Oomycete pathogens of plants secrete effectors to perturb host cell physiology. This study reveals that the P. infestans RXLR-type effector protein PexRD2 interferes with host MAPKKKε immunity-related signaling to the benefit of the pathogen.
Abstract
Mitogen-activated protein kinase cascades are key players in plant immune signaling pathways, transducing the perception of invading pathogens into effective defense responses. Plant pathogenic oomycetes, such as the Irish potato famine pathogen Phytophthora infestans, deliver RXLR effector proteins to plant cells to modulate host immune signaling and promote colonization. Our understanding of the molecular mechanisms by which these effectors act in plant cells is limited. Here, we report that the P. infestans RXLR effector PexRD2 interacts with the kinase domain of MAPKKKε, a positive regulator of cell death associated with plant immunity. Expression of PexRD2 or silencing MAPKKKε in Nicotiana benthamiana enhances susceptibility to P. infestans. We show that PexRD2 perturbs signaling pathways triggered by or dependent on MAPKKKε. By contrast, homologs of PexRD2 from P. infestans had reduced or no interaction with MAPKKKε and did not promote disease susceptibility. Structure-led mutagenesis identified PexRD2 variants that do not interact with MAPKKKε and fail to support enhanced pathogen growth or perturb MAPKKKε signaling pathways. Our findings provide evidence that P. infestans RXLR effector PexRD2 has evolved to interact with a specific host MAPKKK to perturb plant immunity–related signaling.
INTRODUCTION
Plant pathogenic oomycetes and fungi are among the most devastating eukaryotic parasites of socio-economically important food crop species. These filamentous pathogens penetrate plant tissue and extend hyphae into the spaces between cells in search of nutrients. As hyphae extend through plant tissue, they can form intimate contacts with host cells by developing highly specialized structures known as haustoria (Panstruga and Dodds, 2009; Dodds and Rathjen, 2010). These structures are a major interface for exchange of molecules between pathogen and host during infection, including the delivery of pathogen-derived molecules that support infection. These molecules, collectively known as effectors, are thought to be major determinants of pathogenicity, and understanding their molecular functions has become a major theme in the study of plant–microbe interactions. However, our understanding of how filamentous plant pathogen effectors modulate plant immunity is rudimentary.
Plants have developed sophisticated surveillance systems to respond to pathogens and mount defenses against attack (Jones and Dangl, 2006; Dodds and Rathjen, 2010). Defense responses can be initiated at the host cell surface by pattern recognition receptors (PRRs) that detect evolutionarily conserved molecular signatures of pathogens (pathogen-associated molecular patterns (PAMPs) and PRR-mediated immunity) or apoplastic effector proteins (Boller and Felix, 2009; Win et al., 2012b). In turn, pathogens deliver other effector proteins that function within host cells and interfere with PRR-mediated immunity to promote disease (effector-triggered susceptibility). A second layer of plant immunity monitors for the presence of these foreign translocated effector proteins, directly or indirectly (Chisholm et al., 2006; van der Hoorn and Kamoun, 2008; Elmore and Coaker, 2011; Win et al., 2012b). This layer of immunity is intracellular and orchestrated by NB-LRR proteins (plant resistance [R] proteins and NB-LRR–mediated immunity) and can result in localized cell death known as the hypersensitive response (HR). Pathogens also secrete effector proteins that interfere with NB-LRR–mediated immunity. Considerable overlap exists in the downstream signaling cascades activated by PRR- and NB-LRR–mediated immunity, including ion influx, mitogen-activated protein kinase (MAPK) pathways, and increased levels of defense-related hormones. These layers of immunity likely function together as a continuum (Thomma et al., 2011).
In plants, MAPK pathways are important for regulation of development, growth, and the integration of diverse environmental signals, including the response to pathogen attack (Boller and Felix, 2009; Pitzschke et al., 2009; Rodriguez et al., 2010; Meng and Zhang, 2013). MAPK cascades are key players in both PRR- and NB-LRR–mediated immunity (Martin et al., 2003; Boller and Felix, 2009; Segonzac et al., 2011). Phosphorylation of a Thr and a Tyr residue in a Thr-X-Tyr motif, present in the activation loop of MAPKs (Chang and Karin, 2001), regulates diverse plant defense responses, including transcriptional changes. MAPKs are phosphorylated on these residues by MAPK kinases (MAPKKs); in turn, these MAPKKs are regulated through phosphorylation by MAPKK kinases (MAPKKKs) (Chang and Karin, 2001). A major unanswered question is how perception of diverse stimuli signal through these convergent pathways while maintaining the specificity of the signal. MAPKKKs are often multidomain proteins, and one mechanism of achieving specificity may be through differential protein–protein interactions (Rodriguez et al., 2010). Only a limited number of MAPKKKs have been associated with plant immunity signaling. These include MEKK1 that is activated downstream of the PRR receptor for flagellin in Arabidopsis thaliana (Asai et al., 2002; Nicaise et al., 2009); NPK1, a MAPKKK required for triggering an HR response mediated by the N, Bs2, and Rx NB-LRRs (in response to recognition of the p50 protein from tobacco mosaic virus, AvrBs2, and the coat protein of potato virus X, respectively) in Nicotiana benthamiana (Jin et al., 2002); MAPKKKα, which is required for the Pto/Prf-mediated response following AvrPto recognition (del Pozo et al., 2004); and EDR1, a negative regulator of defense responses in Arabidopsis (Frye et al., 2001). Finally, MAPKKKε has been identified as important for resistance in tomato (Solanum lycopersicum) to certain phytopathogenic bacterial strains and for mediating responses downstream of effector/R protein combinations in N. benthamiana (Melech-Bonfil and Sessa, 2010).
Host MAPK signaling pathways are well established as targets of bacterial effector proteins. These effectors have been shown to interfere with signaling from PRR receptors (Göhre et al., 2008; Xiang et al., 2008; Gimenez-Ibanez et al., 2009; Zeng et al., 2012) and coreceptors such as BAK1 (Shan et al., 2008; Cheng et al., 2011). Some of these effectors can covalently modify components of PRR-mediated signaling (Zhang et al., 2010; Feng et al., 2012) and directly inhibit MAPKK and MAPK activity (Zhang et al., 2007, 2012; Wang et al., 2010). However, the extent to which filamentous pathogen effectors target host kinases remains unclear. The Phytophthora infestans CRN8 translocated effector is a functional RD kinase (van Damme et al., 2012), but whether this protein interacts with plant kinases is not known. The Phytophthora sojae effectors Avh238P7076 and Avh331 have been shown to interfere with events dependent on MAPK signaling in Arabidopsis and N. benthamiana, but no association with direct protein–protein interaction was made (Wang et al., 2011; Cheng et al., 2012). Interestingly, no MAPK signaling proteins were identified as effector targets in the plant-pathogen immune network, version 1 (PPIN-1) (Mukhtar et al., 2011).
The hemibiotrophic pathogen P. infestans causes late blight, a devastating disease of potato (Solanum tuberosum) and tomato (Haas et al., 2009). This pathogen is infamous for devastating European potato crops in the mid-nineteenth century and triggering the Irish potato famine. It still is a critical threat to global food security, causing recurrent epidemics in potato, the third most important food crop (Haas et al., 2009; Cooke et al., 2012; Yoshida et al., 2013). P. infestans is a model organism for the study of oomycete pathology (Oliva et al., 2010; Bozkurt et al., 2012) and can infect the model plant N. benthamiana (Chaparro-Garcia et al., 2011). To manipulate its hosts, P. infestans secretes effector proteins, some of which translocate into host cells (Birch et al., 2006; Kamoun, 2006; Whisson et al., 2007; Schornack et al., 2010). RXLR-type effectors are the largest class of host-translocated proteins in Phytophthora (Haas et al., 2009; Raffaele et al., 2010). The biochemical function of RXLR-type effectors has been localized to their C-terminal domains (Win et al., 2007). Although recent studies have identified the first virulence targets of these proteins in plant cells and provided insight into how they are recognized by NB-LRRs (Bos et al., 2010; Bozkurt et al., 2011; Dong et al., 2011; Saunders et al., 2012), functions for the vast majority of RXLR effectors are not yet known. Despite the diversity in protein sequence displayed within their C-terminal domains, structural studies of RXLR effectors have identified a conserved fold (the WY-domain), predicted to be adopted by many, but not all, of these proteins (Boutemy et al., 2011; Win et al., 2012a).
One P. infestans RXLR-type effector that adopts the WY-fold is PexRD2 (Boutemy et al., 2011). PexRD2 is a 121–amino acid protein comprising a signal peptide followed by an RXLR motif (residues 38 to 41, with a degenerate dEER motif spanning residues 48 to 56). The C-terminal effector domain comprises residues 57 to 121. Originally identified in P. infestans isolate 88069, PexRD2 is a member of Tribe 6, an 18-member RXLR effector family in P. infestans (Haas et al., 2009). Members of this family are predicted, at least in part, to adopt the same WY-domain fold, and some of this family can be modeled on the PexRD2 structure with high confidence scores using IntFOLD (Roche et al., 2011). As such, we predict that this family may have evolved as interaction modules, potentially targeting host proteins of similar structure and function. Typical of most RXLR-type effectors, PexRD2 does not share significant sequence similarity to any protein of known function, making prediction of biochemical activity a challenge. Expression of PexRD2 (lacking the signal peptide) in N. benthamiana triggers a weak and delayed dose-dependent cell death response (Oh et al., 2009); a nonspecific cell death response was also recorded on expression in several Solanum species (Vleeshouwers et al., 2008). The crystal structure of PexRD2 revealed a homodimeric state with intimate hydrophobic contacts at the interface (Boutemy et al., 2011). Self-association was confirmed in planta, suggesting the dimer is the unit of function.
In this study, we used a yeast two-hybrid (Y2H) screening approach to identify host targets of PexRD2. PexRD2 was shown to interact with MAPKKKε, a positive regulator of plant immunity–related cell death signaling pathways (Melech-Bonfil and Sessa, 2010). PexRD2 interacts with the kinase domain of MAPKKKε in yeast and in planta. Overexpression of PexRD2 and virus-induced gene silencing (VIGS) of MAPKKKε in N. benthamiana promoted growth of P. infestans. Expression of PexRD2 suppresses both MAPKKKε-triggered cell death and cell death elicited by combinations of effector/R protein pairs dependent on MAPKKKε. MAPKKKε-independent cell death, cell death triggered by MAPKKKα, and cell death triggered by a constitutively active mutant of MAPKK are not affected by expression of PexRD2. Structure-led mutagenesis of PexRD2 revealed the importance of the WY-domain fold and oligomerization of PexRD2 for MAPKKKε interaction and effector activities. Mutations that disrupt MAPKKKε interaction result in the loss of all PexRD2 phenotypes. Our results reveal a P. infestans RXLR-type effector protein that has evolved to interact with a host MAPKKK to perturb plant immunity–related signaling.
RESULTS
Expression of PexRD2 Promotes in Planta Growth of P. infestans
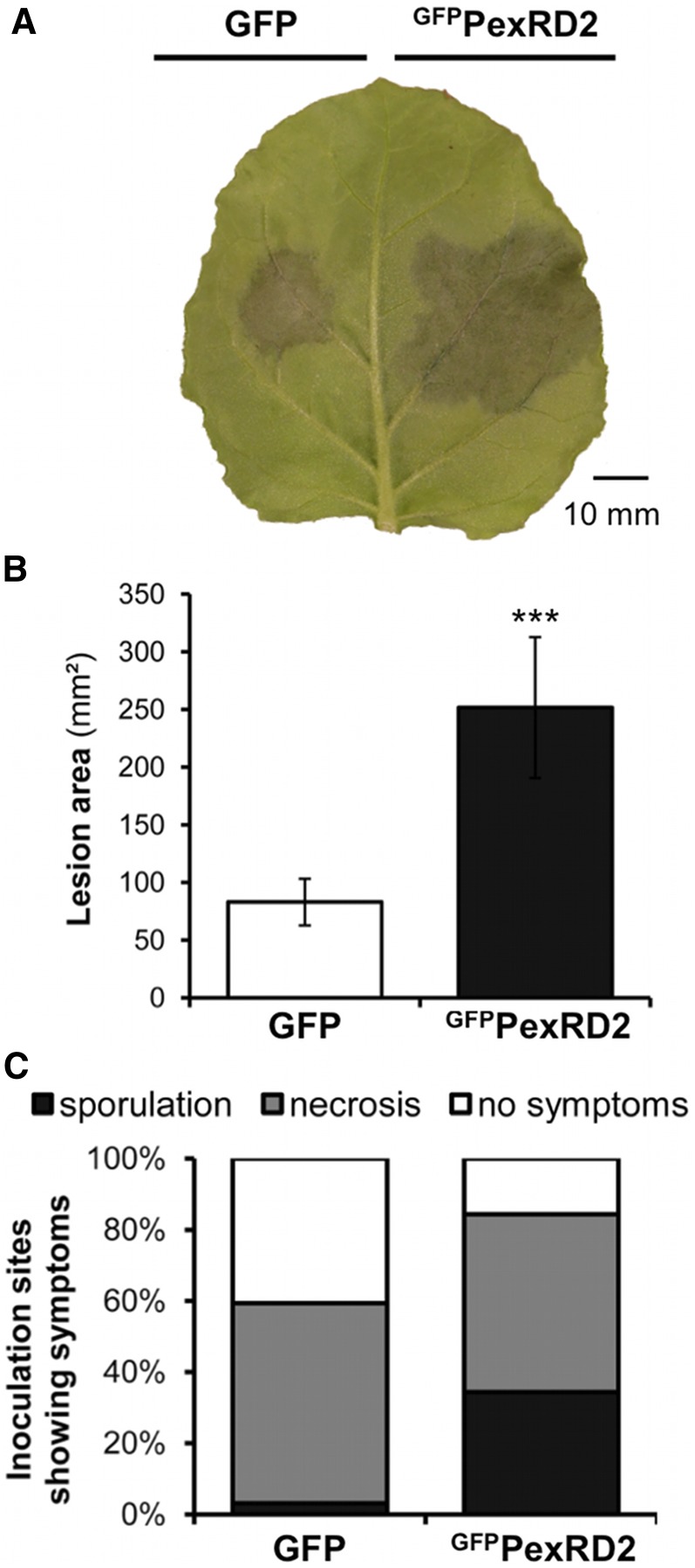
N. benthamiana is a host for P. infestans and a model for functional studies in the Solanaceae. As such, it has been extensively used to investigate host and pathogen gene functions in P. infestans–plant interactions (Whisson et al., 2007; Oh et al., 2009; Bos et al., 2010; Bozkurt et al., 2012; Saunders et al., 2012). To investigate whether PexRD2 could benefit P. infestans during infection, we used an in planta transient assay (McLellan et al., 2013). In this assay, GFPPexRD2 (GFPPexRD221-121, where a green fluorescent protein tag is substituted for the N-terminal signal peptide) was expressed in one half of N. benthamiana leaves via Agrobacterium tumefaciens–mediated transient transformation (agroinfiltration) 24 h prior to inoculation with P. infestans 88069 zoospores; the second half of the leaves expressed the empty vector control (GFP alone). P. infestans 88069 is a standard laboratory strain, particularly appropriate for studying infection of N. benthamiana (Chaparro-Garcia et al., 2011). Five days postinoculation, the mean lesion size and mean percentage of inoculation sites sporulating were recorded (Figure 1). The results clearly demonstrate that leaf areas expressing GFPPexRD2 support enhanced P. infestans growth compared with the control. This suggests that PexRD2 can act within plant cells to promote effector-triggered susceptibility, supporting P. infestans virulence.
Figure 1.
Expression of PexRD2 Promotes in Planta Growth and Sporulation of P. infestans.
(A) Transient expression of GFPPexRD2 in one half of a N. benthamiana leaf by agroinfiltration, followed by inoculation with P. infestans 88069 zoospores, enhances the growth of the pathogen, compared with expression of GFP in the second half. Image taken at 5 d after zoospore inoculation.
(B) Lesion area achieved at 5 d after zoospore inoculation following infection of GFP- or GFPPexRD2-expressing leaf tissue with P. infestans 88069. Results are the mean ± se of infections from two independent experiments using at least eight leaves each. The asterisk indicates a value significantly different from the GFP control (P < 0.001) as determined by t test.
(C) Percentage of total inoculation sites showing necrosis and/or sporulation for the same infections.
PexRD2 Interacts with the Kinase Domain of MAPKKKε
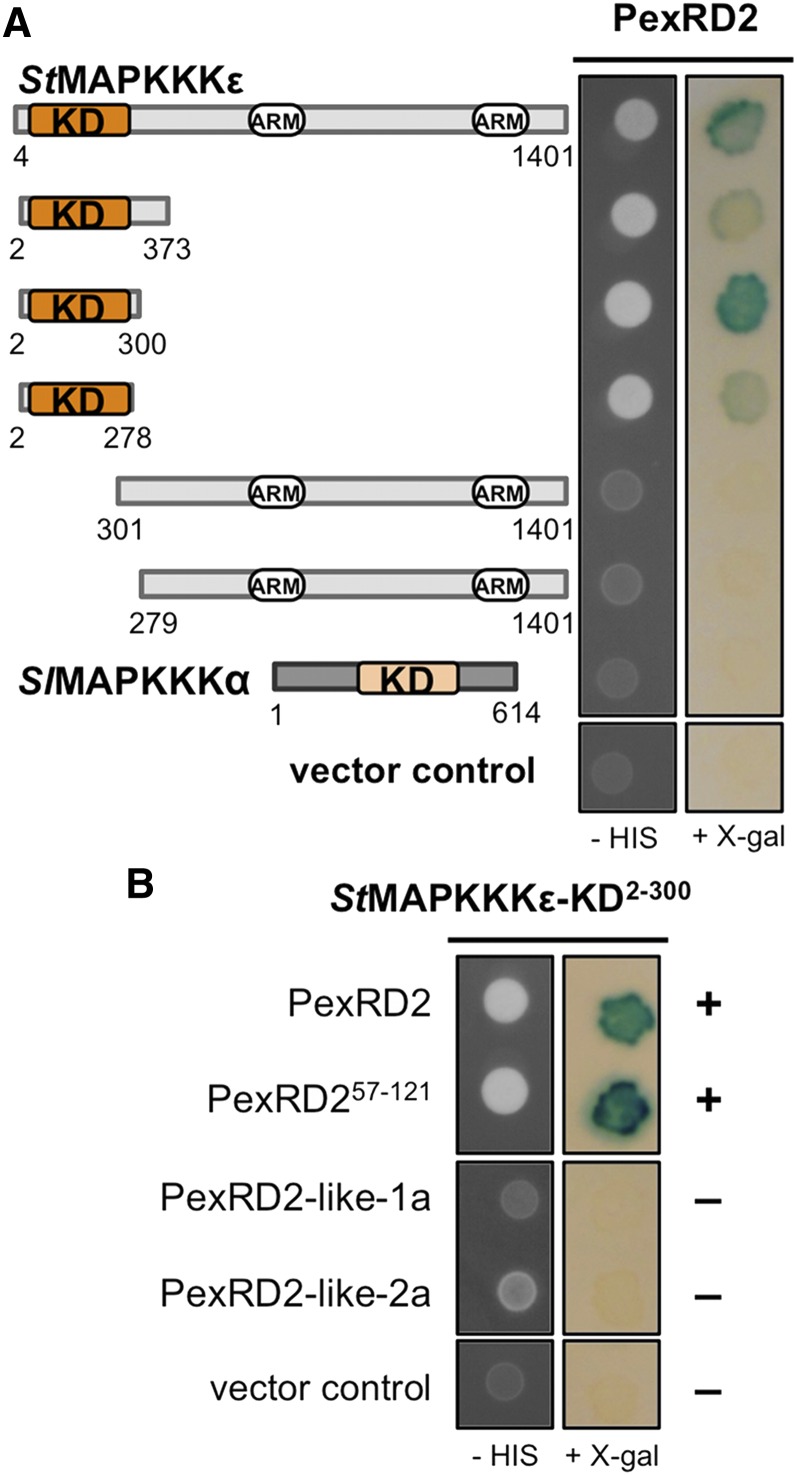
To identify putative host targets of PexRD2, we performed a Y2H screen, using a previously described prey library derived from infected potato tissue (Bos et al., 2010), with PexRD221-121 (henceforth PexRD2) as bait. The screen involved 5.9 × 106 yeast cotransformants. Three transformants were selected from the screen, and each comprised an essentially full-length copy of potato MAPKKKε (St-MAPKKKε residues 4 to 1401) (Figure 2A). St-MAPKKKε is a 154-kD, 1401–amino acid protein with a kinase domain at the N terminus (residues 20 to 274) and two ARM repeat regions (residues 694 to 738 and 1135 to 1220, domains defined by SMART; Letunic et al., 2012). The tomato MAPKKKε homolog (Sl-MAPKKKε, 98% amino acid sequence identity to St-MAPKKKε, 100% in the kinase domain) is a positive regulator of plant immunity signaling pathways (Melech-Bonfil and Sessa, 2010). MAPKKKε homologs can be identified in a range of dicot plants, including two in N. benthamiana (Nb-MAPKKKε1 and Nb-MAPKKKε2, 90 and 93% identical to St-MAPKKKε, respectively; 99% identical in the kinase domain) (Melech-Bonfil and Sessa, 2010; Hashimoto et al., 2012). The interaction between PexRD2 and St-MAPKKKε in yeast was confirmed by retransformation alongside relevant controls.
Figure 2.
PexRD2 Interacts with the Kinase Domain of MAPKKKε.
(A) Y2H assay showing that PexRD2 interacts with the kinase domain (KD) of St-MAPKKKε, as evidenced by growth on selective media lacking His (-HIS) and the development of blue coloration in the presence of X-Gal. No interaction was detected between PexRD2 and a second MAPKKK, Sl-MAPKKKα.
(B) The N-terminal region of PexRD2 (residues 21 to 56, containing the translocation motifs) is not required for the interaction with StMAPKKKε-KD. Furthermore, the two PexRD2-like proteins, PexRD2-like-1a and PexRD2-like-2a, do not interact with StMAPKKKKε-KD. The symbols + and – denote interaction and absence of interaction, respectively.
To map the region of St-MAPKKKε that interacts with PexRD2, we generated a series of truncations that were tested for interaction with the effector by Y2H. Only the constructs expressing an intact kinase domain enabled growth of yeast on selective plates and the development of blue coloration in the presence of X-Gal (Figure 2A). These results show that the kinase domain of St-MAPKKKε (StMAPKKKε-KD) is both necessary and sufficient for the interaction with PexRD2. As a negative control, we used a second related MAPKKK, Sl-MAPKKKα (del Pozo et al., 2004) (which shares 42% sequence identity with St-MAPKKKε in the kinase domain). No interaction with this kinase was detected (Figure 2A). We also showed that only the WY-domain region of PexRD2 (PexRD257-121) is required for the interaction; the RXLR translocation sequence is dispensable (Figure 2B). This validates the use of the PexRD2 structure (which comprised the effector domain only) as a tool for understanding the interaction between this effector and host proteins.
Members of the PexRD2-Like Family Do Not Interact with StMAPKKKε-KD in Yeast
PexRD2 is a member of a family of 18 RXLR-type effector proteins in P. infestans (Haas et al., 2009). To test whether other members of this family might interact with MAPKKKε, we cloned PexRD2-like-1a and PexRD2-like-2a (PITG_14984 and PITG_14787, 47 and 29% sequence identity, respectively, with PexRD2 in the effector domain) into the Y2H bait vector and assayed for interaction with StMAPKKKε-KD. We found no evidence of interaction (Figure 2B). Immunoblotting confirmed the two PexRD2-like baits were expressed and stable (Supplemental Figure 1).
PexRD2 and St-MAPKKKε Localize to, and Interact, in the Plant Cell Cytoplasm
Confocal microscopy with fluorescent-labeled proteins showed PexRD2 locates to the plant cell cytoplasm when expressed on its own, together with St-MAPKKKε, or in the presence of P. infestans infection (Supplemental Methods and Supplemental Figure 2). St-MAPKKKε is also located to the plant cell cytoplasm.
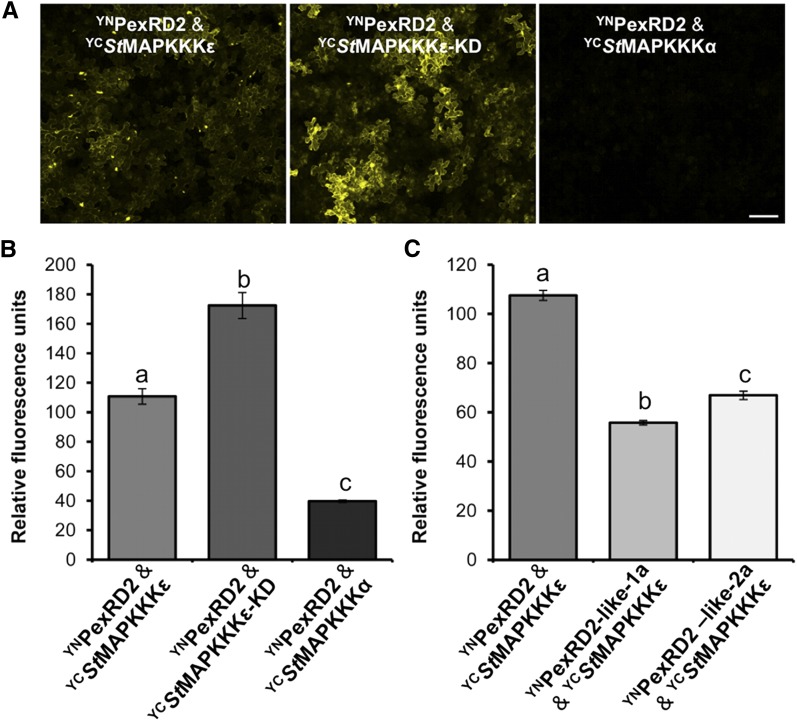
We used a bimolecular fluorescence complementation (BiFC) approach to examine whether interaction between PexRD2 and St-MAPKKKε occurs in the cytoplasm. An N-terminal fragment of YFP (YN) was fused to PexRD2 (YNPexRD2) and coexpressed, in turn, with a C-terminal fragment of YFP (YC) fused to St-MAPKKKε, StMAPKKKε-KD, or Sl-MAPKKKα. Cytoplasmic fluorescence, indicative of YFP reconstitution, was observed with YCStMAPKKKε and YCStMAPKKKε-KD, but not with YCSlMAPKKKα (Figures 3A and 3B). Levels of fluorescence were quantified in leaf disks and compared. Significantly increased fluorescence (one way ANOVA) was observed when coexpressing YCStMAPKKKε or YCStMAPKKKε-KD with YNPexRD2, when compared with YCSlMAPKKKα (the negative control; Figure 3B).
Figure 3.
PexRD2 and MAPKKKε Interact in the Plant Cell Cytoplasm.
(A) BiFC of YNPexRD2 with YCStMAPKKKε (left), YCStMAPKKKε-KD (middle), and YCSlMAPKKKα (right). Bar = 100 µm.
(B) Fluorimeter measurements of BiFC shown in (A), each from 36 independent samples.
(C) Fluorimeter measurements of BiFC between YN and YC fusion proteins as indicated, each from 36 independent samples. Results in (B) and (C) are the mean ± se, with the letters indicating values significantly different to each other (P < 0.001) as determined by one-way ANOVA.
As these results mirrored interactions observed using Y2H, we extended the analysis to PexRD2-like proteins. Consistent with the Y2H results, significantly less fluorescence (one way ANOVA) was detected following coexpression of YCStMAPKKKε with YNPexRD2-like-1a or YNPexRD2-like-2a (Figure 3C). Immunoblots of each construct used for BiFC show that fusion proteins are stable in planta (Supplemental Figure 3).
Variants of PexRD2 Perturb Interactions with StMAPKKKε-KD in Yeast and Disrupt Effector Oligomerization in Planta
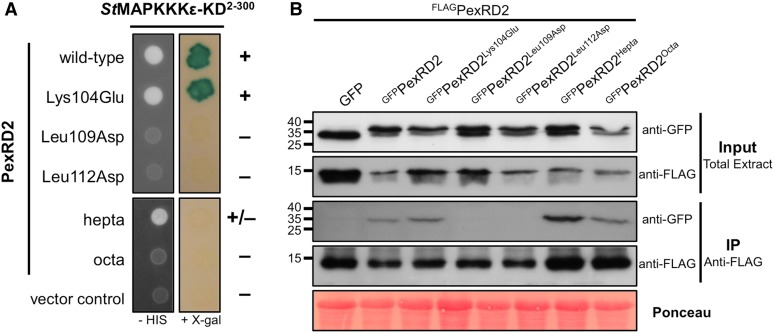
Following the in planta confirmation of interaction between PexRD2 and St-MAPKKKε, we exploited the structure of PexRD2 to design point mutations that might disrupt this interaction (Supplemental Figure 4A) and tested these using the Y2H assay. Two mutations, Leu109Asp and Leu112Asp, were designed to introduce charged residues into the hydrophobic dimerization interface of PexRD2. An additional 10 mutations, Glu61Ala, Asp74Ala, Asp75Ala, Lys79Glu, Lys81Glu, Lys85Glu, Glu97Gln, Glu101Gln, Lys104Glu, and Lys107Glu, were introduced to charged surface residues and were designed to minimize any impact on the WY-domain fold. Only Leu109Asp and Leu112Asp prevented the interaction with StMAPKKKε-KD in yeast (Figure 4A; Supplemental Figure 4B). Immunoblots confirmed the expression of intact fusion proteins for both noninteracting mutants in yeast (Supplemental Figure 4C). Using coimmunoprecipitation from plant tissue, we confirmed that, as expected, the Leu109Asp and Leu112Asp mutations perturb PexRD2 oligomerization in planta (Figure 4B).
Figure 4.
Specific Mutants of PexRD2 Show Perturbation in Interaction with MAPKKKε and in Oligomerization.
(A) In Y2H, point mutations in two Leu residues within PexRD2’s dimerization interface (Leu109Asp and Leu112Asp), but not the surface presented Lys (Lys104Glu), abolishes the interaction with StMAPKKKε-KD. Point mutations at nine other individual surface exposed residues also had no effect on the interaction (Supplemental Figure 4). Mutation of seven surface presented residues within a variable region of the WY-domain fold (PexRD2hepta) weakens the interaction with StMAPKKKε-KD. The mutation of an additional surface residue (Ala90Glu) within this region (PexRD2octa) leads to a loss of interaction with StMAPKKKε-KD. The symbols + and – denote interaction and absence of interaction, respectively.
(B) In coimmunoprecipitation (IP) from plant tissue, the GFPPexRD2Leu109Asp and GFPPexRD2Leu112Asp variants do not interact with FLAGPexRD2, suggesting they prevent dimerization. All other GFP-tagged variants tested, that targeted surface-presented residues, retain interaction with FLAGPexRD2.
We then used the sequence variation between PexRD2 and PexRD2-like-1a, and the structure of PexRD2, to generate mutants within a region of the WY-fold previously shown to accommodate different structures (Boutemy et al., 2011). One of these variants comprised mutations in seven surface-presented residues (Supplemental Figures 5A and 5B). This mutant, PexRD2hepta, still supported very weak yeast growth on selective media but did not promote development of blue coloration in the presence of X-Gal (Figure 4A). A second mutant, PexRD2octa, that comprised the PexRD2hepta background with an additional surface-presented mutation (Ala90Glu), did not interact in the Y2H assays (Figure 4A). These constructs expressed stably in yeast (Supplemental Figure 5C). PexRD2hepta and PexRD2octa were still able to interact with wild-type PexRD2 in planta, as observed by coimmunoprecipitation (Figure 4B), suggesting these variants retain the ability to oligomerize.
Noninteracting Mutants of PexRD2 Fail to Enhance Pathogen Growth
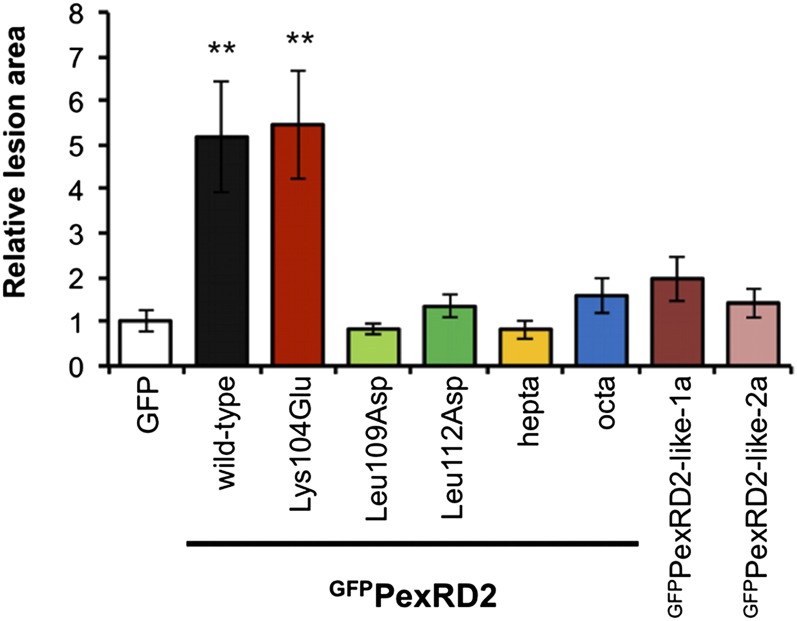
To link the observed interaction of PexRD2 and MAPKKKε with any putative virulence activity, we tested whether PexRD2-like-1a, PexRD2-like-2a, or any of the PexRD2 variants that did not interact with StMAPKKKε-KD, enhanced the growth of P. infestans 88069 on N. benthamiana. PexRD2Lys104Glu, which still interacts with StMAPKKKε-KD, was included as a positive control. All of the noninteracting variants failed to significantly enhance pathogen growth, as measured by the total lesion area relative to the GFP control on the same leaf (Figure 5). By contrast, GFPPexRD2Lys104Glu enhanced pathogen growth to the same level as the wild type. This implicates the interaction between PexRD2 and MAPKKKε as critical to this effector’s virulence activity. All fusion proteins are expressed in planta (Supplemental Figure 6).
Figure 5.
PexRD2 Variants Showing Perturbed Interactions with MAPKKKε, or PexRD2-Like Effectors, Fail to Enhance Pathogen Growth.
Transient expression of wild-type GFPPexRD2 in one half of a N. benthamiana leaf by agroinfiltration, followed by inoculation with P. infestans 88069 zoospores, enhances the lesion area achieved at 5 d after zoospore inoculation, relative to the GFP control. A similar level of enhancement was observed for PexRD2Lys104Glu. No significant enhancement of pathogen growth was observed for the four variants of PexRD2 that show perturbed interactions with MAPKKKε (Leu109Asp, Leu112Asp, PexRD2hepta, or PexRD2octa) or the two PexRD2-like effectors. Results are the mean ± se of infections from two independent experiments using at least eight leaves each time. The asterisks indicate values significantly different from the GFP control (P < 0.01) as determined by one-way ANOVA; all other values were not significantly different (P > 0.05).
Silencing of Nb-MAPKKKε Enhances Pathogen Growth
Having shown that expression of PexRD2 in N. benthamiana enhances pathogen growth, and this effector interacts with MAPKKKε homologs, we used VIGS of Nb-MAPKKKε to determine whether the product of this gene has a role in restricting growth of P. infestans during infection. We used two VIGS constructs, TRV:5′-MAPKKKε (Melech-Bonfil and Sessa, 2010) and TRV:3′-NbMAPKKKε (a construct based on TRV:3′-MAPKKKε; Melech-Bonfil and Sessa, 2010), alongside a TRV:GFP control. We observed an ∼70% reduction in endogenous transcripts of Nb-MAPKKKε in VIGS plants and confirmed that levels of Nb-MAPKKKα were not significantly altered (Supplemental Figure 7). To further assess the potential for off-target silencing, we examined the draft genome of N. benthamiana (see Supplemental Methods for further details) (Bombarely et al., 2012). As observed by Melech-Bonfil and Sessa (2010), Nb-MAPKKKε silencing resulted in a moderate retardation of plant growth.
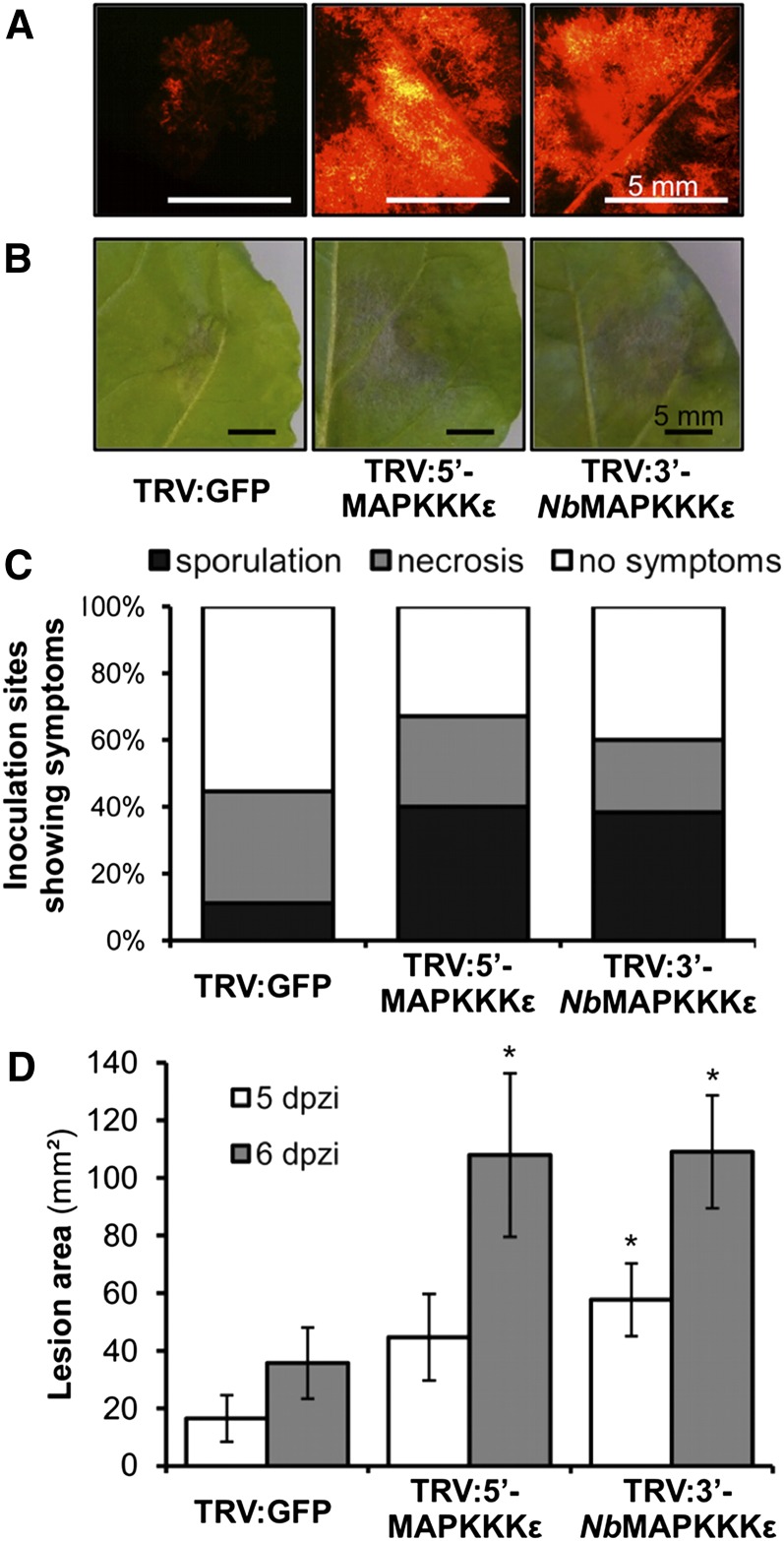
Leaves detached from plants 14 d postinoculation with TRV constructs were inoculated with P. infestans 88069td (P. infestans 88069 expressing a tandem-dimer red fluorescent protein allowing visualization of hyphal growth). Plants silenced with either TRV:5′-MAPKKKε or TRV:3′-NbMAPKKKε repeatedly showed enhanced growth of pathogen hyphae, visible 3 to 4 d postinoculation (Figure 6A). Furthermore, the growth of necrotic lesions (Figure 6B) and progression to sporulation were accelerated in these plants compared with the TRV:GFP control (Figures 6C and 6D). These results indicate that Nb-MAPKKKε is likely involved in a plant immune response to P. infestans 88069 that can limit infection by the pathogen.
Figure 6.
VIGS of Nb-MAPKKKε Enhances Pathogen Growth.
Silencing of MAPKKKε in N. benthamiana, with either TRV:5′-MAPKKKε or TRV:3′-NbMAPKKKε, followed by inoculation with zoospores of the red fluorescent P. infestans 88069td enhances the growth of the pathogen compared with control silencing with TRV:GFP.
(A) Fluorescence microscopy images taken at 4 d after zoospore inoculation indicate more hyphal growth on the MAPKKKε-silenced plants.
(B) and (C) Silencing MAPKKKε also enhances the levels of necrosis and sporulation at 5 d after zoospore inoculation.
(D) Lesion area achieved at 5 and 6 d after zoospore inoculation following infection of TRV-treated plants with P. infestans 88069td. Results are the mean ± se of 40 inoculations from two independent experiments. Asterisks indicate values significantly different from the TRV:GFP control at the same time point (P < 0.05) as determined by one-way ANOVA; all other values were not significantly different (P > 0.05).
MAPKKKε-Triggered Cell Death Is Suppressed by PexRD2
Expression of either full-length Sl-MAPKKKε or SlMAPKKKε-KD1-332 triggers pathogen-independent cell death in N. benthamiana and requires an active kinase domain [the P-loop mutant SlMAPKKKε-KD1-332(Lys49Arg) prevents activity; Melech-Bonfil and Sessa, 2010]. To test whether StMAPKKKε-KD1-332 and StMAPKKKε-KD1-300 also trigger cell death on expression, we cloned these domains into the estradiol-inducible pER8 vector (Zuo et al., 2000) and delivered these into plant cells by agroinfiltration. Two days postagroinfiltration, expression was induced with β-estradiol. All wild-type kinase domains triggered cell death. No cell death was observed following expression of SlMAPKKKε-KDLys49Arg (Supplemental Figure 8).
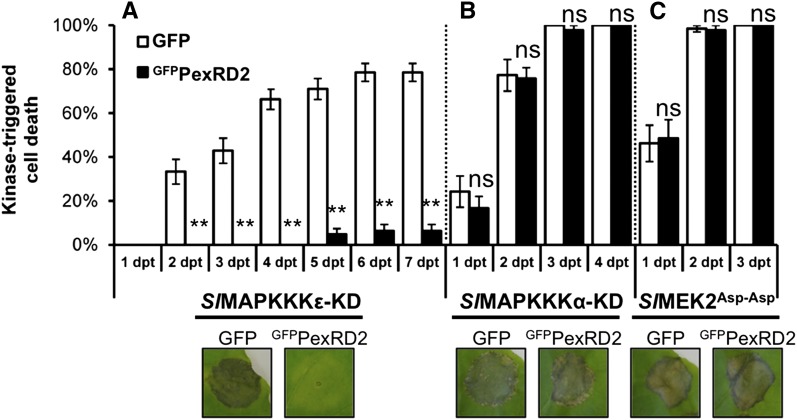
We tested the effect of coexpressing GFPPexRD2 on the cell death triggered by SlMAPKKKε-KD1-332. Two days after coagroinfiltration, expression of Sl-MAPKKKε was induced with β-estradiol. Cell death was then scored for 7 d after β-estradiol treatment (Figure 7A). These assays reveal that PexRD2 is a potent suppressor of the cell death triggered by SlMAPKKKε-KD. The protein levels of SlMAPKKKε-KD in the presence or absence of GFPPexRD2 were confirmed to be similar by immunoblot, indicating that this inhibition is not achieved through any effect on protein expression or stability (Figure 8C). GFPPexRD2 also suppressed cell death when coexpressed with StMAPKKKε-KD1-332 and StMAPKKKε-KD1-300 (Supplemental Figure 8). We also tested the effect of GFPPexRD2 on the cell death triggered by SlMAPKKKα-KD (del Pozo et al., 2004) and a constitutively active variant of the proposed downstream MAPKK, Sl-MEK2 (Melech-Bonfil and Sessa, 2010) or SlMEK2Asp-Asp (Oh and Martin, 2011). No significant difference was observed in the mean level of cell death observed in GFPPexRD2 coagroinfiltrated sites compared with GFP control for either SlMAPKKKα-KD (Figure 7B) or SlMEK2Asp-Asp (Figure 7C). These results show that PexRD2 specifically suppresses the macroscopic cell death triggered by MAPKKKε kinase domains.
Figure 7.
MAPKKKε-Triggered Cell Death Is Suppressed by PexRD2.
Coexpression of GFPPexRD2 with SlMAKKKε-KD in N. benthamiana reduces the percentage of infiltration sites with >50% confluent cell death at 2 to 7 d post-β-estradiol treatment (dpt) compared with coexpression with the GFP control (A). By contrast, PexRD2 did not suppress the cell death triggered by either the kinase domain of SlMAPKKKα-KD (B) or a constitutively active mutant of the proposed downstream MAPKK, SlMEK2Asp-Asp (C). Results are the mean ± se of three independent experiments, each using at least three plants. Asterisks indicate values significantly different from the GFP control at the same time point (P < 0.01) as determined by one-way ANOVA; all other values were not significantly different (ns; P > 0.05). Images indicate representative infiltration sites at 7 dpt.
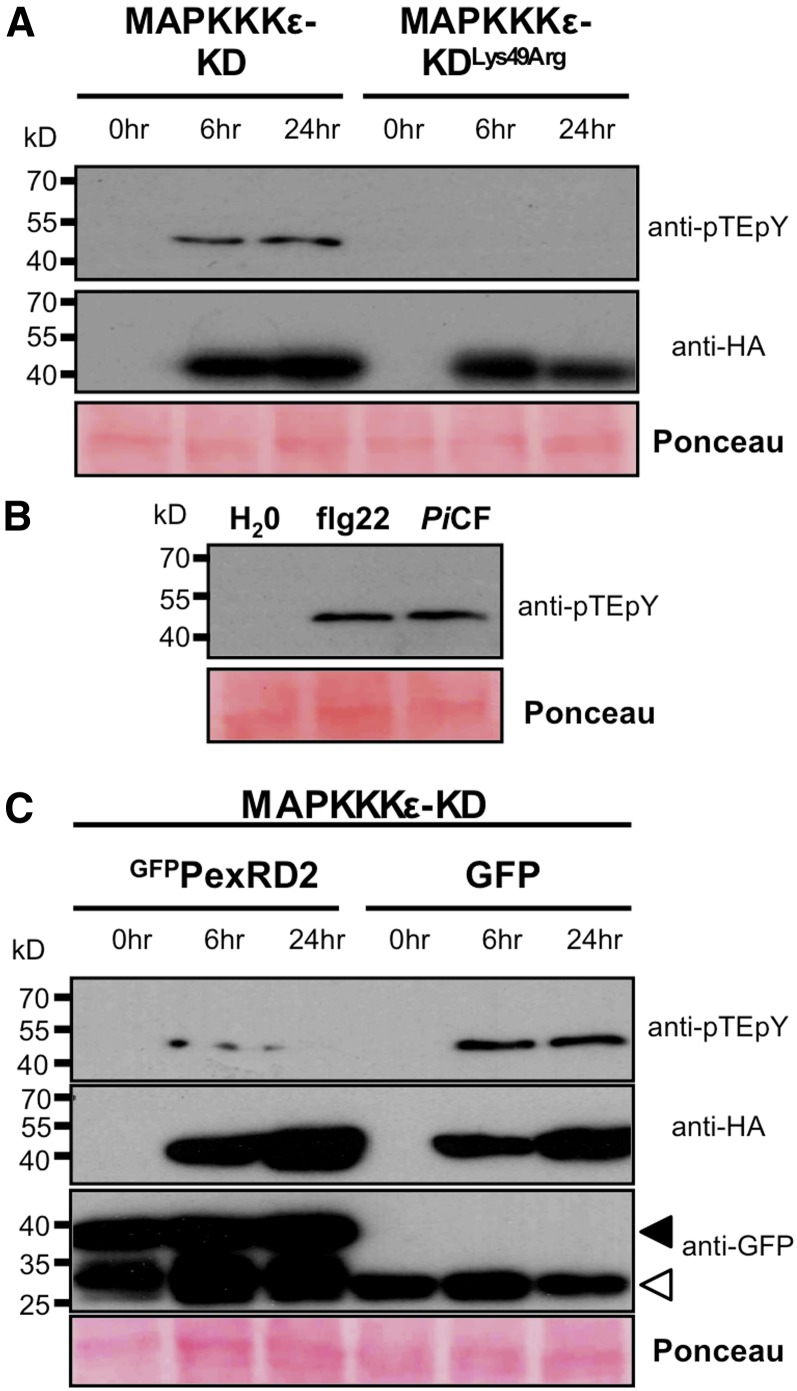
Figure 8.
MAPKKKε-Mediated Activation of MAPKs Is Suppressed by PexRD2.
(A) Immunoblot analysis shows that overexpression of SlMAPKKKε-KD causes activation of an endogenous MAPK in N. benthamiana. Overexpression of the kinase-inactive mutant (MAPKKKε-KDLys49Arg) did not phosphorylate MAPK. Protein levels of both kinases at 6 and 24 h post-β-estradiol treatment were confirmed using an anti-HA antibody (also in [C]).
(B) Treatment of N. benthamiana leaf disks with peptide corresponding to the epitope of the bacterial PAMP flagellin, flg22 (100 nM), or 100-fold diluted P. infestans 88069 culture filtrate (PiCF) results in activation of MAPK.
(C) Coexpression of GFPPexRD2, but not GFP, suppresses the activation of MAPK mediated by SlMAPKKKε-KD. Expression of GFPPexRD2 and SlMAPKKKε-KD was confirmed using anti-GFP and anti-HA antibodies, respectively. Black arrow indicates expected size of the GFPPexRD2 fusion protein and the white arrow free GFP. Similar results were observed in three independent experiments. In all cases, protein loading is confirmed by Ponceau staining.
MAPKKKε Promotes Phosphorylation of MAPKs and PexRD2 Suppresses This Activity
We investigated whether overexpression of MAPKKKε in plant cells leads to phosphorylation of downstream MAPKs, prior to development of macroscopic cell death, and whether PexRD2 could suppress this. First, we expressed SlMAPKKKε-KD, or the inactive SlMAPKKKε-KDLys49Arg, in N. benthamiana, prepared leaf tissue at 6 and 24 h after treatment with β-estradiol and looked for the presence of activated MAPKs using an antibody specific to the phosphorylated Thr-X-Tyr motif of these proteins. We observed a single band in the samples expressing SlMAPKKKε-KD that was absent on expression of SlMAPKKKε-KDLys49Arg (Figure 8A). Following elicitation with flg22, N. benthamiana leaf extracts can show single and multiple phosphorylated MAPKs (Segonzac et al., 2011). In our controls, we observed a single band following treatment with flg22 or P. infestans culture filtrate (Figure 8B). Coagroinfiltration of GFPPexRD2 led to a clear reduction in the accumulation of this phosphorylated MAPK band compared with the GFP control (Figure 8C).
MAPKKKε-Dependent Cell Death Is Suppressed by PexRD2
VIGS of MAPKKKε in N. benthamiana reduces the cell death activated by coexpression of specific fungal and bacterial effectors with their cognate R proteins (Melech-Bonfil and Sessa, 2010). We confirmed these results for the Avr4/Cf4 (Cladosporium fulvum effector) and AvrPto/Pto (Pseudomonas syringae effector) pairs (Supplemental Figures 9A and 9B). We also tested whether silencing of Nb-MAPKKKε reduced cell death in response to the P. infestans effector/R protein pair AVR3aKI/R3a (Armstrong et al., 2005) or expression of the P. infestans effector CRN8 (van Damme et al., 2012) or P. infestans elicitin INF1 (Kamoun et al., 1998). No effect on the mean percentage of cell death mediated by these elicitors was observed (Supplemental Figures 9C to 9E), indicating that they are independent of the MAPKKKε signaling cascade.
We investigated whether PexRD2 interferes with MAPKKKε-dependent and MAPKKKε-independent cell death. When coexpressed with either Avr4/Cf4 (Figure 9) or AvrPto/Pto (Supplemental Figure 10A), PexRD2 strongly suppressed macroscopic MAPKKKε-dependent cell death compared with the GFP control. By contrast, coagroinfiltration of PexRD2 with AVR3aKI/R3a, INF1, or CRN8 did not affect the cell death induced by any of these elicitors (Supplemental Figures 10B to 10D). These experiments reveal a direct correlation in cell death suppressed by either silencing of Nb-MAPKKKε or coexpression of PexRD2 with the elicitors.
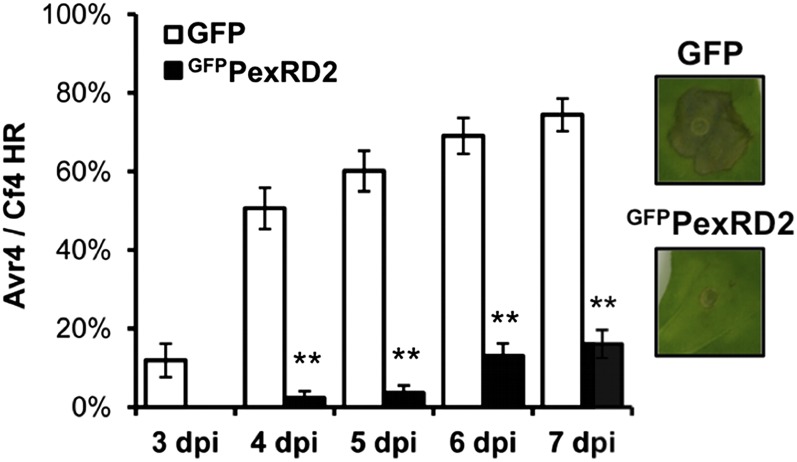
Figure 9.
PexRD2 Suppresses the Cell Death Associated with the Avr4/Cf4 HR.
Coexpression of GFPPexRD2 with the both the C. fulvum effector Avr4 and tomato resistance protein Cf4 in N. benthamiana reduces the percentage of infiltration sites with >50% confluent cell death at 3 to 7 d postagroinfiltration (dpi) compared with the GFP control. Results are the mean ± se of three independent experiments, each using at least four plants. Asterisks indicate a value significantly different from the GFP control at the same time point (P < 0.01) as determined by one-way ANOVA; all other values were not significantly different (P > 0.05). Images indicate representative infiltration sites at 7 dpi.
PexRD2 Variants That Do Not Interact with MAPKKKε Do Not Suppress MAPKKKε-Dependent Cell Death
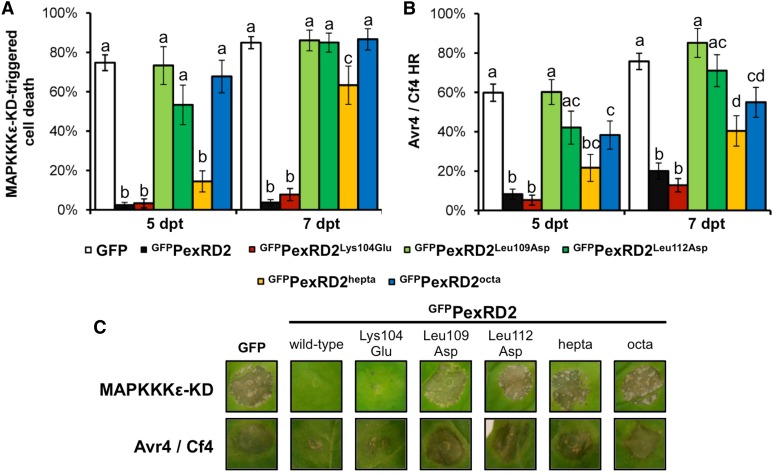
We tested whether variants of PexRD2 that no longer interact with StMAPKKKε-KD have lost their ability to suppress MAPKKKε-triggered or MAPKKKε-dependent cell death. We used the noninteracting mutants PexRD2Leu109Asp, PexRD2Leu112Asp, and PexRD2octa, the weakly interacting mutant PexRD2hepta, and one interacting mutant PexRD2Lys104Glu (as a control) alongside the wild-type protein in coagroinfiltration assays with either SlMAPKKKε-KD or Avr4/Cf4. All the mutants that failed to interact with StMAPKKKε-KD in the Y2H assay failed to suppress both MAPKKKε-triggered and MAPKKKε-dependent cell death activities (Figure 10). Interestingly, the PexRD2hepta mutant showed an intermediate phenotype as it was able to delay, but not prevent, cell death development. This observation corresponds to the strength of interaction between this mutant and StMAPKKKε-KD in the Y2H (Figure 4). However, this mutant did not demonstrate an enhanced infection phenotype (Figure 5).
Figure 10.
PexRD2 Variants That Are Perturbed in MAPKKKε Interaction Also Show Reduced Suppression of MAPKKKε-Triggered or MAPKKKε-Mediated Cell Death.
(A) Percentage of infiltration sites with >50% confluent cell death at 5 and 7 d post-β-estradiol treatment (dpt), following coexpression of the respective PexRD2 mutants with SlMAPKKKε-KD.
(B) Percentage of infiltration sites with >50% confluent cell death at 5 and 7 d after agroinfiltration (dpi), following coexpression of the respective PexRD2 mutants with Avr4 and Cf4. Results are the mean ± se of three independent experiments, each using at least four plants. The same letter above two bars indicates values that are not significantly different from one another at the same time point (P > 0.05) as determined by one-way ANOVA.
(C) Images indicate representative infiltration sites at 7 dpt (MAPKKKε-KD) and 7 dpi (Avr4/Cf4).
DISCUSSION
MAPK cascades are well established as key signaling systems in plants, transducing perception of abiotic and biotic stresses to downstream targets, in addition to playing roles in growth and development. In the defense against pathogens, plant MAPK cascades orchestrate changes in defense-related gene expression and other antimicrobial responses (Pitzschke et al., 2009; Meng and Zhang, 2013). Highlighting their importance, bacterial pathogens of plants have evolved translocated effector proteins that interfere with MAPK signaling pathways (Pitzschke et al., 2009; Meng and Zhang, 2013). In this study, we directly link a filamentous plant pathogen effector with components of MAPK signaling in plants. We identify an RXLR-type effector from the late blight pathogen P. infestans that interacts with MAPKKKε and perturbs plant defense–related processes either triggered by or dependent on the activity of this kinase. This establishes MAPK cascades as direct targets of oomycete RXLR-type effectors.
PexRD2 is one of a limited number of RXLR-type effectors for which structural information is available. Here, we used structure-informed mutagenesis of both surface- and dimerization interface-presented residues of PexRD2 to generate variants with differing interaction strengths. These mutants have demonstrated a positive correlation between the ability to interact with MAPKKKε and the ability to suppress MAPKKKε signaling–dependent cell death. Furthermore, PexRD2 mutants with reduced ability to bind MAPKKKε or inhibit its signaling were no longer able to enhance the growth of P. infestans in planta. The PexRD2-like effectors tested were unable to interact with MAPKKKε. We hypothesize that these and other PexRD2-like effectors, that are predicted to adopt the same overall WY-domain protein fold, may have evolved as kinase interacting modules. Although whether members of the PexRD2 family, other than PexRD2, do indeed target other host kinases remains to be determined. If they do, PexRD2-like effectors may provide useful molecular tools to dissect the role of MAPK signaling in plants. Specificity for different kinases may be encoded in the amino acid residues presented on the surface of the proteins. In support of this, our data showing that PexRD2-like-1a and PexRD2-like-2a do not interact with MAPKKKε and do not display any of the PexRD2 phenotypes in planta suggest they have a different function.
Detailing the roles of MAPK cascades in plant defense responses is frequently challenging due to genetic redundancy and/or crosstalk associated with plant development (Wang et al., 2007; Qiu et al., 2008; Rodriguez et al., 2010). The best-studied plant MAPKs are Arabidopsis MPK3 and MPK6, their orthologs in solanaceous plants, SIPK and WIPK (positive regulators of plant immune pathways), and MPK4 (a negative regulator of plant immune pathways) (Pitzschke et al., 2009; Meng and Zhang, 2013). Double loss-of-function mutations in MPK3 and MPK6 are embryonic lethal, and MPK4 mutants show a severely dwarfed phenotype (Wang et al., 2007; Qiu et al., 2008). Further upstream, double mutation of MAPKKKε1 and MAPKKKε2 in Arabidopsis is pollen-lethal, but plants containing individual mutations to MAPKKKε1 or MAPKKKε2 show no obvious phenotype (Chaiwongsar et al., 2006). The use of an inducible At-MAPKKKε1 construct in the mapkkkε1 mapkkkε2 background rescues pollen unviability, but the plants show reduced root length and reduced cell expansion in rosette leaves (Chaiwongsar et al., 2012). Knocking down MAPKKKε by VIGS in N. benthamiana results in a degree of growth inhibition (Melech-Bonfil and Sessa, 2010). Interestingly, we observe that high levels of PexRD2 expression in young leaves can also lead to developmental phenotypes in N. benthamiana consistent with retardation of plant growth. Defining whether this is directly linked to PexRD2 interaction with Nb-MAPKKKε, and whether the effects on growth are linked to or independent of plant immunity, is beyond the scope of this study.
Epistasis analysis has placed MAPKKKε upstream of MEK2 and SIPK/WIPK in N. benthamiana (Melech-Bonfil and Sessa, 2010). Here, we show that overexpression of MAPKKKε in N. benthamiana results in phosphorylation of MAPKs, and this phosphorylation can be suppressed by coexpression with PexRD2. Moreover, whereas PexRD2 suppresses cell death triggered by overexpression of the MAPKKKε kinase domain, it fails to suppress cell death triggered by MAPKKKα or MEK2Asp-Asp. We find that PexRD2 suppresses all readouts of cell death tested that are either triggered by or dependent on MAPKKKε, while all readouts of cell death tested that are independent of MAPKKKε are not affected by PexRD2. This suggests a high level of specificity for PexRD2 in interfering with plant immunity–related signaling cascades involving MAPKKKε.
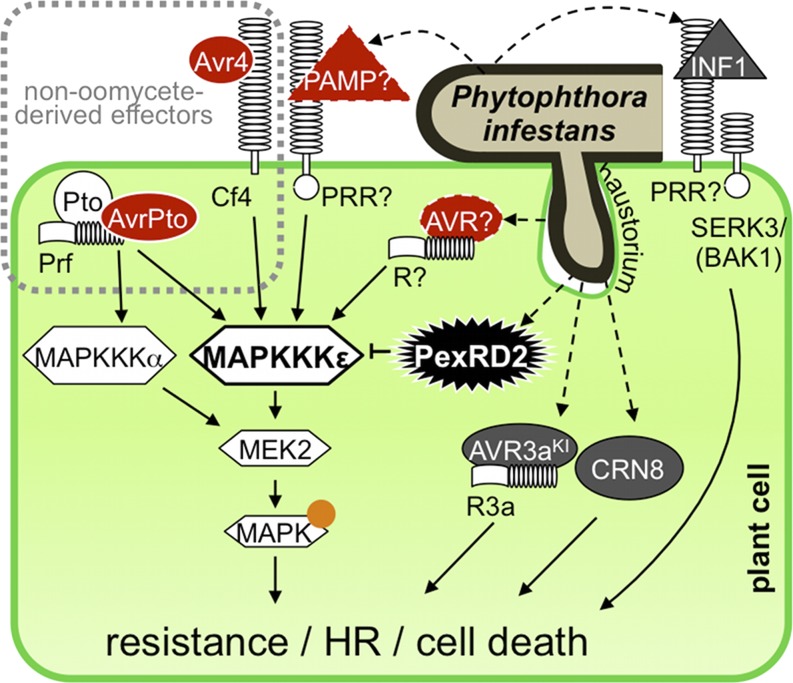
Our data support the hypothesis that P. infestans delivers PexRD2 into host cells to interact with and suppress MAPKKKε activity, and this is of benefit to the pathogen during infection. In our model for PexRD2 function (Figure 11), this effector interacts with MAPKKKε to suppress immunity-related signaling by this kinase. We suggest that MAPKKKε is a target of PexRD2, although we cannot rule out that MAPKKKε acts as a helper (Win et al., 2012b) to modify PexRD2 following delivery and this modified protein then perturbs the function of other molecules involved in MAPK cascade signaling.
Figure 11.
Model of PexRD2 Function during Infection.
Schematic diagram illustrating P. infestans delivering PexRD2 into a haustoriated plant cell. PexRD2 interacts with MAPKKKε in the plant cytosol and inhibits signaling by this protein. The cell death triggered by overexpression of MAPKKKε is inhibited by the presence of PexRD2, but not the cell death triggered by MAPKKKα or MEK2. PexRD2 also suppresses the phosphorylation of MAPKs (orange circle), which occurs following overexpression of MAPKKKε. The HR following recognition of the C. fulvum effector Avr4 by Cf4 and the P. syringe effector AvrPto mediated by Pto/Prf, which are known to be MAPKKKε dependent (red, solid white border), are suppressed by the presence of PexRD2. The HR resulting from the recognition of AVR3aKI by R3a, the cell death following recognition of the PAMP-like INF1, and the cell death mediated by CRN8 are all MAPKKKε independent (dark gray) and are not suppressed by PexRD2. We propose that, in planta, MAPKKKε confers enhanced resistance against P. infestans following recognition of an unidentified PAMP (red triangle, dashed outline), by a PRR or recognized effector/avirulence protein (AVR, red oval, dashed outline) by an R protein. The pathogen delivers PexRD2 inside the host cell to specifically inhibit this MAPKKKε-dependent resistance and promote proliferation.
Both reduction of MAPKKKε activity (through VIGS) and expression of PexRD2 resulted in enhancing P. infestans 88069 growth and suppressing a variety of cell death readouts in N. benthamiana. However, we have yet to establish a direct link between MAPKKKε activity and a specific signaling pathway downstream of oomcyete PAMP perception (PRR-mediated immunity) or recognition of an oomycete effector protein (NB-LRR–mediated immunity). Potentially, MAPKKKε could be important for signaling events following recognition of oomycete PAMPs such as Pep-13 (Halim et al., 2004) and CBEL (Gaulin et al., 2006), as we observed that at least one MAPK is activated by application of P. infestans culture filtrate. Alternatively, MAPKKKε may be required for signaling mediated by one or a subset of the 68 Solanum NB-LRRs documented against P. infestans (Rodewald and Trognitz, 2013). It is also possible that interfering with MAPKKKε activity induces a more general perturbation in immunity signaling that is of benefit to the pathogen.
In summary, we identified a P. infestans RXLR-WY–type effector, PexRD2, which interacts with MAPKKKε and perturbs plant immunity associated signaling pathways dependent on this kinase. Either overexpression of PexRD2 or knockdown of MAPKKKε supports enhanced pathogen growth and suppression of MAPKKKε-triggered or -dependent cell death readouts in N. benthamiana. This study represents a step toward understanding how oomycete RXLR-type effectors directly interact with MAPK cascades, which are well established as key regulators of plant immunity. The next challenge is to better understand the role of PexRD2 and PexRD2-like effectors, and their targets, in the progression of disease in important host crop plants, such as tomato and potato. The ultimate aim of this would be to manipulate these interactions to tip the balance in the coevolutionary arms race between pathogen and host in favor of the plant.
METHODS
Bacterial Strains and Growth Conditions
Escherichia coli, used for molecular biology, was cultivated in Luria-Bertani medium at 37°C. Agrobacterium tumefaciens was grown at 28°C in LB-Lennox broth medium (Lennox, 1955). All binary constructs generated in this study were introduced into Agrobacterium strain GV3101 by electroporation.
Plant Material and Agroinfiltration
Nicotiana benthamiana plants were grown in controlled environment rooms at 22°C with 55% humidity and 16 h light or in controlled glasshouses under similar conditions. Agrobacteria were incubated in infiltration medium (10 mM magnesium chloride, 10 mM MES, pH 5.6, and 150 μM acetosyringone) for at least 1 h prior to infiltration into leaves as described (Van der Hoorn et al., 2000).
Phytophthora infestans Infection Assays
P. infestans 88069 or P. infestans 88069td (a stable transformant of P. infestans 88069 expressing a tandem-dimer red fluorescent protein) strains were grown on rye Suc agar at 18°C in the dark (Kamoun et al., 1998). Two leaves of 4- to 5-week old N. benthamiana plants were agroinfiltrated at an OD600 of 0.3 with the binary vector pK7WGF2 (Karimi et al., 2002) (expressing free GFP) on one half of the mid-vein and an effector cloned into pK7WGF2 (expressing N-terminal GFP fusions) into the other half of the same leaf. Phytophthora spores were harvested and diluted to 100,000 spores/mL (Kamoun et al., 1998; Schornack et al., 2010). Droplets (10 μL) of zoospores were applied onto the abaxial side of detached leaves 24 h postagroinfiltration and incubated for several days on wet paper towels in 100% relative humidity. Lesion areas were determined with GIMP (v2.8) software from white-light photographs. Mycelial growth of P. infestans 88069td was visualized as described (Chaparro-Garcia et al., 2011). Relative lesion area was calculated by dividing the lesion area at 5 d after zoospore inoculation of leaf tissue transiently expressing the appropriate effector/mutant, by the lesion area achieved on tissue from the same leaf expressing the GFP control at the same time point. Data graphs present the mean lesion area/relative lesion area per infected leaf with error bars representing ±se. Either t test or one-way ANOVA and Tukey honest significant difference tests were performed to identify statistically significant differences.
Y2H Screening and Assays
The Proquest two-hybrid system (Invitrogen) was used to detect protein–protein interactions. The original prey library and screening were as described (Bos et al., 2010). PexRD221-121 and PexRD257-121 were amplified from previously generated plasmid DNA (Boutemy et al., 2011) using primers listed in Supplemental Table 1. PexRD2-like effectors were amplified from P. infestans strain T30-4 genomic DNA using primers listed in Supplemental Table 1. The resultant products were cloned into pENTR/D-TOPO and transferred into pDEST 32 using LR clonase. St-MAPKKKε truncations and Sl-MAPKKKα sequences were amplified using primers listed in Supplemental Table 1 and cloned into pENTR/D-TOPO and transferred to pDEST 22.
Mutagenesis
Individual point mutants of PexRD221-121 and the PexRD2octa mutant were either introduced into the wild-type entry clone or synthesized and flanked by the attB recombination sites in a pUC57 vector by Genscript. Mutants in the pUC57 background were transferred into pDONR 201 using BP clonase. The PexRD2hepta mutant was generated by whole-plasmid mutagenesis using Velocity DNA polymerase (BIOLINE) and pDONR 201:PexRD2octa as a template. Methylated parental DNA was removed by DpnI treatment. The resultant Glu-90 to Ala mutation was verified by sequencing. Mutant PexRD2 effectors were moved using LR clonase into pDEST 32 for Y2H and pK7WGF2 for in planta assays.
BiFC
For split-YFP assays, leaves of 4- to 5-week-old N. benthamiana plants were infiltrated with agrobacteria containing constructs expressing the YN fragment fused to PexRD2, PexRD2-like-1a, or PexRD2-like-2a and the YC fragment fused to the various MAPKKK forms. For each combination, the final OD600 for each strain was 0.01 for confocal imaging and 0.1 for fluorimeter measurements. Leaf pieces were imaged on a Zeiss 710 microscope 2 d after agroinfiltration using the 514-nm laser line to excite the YFP and a window of 525 to 560 nm to collect emissions. Quantification of fluorescence was performed using a SpectraMax M5 fluorimeter (Molecular Devices). Leaf disks were cut at 2 d postagroinfiltration and floated abaxial side up on water in 24-well plates. Measurements were made using a well scanning setting taking reads from the top surface. YFP fluorescence was excited at 485 nm and measured at 525 nm. Softmax Pro software (Molecular Devices) was used to collect data that were then exported to Excel for analysis.
Coimmunoprecipitation of PexRD2
FLAG-epitope tagged PexRD2 in pJL-TRBO (Boutemy et al., 2011) and a GFP-tagged mutant PexRD2 were expressed together in N. benthamiana leaves by agroinfiltration. FLAG-epitope tagged proteins were immunoprecipitated by anti-FLAG M2 affinity gel (Sigma-Aldrich) from total protein extracts harvested from leaves 3 d postinoculation (Win et al., 2011). Interacting GFP fusion proteins were detected by immunoblotting of SDS-PAGE separated proteins using anti-GFP antibodies (Invitrogen) as a probe followed by anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Sigma-Aldrich). The expression of effectors in total extract was confirmed using either the procedure above (for GFP-fused effectors) or an anti-FLAG monoclonal antibody conjugated to horseradish peroxidase (Sigma-Aldrich). Chemiluminescent substrate (Thermo Scientific) was added to the blots to visualize positive protein bands after exposure to x-ray films.
VIGS of N. benthamiana
pTRV2:5′-MAPKKKε used in this study has been described (Melech-Bonfil and Sessa, 2010). A second construct was generated by amplifying a 336-bp fragment of the 3092 to 3427 region of MAPKKKε2 from N. benthamiana cDNA with the following primers (5′-GGGGgaattcCATCACAGACTGCGTCAGGT-3′ and 5′-GGGGGgttaacTGATTGCATCTGCTCTCTGG-3′). The resultant PCR product was then ligated into EcoRI and HpaI digested pTRV2, to yield pTRV2:3′-NbMAPKKKε. VIGS was performed as described previously (Valentine et al., 2004). Briefly, Agrobacterium strains harboring pTRV1 vector and pTRV2:GFP, pTRV2:5′-MAPKKKε or pTRV2:3′-NbMAPKKKε were mixed in a 1:1 ratio to achieve final OD600 for each strain of 0.5. The cocultures were then infiltrated into the two largest leaves of 2-week-old plants. The plants were then grown for 2 weeks before using for P. infestans infection or cell death assays.
Cell Death Assays
Agrobacterium strains harboring R protein constructs (Cf4, Pto, or R3a) were mixed with those harboring effector protein constructs (Avr4, AvrPto, or AVR3aKI, respectively) to achieve final OD600 values of 1.0 and 0.5, respectively (Gilroy et al., 2011). Agrobacterium strains harboring INF1 (Gilroy et al., 2011) or CRN8 (residues 118 to 699) ligated into XmaI- and StuI-digested pEAQ-HT vector (Sainsbury et al., 2009) were diluted to achieve final OD600 values of 0.5 and 0.3, respectively. Agrobacterium mixtures were infiltrated into leaves of VIGS-treated plants, and progression of HR/programmed cell death (PCD) was monitored daily up to 7 d postagroinfiltration. An agroinfiltration site was scored positive for HR/PCD following the development of clear necrosis occupying >50% of the agroinfiltrated area. Data graphs present the mean percentage of total inoculations per plant developing a clear HR with error bars representing ±se of combined data from at least three independent experiments. One-way ANOVA and Tukey honest significant difference tests were performed to identify statistically significant differences.
Cell Death Suppression Assays
The β-estradiol–inducible SlMAPKKKε-KD and SlMAPKKKε-KDLys49Arg constructs used in this study are the same as described (Melech-Bonfil and Sessa, 2010). StMAPKKKε-KD1-332 and StMAPKKKε-KD1-300 were amplified using primers listed in Supplemental Table 1, without a stop codon and ligated into XhoI- and PacI-digested pER8 vector, which already had a double hemagglutinin tag cloned into the PacI and SpeI sites. The SlMAPKKKα-KD and SlMEK2Asp-Asp (previously referred to as SlMEK2DD) constructs are the same as described those described by del Pozo et al. (2004) and Oh and Martin (2011), respectively.
Cell death activity was confirmed by agroinfiltration of strains harboring the β-estradiol–inducible kinase constructs, and the P19 suppressor of silencing (Lindbo, 2007), mixed to achieve final OD600 values of 0.25 and 0.1, respectively. Expression of the kinases was induced by application of 10 μM β-estradiol (Sigma-Aldrich) to the leaves at 48 h postagroinfiltration, with additional spray treatments at 48-h intervals when necessary. Progression of cell death was monitored daily up to 7 d after β-estradiol treatment. Effector-mediated suppression of kinase-triggered cell death was assessed in the same way, but with the addition of GFPPexRD2 (wild type/mutant) or GFP vector control (in pK7WGF2) at a final OD600 of 0.3, coagroinfiltrated with the kinase.
Agrobacterium strains harboring R protein/effector protein combinations were diluted with Agrobacterium harboring the wild type or mutant GFPPexRD2 or GFP vector control (in pK7WGF2, except AvrPto/Pto HR suppression, which used pB7WGF2) to achieve final OD600 values of 0.6, 0.3, and 0.3, respectively. Agrobacterium strains harboring INF1 or CRN8 were diluted to achieve final OD600 values, as above, but with the wild type or mutant GFPPexRD2 or GFP vector control at a 1:1 ratio. Following agroinfiltration of these mixtures into N. benthamiana plants, progression of HR/PCD was monitored, scored, and analyzed as described above.
MAPK Activation Assay
The activation of endogenous MAPKs in N. benthamiana following induction of MAPKKKε-KD expression was assessed using a procedure described (Segonzac et al., 2011). Following agroinfiltration and induction of kinase expression as described above, total protein extracts were separated on a SDS-PAGE gel and transferred to polyvinylidene fluoride membrane. The membrane was probed using anti-pTEpY primary antibodies (Cell Signaling), anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Sigma-Aldrich), and a chemiluminescent substrate (Thermo Scientific) in accordance with the manufacturer’s instructions.
For PAMP treatment, eight leaf disks of 5 mm in diameter were harvested from 4-week old N. benthamiana plants and added to wells of a 96-well microtiter plate, containing 100 μL of either MQ-water, 100 nM flg22 peptide, or 100-fold diluted, crude P. infestans 88069 culture filtrate (prepared as described in Chaparro-Garcia et al., 2011). These leaf disks were then pooled, and total protein extraction and assessment of the phosphorylation status of MAPKs were conducted as described above.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL databases under the following accession numbers: PexRD2 (PITG_21422, EEY62542), PexRD2-like-1a (PITG_14984, EEY62514), PexRD2-like-2a (PITG_14787, EEY62361), potato MAPKKKε (KJ504180), S. lycopersicum MAPKKKε (ADK36642), N. benthamiana MAPKKKε (ADK36643 and BAM36969), tomato MAPKKKα (AAS78640), and tomato MEK2 (AAU04434).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PexRD2-Like Bait Proteins Are Expressed and Stable in Yeast.
Supplemental Figure 2. PexRD2 and MAPKKKε Colocalize in the Plant Cell Cytoplasm.
Supplemental Figure 3. Localization and Split YFP Fusion Proteins Are Expressed and Stable in Planta.
Supplemental Figure 4. Structure-Informed Point Mutants of PexRD2 Can Disrupt the Interaction with MAPKKKε.
Supplemental Figure 5. Mutations within the Structurally Variable Region of the WY-Fold of PexRD2 Reduce the Interaction with MAPKKKε.
Supplemental Figure 6. GFP Fusion Proteins Used for in Planta Assays Are Expressed and Stable.
Supplemental Figure 7. Real-Time Quantitative RT-PCR Confirms Specific Silencing of MAPKKKε.
Supplemental Figure 9. VIGS Experiments to Investigate MAPKKKε-Dependent and MAPKKKε-Independent Cell Death Events.
Supplemental Figure 10. PexRD2 Suppresses MAPKKKε-Dependent AvrPto/Pto Hypersensitive Response, but Not MAPKKKε-Independent Cell Death.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the Biotechnology and Biological Science Research Council (BBSRC) (UK; Grants BB/J00453/1, BB/I019557, and BB/G015244), the European Research Council (proposal 294608), the Gatsby Charitable Foundation, the John Innes Foundation, and the Scottish Government Rural and Environment Science and Analytical Services Division for funding in areas relevant to this work. S.R.F.K. is funded by a BBSRC-Doctoral Training Grant (studentship) and the Gatsby Charitable Foundation. We thank Richard Hughes, Angela Chaparro-Garcia, and Liliana Cano for discussion and provision of reagents.
AUTHOR CONTRIBUTIONS
S.R.F.K., H.M., P.C.B., S.K., P.R.J.B., and M.J.B. designed the research. S.R.F.K., H.M., P.C.B., M.R.A., T.B., O.S., and J.W. performed the research. S.R.F.K., H.M., P.C.B., M.R.A., S.K., P.R.J.B., and M.J.B. analyzed data. S.R.F.K. and M.J.B. wrote the article with contributions/comments from all authors.
Glossary
- PRR
pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- HR
hypersensitive response
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- MAPKKK
MAPKK kinase
- Y2H
yeast two-hybrid
- VIGS
virus-induced gene silencing
- BiFC
bimolecular fluorescence complementation
- PCD
programmed cell death
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Armstrong M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102: 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Birch P.R., Rehmany A.P., Pritchard L., Kamoun S., Beynon J.L. (2006). Trafficking arms: Oomycete effectors enter host plant cells. Trends Microbiol. 14: 8–11. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bombarely A., Rosli H.G., Vrebalov J., Moffett P., Mueller L.A., Martin G.B. (2012). A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 25: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Bos J.I., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107: 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutemy L.S., King S.R., Win J., Hughes R.K., Clarke T.A., Blumenschein T.M., Kamoun S., Banfield M.J. (2011). Structures of Phytophthora RXLR effector proteins: A conserved but adaptable fold underpins functional diversity. J. Biol. Chem. 286: 35834–35842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T.O., Schornack S., Banfield M.J., Kamoun S. (2012). Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15: 483–492. [DOI] [PubMed] [Google Scholar]
- Bozkurt T.O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., Cano L.M., Jones A.M., Huitema E., van der Hoorn R.A., Kamoun S. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 108: 20832–20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwongsar S., Otegui M.S., Jester P.J., Monson S.S., Krysan P.J. (2006). The protein kinase genes MAP3K epsilon 1 and MAP3K epsilon 2 are required for pollen viability in Arabidopsis thaliana. Plant J. 48: 193–205. [DOI] [PubMed] [Google Scholar]
- Chaiwongsar S., Strohm A.K., Su S.H., Krysan P.J. (2012). Genetic analysis of the Arabidopsis protein kinases MAP3Kε1 and MAP3Kε2 indicates roles in cell expansion and embryo development. Front. Plant Sci. 3: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Karin M. (2001). Mammalian MAP kinase signalling cascades. Nature 410: 37–40. [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A., Wilkinson R.C., Gimenez-Ibanez S., Findlay K., Coffey M.D., Zipfel C., Rathjen J.P., Kamoun S., Schornack S. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE 6: e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B.P., Yu X.L., Ma Z.C., Dong S.M., Dou D.L., Wang Y.C., Zheng X.B. (2012). Phytophthora sojae effector Avh331 suppresses the plant defence response by disturbing the MAPK signalling pathway. Physiol. Mol. Plant Pathol. 77: 1–9. [Google Scholar]
- Cheng W., Munkvold K.R., Gao H., Mathieu J., Schwizer S., Wang S., Yan Y.B., Wang J., Martin G.B., Chai J. (2011). Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe 10: 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Cooke D.E., et al. (2012). Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog. 8: e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O., Pedley K.F., Martin G.B. (2004). MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23: 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Dong S., et al. (2011). Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 7: e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore J.M., Coaker G. (2011). The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol. Plant 4: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J.M. (2012). A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118. [DOI] [PubMed] [Google Scholar]
- Frye C.A., Tang D., Innes R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18: 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy E.M., Taylor R.M., Hein I., Boevink P., Sadanandom A., Birch P.R. (2011). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190: 653–666. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19: 423–429. [DOI] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Haas B.J., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- Halim V.A., Hunger A., Macioszek V., Landgraf P., Nurnberger T., Scheel D., Rosahl S. (2004). The oligopeptide elicitor Pep-13 induces salicylic acid-dependent and -independent defense reactions in potato. Physiol. Mol. Plant Pathol. 64: 311–318. [Google Scholar]
- Hashimoto M., Komatsu K., Maejima K., Okano Y., Shiraishi T., Ishikawa K., Takinami Y., Yamaji Y., Namba S. (2012). Identification of three MAPKKKs forming a linear signaling pathway leading to programmed cell death in Nicotiana benthamiana. BMC Plant Biol. 12: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Axtell M.J., Dahlbeck D., Ekwenna O., Zhang S., Staskawicz B., Baker B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3: 291–297. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44: 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun S., van West P, Vleeshouwers V.G., de Groot K.E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Lennox E.S. (1955). Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1: 190–206. [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo J.A. (2007). TRBO: A high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 145: 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.B., Bogdanove A.J., Sessa G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. [DOI] [PubMed] [Google Scholar]
- McLellan H., Boevink P.C., Armstrong M.R., Pritchard L., Gomez S., Morales J., Whisson S.C., Beynon J.L., Birch P.R. (2013). An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melech-Bonfil S., Sessa G. (2010). Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 64: 379–391. [DOI] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Mukhtar M.S., et al. European Union Effectoromics Consortium (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V., Roux M., Zipfel C. (2009). Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C.S., Martin G.B. (2011). Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J. Biol. Chem. 286: 14129–14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.K., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21: 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R., et al. (2010). Recent developments in effector biology of filamentous plant pathogens. Cell. Microbiol. 12: 705–715. [DOI] [PubMed] [Google Scholar]
- Panstruga R., Dodds P.N. (2009). Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426. [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., et al. (2010). Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330: 1540–1543. [DOI] [PubMed] [Google Scholar]
- Roche D.B., Buenavista M.T., Tetchner S.J., McGuffin L.J. (2011). The IntFOLD server: An integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 39: W171–W176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald J., Trognitz B. (2013). Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol. Plant Pathol. 14: 740–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E.C., Lomonossoff G.P. (2009). pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Saunders D.G., Breen S., Win J., Schornack S., Hein I., Bozkurt T.O., Champouret N., Vleeshouwers V.G., Birch P.R., Gilroy E.M., Kamoun S. (2012). Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24: 3420–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S., van Damme M., Bozkurt T.O., Cano L.M., Smoker M., Thines M., Gaulin E., Kamoun S., Huitema E. (2010). Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA 107: 17421–17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C., Feike D., Gimenez-Ibanez S., Hann D.R., Zipfel C., Rathjen J.P. (2011). Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 156: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nürnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Nürnberger T., Joosten M.H. (2011). Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine T., Shaw J., Blok V.C., Phillips M.S., Oparka K.J., Lacomme C. (2004). Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol. 136: 3999–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme M., Bozkurt T.O., Cakir C., Schornack S., Sklenar J., Jones A.M., Kamoun S. (2012). The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. PLoS Pathog. 8: e1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R.A., Kamoun S. (2008). From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn R.A., Laurent F., Roth R., De Wit P.J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13: 439–446. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V.G., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3: e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23: 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Hou S., Wang X., Li Y., Ren D., Chen S., Tang X., Zhou J.M. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22: 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S.C., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- Win J., Kamoun S., Jones A.M. (2011). Purification of effector-target protein complexes via transient expression in Nicotiana benthamiana. Methods Mol. Biol. 712: 181–194. [DOI] [PubMed] [Google Scholar]
- Win J., Krasileva K.V., Kamoun S., Shirasu K., Staskawicz B.J., Banfield M.J. (2012a). Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 8: e1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J., Chaparro-Garcia A., Belhaj K., Saunders D.G., Yoshida K., Dong S., Schornack S., Zipfel C., Robatzek S., Hogenhout S.A., Kamoun S. (2012b). Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77: 235–247. [DOI] [PubMed] [Google Scholar]
- Win J., Morgan W., Bos J., Krasileva K.V., Cano L.M., Chaparro-Garcia A., Ammar R., Staskawicz B.J., Kamoun S. (2007). Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19: 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., Li Y., Tang X., Zhu L., Chai J., Zhou J.M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18: 74–80. [DOI] [PubMed] [Google Scholar]
- Yoshida K., et al. (2013). The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. Elife 2: e00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Velásquez A.C., Munkvold K.R., Zhang J., Martin G.B. (2012). A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 69: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.