Abstract
Macrophages are cellular targets for infection by bacteria and viruses. The fate of infected macrophages plays a key role in determining the outcome of the host immune response. Apoptotic cell death of macrophages is considered to be a protective host defense that eliminates pathogens and infected cells. In this study, we investigated the involvement of Notch signaling in regulating apoptosis in macrophages treated with tuberculin purified protein derivative (PPD). Murine bone marrow-derived macrophages (BMMs) treated with PPD or infected with Mycobacterium bovis Bacillus Calmette-Guérin (BCG) induced upregulation of Notch1. This upregulation correlated well with the upregulation of the anti-apoptotic gene mcl-1 both at the transcriptional and translational levels. Decreased levels of Notch1 and Mcl-1 were observed in BMM treated with PPD when a gamma secretase inhibitor (GSI), which inhibits the processing of Notch receptors, was used. Moreover, silencing Notch1 in the macrophage-like cell line RAW264.7 decreased Mcl-1 protein expression, suggesting that Notch1 is critical for Mcl-1 expression in macrophages. A significant increase in apoptotic cells was observed upon treatment of BMM with PPD in the presence of GSI compared to the vehicle-control treated cells. Finally, analysis of the mcl-1 promoter in humans and mice revealed a conserved potential CSL/RBP-Jκ binding site. The association of Notch1 with the mcl-1 promoter was confirmed by chromatin immunoprecipitation. Taken together, these results indicate that Notch1 inhibits apoptosis of macrophages stimulated with PPD by directly controlling the mcl-1 promoter.
Keywords: apoptosis, BCG, macrophage, Mcl-1, Notch signaling, PPD
Introduction
Macrophages play critical roles in the first line of host defense as well as in the elimination of pathogens. They are, however, the main targets for infection by various pathogens, including Mycobacterium tuberculosis (MTB), which is the causative agent of tuberculosis. MTB infects macrophages and escapes macrophage-mediated elimination by various mechanisms.1 Apoptosis of macrophages is one of the defense mechanisms of the host that eliminates both the infected cells and the bacteria residing inside these cells.2,3 Furthermore, apoptotic macrophages are critical for inducing adaptive immune responses, as apoptotic vesicles containing mycobacteria are phagocytized by nearby antigen-presenting cells and presented to CD8+ T cells.4,5 Apoptosis of macrophages in this setting is partially tumor-necrosis factor-dependent.6 Pathogenic MTB is reported to combat apoptosis through various mechanisms, including switching the cell death mode to necrosis and inactivating tumor-necrosis factor signaling.7,8
Myeloid cell leukemia sequence-1 (Mcl-1) is an anti-apoptotic member of the B-cell lymphoma 2 (Bcl-2) family that contains three putative BH domains that participate in protein-protein interactions between members of the protein family.9 Mcl-1 is highly expressed in hematopoietic stem cells and myeloid lineage cells and helps sustain cell viability.10 In macrophages, Mcl-1 is essential for survival, and its expression is mediated through the STAT3 and Akt-1 pathways.11 Expression of Mcl-1 is controlled at the transcriptional, post-transcriptional and post-translational levels. At the transcriptional level, numerous extracellular stimuli, including IL-3, IL-6, IL-15 and growth factors such as vascular endothelial growth factor and epidermal growth factor, are reported to induce mcl-1 expression.12,13 These extracellular stimuli trigger the activation of the JAK/STAT, MAPK and PI3K/AKT pathways. These pathways induce the transcription of mcl-1 mRNA.11,14,15 The promoter region of the mcl-1 gene contains multiple putative transcription factor-binding sites, including those for cAMP response element-binding protein, PU.1, STAT3 and HIF1α.14,15,16 Mcl-1 promotes cell survival by suppressing cytochrome c release from the mitochondria via heterodimerization with the pro-apoptotic Bcl-2 family members Bak and Bax.9,17 During pathogenic infection, Mcl-1 plays an important role in the apoptosis of infected cells. For example, survival of macrophages is regulated by Mcl-1, which promotes the transition from resistance to susceptibility and finally to apoptosis during pneumococcal infection.18 In mycobacterial infection, infected macrophages delay apoptosis by upregulating Mcl-1 and anti-apoptotic proteins that are similar to Mcl-1, including Bfl-1/A1, which promotes mycobacterial survival and chronic intracellular persistence.19,20 Furthermore, a highly virulent clinical isolate of MTB significantly upregulates Mcl-1 and Bfl-1 in the human monocyte cell line THP-1.21
Previously, we and others have reported that Notch signaling is activated in Toll-like receptor (TLR)-stimulated macrophages.22,23 Activation of Notch signaling regulates pro-inflammatory responses.22,24 Furthermore, it has been reported that in macrophages infected with Bacillus Calmette-Guérin (BCG), Notch1 is upregulated and SOCS3 expression is induced.25 Mycobacterial infection also induces the expression of dll4, a Notch ligand, via TLR9 and promotes TH17 activity during granuloma formation.26 Notch signaling regulates apoptosis in various types of cells, and aberrations in Notch signaling have been linked to cancer formation.27 Recently, it was reported that GSI treatment induces apoptosis of chronic lymphocytic leukemia cells while the accumulation of Mcl-1 was observed, suggesting that Mcl-1 accumulation is not sufficient for protecting cells from apoptosis.28 The involvement of Notch signaling in regulating apoptosis in macrophages, however, has not been determined.
In this study, we investigated the roles of Notch signaling in regulating Mcl-1 expression in macrophages that were treated with purified protein derivative (PPD) or infected with BCG. We also studied the effects of inhibition of Notch signaling on Mcl-1 expression and apoptosis in macrophages. Finally, we present evidence linking Notch signaling and mcl-1 gene expression at the transcriptional level.
Materials and methods
Bone marrow-derived macrophage (BMM) and macrophage cell line
BMM derived from female C57BL/6 mice (National Laboratory Animal Center, Mahidol University, Salaya, Thailand) or mice with a MyD88−/− targeted deletion (The Max Planck Institute for Infection Biology, Berlin, Germany) were prepared as described previously.22 All procedures involving laboratory animals were conducted according to the guidelines issued by Chulalongkorn University. The murine macrophage-like cell line RAW264.7 was obtained from Cell Lines Service (Eppelheim, Germany). BMM and RAW264.7 cells were maintained in DMEM (HyClone, USA) supplemented with 10% FBS (v/v) (HyClone), 100 U/ml penicillin (General Drug House, Bangkok, Thailand), 0.4 mg/ml streptomycin (M&H Manufacturing, Samutprakarn, Thailand), 1% sodium pyruvate (Hyclone, Logan, UT, USA) and 1% HEPES (Hyclone, Logan, UT, USA). Expression of F4/80 and CD11b were used as markers to confirm that the BMM were macrophages. Human monocytic leukemia cell line (ATCC TIB-202) was maintained in RPMI1640 (Hyclone, Logan, UT, USA) supplemented with reagents similar to those of DMEM described above.
Reagents
Gamma secretase inhibitor (GSI), IL-CHO (a kind gift of Professor Todd E Golde, University of Florida College of Medicine, University of Florida College of Medicine, Gainesville, FL, USA), were dissolved in dimethyl sulfoxide (DMSO) and used as inhibitors of Notch signaling as described previously.22 PPD was a kind gift from Mrs Thipchuta Bharnthong (Queen Saovabha Memorial Institute, Bangkok, Thailand). To investigate the effects of IL-CHO, cells were pre-treated with the indicated concentration of IL-CHO or vehicle control (DMSO) for 30 min prior to PPD treatment or infection.
Infection of BMM with M. bovis BCG
M. bovis BCG (Copenhagen strain) was grown to a log phase in Middlebrook 7H9 broth (Difco, DifcoTM, BD Biosciences, San Jose, CA, USA) supplemented with 10% albumin-dextrose-catalase (Difco) and 0.05% Tween 80 (Research Organics, Cleveland, OH, USA) at 37 °C under static conditions. Bacteria were harvested and resuspended in DMEM without antibiotics. To eliminate clumping, the bacteria were passed through a 25-gauge needle 10 times before infection. For the infection of BMM, BMM were seeded in 12-well plates and allowed to adhere overnight. The infection was carried out at a multiplicity of infection (MOI) of 1∶10 for 4 h, and the extracellular bacteria were removed by washing one time with phosphate-buffered solution. Cells were incubated in DMEM with antibiotics as indicated.
Silencing of Notch1 in RAW264.7 cell line
To silence the expression of Notch1, the plasmid pShNotch1 and the control plasmid pShLuc were constructed from pBAsi-mU6 Neo (TaKaRa Bio Inc., Shiga, Japan) and transfected into RAW264.7 cells. The hairpin loop sequences targeted nucleotides at positions 1529–1547 of murine Notch1 (accession no. NM_008714.2). To construct the plasmid targeting Notch1, the sense strand (5′-GATCCGGTGTATACTGTGAAATCAGTGTGCTGTCCTGATTTCACAGT ATACACCTTTTTTA-3′) and the antisense strand (5′-AGCTTAAAAAAGGTGTATACTG TGAAATCAGGACAGC ACACTGATTTCACAGTATACACCG-3′) were synthesized. To construct the control plasmid targeting luciferase, the sense strand (5′-GATCCAGCAATAGT TCACG CTGAAGTGTGCTGTCCTTCAGCGTGAACTATTGCTTTTTTTA-3′) and the antisense strand (5′-AGCTTAAAAAAAGCAATAGTTCACGC1TGAAGGACAGCACAC TTCAGCGTGAACATTGCTG-3′). The underlined sequences indicated the hairpin loop regions. Each pair of oligonucleotides were annealed as recommended by the manufacturer. The plasmid and the annealed oligonucleotides were digested by BamHI and HindIII before ligation using T4 DNA ligase (New England Biolab, Ipswich, MA, USA). All plasmids were prepared using the Endo-free Plasmid Maxi kit (Qiagen, Hilden, Germany). Transient transfections were performed using the FuGeneHD transfection reagent (Roche, Germany) according to the manufacturer's protocol. The extent of silencing was confirmed by either quantitative real-time PCR (qPCR) or western blot. To verify that non-overlapping targeting Notch1 for silencing yields similar result, synthesized siRNA for Notch1 or the scramble siRNA as a control (Qiagen) was used to transfect RAW264.7 cells using SuperFect Transfection Reagents (Qiagen) according to the manufacturer's instruction.
Retroviral transduction
The retroviral plasmid vector for the expression of dominant negative Mastermind-like protein (MSCV–Mam (12–74)–EGFP) was a kind gift from Dr Warren Pear (University of Pennsylvania, Philadelphia, PA, USA) and the vector for the expression of truncated Notch1, corresponding to amino-acid residues 1759–2556 of human Notch1 (MSCV–GFP–Myc–Nic) was a kind gift from Dr Barbara Osborne (University of Massachusetts at Amherst, Amherst, MA, USA), respectively. A control empty vector, MSCV–IRES–GFP (Addgene plasmid 20672), was obtained from Addgene (Cambridge, MA, USA). The retroviral vectors and packaging construct pCL-Ampho (Imgenex, San Diego, CA, USA) were co-transfected into 293T cells using FuGene HD transfection reagent (Roche, Mannheim, Germany), according to the manufacturer's instructions. Culture supernatant containing retroviruses were harvested twice, at 48 and 72 h after transfection, and used to transduce THP-1. The transduced cells were sorted using GFP as a marker to obtain more than 90% GFP+ cells using BD FACSAria (BD Biosciences, San Jose, CA, USA).
qPCR
Cells were treated as indicated and total RNA was isolated using the TriZol reagent (Invitrogen Life Technologies, Grand Island, NY, USA) according to the manufacturer's protocol. RNA was converted to cDNA using random hexamers (Qiagen) and RevertAid Reverse Transcriptase (Fermentas, Glen Burnie, MD, USA). qPCR for mcl-1 (accession no. NM_008562.3) was carried out using the mcl-1 forward primer (5′-GACCGGCTCCAAGGACTC-3′) and reverse primer (5′-TGTCCAGT TTCCGGAGCAT-3′). β-actin (accession no. NM_007393) (forward primer 5′-ACCAACTGGG ACGACATGGAGAA-3′ and reverse primer 5′-GTGGTGGTGAAGCTGTAGCC-3′) was used as a reference gene. The reaction was carried out using the 1×Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) in an MJ Mini personal thermal cycler (BioRad, Richmond, VA, USA). The relative expression levels were calculated by 2−ΔΔCTand analyzed.29
Western blot
Cells were treated as indicated and washed twice in cold phosphate-buffered solution. To lyse the cells, cold buffer A (1 mM EGTA, 1 mM DTT, 50 mM Tris-HCl (pH 7.2), 0.14 M KCl, 2.5 mM MgCl2 and protease inhibitor cocktails) was added directly to the cells to rinse away the remaining media. This was followed by the addition of buffer B (Buffer A, 1% nonidet P-40) to lyse the cells. The lysate was collected and centrifuged at 5000 r.p.m. at 4 °C for 5 min. The clear supernatant was used for western blot analysis. Proteins were quantified using the BCA protein assay kit (Pierce, Rockford, IL, USA). The primary antibodies used in this study are as follows: rabbit anti-Notch 1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-Mcl-1 (Santa Cruz Biotechnology), rabbit anti-cleaved Notch1 (Val1744) (Cell Signaling Technology, Beverly, MA, USA), mouse anti-GAPDH (Santa Cruz Biotechnology) and mouse anti-β-Actin (Chemicon International, Temecula, CA, USA). Donkey anti-rabbit IgG or sheep anti-mouse IgG conjugated with horseradish peroxidase (both from GE Healthcare Lifescience, Piscataway Township, NJ, USA) were used as secondary antibodies. The signal was detected by chemiluminescence using Hyperfilm ECL X-ray film (GE Healthcare).
Apoptotic assay
To detect apoptosis, cells were treated as indicated and harvested by gentle scraping in cold phosphate-buffered solution using a rubber policeman. The cells were stained for AnnexinV and propidium iodide using the Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit (Invitrogen, Paisley, UK) according to the manufacturer's protocol. Cells were immediately analyzed by flow cytometry using the FC500 (Beckman Coulter, Indianapolis, IN, USA). The acquired data were analyzed using CXP software (Beckman Coulter). To analyze apoptosis in BMM infected with BCG, the cells were infected with BCG at an MOI of 10 for 5 days, and apoptotic cells were detected using Cell Death Detection ELISA (Roche, Mannheim, Germany) according to the manufacturer's protocol. The specific enrichment of mono- and oligonucleosomes released into the cytoplasm was calculated as the enrichment factor.
Chromatin immunoprecipitation (ChIP) assay
BMMs (1×107 cells/plate) were activated by PPD (5 µg/ml) for 24 h, and ChIP using the rabbit anti-Notch1 antibody (Santa Cruz Biotechnology) was carried out using the EZ Magna ChIP Chromatin Immunoprecipitation Kit (Upstate, Upstate Millipore, Billerica, MA, USA) according to the manufacturer's protocol and as described previously.30 The region with the potential CSL binding site in the murine mcl-1 promoter was detected by PCR using the following primer set: forward primer 5′-CCGGGCTGAGAGTTGTACC-3′ and reverse primer 5′-CGGAAGTCAGGAGTGAGG AA-3′. The PCR reactions were carried out using the Bioer Life Express (Bioer, Bioer Technology, Hangzhou, P.R. China), and the PCR product was visualized on 2% agarose gel.
Statistic analysis
To calculate the significant differences between samples, Student's paired independent t-test was used (SPSS 15.0). Differences with P<0.05 were considered statistically significant.
Results
BCG infection and PPD treatment induces Notch1 and Mcl-1 expression in macrophages
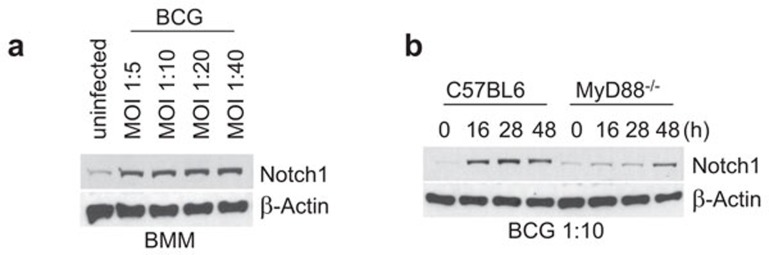
Because it has been previously reported that stimulation of macrophages upregulates Notch receptors, we first investigated whether BCG infection of macrophages induces an upregulation of Notch1. BCG infection with MOIs ranging from 1∶5 to 1∶40 induced the upregulation of Notch1. This upregulation could be readily detected 6 h post infection (Figure 1a). However, the expression levels of Notch1 did not surpass baseline levels in MyD88−/− macrophages at various times post infection (Figure 1b). Therefore, upregulation of Notch1 during BCG infection is dependent on MyD88, which is an adaptor molecule for TLR signaling. Furthermore, this result indicates that macrophages recognize BCG using TLRs in a MyD88-dependent manner. Similar upregulation of Notch1 was observed when macrophages were treated with PPD. This upregulation can be observed as early as 1 h post treatment (Figure 2a).
Figure 1.
Infection with BCG induces Notch1 upregulation in BMM. (a) BMM from C57/BL6 mice was infected with BCG at various MOIs for 28 h, and the expression of Notch1 was detected by western blot. (b) BMM from C57/BL6 mice or MyD88−/− mice were infected with BCG at an MOI of 1∶10, and the cell lysate samples that were collected at different time points were analyzed by western blot. β-actin was used as a loading control. BCG, Bacillus Calmette-Guérin; BMM, bone marrow-derived macrophage; MOI, multiplicity of infection.
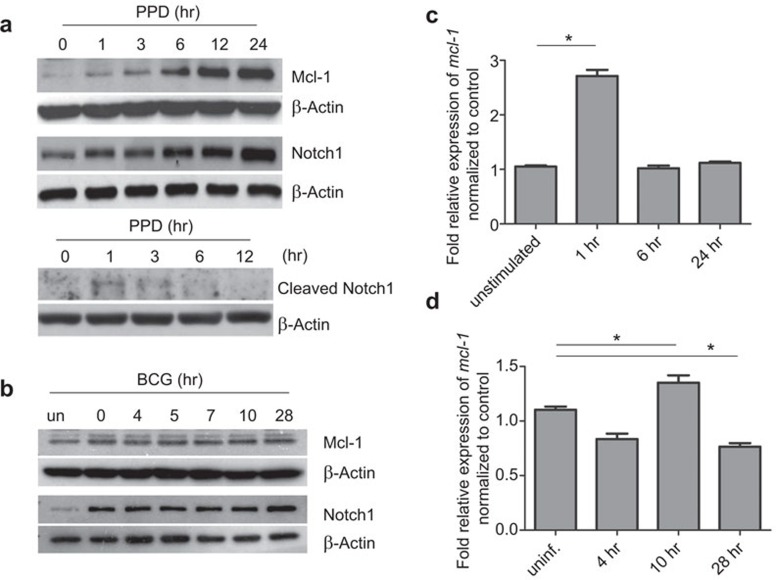
Figure 2.
PPD treatment and BCG infection induces Mcl-1 expression in BMM. (a) BMM were treated with PPD (5 µg/ml) for the indicated times, and the expression of Mcl-1, Notch1 and cleaved Notch1 (Val1744) were detected by western blot. (b) BMM were infected with BCG (MOI 1∶10) for the indicated times, and the expression of Mcl-1 and Notch1 were detected by western blot. (c, d) BMM were treated with PPD (c) or infected with BCG (d) for the indicated times, and the levels of mcl-1 mRNA were detected by qPCR. Cells without any stimulation was labeled as ‘unstimulated' and cells without BCG infection was labeled ‘uninf'. Cells The results represent two independent experiments carried out in triplicate. * indicates where statistical significance (P<0.05) was observed between the levels in uninfected cells and cells infected for 10 and 28 h, respectively. BCG, Bacillus Calmette-Guérin; BMM, bone marrow-derived macrophage; Mcl-1, myeloid cell leukemia sequence-1; MOI, multiplicity of infection; PPD, purified protein derivative; qPCR, quantitative real-time PCR.
In an attempt to identify the target genes of Notch signaling in activated macrophages, a genome wide chromatin immunoprecipitation assay using the anti-Notch1 antibody followed by microarray analysis was conducted, and this experiment yielded various potential target genes of Notch signaling, including mcl-1 (manuscript in preparation). Therefore, we investigated whether the levels of Mcl-1 correlated with the levels of Notch1 in PPD-activated or BCG-infected macrophages. As shown in Figure 2a and b, PPD treatment led to a significant increase in Mcl-1 expression levels. This increase correlated with Notch1 expression levels and the appearance of cleaved Notch1. Mcl-1 expression in BCG-infected macrophages increased slightly compared to uninfected cells, but the levels remained almost constant throughout the time course of the infection. There was no detectable change in the protein levels of Notch1 and Mcl-1 at each time point studied in untreated/unstimulated cells (data not shown). To determine whether this increase in Mcl-1 protein correlated with mcl-1 mRNA, qPCR was performed. As shown in Figure 2c, PPD treatment significantly upregulated mcl-1 mRNA 1 h post treatment. After 1 h, the mRNA levels decreased. In BCG-infected cells, the kinetics of mcl-1 mRNA expression differed from that in PPD-treated cells. The mRNA expression levels in BCG-infected macrophages increased at 10 h post infection but then dropped down to basal levels at 28 h post infection (Figure 2d). Taken together, mcl-1 mRNA and protein expression increases after BCG infection and PPD treatment, but this induction is immediately followed by downregulation.
Effects of Notch signaling inhibition on the levels of Notch1 and Mcl-1
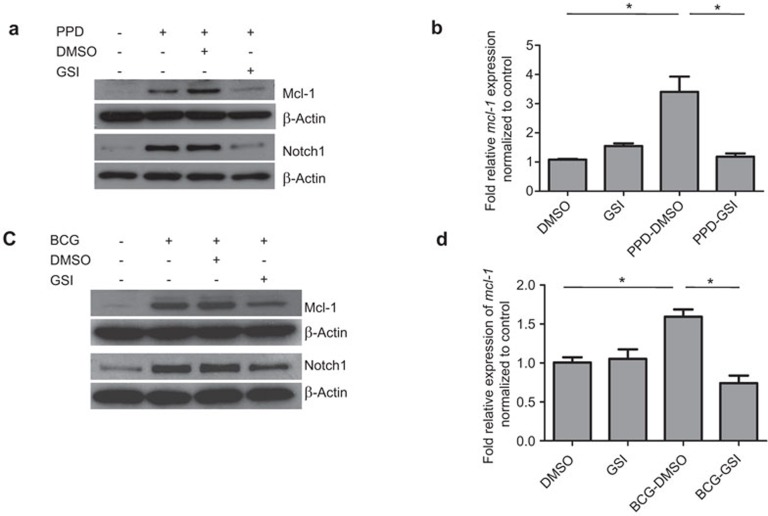
To address whether Notch signaling is involved in the regulation of Mcl-1 expression, we used GSI to inhibit Notch signaling in activated or infected macrophages and investigated whether this treatment affected the levels of Notch1, Mcl-1 protein and mRNA. As shown in Figure 3a and c, treatment with GSI (25 µM) significantly diminished the upregulation of Notch1 in PPD-treated and BCG-infected macrophages compared to the vehicle control. This result is consistent with previously reported results.22,31 This defect in mRNA upregulation correlates with a decrease in mcl-1 mRNA expression (Figure 3b and d).
Figure 3.
GSI treatment decreases Mcl-1 expression in BMM treated with PPD or infected with BCG. (a, b) BMM were pre-treated with GSI (25 µM) or a vehicle control (DMSO) for 1 h prior to stimulation with PPD (5 µg/ml). The expression of Mcl-1 and Notch1 were detected by western blot at 24 h post PPD treatment (a). The levels of mcl-1 mRNA were measured by q-PCR 1 h post PPD treatment (b). The results represent two independent experiments carried out in triplicate. * indicates where statistical significance (P<0.05) is observed. (c, b) BMM were pre-treated with GSI (25 µM) or a vehicle control (DMSO) for 1 h prior to infection with BCG (MOI 1∶10). The expression of Mcl-1 and Notch1 was detected by western blot 24 h post PPD treatment (c). The levels of mcl-1 mRNA were measured by q-PCR 10 h post infection (d). The results represent two independent experiments carried out in triplicate. * indicates where statistical significance (P<0.05) is observed. BCG, Bacillus Calmette-Guérin; BMM, bone marrow-derived macrophage; DMSO, dimethyl sulfoxide; GSI, gamma secretase inhibitor; Mcl-1, myeloid cell leukemia sequence-1; MOI, multiplicity of infection; PPD, purified protein derivative; qPCR, quantitative real-time PCR.
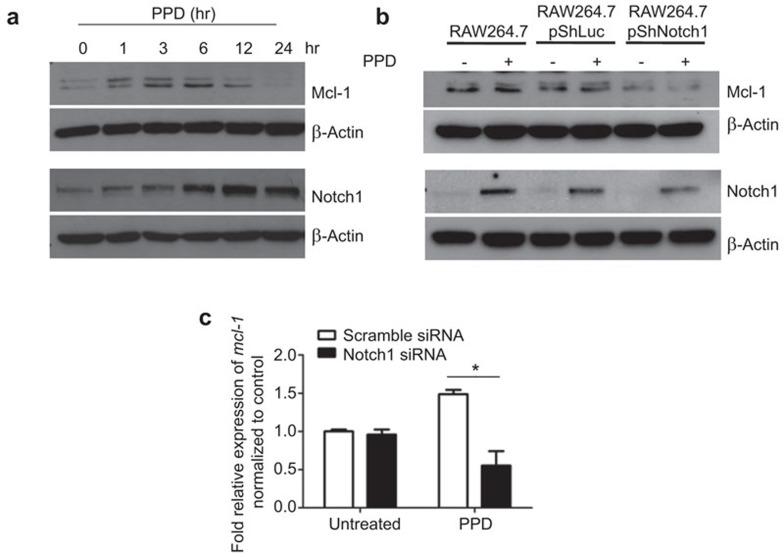
To investigate whether Notch1 regulates Mcl-1 expression, we silenced Notch1 expression in RAW264.7 cells and determined the effect on Mcl-1 expression. We first confirmed that Notch1 and Mcl-1 were upregulated in response to PPD treatment, which is similar to what was observed in BMM (Figure 4a). The kinetics of Mcl-1 expression in RAW264.7 cells differed from those in BMM; PPD treatment induced Mcl-1for a shorter duration, and the levels decreased to basal levels by 24 h. In contrast, the levels of Notch1 remained high. When Notch1 expression was silenced, the level of Mcl-1 remained at basal levels even after PPD treatment (Figure 4b). Using non-overlapping siRNA approach for Notch1 silencing in RAW264.7 cells yielded similar results (Supplementary Figure 1). To investigate whether silencing Notch1 affects the transcription of mcl-1, the relative expression of mcl-1 mRNA in RAW264.7 cells with Notch1 silencing was measured by qPCR upon PPD treatment. As shown in Figure 4c, silencing Notch1 significantly reduced the expression level of mcl-1. Silencing Notch1 in RAW264.7 cells did not affect cell viability up to 72 h after transfection (data not shown). Therefore, Notch1 is indispensable for Mcl-1 upregulation in response to PPD treatment.
Figure 4.
Silencing Notch1 in RAW264.7 decreases Mcl-1 expression. (a) RAW264.7 cells were treated with PPD (5 µg/ml) for the indicated times, and the expression of Mcl-1 and Notch1 were detected by western blot. (b) RAW264.7 cells were transiently transfected with the indicated plasmids for 36 h and then treated with PPD (5 µg/ml) for 12 h. Expression of Notch1 and Mcl-1 were detected by western blot. (c) RAW264.7 cells were transfected with indicated siRNA for 24 h. After the transfection, cells were treated with PPD (5 µg/ml) for 3 h. The levels of mcl-1 mRNA were measured by qPCR. * indicates where statistical significance (P<0.05) is observed. Mcl-1, myeloid cell leukemia sequence-1; PPD, purified protein derivative; qPCR, quantitative real-time PCR.
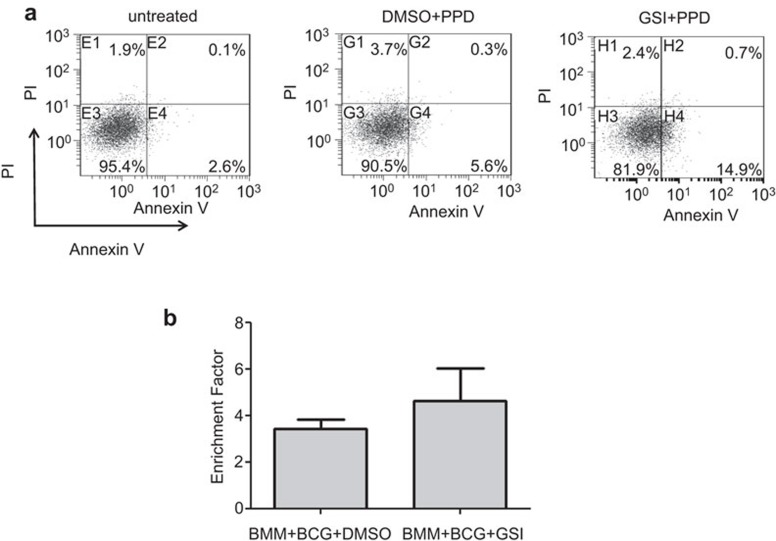
Mcl-1 functions as an anti-apoptotic protein and has been shown to regulate apoptosis in macrophages. We therefore investigated the effect of GSI treatment on apoptosis in PPD-treated BMM. As shown in Figure 5a, treatment of BMM with PPD slightly increased AnnexinV+ apoptotic cells. Pre-treating cells with GSI before PPD treatment significantly increased the percentage of apoptotic cells, suggesting that Notch signaling reduces apoptosis in PPD-treated BMM. We also determined if GSI treatment enhanced apoptosis in BCG-infected BMM, but no significant difference between DMSO-treated and GSI-treated cells was found (Figure 5b).
Figure 5.
GSI treatment enhances apoptosis of BMM. (a) BMM were pre-treated with GSI or DMSO for 1 h and then stimulated with PPD (5 µg/ml) for 18 h. The results represent two independent experiments. (b) BMM were pre-treated with GSI or DMSO for 1 h and infected with BCG (MOI 1∶10). On day 5 post infection, apoptotic cells were measured. The results represent two independent experiments performed in triplicate. BCG, Bacillus Calmette-Guérin; BMM, bone marrow-derived macrophage; DMSO, dimethyl sulfoxide; GSI, gamma secretase inhibitor; PPD, purified protein derivative.
The effect of activation of Notch signaling on the expression of Mcl-1 was investigated using human monocytic cell line, THP-1. Cells were transduced with the retroviral construct containing truncated intracellular Notch1, the form that bypasses the requirement of ligand binding for activation. As shown in Supplementary Figure 2, Mcl-1 is expressed at detectable level even in unstimulated THP-1 cells. Overexpression of activated form of Notch1 slightly increased the level of Mcl-1, but PPD stimulation did not increase the level of Mcl-1. In contrast, inhibiting Notch signaling using dominant negative Mastermind-like protein to suppress the transcriptional activity of Notch signaling,32 decreased Mcl-1 was observed. This result is consistent with the results obtained from murine cells (Supplementary Figure 2).
Direct association of Notch1 with the mcl-1 promoter
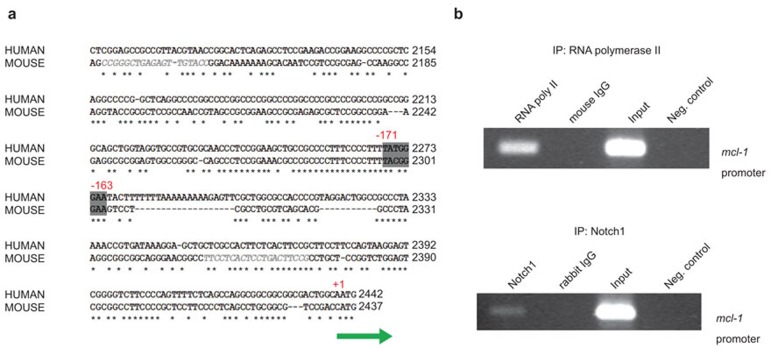
In a preliminary study using the ChIP-seq method, mcl-1 was identified as one of the genes regulated by Notch1 binding. Therefore, the promoter regions of human and murine mcl-1 were analyzed to identify potential CSL/RBP-Jk binding sites. As shown in Figure 6a, there is one region (5′-TATGGGAA-3′) which contains a well conserved consensus CSL/RBP-Jk binding site. To determine if Notch1 is associated with the identified region on the mcl-1 promoter, chromatin immunoprecipitation was performed on the genomic DNA from BMM using either an anti-Notch1 or anti-RNA polymerase II antibody. The mcl-1 promoter region containing the consensus binding site was then amplified by PCR. As shown in Figure 6b, the promoter region of mcl-1 was readily detected when the antibody against RNA polymerase II was used as a control. The PCR product corresponding to the mcl-1 promoter was also detected when the antibody against Notch1 was used. These results strongly suggest that Notch1 is associated with the mcl-1 promoter and may play an important role in the transcriptional regulation of mcl-1.
Figure 6.
Direct association of Notch1 with mcl-1 promoter. (a) The consensus sequence of a potential CSL/RBP-Jκ binding site on the regulatory region in the human and murine mcl-1 promoter is shown. The area highlighted in grey is the conserved binding site. +1 indicates the translation start site. A potential binding site for CSL/RBP-Jκ was found between −171 and −163 bp upstream of transcription initiation site. * indicates conserved nucleotide sequences between human and murine species. (b) BMM were treated with PPD (5 µg/ml) for 24 h, and the genome of treated cells was immunoprecipitated using either an anti-RNA polymerase II or anti-Notch1 antibody. The resulting products were used to PCR amplify the promoter region of mcl-1 containing the potential CSL/RBP-Jκ binding site. Rabbit or mouse normal IgG were used as a control. The PCR reaction performed without addition of the precipitated DNA was used as a negative control, as indicated ‘Neg. control' in the figure. Input indicated the results when the PCR reaction was performed using genomic DNA for ChIP assay as a template. BMM, bone marrow-derived macrophage; ChIP, chromatin immunoprecipitation; GSI, gamma secretase inhibitor; Mcl-1, myeloid cell leukemia sequence-1; PPD, purified protein derivative.
Discussion
In this study, we investigated the involvement of Notch signaling in regulating the levels of Mcl-1, which is an anti-apoptotic protein, and the possible role Notch signaling plays in regulating apoptosis in PPD-treated and BCG-infected macrophages. Notch signaling regulates pro-inflammatory responses in macrophages stimulated with TLR agonists by interacting with other signaling modulators, such as NF-κB, IRF and MAPK.22,23,33 In an effort to identify the potential target genes induced by Notch signaling in inflammatory macrophages, a ChIP-on-chip assay was performed, and mcl-1 was found to be one of the target genes.
Mycobacteria mainly infect macrophages and can escape killing mediated by innate immune cells. These bacteria evade the immune response and survive in the host by controlling the type of cell death in infected macrophages.2,3 While apoptosis of infected macrophages is considered beneficial for controlling bacterial growth and stimulating the adaptive immune response, necrosis of macrophages helps disseminate bacteria.3,34 Virulent Mycobacterium tuberculosis manipulates host cell death mechanisms by inducing the expression of Mcl-1.19
We used PPD treatment and BCG infection to study the involvement of Notch signaling and Mcl-1 in macrophages. PPD treatment of dendritic cells triggers induction of pro-inflammatory mediators via TLR2 in a MyD88-dependent manner, suggesting that PPD signals partially through the TLR pathway.35 Furthermore, BCG infection of macrophages induces SOCS3 expression via TLR2 and Notch1.25 Our results further support the reports that BCG and PPD signal through TLR2 in a MyD88-dependent manner and trigger the upregulation of Notch1. Because Notch signaling can regulate its own expression, which can be observed in activated T lymphocytes, TLR-mediated upregulation of Notch receptors can stimulate Notch signaling in a positive feedback manner.31
Notch signaling has been previously linked to apoptosis and Mcl-1. Notch receptors have been directly linked to lymphoblastic leukemia and other types of cancers.27,36 In T cells, Notch signaling directly suppresses apoptosis by either inhibiting the cell death promoting protein Nur77 or inducing the anti-apoptotic proteins Bcl-2 and Bcl-xL.37,38 In addition, activation of Notch signaling using the Notch receptor that contains the RAM domain promotes the survival of neural precursor cells by regulating the expression of Mcl-1 and Bcl-2.39 Therefore, Notch signaling positively regulates anti-apoptotic proteins, including Mcl-1. However, the mechanism behind this regulation is not known.
Recently, it was reported that GSI treatment induced apoptosis in chronic lymphocytic leukemia cells.28 This report investigated the effect of GSI on Mcl-1 and found the accumulation of Mcl-1 while cells were undergoing apoptosis. Silencing Mcl-1 accelerated cell death in the presence of GSI, suggesting that targeting Notch signaling pathway using GSI and silencing Mcl-1 may be effective in inducing apoptosis of cancer cells.28 The contradictory results on the effect of GSI on Mcl-1 level in our study and this report may be due to differences in cell types used and the stimuli used to induce Mcl-1 expression.
In our study, GSI treatment was employed to inhibit the processing of Notch receptors. This treatment also decreased the expression of Notch1 (Figure 3a and c). This is because activation of Notch induces new transcription of Notch in several systems, including T cells and Drosophila, inhibition of Notch activation should decrease Notch protein expression itself.31,40
A well-conserved consensus binding site for CSL/RBP-Jk was identified in the human and mouse mcl-1 promoter region. Furthermore, the results from the ChIP assay confirmed an association between Notch1 and the mcl-1 promoter region, indicating that Notch/CSL/RBP-Jk directly regulates the mcl-1 promoter. We cannot exclude the possibility that Notch1 may associate with regions of the mcl-1 promoter other than the one we identified in this study. But no potential binding site for CSL was found within at least the region 1400 bp upstream of the transcription start site of mcl-1. Silencing Notch1 or treating macrophages with GSI abrogated the upregulation of mcl-1 mRNA, suggesting that Mcl-1 is regulated by Notch1 at the transcriptional level. However, other studies have reported that Mcl-1 is also regulated post-transcriptionally by other signaling pathways, such as PI3K/AKT, MAPK and JAK/STAT.11,15 Because these pathways are also affected by Notch signaling, it is possible that Notch signaling may regulate Mcl-1 indirectly via these pathways.
Apoptosis of macrophages treated with PPD in the presence of GSI increased significantly when compared to cells receiving PPD and DMSO. Apoptosis of BCG-infected macrophages, however, was not significantly enhanced by GSI treatment, although there was a slight increase (Figure 5b). This result suggests that BCG uses different pathways to suppress apoptosis. Because PPD is derived from the pathogenic MTB and infection with these bacteria also upregulate Notch1 (unpublished observation), the ability of PPD treatment to affect apoptosis in the host may reflect the mechanisms used by MTB.
In conclusion, we provide evidence that links Notch signaling to Mcl-1 expression upon the recognition of Mycobacteria in macrophages. The direct regulation of Mcl-1 by Notch signaling may provide a new way to manipulate apoptosis in macrophages to benefit the host immune response to MTB infection.
Acknowledgments
The authors are grateful to Dr. Stefan H. E. Kaufmann, Dr Barbara Osborne and Dr Todd Golde for sharing reagents. The authors thank Dr Jorg Schriber and Dr Masami Nakatsu for their help with ChIP and ChIP-on-chip analysis. This work was partly supported by the Thailand Research Fund Grant No. RSA5280014, Research Foundation Enhancement from Chulalongkorn University (Ratchadaphiseksomphot Endowment Fund) and by the Thai Government Stimulus Package 2 (TKK2555) under the Project for Establishment of Comprehensive Center for Innovative Food, Health Products and Agriculture and the Higher Education Research Promotion and National Research University Project of Thailand, the Office of the Higher Education Commission (AS613A) and The Fogarty International Research Collaborative Award (Grant No. R03 TW008420-01A1, NIH, Bethesda, MD USA). W. Wongchana was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (PHD/0337/2551) (NIH, Bethesda, MD, USA).
Footnotes
Supplementary Information accompanies the paper on Cellular & Molecular Immunology website.
Supplementary Information
References
- Pieters J. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe. 2008;3:399–407. doi: 10.1016/j.chom.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Lee J, Hartman M, Kornfeld H. Macrophage apoptosis in tuberculosis. Yonsei Med J. 2009;50:1–11. doi: 10.3349/ymj.2009.50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld H, Mancino G, Colizzi V. The role of macrophage cell death in tuberculosis. Cell Death Differ. 1999;6:71–78. doi: 10.1038/sj.cdd.4400454. [DOI] [PubMed] [Google Scholar]
- Fairbairn IP. Macrophage apoptosis in host immunity to mycobacterial infections. Biochem Soc Trans. 2004;32:496–498. doi: 10.1042/BST0320496. [DOI] [PubMed] [Google Scholar]
- Winau F, Weber S, Sad S, de Diego J, Hoops SL, Breiden B, et al. Apoptotic vesicles crossprime CD8 T cells and protect against tuberculosis. Immunity. 2006;24:105–117. doi: 10.1016/j.immuni.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Keane J, Shurtleff B, Kornfeld H. TNF-dependent BALB/c murine macrophage apoptosis following Mycobacterium tuberculosis infection inhibits bacillary growth in an IFN-gamma independent manner. Tuberculosis (Edinb) 2002;82:55–61. doi: 10.1054/tube.2002.0322. [DOI] [PubMed] [Google Scholar]
- Gan H, Lee J, Ren F, Chen M, Kornfeld H, Remold HG. Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat Immunol. 2008;9:1189–1197. doi: 10.1038/ni.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Pathogenic Mycobacterium tuberculosis evades apoptosis of host macrophages by release of TNF-R2, resulting in inactivation of TNF-alpha. J Immunol. 1998;161:2636–2641. [PubMed] [Google Scholar]
- Michels J, Johnson PW, Packham G. Mcl-1. Int J Biochem Cell Biol. 2005;37:267–271. doi: 10.1016/j.biocel.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Zhou P, Qian L, Bieszczad CK, Noelle R, Binder M, Levy NB, et al. Mcl-1 in transgenic mice promotes survival in a spectrum of hematopoietic cell types and immortalization in the myeloid lineage. Blood. 1998;92:3226–3239. [PubMed] [Google Scholar]
- Liu H, Ma Y, Cole SM, Zander C, Chen KH, Karras J, et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- Le Gouill S, Podar K, Amiot M, Hideshima T, Chauhan D, Ishitsuka K, et al. VEGF induces Mcl-1 up-regulation and protects multiple myeloma cells against apoptosis. Blood. 2004;104:2886–2892. doi: 10.1182/blood-2004-05-1760. [DOI] [PubMed] [Google Scholar]
- Le Gouill S, Podar K, Harousseau JL, Anderson KC. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle. 2004;3:1259–1262. doi: 10.4161/cc.3.10.1196. [DOI] [PubMed] [Google Scholar]
- Wang JM, Lai MZ, Yang-Yen HF. Interleukin-3 stimulation of mcl-1 gene transcription involves activation of the PU.1 transcription factor through a p38 mitogen-activated protein kinase-dependent pathway. Mol Cell Biol. 2003;23:1896–1909. doi: 10.1128/MCB.23.6.1896-1909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Yu EZ, Li YY, Kagan E. HIF-1alpha has an anti-apoptotic effect in human airway epithelium that is mediated via Mcl-1 gene expression. J Cell Biochem. 2006;97:755–765. doi: 10.1002/jcb.20683. [DOI] [PubMed] [Google Scholar]
- Thomas LW, Lam C, Edwards SW. Mcl-1: the molecular regulation of protein function. FEBS Lett. 2010;584:2981–2989. doi: 10.1016/j.febslet.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Marriott HM, Bingle CD, Read RC, Braley KE, Kroemer G, Hellewell PG, et al. Dynamic changes in Mcl-1 expression regulate macrophage viability or commitment to apoptosis during bacterial clearance. J Clin Invest. 2005;115:359–368. doi: 10.1172/JCI21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sly LM, Hingley-Wilson SM, Reiner NE, McMaster WR. Survival of Mycobacterium tuberculosis in host macrophages involves resistance to apoptosis dependent upon induction of antiapoptotic Bcl-2 family member Mcl-1. J Immunol. 2003;170:430–437. doi: 10.4049/jimmunol.170.1.430. [DOI] [PubMed] [Google Scholar]
- Kausalya S, Somogyi R, Orlofsky A, Prystowsky MB. Requirement of A1-a for bacillus Calmette-Guerin-mediated protection of macrophages against nitric oxide-induced apoptosis. J Immunol. 2001;166:4721–4727. doi: 10.4049/jimmunol.166.7.4721. [DOI] [PubMed] [Google Scholar]
- Sohn H, Lee KS, Kim SY, Shin DM, Shin SJ, Jo EK, et al. Induction of cell death in human macrophages by a highly virulent Korean Isolate of Mycobacterium tuberculosis and the virulent strain H37Rv. Scand J Immunol. 2009;69:43–50. doi: 10.1111/j.1365-3083.2008.02188.x. [DOI] [PubMed] [Google Scholar]
- Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SH, et al. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur J Immunol. 2008;38:174–83. doi: 10.1002/eji.200636999. [DOI] [PubMed] [Google Scholar]
- Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldi J, Chung AY, Xu H, Zhu J, Outtz HH, Kitajewski J, et al. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J Immunol. 2010;185:5023–5231. doi: 10.4049/jimmunol.1001544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana Y, Balaji KN. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J Biol Chem. 2008;283:12501–12511. doi: 10.1074/jbc.M709960200. [DOI] [PubMed] [Google Scholar]
- Ito T, Schaller M, Hogaboam CM, Standiford TJ, Sandor M, Lukacs NW, et al. TLR9 regulates the mycobacteria-elicited pulmonary granulomatous immune response in mice through DC-derived Notch ligand delta-like 4. J Clin Invest. 2009;119:33–46. doi: 10.1172/JCI35647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L, Osborne B. Arbiter of differentiation and death: Notch signaling meets apoptosis. J Cell Physiol. 1999;181:393–409. doi: 10.1002/(SICI)1097-4652(199912)181:3<393::AID-JCP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Rosati E, Sabatini R, de Falco F, del Papa B, Falzetti F, di Ianni M, et al. gamma-Secretase inhibitor I induces apoptosis in chronic lymphocytic leukemia cells by proteasome inhibition, endoplasmic reticulum stress increase and notch down-regulation. Int J Cancer. 2013;132:1940–1953. doi: 10.1002/ijc.27863. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta CT) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Wongchana W, Palaga T. Direct regulation of interleukin-6 expression by Notch signaling in macrophages. Cell Mol Immunol. 2011;8:1–8. doi: 10.1038/cmi.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- Wu L, Griffin JD. Modulation of Notch signaling by mastermind-like (MAML) transcriptional co-activators and their involvement in tumorigenesis. Semin Cancer Biol. 2004;14:348–356. doi: 10.1016/j.semcancer.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Repasy T, Papavinasasundaram K, Sassetti C, Kornfeld H. Mycobacterium tuberculosis induces an atypical cell death mode to escape from infected macrophages. PLoS One. 6:e18367. doi: 10.1371/journal.pone.0018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AD, Manickasingham SP, Sporri R, Diebold SS, Schulz O, Sher A, et al. Microbial recognition via Toll-like receptor-dependent and -independent pathways determines the cytokine response of murine dendritic cell subsets to CD40 triggering. J Immunol. 2002;169:3652–3660. doi: 10.4049/jimmunol.169.7.3652. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Jehn BM, Bielke W, Pear WS, Osborne BA. Cutting edge: protective effects of notch-1 on TCR-induced apoptosis. J Immunol. 1999;162:635–638. [PubMed] [Google Scholar]
- Deftos ML, He YW, Ojala EW, Bevan MJ. Correlating notch signaling with thymocyte maturation. Immunity. 1998;9:777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Kamakura S, Isazawa Y, Yoshimatsu T, Kuida K, Nakafuku M, et al. Notch promotes survival of neural precursor cells via mechanisms distinct from those regulating neurogenesis. Dev Biol. 2004;276:172–184. doi: 10.1016/j.ydbio.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, et al. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.