Abstract
Aim:
To explore the role of Med19, a component of the Mediator complex that coactivates DNA-binding transcription factors, in the proliferation and tumorigenesis of human hepatocellular carcinoma cells.
Methods:
The human hepatocellular carcinoma cell lines HepG2 and Hep3B were infected with lentiviral vectors encoding interfering RNA (RNAi) targeting the Med19 gene. To further confirm the inhibitory effects of RNAi vectors on Med19 gene expression, quantitative real-time RT-PCR and Western blotting assays were used. The proliferation of HepG2 and Hep3B cells after transduction with the Med19-RNAi-Lentivirus vector was evaluated by MTT conversion, BrdU incorporation, colony formation, and cell-cycle assays in vitro. In addition, the ability of the Med19-RNAi-Lentivirus vector-infected Hep3B cells to form tumors after inoculation into nude mice was determined.
Results:
Recombinant lentiviral vectors expressing small interfering RNA (siRNA) against Med19 were constructed and were found to efficiently downregulate Med19 mRNA and protein levels in HepG2 and Hep3B cells. Furthermore, the inhibition of Med19 by RNAi dramatically reduced hepatocellular carcinoma cell proliferation, induced cell-cycle arrest in the G0/G1 phase, and suppressed tumor formation.
Conclusion:
These results provide new evidence of an important role for Med19 in the development of hepatocellular carcinomas, suggesting that lentivirus-mediated RNAi to target Med19 is a potential tool for inhibiting cancer cell proliferation and tumorigenesis.
Keywords: hepatocellular carcinoma, Med19, RNAi, lentivirus vector, proliferation, tumorigenesis
Introduction
Hepatocellular carcinoma (HCC) is a malignant tumor type with a high rate of relapse and metastasis; it is the third leading cause of cancer-related deaths worldwide. The incidence of HCC has been increasing at an alarming rate on a global scale over the past two decades1, 2, 3, 4, 5. Despite a recent improvement in long-term survival rates, the prognosis of HCC is still poor. Therefore, it is necessary to find new therapeutic molecular targets and therapeutic strategies for the prevention and treatment of HCC. The recent discovery of RNA interference (RNAi), a powerful tool to induce loss-of-function phenotypes through the posttranscriptional silencing of gene expression, has provided new possibilities for cancer therapy6, 7, 8.
As a multi-protein coactivator, the Mediator (Med) complex is an essential component of the RNA polymerase II (pol II)-mediated transcription complex and is required for the transcriptional activation of pol II-transcribed genes by transcription factors9, 10. In addition to stimulating basal transcription, the Mediator complex plays a crucial role in the activation and repression of eukaryotic mRNA synthesis11, 12. The Mediator complex, which consists of at least 20 subunits with multiple activities, including Rox3 (Med19), was first characterized through biochemical and genetic studies in Saccharomyces cerevisiae in the early 1990s13, 14.
In the human genome, the Mediator complex subunit 19 (Med19) gene, also known as lung cancer metastasis-related protein 1 (LCMR1), is located at chr11q12.1. The Med19 protein can modulate both basal and heat-shock gene transcription, which are capabilities of a distinct functional module (head) within the Mediator complex. The Med19 gene was first cloned from a human lung large-cell carcinoma cell line through differential display reverse-transcription PCR by researchers at the Chinese General Hospital of the People's Liberation Army, Beijing, China, in 2003. However, the function of Med19 in the progression of hepatocellular carcinomas has not been investigated.
To study the role of Med19 in HCC cells, Med19 was silenced in HepG2 and Hep3B cells by small interfering RNA (siRNA), expressed by recombinant lentiviral vectors15. The effect of Med19 knockdown on cancer cell proliferation, cell-cycle arrest, and tumorigenesis was investigated. In addition, the expression of cell-cycle-related proteins in Med19-RNAi-Lentivirus vector-infected cells was analyzed to elucidate the possible mechanism of HCC cell proliferation.
Materials and methods
Lentiviral vectors encoding small interfering RNAs targeting Med19
The small interfering RNA (siRNA) sequences targeting the human Med19 gene (GenBank accession number NM_153450) were designed by Genechem Co, Ltd, Shanghai, China. Lentiviral vectors encoding Med19 siRNA and green fluorescent protein (GFP) (Genechem Co, Ltd, Shanghai, China) were constructed. After screening to validate potential siRNAs, the Med19 siRNA target sequence (5′-GGTGAAGGAGAAGCTAAGT-3′) was selected. The recombinant lentivirus particles encoding siRNA targeting Med19 (Med19-RNAi-Lentivirus) and the negative-control lentivirus (pGC-SIL-GFP) were packaged. To confirm the specificity of the RNAi experiment, Med19 gene expression was rescued with a cDNA containing silent third-codon point mutations in the targeted region, which made the cDNA resistant to silencing by the siRNA targeting Med19.
Cell culture and lentivirus infection
The human hepatocellular carcinoma cell lines HepG2 and Hep3B were purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco BRL, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Gibco BRL, Grand Island, NY, USA) at 37 °C in a humidified atmosphere of 5% CO2. HepG2 and Hep3B cells were seeded in 6-well plates (at a density of 5×104cells/well) and infected with lentivirus at an MOI (multiplicity of infection) of 20 when the cells reached 30% confluency. After 24 h of infection at 37 °C, the medium was replaced by fresh DMEM and incubated for another 48 h.
Real-time RT-PCR
Total RNA was extracted from naïve cells and lentivirus-infected cells with Trizol (Life Technologies, Inc, Grand Island, NY, USA) according to the manufacturer's instructions. The first-strand cDNA was synthesized using 2 μg of total RNA, 10 units of M-MLV reverse transcriptase (Promega, Madison, WI, USA), and 0.5 μg of oligo (dT) primer in a reaction volume of 20 μL.
Next, 1 μL of the cDNA reaction product was used for quantitative real-time PCR with SYBR Master Mixture (Takara, Otsu, Japan) on IQ5 (BioRad, Hercules, CA, USA), with reaction conditions consisting of an initial denaturation at 95 °C for 15 s, followed by 45 cycles at 95 °C for 5 s and 60 °C for 30 s. The PCR primer sequences for the Med19 target gene and the beta-actin internal control gene are listed in Table 1. The relative quantity of Med19 mRNA, normalized to beta-actin and relative to a calibrator, was calculated as 2−ΔΔCT, where ΔΔCT=(CTMed19 − CTbeta-actin) sample − (CTMed19 − CTbeta-actin) calibrator16.
Table 1. Base sequence of primers for quantitative real-time RT-PCR.
| Primers | Base sequence | |
|---|---|---|
| Med19 | Forward Reverse | 5′-TGACAGGCAGCACGAATC-3′ 5′-CAGGTCAGGCAGGAAGTTAC-3′ |
| Beta-actin | Forward Reverse | 5′-GGCGGCACCACCATGTACCCT-3′ 5′-AGGGGCCGGACTCGTCATACT-3′ |
Western blotting
Cells from each group were lysed on d 7 after infection by using pre-cooled lysis buffer. Next, 30 μg of lysates were loaded into each well and run on a 10% polyacrylamide gel at 30 mA for 2 h, followed by semidry transfer for a further 2 h. After being blocked with 5% non-fat dry milk for 1 h at room temperature, the membrane was probed with protein-specific antibodies overnight at 4 °C. Next, the membrane was washed with TBST three times and incubated for 2 h with the secondary antibody at room temperature. Following another round of washing with TBST, the membrane was developed using enhanced chemiluminescence (ECL) (Amersham Life Sciences, Amersham, UK). The primary antibodies for Med19 (Abcam, Cambridge, UK) and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were employed at dilutions of 1:200 and 1:6000, and the secondary antibodies were utilized at working concentrations of 1:5000 and 1:6000 for anti-goat IgG and anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), respectively.
Cell proliferation assay
Cell viability was assessed by methyl thiazol tetrazolium (MTT) assay, as in a previous report17, and the absorbance was measured at 570 nm (Sigma-Aldrich Corp, St Louis, MO, USA). Analysis of DNA synthesis, as measured by 5′-bromodeoxyuridine (BrdU, Sigma-Aldrich Corp, St Louis, MO, USA) incorporation, was performed according to a previous report18. Results of the BrdU assay were recorded at 490 nm at 24 and 48 h after the seeding of cells.
Colony-forming assay
Cells from the normal control, infected with Med19-RNAi-Lentivirus vector and negative-control lentivirus vector, were collected by trypsinization and suspended in complete growth medium, and 200 cells from each group were then seeded onto 6-well plates for each well. After being cultured at 37 °C for 2 weeks, visible colonies were detected with a fluorescence microscope. After fixation in paraformaldehyde, the colonies were stained with Giemsa for 10 min and photographed with a digital camera. Finally, the number of colonies formed by each group of cells was counted.
Cell-cycle assay and analysis of cell-cycle-related proteins
Hep3B cells were cultured on 6-well plates and infected with the Med19-RNAi-Lentivirus vector and the negative-control lentivirus vector. After reaching 90% confluency, cells were harvested for cell-cycle detection by propidium-iodide staining and were subsequently analyzed by flow cytometry.
The expression of the cell-cycle-related proteins CDKN1A (cyclin-dependent kinase inhibitor 1A, p21), CDC25A (cell division cycle 25A), CDC25B (M-phase inducer phosphatase 2), and CDC45L (cell division control protein 45 homolog) in Med19-RNAi-Lentivirus-infected Hep3B cells was detected by Western blotting analysis. The following antibodies were used: rabbit polyclonal to p21 (Abcam, Cambridge, UK; ab7960), rabbit polyclonal to CDC25A (Novus Biologicals, Littleton, CO, USA; NB100-213), rabbit polyclonal to CDC25B (Novus Biologicals, Littleton, CO, USA; NB100-91690), and mouse monoclonal to CDC45L (Abcam, Cambridge, UK; ab56476).
Tumor formation in nude mice
To assay tumor formation in nude mice, parental cells and Hep3B cells infected with the Med19-RNAi-Lentivirus vector and the negative-control lentivirus vector were trypsinized, counted, and resuspended in PBS. One hundred microliters of PBS containing 1×106 cells were then injected into 4- to 5-week-old female nude mice (n=8 per group). Tumor volumes were assessed every other day by measuring two perpendicular diameters with vernier calipers and was calculated using the formula (L×W2)/2, where L is the long diameter and W is the short diameter. The mice were sacrificed 30 d after injection and were examined for subcutaneous tumor growth.
Statistical analysis
The expression of Med19 was considered reduced if its relative mRNA level (2−ΔΔCT) was less than 1.0; otherwise, Med19 expression was determined to be preserved. When the 95% confidence interval (CI) of the sample did not overlap that of its calibrator, we considered the result statistically significant. When the P-value was less than 0.05, we considered the difference statistically significant.
Results
GFP expression of cells infected with recombinant lentivirus
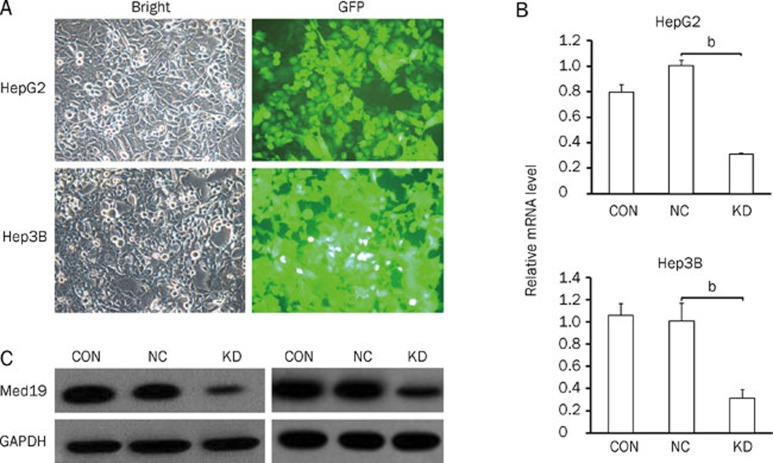
We constructed a lentiviral vector system to express siRNA targeting Med19 and to express GFP as a reporter gene. To determine whether the recombinant lentiviruses were able to infect human HCC cell lines, cells infected with the Med19-RNAi-Lentivirus (pKD) vector and the negative-control lentivirus (pNC) vector were observed under a fluorescence microscope. Seventy-two hours postinfection, more than 80% of the cells expressed GFP (Figure 1A), which indicated high-efficiency infection by the Med19-RNAi-Lentivirus vector.
Figure 1.
Determination of infection efficiency in the human hepatocellular carcinoma cell lines HepG2 and Hep3B. (A) Images recorded under a fluorescence microscope. Expression analyses of Med19 mRNA (B) and protein (C) levels by quantitative real-time RT-PCR and Western blot analysis in uninfected HepG2 and Hep3B cells (CON), cells infected with the pNC vector (NC), and cells infected with the pKD vector (KD). The beta-actin gene and the GAPDH protein are the internal controls for quantitative real-time RT-PCR and Western blot analysis, respectively. Significant difference from NC (bP<0.05).
Silencing of the Med19 gene by RNAi
To verify that the Med19 gene was silenced by the pKD vector's siRNA, we determined the mRNA levels in uninfected, pNC-infected, and pKD-infected cells through real-time RT-PCR. Cells infected with pKD exhibited significantly reduced levels of Med19 mRNA compared with pNC-infected cells and uninfected cells (P<0.05, Figure 1B).
Inhibition of Med19 protein levels by RNAi
After verifying the reduction in Med19 mRNA levels in pKD-infected cells, we detected Med19 protein levels in cell lysates using Western blot analysis with GAPDH as an internal control. Compared with that of uninfected and pNC-infected cells, the Med19 protein levels in cells infected with the pKD vector decreased greatly (Figure 1C).
Effects of Med19 knockdown on cell proliferation
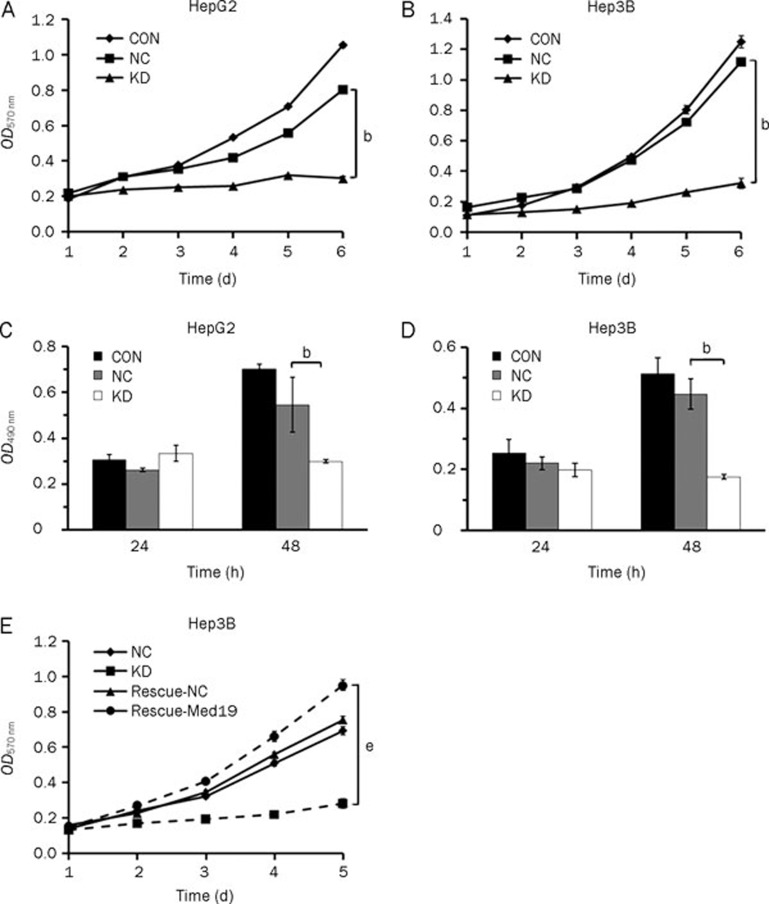
To investigate the effect of Med19 knockdown on cell proliferation, MTT and BrdU assays were performed. The growth curve obtained from the MTT assay indicated that the proliferative ability of pKD-infected cells was significantly decreased when compared with that of pNC-infected cells (P<0.05, Figure 2A and 2B). In the BrdU incorporation assay, the DNA synthesis capacity of pKD-infected HepG2 and Hep3B cells was significantly reduced (P<0.05 vs pKD infected, Figure 2C and 2D). The results of these experiments suggest that the depletion of endogenous Med19 expression in HCC cells correlates with impaired cell growth and DNA synthesis and demonstrate that Med19 is a key factor in cancer cell proliferation. More importantly, when Hep3B cells was treated with a vector encoding a Med19 wobble mutant (Rescue-Med19), which preserves the same amino acid sequence as pKD but contains three point mutations in the target nucleotide sequence, cell proliferation was substantially rescued (P<0.05 vs pKD infected, Figure 2E). The above results indicate that the knockdown of endogenous Med19 by RNAi inhibited the proliferation in HCC cells, and the rescued Med19 expression validated the specificity of the siRNA targeting.
Figure 2.
In vitro proliferation assays of HepG2 and Hep3B cells without infection (CON), cells infected with the negative control lentivirus vector (NC), and cells infected with the Med19-RNAi-Lentivirus vector (KD). Cell proliferation in the KD groups was significantly inhibited, as demonstrated by the MTT assay (A and B) and the BrdU incorporation assay (C and D). Significant difference from NC (bP<0.05). (E) The proliferation of Med19 siRNA-rescued cells (Rescue-KD) was enhanced significantly when compared with the KD group (eP<0.05), as demonstrated by the MTT assay.
Effect of Med19-RNAi on the colony-forming ability of HCC cells
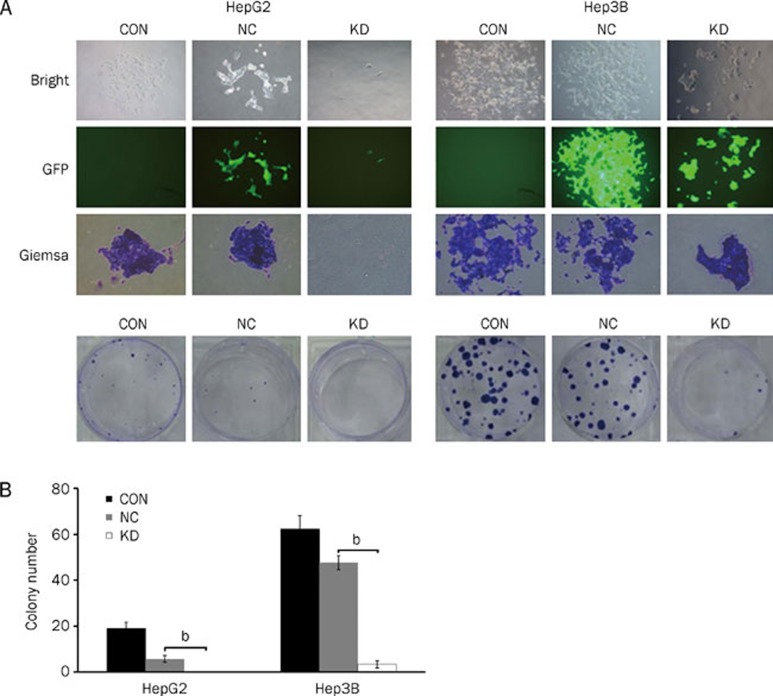
To detect whether Med19 has a significant effect on the colony-forming capacity of human HCC cells, a colony-forming experiment was performed on HepG2 and Hep3B cells. Our data indicated that the number and size of colonies formed from pKD-infected cells were strongly decreased when compared with those of uninfected and pNC-infected cells (Figure 3A and 3B), suggesting that the reduced expression of Med19 could significantly inhibit colony formation in human HCC cells.
Figure 3.
Med19-RNAi-Lentivirus infection inhibits the colony-forming ability of HepG2 and Hep3B cells. (A) Images recorded under a fluorescence microscope, representing the size and number of colonies in each group of cells; (B) Statistical analysis of the number of colonies with Giemsa staining. CON: cells without infection; NC: cells infected with the negative-control vector; KD: cells infected with the Med19-RNAi-Lentivirus vector. Significant difference from NC (bP<0.05).
Effect of Med19 knockdown on cell-cycle distribution
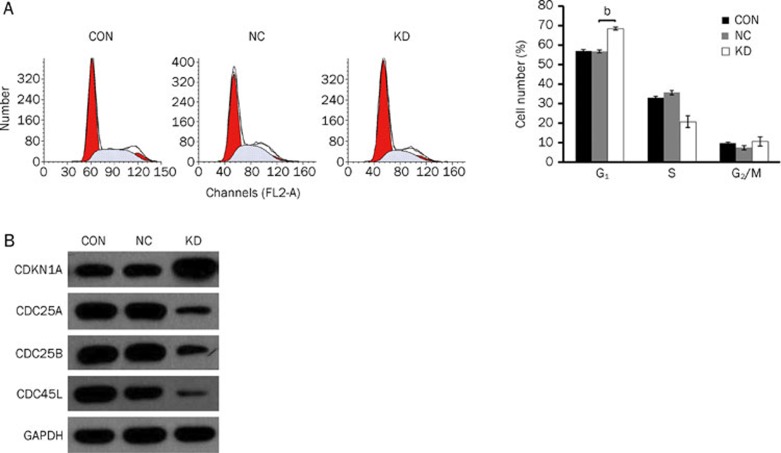
To elucidate whether the pKD-mediated knockdown of Med19 has any impact on the cell-cycle status of HCC cells, all three groups of Hep3B cells were subjected to a flow cytometry assay 7 d after infection. pKD-infected Hep3B cells exhibited a greater proportion of cells in the G0/G1 phase when compared with pNC-infected cells (P<0.05, Figure 4A). We next explored the expression of cell cycle-related proteins to investigate the possible role of Med19 in the control of mitotic cellular process. As shown in Figure 4B, the expression of CDKN1A was increased, whereas the expression of CDC25A, CDC25B, and CDC45L was diminished, in pKD-infected Hep3B cells.
Figure 4.
Med19-RNAi-Lentivirus infection induces cell-cycle arrest at G0/G1 phase and affects the expression of cell-cycle-related proteins in Hep3B cells. (A) Increased cell population in G0/G1 phase in the Med19-RNAi-Lentivirus-infected group (KD); (B) Increased expression of CDKN1A (cyclin-dependent kinase inhibitor 1A) and diminished expression of CDC45L (cell division control protein 45 homolog), CDC25A (cell division cycle 25A), and CDC25B (M-phase inducer phosphatase 2) in Med19-RNAi-Lentivirus-infected Hep3B cells. Significant difference from NC (bP<0.05).
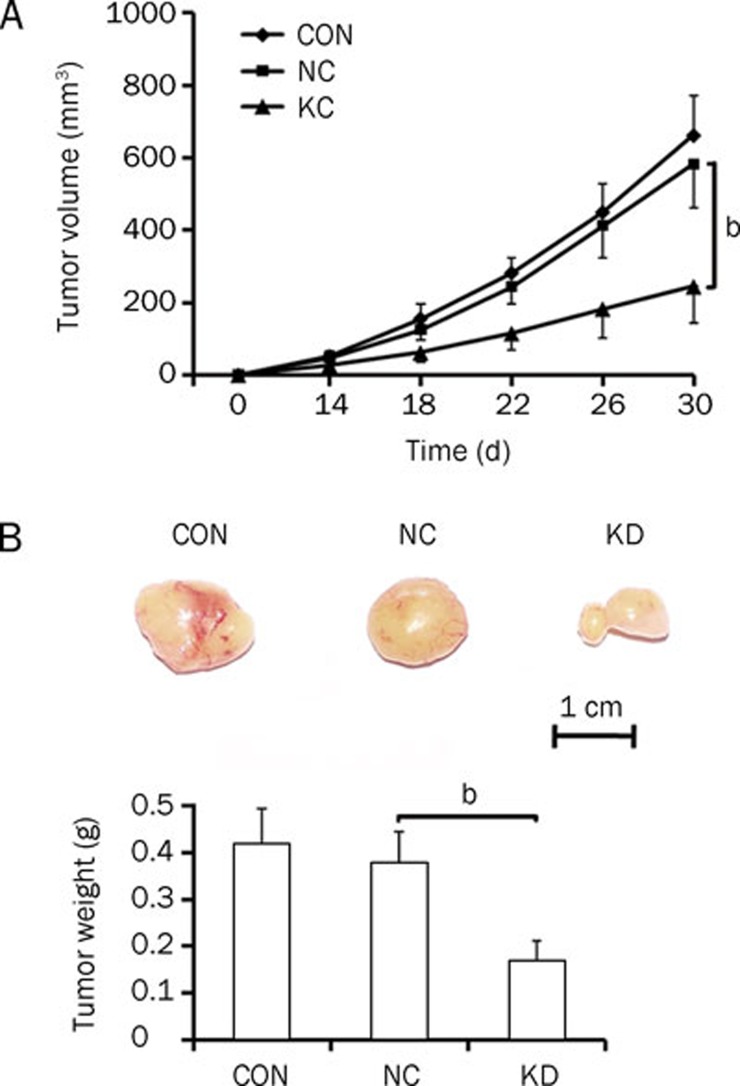
Effects of Med19 knockdown on tumorigenesis
Tumor formation in Med19-silenced Hep3B cells was then evaluated in vivo, and the results are shown in Figure 5. With regard to changes in tumor volume and tumor weight, no significant difference was found between the pNC-infected cells and uninfected cells. However, pKD infection had a significant inhibitory effect on tumor volume (P<0.05 vs pNC infected) and tumor weight (P<0.05 vs pNC infected). Representative pictures of tumors on d 30 can be seen in Figure 5B. The tumors in the uninfected and pNC-infected groups were large, whereas the tumors in the pKD-infected group exhibited a reduced tumorigenic capacity.
Figure 5.
In vitro tumorigenesis assay in mice inoculated with uninfected Hep3B cells (CON), cells infected with the negative-control lentivirus vector (NC), and cells infected with the Med19-RNAi-Lentivirus vector (KD). (A) Tumor volume growth curve. (B) Picture of excised tumor in each group and quantitative measurement of tumor weights. Significant difference from NC (bP<0.05).
Discussion
Conventional gene-suppression technologies, such as chemical inhibitors, antisense technology, ribozymes, and deoxyribozymes, often evoke nonspecific side effects and/or offer only transient and partial suppression of the gene of interest19, 20. Recently emerging as an important tool for silencing specific genes, RNAi is more potent in inhibiting gene expression than other methods and carries less risk of toxicity21, 22, 23. More importantly, with the effectiveness and relative ease with which dsRNA can be introduced into cells and tissues, RNAi has a significant therapeutic potential for targeting human carcinomas. Therefore, RNAi-mediated gene silencing, as a novel tool to arrest tumor growth and kill cancer cells, may open new avenues in cancer therapy24, 25, 26, 27.
Malignant tumor cells have the ability to migrate into surrounding tissue and/or distal organs — a phenomenon known as metastasis. In the complex and multi-step process of metastasis, cancer cells first detach from the primary tumor position, then invade adjacent lymph nodes and tissue, enter the circulatory system, migrate through the vasculature, and, finally, proliferate from microscopic growths into macroscopic secondary tumors in a distant tissue7. To date, several studies have revealed that many genes, such as oncogenes and tumor-suppressor genes, could be involved in complex metastatic processes, including tumor cell proliferation. For example, a few transcription factors that program many biological cell changes are required for executing the initial steps of the tumor invasion-metastasis cascade28, 29, 30, 31, 32, 33. Med19 is a component of the Mediator complex, which is a coactivator for DNA-binding factors that activate transcription via RNA polymerase II9. Med19 is also known as “lung cancer metastasis-related protein 1” (LCMR1) in the human genome. However, there is no report on the effect of Med19 on proliferation and tumorigenesis in HCC.
In our study, lentivirus-mediated RNAi was used to silence Med19 in the human HCC cell lines HepG2 and Hep3B. The siRNA targeting Med19, expressed from the recombinant lentivirus, induced efficient and specific inhibition of endogenous Med19 mRNA and protein expression in both cell lines. Simultaneously, inhibition of Med19 expression led to decreased colony formation and tumorigenic capacity in these cells. Thus, this study is to observe and confirm a crucial role of Med19 in the progression of HCC, showing that the Med19 gene may act as an oncogene to promote HCC development.
Moreover, our results indicated that the downregulation of Med19 expression significantly inhibits cell proliferation and induces G1-phase delay in HCC cells, indicating that Med19 is a proliferation regulator in HCC and may be involved in the control of mitotic cellular processes. Next, the detection of cycle-related proteins was performed on Med19-silenced Hep3B cells. As shown in Figure 4B, the expression of CDKN1A was increased, whereas the expression levels of CDC25A, CDC25B, and CDC45L were diminished in Med19-RNAi-Lentivirus vector-infected Hep3B cells. The cyclin-dependant kinase inhibitors are well known to prevent cyclin D/cdk4, 6, and cyclin E/cdk2-mediated phosphorylation of the retinoblastoma protein (Rb) and to block progression of the cell cycle in the G1 phase. However, CDC25A is required for progression from the G1 to the S phase of the cell cycle, whereas CDC25B activates the cyclin-dependent kinase CDC2 and is required for entry into mitosis. In addition, CDC45L is required for the initiation of chromosomal DNA replication (which occurs during the S phase). Taken as a whole, the enhanced CDKN1A expression and suppressed CDC25A, CDC25B, and CDC45L expression in Med19-silenced HCC cells contributed to arrested cell cycling at G0/G1 phase. These data support the hypothesis that Med19 acts as an oncogene to promote human hepatocellular carcinoma growth by regulating cell-cycle progression. However, further experiments are needed to elucidate the detailed mechanism of cell proliferation.
In conclusion, knockdown of human Med19 gene expression with RNAi successfully reduced tumor-cell proliferation and suppressed tumor formation in HepG2 and Hep3B cells in vitro and in vivo. These results provide new evidence for the involvement of Med19 in carcinogenesis and the development of HCC and suggest that RNAi-directed Med19 silencing may be a potent therapeutic tool for the treatment of HCC, especially in inhibiting or preventing cancer cell proliferation and tumorigenesis.
Author contribution
Shao-wu ZOU, Kai-xing AI, and Qi ZHENG conceived and designed the experiments; Shao-wu ZOU, Kai-xing AI, Zhi-gang WANG, and Zhou YUAN performed the experiments; Shao-wu ZOU, Kai-xing AI, and Qi ZHENG analyzed the data; Zhou YUAN and Jun YAN contributed reagents, materials, and analytical tools; and Shao-wu ZOU and Qi ZHENG wrote the manuscript.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (No 30872512).
References
- Okuda K. Natural history of hepatocellular carcinoma including fibrolamellar and hepato-cholangio carcinoma variants. J Gastroenterol Hepatol. 2002;17:401–5. doi: 10.1046/j.1440-1746.2002.02734.x. [DOI] [PubMed] [Google Scholar]
- Bosch FX, Ribes J, Cleries R, Diaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191–211. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma: Recent trends in the United States. Gastroenterology. 2004;127:S27–34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- McKillop IH, Schrum LW. Alcohol and liver cancer. Alcohol. 2005;35:195–203. doi: 10.1016/j.alcohol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Farazi PA, De Pinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–87. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nature Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Westhof E, Filipowicz W. From RNAi to epigenomes: how RNA rules the world. Chembiochem. 2005;6:441–3. doi: 10.1002/cbic.200400418. [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–91. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–9. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–5. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Casamassimi A, Napoli C. Mediator complexs and eukaryotic transcription regulation: An overview. Biochimie. 2007;89:1439–46. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–49. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- Baidoobonso SM, Guidi BW, Myers LC. Med19 (Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae mediator complex. J Biol Chem. 2007;282:5551–9. doi: 10.1074/jbc.M609484200. [DOI] [PubMed] [Google Scholar]
- Naldini L, Gallay P, Gallay P. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang Z, Wang J, Li J, Wang H, Yue W. Lentivirus-mediated knockdown of cyclin Y (CCNY) inhibits glioma cell proliferation. Oncol Res. 2010;18:359–64. doi: 10.3727/096504010x12644422320582. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK+ yeast strains. Nucleic Acids Res. 2001;29:1433–42. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–9. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–10. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Schweinitz A, Steinmetzer T, Banke IJ, Arlt MJ, Stürzebecher A, Schuster O, et al. Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents. J Biol Chem. 2004;279:33613–22. doi: 10.1074/jbc.M314151200. [DOI] [PubMed] [Google Scholar]
- D'Alessio S, Margheri F, Pucci M, Del Rosso A, Monia BP, Bologna M, et al. Antisense oligodeoxynucleotides for urokinase plasminogen activator receptor have anti-invasive and anti-proliferative effects in vitro and inhibit spontaneous metastases of human melanoma in mice. Int J Cancer. 2004;110:125–33. doi: 10.1002/ijc.20077. [DOI] [PubMed] [Google Scholar]
- Arens N, Gandhari M, Bleyl U, Hildenbrand R. In vitro suppression of urokinase plasminogen activator in breast cancer cells — a comparison of two antisense strategies. Int J Oncol. 2005;26:113–9. [PubMed] [Google Scholar]
- Woessmann W, Damm-Welk C, Fuchs U, Borkhardt A. RNA interference: new mechanisms for targeted treatment. Rev Clin Exp Hematol. 2003;7:270–91. [PubMed] [Google Scholar]
- Jiang M, Rubbi CP, Milner J. Gel-based application of siRNA to human epithelial cancer cells induces RNAi-dependent apoptosis. Oligonucleotides. 2004;14:239–48. doi: 10.1089/oli.2004.14.239. [DOI] [PubMed] [Google Scholar]
- Rye PD, Stigbrand T. Interfering with cancer: a brief outline of advances in RNA interference in oncology. Tumour Biol. 2004;25:329–36. doi: 10.1159/000081403. [DOI] [PubMed] [Google Scholar]
- Wilda M, Fuchs U, Wossmann W, Borkhardt A. Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi) Oncogene. 2002;21:5716–24. doi: 10.1038/sj.onc.1205653. [DOI] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc Natl Acad Sci USA. 2006;103:18969–74. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–74. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]