SUMMARY
Soluble guanylate cyclase (sGC) is the primary mediator of nitric oxide (NO) signaling. NO binds the sGC heme cofactor stimulating synthesis of the second messenger cyclic-GMP (cGMP). As the central hub of NO/cGMP signaling pathways, sGC is important in diverse physiological processes such as vasodilation and neurotransmission. Nevertheless, the mechanisms underlying NO-induced cyclase activation in sGC remain unclear. Here, hydrogen/deuterium exchange mass spectrometry (HDX-MS) was employed to probe the NO-induced conformational changes of sGC. HDX-MS revealed NO-induced effects in several discrete regions. NO binding to the heme-NO/O2-binding (H-NOX) domain perturbs a signaling surface implicated in Per/Arnt/Sim (PAS) domain interactions. Furthermore, NO elicits striking conformational changes in the junction between the PAS and helical domains that propagate as perturbations throughout the adjoining helices. Ultimately, NO-binding stimulates the catalytic domain by contracting the active site pocket. Together, these conformational changes delineate an allosteric pathway linking NO-binding to activation of the catalytic domain.
INTRODUCTION
Nitric oxide (NO) is a diatomic free radical gas that serves as a signaling molecule with diverse physiological roles. NO signaling controls processes in the cardiovascular, neuronal, and gastrointestinal systems (Friebe and Koesling, 2009; Garthwaite, 2008). The primary receptor for NO is soluble guanylate cyclase (sGC). NO binds to the heme cofactor of the sGC sensor domain, stimulating cyclization of GTP to the second messenger cyclic-GMP (cGMP) (Derbyshire and Marletta, 2012; Underbakke et al., 2013b). Elevated cGMP levels trigger complex downstream signaling cascades. Disruptions in the NO/sGC/cGMP signaling pathway have been linked to heart disease, stroke, hypertension, erectile dysfunction, and neurodegeneration (Bredt, 1999; Friebe et al., 2007). Accordingly, sGC is a notable therapeutic target for drug development (Follmann et al., 2013; Stasch et al., 2011).
The predominant sGC isoform is a heterodimer of homologous α1 and β1 subunits (Figure 1). The β1 subunit senses NO via the N-terminal “heme-NO/O2-binding” (H-NOX) domain. The pseudo-H-NOX of the α1 subunit does not bind heme, and its function remains unknown. Both subunits contain Per/Arnt/Sim (PAS) and helical domains responsible for heterodimerization and signal transmission (Ma et al., 2008; Rothkegel et al., 2007; Underbakke et al., 2013a). The C-termini of the α1 and β1 subunits constitute the catalytic domain with the cyclase active site formed at the subunit interface (Winger and Marletta, 2005). While the structure of the sGC holoenzyme remains unknown, structures have been reported for many sGC domain truncations and homologs (Allerston et al., 2013; Ma et al., 2010; Ma et al., 2007; Purohit et al., 2013). Significant progress toward reconstructing the sGC architecture has recently emerged from diverse approaches (Campbell MG, ESU, Potter CS, Carragher B, MAM, submitted)(Fritz et al., 2013; Underbakke et al., 2013a).
Figure 1. Domain organization of sGC subunits and truncations.
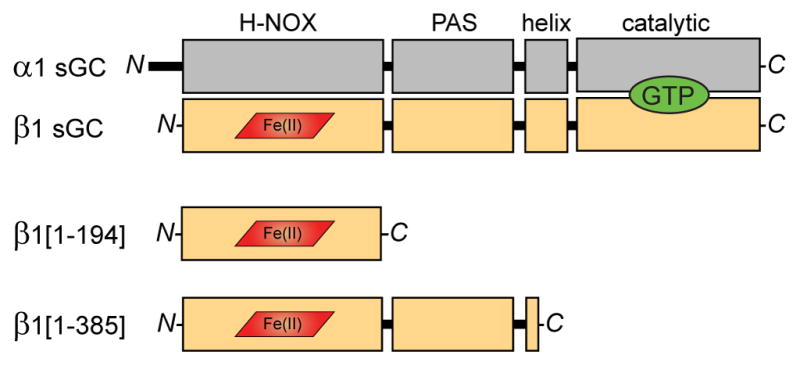
The sGC holoenzyme consists of two homologous subunits, α1 (gray) and β1 (tan), each composed of four domains. NO binds to the ferrous heme cofactor (red) of the β1 H-NOX domain. Cyclization of GTP (green) to cGMP occurs in the C-terminal catalytic domains. Truncations of the β1 sGC subunit preserve NO-binding. β1[1-194] comprises the minimal heme-binding H-NOX domain. β1[1-385] includes the associated PAS domain and a portion of the helical domain.
The molecular mechanisms underlying NO activation of sGC remain a central question in NO/cGMP signaling. NO binds with high affinity (pM) to the ferrous heme of the H-NOX domain. Formation of the Fe2+–NO bond induces cleavage of the bond between the heme iron and the axial histidine ligand, initiating a conformational change that ultimately stimulates cyclase activity within the catalytic domain. In addition, extra equivalents of NO are known to super-activate sGC though cysteine interaction (Cary et al., 2005; Fernhoff et al., 2009; Russwurm and Koesling, 2004). However, the mechanism of sGC allosteric control is largely unknown.
Hydrogen/deuterium exchange mass spectrometry (HDX-MS) is a powerful approach for mapping protein folding, interactions, and conformational changes (Engen, 2009; Englander, 2006; Konermann et al., 2011). HDX-MS reports on the local chemical environment of the protein backbone by monitoring the exchange rate of peptide bond amide protons with the deuterons of a D2O solvent. Exchange rates are particularly sensitive to changes in hydrogen bonding, secondary structure, solvent accessibility, and dynamics. As such, HDX-MS is ideally-suited to interrogating conformational changes (Iacob et al., 2009; Landgraf et al., 2013; Pan et al., 2011; West et al., 2013). Here, we report the results of HDX-MS studies exploring the NO-induced conformational changes of sGC. NO-binding perturbs a contiguous H-NOX signaling surface implicated in PAS-domain interactions. Furthermore, strong NO-induced effects in the PAS and helical domains reveal a central role for this junction in allosteric communication. Ultimately, the conformational change stimulates the catalytic domain by condensing the active site pocket.
RESULTS
NO-induced conformational changes in the H-NOX sensor domain
NO stimulation of sGC cyclase activity originates in the H-NOX sensor domain. Formation of the heme Fe2+–NO bond releases the axial histidine ligand, instigating the conformational change that is ultimately communicated to the catalytic domain. Accordingly, we began our investigations into the local conformational effects of axial histidine release by focusing on the isolated H-NOX domain. Two well-characterized sGC H-NOX truncations preserve important ligand-binding and spectroscopic features of full-length sGC (Figure 1). β1[1-194] comprises the minimal, monomeric H-NOX domain (Karow et al., 2005). The homodimeric β1[1-385] construct includes the PAS domain and a segment of the helical domain (Zhao and Marletta, 1997).
To assess NO-induced conformational changes in the minimal H-NOX domain, HDX-MS was employed to compare differences in dynamics and solvent accessibility between unligated and NO-bound β1[1-194]. Loss of secondary structure (i.e., hydrogen bonding), greater dynamics, and increased solvent exposure manifest as increased exchange rates. Conversely, regional stabilization or burial exhibit local decreases in exchange rates. NO-induced changes in exchange rate were color-coded and mapped to the β1[1-194] primary sequence (Figure 2A).
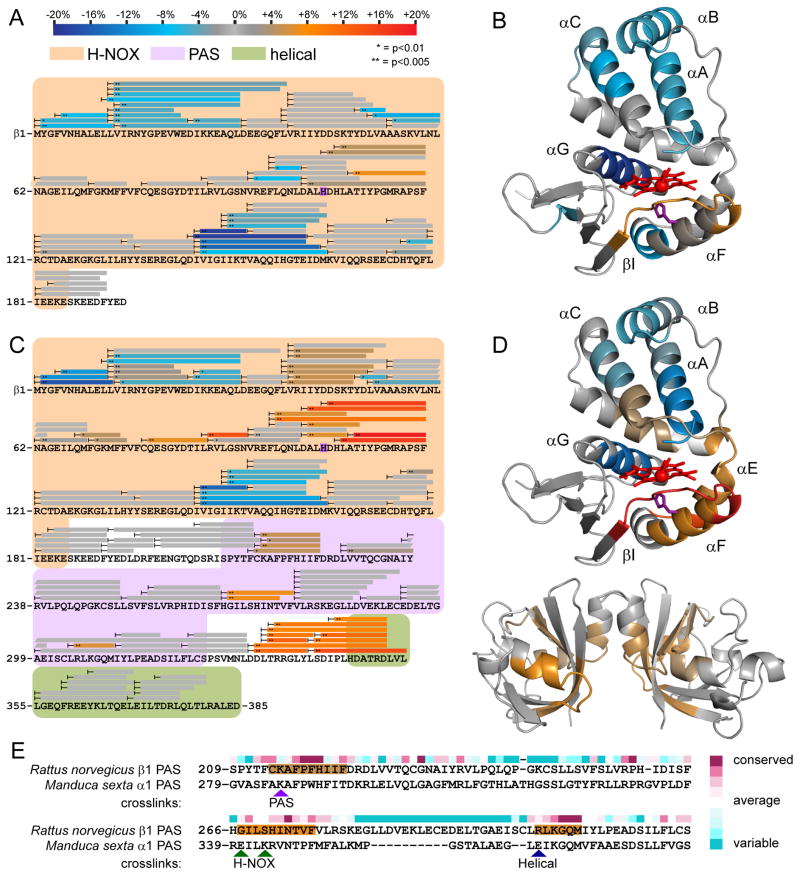
Figure 2. NO-induced conformational changes of sGC H-NOX domain constructs.
Peptide coverage from HDX-MS analysis is represented by bars overlaying sequence. Peptides exhibiting significant NO-induced exchange rate changes are color-coded according to the scale bar. Exchange rate changes are reported as the averaged difference in percent deuterium incorporation (Δ%D) over the actively exchanging portion of the time course. Asterisks denote the results of two-tailed, unpaired Student’s t-tests. Peptides exhibiting non-significant or undetectable changes are colored gray.
(A) Changes in HD exchange kinetics induced by NO-binding to β1[1-194].
(B) Color-coded HDX-MS results for NO-induced changes in β1[1-194] were mapped to a homology model derived from the structure of a bacterial H-NOX (PDB: 2o09) (Ma et al., 2007). Heme (red) and the axial heme ligand His-105 (purple) are represented as sticks.
(C) Changes in HD exchange kinetics induced by NO-binding to β1[1-385].
(D) Color-coded HDX-MS results for NO-induced changes in β1[1-385] were mapped to the sGC β1 H-NOX model (top) and a β1 PAS domain homodimer model (bottom). The β1 PAS domain was modeled using the Robetta server and dimerized by alignment to the structure of a homologous bacterial PAS dimer (PDB: 2P04) (Kim et al., 2004; Ma et al., 2008).
(E) Comparison of NO-induced PAS domain changes with inter-domain contacts identified by crosslinking. Crosslinks identified between the α1 PAS domain of M. sexta sGC and the indicated domains are highlighted with arrowheads(Fritz et al., 2013). NO-induced perturbations identified by HDX-MS are color-coded as in (A) and mapped to the sequence of the rat β1 PAS. Residue conservation (above sequence) of β1 sGC was scored and color-coded using the ConSurf server (Ashkenazy et al., 2010).
Several regions of β1[1-194] exhibited significant differences in exchange rate upon NO-binding (Figure 2B). The N-terminal helices (αA-C) forming the H-NOX lobe above the heme pocket exhibited pronounced decreases in exchange rate with NO-bound. The observed decreases in exchange rates are consistent with a general stabilization of the N-terminal lobe upon NO-binding. Likewise, the α-helix (αG) at the back of the heme pocket was much more resistant to exchange with NO bound. Again, NO-binding appears to confer greater stability to the helical structure of this element of the heme pocket wall.
NO-binding also impacts the heme-associated helix (αF) that harbors His-105, the axial heme ligand freed by NO-binding. The core of the heme-associated helix undergoes little net perturbation. The N-terminal periphery of the helix, however, experiences slower exchange near the rear of the heme pocket. In contrast, the linker (αF–βI) forming the lip of the heme pocket undergoes significant increased exchange, consistent with increased dynamics. Generally, the NO-induced perturbations to the heme-associated helix are subtle in the isolated H-NOX.
PAS domain mediates NO-induced H-NOX conformational changes
Previous investigations focusing on sGC domain interactions revealed close associations between the H-NOX heme-associated helix and PAS domains (Underbakke et al., 2013a). We reasoned that inclusion of the PAS domain may modulate the NO-induced conformational effects on the heme-associated helix. Accordingly, HDX-MS was used to analyze the NO-induced conformational changes of the sGC H-NOX—PAS truncation, β1[1-385]. NO-induced exchange rate differences were color-coded and mapped to the β1[1-385] sequence (Figure 2C).
Several regions of β1[1-385] exhibited effects comparable to β1[1-194] upon NO-binding. The helices (αA-C) of the N-terminal lobe are similarly stabilized in both the isolated H-NOX and H-NOX—PAS truncations (Figure 2D). In addition, the helix constituting the back of the heme pocket (αG) exchanges more slowly with NO bound in both truncations.
Interestingly, inclusion of the PAS domain amplifies several other effects of NO binding, especially around the H-NOX heme pocket. NO-binding elicited modest increases in exchange rate in the “upper” lip (αB-αC linker) of the heme-binding pocket of β1[1-385], whereas no change was evident in β1[1-194] (Figure 2B, D). The heme-associated helix (αF) and neighboring heme pocket lip (αF—BI linker) undergo dramatic NO-induced exchange rate increases in β1[1-385]. In addition, the “hinge” (αE) connecting the N- and C-terminal lobes of the β1[1-385] H-NOX exhibited significantly greater exchange with NO bound. These NO-induced increases in exchange rate are also unique to the PAS-containing truncation, as evidenced by the muted effects in β1[1-194]. Notably, previous work implicated the contiguous surface formed by the signaling helix, heme pocket lip, and hinge as the primary region exhibiting slowed exchange (i.e., burial and hydrogen-bonding) due to PAS domain interactions (Underbakke et al., 2013a). Taken together, NO-binding increases accessibility and dynamics of the heme-associated helix and neighboring surfaces. The strong influence of the PAS domain implies that this conformational change involves opening or relaxation of the H-NOX—PAS domain interface.
HDX-MS analysis of the effects of NO binding on β1[1-385] also revealed conformational changes within the PAS and helical domains. The β1[1-385] PAS domain mediates non-physiological homodimerization, likely due to its homology to the α1 PAS. Here, the HDX-MS results were mapped to a model of a β1 PAS homodimer (Figure 2D). However, the NO-induced PAS effects may be asymmetric in the full-length α1β1 heterodimer. Three discrete regions of the PAS domain exhibit increased exchange rates in NO-bound β1[1-385], each corresponding to relatively conserved segments (Figure 2C, E). Comparisons with a recent crosslinking study using Manduca sexta (tobacco hornworm) sGC provide intriguing context for the localization of the NO-induced effects (Fritz et al., 2013). Indeed, the three NO-influenced PAS regions correspond to locations of inter-domain crosslinks in the M. sexta PAS (Figure 2E). Interestingly, the PAS surface exhibiting the largest NO-induced increases in exchange encompasses the PAS residues (M. sexta α1 E341 and K343) identified as H-NOX contacts in the crosslinking study (Fritz et al., 2013). NO-induced exchange rate increases in complementary surfaces of both PAS and H-NOX domains indicate that NO-binding relaxes contacts and hydrogen bonding networks between the two domains.
The β1[1-385] truncation also includes a segment of the helical domain. Interestingly, NO also stimulates prominent increases in exchange rates in the linker between the PAS domain and helical elements (Figure 2C). The strong linker perturbations upon NO-binding were examined further in the context of the full-length sGC heterodimer.
NO-induced conformational changes in full-length α1β1 sGC
H-NOX truncations revealed the proximate conformational changes associated with NO-occupancy of the heme. Next, we sought to identify the allosteric effects that communicate NO occupancy from the H-NOX domain to the active site of the catalytic domain. To that end, semi-automated HDX-MS was used to analyze the NO-induced changes in conformation and dynamics in the full-length α1β1 sGC heterodimer (Chalmers et al., 2006).
To mimic physiological substrate availability, sGC was pre-incubated with a noncyclizable GTP analog, guanosine-5′-[(α,β)-methylene]triphosphate (GPCPP). (Cary et al., 2005; Russwurm and Koesling, 2004). Unliganded and NO-bound sGC samples were then subjected to parallel H/D exchange time courses and analyzed by MS. NO-induced differences in exchange rates were color-coded and mapped to the primary sequence of each subunit (Figure 3). For a global view, exchange rate differences were condensed and mapped to a domain representation of the sGC heterodimer (Figure 4A).
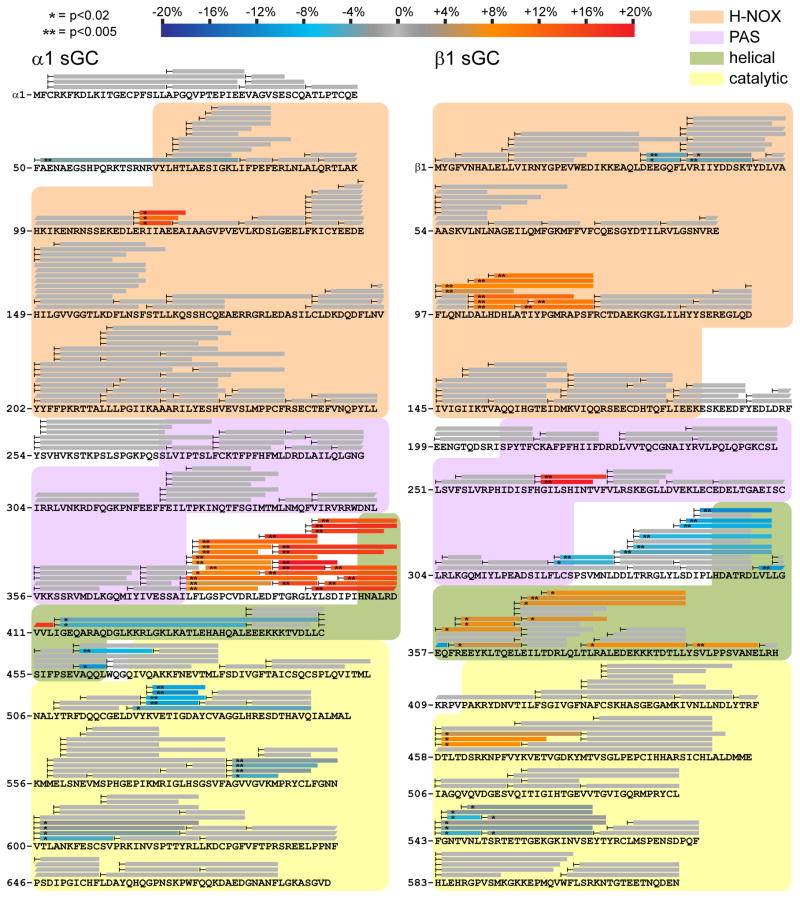
Figure 3. NO-induced conformational changes of the full-length sGC heterodimer.
Changes in HD exchange kinetics induced by NO-binding in full length α1β1 sGC with noncyclizable GTP analog GPCPP. Peptide coverage from HDX-MS analysis is represented by bars overlaying primary sequence of α1 (left) and β1 (right) sGC. Peptides exhibiting significant NO-induced exchange rate changes are color-coded according to the scale bar. Peptides exchanging more rapidly upon NO-binding are colored in red hues, whereas peptides exchanging more slowly are in blue hues. Asterisks denote the results of two-tailed, unpaired Student’s t-tests. Peptides exhibiting non-significant or undetectable changes are colored gray.
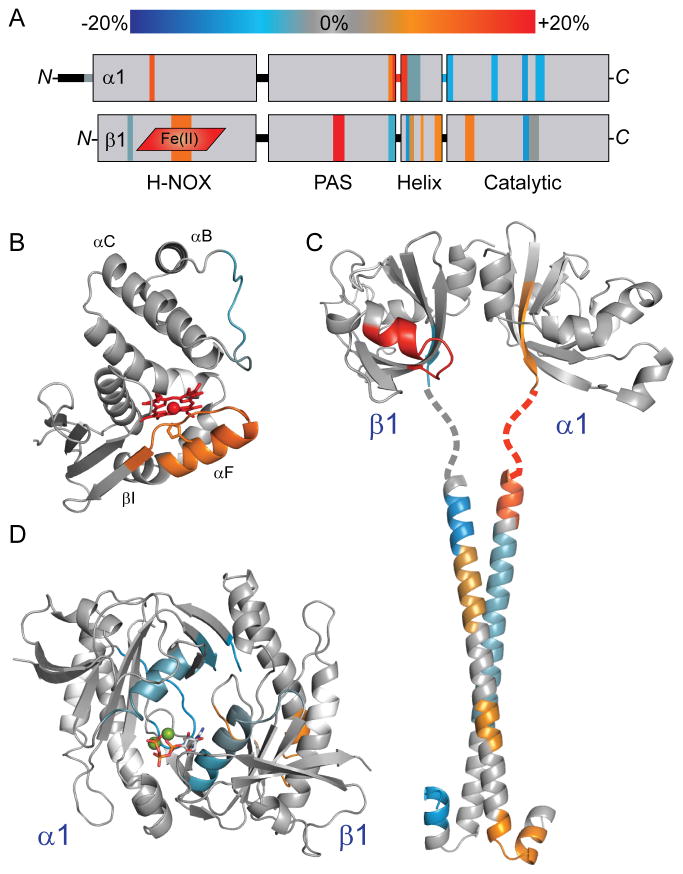
Figure 4. NO-induced conformational changes of full-length sGC mapped to domain structural models.
(A) Results from HDX-MS analysis of the NO-induced changes in α1β1 sGC (Figure 3) were condensed, color-coded according to the scale bar, and mapped to the representation of sGC domain organization.
(B) HD exchange rate perturbations induced by NO-binding were color-coded (as in Figure 3) and mapped to a structural model of the β1 H-NOX, as in Figure 2.
(C) NO-induced changes were mapped to a model of the α1β1 PAS-linker-helical domains. The α1 and β1 PAS domains were modeled independently using the Robetta server, based on homology with the M. sexta α1 PAS domain structure (PDB: 4GJ4) (Kim et al., 2004; Purohit et al., 2013). PAS domain monomers were dimerized by alignment with the structure of the bacterial PAS dimer (PDB: 2P04) (Ma et al., 2008). The structure of the rat sGC β1 helix (PDB: 3HLS) was used to reconstruct the coiled-coil of the helical domain (Ma et al., 2010). The α1 helical subunit was modeled using the Robetta server. The α1β1 coiled-coil was docked using the ClusPro 2.0 server, selecting the highest populated class exhibiting parallel orientation (Comeau et al., 2004). The intervening linker sequence is represented as a dotted line.
(D) NO-induced changes were mapped to the structure of the sGC α1β1 catalytic domain heterodimer (PDB: 3UVJ) (Allerston et al., 2013). For reference, GTP:Mg2+ substrate (sticks and balls) was docked into the active site.
NO elicits conformational changes in the β1 H-NOX of full-length sGC similar to the H-NOX truncations, β1[1-194] and β1[1-385]. Specifically, the heme-associated helix (αF) and adjacent heme pocket loop (αF-βI) demonstrate significant increases in exchange rates, consistent with increased accessibility and dynamics (Figure 4B). The stabilization of α-helices of the N-terminal lobe observed in NO-bound H-NOX truncations is not evident in the full-length sGC, possibly due to the narrower kinetic regime analyzed. Nevertheless, the rapidly exchanging αB-αC loop of the N-terminal lobe does exhibit subtle decreases in exchange rate.
The α1 pseudo-H-NOX was largely unaffected. One short segment increased exchange rates with NO bound. However, the lack of structural characterization of the α1 pseudo-H-NOX precludes interpretation of the effect.
The PAS domains exhibited asymmetric NO-induced effects localized to two regions (Figure 4C). Prominent increases in exchange were evident at the helix and loop implicated in H-NOX interactions by previous crosslinking studies (Fritz et al., 2013). The PAS domain of β1[1-385] also exhibited increased exchange at this H-NOX-associated helix, however the change was more pronounced in full-length sGC. Interestingly, this increased exchange was not observed in the corresponding region of the α1 PAS subunit. The other PAS domain elements perturbed by NO-binding are the C-terminal β-sheets that adjoin the linker to the helical domains. The terminal sheet of the β1 PAS exchanges more slowly with NO bound, while the equivalent sheet of the α1 PAS exchanges much more rapidly. Furthermore, this exchange rate increase extends through the linker into the α1 helical domain.
The NO-induced conformational changes continue in both subunits of the helical domains (Figure 4C). The helical domains exhibit asymmetric stretches of increased and decreased exchange. Limited peptide coverage may obscure the details of the effects in the α1 helical domain. Nevertheless, both helical domains likely participate in transducing the NO-induced conformational change to the catalytic domain.
Ultimately, NO-occupancy of the sGC heme controls the activity of the catalytic domains. The active site pocket is formed by the interface between the α1 and β1 catalytic domain, with residues from both subunits contributing substrate recognition residues (Allerston et al., 2013; Winger et al., 2008). HDX-MS reveals that the catalytic domain responds to NO-binding with conformational changes localized to several discrete surfaces (Figure 4D). The dorsal surface (opposite the active site) exhibits inverse effects in the β1 and α1 halves of the catalytic domain, with increases in exchange rates in the β1 subunit and decreases in the α1 subunit (Figure 5). The other primary NO-induced changes involve the active site itself. Pseudo-symmetric helices flanking the α1 and β1 halves of the substrate pocket exhibit NO-induced decreases in exchange rates. The loop comprising the interior surface of the active site pocket also undergoes slower exchange upon NO-binding. On the whole, the structural elements constituting the active site pocket undergo uniform decreases in exchange, consistent with NO-induced stabilization or “closing” of the active site.
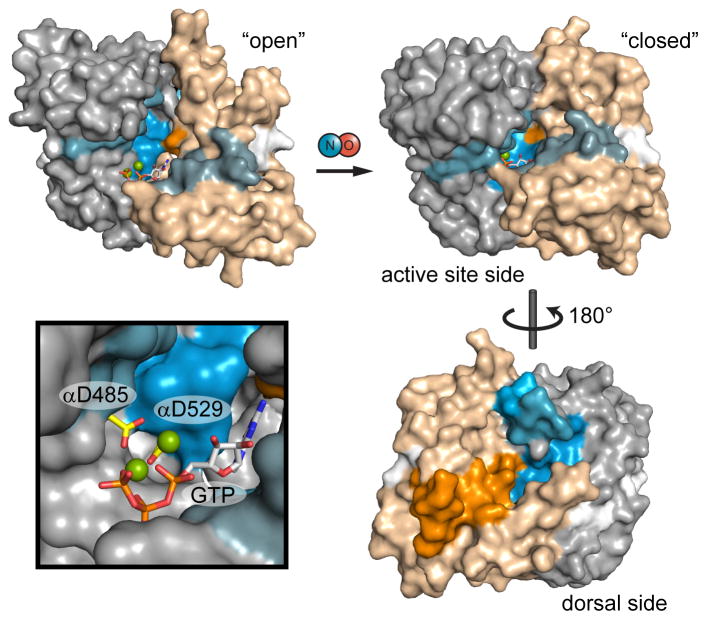
Figure 5. Mapping NO-induced conformational changes to models of catalytic domain activation.
Catalytic domain exchange rate differences induced by NO were mapped to models of the inactive, “open” conformation and active “closed” conformation. Exchange rate differences are color-coded as in Figure 3; here, α1 and β1 subunits are distinguished as gray and tan, respectively. The open conformation is represented by the structure of the human sGC catalytic domain (PDB: 3UVJ) with GTP docked. The closed conformation is a model based on the structure of a putatively active adenylate cyclase (PDB: 1AZS). Active site aspartate residues, α1 Asp-485 and α1 Asp-529, are highlighted (yellow) in the inset.
DISCUSSION
The mechanism underlying NO activation of sGC is a central question in NO/sGC/cGMP signaling. HDX-MS revealed that NO induces several discrete conformational changes in sGC, spanning from the N-terminal H-NOX sensor domain to the C-terminal catalytic output domain. Generally, these conformational effects localize to five regions: (1) the H-NOX heme-associated helix surface, (2) an H-NOX-associated surface of the PAS domain, (3) The PAS—helical domain linker, (4) the helices themselves, and (5) the active site of the catalytic domain.
The proximate effect of NO-binding is the cleavage of the Fe2+-His bond that connects the heme to the αF helix. As hypothesized, HDX-MS analysis implicated the heme-associated helix as a focal point of the NO-induced conformational change. NO induces increased exchange rates in the heme-associated helix and adjacent surfaces, consistent with a helix displacement that increases accessibility and dynamics. The heme-associated helix effects in sGC parallel observations in stand-alone bacterial H-NOX proteins in states mimicking activation (Plate and Marletta, 2013). A Shewanella oneidensis H-NOX variant lacking the His ligand exhibits a helix shift away from the heme (Erbil et al., 2009). Similarly, replacement of heme with the small molecule H-NOX activator cinaciguat caused subtle displacement of the heme-associated helix in the Nostoc sp. PCC7120 H-NOX (Martin et al., 2010). Like the bacterial H-NOX proteins, the isolated sGC H-NOX (β1[1-194]) exhibits significant, albeit subtle, effects on the heme-associated helix with NO bound.
Interestingly, the NO-induced effects on the heme-associated helix are markedly accentuated in the presence of the PAS domain. Previous work mapping sGC inter-domain interactions implicates the same contiguous heme-associated helix surfaces as an interface with the PAS domains (Underbakke et al., 2013a). To account for the influence of the PAS domain, we propose that NO-induced heme-associated helix movement elicits concomitant disassociation or weakening of H-NOX—PAS interactions. Minimally, the PAS domain is a key mediator in transducing the conformational signals from the H-NOX along to the catalytic domain.
The dominant activation-associated conformational changes within the PAS domain localize to two regions. The first is a surface patch of the β1 PAS that undergoes marked increases in exchange with NO bound, an effect common to both the β1[1-385] homodimer and the α1β1 sGC heterodimer. The surface exhibiting increased exchange may be associated with the putative disassociation of H-NOX—PAS contacts. Notably, the corresponding surface of the α1 PAS crosslinks with the β1 H-NOX in M. sexta sGC truncations (Fritz et al., 2013). The β1 H-NOX may bridge both PAS subunits, constitutively interfacing with α1 PAS and shifting with respect to the β1 PAS when NO binds.
NO also strongly affects the C-termini of the PAS domains, a perturbation that continues into the helical domains. The opposing α1 increases and β1 decreases in exchange rates are among the largest magnitude changes stimulated by NO. The structure of the inter-domain linker remains ambiguous, but the strong changes in exchange rates are consistent with a loss of secondary structure or an order-to-disorder transition due to NO-binding. Similar conformational changes in PAS—helical domain linkers have been proposed to dictate activity in bacterial receptors (see below).
The sGC helical domain is thought to assemble into a parallel coiled-coil with functional importance for signal transmission (Ma et al., 2010; Rothkegel et al., 2007). As such, the helical domain is a member of a broader family of signaling coiled-coils dubbed the S-helix (Anantharaman et al., 2006). The S-helix family is ubiquitous in prokaryotic signaling systems, yet relatively uncommon in eukaryotes. S-helices bridge a diverse array of signaling input and output domains. While the S-helix is thought to serve as a conformational switch controlling signaling output, the allosteric mechanism remains unclear. The diversity and modularity of the associated signaling domains argues for a general signaling mechanism. Proposed mechanisms include helical rotation (Möglich et al., 2009), scissoring (Ramamurthy et al., 2001), or dynamic changes to coil packing and stability (Stewart and Chen, 2010; Winkler et al., 2012). The sGC helical domains exhibit patterns of increased and decreased protection along the length of the coiled-coil. Notably, HDX monitors amide protons of the polypeptide backbone, which should be engaged in hydrogen bonds along the long axis in a predominantly α-helical domain. The NO-sensitivity of the helical domains suggests perturbations to secondary structure may be involved in signal transmission. Nevertheless, many of the proposed mechanisms could also account for the observed effects. Indeed, the general signaling mechanism(s) of the S-helix family may rely on multiple aspects of the continuum of conformational changes accessible to coiled-coils.
The NO-activated catalytic domains exhibit conformational effects remarkably consistent with the activation mechanism proposed for adenylate cyclases (AC) (Figure 5). The homologous catalytic domains of sGC and AC share similar C2-pseudosymmetric wreath shapes. Structural investigations focused on AC catalytic domain truncations have advanced a compelling activation mechanism hypothesis (Hurley, 1999; Linder and Schultz, 2008; Tesmer et al., 1997). Binding of a stimulatory G protein activates the AC catalytic domain by inducing an ~10° rotation between the subunits that contracts the active site pocket (Tesmer et al., 1997). This “closed” active site conformation is thought to shift two catalytic aspartate residues into coordination with the Mg2+-triphosphate of ATP, thereby activating the ribose 3′-hydroxyl while stabilizing the pyrophosphate leaving group. HDX-MS revealed that the sGC catalytic domain likely undergoes a similar active site closure upon activation (Figure 5). The decreased exchange rates of the active site pocket and flanking helices are consistent with the solvent-protected, “closed” conformation proposed to align catalytic aspartates with GTP substrate. In fact, the strongly-protected active site wall includes α1 Asp-529, one of catalytic aspartate residues. Notably, substrate binding is not sufficient to induce active site closure, as HDX-MS analysis showed no GPCPP-dependent exchange rate changes in sGC (data not shown).
Active site contraction is accompanied by conformational effects on the “dorsal” surface, opposite the active-site cleft (Figure 5). Dorsal surfaces in the α1 and β1 subunit undergo asymmetric exchange rate increases and decreases, respectively. The proximity to the subunit interface suggests that the dorsal conformational effects are a consequence of flexing and interfacial rotation of the subunits. Interestingly, the affected dorsal surfaces are also close to the linker connecting to the helical domains, despite occurring much farther into the primary sequence. The dorsal surface effects may therefore reflect helical domain shifts promoting the inter-domain torquing.
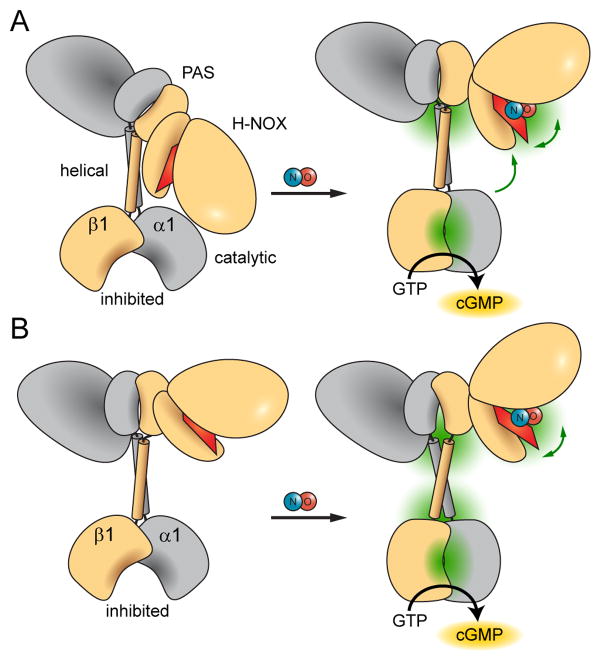
The various conformational changes observed upon NO-binding suggest possible allosteric pathways for cyclase activation. Prior studies indicated that direct interactions between the H-NOX and catalytic domain repress sGC activity (Winger and Marletta, 2005). This proposed mechanism for cyclase regulation is supported by a FRET study that revealed close proximity between the N- and C-termini of sGC (Haase et al., 2010). In addition, previous HDX-MS mapping of sGC inter-domain interactions identified a surface of the catalytic domain protected from exchange by the presence of the H-NOX domain (Underbakke et al., 2013a). The effects of NO-binding observed here indicate that the H-NOX—catalytic domain regulatory interface may be controlled by conformational switching in the linker between PAS and helical domains (Figure 6A). The PAS—helical junction may serve as a flexible pivot point for releasing the inhibitory inter-domain contacts. In support of this model, a recent study employing Förster resonance energy transfer revealed NO-induced proximity changes between the sGC H-NOX and catalytic domains (Busker et al., 2013). On the other hand, the possibility that the PAS—helical junction exerts long-range effects through helical domain conformational changes cannot be ruled out (Figure 6B).
Figure 6. Proposed NO-induced activation mechanisms of sGC.
NO-binding releases and opens the heme-associated helix of the H-NOX domain while and condensing and closing the active site pocket of the catalytic domain. Dominant points of conformational articulation are highlighted in green. Higher-order domain architecture is based on an emerging single particle electron microscopy study (Campbell MG, ESU, Potter CS, Carragher B, MAM, submitted). The allosteric pathway bridging the sensor and output domains may involve two different mechanisms.
(A) Large conformational changes at the junction between PAS and helical domains may indicate inter-domain pivoting that relieves inhibitory contacts between H-NOX and catalytic domains.
(B) Alternatively, the PAS—helical junction may mediate remote allosteric effects via long-range conformational changes propagated through the helical domains.
Ultimately, both mechanisms share a key feature: changes in the conformational flexibility of the junction between the PAS and helical domains modulate the signaling state of the associated output cyclase domain. Signal transmission via PAS—helical signaling modules has emerged as an important theme in prokaryotic signaling systems (Anantharaman et al., 2006; Moglich et al., 2009). PAS domains often function directly as sensor domains, binding ligands or signal-sensitive cofactors (Henry and Crosson, 2011). Unfolding at the helical termini of model PAS domains is thought to be important for communicating PAS signals (Harper et al., 2003; Lee et al., 2001). Indeed, the large-scale increases in exchange rates at the sGC α1 PAS terminus are consistent with local unfolding. Recent structural investigations also offer new insights into PAS sensor signaling mechanisms, highlighting the importance of conformational transitions within PAS—helix linkers (Diensthuber et al., 2013; Wang et al., 2013). While a consensus signaling mechanism has yet to emerge, these diverse prokaryotic PAS modules exhibit conformational transitions with intriguing parallels to sGC signaling mechanisms. The apparent wide evolutionary conservation of the PAS—helical signaling mechanisms highlights the utility of this signaling module in connecting disparate sensor and effector domains.
The HDX approach is useful for probing dynamic proteins in solution, especially complex signaling proteins that resist crystallization. Accordingly, these HDX-MS studies offer valuable insights into the conformational dynamics of the NO-activated sGC holoenzyme, advancing the understanding sGC signaling mechanisms. We anticipate that a comprehensive model of the architecture and signaling mechanisms of sGC will fuel development of therapeutics targeting cGMP-dependent physiological processes.
EXPERIMENTAL PROCEDURES
Expression and purification of sGC constructs
H-NOX truncations, β1[1-194] and β1[1-385] were expressed and purified from Escherichia coli, as previously described (Underbakke et al., 2013a). Full-length Rattus norvegicus α1β1 sGC was expressed and purified from Sf9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen), as previously described (Underbakke et al., 2013a).
HDX-MS on NO-treated β1 H-NOX truncations
H/D exchange time courses were performed as previously described, with modifications (Smith et al., 2013; Underbakke et al., 2013a). Briefly, β1[1-194] or β1[1-385] was reduced with sodium dithionite under anaerobic conditions and buffer-exchanged into anaerobic Assay Buffer (50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH 7.5) using a MiniTrap G-25 desalting column (GE Healthcare). The NO-bound H-NOX truncation was formed by incubating with excess NO-donor diethylamine NONOate (DEA/NO, Cayman Chemical). Unliganded samples were treated with buffer vehicle (1 mM NaOH). NO-bound and unliganded H-NOX samples were both subjected to parallel HDX time courses in triplicate. Exchange was initiated by diluting H-NOX stocks into buffered D2O (50 mM HEPES pD 8.0, 50 mM NaCl, 5 mM DTT). NO-bound exchange reactions were supplemented with DEA/NO to maintain NO-occupancy of the heme. At various time points (6, 10, 30, 60, 150, 300, 1,200, and 7,200 s) exchange was quenched by addition of 5% trifluoroacetic acid (TFA) to final pH ~2.5, followed by immediate freezing in liquid N2. Time point samples were quickly thawed and digested (3 min, 4 °C) into uniquely identifiable peptides with agarose-immobilized pepsin resin (Pierce) in aqueous 0.025% TFA pH 2.5. Pepsin resin was removed by brief centrifugation, and samples were immediately frozen in liquid N2 until analysis. Mass spectrometry analysis of β1[1-194] and β1[1-385] HDX-MS samples was performed as previously described (Underbakke et al., 2013a). Briefly, peptide products of pepsin digestion were analyzed by MS/MS using a quadrupole time-of-flight MS (Q-TOF Premier, Waters) and identified using ProteinLynx Global Server (Waters). Deuterium incorporation for each time point sample was analyzed by LC-MS using an Agilent 1200 HPLC coupled in-line with an LTQ Orbitrap XL MS (Thermo Fisher Scientific). Samples were quickly thawed immediately prior to analysis and separated over a C8 (30 mm × 1 mm, 5 μm particle) reversed-phase column (Restek) in aqueous 0.1% v/v formic acid maintained at 4 °C. The elution profile consisted of linear gradients of 5–10% v/v acetonitrile over 1 min, 10–40% over 5 min, and 40–100% over 4 min (300 μL/min). Mass spectra were collected in positive ion mode with mass range m/z 350 – 1500 using resolution set to 100,000 at m/z 400.
HDX-MS on NO-treated α1β1 sGC holoenzyme
Full-length α1β1 sGC in Assay Buffer was pre-equilibrated with 300 μM GPCPP. NO-bound sGC was formed by incubating with excess DEA/NO (300 μM). Unliganded samples were treated with an equivalent volume of buffer vehicle (1 mM NaOH). Time courses were performed in triplicate, with each time course prepared independently, immediately prior to deuterium exchange initiation.
Exchange reactions and mass spectrometry analysis were performed with a custom-built automated HDX-MS system implementing dual HTS PAL liquid-handling robots (LEAP Technologies), with some adaptations (Chalmers et al., 2006). To initiate exchange time point, 40 pmol sGC was diluted into D2O (20 μL final volume) and exchanged for 10, 30, 60, 300, 900, or 3,600 s at 4 °C. Exchange was quenched with 30 μL 3 M urea, 1% v/v TFA. Pepsinization was performed with an in-line pepsin column (1 mm × 20 mm, 50 μL/min flow rate) at 15 °C and desalted on a C8 (1 mm × 10 mm) trap column (Thermo Scientific). Peptides were separated over a C18 column (1 mm × 50 mm, 5 μm particle, Hypersil Gold, Thermo Scientific) in aqueous 0.3% v/v formic acid maintained at 1 °C. The elution profile was a linear gradient of 5–50% v/v acetonitrile over 5 min. Mass spectra were collected in positive ion mode with an LTQ Orbitrap XL (Thermo Scientific) using mass range m/z 300 – 1700 with mass resolution set to 100,000 at m/z 400.
HDX data analysis
Mass spectra were analyzed using HDX Workbench software to calculate the centroid of the isotopic envelope of each peptide (Pascal et al., 2012). Percent deuterium exchange (%D) and exchange rate differences (Δ%D) were calculated as previously described (Underbakke et al., 2013a).
Highlights.
HDX-MS reveals an allosteric pathway communicating NO-binding through sGC
NO-binding to the H-NOX displaces heme-associated helix and PAS domain contacts
Activation signal transmitted via key linker joining PAS domain and signaling helix
NO induces active site contraction in the catalytic domain for cyclase stimulation
Acknowledgments
We thank Dr. N. Basak Surmeli for experimental assistance. LC-MS/MS instrumentation was acquired with National Institutes of Health (NIH) support (1S10RR022393-01). ESU received financial support from NIH Postdoctoral Fellowship 5F32GM093564.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allerston CK, von Delft F, Gileadi O. Crystal structures of the catalytic domain of human soluble guanylate cyclase. PLoS One. 2013;8:e57644. doi: 10.1371/journal.pone.0057644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Balaji S, Aravind L. The signaling helix: a common functional theme in diverse signaling proteins. Biol Direct. 2006;1 doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS. Endogenous nitric oxide synthesis: Biological functions and pathophysiology. Free Radic Res. 1999;31:577–596. doi: 10.1080/10715769900301161. [DOI] [PubMed] [Google Scholar]
- Busker M, Neidhardt I, Behrends S. Nitric Oxide Activation of Guanylate Cyclase Pushes the α1 Signaling Helix and the β1 Heme Binding Domain closer to the Substrate Binding Site. J Biol Chem. 2013 doi: 10.1074/jbc.M113.504472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary SPL, Winger JA, Marletta MA. Tonic and acute nitric oxide signaling through soluble guanylate cyclase is mediated by nonheme nitric oxide, ATP, and GTP. Proc Natl Acad Sci USA. 2005;102:13064–13069. doi: 10.1073/pnas.0506289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire ER, Marletta MA. Structure and regulation of soluble guanylate cyclase. Annu Rev Biochem. 2012;81:533–559. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- Diensthuber RP, Bommer M, Gleichmann T, Moglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbil WK, Price MS, Wemmer DE, Marletta MA. A structural basis for H-NOX signaling in Shewanella oneidensis by trapping a histidine kinase inhibitory conformation. Proc Natl Acad Sci USA. 2009;106:19753–19760. doi: 10.1073/pnas.0911645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernhoff NB, Derbyshire ER, Marletta MA. A nitric oxide/cysteine interaction mediates the activation of soluble guanylate cyclase. Proc Natl Acad Sci USA. 2009;106:21602–21607. doi: 10.1073/pnas.0911083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmann M, Griebenow N, Hahn MG, Hartung I, Mais FJ, Mittendorf J, Schafer M, Schirok H, Stasch JP, Stoll F, et al. The chemistry and biology of soluble guanylate cyclase stimulators and activators. Angew Chem Int Ed. 2013;52:9442–9462. doi: 10.1002/anie.201302588. [DOI] [PubMed] [Google Scholar]
- Friebe A, Koesling D. The function of NO-sensitive guanylyl cyclase: What we can learn from genetic mouse models. Nitric Oxide. 2009;21:149–156. doi: 10.1016/j.niox.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Friebe A, Mergia E, Dangel O, Lange A, Koesling D. Fatal gastrointestinal obstruction and hypertension in mice lacking nitric oxide-sensitive guanylyl cyclase. Proc Natl Acad Sci USA. 2007;104:7699–7704. doi: 10.1073/pnas.0609778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz BG, Roberts SA, Ahmed A, Breci L, Li W, Weichsel A, Brailey JL, Wysocki VH, Tama F, Montfort WR. Molecular model of a soluble guanylyl cyclase fragment determined by small-angle X-ray scattering and chemical cross-linking. Biochemistry. 2013;52:1568–1582. doi: 10.1021/bi301570m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthwaite J. Concepts of neural nitric oxide-mediated transmission. Eur J Neurosci. 2008;27:2783–2802. doi: 10.1111/j.1460-9568.2008.06285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase T, Haase N, Kraehling JR, Behrends S. Fluorescent fusion proteins of soluble guanylyl cyclase indicate proximity of the heme nitric oxide domain and catalytic domain. PLoS One. 2010;5:e11617. doi: 10.1371/journal.pone.0011617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. Structure, mechanism, and regulation of mammalian adenylyl cyclase. J Biol Chem. 1999;274:7599–7602. doi: 10.1074/jbc.274.12.7599. [DOI] [PubMed] [Google Scholar]
- Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc Natl Acad Sci USA. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow DS, Pan D, Davis JH, Behrends S, Mathies RA, Marletta MA. Characterization of functional heme domains from soluble guanylate cyclase. Biochemistry. 2005;44:16266–16274. doi: 10.1021/bi051601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chemical Society Reviews. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- Landgraf Rachelle R, Goswami D, Rajamohan F, Harris Melissa S, Calabrese MF, Hoth Lise R, Magyar R, Pascal Bruce D, Chalmers Michael J, Busby Scott A, et al. Activation of AMP-activated protein kinase revealed by hydrogen/deuterium exchange mass spectrometry. Structure. 2013 doi: 10.1016/j.str.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Pandit A, Croonquist PA, Hoff WD. Folding and signaling share the same pathway in a photoreceptor. Proc Natl Acad Sci USA. 2001;98:9062–9067. doi: 10.1073/pnas.111153598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder JU, Schultz JE. Versatility of signal transduction encoded in dimeric adenylyl cyclases. Curr Opin Struct Biol. 2008;18:667–672. doi: 10.1016/j.sbi.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Ma X, Beuve A, van den Akker F. Crystal structure of the signaling helix coiled-coil domain of the beta1 subunit of the soluble guanylyl cyclase. BMC Struct Biol. 2010;10:2. doi: 10.1186/1472-6807-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Sayed N, Baskaran P, Beuve A, van den Akker F. PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J Biol Chem. 2008;283:1167–1178. doi: 10.1074/jbc.M706218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Baskaran P, Ma X, Dunten PW, Schaefer M, Stasch JP, Beuve A, van den Akker F. Structure of cinaciguat (BAY 58-2667) bound to Nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase. J Biol Chem. 2010;285:22651–22657. doi: 10.1074/jbc.M110.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Brown L, Konermann L. Hydrogen Exchange Mass Spectrometry of Bacteriorhodopsin Reveals Light-Induced Changes in the Structural Dynamics of a Biomolecular Machine. J Am Chem Soc. 2011;133:20237–20244. doi: 10.1021/ja206197h. [DOI] [PubMed] [Google Scholar]
- Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin P. HDX Workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate L, Marletta MA. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci. 2013;38:566–575. doi: 10.1016/j.tibs.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit R, Weichsel A, Montfort WR. Crystal structure of the alpha subunit PAS domain from soluble guanylyl cyclase. Protein Sci. 2013;22:1439–1444. doi: 10.1002/pro.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V, Tucker C, Wilkie SE, Daggett V, Hunt DM, Hurley JB. Interactions within the Coiled-coil Domain of RetGC-1 Guanylyl Cyclase Are Optimized for Regulation Rather than for High Affinity. J Biol Chem. 2001;276:26218–26229. doi: 10.1074/jbc.M010495200. [DOI] [PubMed] [Google Scholar]
- Rothkegel C, Schmidt PM, Atkins DJ, Hoffmann LS, Schmidt HHHW, Schröder H, Stasch JP. Dimerization region of soluble guanylate cyclase characterized by bimolecular fluorescence complementation in vivo. Mol Pharmacol. 2007;72:1181–1190. doi: 10.1124/mol.107.036368. [DOI] [PubMed] [Google Scholar]
- Russwurm M, Koesling D. NO activation of guanylyl cyclase. EMBO J. 2004;23:4443–4450. doi: 10.1038/sj.emboj.7600422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Underbakke ES, Kulp DW, Schief WR, Marletta MA. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci USA. 2013;110:E3577–E3586. doi: 10.1073/pnas.1313331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasch JP, Pacher P, Evgenov OV. Soluble guanylate cyclase as an emerging therapeutic target in cardiopulmonary disease. Circulation. 2011;123:2263–2273. doi: 10.1161/CIRCULATIONAHA.110.981738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V, Chen LL. The S Helix Mediates Signal Transmission as a HAMP Domain Coiled-Coil Extension in the NarX Nitrate Sensor from Escherichia coli K-12. J Bacteriol. 2010;192:734–745. doi: 10.1128/JB.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJG, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with G(s alpha).GTP gamma S. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- Underbakke ES, Iavarone AT, Marletta MA. Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase. Proc Natl Acad Sci USA. 2013a;110:6777–6782. doi: 10.1073/pnas.1301934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underbakke ES, Surmeli NB, Smith BC, Wynia-Smith SL, Marletta MA. 3.10 - Nitric oxide signaling. In: Jan R Editors-in-Chief, Kenneth P, editors. Comprehensive Inorganic Chemistry II. 2. Amsterdam: Elsevier; 2013b. pp. 241–262. [Google Scholar]
- Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, et al. Mechanistic Insights Revealed by the Crystal Structure of a Histidine Kinase with Signal Transducer and Sensor Domains. PLoS Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West Graham M, Pascal Bruce D, Ng LM, Soon FF, Melcher K, Xu HE, Chalmers Michael J, Griffin Patrick R. Protein conformation ensembles monitored by HDX reveal a structural rationale for abscisic acid signaling protein affinities and activities. Structure. 2013;21:229–235. doi: 10.1016/j.str.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger JA, Derbyshire ER, Lamers MH, Marletta MA, Kuriyan J. The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase. BMC Struct Biol. 2008;8 doi: 10.1186/1472-6807-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger JA, Marletta MA. Expression and characterization of the catalytic domains of soluble guanylate cyclase: interaction with the heme domain. Biochemistry. 2005;44:4083–4090. doi: 10.1021/bi047601d. [DOI] [PubMed] [Google Scholar]
- Winkler K, Schultz A, Schultz JE. The S-Helix Determines the Signal in a Tsr Receptor/Adenylyl Cyclase Reporter. J Biol Chem. 2012;287:15479–15488. doi: 10.1074/jbc.M112.348409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marletta MA. Localization of the heme binding region in soluble guanylate cyclase. Biochemistry. 1997;36:15959–15964. doi: 10.1021/bi971825x. [DOI] [PubMed] [Google Scholar]