Abstract
The fate of plastid DNA (ptDNA) during leaf development has become a matter of contention. Reports on little change in ptDNA copy number per cell contrast with claims of complete or nearly complete DNA loss already in mature leaves. We employed high-resolution fluorescence microscopy, transmission electron microscopy, semithin sectioning of leaf tissue, and real-time quantitative PCR to study structural and quantitative aspects of ptDNA during leaf development in four higher plant species (Arabidopsis thaliana, sugar beet [Beta vulgaris], tobacco [Nicotiana tabacum], and maize [Zea mays]) for which controversial findings have been reported. Our data demonstrate the retention of substantial amounts of ptDNA in mesophyll cells until leaf necrosis. In ageing and senescent leaves of Arabidopsis, tobacco, and maize, ptDNA amounts remain largely unchanged and nucleoids visible, in spite of marked structural changes during chloroplast-to-gerontoplast transition. This excludes the possibility that ptDNA degradation triggers senescence. In senescent sugar beet leaves, reduction of ptDNA per cell to ∼30% was observed reflecting primarily a decrease in plastid number per cell rather than a decline in DNA per organelle, as reported previously. Our findings are at variance with reports claiming loss of ptDNA at or after leaf maturation.
In vascular plants, copy numbers of plastid genomes (plastomes) frequently range from <100 per cell in meristematic cells to several thousand per cell in fully developed diploid leaf parenchyma cells. Microscopy studies have shown that the multicopy organelle genomes are usually condensed in more or less distinct DNA regions (nucleoids) within the organelle matrix or stroma.
During development, the ratio of nuclear to organelle genomes appears to be relatively stringently regulated (Herrmann and Possingham, 1980; Rauwolf et al., 2010). Disregarding greatly varying absolute values (summarized in Rauwolf et al., 2010; Liere and Börner, 2013), there is little dispute that the number of plastid genomes and nucleoids per organelle and cell increase during early leaf development in higher plants (Kowallik and Herrmann, 1972; Selldén and Leech, 1981; Baumgartner et al., 1989; Fujie et al., 1994; Li et al., 2006; Rauwolf et al., 2010). This increase is usually accompanied by an increase in both size and number of plastids per cell (Butterfass, 1979). By contrast, data about plastid DNA (ptDNA) amounts in chloroplasts and cells of mature, ageing, and senescent tissue differ and are highly controversial. Basically two patterns have been described: the maintenance of more or less constant amounts of ptDNA per cell and/or organelle (Li et al., 2006; Zoschke et al., 2007; Rauwolf et al., 2010; Udy et al., 2012) or a significant decrease in copy number brought about by either continued organelle and cell division without ptDNA replication (Lamppa and Bendich, 1979; Scott and Possingham, 1980; Tymms et al., 1983) or by ptDNA degradation (Baumgartner et al., 1989; Sodmergen et al., 1991). In a series of communications, Bendich and coworkers recently reported that ptDNA levels decline drastically before leaf maturation in several plant species. In Arabidopsis thaliana and maize (Zea mays), ptDNA levels were reported to decrease early and precipitously as leaves mature. It was concluded that, in fully expanded leaves, most chloroplasts contain no or only insignificant amounts of DNA long before the onset of leaf senescence (Oldenburg and Bendich, 2004; Rowan et al., 2004; Oldenburg et al., 2006; Shaver et al., 2006; Rowan et al., 2009). Retention of ptDNA was proposed to be dispensable after the photosynthetic machinery was established in that the plastome-encoded photosynthesis genes were no longer needed in adult leaves. Degradation or even entire loss of ptDNA was considered as an event during plastid and leaf development, common to all plants (Rowan et al., 2009). ptDNA degradation was also suggested to act as a signal inducing senescence (Sodmergen et al., 1991).
A priori, there is no reason why different ptDNA patterns should not occur, and there is indeed evidence that organelle DNA can behave differently in different materials, both quantitatively and structurally (e.g., Selldén and Leech, 1981; Baumgartner et al., 1989). However, since contradictory data were reported for the same species that were grown under comparable, if not identical, conditions (Rowan et al., 2004, 2009; Li et al., 2006; Oldenburg et al., 2006; Shaver et al., 2006; Zoschke et al., 2007; Evans et al., 2010; Udy et al., 2012), it is apparent that some of them must reflect methodological insufficiencies of the experimental approaches employed.
From a physiological point of view, the existence of DNA-deficient plastids in photosynthetically competent tissue seems unlikely. For instance, due to its susceptibility to photooxidative damage, the D1 protein (PsbA), a plastome-encoded core subunit of photosystem II, must be replaced continuously by a complex repair system to maintain photosynthesis (Prasil et al., 1992). This replacement requires de novo synthesis of the short-lived D1. There are no data available supporting an extreme mRNA stability, protein stability, or for another compensating biochemistry, preserving organelle functions for weeks or even months. The maximum mRNA half-life reported for psbA is in the range of 40 h (Kim et al., 1993).
Resolving this controversy is of considerable scientific interest, both from a theoretical and an applied perspective. We therefore analyzed the fate of ptDNA in mature, ageing, and senescent leaves of four commonly studied higher plant species (Arabidopsis, sugar beet [Beta vulgaris], tobacco [Nicotiana tabacum], and maize; Figure 1) for which conflicting data have been reported. Four complementary methods were used for assessing the presence of ptDNA as well as its quantitative and morphological changes during leaf development: an improved 4′,6-diamidino-2-phenylindole (DAPI)–based fluorescence microscopy approach including deconvolution of fluorescence images, electron microscopy, semithin sectioning across leaf laminas, and real-time quantitative PCR (see Methods).
Figure 1.
Developmental Leaf Series of Sugar Beet, Tobacco, and Arabidopsis.
(A) Sugar beet leaves, developmental stages II to VI (left to right; see text). Inset: leaf stages y1 and y3. Arrows indicate necrotic areas. Bar = 5 cm.
(B) Tobacco leaves, developmental stages II and IV to VI. Inset: leaf stages y1 and y2. Bar = 5 cm; bar in inset = 1 cm.
(C) Arabidopsis plants (left) from which leaves of developmental stages I to VI were taken. Bar = 4 cm.
Figure 2, Supplemental Methods, and Supplemental Data Sets 1 to 4 present representative micrographs of developmental series of DAPI-stained chloroplasts in leaf spongy parenchyma cells of late ontogenetic stages from sugar beet, Arabidopsis, tobacco, and maize displaying clearly discernible nucleoid patterns. Figures 1A to 1C document some of the leaves from which samples were taken. Mesophyll cells of juvenile leaves investigated in our previous work (Li et al., 2006; Zoschke et al., 2007; Rauwolf et al., 2010) were included for comparison (Supplemental Data Sets 1 to 4, panels 1 to 37, 84 to 94, 112 to 117, and 123 to 128). The staining specificity of the fluorochrome was confirmed enzymatically. Treatment with DNase, but not DNase-free RNase or Proteinase K, either before or after staining with the fluorochrome, abolished the fluorescence but did not significantly affect chloroplast structure (compare with Rauwolf et al., 2010; see Methods).
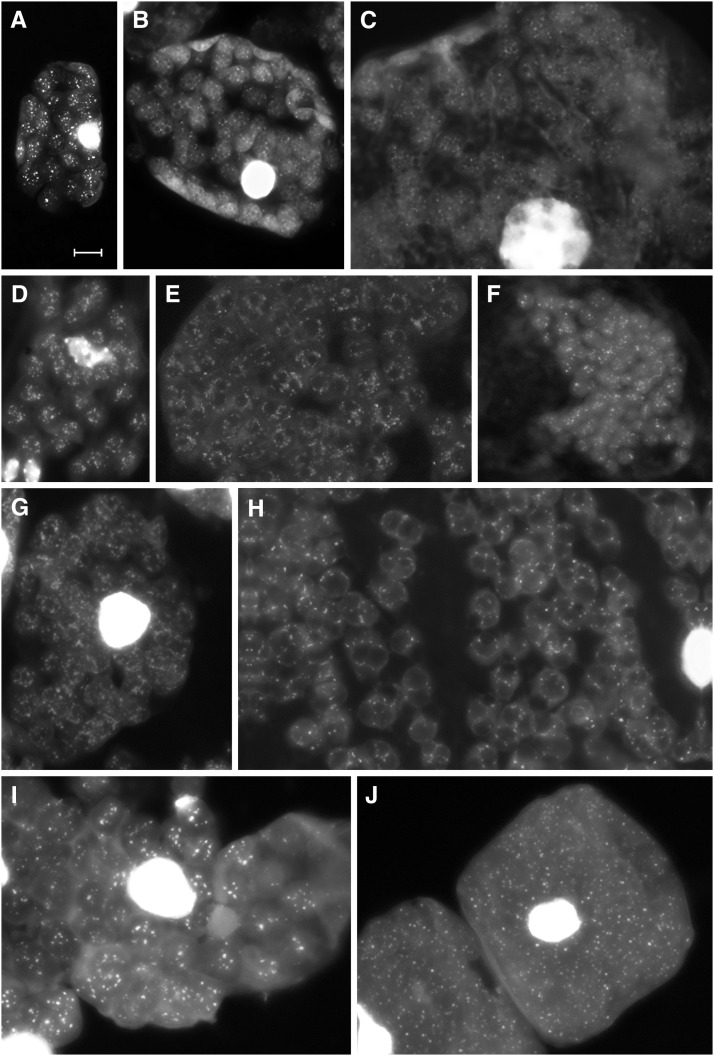
Figure 2.
DAPI-DNA Fluorescence of Mature, Senescent, and Prenecrotic Leaf Mesophyll Cells or Cell Segments.
Representative DAPI-stained squashed mesophyll cells of sugar beet ([A] to [C]), Arabidopsis ([D] to [F]), tobacco ([G] and [H]), and maize ([I] and [J]) leaflets or leaves (cell detail in [C], [E], [F], and [H]) of the developmental stages III/IV (I), IV ([A] and [D]), V ([B], [E], and [G]), and VI ([C], [F], [H], and [J]). Note that (E) represents a cell fragment of Supplemental Data Set 2, panel 102. Bar = 5 μm in (A), also for (B) to (J).
DAPI-DNA FLUORESCENCE PATTERNS IN YOUNG AND MATURE LEAVES
As expected, the images obtained from individual organelles or cells of juvenile material were heterogeneous. During early organelle and cell development, nucleoid numbers and DNA quantity per organelle and cell increased significantly. Nucleoids also varied in DNA content, texture, and location within the chloroplast (Figures 2A, 2D, and 2I; Supplemental Data Sets 1 to 4, panels 1 to 37, 84 to 94, 112 to 117, and 123 to 128; Rauwolf et al., 2010). In mature sugar beet leaves, scattered nucleoids per organelle varied between 15 and >30, the average (based on 782 determinations) being 23.3 ± 5.4. A visual inspection suggests that their fluorescence intensities remained comparable from fully expanded leaves with a glossy surface to older leaves with matt surfaces, indicating unaltered DNA contents per organelle. Similar patterns were observed for Arabidopsis, tobacco, and maize (Supplemental Data Sets 1 to 4, panels 38 to 60, 95 to 101, 118, 119, and 129 to 131; see also Supplemental Methods). In general, there was no significant intensity difference between DNA regions in individual organelles of different sizes or in chloroplasts of cells differing in nuclear ploidy (compare with Supplemental Methods). No indication for a notable loss of DNA in chloroplasts of premature or mature leaves was observed.
DAPI-DNA FLUORESCENCE PATTERNS IN AGEING, SENESCING, AND SENESCENT LEAVES
With the onset of senescence, chloroplasts convert to gerontoplasts. The thylakoid system begins to distort and globular particles, either plastoglobules (in sugar beet and Arabidopsis: Figures 3A and 3B) or starch grains (in tobacco: Figure 3C) appeared (compare with Figures 2E, 2H, and 2I; Supplemental Data Sets 1 to 3, panels 62, 65, 68, 71-83, 96 to 99, and 119 to 122). In spite of profound changes of the organelle architecture, in all four species studied, well-spread nucleoid patterns resembling those of mesophyll chloroplasts of mature and ageing leaves were clearly present in plastids of senescing leaves and of yellow sectors in old leaves (Figures 2B, 2C, 2E to 2H, and 2J; Supplemental Data Sets 1 to 4, panels 61 to 77, 79, 80, 103 to 111, 120 to 122, and 132 to 134). In gerontoplasts of advanced senescent or prenecrotic tissue, DNA fluorescence became more and more diffuse and more difficult to visualize, presumably because of changes in DNA conformation and/or destructive processes in the stroma (Figures 2C, 2H, and 2J; e.g., for sugar beet Supplemental Data Set 1, panels 61, 67 to 71, and 73 to 80). Even in such stages, at least some nucleoids or residual nucleoid structures could still unambiguously be recognized in a substantial fraction of organelles, often surrounding globular particles. In late senescent and near-necrotic leaf tissues, gerontoplasts often decreased in size. In the case of Arabidopsis, plastids finally contained only a single or a few relatively brightly fluorescing nucleoids (Figure 2F; Supplemental Data Set 2, panels 106 to 111). Only in highly necrotic tissue, such as from lamina regions adjacent to brown sectors (Figures 1A and 1C), DAPI staining of plastids frequently failed to produce a distinct spotted pattern (Supplemental Data Set 1, panels 81 to 83). Somewhat diffuse nucleoid-like areas and relatively high diffuse background fluorescence could often be detected, probably reflecting the presence of DNA. The fluorescence was absent from unstained preparations verifying that it was not due to autofluorescence.
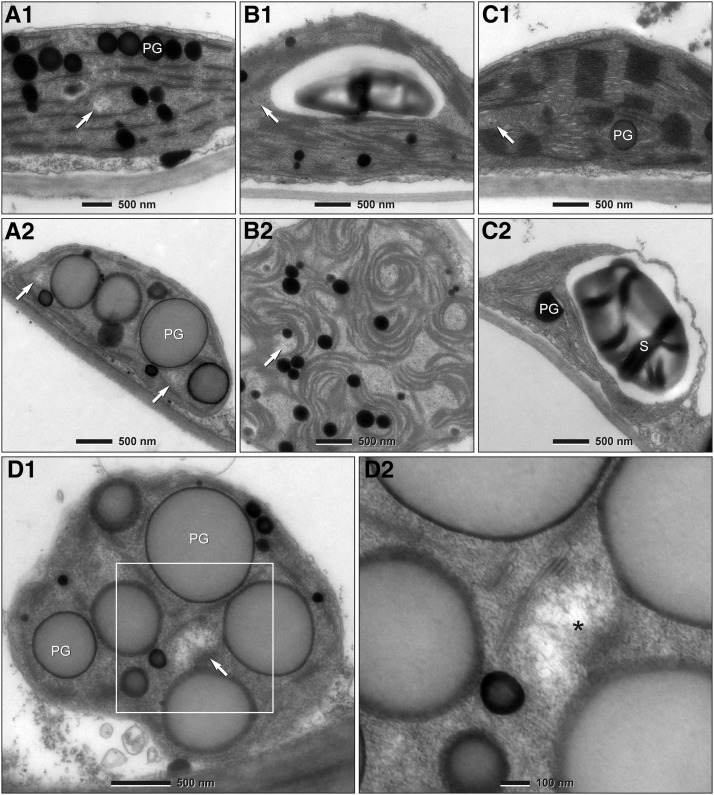
Figure 3.
Transmission Electron Microscopy of Mature and Senescent Leaf Mesophyll Plastids.
Transmission electron micrographs of ultrathin sections of mesophyll chloroplasts of mature ([A1] to [C1]) and gerontoplasts of senescent ([A2] to [D]) leaves of sugar beet ([A] and [D]), Arabidopsis (B), and tobacco (C). The gerontoplast shown in (A2) is from a leaf sector at a relatively early stage of senescence. (D1) shows a gerontoplast of highly senescent mesophyll tissue of sugar beet. Detail at higher magnification ([D2], boxed area of [D1]) demonstrates a typical nucleoplasm (nucleoid; asterisk) in which DNA fibrils are clearly apparent. Arrows indicate DNA-containing areas (nucleoids). PG, plastoglobule; S, starch grain.
Ultrastructural Analysis
In ultrathin sections of chloroplasts, nucleoids with tangled 2.5-nm DNA fibrils usually appear in localized electron-lucid areas within the dense stroma between photosynthetic or near envelope membranes. The ultrastructural analysis of ptDNA in senescent leaves confirmed the data from DAPI-ptDNA fluorescence (Figures 3A to 3C). DNA-containing areas were often found close to thylakoid remnants that frequently formed sheath-like structures around the globules (Figure 3D1). At higher magnifications, the typical 2.5-nm fibrillar structures and clumps of larger diameter became clearly visible in these areas, even within organelles of advanced senescent tissue (Figure 3D). The identification of finely fibrillar nucleoplasms of characteristic appearance in size, shape, and location as well as their abundance in plastid sections with altered thylakoid systems and globular structures provides independent proof of the maintenance of ptDNA until leaf necrosis.
Real-Time Quantitative PCR
The presence and abundance of DNA in plastids were quantitatively evaluated by real-time quantitative (qPCR). In silico analysis was employed to estimate whether the ptDNA copy numbers obtained by real-time qPCR could be adulterated by nuclear copies of plastid DNA (NUPTs). To identify similarities between plastid und NUPT sequences of maize, the amplicon sequences of the three plastid gene fragments used were aligned against the total maize nuclear genome sequence (http://www2.genome.arizona.edu/genomes/maize) using the BLASTN program. Three and six near-exact nuclear copies were found for clpP and psbA, respectively, which are expected to be coamplified with the ptDNA. However, the six NUPTs identified for ndhH are unlikely to be coamplified since their target sequence of the reverse primer is truncated (by seven nucleotides). The ptDNA copy numbers of clpP and psbA gene fragments obtained by real-time qPCR did not differ significantly from those of ndhH. We conclude that promiscuous DNA does not falsify the signal from authentic ptDNA and that the method provides valid data.
In agreement with the histological data, in all samples spanning the developmental stages from premature to advanced senescent tissue, significant quantities of ptDNA could be detected by pPCR. The ploidy-normalized ptDNA levels increased in juvenile material from between 700 and 1200 plastome copies per diploid cell to maximum levels in the order of 2000 copies. This increase happens relatively early in development, i.e., usually before full leaf maturation and before organelles reached their final volume (Figure 4; compare with Li et al., 2006; Zoschke et al., 2007; Rauwolf et al., 2010). It is important to note that real-time qPCR determines average copy numbers over all cell types present in a leaf including green and nongreen cells. Thus, somewhat higher ptDNA copy numbers are obtained when only green (mesophyll cells) are examined (Rowan et al., 2009; Rauwolf et al., 2010; Liere and Börner, 2013).
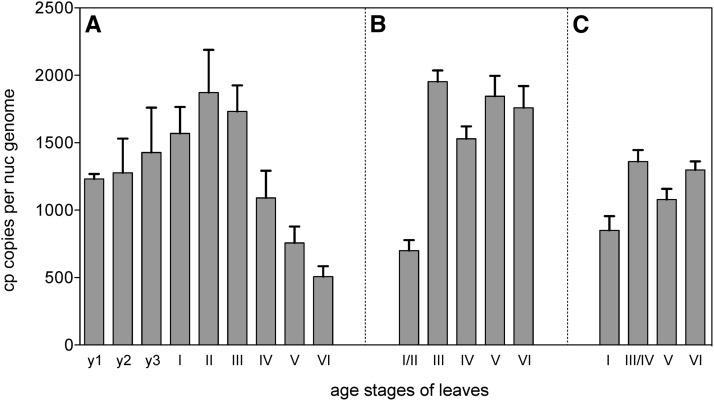
Figure 4.
Plastome Copy Numbers per Cell during Leaf Development.
Sugar beet (A), tobacco (B), and maize (C). For determination of copy numbers, three chloroplast genes (clpP, ndhH, and psbA) were measured by real-time qPCR using total cellular DNA as template against nuclear reference genes (see Methods). The graphs show means and standard deviations from nine measurements per leaf sample (three replicates per gene). To obtain total chloroplast genome copy numbers, average ploidy levels of 3C ([A] and [C]) and 4C (B) were assumed. For Arabidopsis, see Zoschke et al. (2007).
The patterns in ageing and senescent material differed between the species investigated. Consistent with our previous findings (Li et al., 2006; Zoschke et al., 2007), ptDNA levels remained relatively constant until leaf necrosis in Arabidopsis, tobacco, and maize (Figures 4B and 4C), but dropped to ∼700 plastome copies per cell during ageing of sugar beet leaves (Figure 4A). At first glance, this decline contrasts with our fluorescence microscopic data that indicated little change in both organelle sizes and DNA quantity per organelle during late leaf development (Figures 2B, 2C, and 3A1 versus 3A2; Supplemental Data Set 1, panels 61 to 74). Semithin sections of appropriate leaf tissue samples and inspection of cellular plastid numbers in several hundred sections resolved this seeming contradiction in sugar beet. It appeared that the ptDNA decline during senescence was correlated with an ∼60% reduction of plastid numbers per cell (Supplemental Figures 1A, 2, and 3) and not caused primarily by DNA degradation in organelles as reported by Tymms et al. (1983). In Arabidopsis, tobacco, and maize, a significant decrease of ptDNA was found only very late, in tissue neighboring necrotic leaf sectors (Figures 1A, 1C, and 2F; compare with real-time qPCR of Rowan et al., 2009; Evans, et al., 2010).
ptDNA Is Ontogenetically Stable
With the combination of independent approaches employed in this study, we find consistently and unmistakably that the organelle DNA is readily detectable during late stages of leaf development until necrosis. Complete or near-complete loss of ptDNA was not observed in any of the species studied. Virtually all gerontoplasts of senescent and even near-necrotic cells possessed significant amounts of DNA dispersed in scattered nucleoids, which appears as the predominant kind of plastome arrangement in the organelle (Figures 2B, 2C, 2E, 2F to 2H, and 2J; Supplemental Data Sets 1 to 4, panels 61 to 80, 103 to 111, 120 to 122, and 132 to 134). The enormous conversion of plastid structures during gerontoplast formation (Figure 3) clearly preceded the degradation of ptDNA and not vice versa, which excludes the possibility that DNA degradation triggers senescence (compare with Sodmergen et al., 1991).
Our findings cannot be reconciled with those of Bendich and coworkers, who have claimed that ptDNA sharply declines or even vanishes completely from chloroplasts long before the onset of senescence in several species, notably including Arabidopsis and maize (Oldenburg and Bendich, 2004; Rowan et al., 2004, 2009; Oldenburg et al., 2006; Shaver et al., 2006). The reasons for these differences are not entirely clear, but in our opinion, the reported findings are not conclusive because of artifact-prone methods and the lack of appropriate controls checking the biochemistry used. In addition to the concerns previously raised by Li et al. (2006), three examples may illustrate this. First, undetectability of stainable DNA, even if the plastids were not treated with DNase during the isolation procedure, is not a valid criterion per se to postulate the absence of DNA or to assess nature and impact of changes of in-gel DNA structures remaining after lysis of embedded chloroplasts (Oldenburg and Bendich, 2004; Rowan et al., 2004, 2009; Oldenburg et al., 2006; Shaver et al., 2006). Besides possible technical problems with insufficient dye penetration (Selldén and Leech, 1981; Evans et al., 2010), leaf tissue, especially if more mature, is known to contain endogenous nucleases that can be sufficiently active to destroy accessible DNA in organelle preparations (Selldén and Leech, 1981). This can affect not only nuclear DNA and DNA of broken chloroplasts, but also DNA of morphologically intact plastids with leaky envelopes (Atchison et al., 1976). Moreover, apart from the fact that mature and aging tissues often accumulate secondary metabolites that are released during homogenization and may adversely affect envelopes, the two kinds of homogenization buffers used by the authors are physiologically imbalanced. The added polyvinyl pyrrolidone (Oldenburg and Bendich, 2004) is known to act as a weak detergent and bears the risk of altering envelopes that may become leaky. High-salt buffer >100 mM (1.25 M NaCl was used by Shaver et al., 2006), in turn, injures and even destroys envelope membranes. Second, ptDNA in senescing and necrotic tissue appears to be more difficult to visualize by DAPI. The failure of Bendich and coworkers to observe DNA staining in most chloroplasts of mature and senescent mesophyll cells in situ, in addition to possible dye penetration problems (Selldén and Leech, 1981; Evans et al., 2010), could also be caused by ploidy reduction of nucleoids with leaf ageing (Figures 2C and 2J; Supplemental Data Sets 1, 2, and 4, panels 42, 52 to 55, 61, 63, 100, 103, 133, and 134, and Supplemental Methods) and DNA dispersion with progressing senescence (Figures 2C, 2E, and 2H; Supplemental Data Set 1, panels 61 and 65 to 80). Although DAPI is capable of tracing relative DNA quantities down to a single plastid chromosome (James and Jope, 1978), this sensitivity may only be reached when the DNA is in a sufficiently condensed state. Original prints of an appropriate DAPI-stained leaf control in situ were not included in Rowan et al. (2004), and those presented in Oldenburg and Bendich (2004), Shaver et al. (2006), or Rowan et al. (2009) do not resolve sufficient detail (compare with James and Jope, 1978; Selldén and Leech, 1981; Fujie et al., 1994; Rauwolf et al., 2010). To avoid artifacts, we have taken various precautions. In addition to specific fixation (see Methods) and tissue maceration (Rauwolf et al., 2010), the applied deconvolution of fluorescence images is equivalent to confocal imaging and takes advantage of a fast combination of records from different focal planes. It is superior in resolution to conventional pictures taken from only a single plane. The fluorescence microscopy images of macerated cells and flattened organelles taken with the advanced technology clearly displayed the stained nucleoid patterns as intrinsic elements of the organelle architecture (Figure 2; Supplemental Data Sets 1 to 4). Third, the real-time qPCR data reported by Bendich and coworkers are at variance with their findings obtained by DAPI staining, in that these did not indicate as dramatic a ptDNA loss during chloroplast development (compare with Rowan et al., 2009). This discrepancy has been attributed to the coamplification of NUPTs (Kumar and Bendich, 2011; Zheng et al., 2011). However, in the case of maize, for which total genomic sequence data are available, our analyses show that a few NUPTs (versus more than 1000 authentic ptDNA copies) have no significant influence on the estimated copy numbers per cell. Recent findings about the role of the maize white2 gene mutation on plastid genome copy numbers, which showed that the fraction of the total signal in real-time qPCR measurements that could arise from NUPTs is in the order of 1% (Udy et al., 2012), corroborate our findings. In Arabidopsis, the impact of coamplification of NUPTs is expected to be even lower because of the significantly lower abundance of NUPTs in the genome (Smith et al., 2011) and the possibility of selecting primers (on the basis of complete genome sequence information) that will not coamplify nuclear DNA sequences (Zoschke et al., 2007).
In summary, the observation that all four studied species (spanning a reasonably wide range of angiosperm evolution) were found to retain substantial amounts of ptDNA even in senescent tissue demonstrates that the DNA of the organelle is ontogenetically stable. These results suggest that its preservation during late development reflects a general phenomenon, at least in seed plants.
METHODS
Plant Material
Field-grown diploid sugar beet (Beta vulgaris var Felicita) samples were collected shortly before harvest early October. The plants studied carried between 20 and 30 rosette leaves with >1.5-cm lamina length. Approximately half of them could be classified as developing leaves and 30 to 40% as fully developed leaves, which, in turn, can operationally be grouped into three categories: leaves with glossy surface, matt surface, and beginning senescent areas, respectively (Figure 1A). About 15% of the leaves were senescent or necrotic (Figure 1A). The beet leaves were grouped into six distinct developmental classes ranging in sizes between 6 and 8 cm (fraction I) and 10 to 30 cm (II to VI), depending on the individual plant. Fraction II consisted of maturing leaves, fraction III of nearly mature leaves, both with a glossy surface, fraction IV of older leaves usually with a matt surface, and fractions V and VI contained leaves with varying degrees of senescence and necrotic sectors (Figure 1A). In some instances, additional samples were taken, including younger leaves from 2.5 to 6 cm in size (designated y1 to y4; Figure 1A, inset) with developmental stages overlapping with those studied previously (y1 to y4 correspond to leaf fractions III and IV in Rauwolf et al., 2010).
Tobacco (Nicotiana tabacum var Petit Havana) and maize (Zea mays var H99) were grown in a greenhouse as previously described (Li et al., 2006). The parenchymous tissue was largely depleted of transitory starch by placing plants in darkness for 24 to 48 h before use. Samples were taken from leaves of six developmental stages from 8- to 10-week-old plants with up to ∼20 leaves (Figure 1B); leaves between numbers 11 and 14 with ∼30-cm lamina length were usually fully expanded. Four developmentally different sectors (fractions I/II and III/IV to VI), with sample VI taken from yellow-green mottled, (i.e., senescent) areas, were excised from maize leaves.
Arabidopsis thaliana (ecotype Columbia-0) was grown on Vermiculite/soil mix (1:4) under an 8-h/16-h light/dark cycle at 20°C (Zoschke et al., 2007). After 14 d, the seedlings were exposed to a light regime of 16-h light/8-h darkness. The light intensity was 150 μE s−1 m−2. Samples varying in age and progression of senescence were taken from leaves of six developmental stages from 15-d-old seedlings (I), 30-d-old (II and III, young and old, respectively), and 55-d-old (IV to VI, young, old, and highly senescent) plants (Figure 1C).
Microscopy
Different fixatives were employed for light microscopy and electron microscopy work. The fixative chosen for fluorescence microscopy locations of nucleoids enhanced DNA compactness to generate spot-like emission sources (Herrmann, 1970) because at a given base composition, the visibility of nucleoids is influenced not only by DNA content, but also by DNA conformation (Ris and Plaut, 1962; Coleman, 1978). On the other hand, the fixatives used for ultrastructural investigations revealed the finely fibrillar histone-free nucleoplasm of organelles (Figure 3), which is only preserved under certain conditions (Ryter et al., 1958).
For light microscopy, explants of ∼0.3 × 0.3 cm from central laminal leaf regions (sugar beet, tobacco, and maize) or entire leaves (Arabidopsis) varying in age and degree of senescence or necrosis were fixed for at least 20 h after a brief vacuum infiltration in either 5% buffered, weakly hypotonic formaldehyde (0.1 M Suc, 20 mM Tris/HCl, and 2 mM EDTA, pH 7.2) or 2.5% glutaraldehyde in 50 mM cacodylate buffer, pH 7.2 (Rauwolf et al., 2010). The fixative was replaced by several changes with plain buffer. Enzyme treatments of explants, DAPI staining, and visualization of stained ptDNA were performed essentially as described by Rauwolf et al. (2010). DNA staining specificity of DAPI was enzymatically verified. DNase treatments (10 μg/mL for 2 h at room temperature), either before or after staining, removed fluorescence. DNase-free RNase (20 μg/mL for 2 h at room temperature) or Proteinase K (1 mg/mL for 3 h at 25°C) had no effect on DAPI fluorescence. All treatments did not significantly affect chloroplast structure (compare with Rauwolf et al., 2010).
For electron microscopy, pieces of leaf tissue were fixed immediately with 2.5% glutaraldehyde in 75 mM sodium cacodylate and 2 mM MgCl2, pH 7.0, for 1 h at room temperature, rinsed several times in fixative buffer, and postfixed for 2 h with 1% osmium tetroxide in fixative buffer at room temperature. After two washing steps in distilled water, the cells were stained en bloc with 1% uranyl acetate in 20% acetone for 30 min. Dehydration was performed with a graded acetone series. Samples were then infiltrated and embedded in Spurr’s low-viscosity resin (Spurr, 1969). After polymerization, ultrathin sections of thicknesses between 50 and 70 nm were cut with a diamond knife and mounted on uncoated copper grids. The sections were poststained with aqueous lead citrate (100 mM, pH 13.0). Micrographs were taken with an EM 912 electron microscope (Zeiss) equipped with an integrated OMEGA energy filter operated in the zero loss mode. Semithin sections were cut with a diamond knife at a thickness of 500 nm and mounted on glass slides. Phase contrast micrographs were taken with a Zeiss Axiophot microscope and a Nikon D1000 digital camera.
Real-Time qPCR
qPCR was conducted as described by Zoschke et al. (2007). Three chloroplast genes, clpP, psbA, and ndhH, topographically well spread on the plastid chromosome, were chosen to determine plastome copy numbers per cell. As nuclear reference genes, PhyA (accession number GU057342.1) was chosen for sugar beet, RpoT1 (RpoTm) (accession number AF127021.1) for maize, and RpoT2-syl (accession number AJ416574.1) for tobacco. The primers used in real-time qPCR analyses are listed in Supplemental Table 1. Copy numbers of ptDNA per cell were normalized by multiplying the relative copy numbers of ptDNA as obtained by real-time qPCR by the nuclear ploidy level. An average ploidy level of 3C was assumed for maize (Oldenburg et al., 2006) and sugar beet (Butterfass, 1979) and of 4C for tobacco.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Representative Examples of Phase Contrast Light Micrographs of Semithin Cross Sections of Leaf Sectors of Sugar Beet, Arabidopsis thaliana, and Tobacco.
Supplemental Figure 2. Representative Examples of Phase Contrast Micrographs of Semithin Cross Sections of Sugar Beet Leaves.
Supplemental Figure 3. Representative Examples of Nomarski Interference Contrast Micrographs of Leaf Sectors of Sugar Beet.
Supplemental Table 1. Oligonucleotides Used for qPCR.
Supplemental Methods. Aspects of the Enormous Structural/Quantitative Complexity and Variability of Plastids and Their DNA during Leaf Development.
Supplemental Data Set 1. DAPI-Stained Leaf Mesophyll Cells of Beta vulgaris (Sugar Beet).
Supplemental Data Set 2. DAPI-Stained Leaf Mesophyll Cells of Arabidopsis thaliana.
Supplemental Data Set 3. DAPI-Stained Leaf Mesophyll Cells of Nicotiana tabacum (Tobacco).
Supplemental Data Set 4. DAPI-Stained Leaf Mesophyll Cells of Zea mays (Maize).
Supplementary Material
Acknowledgments
We thank Silvia Dobler and Jennifer Grünert for excellent technical assistance. We are grateful to the Max-Planck-Institut für Molekulare Pflanzenphysiologie Green Team for plant cultivation and to Josef Bergstein (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for expert photographic service. This work was supported by the Deutsche Forschungsgemeinschaft to R.G.H. (SFB TR1 and He 693), to T.B., R.B., and A.W. (SFB 429), by the Catholic University of Lublin to H.G., and by the Max Planck Society to R.B. and S.G.
AUTHOR CONTRIBUTIONS
R.G.H., T.B., and S.G. designed research. H.G., G.W., A.W., and S.G. performed experiments. R.G.H., H.G., G.W., A.W., S.G., T.B., and R.B. analyzed data. R.G.H. wrote most of the article. S.G., T.B., and R.B. participated in writing.
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Atchison B.A., Whitfeld P.R., Bottomley W. (1976). Comparison of chloroplast DNA by specific fragmentation with EcoRI endonuclease. Mol. Gen. Genet. 148: 263–269. [Google Scholar]
- Baumgartner B.J., Rapp J.C., Mullet J.E. (1989). Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol. 89: 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfass T. (1979). Patterns of chloroplast reproduction. A developmental approach to protoplasmic plant anatomy. In Cell Biology Monographs. Continuation of Protoplasmatologie, Vol. 6. L.V. Heilbrunn, W. Beermann, and G. Rudkin, eds (Wien, New York: Springer), pp. 1–205. [Google Scholar]
- Coleman A.W. (1978). Visualization of chloroplast DNA with two fluorochromes. Exp. Cell Res. 114: 95–100. [DOI] [PubMed] [Google Scholar]
- Evans I.M., Rus A.M., Belanger E.M., Kimoto M., Brusslan J.A. (2010). Dismantling of Arabidopsis thaliana mesophyll cell chloroplasts during natural leaf senescence. Plant Biol. (Stuttg.) 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie M., Kuroiwa H., Kawano S., Mutoh S., Kuroiwa T. (1994). Behaviour of organelles and their nucleoids in the shoot apical meristem during leaf development in Arabidopsis thaliana L. Planta 194: 395–405. [DOI] [PubMed] [Google Scholar]
- Herrmann R.G. (1970). Multiple amounts of DNA related to the size of chloroplasts : I. An autoradiographic study. Planta 90: 80–96. [DOI] [PubMed] [Google Scholar]
- Herrmann R.G., Possingham J.V. (1980). Plastid DNA: The plastome. In Results and Problems in Cell Differentiation. Chloroplasts, Vol. 10, J. Reinert, ed (Berlin, Heidelberg, New York: Springer), pp. 45–96. [DOI] [PubMed] [Google Scholar]
- James T.W., Jope C. (1978). Visualization by fluorescence of chloroplast DNA in higher plants by means of the DNA-specific probe 4′6-diamidino-2-phenylindole. J. Cell Biol. 79: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Christopher D.A., Mullet J.E. (1993). Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol. Biol. 22: 447–463. [DOI] [PubMed] [Google Scholar]
- Kowallik K.V., Herrmann R.G. (1972). Variable amounts of DNA related to the size of chloroplasts. IV. Three-dimensional arrangement of DNA in fully differentiated chloroplasts of Beta vulgaris L. J. Cell Sci. 11: 357–377. [DOI] [PubMed] [Google Scholar]
- Kumar R.A., Bendich A.J. (2011). Distinguishing authentic mitochondrial and plastid DNAs from similar DNA sequences in the nucleus using the polymerase chain reaction. Curr. Genet. 57: 287–295. [DOI] [PubMed] [Google Scholar]
- Lamppa G.K., Bendich A.J. (1979). Changes in chloroplast DNA levels during development of pea (Pisum sativum). Plant Physiol. 64: 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ruf S., Bock R. (2006). Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol. Genet. Genomics 275: 185–192. [DOI] [PubMed] [Google Scholar]
- Liere K., Börner T. (2013). Development-dependent changes in the amount and structure of plastid DNA. In Chloroplast Development during Leaf Growth and Senescence. Advances in Photosynthesis and Respiration, Vol. 36, B. Biswal, K. Krupinska, and U.C. Biswal, eds (Dordrecht, The Netherlands: Springer), pp. 215–237. [Google Scholar]
- Oldenburg D.J., Bendich A.J. (2004). Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J. Mol. Biol. 344: 1311–1330. [DOI] [PubMed] [Google Scholar]
- Oldenburg D.J., Rowan B.A., Zhao L., Walcher C.L., Schleh M., Bendich A.J. (2006). Loss or retention of chloroplast DNA in maize seedlings is affected by both light and genotype. Planta 225: 41–55. [DOI] [PubMed] [Google Scholar]
- Prasil O., Adir N., Ohad I. (1992). Dynamics of photosystem II: Mechanism of photoinhibition and recovery processes. In Topics in Photosynthesis, Vol. 11, J. Barber, ed (Amsterdam: Elsevier), pp. 295–348. [Google Scholar]
- Rauwolf U., Golczyk H., Greiner S., Herrmann R.G. (2010). Variable amounts of DNA related to the size of chloroplasts III. Biochemical determinations of DNA amounts per organelle. Mol. Genet. Genomics 283: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris H., Plaut W. (1962). Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 13: 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan B.A., Oldenburg D.J., Bendich A.J. (2004). The demise of chloroplast DNA in Arabidopsis. Curr. Genet. 46: 176–181. [DOI] [PubMed] [Google Scholar]
- Rowan B.A., Oldenburg D.J., Bendich A.J. (2009). A multiple-method approach reveals a declining amount of chloroplast DNA during development in Arabidopsis. BMC Plant Biol. 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter A., Kellenberger E., Birchandersen A., Maaløe O. (1958). Etude au microscope électronique de plasmas contenant de l’acide désoxyribonucliéique. I. Les nucléoides des bactéries en croissance active. Z. Naturforsch. B 13B: 597–605. [PubMed] [Google Scholar]
- Scott N.S., Possingham J.V. (1980). Chloroplast DNA in expanding spinach leaves. J. Exp. Bot. 31: 1081–1092. [Google Scholar]
- Selldén G., Leech R.M. (1981). Localization of DNA in mature and young wheat chloroplasts using the fluorescent probe 4′-6-diamino-2-phenylindole. Plant Physiol. 68: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver J.M., Oldenburg D.J., Bendich A.J. (2006). Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224: 72–82. [DOI] [PubMed] [Google Scholar]
- Smith D.R., Crosby K., Lee R.W. (2011). Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol. Evol. 3: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodmergen X.X., Kawano S., Tano S., Kuroiwa T. (1991). Degradation of chloroplast DNA in second leaves of rice (Oryza sativa) before leaf yellowing. Protoplasma 160: 89–98. [Google Scholar]
- Spurr A.R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26: 31–43. [DOI] [PubMed] [Google Scholar]
- Tymms M.J., Scott N.S., Possingham J.V. (1983). DNA content of Beta vulgaris chloroplasts during leaf cell expansion. Plant Physiol. 71: 785–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy D.B., Belcher S., Williams-Carrier R., Gualberto J.M., Barkan A. (2012). Effects of reduced chloroplast gene copy number on chloroplast gene expression in maize. Plant Physiol. 160: 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Oldenburg D.J., Bendich A.J. (2011). Independent effects of leaf growth and light on the development of the plastid and its DNA content in Zea species. J. Exp. Bot. 62: 2715–2730. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Liere K., Börner T. (2007). From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50: 710–722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.