Abstract
Mutation of the gene PARK2, which encodes an E3 ubiquitin ligase, is the most common cause of early-onset Parkinson's disease1, 2, 3. In a search for multisite tumor suppressors, we identified PARK2 as a frequently targeted gene on chromosome 6q25.2–q27 in cancer. Here we describe inactivating somatic mutations and frequent intragenic deletions of PARK2 in human malignancies. The PARK2 mutations in cancer occur in the same domains, and sometimes at the same residues, as the germline mutations causing familial Parkinson's disease. Cancer-specific mutations abrogate the growth-suppressive effects of the PARK2 protein. PARK2 mutations in cancer decrease PARK2's E3 ligase activity, compromising its ability to ubiquitinate cyclin E and resulting in mitotic instability. These data strongly point to PARK2 as a tumor suppressor on 6q25.2–q27. Thus, PARK2, a gene that causes neuronal dysfunction when mutated in the germline, may instead contribute to oncogenesis when altered in non-neuronal somatic cells.
Main
Parkinson's disease (PD) is the most common neurodegenerative movement disorder4. The familial, autosomal recessive form of PD is caused by germline mutations in the PARK2 gene, which result in early-onset loss of dopaminergic neurons in the substantia nigra1, 2, 3, 5. PARK2 is widely expressed in a variety of tissues, including the brain (in neurons and astrocytes), lung, colon and testes6, 7, 8.
PARK2 associates with ubiquitin-conjugating enzymes, including UBCH7 and UBCH8, and is capable of promoting mono- and polyubiquitination of target proteins8, 9, 10. In neuronal model systems, these activities can regulate proteasome-mediated degradation11, 12. PARK2 can target a number of protein substrates, which have been identified primarily using systems focused on studying neuronal cytoprotection13, 14, 15, 16, 17. Notably, PARK2-mediated degradation of cyclin E is important for preventing excitotoxicity in postmitotic neurons14. In neuronal model systems, PARK2 mutations that cause juvenile PD disrupt the ubiquitination activity of PARK2 and the regulation of proteasome-mediated degradation8, 9, 12, 14, 18, 19, 20, 21. How PARK2 loss leads to PD is not entirely clear.
Chromosome 6q25.2–27 spans a large genomic region and undergoes frequent loss in a number of human cancers22, 23, 24, 25, 26. PARK2 is a potential candidate for a tumor suppressor gene at this locus, but intragenic mutations of this gene have not been reported 25, 27, 28. Furthermore, copy number loss within this region varies greatly in size from one tumor to another, and the identity of a common target of deletion remains unclear29. PARK2 maps near FRA6E, a common fragile site in the human genome, which displays complicated copy number variants. Like the locations of FHIT (3p14.2) and WWOX (16q23.3), this site is hypothesized to contain a tumor suppressor gene30, 31.
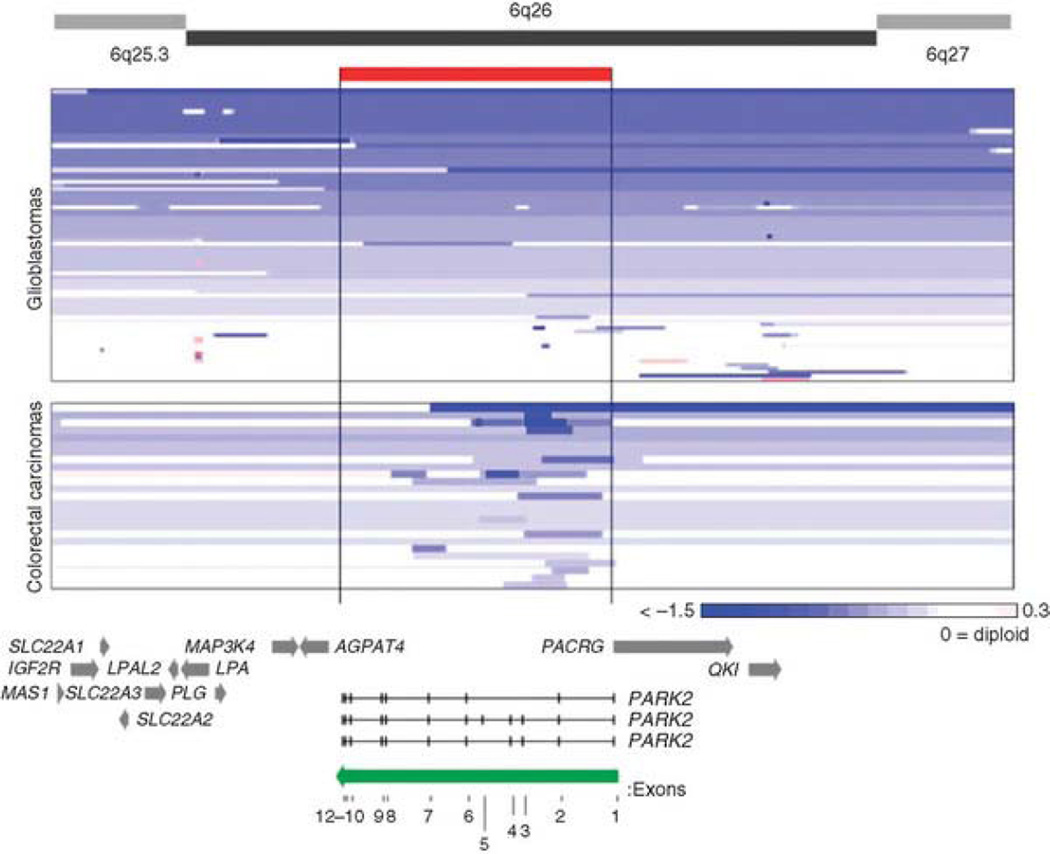
Here we present mutational and functional data that identify the ubiquitin E3 ligase PARK2 as a chromosome 6q tumor suppressor in glioblastoma multiforme (GBM), colon cancer and lung cancer. To identify tumor suppressors that are targeted in multiple tumor types, we examined array comparative genomic hybridization (aCGH) results from 98 colon cancer samples and 216 GBM tumor samples. For colon cancer, analysis of loci that were recurrently deleted demonstrated a focal region on chromosome 6q (Supplementary Fig. 1). The GBM dataset is from the Cancer Genome Atlas (TCGA)23. Copy number alterations (CNA) at the PARK2 locus for both GBM and colon cancers are shown in Figure 1. As expected, we observed frequent heterozygous and homozygous loss of variable size on 6q in GBM (see Online Methods)23. In GBM samples, 85% (53 out of 62) of samples with loss on 6q showed loss of the PARK2 gene within the area of CNA. In colon cancer samples, 100% (24 out of 24) of samples with loss on 6q showed loss of PARK2 within the CNA region. Loss of regions of various size encompassed PARK2 and were found in a substantial portion of tumors (Fig. 1, Table 1). Notably, in both tumor types, intragenic homozygous deletions were found in the PARK2 gene that removed exons but not any surrounding genes, thus pointing to PARK2 as a targeted gene on chromosome 6q. Genomic loss in GBM samples tended to encompass broad regions including the PARK2 gene, and intragenic microdeletions in PARK2 occurred in 2.3% of samples (Fig. 1, Table 1). This pattern is also seen in lung cancer, which is thought to be associated with deletion of a putative tumor suppressor at 6q25?27. The identity of the gene of interest in this region is difficult to determine due to the variable nature of copy number loss in this tumor type24. In contrast, the majority of the copy number loss on 6q25.2–q27 in our colon cancer samples occurred via focal events that affect PARK2 but not surrounding genes (Fig. 1, bottom). Such focal losses occurred in approximately 25% of all colon cancer samples we examined (Table 1), robustly identifying the PARK2 gene as the target of CNA. These results demonstrate the diversity of deletions at the PARK2 locus. Focal deletions that target PARK2 occur in both GBM and colon cancer, and PARK2 constitutes a specifically targeted gene on 6q for both tumor types. It is well known that different tumor types have an intrinsic variation in the sizes of, and the tendency to undergo, this type of genetic abnormality, which may be dependent on chromatin state32. However, at least in the case of GBM, we cannot rule out the possibility that there exist other 6q tumor suppressors.
Figure 1. Diversity of deletions at the PARK2 locus in colon cancer and GBM.
Array CGH segmentation map showing GBM (TCGA) and colon cancers (Memorial Sloan-Kettering Cancer Center) for the area surrounding PARK2 on chromosome 6. Analysis and scores were calculated as previously described57. Tumors are sorted by amount of loss at the PARK2 locus for convenient viewing. Only tumors showing loss on 6q are shown. The color gradient depicts the extent of copy number loss. The position and boundaries of the PARK2 gene (red bar) are indicated. PARK2 direction and individual exons are labeled (green arrow). Surrounding genes are indicated with gray arrows.
Table 1.
Frequencies of PARK2 Copy Number Loss
| Cancer type | Total samples with alterations |
No. heterozygous loss |
No. homozygous loss |
Total samples |
|---|---|---|---|---|
| Glioblastoma | 53 (24.5%) | 48 (22.2%) | 5 (2.3%) | 216 |
| Colon | 24 (24.4%) | 18 (18.4%) | 6 (6.1%) | 98 |
| Total | 77 | 66 | 11 | 314 |
No somatic mutations in PARK2 have been reported to date. To determine whether PARK2 mutations are present in GBM and other human tumors, we sequenced all exons of the gene in 242 human cancers (Supplementary Table 1). Whenever a presumptive mutation was identified in a primary tumor, we verified that the change did not correspond to a known SNP and determined whether it was somatically acquired (that is, tumor specific) by examining the sequence of the gene in genomic DNA from normal tissue of the same individual. No study participant had a history of early-onset PD, nor did any carry germline PD-associated alleles. Using this strategy, we identified PARK2 somatic mutations in human cancers for the first time, to our knowledge (Table 2, Supplementary Table 1 and Supplementary Fig. 2).
Table 2.
Somatic Mutations of PARK2 in Human Cancers
| Cancer type | Genomic position |
Normal genotype |
Tumor genotype |
Amino acid change |
Zygosity | Domain |
|---|---|---|---|---|---|---|
| Glioblastoma | 161889928 | GAG | GGG | E344G | het | InterPro IPR002867 IBR domain |
| Glioblastoma | 162126841 | CGG | CAG | R275Q | het | Ring finger domain |
| Glioblastoma | 162542170 | ACG | GCG | T173A | het | SH2-like domain |
| Glioblastoma | 162784379 | CGT | TGT | R42C | het | InterPro IPR000626 Ubiquitin domain |
| Glioblastoma | 163068687 | ATA | GTA | I2V | het | InterPro IPR000626 Ubiquitin domain |
| Glioblastoma | 161890026 | C | T | eliminates 3’ splice site (position 0) | het | Exon 8 |
| Glioblastoma cell line – T98G | 161701212 | GAA | TAA | E395STOP | het | truncation |
| Lung | 162126904 | AAC | AGC | N254S | het | Ring finger domain |
| Lung | 162314331 | GAC | AAC | D243N | het | Ring finger domain |
| Lung | 162126829 | CAC | CCC | H279P | het | Ring finger domain |
| Lung | 162784367 | GCA | ACA | A46T | hom | InterPro IPR000626 Ubiquitin domain |
| Colon cell line – DLD1 (both alleles mut.) | 161701136 161727847 |
CGC GCC |
CAC GTC |
R420H A379V |
het het |
Ring finger domain |
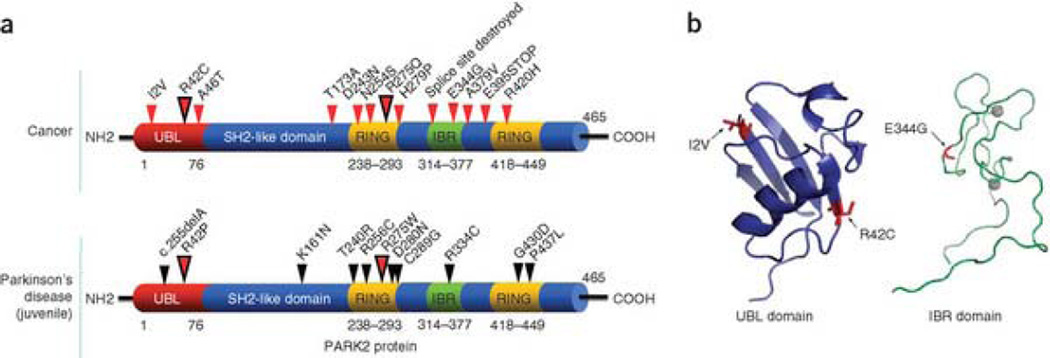
Figure 2 shows the distribution of our newly discovered mutations of PARK2 in cancers (Fig. 2a, top diagram). The bottom diagram shows the most common point mutations that cause early-onset PD. Notably, somatic PARK2 mutations in cancer occur in the same domains as the germline PD mutations. In both cases, mutations cluster in the ubiquitin-like domain (UBL), the RING finger domain and the in-between RING fingers domain (IBR). Two residues, Arg42 and Arg275, are mutated both in colon cancer and GBM and in PD. However, the resultant amino acids differ between the somatic cancer mutations and the germline PD mutations. We have mapped several of the cancer-specific mutations that lie within domains whose structures have been solved (UBL and IBR)33, 34. The UBL domain is required for interactions with proteasomes and ligands. Disruption of Arg42 is predicted to disturb these interations33. Ile2 is located on the surface of the conserved β sheet of the UBL domain. The E344G mutation is located in the IBR domain, a region crucial for interaction with E2 and other members of the ubiquitination machinery35, 36, 37. This mutation lies adjacent to the zinc-binding core and resides within a region predicted to be critical for proper ubiquitination (Fig. 2b)34.
Figure 2. Somatic mutations of PARK2 in human cancers.
(a) Summary of PARK2 mutations found in cancer (top) and early-onset Parkinson's disease (bottom). Small arrows show the location of mutations and corresponding amino acid changes. Larger dual-color arrows indicate amino acids that are affected in both cancer and PD; resultant amino acids are different. Mutations cluster in similar regions in both cancer and PD. (b) Structural analysis of cancer-specific mutations in the UBL (left) and IBR (right) domains. Ribbon diagram is shown (with alpha helices and beta sheets). Mutations are shown in red and labeled. Gray circles represent zinc atoms.
Examination of the copy number and mutation data (Fig. 1, Table 2) shows that although homozygous alterations do occur, most changes were heterozygous in nature. Thus, it may be that inactivation of a single copy of PARK2 is sufficient to impart a clonal growth advantage during tumor development. It is interesting to note that, in the literature, there is well-described precedent for haploinsufficiency of another cyclin E–targeting E3 ligase—encoded by the tumor suppressor FBXW7, also known as hCDC4 (ref. 38).
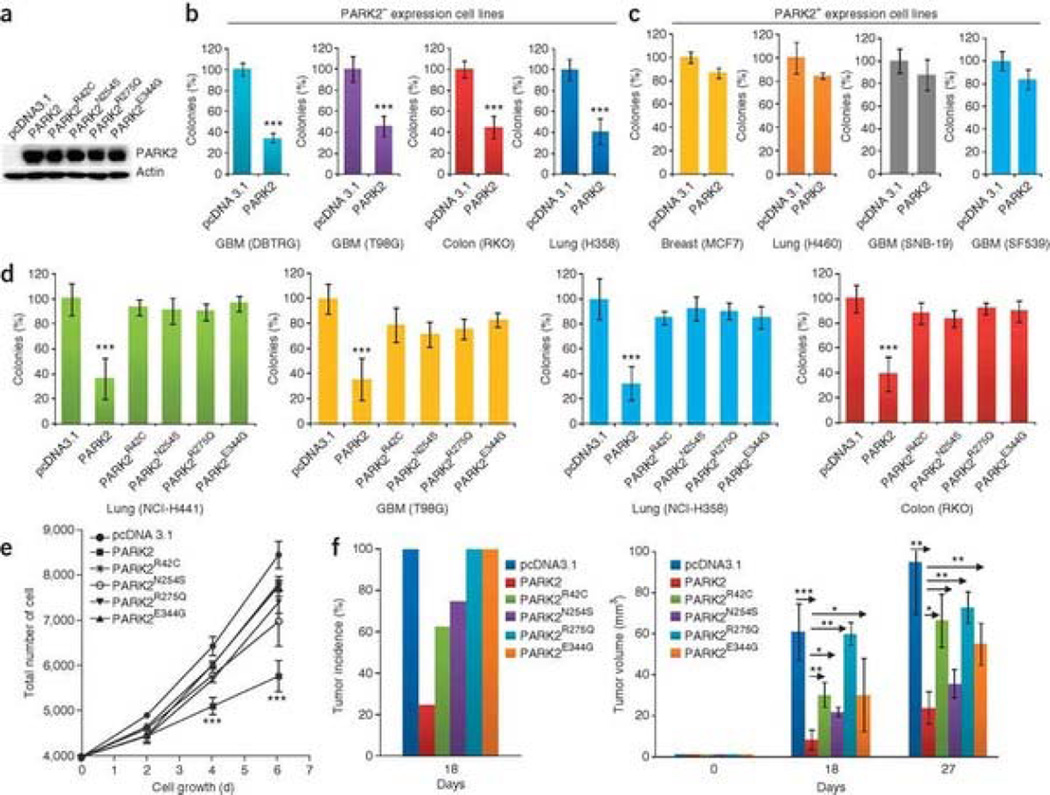
The molecular function of PARK2 in neurons is a subject of considerable investigation, and it is still not clear how PARK2 mutations cause PD. Less is known about the biological function of PARK2 in human cancers. We first sought to determine if PARK2 possesses growth-suppressive properties. PARK2 protein expression was determined in several cell lines (Supplementary Fig. 3). We cloned wild-type (WT) PARK2 cDNA and four PARK2 mutants. Transfection of all cDNAs resulted in production of PARK2 protein (Fig. 3a). To examine the functional consequences of reconstituting PARK2 expression in cancer cells, we transfected WT PARK2 into human cancer cell lines. PARK2 potently inhibited colony-forming activity in cell lines lacking PARK2 protein expression (Fig. 3b, Supplementary Fig. 4a) but not in cell lines that retained PARK2 expression (Fig. 3c, Supplementary Fig. 4b). We next sought to determine whether the cancer-specific mutations in PARK2 altered the protein's growth-suppressive properties. Expression of PARK2 with tumor-derived mutations resulted in substantially decreased colony-forming activity as compared to WT PARK2 (Fig. 3d, Supplementary Fig. 4c). Transfection of WT but not mutant PARK2 into cells from the DBTRG line resulted in a reduction in the rate of cell growth (Fig. 3e). WT PARK2 decreased tumor growth in vivo, a property that was reduced by the cancer-specific mutations (Fig. 3f). These data demonstrate that the PARK2 mutations in cancers have clear functional consequences.
Figure 3. Functional analysis of somatic PARK2 mutations in human cancer cell lines.
(a) Protein blot showing expression of WT PARK2 and PARK2 with four cancer-specific mutations. Representative data for transfection into T98G are shown. pcDNA3.1, vector-only control. (b) Reconstitution of WT PARK2 suppresses colony-forming ability of human cancer cells lacking PARK2 expression. All assays performed in triplicate. Error bars, ± 1 s.d. ***P < 0.001 (Student's t-test) in all cases. (c) Specificity of PARK2 suppressive effects on colony formation. WT PARK2 was transfected into PARK2-expressing cell lines. Suppressive effects on colony formation are minimal in PARK2+ lines. P > 0.1 (Student's t-test) for all experiments. Error bars, ± 1 s.d. (d) Tumor-derived mutations compromise the colony-forming ability of PARK2 in cancer cells. WT or mutant PARK2 was transfected into the cells indicated. All experiments performed in triplicate. ***P < 0.001 (ANOVA) for all mutants. Error bars, ± 1 s.d. (e) Reconstitution of PARK2 reduces growth rate in cancer cells. DBTRG cells were transfected with each of the constructs shown. All experiments were performed in triplicate. ***P < 0.0001 (ANOVA) for WT PARK2 compared to all others. Error bars, ± 1 s.d. (f) PARK2 reconstitution results in decreased tumor growth in vivo. DBTRG glioma cells stably transfected with vector alone, WT PARK2 and four PARK2 mutants were injected as xenografts. Tumor incidence (left) and tumor size (right) are shown (n= 16). Days on x-axis refer to days following injection of cells into animals. All experiments were performed in duplicate. Arrows indicate comparisons made. *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA). Error bars, ± 1 s.d.
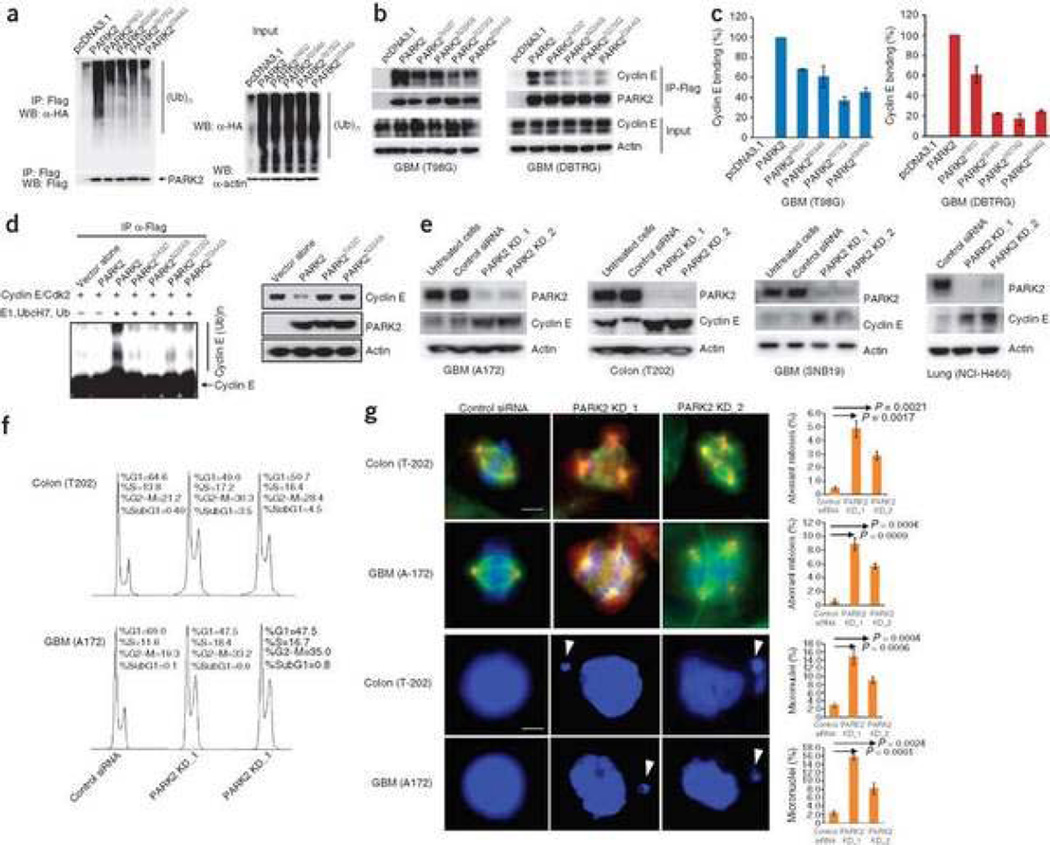
What are the molecular mechanisms underlying PARK2 tumor suppression? PARK2 has previously been shown to be a ubiquitin E3 ligase that facilitates the ubiquitination of target proteins, leading to proteasome-mediated degradation8, 13, 39, 40. We first wanted to determine whether cancer-specific mutations in PARK2 altered its ubiquitin ligase activity. We used a well-established assay to measure the ubiquitination function of PARK2 mutants in cells8, 11, 41. Cancer-specific mutations in PARK2 were found to substantially compromise the association of PARK2 with ubiquitinated target proteins in cancer cells (Fig. 4a). The mutations appeared to substantially decrease, but not completely abolish, the E3 ligase function. It is widely known that cyclin E is a fundamental component of the cell cycle machinery and is encoded by an oncogene42, 43, 44. We found that all PARK2 cancer mutations we analyzed resulted in a decreased ability of PARK2 to interact with cyclin E (Fig. 4b,c). Furthermore, the cancer-specific mutations compromised PARK2's ability to ubiquitinate cyclin E in vitro and degrade it (Fig. 4d). PARK2 mutations did not alter the protein's ability to regulate phosphorylation of c-Jun (Supplementary Fig. 5), another candidate effector of PARK2 function identified in neuronal systems19, 45. Thus, the cancer-specific mutations in PARK2 abrogate the protein's ability both to block tumor cell growth and to ubiquitinate cyclin E, establishing a mechanistic link for the loss-of-function mutations.
Figure 4. PARK2 cancer-specific mutations compromise ubiquitination activity.
(a) Tumor-derived mutations disrupt PARK2-mediated ubiquitination in cancer cells. T98G cells were transfected with hemagglutinin-ubiquitin (HA-Ub), vector only (pcDNA3.1), WT PARK2 (Flag-tagged) or one of four mutant PARK2 cDNAs (Flag-tagged). Assay was performed as previously described8. (b) Cancer-derived mutations of PARK2 decrease association with cyclin E. Indicated cells were treated as above, immunoprecipitated with Flag and detected by protein blot. (c) Quantitation of cyclin E binding efficiency by densitometry. Representative plots shown. For each mutant versus WT, P < 0.05 (Student's t-test). Error bars, ± 1 s.d. (d) Protein blot showing cancer-derived mutations that compromise PARK2-mediated cyclin E ubiquitination in vitro (left). Expression of WT PARK2 but not mutant PARK2 decreases cyclin E levels (right). (e) Knockdown of PARK2 results in increased cyclin E levels. Cells indicated were transfected with PARK2 siRNAs or scrambled siRNA controls and protein blots were performed. (f) Flow cytometry analysis of the indicated cells following PARK2 knockdown. Experiments were performed in triplicate. Representative results are shown. (g) Knockdown of PARK2 results in multipolar spindles and increased frequency of abnormal mitoses (top two rows) and the development of micronuclei (bottom two rows, white arrows). Examples for indicated cells shown using siRNAs targeting PARK2 and scrambled siRNA controls. Red, γ-tubulin; green, α-tubulin. Graphs show quantitation of experiments. Black arrows indicate comparisons made and corresponding P values (Student's t-test). White scale bar for top two rows, 15 µm; bottom two rows, 5 µm. Error bars, ± 1 s.d.
If PARK2 normally targets cyclin E for ubiquitination and degradation in cancer cells, then depletion of PARK2 should result in an increase of cyclin E levels. We knocked down PARK2 in four cancer cell lines that show PARK2 expression. In all cell lines examined, knockdown of PARK2 with two independent short interfering RNAs (siRNAs), but not with scrambled-sequence siRNAs, resulted in an accumulation of cyclin E levels (Fig. 4e). Thus, PARK2 mutation and inactivation disrupts the ability of PARK2 to ubiquitinate cyclin E.
Fluorescence-activated cell sorting analysis revealed that PARK2 knockdown increased the proportion of cells in the S and the G2-M phases (Fig. 4f). Immunofluorescence staining showed a significant increase in the frequency of multipolar spindles and abnormal mitoses (Fig. 4g). Furthermore, PARK2 knockdown cells showed a marked increase in nuclear atypia characterized by micronuclei. Notably, this is what one observes when cyclin E is overexpressed46 or when FBXW7 (hCDC4), another protein that targets cyclin E for degradation, is inactivated47, 48. These data show that PARK2 inactivation can lead to impaired mitosis.
The genetic and functional data we have presented demonstrate that the PD-associated gene PARK2 is a bona fide tumor suppressor gene that is inactivated and mutated in GBM, colon cancer and lung cancer. Genetic loss or mutational inactivation of PARK2 abrogates the ability of PARK2 to promote ubiquitination and results in cyclin E dysregulation, which can promote tumor cell growth 49. Although the gene encoding cyclin E is an oncogene that has been strongly linked to tumorigenesis, we nevertheless cannot rule out the possibility that regulation of other targets is important. In addition, our study reveals several important points. First, the finding of somatic mutations and high frequency intragenic copy-number loss provides the strongest evidence yet that PARK2 is the (or at least one of the) 'long-sought' tumor suppressors on chromosome 6q. PARK2 may be one of a select group of tumor suppressors inactivated in a wide range of human malignancies23, 24, 32, 50. Second, we determined that PARK2 mutations in cancer can decrease the E3 ligase's ability to ubiquitinate cyclin E. Many human tumors have increased cyclin E levels 42, but to date, the mechanisms underlying this increase are unclear. Our study suggests that PARK2 can target cyclin E for ubiquitination, which, together with other factors such as FBXW7 (hCDC4), helps regulate cyclin E levels48, 51, 52. Because PARK2 is mutated in both PD and cancer, it is tempting to hypothesize that alterations of this gene may result in very different phenotypes depending on cellular context (Supplementary Fig. 6).
The finding of somatic mutations of PARK2, a PD-causing gene, in cancer is noteworthy from a pathophysiologic standpoint. It seems that inactivation of certain genes, such as PARK2, results in distinct physiological outcomes depending on cellular context. Indeed, the PTEN and ATM genes function as tumor suppressors, but their inactivation also leads to neuronal loss when the mutations are in the germline53, 54. Unlike in these two cases, PARK2 germline mutation gives rise to a neurological disease but not also to a cancer predisposition syndrome. It is possible that PARK2 function may result in biological outcomes that are very different depending on whether the affected cell is a neuron or a dividing cell such as an astrocyte or epithelial cell; this seems more true for PARK2 than for PTEN or ATM. Notably, cohorts of individuals with PD do reveal a small but significant increase in the risk of malignancies such as brain and lung cancers55, 56. We believe our study has wide implications for understanding oncogenesis for a number of tumor types.
Methods
Tumor samples, array CGH analysis and bioinformatics
Colon tumor samples (n=98) from the Memorial Sloan-Kettering Cancer Center were obtained following participant consent and with institutional review board approval (Supplementary Note). Source DNAs were extracted from primary tumors for the aCGH study. The GBMs (n= 216) in the aCGH study were part of the TCGA initiative (4/14/2008 data freeze) (see URL section). aCGH was performed using the Agilent 244K microarray according to the manufacturer's instructions (Agilent Technologies). In our examination of both the colon cancer and GBM datasets, analysis of data for the CNAs observed was performed using the RAE method. The status of genomic loss at the PARK2 locus in colon cancer samples was assigned as either likely heterozygous loss (D0 ≥ 0.9) or homozygous deletion (D0 ≥ 0.9 and D1 ≥ 0.5), and in GBM as previously described per the multi-component model in RAE23, 57. Cell lines sequenced were T98G, DBTRG, RKO, H441 and H358.
PCR amplification and sequencing
Exonic regions for the PARK2 gene (NCBI Human Genome Build 36.1) were broken into 16 amplicons of 500 bp or less, and specific primers were designed using Primer3. Primers are listed in Supplementary Table 2. Standard M13 tails were added to the primers to facilitate Sanger sequencing. PCR reactions were carried out in 384-well plates in a Duncan DT-24 water bath thermal cycler with 10 ng of whole-genome amplified DNA (REPLI-g Midi, Qiagen) as a template, using a touchdown PCR protocol with KAPA Fast HotStart (Kapa Biosystems). The touchdown PCR method consisted of: 1 cycle of 95 °C for 5 min; 3 cycles of 95 °C for 30 s, 64 °C for 15 s, 72 °C for 30 s; 3 cycles of 95 °C for 30 s, 62 °C for 15 s, 72 °C for 30 s; 3 cycles of 95 °C for 30 s, 60 °C for 15 s, 72 °C for 30 s; 37 cycles of 95 °C for 30 s, 58 °C for 15 s, 72 °C for 30 s; 1 cycle of 70 °C for 5 min. Templates were purified using AMPure (Agencourt Biosciences). The purified PCR reactions were split into two and sequenced bidirectionally with M13 forward and reverse primer and the Big Dye Terminator Kit v.3.1 (Applied Biosystems) at Agencourt Biosciences. Dye terminators were removed using the CleanSEQ kit (Agencourt Biosciences), and sequence reactions were run on ABI PRISM 3730xl sequencing apparatus (Applied Biosystems).
Mutation detection
Passing reads were assembled against the PARK2 reference sequence, which contains all coding exons in PARK2 including those 5 kb upstream and downstream of the gene, using command line Consed 16.0 (ref. 58). Assemblies were passed on to Polyphred 6.02b59, which generated a list of putative candidate mutations, and to Polyscan 3.0 (ref. 60), which generated a second list of putative mutations. The lists were merged together into a combined report, and the putative mutation calls were normalized to '+' genomic coordinates and annotated using the genomic mutation consequence calculator61. The resulting list of annotated putative mutations was loaded into a Postgres database along with select assembly details for each mutation call (assembly position, coverage and methods supporting mutation call). To reduce the number of false positives generated by the mutation detection software packages, only point mutations that are supported by at least one bidirectional read pair and at least one sample mutation called by Polyphred were considered, and only the putative mutations that are annotated as having nonsynonymous coding effects, occur within 11 bp of an exon boundary, or have a conservation score >0.699 (see URL section) were included in the final candidate list. Indels were manually reviewed and included in the candidate list if found to hit an exon. All putative mutations were confirmed by a second PCR and sequencing reaction in parallel with amplification and sequencing of matched normal tissue DNA.
Cell culture
All cell lines were obtained from American Type Tissue Culture and cultured using the recommended media (Invitrogen) + 10% FBS (Invitrogen) and penicillin plus streptomycin at 37 °C in 5% CO2. HEK 293T cells were cultured in Dulbecco's modified eagle's medium (DMEM) + 10% FBS (Invitrogen). Expression of PARK2 was accomplished by cloning the gene into the vector pcDNA 3.1 with a Flag tag (Invitrogen). Transfection was performed using Lipofectamine reagent according to the manufacturer's protocol (Invitrogen). Selection was performed using G418 or hygromycin. Cells used in colony formation assays were stained with crystal violet. Growth curve assays were quantified by manual counting with a Motic inverted microscope. All experiments were performed in triplicate.
Protein blot, immunoprecipitation and immunostaining
Protein blot analysis was performed using standard methods. Antibody against the Flag epitope, the hemagglutinin tag and beta-actin were obtained from Sigma. PARK2 and cyclin E1 antibodies were obtained from Cell Signaling. Immunoprecipitation was performed using the Flag immunoprecipitation kit (Sigma). In vivo ubiquitination assay was performed as previously described8. The in vitro PARK2 ubiquitination assay was performed using the Parkin ubiquitination kit (Boston Biochem) per the manufacturer's protocol. Immunostaining was performed with antibodies to α-tubulin and γ-tubulin as previously described47. Staining for atypical nuclei and micronuclei was performed as previously described 48 h after siRNA transfection47.
Knockdown of PARK2
PARK2 siRNAs were obtained from Invitrogen. PARK2 targeted sequences are listed in Supplementary Table 2. For siRNA knockdown of PARK2, cells were transfected using Lipofectamine RNAiMAX system (Invitrogen).
Retrovirus production
For retrovirus production, WT PARK2 and mutants were cloned into the vector pQCXIP (Clontech). HEK 293T cells were seeded in 10-cm-diameter dishes. The HEK 293T packaging cells (at 30–50% confluency) were co-transfected using Lipofectamine (Invitrogen) with pE-ampho vector (Takara Bio) and pQCXIP-PARK2. Retroviral particles were collected, filtered through the 0.45-µm syringe filter and used in the presence of polybrene (8 µg/ml final concentration) to infect cells for 12 h.
Site-directed mutagenesis
Mutations identified were engineered into pcDNA3.1-PARK2 using the QuikChange II XL kit (Stratagene). All changes were verified by Sanger sequencing.
Flow cytometry
Cells were trypsinized, fixed and stained using the standard propidium iodide method 48 h after transfection. Cell cycle analysis was performed on stained cells using a MoFlo cell sorter (Cytomation).
Mouse xenograft studies
1 × 106 cells were suspended in 50% Matrigel and injected into the flanks of severe combined immunodeficiency mice. Growth was followed over time by taking caliper measurements. Eight mice were injected and 16 tumors were assessed for each condition.
Statistical analysis
Two-tailed Student's t-test analysis was performed using GraphPad Prism software.
URLs
TCGA initiative, http://cancergenome.nih.gov/index.asp; Primer3, http://frodo.wi.mit.edu/primer3/; Postgres, http://www.postgresql.org/; 17- way Cons Track Settings, http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=108554407&g=multiz17way; GraphPad Prism, http://www.graphpad.com/prism/Prism.htm.
Accession numbers
Colon cancer datasets are deposited in the Gene Expression Omnibus via accession number GSE18638. All GBM datasets are publically available at http://cancergenome.nih.gov.
Accession codes
Referenced accessions
Gene Expression Omnibus
Supplementary Material
Acknowledgments
We thank J. Wongvipat, K. Huberman, I. Dolgalev and S. Thomas for exceptional technical expertise. We thank R. Levine for helpful discussions and comments. B.S.T. is the David H. Koch Fellow in cancer genomics. This work was supported in part by The Brain Tumors Funders' Collaborative Fund (I.K.M.), The Cancer Genome Atlas Project (N.S, C.S.), the Flight Attendants Medical Research Institute (T.A.C.), the Louis Gerstner Foundation (T.A.C.), the Memorial Sloan-Kettering Society (T.A.C.), the Elsa U. Pardee Foundation (T.A.C.) and the Doris Duke Charitable Foundation (T.A.C.).
Footnotes
Contributions
T.A.C. and S.V. designed the experiments. S.V., S.M., F.F., E.Y. and M.J. performed the experiments. S.V., B.S.T., N.S., C.S. and T.A.C. analyzed the data. W.P., M.L., E.C.H., I.V., P.B.P., L.L., P.S.M., A.H., T.F.C., A.J.H., I.K.M. and D.B.S. contributed new reagents and analytic tools. T.A.C. and S.V. wrote the paper.
Contributor Information
Selvaraju Veeriah, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Barry S Taylor, Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Shasha Meng, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Fang Fang, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Emrullah Yilmaz, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Igor Vivanco, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Manickam Janakiraman, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Nikolaus Schultz, Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Aphrothiti J Hanrahan, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
William Pao, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.; Department of Medicine, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Marc Ladanyi, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.; Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Chris Sander, Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Adriana Heguy, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Eric C Holland, Department of Neurosurgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Philip B Paty, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA..
Paul S Mischel, David Geffen School of Medicine, University of California, Los Angeles, California, USA..
Linda Liau, David Geffen School of Medicine, University of California, Los Angeles, California, USA..
Timothy F Cloughesy, David Geffen School of Medicine, University of California, Los Angeles, California, USA..
Ingo K Mellinghoff, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.; Department of Neurology, University of California, Los Angeles, California, USA.
David B Solit, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.; Department of Medicine, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Timothy A Chan, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.; Department of Radiation Oncology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
References
- 1.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 2.Lücking CB, et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N. Engl J. Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 3.Fearnley JM, Lees AJ. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114:2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 4.Samii A, Nutt JG, Ransom BR. Parkinson's disease. Lancet. 2004;363:1783–1793. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 5.Abbas N, et al. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. French Parkinson's Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson's Disease. Hum. Mol. Genet. 1999;8:567–574. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- 6.Ledesma MD, et al. Astrocytic but not neuronal increased expression and redistribution of parkin during unfolded protein stress. J. Neurochem. 2002;83:1431–1440. doi: 10.1046/j.1471-4159.2002.01253.x. [DOI] [PubMed] [Google Scholar]
- 7.Kitada T, et al. Molecular cloning, gene expression, and identification of a splicing variant of the mouse parkin gene. Mamm. Genome. 2000;11:417–421. doi: 10.1007/s003350010080. [DOI] [PubMed] [Google Scholar]
- 8.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 9.Hampe C, et al. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- 10.Corti O, et al. Parkinson's disease: from causes to mechanisms. C. R. Biol. 2005;328:131–142. doi: 10.1016/j.crvi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Shimura H, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 12.Sriram SR, et al. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- 13.Corti O, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 14.Staropoli JF, et al. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- 15.Moore DJ, et al. Parkin mediates the degradation-independent ubiquitination of Hsp70. J. Neurochem. 2008;105:1806–1819. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith WW, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc. Natl. Acad. Sci. USA. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai Y, et al. CHIP is associated with Parkin a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa T, et al. Parkin protects against tyrosinase-mediated dopamine neurotoxicity by suppressing stress-activated protein kinase pathways. J. Neurochem. 2008;105:1700–1715. doi: 10.1111/j.1471-4159.2008.05277.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, et al. Parkin regulates Eg5 expression by Hsp70 ubiquitination-dependent inactivation of c-Jun NH2-terminal kinase. J. Biol. Chem. 2008;283:35783–35788. doi: 10.1074/jbc.M806860200. [DOI] [PubMed] [Google Scholar]
- 20.Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cesari R, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc. Natl. Acad. Sci. USA. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toma MI, et al. Loss of heterozygosity and copy number abnormality in clear cell renal cell carcinoma discovered by high-density Affymetrix 10K single nucleotide polymorphism mapping array. Neoplasia. 2008;10:634–642. doi: 10.1593/neo.08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denison SR, et al. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22:8370–8378. doi: 10.1038/sj.onc.1207072. [DOI] [PubMed] [Google Scholar]
- 28.Wang F, et al. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosom. Cancer. 2004;40:85–96. doi: 10.1002/gcc.20020. [DOI] [PubMed] [Google Scholar]
- 29.Yin D, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol. Cancer Res. 2009;7:665–677. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 30.Ohta M, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 31.Sozzi G, et al. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 32.Cox C, et al. A survey of homozygous deletions in human cancer genomes. Proc. Natl. Acad. Sci. USA. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata E, et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 2003;4:301–306. doi: 10.1038/sj.embor.embor764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beasley SA, Hristova VA, Shaw GS. Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2007;104:3095–3100. doi: 10.1073/pnas.0610548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dächsel JC, et al. Parkin interacts with the proteasome subunit alpha4. FEBS Lett. 2005;579:3913–3919. doi: 10.1016/j.febslet.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Rankin CA, Joazeiro CA, Floor E, Hunter T. E3 ubiquitin-protein ligase activity of Parkin is dependent on cooperative interaction of RING finger (TRIAD) elements. J. Biomed. Sci. 2001;8:421–429. doi: 10.1007/BF02255952. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Noel G, Müller U, Harbers K. Identification of molecular determinants required for interaction of ubiquitin-conjugating enzymes and RING finger proteins. Eur J. Biochem. 2001;268:5912–5919. doi: 10.1046/j.0014-2956.2001.02541.x. [DOI] [PubMed] [Google Scholar]
- 38.Mao JH, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 39.Betarbet R, Sherer TB, Greenamyre JT. Ubiquitin-proteasome system and Parkinson's diseases. Exp. Neurol. 2005;191(Suppl. 1):S17–S27. doi: 10.1016/j.expneurol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Dohm CP, Kermer P, Bahr M. Aggregopathy in neurodegenerative diseases: mechanisms and therapeutic implication. Neurodegener. Dis. 2008;5:321–338. doi: 10.1159/000119459. [DOI] [PubMed] [Google Scholar]
- 41.Schlossmacher MG, Shimura H. Parkinson's disease: assays for the ubiquitin ligase activity of neural Parkin. Methods Mol. Biol. 2005;301:351–369. doi: 10.1385/1-59259-895-1:351. [DOI] [PubMed] [Google Scholar]
- 42.Donnellan R, Chetty R. Cyclin E in human cancers. FASEB J. 1999;13:773–780. doi: 10.1096/fasebj.13.8.773. [DOI] [PubMed] [Google Scholar]
- 43.Courjal F, et al. Cyclin gene amplification and overexpression in breast and ovarian cancers: evidence for the selection of cyclin D1 in breast and cyclin E in ovarian tumors. Int J. Cancer. 1996;69:247–253. doi: 10.1002/(SICI)1097-0215(19960822)69:4<247::AID-IJC1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 44.Kitagawa K, Kotake Y, Kitagawa M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci. 2009;100:1374–1381. doi: 10.1111/j.1349-7006.2009.01196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Um JW, et al. Molecular interaction between parkin and PINK1 in mammalian neuronal cells. Mol. Cell. Neurosci. 2009;40:421–432. doi: 10.1016/j.mcn.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Keck JM, et al. Cyclin E overexpression impairs progression through mitosis by inhibiting APC (Cdh1) J. Cell Biol. 2007;178:371–385. doi: 10.1083/jcb.200703202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopalan H, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 48.Ekholm-Reed S, et al. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004;64:795–800. doi: 10.1158/0008-5472.can-03-3417. erratum 64, 2939 (2004) [DOI] [PubMed] [Google Scholar]
- 49.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 50.Mhawech-Fauceglia P, et al. Array-comparative genomic hybridization analysis of primary endometrial and ovarian high-grade neuroendocrine carcinoma associated with adenocarcinoma: mystery resolved? Int J. Gynecol. Pathol. 2008;27:539–546. doi: 10.1097/PGP.0b013e31816bcda4. [DOI] [PubMed] [Google Scholar]
- 51.Sangfelt O, et al. Both SCF(Cdc4α) and SCF(Cdc4γ) are required for cyclin E turnover in cell lines that do not overexpress cyclin E. Cell Cycle. 2008;7:1075–1082. doi: 10.4161/cc.7.8.5648. [DOI] [PubMed] [Google Scholar]
- 52.Strohmaier H, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 53.Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 54.Shiloh Y, Rotman G. Ataxia-telangiectasia and the ATM gene: linking neurodegeneration, immunodeficiency, and cancer to cell cycle checkpoints. J. Clin. Immunol. 1996;16:254–260. doi: 10.1007/BF01541389. [DOI] [PubMed] [Google Scholar]
- 55.Olsen JH, et al. Atypical cancer pattern in patients with Parkinson's disease. Br J. Cancer. 2005;92:201–205. doi: 10.1038/sj.bjc.6602279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Møller H, Mellemkjaer L, McLaughlin JK, Olsen JH. Occurrence of different cancers in patients with Parkinson's disease. Br. Med. J. 1995;310:1500–1501. doi: 10.1136/bmj.310.6993.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor BS, et al. Functional copy-number alterations in cancer. PLoS One. 2008;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 59.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K, et al. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–666. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Major JE. Genomic mutation consequence calculator. Bioinformatics. 2007;23:3091–3092. doi: 10.1093/bioinformatics/btm339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.