be considered that SNX2a does not participate directly in iron uptake. The phenotypes described in this study can be observed in single snx1 mutants, and the snx triple mutant shows the same tendency; however, in some cases, the effects were less pronounced. Thus, in the absence of SNX1, the accumulation of SNX2b in the cytoplasm may cause additional negative effects diminishing the performance of the plants under iron deficiency. In the triple mutant, these additional effects are not present due to the absence of SNX2b. Therefore, SNX2b needs to be loaded on the endosomal membrane for correct IRT1 regulation, suggesting that in the root hair cells the iron uptake efficiency depends on the formation of a complex containing at least SNX1 and SNX2b proteins. At present, we cannot exclude also an alternative hypothesis, where the lack of all three SNX proteins leads to activation of compensation mechanisms, suppressing part of the phenotype severity. The fact that SNX2a probably does not participate in this process is not surprising. Despite the high amino acid identity between SNX2a and SNX2b, a functional diversification between them has already been shown with regard to the 12S globulin maturation during seed development (Pourcher et al., 2010).
SNX Proteins and Iron Deficiency Signaling
In snx mutants grown under iron deficiency, we observed overall stronger upregulation of iron-related gene expression. This is consistent with the reported phenotypes and suggests the action of a feedback signaling mechanism in order to compensate for the enhanced deficiency. Unexpectedly, BHLH101 and FIT expression levels remained similar to the wild type, suggesting that not FIT but specifically some of its partners are targets of this extra signaling event. While it is not yet clear what may be the biological reason for the difference between BHLH039 (upregulated) and BHLH101 (unchanged), these two have been recently shown to have higher importance for iron deficiency responses than BHLH038 and BHLH100 (Wang et al., 2013). However, it has to be noted that among the tested transcriptional regulator genes BHLH101 shows the weakest expression and upregulating it under conditions of more pronounced iron deficiency might not be a priority.
An interesting result, which offers many possibilities for further research, is the change in the expression pattern of the IRT1 promoter in the snx1-2 mutant. The RT-PCR results show that the net outcome of this reprogramming is an increase of the total IRT1 mRNA levels. This suggests that under stress, iron deficiency response becomes centralized and induced only in localized zones, where a certain combination of factors might favor an efficient iron uptake. In this respect, further analysis of gene expression needs to take into consideration the topology of expression of genes involved in iron uptake responses.
Our analysis reveals the participation of the intracellular trafficking machinery in the regulation of the iron acquisition from the rhizosphere. SNX family proteins are responsible for the recycling and preventing the premature degradation of the iron transporter IRT1. In the absence of SNX1, the iron uptake efficiency of the root is compromised, resulting in the strong local upregulation of the IRT1 promoter through a feedback signaling mechanism acting specifically via subgroup Ib bHLH transcriptional regulators.
METHODS
Plant Lines
Arabidopsis thaliana snx1-1, snx1-2, and snx1-2 snx2a-2 snx2b-1 (snx triple), SNX1pro:SNX1-GFP in snx1-1, SNX2apro:SNX2a-GUS in snx2a-1, and SNX2bpro:SNX2b-GUS in snx2b-1 are a gift from Thierry Gaude (Jaillais et al., 2006; Pourcher et al., 2010); irt1-1 and IRT1pro:GUS are a gift from Catherine Curie (Vert et al., 2002). The FITox line was described previously (Jakoby et al., 2004). The snx1-2 allele was introduced into the IRT1pro:GUS line by crossing. Homozygous snx1-2 lines were selected by genotyping PCR (Supplemental Figure 4).
Molecular Cloning and Plant Transformation
An IRT1 fragment was amplified from genomic DNA using the primers I1B1 and FLI1B2, an SNX1 fragment was amplified from cDNA using primers SNX1B1 and SNX1nsB2, and an SKD1 fragment was amplified from cDNA using primers AtVPS4B1 and AtVPS4B2 (see Supplemental Table 3 for primer sequences and Arabidopsis gene identification numbers). All fragments were introduced into pDONR207 (Invitrogen) by recombination. RFP-ARA7 and TLG2a pDONR207 clones were provided by Thierry Gaude. For generating GFP or mCherry fusions, a second recombination step using either pMDC83 (GFP) (Curtis and Grossniklaus, 2003) or pJNC1 (a derivative of pMDC83, where the mCherry gene is inserted in the AscI site) were used.
Transformation of Nicotiana benthamiana leaf epidermis cells was performed as described previously (Hötzer et al., 2012).
Plant Growth and Treatments
Seeds were surface sterilized, germinated, and grown upright on Hoagland medium agar plates supplemented with 50 μM ferric iron in the form of FeNaEDTA (Jakoby et al., 2004). After 14 d, seedlings were transferred to new Hoagland medium plates containing either 50 μM ferric iron (50 μM Fe) or no iron (0 μM Fe). Two days later, some plants were transferred from 0 μM Fe back to 50 μM Fe (resupply). After two more days, the roots from all three conditions were harvested for further analysis. This growth regime is referred to as the 2-week growth system in the article. Alternatively, following sterilization, seeds were directly germinated and grown on 50 μM Fe or 0 μM Fe Hoagland plates for the indicated time periods. For IRT1 protein stability, plants were grown for 2 weeks under sufficient iron supply followed by 4 d under iron deficiency. Roots were then placed for 3 h in liquid iron-deficient Hoagland medium in the presence of 30 μM CHX or in identical medium containing only the solvent DMSO, instead of CHX, as a mock treatment.
Total Chlorophyll Content Determination
Approximately 500 mg of plant green tissues was ground, and pigments were extracted in 80% aqueous acetone. Tissue remains were removed by centrifugation and were then dried and used for dry weight determination. For the extracts, absorption was measured at 646, 663, and 750nm. Calculations were made according to Arnon (1949).
Iron Content Determination
Ten milligrams of dried plant material of 2-week-old plants grown on Hoagland agar medium, containing 50 or 0 µM Fe as described above, was used for direct solid sampling graphite furnace absorption spectrometry (GF AAS 6; Analytik Jena) for determination of Fe. Reference standards were used for quantification. Each biological sample was measured five times, and a mean value was calculated.
Root Length Calculations
Images of upright-grown plants on plates were taken and root lengths were measured using the JMicroVision software, version 1.2.7 (http://www.jmicrovision.com). Spatial calibration was first performed for converting pixel values into centimeters. The 1D measurements were made of freehand lines drawn upon the root image. The line lengths were used for calculating average and sd, as well as making the comparisons.
Histochemical GUS Activity Detection
GUS activity was visualized by the incubation of GUS-expressing plants in staining solution (100 mM Na2HPO4, 100 mM NaH2PO4, 2 mM K4[Fe(CN)6], 2 mM K3[Fe(CN)6], 0.2 Triton X-100, and 2 mM 5-bromo-4-chloro-3-indolyl glucuronide, pH 7.0). After a 1-h incubation at 37°C, the reaction was stopped by fixing the samples in 70% ethanol. Samples were observed with a Keyence BZ 9000 microscope. Images were taken at ×20 magnification and assembled using the Keyence BZ Analyzer software.
Confocal Microscopy
An LSM 510 Meta (Zeiss) confocal microscope was employed for the study. For GFP/AlexaFluor 488, excitation at 488 nm and detection between 500 and 530 nm were used. For mRFP/mCherry, excitation at 563 nm and detection 560 to 615 nm were used. Pinholes for both channels were set at 1 Airy Unit. Images were recorded in either 1024 × 1024 or 512 × 512 pixel format, resulting in pixel sizes (xy resolution) between 120 and 200 nm. Colocalization analysis was performed on 8-bit gray-scale image pairs, representing the two channels. Images were loaded in ImageJ software (http://rsb.info.nih.gov/ij) and analyzed using the JACoP v2.0 plug-in (Bolte and Cordelières, 2006). Threshold values were manually adjusted for each image and fell in the range of 21 to 40. Object-based analysis was performed, and the colocalization was calculated based on the distance between the geometrical centers of signals with size between 10 and 5000 pixels. Reference distance was automatically calculated for each comparison and equaled the xy resolution of the image, varying between 120 and 180 nm. In Supplemental Figure 5K, colocalization is presented for both channels. It represents the percentage of the detected objects in one channel colocalizing with objects from the other channel.
Histochemical Detection of Iron (Perls Stain)
Whole seedlings were fixed in methanol/chloroform/glacial acetic acid (6:3:1) solution for 1 h and washed three times, 5 min each, in distilled water. Samples were then incubated with Perls stain solution (2% K4[Fe(CN)6] and 2% HCl) for 30 min and washed for three more rounds in distilled water. The 3,3′-diaminobenzidine tetrahydrochloride intensification was then applied: 1-h incubation in solution containing 0.01 M NaN3 and 0.3% H2O2, followed by three washes in phosphate buffer (100 mM Na2HPO4 and 100 mM NaH2PO4, pH 7.0) and a 5- to 15-min incubation in staining solution (100 mM Na2HPO4, 100 mM NaH2PO4, 0.025% 3,3′-diaminobenzidine tetrahydrochloride [Sigma-Aldrich]), 0.005% H2O2, and 0.005% CoCl2, pH 7.0). Samples were observed under Keyence BZ 9000 microscope. Images were taken at ×20 magnification and assembled using the Keyence BZ Analyzer software.
Gene Expression Analysis by Quantitative RT-PCR
Total RNA was isolated from roots using the Spectrum Plant Total RNA kit (Sigma-Aldrich). Reverse transcription with oligo(dT) primer was performed using the RevertAid first-strand cDNA synthesis kit (Fermentas). Quantitative RT-PCR was performed on an iCycler/MyIQ single-color real-time PCR detection system (Bio-Rad), and the results were processed with the Bio-Rad iQ5-Standard Edition software (version 2.0) as described previously (Wang et al., 2007; Klatte and Bauer, 2009). Normalized gene expression was calculated by mass standard curve analysis, normalized to elongation factor EF1Bα signals. Primer pairs used for this study are listed in Supplemental Table 3. The experiment was performed in three biological repetitions with two technical replicates each. For statistical analysis, a t test was performed.
Generation of Gene Coexpression Networks
SNX1 (At5g06140) and SNX2a (At5g58440) were each used as a query to generate a coexpression network using the ATTED-II tool (Obayashi et al., 2009). The output was processed as previously described (Ivanov et al., 2012a) to generate image representations.
SDS-PAGE and Immunoblotting
Total protein from plant root tissues was extracted with SDG buffer (62 mM Tris-HCl, pH 8.6, 2.5% SDS, 2% DTT, and 10% glycerol). Protein amount was measured using the 2-D Quant Kit (GE Healthcare) according to the manufacturer’s instructions. Samples containing 5 µg protein were separated on 12% (w/v) SDS-polyacrylamide gel. Electrophoresis was performed in Tris-Gly buffer (25 mM Tris, 192 mM Gly, and 0.1% SDS, pH 8.3) using standard mini gel equipment (Bio-Rad). After electrophoresis, the proteins were transferred in blotting buffer (25 mM Tris, 192 mM Gly, and 20% ethanol, pH 8.3) to a Protran nitrocellulose membrane (Schleier and Schuell). The nitrocellulose filter was blocked by a 1-h incubation in 5% milk solution (Roth), dissolved in TBST (20 mM Tris-HCl, pH 7.4, 180 mM NaCl, and 0.1% Tween 20), to prevent nonspecific binding of the antibody, followed by a 1-h incubation in a dilution of the primary antibody in TBST containing 2.5% milk. After three washes in TBST, 10 min each, the membrane was incubated in a dilution of the secondary antibody in TBST containing 2.5% milk. The membrane was washed four times in TBST, 10 min each, before detection using the ECL system (GE Healthcare).
Antibody dilutions were as follows: rabbit anti-GFP (11814460001; Roche) 1:1000; rabbit anti-IRT1 (AS11 1780; Agrisera) 1:5000; rabbit anti-BiP (AS09 481; Agrisera) 1:2500; rabbit anti-PM (H+)-ATPase (AS07 260; Agrisera) 1:1000; goat anti-mouse IgG horseradish peroxidase (W4011; Promega) 1:5000; goat anti-rabbit IgG horseradish peroxidase (AS09 602; Agrisera) 1:5000.
Quantitation of Image Signal Intensity
Color images were converted to 8-bit grayscale prior to the analysis and subsequently loaded to ImageJ (http://rsb.info.nih.gov/ij). The evaluation of immunoblots was performed as described previously (Ivanov et al., 2012b). For the quantification of signal intensity of confocal microscopy images, sequential manual selection of zones was performed as shown in Supplemental Figure 6. The total signal intensity values were recorded for each zone, and the percentage of endosomal and PM signal was calculated in relation to the total signal intensity for every cell.
Immunolocalization
Roots from 2-week-old plants grown for 4 d under iron deficiency were used. Material was embedded in 3% agarose before 50-µm cross sections were made using a Leica VT-1000P vibratome. Immunolocalizations were performed as described previously (Ivanov and Gaude, 2009). Rabbit anti-IRT1 (AS11 1780; Agrisera) was used as a primary antibody in a 1:200 dilution and Alexa488 goat anti-rabbit IgG (A11034; Invitrogen) as a secondary in a 1:500 dilution.
PM Isolation
PMs were purified from iron-deficient root extracts by aqueous two-phase partitioning as described by Alexandersson et al. (2008). The starting tissue quantity was 400 mg. High-speed centrifugation was performed in a Beckman L8M ultracentrifuge with an SW40Ti swinging rotor. After the last centrifugation, membranes were dissolved in 20 μL SDG buffer (see SDS-PAGE and immunoblotting section above). The samples loaded on the protein gel represented 10% of the total protein fraction, 50% of the PM-depleted fraction, and 50% of the PM-enriched fraction. The signal in the PM-enriched fraction was divided by the sum of the signals in the PM-depleted and PM-enriched fractions and multiplied by 100.
Statistical Analysis
Student’s unpaired t test was used to calculate statistical significance of observed differences. P values of 0.05 or less were considered as statistically significant.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: FIT, At2g28160; FRO2, At1g01580; IRT1, At4g19690; BHLH038, At3g56970; BHLH039, At3g56980; BHLH100, At2g41240; BHLH101, At5g04150; SNX1, At5g06140; SNX2a, At5g58440; SNX2b, At5g07120, ARA7 At4g19640; SKD1, At2g27600; and TLG2a, At5g26980.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Coexpression Networks for SNX1 and SNX2a.
Supplemental Figure 2. Expression Pattern of SNX in the Root.
Supplemental Figure 3. SNX1 Expression and Protein Abundance under Iron Deficiency.
Supplemental Figure 4. Verification of the IRT1pro:GUS/snx1-2 Line.
Supplemental Figure 5. IRT1 Colocalization Analysis.
Supplemental Figure 6. Quantification of IRT1 Localization in Root Epidermis Cells.
Supplemental Table 1. Genes Coexpressed with SNX1.
Supplemental Table 2. Genes Coexpressed with SNX2a.
Supplemental Table 3. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Jonas Chodorsky for help in preparing pJNC1, Angelika Anna for technical assistance, and all the members of the Plant Biology team for discussions. This research was supported by the Saarland University and Heinrich-Heine University.
AUTHOR CONTRIBUTIONS
R.I. designed the research. R.I., T.B., A.B., C.F.-S., and A.-M.J. performed research. R.I., T.B., and P.B. analyzed data. R.I. wrote the article.
Glossary
- PM
plasma membrane
- bHLH
basic helix-loop-helix
- Col-0
Columbia-0
- TGN
trans-Golgi network
- CHX
cycloheximide
Footnotes
Online version contains Web-only data.
References
- Alexandersson E., Gustavsson N., Bernfur K., Karlsson A., Kjellbom P., Larsson C. (2008). Purification and proteomic analysis of plant plasma membranes. Methods Mol. Biol. 432: 161–173. [DOI] [PubMed] [Google Scholar]
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L.M., Catching A., Kirik V., Marc J., Overall R., Wasteneys G.O. (2013). CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 24: 649–659. [DOI] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224: 213–232. [DOI] [PubMed] [Google Scholar]
- Brumbarova T., Bauer P. (2005). Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol. 137: 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T., Matros A., Mock H.P., Bauer P. (2008). A proteomic study showing differential regulation of stress, redox regulation and peroxidase proteins by iron supply and the transcription factor FER. Plant J. 54: 321–334. [DOI] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly E.L., Campbell N.H., Grotz N., Prichard C.L., Guerinot M.L. (2003). Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 133: 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly E.L., Fett J.P., Guerinot M.L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., Korswagen H.C. (2012). Sorting nexins provide diversity for retromer-dependent trafficking events. Nat. Cell Biol. 14: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Donnini S., Prinsi B., Negri A.S., Vigani G., Espen L., Zocchi G. (2010). Proteomic characterization of iron deficiency responses in Cucumis sativus L. roots. BMC Plant Biol. 10: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett T.P., Connolly E.L., Rogers E.E. (2006). Arabidopsis cpFtsY mutants exhibit pleiotropic defects including an inability to increase iron deficiency-inducible root Fe(III) chelate reductase activity. Plant J. 47: 467–479. [DOI] [PubMed] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93: 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang A.T., Guo Y., Cuin T.A., Qiu Q., Song C., Kristiansen K.A., Bych K., Schulz A., Shabala S., Schumaker K.S., Palmgren M.G., Zhu J.K. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Robertson F.C., Soares D.C., Ingram G.C. (2005). ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17: 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Burch H.L., Nelson R.B., Barrett-Wilt G., Kline K.G., Mohsin S.B., Young J.C., Otegui M.S., Sussman M.R. (2010). Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285: 17918–17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Henriques R., Jásik J., Klein M., Martinoia E., Feller U., Schell J., Pais M.S., Koncz C. (2002). Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol. Biol. 50: 587–597. [DOI] [PubMed] [Google Scholar]
- Hötzer B., Ivanov R., Brumbarova T., Bauer P., Jung G. (2012). Visualization of Cu2+ uptake and release in plant cells by fluorescence lifetime imaging microscopy. FEBS J. 279: 410–419. [DOI] [PubMed] [Google Scholar]
- Ivanov R., Gaude T. (2009). Endocytosis and endosomal regulation of the S-receptor kinase during the self-incompatibility response in Brassica oleracea. Plant Cell 21: 2107–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R., Brumbarova T., Bauer P. (2012a). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5: 27–42. [DOI] [PubMed] [Google Scholar]
- Ivanov R., Tiedemann J., Czihal A., Baumlein H. (2012b). Transcriptional regulator AtET2 is required for the induction of dormancy during late seed development. J. Plant Physiol. 169: 501–508. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Gaude T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 53: 237–247. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109. [DOI] [PubMed] [Google Scholar]
- Jakoby M., Wang H.Y., Reidt W., Weisshaar B., Bauer P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577: 528–534. [DOI] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeb L., Mukherjee I., Chatterjee I., Lahner B., Salt D.E., Connolly E.L. (2008). Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol. 146: 1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte M., Bauer P. (2009). Accurate real-time reverse transcription quantitative PCR. Methods Mol. Biol. 479: 61–77. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W., Wen T.N., Shiau J.Y., Wu Y.C., Lin W., Schmidt W. (2011). iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol. 155: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemes S., Langhans M., Viotti C., Scheuring D., San Wan Yan M., Jiang L., Hillmer S., Robinson D.G., Pimpl P. (2010). Retromer recycles vacuolar sorting receptors from the trans-Golgi network. Plant J. 61: 107–121. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcher M., Santambrogio M., Thazar N., Thierry A.M., Fobis-Loisy I., Miège C., Jaillais Y., Gaude T. (2010). Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 22: 3980–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Alvarez R., Andaluz S., Rodríguez-Celma J., Wohlgemuth G., Zocchi G., Alvarez-Fernández A., Fiehn O., López-Millán A.F., Abadía J. (2010). Changes in the proteomic and metabolic profiles of Beta vulgaris root tips in response to iron deficiency and resupply. BMC Plant Biol. 10: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697. [DOI] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S., Schmidt W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183: 1072–1084. [DOI] [PubMed] [Google Scholar]
- Schuler M., Keller A., Backes C., Philippar K., Lenhof H.P., Bauer P. (2011). Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biol. 11: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla M., Briat J.F., Vert G., Curie C. (2008). Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J. 55: 289–300. [DOI] [PubMed] [Google Scholar]
- Shin L.J., Lo J.C., Chen G.H., Callis J., Fu H., Yeh K.C. (2013). IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25: 3039–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A.B., Hermand V., Curie C., Vert G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierhof Y.D., Viotti C., Scheuring D., Sturm S., Robinson D.G. (2013). Sorting nexins 1 and 2a locate mainly to the TGN. Protoplasma 250: 235–240. [DOI] [PubMed] [Google Scholar]
- Strochlic T.I., Setty T.G., Sitaram A., Burd C.G. (2007). Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M.R. (1994). Molecular analysis of proteins in the plant plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45: 211–234. [Google Scholar]
- Vanoosthuyse V., Tichtinsky G., Dumas C., Gaude T., Cock J.M. (2003). Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol. 133: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C., Maiwald D., Pesaresi P., Jahns P., Salamini F., Leister D. (2002). The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 31: 589–599. [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Klatte M., Jakoby M., Bäumlein H., Weisshaar B., Bauer P. (2007). Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908. [DOI] [PubMed] [Google Scholar]
- Wang N., Cui Y., Liu Y., Fan H., Du J., Huang Z., Yuan Y., Wu H., Ling H.Q. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6: 503–513. [DOI] [PubMed] [Google Scholar]
- Yi Y., Guerinot M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10: 835–844. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., Wang D., Ling H.Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18: 385–397. [DOI] [PubMed] [Google Scholar]
- Yuan Y.X., Zhang J., Wang D.W., Ling H.Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15: 613–621. [DOI] [PubMed] [Google Scholar]
- Zelazny, E., Ivanov, R., and Gaude, T. (2012). The plant SNX family and its Role in endocytosis. In Endocytosis in Plants, J. Šamaj, ed (Berlin, Heidelberg: Springer-Verlag), pp. 233–247. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.