Abstract
Background
NOD2 single nucleotide polymorphisms have been associated with increased risk of ileal Crohn’s disease. This exploratory study was conducted to compare ileal mucosal gene expression in Crohn’s disease (CD) patients with and without NOD2 risk alleles.
Methods
Ileal samples were prospectively collected from eighteen non-smoking CD patients not treated with anti-TNFα biologics and nine non-smoking control patients without inflammatory bowel disease undergoing initial resection, and genotyped for the three major NOD2 risk alleles (Arg702Trp, Gly908Arg, Leu1007fs). Microarray analysis was performed in samples from four NOD2R (at least one risk allele) CD patients, four NOD2NR (no risk alleles) CD patients and four NOD2NR controls. Candidate genes selected by significance analysis of microarrays (SAM) were confirmed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays of all the samples.
Results
SAM detected upregulation of 18 genes in affected ileum in NOD2R compared to NOD2NR CD patients, including genes related to lymphocyte activation. SAM also detected altered ileal gene expression in unaffected NOD2NR ileal mucosal CD samples compared to NOD2NR control samples. QRT-PCR conducted on all the samples confirmed that increased CD3D expression in affected samples was associated with NOD2R status, and that increased MUC1, DUOX2, DMBT1 and decreased C4orf7 expression in unaffected samples was associated with CD, independent of NOD2 status.
Conclusions
The results support the concept that NOD2 risk alleles contribute to impaired regulation of inflammation in the ileum. Furthermore, altered ileal gene expression, independent of NOD2 status, is detected in the unaffected proximal margin of resected ileum from CD patients.
Keywords: NOD2, Crohn’s disease, ileum, microarray
INTRODUCTION
Crohn’s disease (CD) is a chronic relapsing inflammatory disorder that can affect any part of the GI tract, but most commonly involves the terminal ileum. The majority of patients with ileal CD eventually undergo resection of diseased ileum [1]. Unfortunately, surgery is not curative. Endoscopic evidence of recurrent disease in the formerly unaffected neo-terminal ileum is often observed within months after resection [2]. Nucleotide-binding oligomerization domain 2 or NOD2 genetic polymorphisms were the first genetic risk alleles identified for Crohn’s disease [3–7]. Three single nucleotide polymorphisms (SNPs), Arg702Trp, Gly908Arg and Leu1007fs, account for about 80% of the NOD2 variants associated with Crohn’s disease. The frequency of these risk alleles is <5% in Caucasian control subjects but approximately 30% in CD patients. NOD2 risk alleles are associated with ileal disease location, younger age of onset and earlier initial surgery [6–8].
NOD2 is an intracellular receptor of bacteria that recognizes muramyl dipeptide (MDP), a component of the bacterial cell wall. NOD2 is expressed in monocytes, macrophages and epithelial cells, particularly in Paneth cells [9, 10]. Expression of α-defensins (DEFA5 and DEFA6), which are anti-microbial peptides secreted by Paneth cells, is reported to be decreased in ileal tissues collected from CD patients harboring NOD2 risk alleles (NOD2R patients) compared to CD patients with only notno risk alleles (NOD2NR) in one study [11], but a subsequent study reports that expression is decreased with inflammation and is not related to NOD2 genotype [12]. While one study detected no significant difference in colonic gene expression between NOD2R and NOD2NR CD patients [13], another study reported that expression of the deleted in malignant brain tumors 1 (DMBT1) gene, is decreased in disease affected colon biopsies from NOD2R compared to NOD2NR CD patients [14]. This gene encodes a member of the scavenger receptor cysteine-rich superfamily that binds bacteria and is implicated in mucosal protection [15]. Increased expression of DMBT1 has been reported to correlate with disease activity in inflamed Crohn’s disease tissues and DMBT1 knockout mice exhibit increased susceptibility to dextran sodium sulfate-induced colitis [16, 17].
To further examine the functional consequences of the NOD2 risk alleles, ileal mucosal gene expression was compared in tissue samples that were prospectively collected from CD patients with and without NOD2 risk alleles undergoing initial ileocolic resection.
MATERIALS AND METHODS
Patients and acquisition of ileal tissue samples
This study was approved by the Washington University-St. Louis Human Research Protection Office. CD patients undergoing initial ileocolic resection (ICR) were prospectively enrolled in a consecutive fashion by the Washington University Digestive Diseases Research Core Center (DDRCC) Tissue Procurement Facility to donate surgically resected tissue samples between April 2005 and October 2007. The indications for surgery included failure of medical management, small bowel obstruction, fistula, abscess or perforation. The diagnosis of Crohn’s disease was established according to conventional clinical and pathological criteria [18]. Patients who were unwilling or unable to give informed consent were excluded. Gender and race were recorded as reported by the patients. The patients were phenotyped using the Montreal classification with respect to age of diagnosis, disease location and disease behavior at the time of surgery as previously described [19, 20]. A detailed smoking history and medication history was obtained by reviewing the medical records and interviewing the patients. Samples from patients who smoked (≥ 7 cigarettes a week ) or received anti-TNFα therapy (e.g. infliximab or adalimumab) within a year of surgery were excluded from this study. Control ileal mucosal biopsies were collected from non-smoking patients without inflammatory bowel disease undergoing initial surgery.
Biopsies of macroscopically disease affected and unaffected areas of ileal mucosa were collected from fresh pathologic specimens using Radial JawTM4 large capacity biopsy forceps (Boston Scientific Corp., Natick, MA). Biopsies of ileal mucosa were also collected from fresh pathologic specimens of patients without inflammatory bowel disease undergoing either a right hemicolectomy or subtotal colectomy for colonic neoplasm or colonic inertia. A minimum of 4 biopsies were taken within a 2 × 2 cm2 area from each region, immediately placed in an RNA stabilization solution (RNAlater, Applied Biosystems/Ambion, Austin, TX), and archived at −80°C. The samples were de-identified and linked to a detailed clinical database by a patient study code.
Genotyping of NOD2 SNPs
Each subject was genotyped for the Leu1007fsInsC (rs2066847, SNP13), R702W (rs2066844, SNP8) and G908R (rs2066845, SNP12) SNPs by direct sequencing or by a Taqman MGB (Applied Biosystems, Foster City, CA) genotyping platform [21] using genomic DNA prepared from peripheral venous blood or tissue. NOD2R patients (R = risk alleles) were defined as patients who carried at least one of the three risk alleles (NOD2R/R and NOD2R/NR). NOD2NR patients (non-risk allele) were defined as patients who carried none of the three risk alleles (NOD2NR/NR). In addition, the patients were genotyped for the ATG16L1T300A (rs2241880) and IL23R381N (rs11109026) risk alleles by the Sequenom Technology Core within the Washington University Division of Human Genetics (http://hg.wustl.edu/info/Sequenom_description.html).
RNA isolation and Microarray Analysis
Total RNA was prepared from ileal biopsies stored in RNAlater using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations. RNA quality was assessed using an Agilent 2100 Bioanalyzer. The test ileal RNA and a common reference ileal RNA (NOD2NR, non-IBD) were labeled using the Agilent low RNA input linear amplification kit. The resulting probes were hybridized to Agilent Whole Human Genome Arrays (Agilent No.G4410A), using established protocols. Each array was globally normalized such that the overall ratio of medians of the test and reference probes was made equal to one. The arrays have been deposited in Gene Expression Omnibus (GEO accession no. GSE17594). A primary statistical analysis of the array data was performed using the significance analysis of microarrays (SAM) procedure [22]. These analyses were performed on log-transformed mean signal intensities. SAM assigns a gene-specific t-test (q-value) based on changes in gene expression relative to the standard deviation of repeated measurements for that gene. The criteria set for differences between groups in the SAM analysis were q < 0.05, fold change < 0.5 or > 2.
Quantitative real time RT-PCR
One µg total RNA was used for first strand cDNA synthesis using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations. cDNA was then diluted and amplified using SYBR Green Master Mix 2× reagent (Applied Biosystems, Foster City, CA) in an Applied Biosystems 7500 Real-time analyzer in a total of 25 µl per reaction, per the manufacturer's instructions. The target gene X was expressed relative to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene as −ΔCT (CT of GAPDH – CT of gene X), and represents a logarithmic transformation of the mRNA levels [23]. Each measurement was carried out in triplicate. The primers (see Table 1) were selected from the Primer Bank database (http://pga.mgh.harvard.edu/primerbank/). In addition to the candidate genes identified by SAM, interleukin-8 (IL-8) mRNA expression was also measured by RT-PCR as a marker of intestinal inflammation. Fold change between categorical groups was calculated as 2−ΔΔCT, where ΔΔCT was the difference between the median ΔCT values for the two groups.
Table 1.
Primer sequences for qRT-PCR assays.
| Primer Name | Primer Sequence | Accession Number |
|---|---|---|
| hIL-8 Forward | 5’-ACT GAG AGT GAT TGA GAG TGG AC-3' | NM_000584 |
| hIL-8 Reverse | 5’-AAC CCT CTG CAC CCA GTT TTC-3' | NM_000584 |
| hGAPDH Forward | 5’-CAT GAG AAG TAT GAC AAC AGC CT-3' | NM_002046 |
| hGAPDH Reverse | 5’-AGT CCT TCC ACG ATA CCA AAG T-3' | NM_002046 |
| hCD3D Forward | 5’-TAC CGT GCA AGT TCA TTA TCG AA-3’ | NM_000732 |
| hCD3D Reverse | 5’-GCC TTC CAG TCT CAT GTC CAG-3’ | NM_000732 |
| hMUC1 Forward | 5’ –AAG CAG CCT CTC GAT ATA ACC T -3’ | NM_002456 |
| hMUC1 Reverse | 5’ –GGT ACT CGC TCA TAG GAT GGT -3’ | NM_002456 |
| hDMBT1 Forward | 5’-TGC ACC TGG TCA TAC GGA GTT GAA-3' | NM_004406 |
| hDMBT1 Reverse | 5’-TGG ATT CTG GGA CGT CAG GTC TAT-3' | NM_004406 |
| hC4orf7 Forward | 5’-AGT GGC TGT TGG TTT CCC AG-3’ | NM_152997 |
| hC4orf7 Reverse | 5’-GGC GAA ATG GAT ATG GGT AAG G-3’ | NM_152997 |
Statistical analysis
Statistical analyses were carried out with the aid of Graphpad Prism 3.0. Differences in the clinical characteristics between two patient groups were compared by the nonparametric Mann-Whitney test. Differences in median levels of mRNA expression between more than two categorical groups were compared by the Kruskal Wallis test, a nonparametric alternative to the one-way analysis of variance test. The Dunn’s post-test was calculated to compare pairs of categorical groups when the Kruskal Wallis test was significant. A P value of <0.05 was considered statistically significant. All P-values were two-tailed.
An empirical procedure was applied to estimate the power in the sample for every gene that is on the microarray and passed data quality control. The power calculation was based on a two-sample t-test with pooled variance similar to the SAM statistic. We assumed that for each gene the test statistic based on the log-transformed expression levels followed an asymptotic t-distribution. The observed variation in the control samples was used to approximate the standard deviation; and various effect sizes were consider for a realistic power analysis. To minimize false positive rates, Bonferroni correction was used to derive a conservative estimate adjusting for multiple testing.
RESULTS
Clinical characteristics of patients
The ileal CD mucosal RNA samples used in this study were prospectively collected from 18 CD patients (median age 36 years, range 18–74 years, 43% female), who underwent an initial ileocolic resection between April 1, 2005 and October 1, 2007 and had never smoked or received anti-TNFα therapy within one year of surgery. The clinical characteristics of the CD patients as well as the 9 NOD2NR control subjects are summarized in Table 2. Seven of the 18 CD patients harbored at least one of the three major NOD2 risk alleles (NOD2R) and eleven patients had no risk alleles (NOD2NR). The median age of diagnosis in the NOD2R group (21 years, range 17–27 years) was significantly younger than that of the NOD2NR group (33 years, range 18–71 years, P <0.01). The median age of surgery in the NOD2R group (28 years, range 24–37 years) was also younger than the NOD2NR group (40 years, range 18–74 years, P < 0.05). However, there was no significant difference in the median duration of disease prior to surgery between NOD2NR and NOD2R CD patients. These observations are consistent with previous reports that NOD2R alleles are associated with younger age of disease onset and initial surgery [6–8]. The CD patients were homogeneous with respect to disease location, since all of the patients had disease located in the ileum with or without disease limited to the cecum (Montreal classification L1). With the exception of one patient, all patients exhibited either striucturing disease behavior (Montreal classification B2) or penetrating disease behavior (Montreal classification B3) at the time of surgery (see Table 2).
Table 2.
| a. Clinical characteristics of NOD2R CD patients. | |||||||
|---|---|---|---|---|---|---|---|
| Patient | NOD2 Genotype |
Age at Surgery (years) |
Disease Duration1 (years) |
Disease Behavior2 |
Sex | Race3 | Medication4 |
| 15* | R702W/WT | 37 | 10 | B2 | F | W | C,D |
| 196* | InsC/R702W | 24 | 7 | B3 | F | W | B |
| 235* | R702W/WT | 28 | 11 | B3 | M | W | B,D |
| 312* | InsC/InsC | 28 | 1 | B3 | F | W | B |
| 382 | InsC/G908R | 24 | 3 | B3 | M | W | A,B,C |
| 404 | R702W/WT | 24 | 0 | B3 | F | W | A,B,C,D |
| 442 | InsC/WT | 33 | 13 | B1 | M | W | C,D |
| b. Clinical characteristics of NOD2NR CD patients. | |||||||
|---|---|---|---|---|---|---|---|
| Patient | NOD2 Genotype |
Age at Surgery (years) |
Disease Duration1 (years) |
Disease Behavior2 |
Sex | Race3 | Medication4 |
| 16 | WT/WT | 40 | 4 | B2 | F | A | |
| 19* | WT/WT | 47 | 19 | B3 | M | W | B,C |
| 22* | WT/WT | 74 | 3 | B3 | M | W | A, D |
| 124* | WT/WT | 36 | 7 | B2 | M | W | A |
| 178 | WT/WT | 20 | 0 | B3 | F | W | B |
| 204 | WT/WT | 48 | 2 | B2 | M | B | A,C,D |
| 163 | WT/WT | 35 | 1 | B3 | F | B | A,C |
| 218 | WT/WT | 38 | 5 | B3 | M | W | B,C,D |
| 256* | WT/WT | 46 | 16 | B3 | M | W | A,D |
| 233 | WT/WT | 69 | 5 | B2 | F | W | A, C, D |
| 576 | WT/WT | 18 | 0 | B2 | M | W | B,C |
| C. Clinical characteristics of NOD2NR non-IBD control subjects. | |||||
|---|---|---|---|---|---|
| Sample | NOD2 Genotype |
Age at Surgery (years) |
Sex | Race1 | Diagnosis |
| N13* | WT/WT | 61 | M | W | Colonic inertia |
| N14 | WT/WT | 72 | M | W | Colon cancer |
| N15* | WT/WT | 71 | M | W | Colon cancer |
| N17* | WT/WT | 56 | F | W | Ileal nodule |
| N19 | WT/WT | 47 | F | W | Fistula, adenomatous polyp |
| N21 | WT/WT | 51 | M | B | Colonic inertia |
| N33* | WT/WT | 55 | F | W | Colonic inertia |
| N34* | WT/WT | 41 | F | W | Colonic inertia |
| N39 | WT/WT | 40 | F | W | Colonic inertia |
Disease duration is defined as Age at Surgery – Age of Diagnosis
Disease behavior at the time of surgery is defined using the Montreal Classification: B1 inflammatory (not stenosing or penetrating); B2, stenosing; B3, penetrating.
Race: W, white; B, black; A, asian.
Medication: A, 5-aminosalicylic acid; B, antibiotics (in addition to perioperative cefoxitin); C steroids; D immunomodulators (azathioprine, 6-mercaptopurine or methotrexate).
Samples selected for microarray analysis
Disease duration is defined as Age at Surgery – Age of Diagnosis
Disease behavior at the time of surgery is defined using the Montreal Classification: B1 inflammatory (not stenosing or penetrating); B2, stenosing; B3, penetrating.
Race: W, white; B, black; A, asian.
Medication: A, 5-aminosalicylic acid; B, antibiotics (in addition to perioperative cefoxitin); C steroids; D immunomodulators (azathioprine, 6-mercaptopurine or methotrexate).
Samples selected for microarray analysis
Race: W, white; B, black; A, asian.
Samples selected for microarray analysis
Comparison of gene expression by microarray
In order to identify candidate genes that exhibited altered expression due to NOD2 risk variants, whole human genome microanalyses were performed using RNA samples isolated from regions of macroscopically unaffected and affected ileal mucosa collected from the first four NOD2R CD patients recruited for the study (see Table 2). Because all the NOD2R CD patients were Caucasian, the four NOD2NR CD patients selected for microarray analysis were also Caucasian. The microarrays probed with RNA prepared from unaffected regions of the resected ileum were analyzed separately from those probed with RNA from affected regions. Significance analysis of microarrays (SAM) detected 18 genes (q < 0.05, fold change >2) that were upregulated in affected regions of the ileum in NOD2R CD patients compared to NOD2NR CD patients (see Table 3). As shown in Table 4, the mean log ratios of the intensities for the test probe relative to the reference control probe for the α-defensin genes, hDEFA5 and hDEFA6, were not significantly decreased in the NOD2R CD patients compared to NOD2NR CD patients.
Table 3.
Upregulated genes in affected ileum from NOD2R vs. NOD2NR CD patients.
| Gene Accession | Gene Description | Fold change | q-value |
|---|---|---|---|
| NM_000732* | T-cell surface glycoprotein CD3 delta chain precursor (CD3D) | 3.6 | <0.001 |
| THC2437822 | Unknown | 2.9 | <0.001 |
| THC2437476 | Unknown | 3.6 | <0.001 |
| NM_001778 | CD 48 antigen (CD48) | 6.6 | <0.001 |
| NM_002514 | Nephroblastoma overexpressed gene (NOV) | 3.3 | <0.001 |
| NM_000397 | Cytochrome b -245, beta polypeptide (CYBB) | 2.4 | <0.001 |
| NM_002828 | Protein tyrosine phosphatase, receptor type C (PTPRC) | 3.9 | <0.001 |
| NM_013351 | T-box expressed in T-cells (TBX21) | 2.9 | <0.001 |
| NM_001037332 | Cytoplasmic FMR1 interacting protein 2 (CYFIP2) | 3.0 | <0.001 |
| NM_004951 | Epstein-Barr virus induced gene 2 (EBI2) | 5.2 | <0.001 |
| ENST00000381961 | Unknown | 3.0 | <0.001 |
| NM_004510 | SP110 nuclear body protein (SP110) | 2.3 | <0.001 |
| NM_002988 | Chemokine (C-C motif) ligand 18 (CCL18) | 4.2 | 0.04 |
| NM_002738 | Protein kinase C, beta 1 (PRKCB1) | 4.7 | 0.04 |
| ENST00000371030 | Unknown | 4.1 | 0.04 |
| NM_024315 | C7 open reading frame 23 | 2.1 | 0.04 |
| A_24_P289043 | Unknown | 2.3 | 0.04 |
| THC2337832 | Unknown | 2.9 | 0.04 |
Verified by real-time RT PCR
Table 4.
Mean hDEFA5 and hDEFA6 log2 ratios of intensities (relative to a control NOD2NR RNA) from NOD2R and NOD2NR CD microarrays
| Gene ID | Tissue | NOD2R log ratio Mean ± std dev |
NOD2NR log ratio Mean ± std dev |
P (t-test) |
|---|---|---|---|---|
| hDEFA5 | Unaffected ileum | −1.6 ± 2.4 | 1.1 ± 0.6 | 0.1 |
| Affected ileum | −1.3 ± 0.6 | −0.6 ± 0.8 | 0.4 | |
| hDEFA6 | Unaffected ileum | −1.3 ± 2.7 | 1.1 ± 0.4 | 0.1 |
| Affected ileum | −1.2 ± 1.2 | 0.0 ± 0.8 | 0.2 |
To identify candidate genes that reflect early pathogenic changes prior to the development of macroscopic disease, we performed SAM analysis to compare four microarrays probed with NOD2NR CD unaffected mucosal RNA with four microarrays probed with NOD2NR control mucosal RNA. SAM detected upregulation of 165 genes (q < 0.05, fold change > 2) and downregulation of 196 genes (q < 0.05, fold change <0.5) in unaffected ileum from NOD2NR CD patients compared to NOD2NR control patients (see supplementary Tables 1 and 2).
Confirmation of candidate genes by qRT-PCR
Altered expression of selected candidate genes were then confirmed by qRT-PCR analysis of samples from all the patients listed in Table 2 (7 NOD2R CD patients, 11 NOD2NR CD patients and 9 NOD2NR control patients). We focused on candidate genes with (q < 0.001 and fold change either <0.5 or >2). The median CD3D mRNA expression was 3-fold higher (P = 0.004) in affected ileal samples from 7 NOD2R CD patients compared to 11 NOD2NR CD patients. While there was a trend towards increased expression of the CD48 and CYBB genes in affected ileal samples from NOD2R CD patients compared to NOD2NR patients, the differences did not reach statistical significance. There was no significant difference in hDEFA5 or hDEFA6 expression between NOD2R and NOD2NR patients detected by qRT-PCR assays (data not shown).
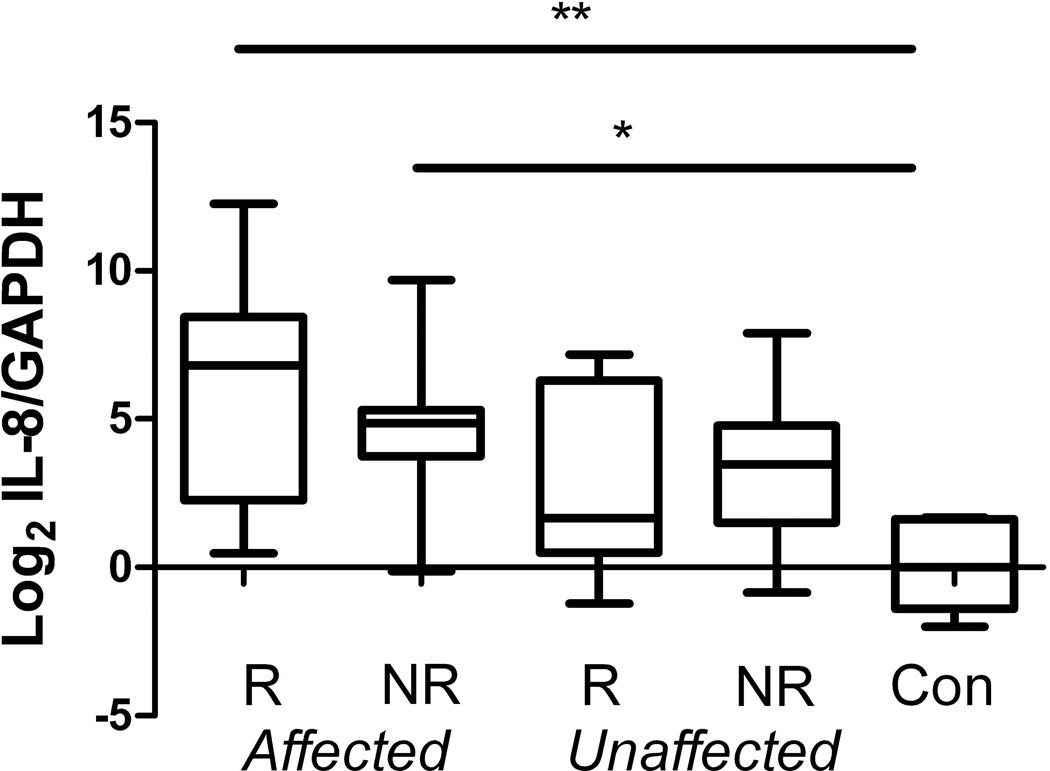
Because of the patchy nature of the inflammatory changes in Crohn’s disease, we measured IL-8 mRNA expression as an internal marker of inflammation rather than by grading the histopathology of adjacent biopsies. While the median IL-8 mRNA expression levels in affected ileum was significantly elevated compared to control ileal samples, both the affected and unaffected ileum from CD patients exhibited a considerable range of IL-8 mRNA expression levels (see Figure 1). Upregulation of MUC1 DUOX2 and DMBT1 expression and downregulation of C4orf7 gene (encodes follicular dendritic cell secreted protein) expression was confirmed by qRT-PCR analysis of a larger set of disease unaffected mucosal RNA from 17 CD patients (both NOD2R and NOD2NR CD patients) and 9 NOD2NR control subjects.The altered expression of these genes could potentially be linked to upregulation of pro-inflammatory genes such as IL-8, even in samples of macroscopically disease unaffected ileal samples. We therefore recalculated the differences in the expression of these genes after excluding CD ileal samples with elevated IL-8 mRNA expression (defined as exceeding the mean control IL-8/GAPDH ΔCT level by greater than one standard deviation or 1.4). Upregulation of the MUC1 and DMBT1 genes, and downregulation of the C4orf7 gene, remained significant at a level of P = 0.03 (uncorrected for multiple comparisons).
Figure 1.
IL-8 mRNA expression is increased in affected ileum from NOD2R(R) CD and NOD2NR(NR) CD patients compared to control subjects (Kruskal Wallis test P = 0.004, * and ** denotes significance in Dunn’s multiple comparison tests between categorical groups P < 0.05). The ends of the box represent the 25th and 75th percentiles respectively. The line in the box represents the median value. The whiskers represent the minimum and maximum values. The results are expressed relative to the median −ΔCT values from control subjects.
DISCUSSION
The major aim of this exploratory study was to identify candidate ileal genes that were affected by NOD2R status by comparing separately, whole human genome expression profiling of disease unaffected (proximal margin) and affected regions of the resected ileum in CD patients undergoing initial ileocolic resection. A secondary aim was to identify candidate genes that were altered in the disease unaffected proximal margin in CD patients compared to control non-IBD patients. In this study, the CD patients were relatively homogeneous with respect to disease location and had advanced disease requiring surgical intervention. Since smoking was linked to ileal disease location [24] and anti-TNFα (e.g. Remicade or Humira) therapy often resulted mucosal healing [25], these patients were excluded from this analysis. However, the patients were still heterogeneous with respect to exposure to other medications (e.g. steroids, immunomodulators). Because of the small sample size, patients with different NOD2 SNPs were combined into a single categorical group, regardless of gene dosage. In addition, the patients were potentially genetically heterogeneous at other CD susceptibility loci [26]. Genotyping at the ATG16L1T300A (rs2241880) revealed, however, that all of the CD patients carried at least one risk allele. Genotyping at the IL23R (rs11209026) revealed that all of the CD patients were homozygous for the risk allele except for two that were heterozygous for the risk allele.
A post-hoc power analysis estimates that 28 and 14 microarrays are were required in each categorical group to reach a median power of 80% for respectively a 2-fold and 3-fold change in expression for the >40, 000 genes present on the microarray. Hence the study is was underpowered to determine whether lack of expression differences in genes (e.g. hDEFA5 or hDEFA6) is was significant. In addition, the sample is was heterogeneous with respect to the three NOD2 risk alleles, which may not have differing effects on gene expression.
Nonetheless, a limited number of candidate genes were confirmed by qRT-PCR on samples in addition to the original set used for the microarrays. Upregulation of CD3D, in the affected ileum of NOD2R CD patients was confirmed by qRT-PCR assays. Upregulation of this gene as well as others [27–32] identified by SAM analysis, supported the concept that NOD2 risk alleles contributed to impaired regulation of inflammation in ileal CD. Both reduced tolerance to bacterial products [33, 34] and impaired clearance of invasive bacteria in NOD2R CD patients [35] were previously proposed to drive deregulated inflammation.
MUC1, DUOX2 and DMBT1 gene expression are were confirmed by qRT-PCR to be significantly upregulated in the disease unaffected proximal margin of CD compared to control non-IBD ileal resection specimens. MUC1 encodes mucin 1, an epithelial glycoprotein that is overexpressed in human adenocarcinoma and inflammatory bowel diseases [36]. This protein is implicated in host defense and in exacerbation of colitis in animal models [37]. DUOX2 encodes dual oxidase 2, which is implicated in the generation of hydrogen peroxide and is expressed along the length of the GI tract [38]. Upregulation of DUOX2 has been previously reported in disease unaffected regions of the colon from CD patients compared to control non-IBD patients [39]. DMBT1 encodes a member of the scavenger receptor cysteine-rich superfamily that binds bacteria and is implicated in mucosal protection [15]. Some of the observed changes could reflect inflammation as detected by elevated IL-8 mRNA expression in samples collected from the CD patients. However, increased expression of MUC1 and DMBT1 was still observed after exclusion of CD samples with elevated IL-8 mRNA expression levels. Because both of these genes are implicated in mucosal defense against bacteria, these results suggest that there are abnormal mucosal-microbial interactions in the region of the ileocolic anastomosis at the time of surgery. Down-regulation of C4orf7 gene expression, which encodes a follicular dendritic cell secreted peptide [40, 41], was also observed after exclusion of CD samples with elevated IL-8 mRNA expression levels. SAM also detected down-regulation of other genes (CCL23, BACH2, LRMP, FCRL4, see Supplementary Table 2) associated with organized lymphoid structures and/or B-cell function in the disease unaffected proximal margin of the resected ileum in the CD patients [42–45]. These changes in gene expression support the hypothesis that changes in lymphoid follicles and the associated epithelium play a role in the pathogenesis of Crohn’s disease [46].
Increased expression of a number of genes related to neuronal differentiation in unaffected regions of the ileum, such as Olig1, DRD4, NPAS3, LBX1 and IRX 1 (see supplementary Table 1), could reflect previously described neural proliferation in mildly affected areas or areas adjacent to severely diseased areas of the intestine in CD [47]. Isolation of a pure cell population by methods such as laser capture microdissection would be required in order to distinguish between changes in gene expression within a cell population from changes reflecting architectural alterations or compositional changes (e.g. influx of inflammatory cells) in the mucosal samples.
Further analysis of the contribution of individual NOD2 risk alleles and other risk alleles to CD pathogenesis will require collection of additional patient samples with detailed annotation of disease phenotype, genetic susceptibility loci and environmental exposures. Nonetheless, in this limited dataset, our results support the concept that NOD2 risk alleles contribute to impaired regulation of inflammation in the ileum. Furthermore, altered ileal gene expression, independent of NOD2 status, is was detected in the disease unaffected proximal margin of the ileocolic resection specimen of CD patients compared to control patients.
Table 5.
qRT-PCR of selected genes with altered regulation in unaffected ileum from CD patients regardless of NOD2 genotype.
| Gene Accession |
Gene Symbol | CD (n=16) vs. Control (n=9) Fold-change |
P -value | CD (n=7)* vs. Control (n=9) Fold-change |
P-value |
|---|---|---|---|---|---|
| NM_002456 | MUC1 | 12 | 0.0007 | 5 | 0.03 |
| NM_014080 | DUOX2 | 10 | 0.008 | 3 | 0.3 |
| NM_00739 | DMBT1 | 4 | 0.004 | 3 | 0.03 |
| NM_152997 | C4orf7 | 0.05 | 0.008 | 0.01 | 0.02 |
Excluded samples with elevated IL-8 mRNA expression
ACKNOWLEDGEMENTS
The authors thank the patients who have contributed their blood, tissue and medical information to the Digestive Diseases Research Core Center (DDRCC) Tissue Procurement Facility, Candace Miller, Michael Lewis and Drs. Elisa Birnbaum, James Fleshman, Ira Kodner, Jennifer Lowney, Matthew Mutch in the Section of Colon and Rectal Surgery, Drs. Matthew Ciorba, Ray Clouse, Nicholas Davidson, Dayna Early, Rodney Newberry, Chandra Prakash, Deborah Rubin and William Stenson in the Division of Gastroenterology at Washington University for their help and support in recruiting patients, the community gastroenterologists including Drs. Barry Abramson, Marc Bernstein, Paul Buse, David Cort, Steve Fern, John J. Kelly, Carl Lyss, Robert Shuman, Eric Thyssen, Janet Todorczuk, David Walden, Michele Woodley, Leonard Weinstock, Michael Zerega for their help and support in recruiting patients. We want to thank Dr. Rodney Newberry for helpful discussions regarding organized lymphoid structures in the intestine. We acknowledge the use of the Washington University Digestive Diseases Research Core Center Tissue Procurement Facility (P30 DK505274), the Human Genetics Division Genotyping Core and the Center for Biomedical Informatics within the Washington University Institute of Clinical and Translational Sciences (UL1 RR024992) in conducting this study.
This work was supported in part by NIH grant P30 DK52574 which supports the Washington University Digestive Diseases Research Core Center (Li), Barnes Jewish Foundation Grant 00434-0805-01 (Dietz/Hunt), Johnson and Johnson Translational Seed Award (Li), Crohn’s Colitis Foundation of America (Li). C.M. was supported by T35 DK074375.
Footnotes
This work was presented in part at the Society of Surgery Alimentary Tract Annual Meeting at San Diego May 19, 2008.
REFERENCES
- 1.Farmer RG, Whelan G, Fazio VW. Long-term follow-up of patients with CD. Relationship between the clinical pattern and prognosis. Gastroenterology. 1985;88:1818–1825. doi: 10.1016/0016-5085(85)90006-x. [DOI] [PubMed] [Google Scholar]
- 2.Rutgeerts P, Geboes K, Vantrappen G, et al. Predictability of the postoperative course of Crohn’s Disease. Gastroenterology. 1990;99:956–963. doi: 10.1016/0016-5085(90)90613-6. [DOI] [PubMed] [Google Scholar]
- 3.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 4.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad T, Armuzzi A, Bunce M, et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology. 2002;122:854–866. doi: 10.1053/gast.2002.32413. [DOI] [PubMed] [Google Scholar]
- 6.Brant SR, Picco MF, Achkar JP, et al. Defining complex contributions of NOD2/CARD15 gene mutations, age at onset, and tobacco use on Crohn's disease phenotypes. Inflamm Bowel Dis. 2003;9:281–289. doi: 10.1097/00054725-200309000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Economou M, Trikalinos TA, Loizou KT, et al. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez-Lobos M, Arostegui JI, Sans M, et al. Crohn's disease patients carrying NOD2/CARD15 gene variants have an increased and early need for first surgery due to stricturing disease and higher rate of surgical recurrence. Ann Surg. 2005;242:693–700. doi: 10.1097/01.sla.0000186173.14696.ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lala S, Ogura Y, Osborne C, et al. Crohn’s disease and the NOD2 gene: a role for Paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 10.Ogura Y, Lala S, Xin W, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wehkamp J, Harder J, Weichenthal M, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simms LA, Doecke JD, Walsh MD, et al. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 13.Csillag C, Nielsen OH, Borup R, et al. CARD15 status and familial predisposition for Crohn's disease and colonic gene expression. Dig Dis Sci. 2007;52:1783–1789. doi: 10.1007/s10620-006-9737-5. [DOI] [PubMed] [Google Scholar]
- 14.Rosenstiel P, Sina C, End C, et al. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- 15.Bikker FJ, Ligtenberg AJ, Nazmi K, et al. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J Biol Chem. 2002;277:32109–32115. doi: 10.1074/jbc.M203788200. [DOI] [PubMed] [Google Scholar]
- 16.Renner M, Bergmann G, Krebs I, et al. DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn's disease. Gastroenterology. 2007;133:1499–1509. doi: 10.1053/j.gastro.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Wu F, Dassopoulos T, Cope L, et al. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13(7):807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 18.Stange EF, Travis SP, Vermeire S, et al. European evidence based consensus on the diagnosis and management of Crohn's disease: definitions and diagnosis. Gut. 2006;55(Suppl 1):i1–i15. doi: 10.1136/gut.2005.081950a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unkart JT, Anderson L, Li E, et al. Risk factors for surgical recurrence after ileocolic resection of Crohn's disease. Dis Colon Rectum. 2008;51:1211–1216. doi: 10.1007/s10350-008-9348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abreu MT, Taylor KD, Lin YC, et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123:679–688. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaparro J, Reeds DN, Wen W, et al. Alterations in thigh subcutaneous adipose tissue gene expression in protease inhibitor-based highly active antiretroviral therapy. Metabolism. 2005;54:561–567. doi: 10.1016/j.metabol.2004.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldhous MC, Drummond HE, Anderson N, et al. Does cigarette smoking influence the phenotype of CD? Analysis using the Montreal classification. Am J Gastroenterol. 2007;102:2577–2588. doi: 10.1111/j.1572-0241.2007.01064.x. [DOI] [PubMed] [Google Scholar]
- 25.Rutgeerts P, Vermeire S, Van Assche G. Mucosal healing in inflammatory bowel disease: impossible ideal or therapeutic target? Gut. 2007;56:453–455. doi: 10.1136/gut.2005.088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer A, de Saint Basile G, Le Deist F. CD3 deficiencies. Curr Opin Allergy Clin Immunol. 2005;5:491–495. doi: 10.1097/01.all.0000191886.12645.79. [DOI] [PubMed] [Google Scholar]
- 28.Abadía-Molina AC, Ji H, Faubion WA, et al. CD48 controls T-cell and antigen-presenting cell functions in experimental colitis. Gastroenterology. 2006;130:424–434. doi: 10.1053/j.gastro.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Hermiston ML, Xu Z, Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol. 2003;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 30.Neurath MF, Weigmann B, Finotto S, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J Exp Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayne M, Moffatt T, Kong H, et al. CYFIP2 is highly abundant in CD4+ cells from multiple sclerosis patients and is involved in T cell adhesion. Eur J Immunol. 2004;34:1217–1227. doi: 10.1002/eji.200324726. [DOI] [PubMed] [Google Scholar]
- 32.Hausmann M, Spöttl T, Andus T, et al. Subtractive screening reveals up-regulation of NADPH oxidase expression in Crohn's disease intestinal macrophages. Clin Exp Immunol. 2001;125:48–55. doi: 10.1046/j.1365-2249.2001.01567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedi M, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strober W, Kitani A, Fuss I, et al. The molecular basis of NOD2 susceptibility mutations in Crohn's disease. Mucosal Immunol. 2008;1(Suppl 1):S5–S9. doi: 10.1038/mi.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franchi L, Warner N, Viani K, et al. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAuley JL, Linden SK, Png CW, et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–2334. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beatty PL, Plevy SE, Sepulveda AR, et al. Cutting edge: transgenic expression of human MUC1 in IL-10−/− mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179:735–739. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 38.El Hassani RA, Benfares N, Caillou B, et al. Dual oxidase2 is expressed all along the digestive tract. Am J Physiol Gastrointest Liver Physiol. 2005;288:933–942. doi: 10.1152/ajpgi.00198.2004. [DOI] [PubMed] [Google Scholar]
- 39.Csillag C, Nielsen OH, Vainer B, et al. Expression of the genes dual oxidase 2, lipocalin 2 and regenerating islet-derived 1 alpha in Crohn's disease. Scand J Gastroenterol. 2007;42:454–463. doi: 10.1080/00365520600976266. [DOI] [PubMed] [Google Scholar]
- 40.Marshall AJ, Du Q, Draves K, et al. FDC-SP, a novel secreted protein expressed by follicular dendritic cells. J Immunol. 2002;169:2381–2389. doi: 10.4049/jimmunol.169.5.2381. [DOI] [PubMed] [Google Scholar]
- 41.Al-Alwan M, Du Q, Hou S, et al. Follicular dendritic cell secreted protein (FDC-SP) regulates germinal center and antibody responses. J Immunol. 2007;178:7859–7867. doi: 10.4049/jimmunol.178.12.7859. [DOI] [PubMed] [Google Scholar]
- 42.Anderle P, Rumbo M, Sierro F, et al. Novel markers of the human follicle-associated epithelium identified by genomic profiling and microdissection. Gastroenterology. 2005;129:321–327. doi: 10.1053/j.gastro.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 43.Muto A, Tashiro S, Nakajima O, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 44.Tedoldi S, Paterson JC, Cordell J, et al. Jaw1/LRMP, a germinal centre-associated marker for the immunohistological study of B-cell lymphomas. J Pathol. 2006;209(4):454–463. doi: 10.1002/path.2002. [DOI] [PubMed] [Google Scholar]
- 45.Ehrhardt GR, Leu CM, Zhang S, et al. Fc receptor-like proteins (FCRL): immunomodulators of B cell function. Adv Exp Med Biol. 2007;596:155–162. doi: 10.1007/0-387-46530-8_14. [DOI] [PubMed] [Google Scholar]
- 46.Gullberg E, Söderholm JD. Peyer's patches and M cells as potential sites of the inflammatory onset in Crohn's disease. Ann N Y Acad Sci. 2006;1072:218–232. doi: 10.1196/annals.1326.028. [DOI] [PubMed] [Google Scholar]
- 47.Geboes K, Collins S. Structural abnormalities of the nervous system in Crohn’s disease and ulcerative colitis. Neurogastroenterol Motil. 1998;10:189–202. doi: 10.1046/j.1365-2982.1998.00102.x. [DOI] [PubMed] [Google Scholar]