Abstract
Aim:
To examine the regulatory crosstalk between the transcription factors Nrf2 and AP-1 in prostate cancer (PCa) by dietary cancer chemopreventive compounds (−)epigallocatechin-3-gallate (EGCG) from green tea and sulforaphane (SFN) from cruciferous vegetables.
Methods:
We performed (i) in vitro studies including luciferase reporter gene assays, MTS cell viability assays, and quantitative real-time PCR (qRT-PCR) in PC-3 AP-1 human PCa cells, (ii) in vivo temporal (3 h and 12 h) microarray studies in the prostate of Nrf2-deficient mice that was validated by qRT-PCR, and (iii) in silico bioinformatic analyses to delineate conserved Transcription Factor Binding Sites (TFBS) in the promoter regions of Nrf2 and AP-1, as well as coregulated genes including ATF-2 and ELK-1.
Results:
Our study shows that AP-1 activation was attenuated by the combinations of SFN (25 μmol/L) and EGCG (20 or 100 μmol/L) in PC-3 cells. Several key Nrf2-dependent genes were down-regulated (3-fold to 35-fold) after in vivo administration of the combination of EGCG (100 mg/kg) and SFN (45 mg/kg). Conserved TFBS signatures were identified in the promoter regions of Nrf2, AP-1, ATF2, and ELK-1 suggesting a potential regulatory mechanism of crosstalk between them.
Conclusion:
Taken together, our present study of transcriptome profiling the gene expression changes induced by dietary phytochemicals SFN and EGCG in Nrf2-deficient mice and in PC-3 cells in vitro demonstrates that the effects of SFN+EGCG could be mediated via concerted modulation of Nrf2 and AP-1 pathways in the prostate.
Keywords: prostate cancer, sulforaphane, EGCG, Nrf2, AP-1, ATF-2, ELK-1, gene expression profiles
Introduction
Prostate cancer (PCa), according to the Centers for Disease Control and Prevention (CDC)1, is the second leading cause of cancer deaths among men in the United States, and the seventh leading cause of deaths overall for men. The incidence of PCa in the United States has increased by 1.1% per year from 1995–20031, 2. In addition, the National Cancer Institute (NCI)'s Surveillance Epidemiology and End Results (SEER) Statistics Fact Sheets3 show that, based on rates from 2002–2004, 16.72% of men born today will be diagnosed with cancer of the prostate at some time during their lifetime, ie, 1 in 6 men in the United States are at a lifetime risk of developing PCa. The prostate-specific antigen (PSA) nadir4 while intermittently taking a testosterone-inactivating pharmaceutical agent has been determined to be the best predictor of prostate cancer-specific mortality. Nevertheless, despite the considerable attention given to PSA as a screening test for prostate cancer, it is needle biopsy, and not the PSA test result, that actually establishes the diagnosis of PCa5.
Pivotal to the antioxidant response6, 7, 8, 9 typical in mammalian homeostasis and oxidative stress is the important transcription factor Nuclear Factor-E2-related factor 2 (Nrf2) that has been extensively studied by many investigators including us, as noted elsewhere10, 11. Nrf2 is indispensable to cellular defense against many chemical insults of endogenous and exogenous origin, which play major roles in the etiopathogenesis of many cancers as well as inflammatory bowel disease12 and Parkinson's disease13. Rushmore et al14 were the first to identify a core antioxidant response element (core ARE or cARE) sequence 5′-RGTGACNNNGC-3′ responsible for transcriptional activation by xenobiotics, that was later expanded by Wasserman and Fahl15 giving rise to an expanded ARE (eARE) sequence described by 5′-TMAnnRTGAYnnnGCRwwww-3′. We observed16 that the induction of antioxidant response element (ARE)-regulated genes in vitro in human prostate cancer PC-3 cells upon treatment with phenethyl isothiocyanate (PEITC) is associated with the activation of extracellular signal-regulated kinase (ERK) and c-jun N-terminal kinase (JNK) resulting in the phosphorylation and nuclear translocation of Nrf2. Watai et al17 showed that endogenous Keap1, which acts as a regulator of Nrf2 activity through an interaction with the Nrf2 Neh2 domain, remains mostly in the cytoplasm, and electrophiles promote nuclear accumulation of Nrf2 without altering the subcellular localization of Keap1. Thus, the Keap1-Nrf2-ARE axis potentially has an important role to play in various forms of cancer including that of the prostate.
AP-1 is a redox-sensitive transcription factor that senses and transduces changes in cellular redox status and modulates gene expression responses to oxidative and electrophilic stresses presumably via sulfhydryl modification of critical cysteine residues found on this protein and/or other upstream redox-sensitive molecular targets10. AP-1 is composed of heterodimeric protein complexes of members of the basic leucine zipper (bZIP) protein families, including the Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra-1, and Fra-2) families, Maf (c-Maf, MafB, MafA, Maf G/F/K, and Nrl), Jun dimerization partners (JDP1 and JDP2) and the closely related activation transcription factor (ATF; ATF2, LRF1/ATF3, and B-ATF) subfamilies18, 19, 20 which recognize either 12-O-tetradecanoylphorbol-13-acetate (TPA) response elements (TRE, 5′-TGAG/CTCA-3′) or cAMP response elements (CRE, 5′-TGACGTCA-3′)21. We have shown that ERK and JNK signaling pathways are involved in the regulation of AP-1 and cell death elicited by three isothiocyanates (sulforaphane, SFN; PEITC; and allyl isothiocyanate, AITC) in human PCa PC-3 cells19.
Evidence derived from epidemiological studies has revealed an inverse correlation between the intake of cruciferous vegetables and the risk of certain types of cancer22. Isothiocyanates are a chemical class of compounds that are not naturally present in cruciferous vegetables, such as broccoli and cauliflower, but are nevertheless generated from hydrolysis of secondary metabolites known as glucosinolates by the enzyme myrosinase during the process of vegetable crushing or mastication23. They may also be produced in the intestines where resident microflora can promote the hydrolysis of glucosinolates to isothiocyanates24. SFN, a dietary phytochemical obtained from broccoli, has been implicated in several physiological processes consistent with anticarcinogenic activity, including enhanced xenobiotic metabolism, cell cycle arrest, and apoptosis25. SFN has been shown26 to retard the growth of human PC-3 xenografts and inhibit HDAC activity in human subjects. We have observed27 that SFN induces hemoxygenase-1 (HO-1) by activating the antioxidant response element (ARE) through the induction of Nrf2 protein in HepG2 cells. It has also been reported28 that SFN-induced cell death in PC-3 and DU145 human PCa cells is initiated by reactive oxygen species and that induction of autophagy29 represents a defense mechanism against SFN-induced apoptosis in PC-3 and LNCaP human PCa cells. In addition, we have observed30 that SFN suppresses the transcriptional activation of NF-κB as well as NF-κB-regulated gene expression in PC-3 cells via inhibition of IKKbeta phosphorylation as well as IkappaBalpha phosphorylation and degradation. Mass spectrometric methods have shown that SFN exists as the SFN-glutathione conjugates in enterocytes whereas the N-acetyl cysteine conjugate is the primary urinary metabolite31.
The water-extractable fraction of green tea contains abundant polyphenolic compounds, in which (−)epigallocatechin-3-gallate (EGCG) is the major constituent (>50% of polyphenolic fraction)32. We have observed33 that EGCG treatment causes damage to mitochondria, and that JNK mediates EGCG-induced apoptotic cell death in HT-29 human colon cancer cells. EGCG is also reported34, 35 to inhibit DNA methyltransferase with demethylation of the CpG islands in the promoters, and to reactivate methylation-silenced genes such as p16INK4a, retinoic acid receptor beta, O6-methylguanine methyltransferase, human mutL homolog 1, and glutathione S-transferase-pi in human colon cancer HT-29 cells, esophageal cancer KYSE 150 cells, and PCa PC-3 cells. It was noted36 that EGCG suppresses early stage, but not late stage, PCa in TRAMP (Transgenic Adenocarcinoma Mouse Prostate) animals without incurring undue toxicity. Besides, EGCG has been reported37 to modulate the phosphatidylinositol-3-kinase/protein kinase B- and MAPK-pathways in DU145 and LNCaP human PCa cells, and to have combined inhibitory effects with selective cyclooxygenase-2 inhibitors38 on the growth of human PCa cells both in vitro and in vivo. We have also shown39 that a greater number of Nrf2-regulated genes are modulated in murine liver on oral administration of EGCG than in small intestine. Mass spectrometric methods have shown that EGCG produces methylated and conjugated metabolites in mice40.
Nrf2 knockout mice are greatly predisposed to chemical-induced DNA damage and exhibit higher susceptibility towards cancer development in several models of chemical carcinogenesis41. In the present study, we investigated via transcriptome profiling the gene expression changes induced by a combination of dietary phytochemicals SFN and EGCG in Nrf2-deficient mice, the in vitro effects of this combination in PC-3 AP-1 cells, and delineated conserved Transcription Factor Binding Sites (TFBS) in the promoter regions of Nrf2 and AP-1, as well as coregulated genes including ATF-2 and ELK-1, by in silico bioinformatic analyses. We demonstrate that the effects of the combination of SFN+EGCG in PCa may be mediated via concerted modulation of Nrf2 and AP-1 pathways.
Materials and methods
Cell culture and reagents
Human PCa PC-3 cells were stably transfected with an Activator Protein (AP-1) luciferase reporter construct, and are referred to as PC-3 AP-1 cells. The cells were cultured in Minimum Essential Medium (MEM) containing 10% Fetal Bovine Serum (FBS) and 1% Penicillin-Streptomycin. Twelve hours prior to experimental treatments, the cells were exposed to MEM containing 0.5% FBS. SFN was obtained from LKT Labs (St Paul, MN); whereas EGCG and superoxide dismutase (SOD) were obtained from Sigma-Aldrich (St Louis, MO). Both SFN and EGCG were dissolved in dimethylsulfoxide (DMSO, Sigma), whereas SOD was dissolved in 1×phosphate-buffered saline (PBS).
Reporter gene assays
PC-3 AP-1 cells were seeded in six-well culture plates and treated in triplicate with 0.1% dimethylsulfoxide (control), 20 μmol/L EGCG, 100 μmol/L EGCG, 25 μmol/L SFN, 20 μmol/L EGCG+25 μmol/L SFN, or 100 μmol/L EGCG+25 μmol/L SFN for 24 h. Thereafter, the supernatant medium was aspirated on ice, cells were washed thrice with ice-cold 1×PBS, treated with 1×Luciferase Reporter Lysis Buffer (Promega) and subjected to one cycle of snap freeze-thaw at −80 °C. Cell lysates were harvested with sterile RNAse-free and DNAse-free cell scrapers into microcentrifuge tubes that were immediately placed on ice. They were then centrifuged at 4 °C for ten minutes at 13000×g and returned to ice. Twenty microliters of supernatant solution was analyzed for relative luciferase activity using a Sirius Luminometer (Berthold Detection Systems). The relative luciferase activities were normalized by protein concentrations of individual samples determined as described below.
Protein assays
Protein concentrations of samples were determined by the bicinchonic acid-based BCA Protein Assay Kit (Pierce) according to the manufacturer's instructions using a 96-well plate. Standard curves were constructed using bovine serum albumin (BSA) as a standard. The sample readings were obtained on a μQuant microplate reader (Bio-tek Instruments Inc) at 560 nm.
Cell viability assays
The cell viability assays were performed in 24-well cell culture plates using MTS Assay Kit (Promega) according to the manufacturer's instructions. Cell viability was determined at both 24 and 48 h after treatment with dietary factors. The absorbance readings were obtained on an μQuant microplate reader (Bio-tek Instruments, Inc) at recommended wavelength of 490 nm.
RNA extraction and assessment of RNA integrity
PC-3 AP-1 cells were subjected to treatment with different dietary factors in triplicate for 6 or 10 h. RNA was harvested using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA integrity was assessed using formaldehyde gels in 1×MOPS buffer and RNA concentration was determined by the 260/280 ratio on a DU 530 UV/Visible spectrophotometer (Beckman).
Quantitative real-time PCR assays
Several genes of interest including luciferase gene as well as genes known to be either under the control of the AP-1 promoter or involved in cell cycle regulation or cellular influx-efflux such as cyclin D1, cMyc, ATF-2, Elk-1, SRF, CREB5, MDR1, SLCO1B3, MRP1, MRP2, and MRP3 were selected for quantitative real-time PCR analyses. Beta-actin served as the “housekeeping” gene. The specific primers for these genes were designed using Primer Express 2.0 software (Applied Biosystems, Foster City, CA) and were obtained from Integrated DNA Technologies, Coralville, IA. The specificity of the primers was examined by a National Center for Biotechnology Information Blast search of the human genome. For the real-time PCR assays, briefly, after the RNA extraction and assessment of RNA integrity, first-strand cDNA was synthesized using 4 μg of total RNA following the protocol of SuperScript III First-Strand cDNA Synthesis System (Invitrogen) in a 40 μL reaction volume. The PCR reactions based on SYBR Green chemistry were carried out using 100 times diluted cDNA product, 60 nmol/L of each primer, and SYBR Green master mix (Applied Biosystems, Foster City, CA) in 10 μL reactions. The PCR parameters were set using SDS 2.1 software (Applied Biosystems, Foster City, CA) and involved the following stages: 50 °C for 2 min, 1 cycle; 95 °C for 10 min, 1 cycle; 95 °C for 15 s→55 °C for 30 s→72 °C for 30 s, 40 cycles; and 72 °C for 10 min, 1 cycle. Incorporation of the SYBR Green dye into the PCR products was monitored in real time with an ABI Prism 7900HT sequence detection system, resulting in the calculation of a threshold cycle (CT) that defines the PCR cycle at which exponential growth of PCR products begins. The carboxy-X-rhodamine (ROX) passive reference dye was used to account for well and pipetting variability. A control cDNA dilution series was created for each gene to establish a standard curve. After conclusion of the reaction, amplicon specificity was verified by first-derivative melting curve analysis using the ABI software; and the integrity of the PCR reaction product and absence of primer dimers was ascertained. The gene expression was determined by normalization with control gene beta-actin.
Promoter analyses for transcription factor binding sites (TFBS)
The promoter analyses were performed using Genomatix MatInspector42, 43. Briefly, human promoter sequences of Nrf2, AP-1, ATF-2, and ELK-1, or corresponding murine promoter sequences, were retrieved from Gene2Promoter (Genomatix). Comparative promoter analyses were then performed by input of these sequences in FASTA format into MatInspector using optimized default matrix similarity thresholds. The similar and/or functionally related TFBS were grouped into “matrix families” and graphical representations of common TFBS were generated. The 'V$' prefixes to the individual matrices are representative of the Vertebrate MatInspector matrix library. We also elucidated common regulatory sequences in promoter regions of human, or murine, Nrf2 and AP-1. The ”core sequence” of a matrix is defined as the (usually four) highest conserved positions of the matrix that is provided in upper case letters in our Tables. The maximum core similarity of 1.0 is only reached when the highest conserved bases of a matrix match exactly in the sequence. Only matches that contain the “core sequence” of the matrix with a score higher than the core similarity are listed in the output.
Animals and dosing
The protocol for animal studies was approved by the Rutgers University Institutional Animal Care and Use Committee (IACUC). Nrf2 knockout mice Nrf2(−/−) (C57BL/SV129) have been described previously44. Nrf2(−/−) mice were backcrossed with C57BL/6J mice (The Jackson Laboratory, ME USA). DNA was extracted from the tail of each mouse and genotype of the mouse was confirmed by polymerase chain reaction (PCR) by using primers (3'-primer, 5′-GGA ATG GAA AAT AGC TCC TGC C-3′ 5′-primer, 5′-GCC TGA GAG CTG TAG GCC C-3′ and lacZ primer, 5′-GGG TTT TCC CAG TCA CGA C-3′). Nrf2(−/−) mice-derived PCR products showed only one band of ∼200 bp, Nrf2 (+/+) mice-derived PCR products showed a band of ∼300 bp while both bands appeared in Nrf2(+/−) mice PCR products. Male C57BL/6J/Nrf2(−/−) mice from third generation of backcross were used in this study. Age-matched male C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice in the age-group of 9–12 weeks were housed at Rutgers Animal Facility with free access to water and food under 12 h light/dark cycles. After one week of acclimatization, the mice were put on AIN-76A diet (Research Diets Inc, NJ) for another week. The mice were then administered both SFN (LKT Labs, St Paul, MN) and EGCG (Sigma-Aldrich, St Louis, MO) at doses of 45 and 100 mg/kg respectively (dissolved in 50% PEG 400 aqueous solution) by oral gavage. The control group animals were administered only vehicle (50% PEG 400 aqueous solution). Each treatment was administered to a group of four animals for both C57BL/6J and C57BL/6J/Nrf2(−/−) mice. Mice were sacrificed at either 3 h or 12 h after dietary factor treatment or vehicle administration (control group). The prostates of the animals were retrieved and stored in RNA Later (Ambion, Austin,TX) solution.
Microarray sample preparation and hybridization
Total RNA from prostate tissues was isolated by using TRIzol (Invitrogen, Carlsbad, CA) extraction coupled with the RNeasy kit from Qiagen (Valencia, CA). Briefly, tissues were homogenized in Trizol and then extracted with chloroform by vortexing. A small volume (1.2 mL) of aqueous phase after chloroform extraction and centrifugation was adjusted to 35% ethanol and loaded onto an RNeasy column. The column was washed, and RNA was eluted following the manufacturer's recommendations. RNA integrity was examined by electrophoresis, and concentrations were determined by UV spectrophotometry. Affymetrix (Affymetrix, Santa Clara, CA) Mouse Genome 430 2.0 array was used to probe the global gene expression profiles in mice following SFN+EGCG treatment. The Mouse Genome 430 2.0 Array is a high-density oligonucleotide array comprised of over 45 101 probe sets representing over 34 000 well-substantiated mouse genes. The library file for the above-mentioned oligonucleotide array is readily available at http://www.affymetrix.com/support/technical/libraryfilesmain.affx. After RNA isolation, the subsequent technical procedures including quality control and estimation of RNA concentration, cDNA synthesis and biotin-labeling of cRNA, hybridization and scanning of the arrays, were performed at CINJ Core Expression Array Facility of Robert Wood Johnson Medical School (New Brunswick, NJ). Each chip was hybridized with cRNA derived from a pooled total RNA sample from four mice per treatment group, per time-point, and per genotype (a total of eight chips were used in this study). Briefly, double-stranded cDNA was synthesized from 5 μg of total RNA and labeled using the ENZO BioArray RNA transcript labeling kit (Enzo Life Sciences Inc, Farmingdale, NY) to generate biotinylated cRNA. Biotin-labeled cRNA was purified and fragmented randomly according to Affymetrix's protocol. Two hundred microliters of sample cocktail containing 15 μg of fragmented and biotin-labeled cRNA was loaded onto each chip. Chips were hybridized at 45 °C for 16 h and washed with fluidics protocol EukGE-WS2v5 according to Affymetrix's recommendation. At the completion of the fluidics protocol, the chips were placed into the Affymetrix GeneChip Scanner where the intensity of the fluorescence for each feature was measured.
Microarray data analyses
The CEL files (intensity DATA fields) were created from the scanned image and analyzed using dChip software45, 46 for further data characterization. Briefly, a gene information file with current annotations and functional gene ontology was generated and the Affymetrix Chip Description File (CDF, information on the location and identity of different probe cells) was specified. The data were then normalized in dChip and the expression value for each gene was determined by calculating the average of differences in intensity (perfect match intensity minus mismatch intensity) between its probe pairs. The expression values were imported into GeneSpring 7.2 (Agilent Technologies Inc, Palo Alto, CA) followed by data filtration based on flags present in at least one of the samples, and a corresponding gene list based on those flags was generated. Lists of genes that were either induced or suppressed more than three fold between treated versus vehicle group of same genotype were created by filtration-on-fold function within the presented flag list. By use of Venn Diagram function, lists of genes that were regulated more than three fold only in prostate of C57BL/6J mice but not in prostate of C57BL/6J/Nrf2(−/−) mice at both 3 and 12 h were generated, and were designated as Nrf2-dependent genes. The Affymetrix Probe Set IDs for the Nrf2-dependent genes were matched against the “all genes” expression values list for these set of samples, and expression values for these Affymetrix Probe Set IDs were retrieved. This “external data” was then imported into dChip whereupon Clustering and Enrichment Analysis was performed to obtain hierarchical tree clustering diagrams for the Nrf2-dependent genes. This clustering provided functional classification of Affymetrix Probe Set IDs and gene descriptions that were then matched with the GeneSpring-generated Nrf2-dependent Affymetrix Probe Set IDs with fold-change values. Quantitative real-time PCR assays as described earlier were performed on several genes to validate the microarray results.
Statistical analyses
Data are expressed as mean ± standard deviation, and comparisons among treatment groups were made using one-way analysis of variance (ANOVA) followed by a post hoc test for multiple comparisons — the Tukey's Studentized Range Honestly Significant Difference (HSD) test. In all these multiple comparisons, P<0.05 was considered statistically significant. In order to validate the microarray results, the correlation between corresponding microarray data and real-time PCR data was evaluated by r2, the statistical “coefficient of determination” (r2=0.96). Statistical analyses were performed using SAS 9.1 software (SAS Institute Inc, NC) licensed to Rutgers University.
Results
Diminished transactivation of AP-1 luciferase reporter by combinations of SFN and EGCG
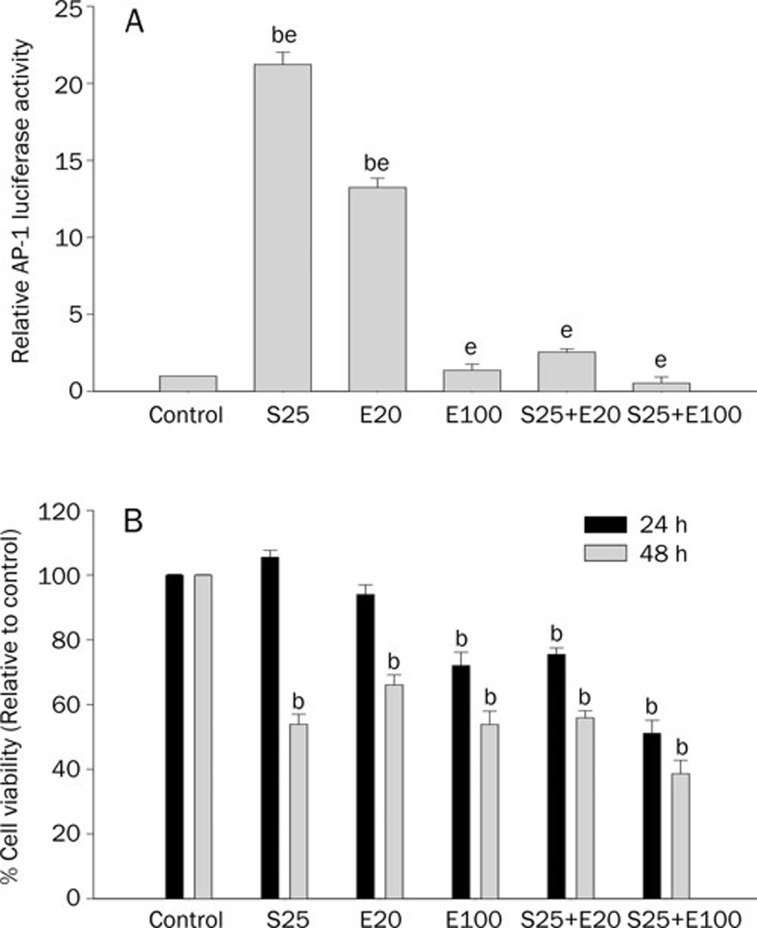
As shown in Figure 1A, treatment of PC-3 AP-1 cells for 24 h with either 25 μmol/L SFN, 100 μmol/L EGCG or 20 μmol/L EGCG individually resulted in variable induction of AP-1 luciferase activity as compared to control cells that were treated with DMSO. Surprisingly, a low-dose combination of 25 μmol/L SFN+20 μmol/L EGCG elicited a diminished induction of AP-1 luciferase activity (less than 5-fold). In addition, a high-dose combination of 25 μmol/L SFN+100 μmol/L EGCG further diminished the induction of the AP-1 luciferase reporter. We also investigated the effects of pre-treatment on the induction of AP-1 luciferase activity. In these experiments (data not shown), we first pre-treated the PC-3 AP-1 cells for 6 h with EGCG (20 μmol/L or 100 μmol/L), then washed off the EGCG thrice with phosphate-buffered saline (PBS) and treated the cells with 25 μmol/L SFN for an additional 18 h before assaying for luciferase activity. Alternatively, we also pre-treated the cells with 25 μmol/L SFN for 6 h before washing with PBS as above and treating with EGCG (20 μmol/L or 100 μmol/L) for an additional 18 h. It was observed that there was no significant difference in induction of AP-1 luciferase activity in these pre-treatment experiments (data not shown) as compared to when the two agents were co-treated as shown in Figure 1A. This enabled us to rule out any physicochemical interaction between the two agents in cell culture, when co-treated, that may have otherwise produced any experimental artifacts in the luciferase assay. Hence, since the effects of the combinations when co-treated were not physicochemical, but potentially modulated at a mechanistic level, we continued co-treating both agents together for a duration of 24 h for ease of experimentation without confounding variables.
Figure 1.
(A) Diminished transactivation of AP-1 luciferase reporter by combinations of SFN and EGCG. PC-3 AP-1 cells were seeded in six-well plates and treated with individual dietary factors, or with combinations of SFN and EGCG, as indicated (S25, sulforaphane 25 μmol/L; E20, EGCG 20 μmol/L; E100, EGCG 100 μmol/L). The AP-1 luciferase activity was measured relative to vehicle control (DMSO) after 24 h of incubation and normalized against protein concentration. Values represent mean± standard deviation for three replicates, and are representative of seven independent experiments. bP<0.05 vs vehicle control; eP<0.05, significantly different from each other. (B) Viability of the PC-3 AP-1 cells with the combinations of SFN and EGCG. PC-3 AP-1 cells were treated with individual dietary factors or with combinations of SFN and EGCG for 24 h or 48 h, as indicated, and treated with MTS assay reagent to ascertain cell viability (S25, sulforaphane 25 μmol/L; E20, EGCG 20 μmol/L; E100, EGCG 100 μmol/L). Values represent mean±standard deviation for six replicates, and are representative of three independent experiments. bP<0.05 vs control.
Viability of the PC-3 AP-1 cells with the combinations of SFN and EGCG
In order to ascertain the effects of the combinations of SFN and EGCG on the cell viability of the PC-3 AP-1 cells, we used the MTS assay with treatment durations of 24 h and 48 h. As shown in Figure 1B, the cell viability at 24 h for the low-dose combination treatment of 25 μmol/L SFN+20 μmol/L EGCG was about 75% to 80%, whereas it was about 60% for the high-dose combination of 25 μmol/L SFN+100 μmol/L EGCG. The low-dose combination of 25 μmol/L SFN+20 μmol/L EGCG may be more appropriate to pursue in longer duration in vitro studies or potential in vivo studies without seemingly toxic effects a priori, and at the same time not compromising on the efficacy elicited by the combination of these two chemopreventive agents.
Temporal gene expression profiles elicited by combinations of SFN and EGCG
We performed quantitative real-time PCR (qRT-PCR) experiments with primers for the luciferase gene to corroborate the synergism elicited with the combinations of SFN and EGCG in the luciferase protein assay with mRNA levels in qRT-PCR. The temporal expression (at 6 h and 10 h) of luciferase gene in qRT-PCR assays (Figure 2A) was lower for the combinations of SFN and EGCG as compared to individual dietary factor treatments in consonance with our data in the luciferase protein assays (Figure 1A). The treatment means for all the treatment groups at a specific time point (6 h or 10 h) were significantly different from each other (P<0.05 by ANOVA and post hoc Tukey's test for multiple comparisons to detect significantly different means). We also determined by qRT-PCR the relative expression levels of transcripts of many genes that were known to be either under the control of the AP-1 promoter or involved in cell cycle regulation or cellular influx-efflux such as cyclin D1, cMyc, ATF-2, ELK-1, SRF, CREB5, SLCO1B3, MRP1, MRP2, and MRP3 (Figure 2).
Figure 2.
Temporal gene expression profiles elicited by combinations of SFN and EGCG. PC-3 AP-1 cells were treated for 6 h or 10 h with individual dietary factors, or combinations of SFN and EGCG, as indicated (S25, sulforaphane 25 μmol/L; E20, EGCG 20 μmol/L; E100, EGCG 100 μmol/L). RNA was extracted, transcribed into cDNA after ascertaining RNA integrity, and quantitative real-time PCR assays were performed for twelve genes at both time-points, as indicated, using beta-actin as the housekeeping gene. Values represent mean±standard deviation for three replicates of each gene, and are representative of two independent experiments.
The low- and high-dose combinations of SFN and EGCG in this study elicited the downregulation of positive cell cycle regulator cyclin D1 expression as compared with individual dietary factors especially at 10 h (Figure 2B). There was, however, no appreciable change in expression of cell proliferation-related cMyc Binding Protein (Figure 2C). In addition, transcription factors/coactivators that are known to be under the control of the AP-1 promoter such as activating transcription factor (ATF-2), Ets-like transcription factor (ELK-1), serum response factor (SRF) and cyclic AMP response element binding protein 5 (CREB5) were also studied. Interestingly, both ATF-2 and ELK-1 were significantly downregulated by the combinations of SFN and EGCG as compared to individual dietary factors (Figure 2D and 2E). Besides, the low-dose combination of SFN and EGCG at 6 h (and the high-dose combination at 10 h) inhibited the expression of SRF as compared to individual dietary factors (Figure 2F). Similarly, the combinations inhibited the expression of CREB5 as compared to individual agents (Figure 2G). Since exogenous stress can potentially stimulate the influx-efflux machinery of cells, we also investigated some key transporter genes. In this study, we observed that the combinations of SFN and EGCG inhibited the SFN-induced expression of the SLCO1B3 gene, which encodes for the organic anion transporter protein OATP1B3, whereas the combinations did not have any clear effect on the expression of MDR1 gene (Figure 2H and 2I). Interestingly, the combinations of SFN and EGCG greatly induced the expression of the efflux transporter MRP2 as compared to individual dietary factors SFN or EGCG (Figure 2K). In addition, the expression of influx transporters MRP1 and MRP3 was not significantly different for the combination-treated cells as compared to the individual agent-treated cells (Figure 2J and 2L).
Comparative promoter analyses of Nrf2 and AP-1, as well as ATF-2 and ELK-1, for conserved Transcription Factor Binding Sites (TFBS)
We performed comparative analyses of Nrf2 and AP-1 human promoter sequences as described in Materials and Methods. We also studied Nrf2 and AP-1 murine promoter sequences similarly. We have alphabetically listed the conserved vertebrate (V$) matrix families between these two transcription factors in Table 1. The major human families included Activator protein 4 and related proteins, cell cycle regulators, E-box binding factors, human and murine ETS1 factors, fork head domain factors, hypoxia inducible factor, myc-associated zinc fingers, nuclear respiratory factor 1, serum response element binding factor, and signal transducer and activator of transcription amongst others. Interestingly, NF-κB was conserved in human sequences of Nrf2 and AP-1. We also performed comparative promoter analyses on ATF-2 and ELK-1 which are AP-1-regulated genes. We have alphabetically listed the conserved vertebrate (V$) matrix families between ATF-2 and ELK-1 in Table 2 and have provided a pictorial representation in Figures 3A and 3B. Some key conserved matrix families included AP-1, cyclic AMP-responsive element binding proteins, estrogen response elements, human and murine ETS1 factors, fork head domain factors, farnesoid-X-activated receptor response elements, human acute myelogenous leukemia factors, Ikaros zinc finger family, myc-associated zinc fingers, nuclear factor of activated T-cells, NF-κB, peroxisome proliferators-activated receptor, ras-responsive element binding protein, serum response element binding factor, signal transducer and activator of transcription, and X-box binding factors amongst others. Interestingly, as is evident from Tables 1 and 2, several key matrix families were conserved not just between Nrf2 and AP-1, or between the AP-1-regulated genes ATF-2 and ELK-1, in either human or murine species, but there was also some degree of overlap between TFBS identified in Tables 1 and 2. Furthermore, as shown in the Supplementary Table, we identified matrices (individual matrix family members) with conserved regulatory sequences in promoter regions of Nrf2 and AP-1 in either human or murine species. Multiple matches that were elicited with the same core sequence have also been grouped together and listed in the Supplementary Table.
Table 1. Human and murine matrix families conserved between Nrf2 and AP-1.
| Matrix family human Nrf2 vs AP-1 | Matrix family murine Nrf2 vs AP-1 | Family information |
|---|---|---|
| V$AP4R | V$AP4R | Activator protein 4 and related proteins |
| V$CDEF | - | Cell cycle regulators: Cell cycle dependent element |
| V$CHRF | - | Cell cycle regulators: Cell cycle homology element |
| - | V$CP2F | CP2-erythrocyte factor related to drosophila Elf1 |
| V$E2FF | - | E2F-myc activator/cell cycle regulator |
| - | V$E4FF | Ubiquitous GLI - Krueppel like zinc finger involved in cell cycle regulation |
| V$EBOX | – | E-box binding factors |
| V$EGRF | V$EGRF | EGR/nerve growth factor induced protein C & related factors |
| – | V$EKLF | Basic and erythroid krueppel like factors |
| – | V$ETSF | Human and murine ETS1 factors |
| V$EVI1 | – | EVI1-myleoid transforming protein |
| – | V$FKHD | Fork head domain factors |
| V$GATA | – | GATA binding factors |
| – | V$GLIF | GLI zinc finger family |
| – | V$GREF | Glucocorticoid responsive and related elements |
| V$HAND | – | bHLH transcription factor dimer of HAND2 and E12 |
| – | V$HESF | Vertebrate homologues of enhancer of split complex |
| – | V$HIFF | Hypoxia inducible factor, bHLH/PAS protein family |
| – | V$HNF6 | Onecut homeodomain factor HNF6 |
| – | V$INSM | Insulinoma associated factors |
| V$MAZF | V$MAZF | Myc associated zinc fingers |
| – | V$MOKF | Mouse Krueppel like factor |
| V$MYBL | – | Cellular and viral myb-like transcriptional regulators |
| V$MYOD | V$MYOD | Myoblast determining factors |
| V$NEUR | – | NeuroD, Beta2, HLH domain |
| V$NFKB | – | NF-κB/c-rel |
| V$Nrf1 | V$Nrf1 | Nuclear respiratory factor 1 |
| V$PAX5 | – | PAX-5 B-cell-specific activator protein |
| V$PAX9 | – | PAX-9 binding sites |
| V$PBXC | – | PBX1 - MEIS1 complexes |
| – | V$PLAG | Pleomorphic adenoma gene |
| – | V$SF1F | Vertebrate steroidogenic factor |
| V$SP1F | V$SP1F | GC-Box factors SP1/GC |
| – | V$SRFF | Serum response element binding factor |
| V$STAF | – | Selenocysteine tRNA activating factor |
| V$STAT | – | Signal transducer and activator of transcription |
| – | V$XBBF | X-box binding factors |
| V$ZBPF | V$ZBPF | Zinc binding protein factors |
| – | V$ZF35 | Zinc finger protein ZNF35 |
Table 2. Human and murine matrix families conserved between ATF-2 and ELK1.
| Matrix family human ATF2 vs ELK1 | Matrix family murine Atf2 vs Elk1 | Family information |
|---|---|---|
| V$AP1F | – | AP1, activator protein 1 |
| – | V$BARB | Barbiturate-inducible element box from pro+eukaryotic genes |
| – | V$BNCF | Basonuclein rDNA transcription factor (PolI) |
| – | V$BRNF | Brn POU domain factors |
| V$CAAT | – | CCAAT binding factors |
| – | V$CLOX | CLOX and CLOX homology (CDP) factors |
| V$COMP | – | Factors which cooperate with myogenic proteins |
| – | V$CP2F | CP2-erythrocyte factor related to drosophila Elf1 |
| V$CREB | – | Camp-responsive element binding proteins |
| – | V$E2FF | E2F-myc activator/cell cycle regulator |
| V$EBOX | – | E-box binding factors |
| V$EGRF | – | EGR/nerve growth factor induced protein C & related factors |
| V$EKLF | – | Basic and erythroid krueppel like factors |
| V$EREF | – | Estrogen response elements |
| V$ETSF | V$ETSF | Human and murine ETS1 factors |
| – | V$EVI1 | EVI1-myleoid transforming protein |
| V$FKHD | V$FKHD | Fork head domain factors |
| – | V$FXRE | Farnesoid X-activated receptor response elements |
| – | V$GATA | GATA binding factors |
| – | V$GCMF | Chorion-specific transcription factors with a GCM DNA binding domain |
| V$GFI1 | – | Growth factor independence transcriptional repressor |
| V$GKLF | V$GKLF | Gut-enriched Krueppel like binding factor |
| V$GLIF | – | GLI zinc finger family |
| – | V$HAML | Human acute myelogenous leukemia factors |
| – | V$HOXC | HOX-PBX complexes |
| – | V$HOXF | Factors with moderate activity to homeo domain consensus sequence |
| – | V$IKRS | Ikaros zinc finger family |
| V$IRFF | V$IRFF | Interferon regulatory factors |
| V$LEFF | V$LEFF | LEF1/TCF, involved in the Wnt signal transduction pathway |
| V$MAZF | – | Myc associated zinc fingers |
| – | V$MEF2 | MEF2, myocyte-specific enhancer binding factor |
| V$MYBL | – | Cellular and viral myb-like transcriptional regulators |
| – | V$MYT1 | MYT1 C2HC zinc finger protein |
| V$MZF1 | – | Myeloid zinc finger 1 factors |
| V$NBRE | – | NGFI-B response elements, nur subfamily of nuclear receptors |
| – | V$NFAT | Nuclear factor of activated T-cells |
| V$NFKB | – | NF-κB/c-rel |
| V$NKXH | V$NKXH | NKX homeodomain factors |
| V$NR2F | V$NR2F | Nuclear receptor subfamily 2 factors |
| – | V$OCT1 | Octamer binding protein |
| V$PARF | V$PARF | PAR/bZIP family |
| V$PAX5 | – | PAX-5 B-cell-specific activator protein |
| V$PAX6 | – | PAX-4/PAX-6 paired domain binding sites |
| V$PBXC | – | PBX1 - MEIS1 complexes |
| V$PERO | – | Peroxisome proliferator-activated receptor |
| – | V$PLZF | C2H2 zinc finger protein PLZF |
| V$RREB | – | Ras-responsive element binding protein |
| V$RXRF | – | RXR heterodimer binding sites |
| V$SNAP | – | snRNA-activating protein complex |
| V$SP1F | – | GC-Box factors SP1/GC |
| – | V$SRFF | Serum response element binding factor |
| V$STAF | – | Selenocysteine tRNA activating factor |
| – | V$STAT | Signal transducer and activator of transcription |
| V$TALE | – | TALE homeodomain class recognizing TG motifs |
| – | V$TBPF | Tata-binding protein factor |
| – | V$TEAF | TEA/ATTS DNA binding domain factors |
| – | V$XBBF | X-box binding factors |
| V$ZBPF | – | Zinc binding protein factors |
Figure 3.
Conserved transcription factor binding sites (TFBS) in promoter regions of ATF-2 and ELK-1. Human promoter sequences of ATF-2 and ELK-1, or corresponding murine promoter sequences, were retrieved from Gene2Promoter (Genomatix). Comparative promoter analyses were then performed by input of these sequences in FASTA format into MatInspector using optimized default matrix similarity thresholds. The similar and/or functionally related TFBS were grouped into 'matrix families' and graphical representations of common TFBS were generated (Figure 3A, human ATF-2 and ELK-1; Figure 3B, murine Atf-2 and Elk-1). The 'V$' prefixes to the individual matrices are representative of the Vertebrate MatInspector matrix library.
Temporal microarray analyses of genes modulated by SFN+EGCG combination in the prostate of Nrf2-deficient mice
We clustered the genes that were downregulated at both 3 h and 12 h by the SFN+EGCG combination in the prostate of Nrf2-deficient mice according to their biological functions and listed them in Table 3. Interestingly, downregulation of genes appeared more important in the prostate of these mice, since the upregulation of genes was negligible (data not shown). Indeed, a strong degree of downregulation ranging from 3 to around 35 fold was observed in vivo. This was also in consonance with our in vitro results in Figure 1A where the combination of SFN+EGCG elicited diminished activation of the luciferase reporter. Furthermore, several genes that were downregulated in our in vivo study were also common to our regulatory comparative promoter analyses between Nrf2 and AP-1, and ATF-2 and ELK-1, as described earlier. These included Ikaros family zinc fingers, forkhead box members, and ATF-2 amongst others. Interestingly, several coactivators and corepressors of Nrf2, as well as Nrf3, and the adenomatosis polyposis coli (Apc) gene, were also shown to be modulated via Nrf2 in response to the combination of SFN+EGCG.
Table 3. Temporal microarray analyses of genes modulated by SFN+EGCG combination in the prostate of Nrf2-deficient mice.
| GenBank accession No | Gene symbol | Gene title | Prostate 3 ha | Prostate 12 hb |
|---|---|---|---|---|
| Apoptosis and cell cycle | ||||
| NM_019816 | Aatf | Apoptosis antagonizing transcription factor | – | 3.09 |
| NM_007466 | Api5 | Apoptosis inhibitor 5 | – | 7.26 |
| NM_007609 | Casp4 | Caspase 4, apoptosis-related cysteine peptidase | – | 4.24 |
| NM_011997 | Casp8ap2 | Caspase 8 associated protein 2 | – | 4.67 |
| NM_026201 | Ccar1 | Cell division cycle and apoptosis regulator 1 | – | 3.31 |
| NM_027545 | Cwf19l2 | CWF19-like 2, cell cycle control (S pombe) | – | 3.48 |
| NM_001037134 | Ccne2 | Cyclin E2 | – | 4.04 |
| NM_009831 | Ccng1 | Cyclin G1 | – | 3.82 |
| NM_028399 | Ccnt2 | Cyclin T2 | – | 3.16 |
| NM_145991 | Cdc73 | Cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S cerevisiae) | – | 6.13 |
| Calcium ion binding | ||||
| NM_009722 | Atp2a2 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 | – | 3.14 |
| NM_009784 | Cacna2d1 | Calcium channel, voltage-dependent, alpha2/delta subunit 1 | – | 4.81 |
| NM_007977 | F8 | Coagulation factor VIII | – | 4.18 |
| NM_007868 | Dmd | Dystrophin, muscular dystrophy | – | 3.21 |
| NM_007943 | Eps15 | Epidermal growth factor receptor pathway substrate 15 | – | 5.26 |
| NM_001039644 | Edem3 | ER degradation enhancer, mannosidase alpha-like 3 | – | 4.54 |
| NM_010427 | Hgf | Hepatocyte growth factor | – | 4.49 |
| XM_001472723 | Macf1 | Microtubule-actin crosslinking factor 1 | – | 3.96 |
| NM_011110 | Pla2g5 | Phospholipase A2, group V | – | 3.89 |
| NM_013829 | Plcb4 | Phospholipase C, beta 4 | – | 3.94 |
| NM_009048 | Reps1 | RalBP1 associated Eps domain containing protein | – | 6.19 |
| NM_021450 | Trpm7 | Transient receptor potential cation channel, subfamily M, member 7 | – | 4.46 |
| Digestion | ||||
| NM_025583 | Ctrb1 | Chymotrypsinogen B1 | – | 4.43 |
| NM_025469 | Clps | Colipase, pancreatic | – | 3.85 |
| NM_009430 | Prss2 | Protease, serine, 2 | – | 28.91 |
| Extracellular space | ||||
| NM_009692 | Apoa1 | Apolipoprotein A–I | – | 9.72 |
| NM_018782 | Calcrl | Calcitonin receptor-like | – | 5.1 |
| NM_001042611 | Cp | Ceruloplasmin | – | 3.77 |
| NM_019919 | Ltbp1 | Latent transforming growth factor beta binding protein 1 | – | 3.46 |
| NM_001039094 | Negr1 | Neuronal growth regulator 1 | – | 3.15 |
| NM_011964 | Psg19 | Pregnancy specific glycoprotein 19 | – | 8.93 |
| NM_009936 | Col9a3 | Procollagen, type IX, alpha 3 | - | 3.48 |
| NM_001081385 | Pcdh11x | Protocadherin 11 X-linked | – | 8.52 |
| NM_053141 | Pcdhb16 | Protocadherin beta 16 | – | 3.17 |
| NM_021289 | Smr2 | Submaxillary gland androgen regulated protein 2 | – | 29.55 |
| Integral to plasma membrane | ||||
| NM_008309 | Htr1d | 5-hydroxytryptamine (serotonin) receptor 1D | 10.02 | – |
| NM_008427 | Kcnj4 | Potassium inwardly-rectifying channel, subfamily J, member 4 | 3.49 | – |
| NM_008422 | Kcnc3 | Potassium voltage gated channel, Shaw-related subfamily, member 3 | 11.48 | – |
| NM_008434 | Kcnq1 | Potassium voltage-gated channel, subfamily Q, member 1 | 3.34 | – |
| NM_001098170 | Pcdh10 | Protocadherin 10 | 4.1 | – |
| BC098457 | Pcdhgc3 | Protocadherin gamma subfamily C, 3 | 3.5 | – |
| AK041751 | Tcrb-J | T-cell receptor beta, joining region | 11.66 | – |
| NM_029975 | Ulbp1 | UL16 binding protein 1 | 3.57 | – |
| Intracellular | ||||
| NM_007478 | Arf3 | ADP-ribosylation factor 3 | – | 4.82 |
| NM_134037 | Acly | ATP citrate lyase | – | 4.49 |
| NM_015802 | Dlc1 | Deleted in liver cancer 1 | 3.23 | – |
| NM_178118 | Dixdc1 | DIX domain containing 1 | 8.24 | 3.65 |
| NM_007961 | Etv6 | Ets variant gene 6 (TEL oncogene) | 4.57 | – |
| NM_080433 | Fezf2 | Fez family zinc finger 2 | – | 3.44 |
| NM_053202 | Foxp1 | Forkhead box P1 | – | 3.09 |
| NM_175143 | Qrich1 | Glutamine-rich 1 | 4.75 | – |
| NM_178888 | Garnl3 | GTPase activating RANGAP domain-like 3 | 3.02 | – |
| NM_001025597 | Ikzf1 | IKAROS family zinc finger 1 | 13.8 | – |
| NM_011771 | Ikzf3 | IKAROS family zinc finger 3 | – | 5.44 |
| NM_016889 | Insm1 | Insulinoma-associated 1 | 4.07 | – |
| NM_029416 | Klf17 | Kruppel-like factor 17 | 3.25 | – |
| NM_053158 | Mrpl1 | Mitochondrial ribosomal protein L1 | – | 7.61 |
| NM_031260 | Mov10l1 | Moloney leukemia virus 10-like 1 | 4.83 | – |
| NM_010823 | Mpl | Myeloproliferative leukemia virus oncogene | 4.77 | – |
| NM_031881 | Nedd4l | Neural precursor cell expressed, developmentally down-regulated gene 4-like | 3.76 | – |
| NM_139144 | Ogt | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine: polypeptide-N-acetylglucosaminyl transferase) | 3.02 | – |
| AY033991 | Plcd4 | Phospholipase C, delta 4 | 4.55 | – |
| NM_008884 | Pml | Promyelocytic leukemia | 4.5 | – |
| NM_009391 | Ran | RAN, member RAS oncogene family | – | 3.96 |
| NM_011246 | Rasgrp1 | RAS guanyl releasing protein 1 | 3.43 | – |
| NM_001081105 | Rhoh | Ras homolog gene family, member H | 5.73 | – |
| NM_145452 | Rasa1 | RAS p21 protein activator 1 | – | 3.31 |
| NM_172525 | Arhgap29 | Rho GTPase activating protein 29 | – | 5.05 |
| NM_015830 | Solh | Small optic lobes homolog (Drosophila) | 4.86 | – |
| NM_028004 | Ttn | Titin | 4.77 | – |
| NM_011529 | Tank | TRAF family member-associated Nf-kappa B activator | – | 6.11 |
| NM_009541 | Zbtb17 | Zinc finger and BTB domain containing 17 | 5.46 | – |
| NM_008717 | Zfml | Zinc finger, matrin-like | 3.36 | – |
| Kinases | ||||
| NM_134079 | Adk | Adenosine kinase | – | 4.07 |
| NM_007561 | Bmpr2 | Bone morphogenic protein receptor, type II (serine/threonine kinase) | – | 6.14 |
| NM_001025439 | Camk2d | Calcium/calmodulin-dependent protein kinase II, delta | – | 4.6 |
| NM_001042634 | Clk1 | CDC-like kinase 1 | – | 3.61 |
| NM_007714 | Clk4 | CDC like kinase 4 | – | 3.33 |
| NM_001109626 | Crkrs | Cdc2-related kinase, arginine/serine-rich | – | 7.33 |
| NM_009974 | Csnk2a2 | Casein kinase 2, alpha prime polypeptide | 6.07 | – |
| XM_979562 | Etnk1 | Ethanolamine kinase 1 | – | 3.19 |
| NM_019827 | Gsk3b | Glycogen synthase kinase 3 beta | – | 4.47 |
| NM_010367 | Magi1 | Membrane associated guanylate kinase, WW and PDZ domain containing 1 | 3.12 | – |
| NM_015823 | Magi2 | Membrane associated guanylate kinase, WW and PDZ domain containing 2 | 6.15 | |
| NM_009157 | Map2k4 | Mitogen activated protein kinase kinase 4 | – | 3.31 |
| NM_011946 | Map3k2 | Mitogen activated protein kinase kinase kinase 2 | – | 5.02 |
| NM_025609 | Map3k7ip1 | Mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | 3.04 | 3.93 |
| NM_008696 | Map4k4 | Mitogen-activated protein kinase kinase kinase kinase 4 | 3.54 | – |
| NM_201519 | Map4k5 | Mitogen-activated protein kinase kinase kinase kinase 5 | – | 3.28 |
| NM_011161 | Mapk11 | Mitogen-activated protein kinase 11 | – | 10.9 |
| NM_172632 | Mapk4 | Mitogen-activated protein kinase 4 | 3.51 | – |
| NM_015806 | Mapk6 | Mitogen-activated protein kinase 6 | 3.53 | – |
| NM_010878 | Nck1 | Non-catalytic region of tyrosine kinase adaptor protein 1 | – | 4.24 |
| NM_010879 | Nck2 | Non-catalytic region of tyrosine kinase adaptor protein 2 | 3.38 | – |
| NM_021605 | Nek7 | NIMA (never in mitosis gene a)-related expressed kinase 7 | – | 3.13 |
| NM_008702 | Nlk | Nemo like kinase | – | 3.99 |
| NM_172783 | Phka2 | Phosphorylase kinase alpha 2 | 3.07 | – |
| NM_011083 | Pik3c2a | Phosphatidylinositol 3-kinase, C2 domain containing, alpha polypeptide | – | 5.18 |
| NM_008839 | Pik3ca | Phosphatidylinositol 3-kinase, catalytic, alpha polypeptide | – | 3.92 |
| NM_008862 | Pkia | Protein kinase inhibitor, alpha | – | 3.28 |
| NM_008855 | Prkcb1 | Protein kinase C, beta 1 | – | 3.34 |
| NM_029239 | Prkcn | Protein kinase C, nu | 3.93 | – |
| NM_001013833 | Prkg1 | Protein kinase, cGMP-dependent, type I | 3.31 | – |
| NM_007982 | Ptk2 | PTK2 protein tyrosine kinase 2 | 3.47 | – |
| NM_009184 | Ptk6 | PTK6 protein tyrosine kinase 6 | 4.66 | – |
| NM_009071 | Rock1 | Rho-associated coiled-coil containing protein kinase 1 | – | 4.71 |
| NM_009072 | Rock2 | Rho-associated coiled-coil containing protein kinase 2 | – | 4.42 |
| NM_148945 | Rps6ka3 | Ribosomal protein S6 kinase polypeptide 3 | – | 8.82 |
| NM_028259 | Rps6kb1 | Ribosomal protein S6 kinase, polypeptide 1 | – | 3.36 |
| NM_019635 | Stk3 | Serine/threonine kinase 3 (Ste20, yeast homolog) | – | 3.03 |
| NM_144825 | Taok1 | TAO kinase 1 | – | 3.93 |
| XM_001474897 | Tnik | TRAF2 and NCK interacting kinase | 4.01 | 8.1 |
| NM_010633 | Uhmk1 | U2AF homology motif (UHM) kinase 1 | 3.19 | – |
| NM_030724 | Uck2 | Uridine-cytidine kinase 2 | 4.1 | – |
| NM_198703 | Wnk1 | WNK lysine deficient protein kinase 1 | – | 6.45 |
| Metal ion binding | ||||
| NM_009530 | Atrx | Alpha thalassemia/mental retardation syndrome X-linked homolog (human) | – | 4.12 |
| NM_013476 | Ar | Androgen receptor | 3.23 | – |
| NM_133738 | Antxr2 | Anthrax toxin receptor 2 | – | 3.15 |
| AB088408 | Atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit D1 | 6.95 | – |
| NM_139294 | Braf | Braf transforming gene | – | 3.67 |
| NM_153788 | Centb1 | Centaurin, beta 1 | – | 6.1 |
| NM_007805 | Cyb561 | Cytochrome b-561 | 5.53 | – |
| NM_010006 | Cyp2d9 | Cytochrome P450, family 2, subfamily d, polypeptide 9 | 3.28 | – |
| NM_027816 | Cyp2u1 | Cytochrome P450, family 2, subfamily u, polypeptide 1 | 5.7 | – |
| NM_011935 | Esrrg | Estrogen-related receptor gamma | 3.85 | – |
| NM_025923 | Fancl | Fanconi anemia, complementation group L | – | 3.38 |
| NM_053242 | Foxp2 | Forkhead box P2 | – | 4.42 |
| NM_026148 | Lims1 | LIM and senescent cell antigen-like domains 1 | – | 4.31 |
| NM_008636 | Mtf1 | Metal response element binding transcription factor 1 | 15.93 | – |
| NM_028757 | Nebl | Nebulette | 4.66 | – |
| NM_152229 | Nr2e1 | Nuclear receptor subfamily 2, group E, member 1 | 11.85 | – |
| NM_030676 | Nr5a2 | Nuclear receptor subfamily 5, group A, member 2 | 4.12 | – |
| NM_010264 | Nr6a1 | Nuclear receptor subfamily 6, group A, member 1 | 10.31 | – |
| NM_026000 | Psmd9 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 | 3.96 | – |
| NM_177167 | Ppm1e | Protein phosphatase 1E (PP2C domain containing) | 3.56 | – |
| NM_009088 | Rpo1-4 | RNA polymerase 1–4 | 4.87 | 4.31 |
| NM_001103157 | Steap2 | Six transmembrane epithelial antigen of prostate 2 | – | 3.76 |
| NM_009380 | Thrb | Thyroid hormone receptor beta | – | 4.67 |
| NM_009371 | Tgfbr2 | Transforming growth factor, beta receptor II | 4.54 | – |
| Phosphatases | ||||
| NM_008960 | Pten | Phosphatase and tensin homolog | – | 3.05 |
| NM_027892 | Ppp1r12a | Protein phosphatase 1, regulatory (inhibitor) subunit 12A | – | 3.77 |
| NM_008913 | Ppp3ca | Protein phosphatase 3, catalytic subunit, alpha isoform | – | 9.33 |
| NM_182939 | Ppp4r2 | Protein phosphatase 4, regulatory subunit 2 | – | 3.39 |
| NM_011200 | Ptp4a1 | Protein tyrosine phosphatase 4a1 | – | 3.11 |
| NM_008979 | Ptpn22 | Protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | – | 3.65 |
| NM_001014288 | Ptprd | Protein tyrosine phosphatase, receptor type, D | – | 6.2 |
| NM_025760 | Ptplad2 | Protein tyrosine phosphatase-like A domain containing 2 | – | 4.42 |
| NM_130447 | Dusp16 | Dual specificity phosphatase 16 | – | 5.97 |
| NM_177730 | Impad1 | Inositol monophosphatase domain containing 1 | – | 3.72 |
| NM_011210 | Ptprc | Protein tyrosine phosphatase, receptor type, C | 4.33 | – |
| Transcription factors and interacting partners | ||||
| AY902311 | Atf2 | Activating transcription factor 2 | – | 8.21 |
| AY903215 | Atf7ip2 | Activating transcription factor 7 interacting protein 2 | – | 9.83 |
| NM_001025392 | Bclaf1 | BCL2-associated transcription factor 1 | – | 3.78 |
| NM_015826 | Dmrt1 | Doublesex and mab-3 related transcription factor 1 | – | 6.96 |
| NM_009210 | Hltf | Helicase-like transcription factor | – | 3.93 |
| NM_033322 | Lztfl1 | Leucine zipper transcription factor-like 1 | – | 4.08 |
| NM_172153 | Lcorl | Ligand dependent nuclear receptor corepressor-like | – | 3.68 |
| NM_183355 | Pbx1 | Pre B-cell leukemia transcription factor 1 | – | 5.26 |
| NM_001018042 | Sp3 | Trans-acting transcription factor 3 | – | 3.65 |
| AM295492 | Sp6 | Trans-acting transcription factor 6 | – | 32.91 |
| NM_013685 | Tcf4 | Transcription factor 4 | – | 3.84 |
| NM_178254 | Tcfl5 | Transcription factor-like 5 (basic helix-loop-helix) | – | 4.6 |
| NM_016767 | Batf | Basic leucine zipper transcription factor, ATF-like | – | 3.89 |
| NM_001109661 | Bach2 | BTB and CNC homology 2 | – | 4.89 |
| NM_133828 | Creb1 | cAMP responsive element binding protein 1 | – | 5.33 |
| NM_001005868 | Erbb2ip | Erbb2 interacting protein | – | 3.66 |
| NM_008031 | Fmr1 | Fragile X mental retardation syndrome 1 homolog | – | 3.96 |
| NM_010431 | Hif1a | Hypoxia inducible factor 1, alpha subunit | – | 8.62 |
| NM_009951 | Igf2bp1 | Insulin-like growth factor 2 mRNA binding protein 1 | 5.27 | – |
| NM_027910 | Klhdc3 | Kelch domain containing 3 | 3.33 | – |
| NM_172154 | Lcor | Ligand dependent nuclear receptor corepressor | – | 3.25 |
| NM_001005863 | Mtus1 | Mitochondrial tumor suppressor 1 | 3.29 | – |
| NM_001081445 | Ncam1 | Neural cell adhesion molecule 1 | 3.08 | – |
| AY050663 | Nfat5 | Nuclear factor of activated T-cells 5 | – | 4.4 |
| NM_010903 | Nfe2l3 | Nuclear factor, erythroid derived 2, like 3 (Nrf3) | – | 3.03 |
| NM_001024205 | Nufip2 | Nuclear fragile X mental retardation protein interacting protein 2 | – | 4.4 |
| NM_010881 | Ncoa1 | Nuclear receptor coactivator 1 | 3.55 | 3.36 |
| NM_172495 | Ncoa7 | Nuclear receptor coactivator 7 | – | 4.46 |
| NM_011308 | Ncor1 | Nuclear receptor co-repressor 1 | – | 5.37 |
| NM_011544 | Tcf12 | Transcription factor 12 | – | 6.06 |
| Transferases | ||||
| NM_144807 | Chpt1 | Choline phosphotransferase 1 | – | 4.13 |
| NM_133869 | Cept1 | Choline/ethanolaminephosphotransferase 1 | – | 5.65 |
| NM_030225 | Dlst | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | – | 4.04 |
| NM_028087 | Gcnt3 | Glucosaminyl (N-acetyl) transferase 3, mucin type | – | 7.23 |
| NM_028108 | Nat13 | N-acetyltransferase 13 | – | 4.05 |
| NM_008708 | Nmt2 | N-myristoyltransferase 2 | – | 4.07 |
| NM_027869 | Pnpt1 | Polyribonucleotide nucleotidyltransferase 1 | – | 3.04 |
| NM_172627 | Pggt1b | Protein geranylgeranyltransferase type I, beta subunit | – | 5.61 |
| NM_183028 | Pcmtd1 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing | – | 7.36 |
| NM_028604 | Trmt11 | tRNA methyltransferase 11 homolog (S cerevisiae) | – | 3.67 |
| NM_144731 | Galnt7 | UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 | – | 3.06 |
| NM_172829 | St6gal2 | Beta galactoside alpha 2,6 sialyltransferase 2 | – | 3.22 |
| NM_009178 | St3gal4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | – | 8.11 |
| NM_011674 | Ugt8a | UDP galactosyltransferase 8A | – | 13.64 |
| Ubiquitination | ||||
| XM_001478436 | Fbxl17 | F-box and leucine-rich repeat protein 17 | – | 3.86 |
| NM_016736 | Nub1 | Negative regulator of ubiquitin-like proteins 1 | 3.57 | – |
| NM_146003 | Senp6 | SUMO/sentrin specific peptidase 6 | – | 4.5 |
| NM_016723 | Uchl3 | Ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) | – | 6.71 |
| NM_173010 | Ube3a | Ubiquitin protein ligase E3A | – | 3.12 |
| NM_009481 | Usp9x | Ubiquitin specific peptidase 9, X chromosome | – | 3.47 |
| NM_152825 | Usp45 | Ubiquitin specific petidase 45 | – | 3.09 |
| NM_023585 | Ube2v2 | Ubiquitin-conjugating enzyme E2 variant 2 | – | 5.5 |
| NM_172300 | Ube2z | Ubiquitin-conjugating enzyme E2Z (putative) | 3.41 | – |
| NM_177327 | Wwp1 | WW domain containing E3 ubiquitin protein ligase 1 | – | 3.9 |
| Others | ||||
| NM_007462 | Apc | Adenomatosis polyposis coli | – | 5.42 |
| NM_021456 | Ces1 | Carboxylesterase 1 | 4.09 | – |
| NM_021369 | Chrna6 | Cholinergic receptor, nicotinic, alpha polypeptide 6 | 4.25 | – |
| NM_028870 | Cltb | Clathrin, light polypeptide (Lcb) | 22.37 | – |
| NM_016716 | Cul3 | Cullin 3 | – | 3.2 |
| NM_010076 | Drd1a | Dopamine receptor D1A | 12.13 | – |
| NM_001033360 | Gpr101 | G protein-coupled receptor 101 | 4.26 | – |
| NM_008211 | H3f3b | H3 histone, family 3B | 3.47 | – |
| NM_008285 | Hrh1 | Histamine receptor H 1 | 10.24 | – |
| NM_133892 | Lao1 | L-amino acid oxidase 1 | 3.39 | – |
| NM_020280 | Magea4 | Melanoma antigen, family A, 4 | 22.06 | – |
| NM_175632 | Oscar | Osteoclast associated receptor | 3.36 | – |
| NM_009419 | Tpst2 | Protein-tyrosine sulfotransferase 2 | 7.76 | – |
| NM_146255 | Slc1a7 | Solute carrier family 1 (glutamate transporter), member 7 | 9.36 | – |
| NM_009206 | Slc4a1ap | Solute carrier family 4 (anion exchanger), member 1, adaptor protein | – | 3.27 |
| NM_022025 | Slc5a7 | Solute carrier family 5 (choline transporter), member 7 | 9.51 | – |
| NM_177909 | Slc9a9 | Solute carrier family 9 (sodium/hydrogen exchanger), isoform 9 | 4.23 | – |
| NM_011506 | Sucla2 | Succinate-Coenzyme A ligase, ADP-forming, beta subunit | 5.02 | – |
| NM_009409 | Top2b | Topoisomerase (DNA) II beta | 3.3 | |
aRelative mRNA expression levels of genes that were suppressed >3-fold by SFN+EGCG combination in prostate of Nrf2 wild-type mice but not in prostate of Nrf2 knockout mice compared with vehicle treatment at 3 h.
bRelative mRNA expression levels of genes that were suppressed >3-fold by SFN+EGCG combination in prostate of Nrf2 wild-type mice but not in prostate of Nrf2 knockout mice compared with vehicle treatment at 12 h.
Discussion
Expression profiling and proteomics have been credited47 with the potential to transform the management of PCa by identifying new markers for screening, diagnosis, prognosis, monitoring and targets for therapy. Several studies have addressed the putative role(s) of either Nrf2 or AP-1 in PCa; however, the potential for putative crosstalk between these two important transcription factors in the pathogenesis of PCa has not been explored so far. In order to better appreciate the regulatory potential for concerted modulation of Nrf2 and AP-1 in PCa on treatment with dietary factors SFN and EGCG in combination, we performed in vitro studies in PC-3 AP-1 PCa cells, in vivo studies in the prostate of Nrf2-deficient mice, and in silico bioinformatic analyses to elucidate conserved motifs in the promoter regions of these transcription factors, as well as genes coregulated by them.
We have previously reported48 that the peak plasma concentration (Cmax) achievable with SFN in rats was 20 μmol/L after oral administration. In addition, we have reported49 that SFN 50 μmol/L was toxic to HepG2 C8 cells, whereas SFN 25 μmol/L was suboptimal in its efficacy. Since a desirable objective of using combinatorial approaches is to reduce the dose of the administered agents thereby reducing toxic side-effects, our dose selection of 25 μmol/L of SFN for the current study was guided by its proximity to the observed Cmax and its suboptimal effectiveness in eliciting transcriptional effects as compared to higher doses of SFN. Besides, in most studies, the concentrations needed to observe the activities of EGCG typically range from 1 to 100 μmol/L; these are, in reality, concentrations that exceed those found in rodent and human plasma by 10- to 100-fold50, 51. However, the uptake of EGCG in HT-29 cells has also been shown to be concentration-dependent in the range of 20–600 μmol/L50. In addition, we have also previously reported33 that EGCG inhibited HT-29 cell growth with an IC50 of approximately 100 μmol/L. Accordingly, we elected to test two doses of EGCG (20 and 100 μmol/L) in the current study in combination with the 25 μmol/L dose of SFN. In support of the rationale for combination regimens, a combination of atorvastatin and celecoxib has been reported to be more potent in inhibiting growth of PC-3 cells in vitro or grown in SCID mice than individual agents52 for the prevention of prostate cancer. Besides, as we discussed earlier in this journal10, our laboratory has been studying two groups of dietary phytochemical cancer-chemopreventive compounds (isothiocyanates and polyphenols), which are effective in chemical-induced, as well as genetically-induced, animal carcinogenesis models. These compounds typically generate “cellular stress” and modulate gene expression of phase II detoxifying/antioxidant enzymes. Some of the most promising members of these two classes of phytochemicals are EGCG (polyphenol) from green tea and sulforaphane (isothiocyanate) from cruciferous vegetables, hence we decided to focus on these two agents in the current study.
Interestingly, from our in vitro data in PC-3 AP-1 cells (Figure 1A), we observed a diminished induction of the luciferase reporter on treatment with a combination of SFN+EGCG that was dose-dependent. We observed similar trends in our in vivo microarray data in Nrf2-deficient mice (Table 3), where downregulation (3-fold to around 35-fold) of key genes identified as Nrf2-dependent appeared to be the dominant response to oral administration of the SFN+EGCG combination at both 3 and 12 h. Quantitative real-time PCR analyses in our in vitro system (Figure 2) confirmed that several genes including ATF-2 and ELK-1 were regulated by AP-1. Bioinformatic analyses of the promoter regions of Nrf2 and AP-1, as well as ATF-2 and ELK-1, revealed an interesting group of conserved TFBS (Tables 1–2 and Figure 3) in both human and murine promoters. Furthermore, we were able to identify genes with conserved regulatory sequences in the promoter regions of human, or murine, Nrf2 and AP-1 as shown in the Supplementary Table. Indeed, microarray analyses in Nrf2-deficient mice (Table 3) confirmed that genes identified as Nrf2-dependent, including ATF-2, were coregulated with genes elicited from our AP-1 in vitro studies as well as the comparative analyses of promoter regions of Nrf2 and AP-1 using bioinformatic approaches. It has been noted47 that a majority of prostate cancers contain fusion genes that result in regulation via ETS family transcription factors. Our in silico results (Tables 1–2 and Supplementary Table) as well as our in vitro qRT-PCR data (Figure 2) also demonstrate a role for ETS family members including ELK-1 which is, thus, in consonance with previous reports.
The identification of conserved TFBS for pro-survival transcription factor NF-κB in the promoter regions of Nrf2 and AP-1 (Tables 1–2, Supplementary Table and Figure 3) raises an important question as to whether there could be any possible crosstalk between Nrf2, AP-1, and NF-κB in concert that may contribute to the overall effects of the SFN+EGCG combination in PCa. Indeed, further studies would be necessary to explore this possibility in greater detail. The identification of several key MAPK genes in our microarray studies (Table 3) as Nrf2-dependent is in congruence with the known role(s) of MAPKs in Nrf2 phosphorylation and activation. Besides, the downregulation of Nrf3, a negative regulator of ARE-mediated gene expression53 with substantial homology to Nrf2, in our microarray data (Table 3) reinforces the putative chemopreventive potential of the SFN+EGCG combination. Furthermore, the elucidation of several coactivators and corepressors as Nrf2-dependent, including nuclear receptor coactivators 1 and 7 (Ncoa1 and Ncoa7) and nuclear receptor corepressor 1 (Ncor1), indicated that these cofactors may have a potentially significant role to play in the ability of Nrf2 to crosstalk with AP-1 in vivo.
Given the biological complexity of PCa and the need for identification of better targets, the various potential biomarkers elicited in this study may be adapted into early discovery screens for chemopreventive or chemotherapeutic intervention in PCa. The current transcriptional regulation study is a first step in understanding putative crosstalk between Nrf2 and AP-1 in PCa that may potentially be exploited by intervention with dietary phytochemicals SFN and EGCG in combination to delay the onset of clinically-evident PCa, or to arrest progression of high-grade prostate intraepithelial neoplasia (HG-PIN) to metastatic hormone-refractory prostate cancer (HRPC). Future goals include dissecting the functional biological networks for cross-talk between Nrf2 and AP-1 that will throw light on specific target hubs that play major role(s) in this cross-talk including a role(s) for potential party hubs and date hubs that might be important in this process54, 55, 56. Indeed, further studies focusing on the specific signaling intermediates, as well as clinical studies, would eventually be necessary to better appreciate the putative role(s) of the combination of dietary factors SFN and EGCG in the management of PCa.
Author contribution
Sujit NAIR, Li CAI, and Ah-Ng KONG conceived and designed the study; Sujit NAIR performed the in vitro studies, processing of in vivo samples and assays on in vivo samples; Avantika BARVE, Tin-Oo KHOR, Guo-xiang SHEN, and Wen LIN dosed the animals with phytochemicals and sacrificed them; Sujit NAIR, Avantika BARVE, Tin-Oo KHOR, Guo-xiang SHEN, and Wen LIN harvested the prostate samples after dissection; Sujit NAIR and Li CAI performed the in silico studies and microarray analyses; Jefferson Y CHAN provided the first generation of Nrf2-knockout mice; Sujit NAIR, Li CAI, and Ah-Ng KONG analyzed the data; Sujit NAIR wrote the manuscript with contributions from Li CAI and Ah-Ng KONG; Li CAI and Ah-Ng KONG edited the manuscript; Ah-Ng KONG provided the funding support for the studies.
Acknowledgments
This work was supported in part by RO1-CA118947 and RO1-CA094828 to Ah-Ng Tony KONG, and R21-CA133675 to Li CAI from the National Institutes of Health (NIH).
Glossary
- Nrf2
Nuclear Factor-E2-related factor 2
- AP-1
activator protein-1
- ATF-2
activating transcription factor 2
- SFN
sulforaphane
- EGCG
(−)epigallocatechin-3-gallate
- MAPK
mitogen-activated protein kinase
- qRT-PCR
quantitative real-time PCR
- TFBS
Transcription Factor Binding Sites
Footnotes
Supplementary table is available at Acta Pharmacologica Sinica website of NPG.
Supplementary Information
Matrices with conserved regulatory sequences in promoter regions of human, or murine, NRF2 and AP-1
References
- Centers for Disease Control and Prevention (CDC). Department of Health and Human Services. Available at http://www.cdc.gov/cancer/prostate/basic_info/
- US Cancer Statistics Working Group. United States Cancer Statistics: 2003 Incidence and Mortality. Atlanta (GA). Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute; 2007
- National Cancer Institute (NCI). Surveillance Epidemiology and End Results (SEER). Available at http://seercancergov/statfacts/html/prosthtml .
- Scholz M, Lam R, Strum S, Jennrich R, Johnson H, Trilling T. Prostate cancer-specific survival and clinical progression-free survival in men with prostate cancer treated intermittently with testosterone inactivating pharmaceuticals. Urology. 2007;70:506–10. doi: 10.1016/j.urology.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Welch HG, Fisher ES, Gottlieb DJ, Barry MJ. Detection of prostate cancer via biopsy in the Medicare-SEER population during the PSA era. J Natl Cancer Inst. 2007;99:1395–400. doi: 10.1093/jnci/djm119. [DOI] [PubMed] [Google Scholar]
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–8. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, et al. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–307. [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- Prochaska HJ, De Long MJ, Talalay P. On the mechanisms of induction of cancer-protective enzymes: a unifying proposal. Proc Natl Acad Sci USA. 1985;82:8232–6. doi: 10.1073/pnas.82.23.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Li W, Kong AN. Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharmacol Sin. 2007;28:459–72. doi: 10.1111/j.1745-7254.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- Yates MS, Kensler TW. Keap1 eye on the target: chemoprevention of liver cancer. Acta Pharmacol Sin. 2007;28:1331–42. doi: 10.1111/j.1745-7254.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Kwon KH, Chan JY, Reddy BS, Kong AN. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66:11580–4. doi: 10.1158/0008-5472.CAN-06-3562. [DOI] [PubMed] [Google Scholar]
- Burton NC, Kensler TW, Guilarte TR. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology. 2006;27:1094–100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–9. [PubMed] [Google Scholar]
- Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci USA. 1997;94:5361–6. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, et al. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5:1918–26. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- Watai Y, Kobayashi A, Nagase H, Mizukami M, McEvoy J, Singer JD, et al. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells. 2007;12:1163–78. doi: 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- Hsu TC, Young MR, Cmarik J, Colburn NH. Activator protein 1 (AP-1)- and nuclear factor kappaB (NF-kappaB)-dependent transcriptional events in carcinogenesis. Free Radic Biol Med. 2000;28:1338–48. doi: 10.1016/s0891-5849(00)00220-3. [DOI] [PubMed] [Google Scholar]
- Xu C, Shen G, Yuan X, Kim JH, Gopalkrishnan A, Keum YS, et al. ERK and JNK signaling pathways are involved in the regulation of activator protein 1 and cell death elicited by three isothiocyanates in human prostate cancer PC-3 cells. Carcinogenesis. 2006;27:437–45. doi: 10.1093/carcin/bgi251. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–52. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Steinmetz KA, Potter JD. Vegetables, fruit, and cancer. I. Epidemiology. Cancer Causes Control. 1991;2:325–57. doi: 10.1007/BF00051672. [DOI] [PubMed] [Google Scholar]
- Leoni O, Iori R, Palmieri S. Hydrolysis of glucosinolates using nylon-immobilized myrosinase to produce pure bioactive molecules. Biotechnol Bioeng. 2000;68:660–4. doi: 10.1002/(sici)1097-0290(20000620)68:6<660::aid-bit9>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Getahun SM, Chung FL. Conversion of glucosinolates to isothiocyanates in humans after ingestion of cooked watercress. Cancer Epidemiol Biomarkers Prev. 1999;8:447–51. [PubMed] [Google Scholar]
- Traka M, Gasper AV, Smith JA, Hawkey CJ, Bao Y, Mithen RF. Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J Nutr. 2005;135:1865–72. doi: 10.1093/jn/135.8.1865. [DOI] [PubMed] [Google Scholar]
- Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232:227–34. [PMC free article] [PubMed] [Google Scholar]
- Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, et al. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–13. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- Singh SV, Srivastava SK, Choi S, Lew KL, Antosiewicz J, Xiao D, et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J Biol Chem. 2005;280:19911–24. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- Herman-Antosiewicz A, Johnson DE, Singh SV. Sulforaphane causes autophagy to inhibit release of cytochrome c and apoptosis in human prostate cancer cells. Cancer Res. 2006;66:5828–35. doi: 10.1158/0008-5472.CAN-06-0139. [DOI] [PubMed] [Google Scholar]
- Xu C, Shen G, Chen C, Gelinas C, Kong AN. Suppression of NF-kappaB and NF-kappaB-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene. 2005;24:4486–95. doi: 10.1038/sj.onc.1208656. [DOI] [PubMed] [Google Scholar]
- Petri N, Tannergren C, Holst B, Mellon FA, Bao Y, Plumb GW, et al. Absorption/metabolism of sulforaphane and quercetin, and regulation of phase II enzymes, in human jejunum in vivo. Drug Metab Dispos. 2003;31:805–13. doi: 10.1124/dmd.31.6.805. [DOI] [PubMed] [Google Scholar]
- Mukhtar H, Ahmad N. Mechanism of cancer chemopreventive activity of green tea. Proc Soc Exp Biol Med. 1999;220:234–8. doi: 10.1046/j.1525-1373.1999.d01-40.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Shen G, Hebbar V, Hu R, Owuor ED, Kong AN. Epigallocatechin-3-gallate-induced stress signals in HT-29 human colon adenocarcinoma cells. Carcinogenesis. 2003;24:1369–78. doi: 10.1093/carcin/bgg091. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (–)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–70. [PubMed] [Google Scholar]
- Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–8S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- Harper CE, Patel BB, Wang J, Eltoum IA, Lamartiniere CA. Epigallocatechin-3-Gallate suppresses early stage, but not late stage prostate cancer in TRAMP mice: mechanisms of action. Prostate. 2007;67:1576–89. doi: 10.1002/pros.20643. [DOI] [PubMed] [Google Scholar]
- Siddiqui IA, Adhami VM, Afaq F, Ahmad N, Mukhtar H. Modulation of phosphatidylinositol-3-kinase/protein kinase B- and mitogen-activated protein kinase-pathways by tea polyphenols in human prostate cancer cells. J Cell Biochem. 2004;91:232–42. doi: 10.1002/jcb.10737. [DOI] [PubMed] [Google Scholar]
- Adhami VM, Malik A, Zaman N, Sarfaraz S, Siddiqui IA, Syed DN, et al. Combined inhibitory effects of green tea polyphenols and selective cyclooxygenase-2 inhibitors on the growth of human prostate cancer cells both in vitro and in vivo. Clin Cancer Res. 2007;13:1611–9. doi: 10.1158/1078-0432.CCR-06-2269. [DOI] [PubMed] [Google Scholar]
- Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, et al. Comparison of (–)-epigallocatechin-3-gallate elicited liver and small intestine gene expression profiles between C57BL/6J mice and C57BL/6J/Nrf2(–/–) mice. Pharm Res. 2005;22:1805–20. doi: 10.1007/s11095-005-7546-8. [DOI] [PubMed] [Google Scholar]
- Sang S, Yang I, Buckley B, Ho CT, Yang CS. Autoxidative quinone formation in vitro and metabolite formation in vivo from tea polyphenol (–)-epigallocatechin-3-gallate: studied by real-time mass spectrometry combined with tandem mass ion mapping. Free Radic Biol Med. 2007;43:362–71. doi: 10.1016/j.freeradbiomed.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Kensler T. Nrf2 as a target for cancer chemoprevention. Mutat Res. 2005;591:93–102. doi: 10.1016/j.mrfmmm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–84. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. Nrf2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci USA. 1996;93:13943–8. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hung Wong W.Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application Genome Biol 20012RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters JR. Clinical applications of expression profiling and proteomics in prostate cancer. Anticancer Res. 2007;27:1273–6. [PubMed] [Google Scholar]
- Hu R, Hebbar V, Kim BR, Chen C, Winnik B, Buckley B, et al. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther. 2004;310:263–71. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- Kim BR, Hu R, Keum YS, Hebbar V, Shen G, Nair SS, et al. Effects of glutathione on antioxidant response element-mediated gene expression and apoptosis elicited by sulforaphane. Cancer Res. 2003;63:7520–5. [PubMed] [Google Scholar]
- Lambert JD, Lee MJ, Diamond L, Ju J, Hong J, Bose M, et al. Dose-dependent levels of epigallocatechin-3-gallate in human colon cancer cells and mouse plasma and tissues. Drug Metab Dispos. 2006;34:8–11. doi: 10.1124/dmd.104.003434. [DOI] [PubMed] [Google Scholar]
- Lambert JD, Lee MJ, Lu H, Meng X, Hong JJ, Seril DN, et al. Epigallocatechin-3-gallate is absorbed but extensively glucuronidated following oral administration to mice. J Nutr. 2003;133:4172–7. doi: 10.1093/jn/133.12.4172. [DOI] [PubMed] [Google Scholar]
- Zheng X, Cui XX, Avila GE, Huang MT, Liu Y, Patel J, et al. Atorvastatin and celecoxib inhibit prostate PC-3 tumors in immunodeficient mice. Clin Cancer Res. 2007;13:5480–7. doi: 10.1158/1078-0432.CCR-07-0242. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–7. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Jin G, Zhang S, Zhang XS, Chen L. Hubs with network motifs organize modularity dynamically in the protein-protein interaction network of yeast. PloS one. 2007;2:e1207. doi: 10.1371/journal.pone.0001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Deane CM, Porter MA, Jones NS. Revisiting date and party hubs: novel approaches to role assignment in protein interaction networks. PLoS Comput Biol. 2010;6:e1000817. doi: 10.1371/journal.pcbi.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Matrices with conserved regulatory sequences in promoter regions of human, or murine, NRF2 and AP-1