Methylation of DNA is an essential epigenetic control mechanism in mammals. Messerschmidt et al. review the current understanding of epigenetic dynamics regulating the molecular processes that prepare the mammalian embryo for normal development.
Keywords: DNA methylation, development, epigenetics, reprogramming
Abstract
Methylation of DNA is an essential epigenetic control mechanism in mammals. During embryonic development, cells are directed toward their future lineages, and DNA methylation poses a fundamental epigenetic barrier that guides and restricts differentiation and prevents regression into an undifferentiated state. DNA methylation also plays an important role in sex chromosome dosage compensation, the repression of retrotransposons that threaten genome integrity, the maintenance of genome stability, and the coordinated expression of imprinted genes. However, DNA methylation marks must be globally removed to allow for sexual reproduction and the adoption of the specialized, hypomethylated epigenome of the primordial germ cell and the preimplantation embryo. Recent technological advances in genome-wide DNA methylation analysis and the functional description of novel enzymatic DNA demethylation pathways have provided significant insights into the molecular processes that prepare the mammalian embryo for normal development.
“When you arrive at a fork in the road, take it.”
—Yogi Berra
Cells in adult higher organisms are either committed to highly specialized tasks or reside in adult stem cell pools with limited potential; while they share the same genetic information, each cell type is defined by a specialized gene expression pattern. During development, the precursors of these cells commit to their fate in a step-by-step differentiation process, which is driven by a multitude of inputs (position, signaling, etc.) and is accompanied by epigenetic measures reinforcing the commitment decisions. These epigenetic changes form barriers to ensure that cell type specification is a one-way street. Such epigenetic modifications are stable and heritable through mitosis, allowing a faithful and directed differentiation process and the propagation of lineage-specific transcription profiles over many cell divisions.
However, these epigenetic barriers also pose a major challenge to sexual reproduction, where preparation for the next generation requires a reset of the (epi)genome to a basic, totipotent state. Particularly in mammals (where germ cells are not defined at fertilization but rather arise from later embryonic tissues), resetting the epigenome is of great importance (Hayashi et al. 2007; Strome and Lehmann 2007). The preparation for sexual reproduction is a three-step process consisting of (in chronological order) erasure of somatic signatures in the germ cell precursors (primordial germ cells [PGCs]) via a comprehensive reprogramming process, establishment of sex-specific and germ cell-specific epigenetic signatures and transcription profiles that enable the highly complex and specialized processes of meiotic maturation and fertilization, and finally, the post-fertilization removal of these signatures to trigger the embryonic developmental program and beginning of a new life cycle.
Although the basic principles of epigenetic reprogramming in embryos and germ cells have been known and studied for many years, major aspects, including the dynamics of these processes, remain enigmatic. In recent years, however, two main advances have propelled the field rapidly forward. Next-generation sequencing has allowed genome-wide epigenetic analysis of ever decreasing amounts of biological material, giving unprecedented insights into the epigenetic states of germ cells and early embryos at multiple stages (Borgel et al. 2010; Popp et al. 2010; Smallwood et al. 2011; Smith et al. 2012; Seisenberger et al. 2012; Hackett et al. 2013; Kobayashi et al. 2013). Furthermore, a long-standing void in our understanding of epigenetic reprogramming was filled by the discovery of the previously elusive enzymatic activities that initiate active erasure of DNA methylation in germ cells and embryos (Ito et al. 2010; He et al. 2011; Inoue and Zhang 2011; Inoue et al. 2011).
At present, our knowledge of epigenetic reprogramming is accumulating at a dizzying pace. Here we give a snapshot of the current understanding of epigenetic dynamics during the mammalian life cycle, with a focus on DNA methylation and genomic imprinting of PGCs and the preimplantation embryo.
The fifth base—5-methylcytosine (5mC) and its functions
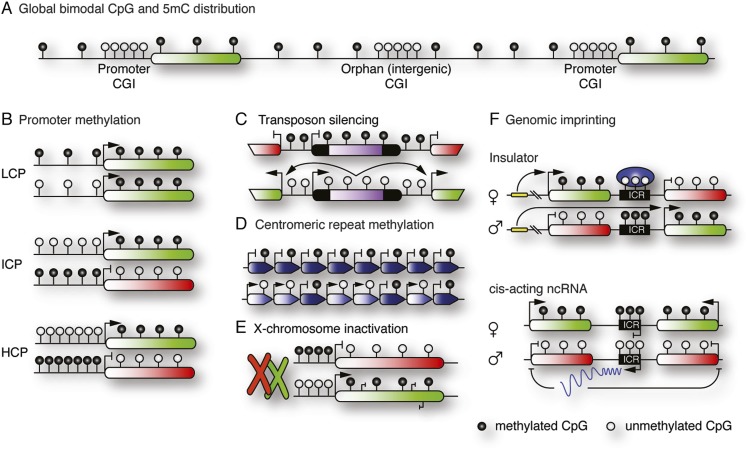
The methylation of the fifth carbon of cytosine produces 5mC, often referred to as the “fifth base” of the DNA code. DNA methylation is common in eukaryotes ranging from fungi to vertebrates, although its significance and function in these organisms varies greatly. While undetectable in certain yeast, nematode, and fly species, DNA methylation is vital for mammalian development and adult homeostasis. Here, DNA methylation is most frequently found in a symmetrical CpG context, and although rare non-CpG (CpH) methylation occurs, its function remains unclear (Ramsahoye et al. 2000; Smallwood et al. 2011; Ziller et al. 2011). The first global methylomes showed that CpG methylation follows a bimodal distribution (Fig. 1A). While “single” CpGs are generally hypermethylated (60%–90%, depending on cell type), CpG islands (CGIs; 500- to 2000-base-pair-long regions with increased CpG density) remain predominantly hypomethylated (Deaton and Bird 2011). Given the instability of 5mC due to its propensity to deaminate to thymidine, this bimodal distribution may account for the evolutionary underrepresentation of CpGs in the mammalian genome outside of CGIs and, conversely, its enrichment within unmethylated CGIs (Weber et al. 2007; Cohen et al. 2011).
Figure 1.
DNA methylation in the mammalian genome. (A) Genomic CpG distribution: CGIs are generally hypomethylated and found at promoters or intergenic regions (orphan CpG). Non-CGI CpGs are generally hypermethylated. (B) Three categories of gene promoters according to CpG density respond differently to methylation. (LCP) Low CpG density promoter; (ICP) intermediate CpG density promoter; (HCP) high CpG density promoter. (C) Retrotransposons are repressed by DNA methylation. Derepression can result in coactivation of neighboring genes. (D) Basal transcription of centromeric repeats interferes with chromosome alignment and is repressed by DNA methylation. (E) DNA methylation reinforces gene silencing on the inactivated X chromosome. Transcribed genes on the active X chromosome (and globally) display gene body methylation, possibly repressing spurious expression from cryptic transcription start sites or aiding RNA processing. (F) Genomic imprinting drives allele-specific gene expression. DNA methylation at imprinting control regions (ICRs) regulates binding of insulator proteins or expression of cis-acting, noncoding RNAs.
Role of DNA methylation in transcriptional control
Promoters
CGIs are often associated with gene promoters, particularly those of developmental or housekeeping genes, where they remain hypomethylated even if the locus is transcriptionally silent (Weber et al. 2007; Meissner et al. 2008; Deaton and Bird 2011). Intragenic or intergenic CGIs behave like promoter CGIs, yet their function in the genome is not fully understood (Fig. 1A). The hypomethylated state of promoter CGIs is associated with transcription factor binding (which prevents the DNA methylation machinery from targeting these regions) (Brandeis et al. 1994; Lienert et al. 2011; Meissner 2011; Stadler et al. 2011) as well as their specialized, histone 3 Lys4 trimethylation (H3K4me3)-heavy chromatin state (Erfurth et al. 2008; Thomson et al. 2010; Smith and Meissner 2013).
Historically, DNA methylation has been linked to transcriptional repression. However, detailed analysis of CGI-associated promoters and their effect on transcriptional activity has allowed for their classification into three categories based on CpG content and sequence length (Fig. 1B; Weber et al. 2007; Meissner et al. 2008). High CpG density promoters (HCPs) are rarely DNA-methylated, which supports the traditional view that DNA methylation represses transcription. In fact, the rare methylation of HCPs does result in efficient gene silencing. Similarly, intermediate CpG density promoters (ICPs) are inactive when methylated (Meissner et al. 2008; Borgel et al. 2010). However, ICPs more frequently acquire differentiation-dependent hypermethylation; i.e., at pluripotency gene promoters, where DNA methylation is thought to act as a safety mechanism to reinforce silencing during differentiation, as well as at promoters of germ cell-specific genes (Maatouk et al. 2006; Weber et al. 2007; Farthing et al. 2008; Meissner et al. 2008; Borgel et al. 2010). In contrast, low CpG density promoters (LCPs), which are generally hypermethylated, remain transcriptionally active regardless of their methylation state (Weber et al. 2007; Meissner et al. 2008).
Enhancers
Transcriptional enhancers physically interact with gene promoters and support tissue-specific differentiation. Like promoters, enhancers have characteristic DNA methylation patterns (Stadler et al. 2011), and hypomethylation is correlated with active gene expression (Carone et al. 2010; Sandovici et al. 2011). Remarkably, such enhancer methylation was shown to be more closely associated with gene expression changes in cancer cells than promoter methylation itself (Aran et al. 2013).
Role of DNA methylation in controlling specialized regions
Transposons
DNA methylation is conserved throughout evolution to repress endogenous transposable elements, which comprise ∼40% of the mammalian genome and thus significantly contribute to the overall hypermethylated state of the genome (Lander et al. 2001; Mouse Genome Sequencing Consortium 2002). In particular, LINEs (long interspersed nuclear elements) and LTR (long terminal repeat)-containing elements carry strong, generally hypermethylated promoters (Fig. 1C). Loss of methylation can cause massive transcriptional activation and, potentially, retrotranspositions (Walsh et al. 1998). Derepression of retrotransposons is also linked to the coactivation of neighboring genes or chimeric transcripts composed of retrotransposable elements and endogenous genes (Peaston et al. 2004). Particularly in oocytes and two-cell embryos, this indirect retrotransposon-mediated gene regulation may be critical for successful development (Peaston et al. 2004; Macfarlan et al. 2012).
Pericentromeric repeats
These elements show latent transcriptional activity, which is repressed by DNA methylation to achieve proper chromosome alignment and segregation (Fig. 1D). The importance of this repression is evident in the rare immunodeficiency, centromeric instability, and facial anomaly (ICF) syndrome, which is caused by missense mutations in the DNA methyltransferase gene DNMT3B. Continued transcription of the pericentromeric repeats causes rearrangements in the vicinity of the centromeres, likely originating from chromosome misalignments during mitosis (Okano et al. 1999; Xu et al. 1999; Bestor 2000; Chen and Li 2004; Jin et al. 2008; Gopalakrishnan et al. 2009).
X-chromosome inactivation
DNA methylation mediates gene dosage control via inactivation of the second X chromosome in females (Fig. 1E). In the case of X-chromosome inactivation, DNA methylation is the outcome of a cascade of events beginning with activation of a cis-acting noncoding RNA, Xist, which coats the X chromosome and therefore triggers displacement of transcription factors, chromatin changes, and, ultimately, CpG methylation at promoter CGIs. Loss of methylation, however, has only moderate effects on X inactivation, suggesting that it functions as long-term insurance rather than an initiator of X inactivation. Studies of DNA methylation in X inactivation led to the interesting observation that active genes display higher methylation levels in gene bodies, which could indicate transcriptional repression from cryptic promoters or merely the greater accessibility of actively transcribed DNA to the methylation machinery (Fig. 1E; Hellman and Chess 2007). Alternatively, DNA methylation at intragenic regions may correlate with RNA processing and alternative splicing, since methylation patterns can mark intron–exon boundaries (Hodges et al. 2009; Anastasiadou et al. 2011).
Imprinted genes
DNA methylation is vital to the controlled expression of imprinted genes (Fig. 1F; Bartolomei 2009; Ferguson-Smith 2011). The hallmark of imprinted genes is their parent-of-origin-specific expression, which is coordinated by differential DNA methylation (on paternal or maternal alleles) at imprinting control regions (ICRs). ICR methylation controls region-specific downstream mechanisms, such as insulator protein binding or antisense noncoding RNA expression. This results in allele-specific expression of gene clusters in which genes are either repressed or activated. Parent-of-origin-specific DNA methylation is introduced during gamete differentiation and maintained throughout life. Genomic imprinting therefore poses the greatest complication to epigenetic reprogramming in the mammalian life cycle, as their methylation patterns must first be erased in PGCs, re-established in an allele-specific manner in gametes, and then preserved during embryonic reprogramming. Genomic imprinting patterns play essential roles in development and growth, and their deregulation impacts grievously on the embryonic phenotype and may well be the chief cause of defects in methylation-deficient mutants.
The machinery
DNA methylation
DNA methyltransferases catalyze the transfer of a methyl group from S-adenosyl-l-methionine to the fifth position of cytosine residues in DNA (Chen and Li 2004). The predominant positioning of 5mC in the symmetrical CpG context led to the early proposal of DNA methylation inheritance through semiconservative DNA replication, which implied two distinct modes of DNA methylation activity: maintenance methylation and de novo methylation (Holliday and Pugh 1975; Riggs 1975).
Methylation maintenance
A requisite for a maintenance mechanism is a high affinity for hemimethylated CpGs (i.e., methylation of the palindromic CpG on only one strand). The first eukaryotic enzyme with DNA methyltransferase activity (DNMT1) (Bestor 1988) displayed this predicted function (Ruchirawat et al. 1987; Hitt et al. 1988; Yoder et al. 1997; Pradhan et al. 1999). Dnmt1 expression is activated by cell cycle-dependent transcription factors in S phase and thus is highly expressed in most mitotic cells (Kishikawa et al. 2003). Attracted by its proliferating cell nuclear antigen (PCNA)-interacting binding partner, NP95, DNMT1 is localized to replication foci, where it restores hemimethylated CpGs to full methylation (Leonhardt et al. 1992; Arand et al. 2012). NP95 specifically attracts DNMT1 to the parental, methylated strand, orienting the enzyme and its activity to the newly synthesized, unmethylated strand (Bostick et al. 2007; Sharif et al. 2007).
In mice, the Dnmt1 locus encodes two functionally identical isoforms: a somatic (Dnmt1s) and an oocyte-specific (thus only maternally expressed) isoform (Dnmt1o) (Rouleau et al. 1992; Gaudet et al. 1998; Mertineit et al. 1998; Ding and Chaillet 2002). Deletion of mouse Dnmt1 is lethal at/after gastrulation, the result of a substantial, global loss of DNA methylation (Ruchirawat et al. 1987; Hitt et al. 1988; Lei et al. 1996; Yoder et al. 1997; Kurihara et al. 2008; Arand et al. 2012). Loss of NP95 causes similar defects (Sharif et al. 2007).
De novo methylation
Dnmt1 deletion in mouse embryonic stem cells (mESCs) causes dramatic DNA hypomethylation but not a complete loss, predicting the presence of further enzymes with methyltransferase activity. Two additional enzymes, DNMT3A and DNMT3B, were identified and shown to act as de novo DNA methyltransferases (Okano et al. 1998a). Dnmt3a is maternally provided and predominates in oocytes and early preimplantation embryos. DNMT3A establishes the differential DNA methylation patterns at ICRs in male and female gametes (Kaneda et al. 2004; Kato et al. 2007). Dnmt3b is transcribed upon zygotic gene activation (ZGA) and is robustly expressed by the blastocyst stage, at which time its presence is predominantly restricted to the epiblast lineage (Watanabe et al. 2002). Deletion of Dnmt3b causes embryonic lethality, while Dnmt3a knockouts are partially viable (Okano et al. 1999). Combined genetic deletion results in earlier embryonic lethality, indicating at least partial functional redundancy of the two enzymes (Okano et al. 1999).
A third de novo DNA methyltransferase, DNMT3L, lacks the characteristic N-terminal catalytic domain but is nonetheless required for DNA methylation, predominantly establishing ICR methylation in gametes (Bourc’his et al. 2001; Bourc’his and Bestor 2004). DNMT3L is a crucial activating cofactor of DNMT3A/B, explaining its methylation effects despite the lack of inherent catalytic activity (Chedin et al. 2002; Gowher et al. 2005; Jia et al. 2007). Dnmt3l knockout mice are viable, but the lack of de novo methylation in the germline causes sterility in males and embryonic lethality of maternal null-derived embryos (Bourc’his et al. 2001; Bourc’his and Bestor 2004).
Last, DNMT2 differs structurally from other DNA methyltransferases, and knockout mice display no phenotype (Okano et al. 1998b). Indeed, DNMT2 is a misnomer, as it shows methylation activity toward RNA (Goll et al. 2006).
DNA demethylation
In contrast to the enzymatically controlled, straightforward methylating mechanism, a direct DNA demethylase capable of breaking carbon–carbon bonds has not yet been identified. Instead, several alternative, active demethylation mechanisms that are passive and indirect have been proposed and demonstrated (Wu and Zhang 2014, 2010).
Passive, replication-dependent dilution
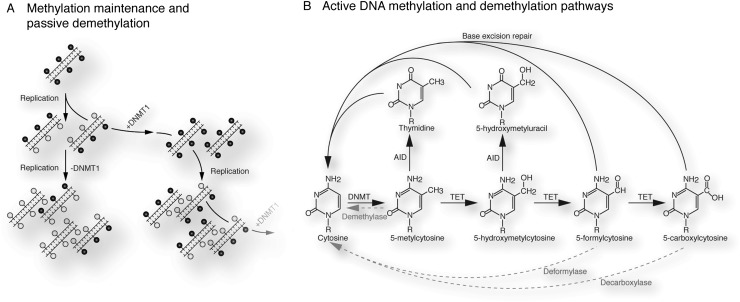
Loss of 5mC in mitotic cells can be achieved by down-regulation or exclusion of the DNA methylation maintenance machinery (DNMT1 or its recruitment factors, such as NP95) from the nucleus (Fig. 2A). Although allowing efficient global DNA demethylation, the passive mechanism depends on repeated DNA replication and therefore cannot account for the rapid loss of DNA methylation in slowly or nondividing cells. Furthermore, this mechanism does not allow locus-specific, but only global, removal of DNA methylation marks.
Figure 2.
DNA methylation and demethylation mechanisms. (A) The palindromic nature of the CpG DNA replication creates hemimethylated DNA. DNMT1 restores hemimethylated DNA to full methylation. Absence of the DNMT1 machinery produces unmethylated DNA after a subsequent round of cell division. (Black filled circles) Methylated CpG; (white filled circles) unmethylated CpG. (B) Possible active demethylation pathways: A direct demethylase converting 5mC to cytosine is, to date, speculative. TET enzymes oxidize 5mC to 5hmC, 5fC, and 5caC. Deamination of 5mC and 5hmC, potentially by AID, produces thymidine or hydroxymethyluracil, respectively. The base excision repair (BER) mechanism may target AID deamination products and possibly 5fC and 5caC directly. Direct deformylation or decarboxylation of 5fC and 5caC has been proposed but remains speculative.
Active demethylation
An indirect, enzyme-catalyzed DNA demethylation mechanism could involve the deamination of 5mC to thymidine by the activation-induced deaminase (AID) or apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (APOBEC1) (Fig. 2B). Deamination of 5mC creates T:G mismatches, recognized by thymine–DNA glycosylase (TDG) or methyl-CpG-binding domain protein 4 (MBD4), which catalyze removal of the thymine base. Abasic sites then trigger the base pair excision repair (BER) machinery to reinstate an unmodified cytosine, effectively resulting in removal of the methyl mark (Fig. 2B; Morgan et al. 2004). The extent of AID-triggered DNA demethylation mechanisms required for the reprogramming in PGCs and early embryos is a matter of debate and is described in detail below.
The existence of a ‘sixth base’
5-hydroxymethylcytosine (5hmC) and the recent discovery of the ten-eleven translocation (TET) family of dioxygenases (Tahiliani et al. 2009) imply new possibilities of active DNA demethylation. TET enzymes catalyze the iterative oxidation of 5mC to 5hmC and further to 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) (Fig. 2B; He et al. 2011; Inoue et al. 2011; Ito et al. 2011). All three derivatives are found in cells displaying TET activity, yet their biological significance is unresolved. Given the existence of a decarboxylating enzyme in the pyrimidine salvage pathway, a similar enzyme, elusive to date, could directly restore cytosine from 5caC (Ito et al. 2010). Alternatively, TDG, MBD4, or a yet unidentified glycosylase may remove oxidized methyl-cytosine derivatives, with the resulting abasic site triggering BER. As recent observations suggest, TDG has direct activity on 5fC and 5caC without requiring a previous deamination step (Fig. 2B; He et al. 2011; Shen et al. 2013).
5hmC may also aid passive DNA demethylation even in the presence of DNMT1, which itself has low affinity for 5hmC hemimethylated DNA (Valinluck and Sowers 2007). Conversely, NP95 interacts with both 5mC and 5hmC. Further studies are required to define this particular mechanism. The observed, divergent affinities of many interaction partners to 5mC and 5hmC (Valinluck and Sowers 2007; Liu et al. 2012) may indicate other functions of 5hmC; i.e., altering the local chromatin environment by recruitment and/or displacement of methylation- or hydroxymethylation-dependent binding proteins.
TET-catalyzed conversion of 5mC to 5hmC is a plausible mechanism to allow cell cycle-independent removal of DNA methylation. In mice, the TET family comprises three members (Tet1–3), which are differentially expressed during development and in adult tissues (Tahiliani et al. 2009). Tet1 and Tet2 are detected in ESCs and PGCs. Tet2 expression is essential in hematopoietic cells; knockouts are viable but develop severe, lethal hematopoietic malignancies at 4–6 mo of age (Li et al. 2011; Moran-Crusio et al. 2011; Quivoron et al. 2011; Koh and Rao 2013). Tet1 knockout mice are viable, displaying somewhat reduced body size, suggesting potential developmental delay, and both knockout males and females are subfertile (Dawlaty et al. 2011; Yamaguchi et al. 2012, 2014). Given their partially overlapping expression patterns, a certain degree of functional redundancy of Tet1 and Tet2 has been suggested. Indeed, double-knockout embryos show more pronounced developmental defects, yet loss of both Tet1 and Tet2 can still be compatible with normal development (Dawlaty et al. 2013). Tet3 expression patterns share little overlap with Tet1/2 expression and are found to be highly expressed in oocytes, spermatozoa, and at early, preimplantation embryonic stages. Maternal loss of Tet3 can cause developmental arrest in a subset of embryos (Gu et al. 2011).
Epigenetic reprogramming in PGCs: the true blank slate
PGCs are either defined by the inherent germplasm in the oocyte (flies, nematodes, frogs, and fish) or are derived from somatic precursors in the embryo by instructive signals (mammals) (Fig. 3; Strome and Lehmann 2007; Saitou and Yamaji 2012). In mice, a small group of PGCs originates from the proximal embryonic ectoderm at embryonic days 6.5–7.5 (E6.5–E7.5) upon receiving instructive signals (the earliest being BMP4 signaling) from the visceral endoderm and the surrounding embryonic ectoderm itself (McLaren and Lawson 2005; Hayashi et al. 2007; Seki et al. 2007; Saitou and Yamaji 2012). As the epiblast rapidly adopts somatic epigenetic features after implantation, the DNA methylation levels and patterns of the embryonic ectoderm in the E6.5 mouse embryo are more closely related to somatic tissues than to the inner cell mass of the blastocyst (Borgel et al. 2010; Popp et al. 2010). In particular, pluripotency genes (i.e., Oct4 and Nanog) and CGI-associated germline-specific genes are tightly repressed by DNA methylation, preventing any deleterious effects that could occur if they were ectopically activated (Maatouk et al. 2006; Shen et al. 2007; Weber et al. 2007; Straussman et al. 2009; Borgel et al. 2010; Arand et al. 2012; Seisenberger et al. 2012; ). Thus, PGCs at this point display the transcription profiles, chromatin modifications, and DNA methylation levels and patterns characteristic of their somatic origin (Ohinata et al. 2005; Maatouk et al. 2006; Hajkova et al. 2010; Popp et al. 2010; Seisenberger et al. 2012).
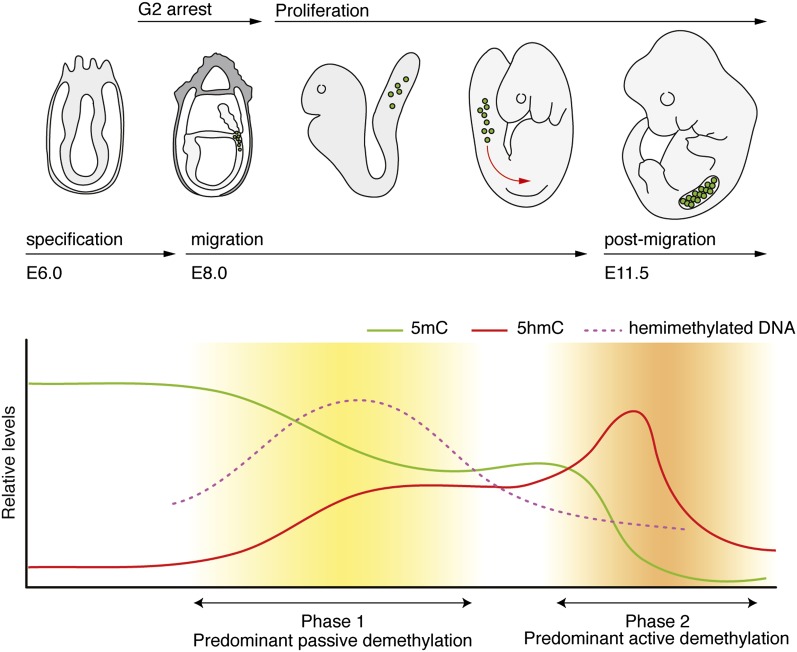
Figure 3.
Biphasic demethylation dynamics in mouse PGCs. PGCs are derived from the embryonic ectoderm of the E6.5 embryo and display high (somatic) 5mC levels (green lines) and low 5hmC levels (red lines). Upon migration, PGCs proliferate, and 5mC levels are passively diluted. Coincidently, hemimethylated DNA strands accumulate transiently and are subsequently lost (purple dashed line). Post-migratory PGCs enter a phase of active DNA demethylation, resulting in an almost complete loss of 5mC and a transient enrichment of 5hmC. At E13.5, both 5mC and 5hmC levels are low.
The major task of epigenetic reprogramming to totipotency that takes place in early PGCs is accomplished by an elaborate transcriptional program driving PGCs away from differentiation and forcing the intimately linked re-expression of key pluripotency markers (for review, see Hayashi et al. 2007; Saitou and Yamaji 2012). During this process, PGCs proliferate and migrate from the proximal epiblast along the hindgut to the genital ridges, where their genomes display very low levels of global DNA methylation (E12.5–E13.5) (Fig. 3; Popp et al. 2010; Guibert and Weber 2012; Seisenberger et al. 2012). This reprogramming process also reverses other somatic epigenetic features: Parental imprints are erased (Hajkova et al. 2002; Sasaki and Matsui 2008), and the silenced X chromosome is reactivated in PGCs of female mouse embryos (Chuva de Sousa Lopes et al. 2008).
DNA demethylation mechanisms in PGCs
It was initially speculated that the proliferation rate of PGCs is insufficient to allow passive demethylation (Fig. 2A), and all active DNA demethylation mechanisms (Fig. 2B) were proposed and explored. However, a reliable, robust, genome-wide removal of nearly all 5mCs by active means seems energetically unlikely and, given the required double-strand breaks involved (if BER-mediated), highly risky. Indeed, recent studies indicate that passive dilution of methylation might well be sufficient for global demethylation, providing an energetically more plausible, robust mechanism. With the help of genome-wide analysis methods applicable to small cell numbers, a combinatorial, almost sequential, and perhaps context-dependent process of active and passive DNA demethylation is emerging (Fig. 3).
Passive DNA demethylation in PGCs
The silencing of key genes of the DNA methylation machinery in PGCs could facilitate passive DNA demethylation. By E9.5, de novo methyltransferases (Dnmt3a/b) are repressed and remain virtually absent throughout the reprogramming period (Seki et al. 2005; Yabuta et al. 2006; Kurimoto et al. 2008; Seisenberger et al. 2012; Kagiwada et al. 2013). In addition, although the maintenance methyltransferase Dnmt1 remains expressed and the protein remains abundant, its essential cofactor, Np95, is repressed at E9.5, and NP95 protein is excluded from the nucleus (Kurimoto et al. 2008; Seisenberger et al. 2012; Kagiwada et al. 2013). Consequently, DNMT1 staining is strongly reduced at S-phase replication foci, suggesting poor methylation maintenance in PGCs. It is only after E12.5 that NP95 protein levels increase slightly, and DNMT1 localization to replication foci is slowly re-established (Kagiwada et al. 2013).
The majority of PGCs are arrested in the G2 phase of the cell cycle, while they migrate toward the hindgut (E7.5–E8.5), but proliferation rapidly ensues at E9.5 (Fig. 3; Seki et al. 2007). In contrast to early data suggesting a 16-h cell cycle in PGCs (Tam and Snow 1981) recent BrdU pulse-chase experiments describe faster (∼12 h per cycle), exponential proliferation until at least E12.5, significantly increasing chances for passive demethylation (Kagiwada et al. 2013). This replication-dependent DNA demethylation creates hemimethylated DNA strands (Fig. 2A), which can indeed be detected by hairpin bisulfite sequencing (Arand et al. 2012), with increased incidence in E9.5–10.5 PGCs (Fig. 3, pink dashed line; Seisenberger et al. 2012).
Bisulfite conversion (BSC) and sequencing of PGC-derived DNA revealed that demethylation indeed affects the global genome, including promoters, CGIs, introns, exons, and intergenic regions. However, a subset of sequences, including imprinted genes, promoters of genes necessary for germ cell formation and meiosis, and CGIs of the inactive X chromosome in females, appears to follow slower kinetics and become fully hypomethylated only after a second wave of demethylation (Fig. 3; Seisenberger et al. 2012). The observed late reprogramming of differentially methylated regions (DMRs) in PGCs is in agreement with some studies showing erasure of imprints after E11.5, when PGCs enter the gonads (Hajkova et al. 2002; Guibert and Weber 2012), yet is in conflict with other studies showing early DMR demethylation, with kinetics paralleling global demethylation (Lee et al. 2002; Kagiwada et al. 2013).
Active DNA demethylation in PGCs
The two-phase demethylation process in PGCs (Guibert and Weber 2012; Seisenberger et al. 2012; Hackett et al. 2013; Yamaguchi et al. 2013) raises the question as to the molecular mechanisms involved in this distinction. It is likely that specific sequences are protected from passive demethylation in early PGCs but are then demethylated by either passive, active, or a combination of both means at later stages. The prevailing expression and nuclear presence of DNMT1 but reduction and nuclear exclusion of NP95 could indicate a noncanonical DNA methylation maintenance mechanism, possibly paralleling methylation protection of imprinted gene loci in the early embryo (see below).
The early assumptions of faster PGC proliferation kinetics have led to an intense investigation of active DNA demethylation mechanisms in these cells (Hajkova et al. 2008, 2010; Feng et al. 2010; Popp et al. 2010; Wu and Zhang 2010; Cortellino et al. 2011; Dawlaty et al. 2011, 2013; Yamaguchi et al. 2012, 2013, 2014; Hackett et al. 2013; Vincent et al. 2013). AID is expressed in E12.5 PGCs (Morgan et al. 2004) and linked to active DNA demethylation. Aid-deficient E13.5 PGCs display a very mild increase in DNA methylation levels over their wild-type counterparts; however, they also undergo evident global DNA demethylation between E8.5 and E13.5 (Popp et al. 2010). General demethylation processes in PGCs are thus either AID-independent or compensated for by other deaminases, possibly APOBEC proteins. However, loss of either APOBEC1, APOBEC2/3, or AID does not severely impact fertility (Muramatsu et al. 2000; Mikl et al. 2005; Popp et al. 2010). AID activity on 5mC is also much lower than on unmethylated cytosine in vitro (Neuberger and Rada 2007), and Aid and Apobec1 expression appears to be very low or absent throughout the PGC reprogramming period (Kagiwada et al. 2013), calling into question whether or to what extent AID and/or other deaminases are involved in PGC reprogramming. Locus-specific re-examination of Aid-deficient PGCs, particularly of imprinted gene loci, using refined analysis methods should resolve these uncertainties.
Judging by mRNA expression levels, Tdg is expressed in PGCs (Kagiwada et al. 2013). However, the protein was not detected by immunofluorescence (IF) (E10.5–13.5) (Hajkova 2010). The biallelic methylation of the Igf2r imprinted region can be detected in Tdg mutant PGCs, suggesting that TDG activity is at least required for maintenance of the unmethylated allele at this locus. However, it is likely that the hypermethylation occurs prior to PGC specification, as de novo DNA methylation is repressed in PGCs (Cortellino et al. 2011). Components of the BER pathway were also detected in PGCs (Hajkova et al. 2002), perhaps a testimony to DNA repair after AID/TDG or even TET activity.
Given the questionable expression of Aid but continued expression of Tet enzymes and Tdg, sequential oxidation of 5mC to 5fC or 5caC, which can be direct targets of TDG, appears to be the most likely scenario for an active demethylation mechanism in PGCs. Preliminary efforts failed to detect drastic changes of 5fC/5caC levels throughout the reprogramming period by IF (Hackett et al. 2013; Yamaguchi et al. 2013). However, until the importance of each of the 5mC oxidation derivatives is thoroughly addressed in vivo, these questions will remain unanswered.
Two recent studies addressed the distribution and dynamics of 5hmC in PGCs, adding another layer of complexity to our understanding of the demethylation process (Hackett et al. 2013; Yamaguchi et al. 2013). These findings show a temporal increase of 5hmC in E9.5/E10.5 PGCs, accompanied by decreasing 5mC levels followed by a reduction of 5hmC levels at E11.5/E12.5 (Fig. 3, red and green lines). Combined knockdown of Tet1 and Tet2 revealed slightly elevated 5mC levels in in vitro generated PGC-like cells, yet in vitro PGC specification itself was unaffected (Hackett et al. 2013).
Detailed analysis of a Tet1 knockout mouse strain revealed a reduction in female germ cells and ovary size. This phenotype is most likely the consequence of the reduced expression of genes affecting meiosis, which retain uncommonly high promoter methylation levels in mutant gametes (Yamaguchi et al. 2012). More recently, TET1 was also shown to be required for the efficient erasure of imprints in the paternal germline (Yamaguchi et al. 2014). However, the low penetrance of the phenotype suggests a redundant function and partial rescue of the phenotype with/by TET2. Tet1/2 double-knockout PGCs completely lack 5hmC (as detected by IF). Interestingly, global 5mC levels are not increased in these cells (also detected by IF), and TET1/2 loss can be compatible with development, with both mutant males and females maintaining fertility (Dawlaty et al. 2013). However, the majority of Tet1/2-null mutants die during embryogenesis or shortly after birth due to various and variable developmental abnormalities (Dawlaty et al. 2013). Crucially, progeny of Tet1/2-null males and females display hypermethylation at several imprinted gene regions, supporting a role for 5hmC removal of imprints in PGCs, probably during the second demethylation wave (Fig. 3). However, these defects are variable, and even in the absence of TET1/2, normal imprint erasure can occur in a fraction of the progeny (Dawlaty et al. 2013), likely by passive means (Lee et al. 2002; Kagiwada et al. 2013). A caveat to these studies is they were performed in mixed genetic backgrounds, which may itself explain the observed phenotypic variability (Dawlaty et al. 2013).
Germ cell specialization and sex-specific remethylation
Sex-specific differentiation of the mouse embryo ensues from E12.5 onward (McLaren 1984; Saitou and Yamaji 2012). The establishment of germ cell-specific methylation marks after PGC reprogramming occurs at different times and in different cellular environments in males and females, ultimately leading to sperm- and oocyte-specific patterns (Sasaki and Matsui 2008; Saitou and Yamaji 2012). In female germ cells, DNA methylation levels remain low at E16.5. In contrast, by the same time, male PGCs have already attained 50% global DNA methylation, as remethylation initiates as early as E14.5 in prospermatogonia, which are arrested in the G1 phase of mitosis (Kota and Feil 2010). The methylation patterns in male germ cells are fully established at birth (days 19–21) and are then maintained throughout many cycles of mitotic divisions before the cells enter meiosis (Davis et al. 2000; Henckel et al. 2009). The chances of accumulating DNA methylation errors and the propagation of mutations occurring due to spontaneous deamination of 5hmC during this extended period of replication and cell division are much greater in males than in females. In females, remethylation of gametes initiates only at birth during the oocyte growth phase, while the eggs are arrested in prophase of meiosis I (Kota and Feil 2010).
Despite divergent development and remethylation dynamics, DNMT3L and DNMT3A are required to establish the sex-specific imprints (Bourc’his et al. 2001; Hata et al. 2002; Bourc’his and Bestor 2004; Kaneda et al. 2004; Kato et al. 2007). Loss of DNMT3A or DNMT3L in males causes catastrophic meiotic arrest in spermatogonia, up-regulation of IAPs and LINE elements, and apoptotic loss of spermatocytes (Walsh et al. 1998; Bourc’his and Bestor 2004). On the other hand, de novo methylation-impaired oocytes can give rise to embryos that die at E10.5 due to the lack of imprinting control (Bourc’his et al. 2001).
Although the central players of ICR methylation have been identified in the differentiating germ cells, it is still unclear how they are targeted to imprinted regions. Several suggestions, such as sequence specificity, underlying chromatin structure and histone modifications, or traversing transcription, have been made and reviewed in excellent studies elsewhere (Smallwood and Kelsey 2012; Strogantsev and Ferguson-Smith 2012; Kelsey and Feil 2013). Recent efforts to decipher oocyte-specific, sperm-specific, and early embryo-specific methylomes, however, have cast a different light on imprinting-specific, DNA methylation targeting mechanisms (Smallwood et al. 2011). In addition to maternal germline ICRs, which are structurally CGIs, more than a thousand nonimprinted CGIs are also differentially methylated in oocytes compared with sperm. However, during post-fertilization reprogramming (see below), <15% of these actually retain their (differential) DNA methylation patterns, inclusive of all maternally imprinted regions (Smallwood et al. 2011). Therefore, a new concept suggesting that maternally methylated CGIs are not targeted by the underlying sequence but rather by histone modifications defined by active transcription enabling access to de novo methylation has emerged (Kelsey and Feil 2013).
Epigenetic reprogramming in preimplantation embryos: selective DNA methylation maintenance
The second wave of global epigenetic reprogramming occurs during early embryogenesis and is crucial to establishing pluripotency. The newly formed embryo undergoes massive, global DNA demethylation such that by the time the early blastocyst stage (32–64 cells) is reached, methylation levels are at their lowest (Fig. 4A). However, the process in embryos differs from that in PGCs. First, demethylation is close to absolute in PGCs, with the exception of a few resistant retroelements, while DNA methylation of imprinted gene regions is preserved in embryos, enabling parent-of-origin-specific gene expression in later tissues. Also, the imprinted paternal X inactivation found in early mouse embryos is not reversed until the late epiblast stage. Second, the genome of the zygote (which contains haploid contributions from the oocyte and sperm genome, each with their own specific chromatin properties) follows different DNA demethylation kinetics after fertilization (Fig. 4A; Mayer et al. 2000; Oswald et al. 2000; Santos et al. 2002; Santos and Dean 2004).
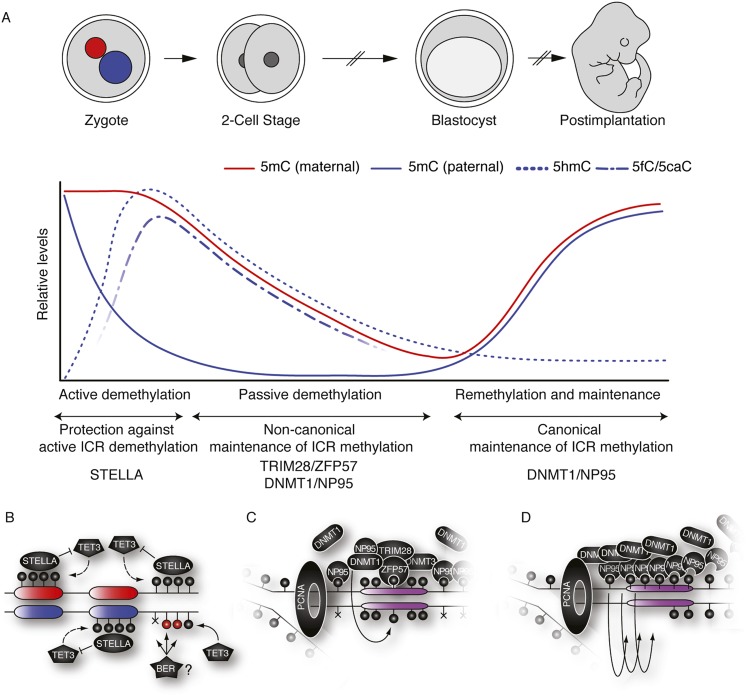
Figure 4.
DNA demethylation dynamics and imprinting maintenance in preimplantation embryos. (A) Distinct characteristics of maternal and paternal genomes impose an epigenetic asymmetry in the zygote. The maternal genome (red pronucleus; red line) undergoes passive DNA demethylation throughout several rounds of DNA replication. The paternal genome (blue pronucleus; blue lines) undergoes active demethylation before DNA replication in the zygote ensues. Concomitant with global loss of paternal 5mC, 5hmC (blue dotted line) and the further oxidation derivatives (5fC and 5caC; blue dashed line) are enriched. Although selected loci are restored to unmodified cytosines, the bulk of paternal 5hmC is passively diluted, paralleling demethylation of the maternal genome. (B) In the zygote, STELLA prevents TET3-dependent oxidation of 5mC through binding to H3K9me2-marked chromatin (maternal genome and paternally imprinted regions) and subsequent active restoration of cytosine by BER (or other pathways). (C) Throughout early cleavage stages, DNMT1 is largely excluded from the nucleus and requires noncanonical targeting to imprinted regions by the ZFP57/TRIM28 complex binding to its methylated consensus sequence found at most ICRs. Nonimprinted regions are efficiently demethylated through replication, while ICRs are maintained by DNMT1 and DNMT3A/B. (D) At later stages of embryogenesis and in adult tissues, high DNMT1/NP95 levels during replication maintain DNA methylation by targeting hemimethylated DNA in a canonical fashion.
Active DNA demethylation of the paternal genome
The mature sperm genome shows 80%–90% overall CpG methylation, the highest global DNA methylation level of any cell in the mouse (Popp et al. 2010), yet the paternal genome is apparently completely demethylated shortly after zygote formation (Fig. 4A, blue line; Mayer et al. 2000; Oswald et al. 2000). This loss must be due to an active demethylation mechanism, as it is completed before the onset of DNA replication at the pronuclear stage 3 (PN3). Conversely, the maternal genome shows lower global methylation levels (∼40%) and undergoes replication-dependent demethylation (Fig. 4A, red line), thereby establishing a significant epigenetic asymmetry in the early embryo (Fig. 4; Mayer et al. 2000; Oswald et al. 2000; Santos et al. 2002).
Conflicting observations made by IF analysis in zygotes and DNA sequencing after BSC created a conundrum regarding DNA demethylation and reprogramming. While the loss of 5mC in the paternal pronucleus was evident by IF, BSC analysis did not wholly support this observation (Hajkova et al. 2008; Wossidlo et al. 2010). It was only after the description of 5hmC, the oxidation product of 5mC by TET enzymes, that a unifying explanation came to hand (Gu et al. 2011; Iqbal et al. 2011; Wossidlo et al. 2011). BSC cannot distinguish between 5mC and 5hmC, whereas oxidation of 5mC removes the epitope recognized in IF.
Indeed, TET3 specifically localizes to the paternal pronucleus (Gu et al. 2011), where it was shown to be responsible for 5mC to 5hmC conversion (Fig. 4A, dotted blue line). In its absence, 5hmC is not detectable (Gu et al. 2011; Wossidlo et al. 2011), thus excluding functional redundancy with other TET proteins in the zygote. Lack of TET3 may result in the delayed activation of paternal alleles of genes required for embryonic development and establishment of the pluripotent epiblast (e.g., Nanog and Oct4), potentially causing the reduced fecundity and partial developmental failure of maternal TET3-null offspring (Gu et al. 2011).
The fate of paternal 5hmC
The bulk of 5mCs in the paternal genome is hydroxylated in the late zygote, but only a few regions were shown to completely revert to unmodified cytosine before the first cleavage division. BER activity could account for this loss of 5mC and the hypomethylation of the Nanog and Oct4 promoters (Gu et al. 2011). Several components of the BER pathway are specifically localized to the paternal pronucleus; i.e., XRCC1 (X-ray repair cross-complementing protein 1) tightly binds to paternal, but not maternal, DNA (Hajkova et al. 2010; Wossidlo et al. 2010). Concomitant with 5hmC, higher oxidation products of 5mC (5fC and 5caC), which can be direct targets for the TDG/BER pathway (He et al. 2011; Maiti and Drohat 2011), were also observed in the zygote (Fig. 4A, dotted/dashed blue lines; Inoue et al. 2011). However, this route of demethylation requires further investigation, since TDG was not detected in zygotes (Hajkova 2010). Certainly, other enzymes with similar activity could perform glycosylation. Alternatively, direct decarboxylation of 5caC has been described in other systems (Schiesser et al. 2012).
However, there is also compelling evidence that paternal 5mC is actively converted to 5hmC (and possibly further to 5fC and 5caC), which then undergoes replication-mediated dilution throughout subsequent cleavage divisions (Fig. 4A, dotted/dashed blue lines; Inoue and Zhang 2011; Inoue et al. 2011). Fittingly, the maintenance methyltransferase DNMT1 shows very limited affinity to oxidized 5mC derivatives (Hashimoto et al. 2012) and is generally excluded from the nucleus of preimplantation embryos (Howell et al. 2001; Hirasawa et al. 2008).
Distinction of parental pronuclei
How is TET3 targeted to the paternal genome without affecting the maternal genome? Prior to description of 5hmC dynamics in the zygote, the protein STELLA (Payer et al. 2003) was found to convey specificity to the, then elusive, active demethylation machinery in the early zygote (Fig. 4B; Nakamura et al. 2006). In its absence, loss of 5mC is observed in both pronuclei, accompanied by 5hmC accumulation in the maternal pronucleus (Wossidlo et al. 2011). Thus, the divergent demethylation dynamics in maternal and paternal genomes in the zygote are the consequence of specific protection of the maternal genome from TET3-mediated 5mC oxidation by STELLA. Although STELLA appeared to localize to both pronuclei in the zygote, its binding to the paternal genome is weak (Nakamura et al. 2012). The protective function is specifically mediated by STELLA’s interaction with dimethylated histone H3 Lys9-marked chromatin (H3K9me2), which is enriched in the maternal but not paternal pronucleus (Santos et al. 2005). This interaction of STELLA and H3K9me2 nucleosomes alters chromatin configuration, preventing TET3 binding and activity (Nakamura et al. 2012). Aside from its global function in the maternal genome, STELLA also protects at least two paternally methylated, imprinted gene loci (Rasgrf1 and H19, but not the IG-DMR) from aberrant demethylation (Nakamura et al. 2006). These loci retain H3K9me2-marked chromatin during spermatogenesis and protamine exchange, which mediates their protection after fertilization (Nakamura et al. 2012). However, not all paternally or maternally imprinted regions are affected equally, and possibly other mechanisms (described below) act in partial redundancy with STELLA (Messerschmidt 2012). Remarkably, in the absence of STELLA, the loss of DNA methylation at imprinted gene loci in the zygote is complete (Nakamura et al. 2006). Thus, at least the imprinted gene regions undergo active removal of 5hmC or rapidly iterated oxidations to 5caC by TET3 (Wu and Zhang 2010). Interestingly, the BER component XRCC1, which ordinarily exclusively localizes to the paternal genome, is found in both pronuclei in STELLA mutants (Hajkova 2010).
Embryos lacking STELLA display severe phenotypes, rarely developing past the four-cell stage, with few embryos surviving to birth. Remarkably, though, STELLA-deficient oocytes show neither methylation nor developmental defects prior to fertilization (Nakamura et al. 2006). This is particularly intriguing, as TET3 is present in the oocyte but only affects (in the absence of STELLA) the maternal genome after fertilization, in the zygotic context (Nakamura et al. 2006; Gu et al. 2011). The interplay of these two antagonistic factors must be addressed in more detail to define whether TET3 is merely prevented from acting on the maternal genome or must be molecularly targeted to its paternal substrates.
Is active DNA demethylation of the paternal genome required?
It is undisputed that global demethylation in the early mouse embryo is required to impose an open, totipotent or pluripotent state in the forming epiblast. However, it is not clear why only the paternal genome is targeted for active demethylation or, indeed, whether active demethylation is at all required. Although TET3-mediated demethylation of Nanog and Oct4 promoters was linked to embryo viability, loss of TET3 is nonetheless compatible with normal development (Gu et al. 2011). In fact, embryos derived from round spermatid-injected oocytes (containing histone-bound paternal DNA, which is not actively demethylated in the zygote) can develop into viable pups (Polanski et al. 2008). In other mammalian species, active demethylation of the paternal genome is followed by immediate de novo remethylation before parallel, passive demethylation of the maternal and paternal genomes occurs (Fulka et al. 2004; Park et al. 2007; Abdalla et al. 2009). Thus, while active demethylation of the paternal genome is beneficial, perhaps it simply provides an additional measure to ensure efficient reprogramming. Another hypothesis is that the oocyte is programmed to remove distinguishing paternal epigenetic features, which may provide a developmental advantage to any individual embryo. It is in the “interest of the mother” to distribute resources equally among her progeny, necessitating the removal of any unique paternal epigenetic marks that would favor a particular embryo (Moore and Haig 1991). More detailed investigations addressing 5hmC and locus-specific demethylation in round spermatid injection-derived embryos, coupled with a base-resolution view of the dynamics of 5mC and its oxidation products, are required to deepen our understanding of these processes.
Passive DNA demethylation and maintenance of parental imprints
The maternal genome is, at least globally, resistant to hydroxylation by TET3 yet nonetheless loses the bulk of its oocyte-specific DNA methylation pattern by replication-mediated 5mC dilution during preimplantation development (Fig. 4A, red line). Here, passive loss of 5mC is achieved by nuclear exclusion of DNMT1 (Howell et al. 2001; Ratnam et al. 2002; Branco et al. 2008; Hirasawa et al. 2008) rather than by down-regulation of NP95, as proposed in PGCs (Kagiwada et al. 2013).
Nuclear exclusion of DNMT1, however, poses a problem for the required maintenance of genomic imprints and other sequences that must retain their DNA methylation patterns throughout development. Although zygotic deletion of DNMT1 was known to cause massive loss of DNA methylation both globally and at imprinted gene loci and to be embryonic-lethal (Li et al. 1992, 1993), the role of DNMT1 in early preimplantation embryos was only recently resolved. The oocyte-specific form of DNMT1 (DNMT1o) was shown to be required for imprint maintenance only during one cell cycle at the eight-cell stage, where DNMT1o was found to transiently translocate into the nucleus (Carlson et al. 1992; Howell et al. 2001; Ratnam et al. 2002). Because the somatic form of DNMT1 (DNMT1s) was not detected until the blastocyst stage, the maintenance of imprints during early cleavage divisions was attributed to unidentified DNA methyltransferases. Re-examination of DNMT3A, DNMT3B, and DNMT1o/s expression and function finally resolved the issue by first excluding the de novo DNA methyltransferases from general imprinting maintenance in the embryo (Hirasawa et al. 2008). Complete (maternal and zygotic) knockout of DNMT1, however, completely abolished DNA methylation at all imprinted gene loci (Cirio et al. 2008; Hirasawa et al. 2008; Kurihara et al. 2008). Thus, both DNMT1o and DNMT1s contribute to DNA methylation maintenance at imprinted regions even though the vast amount of protein is excluded from the nucleus (Branco et al. 2008).
Noncanonical targeting of DNMT1 to imprinted regions in preimplantation embryos
DNMT1 is required to maintain imprints, yet, at the same time, its nuclear protein levels are drastically reduced to allow global demethylation. How is DNMT1 targeted to imprinted gene loci? STELLA is required for imprinting maintenance. However, STELLA’s global binding and protection of the whole maternal genome from active demethylation makes it an unlikely candidate for DNA methylation maintenance at specific loci. ZFP57, a Krueppel-associated box (KRAB) domain zinc finger protein, has also been associated with imprinting maintenance (Fig. 4C; Li et al. 2008; Mackay et al. 2008). Loss of ZFP57 in mouse embryos and ESCs causes hypomethylation of both paternal and maternal ICRs and misregulation of imprinted genes (Li et al. 2008; Quenneville et al. 2011; Zuo et al. 2012). This function of ZFP57 is evolutionarily conserved; in humans, loss-of-function mutations also result in ICR hypomethylation, ultimately causing transient, neonatal diabetes (Mackay et al. 2006, 2008).
KRAB zinc finger proteins often act as epigenetic repressors through their interaction with TRIM28. TRIM28 is a component of a multifunctional repressor complex comprised of, at least in this instance, the nucleosome remodeling and histone deacetylation (NuRD) complex, the H3K9me3-catalyzing histone methyltransferase SETDB1, the heterochromatin protein 1 (HP1), and DNA methyltransferases DNMT1, DNMT3A, and DNMT3B (Fig. 4C; Schultz et al. 2001, 2002; Iyengar and Farnham 2011; Quenneville et al. 2011; Zuo et al. 2012). While the DNA methylation maintenance defects are less pronounced in zygotic ZFP57 mutants and the lack of maternal ZFP57 is rescued by expression of paternal Zfp57 (Li et al. 2008), loss of maternal Trim28 alone is embryonic-lethal (Messerschmidt et al. 2012). The times of death and the embryonic phenotypes of maternal Trim28 mutants are highly variable, as is the occurrence of hypomethylation at several maternal and paternal ICRs (Messerschmidt et al. 2012). DNA methylation analysis of individual blastomeres confirmed that stochastic, random methylation defects (and phenotypes) are based on the mosaic composition of early Trim28 maternal null embryos carrying normally and aberrantly imprinted gene loci (Messerschmidt 2012; Messerschmidt et al. 2012; Lorthongpanich et al. 2013).
Binding of both proteins and the presence of H3K9me3, the product of the TRIM28 complex, were detected at imprinted loci in embryos (Messerschmidt et al. 2012) and mESCs (Quenneville et al. 2011). Being a DNA-binding transcription factor, ZFP57 localization to ICRs opens the exciting prospect of sequence-specific recognition and maintenance of imprinted loci. Indeed, sequence analysis of loci enriched for H3K9me3, TRIM28, and ZFP57, as identified by chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) in mESCs, revealed a hexanucleotide consensus ZFP57 recognition site (TGCCGC), which is highly conserved in 81 of 91 identified (H3K9me3/TRIM28/ZFP57) sites (Quenneville et al. 2011). Remarkably, binding of TRIM28 to the hypomethylated sites was abrogated in Trim28 maternal null embryos and was not restored by TRIM28 re-expression from the paternal allele in the two- to four-cell stage embryo, indicating the DNA methylation-dependent binding of the ZFP57/TRIM28 complex. In fact, ZFP57 displays much higher binding affinity for its methylated consensus binding site in vitro (Quenneville et al. 2011; Liu et al. 2012). Once lost, DNA methylation cannot be restored via the ZFP57/TRIM28 complex in vivo (Messerschmidt et al. 2012) or ectopic re-expression of ZFP57 in Zfp57-deficient ESCs (Zuo et al. 2012). To a point, the interaction of TRIM28 with DNMT1/NP95 suggests the noncanonical, ZFP57-mediated targeting of the DNA methylation maintenance machinery to ICRs in the preimplantation embryo (Fig. 4C). This targeting mode would compensate for the drastic reduction of nuclear DNMT1 during early cleavage divisions and thus enable DNA methylation maintenance at imprinted regions (Messerschmidt 2012). Only at later embryonic stages, after DNMT1 levels are significantly increased in the nuclei, does canonical DNA methylation maintenance ensue (Fig. 4D).
The maintenance of methylation at imprinted regions in preimplantation embryos was thought to be very robust until genome-wide methylation analysis showed that even ICRs are partially demethylated, particularly at peripheral regions (Tomizawa et al. 2011; Kobayashi et al. 2012). Such loss, however, would be of little to no consequence as long as the ZFP57-binding site itself remains methylated and targeted by the ZFP57/TRIM28 complex. De novo methylation enzymes DNMT3A and DNMT3B, which are also found in the ZFP57/TRIM28 complex (Quenneville et al. 2011; Zuo et al. 2012), may account for the recovery of DNA methylation at these peripheral regions at later stages of development (Tomizawa et al. 2011; Kobayashi et al. 2012).
Finally, given that DNA demethylation mechanisms act during early embryonic development, a drastic reduction of methylation levels is expected and in fact found at later preimplantation stages. Surprisingly, in addition to imprinted genes and retrotransposons, a substantial quantity of differentially methylated CGIs in oocyte-specific and a subset of sperm-specific methylated regions retain DNA methylation at much higher levels than expected if unhindered passive demethylation were to take place (Smallwood et al. 2011; Kobayashi et al. 2012). It remains to be seen whether these CGIs, as has been shown for ICRs, are targeted and protected by ZFP57/TRIM28, other KRAB zinc finger proteins, or entirely different mechanisms.
Concluding remarks
The veritable flood of new, genome-wide methylation data and insights into epigenetic reprogramming of embryos and PGCs allows us to view this process from a new angle. It is now clear that passive DNA demethylation is the most parsimonious mechanism of both PGCs and preimplantation embryos and is probably sufficient for that purpose. Moreover, the exact time and concentration of gene products at specific sites in the genome are now clarified. Nevertheless, these new insights also accentuate the limitations of our current understanding of the reprogramming process. Although global, genome-wide methylation and demethylation pathways have been identified, their disruption produces embryos with less than fully penetrant phenotypes, leading to confounding and confusing interpretations (Li et al. 2008; Popp et al. 2010; Dawlaty et al. 2011, 2013; Gu et al. 2011; Messerschmidt et al. 2012; Yamaguchi et al. 2012). Loss of TET3 in the early embryo, for instance, causes developmental arrest in only a subset of embryos, which is likely the result of delayed activation of the paternal pluripotency genes (Gu et al. 2011). Nonetheless, in wild-type embryos, the bulk of paternal 5mC undergoes conversion to 5hmC, which is then passively removed over subsequent cleavages. However, TET3-independent passive demethylation also occurs in the maternal genome, thus calling into question the significance of the conversion of the global paternal genome. Similarly, TET1/2-mediated oxidation in PGCs might have a broader genomic range than merely to ensure robust demethylation of meiotic and imprinted genes. It is therefore prudent not to assume active reversion to unmodified cytosine if TET-mediated oxidation is involved; divergent biological function and relevance of 5mC and 5hmC must also be considered.
We are far from understanding active DNA demethylation. The direct removal of 5mC or its deamination via AID/APOBECs has still not been conclusively addressed in vivo. TET-dependent demethylation can occur via multiple, interconnected pathways (Wu and Zhang 2010, 2014). However, even in the absence of TETs, AID, APOBECs, etc., PGCs and early embryos can develop into functional gametes or give rise to live pups, although often inefficiently. The long-term transgenerational effects of epigenetic defects in these surviving pups have yet to be addressed. Therefore, it appears that several redundant, fail-safe proofing mechanisms have evolved to act in parallel to ensure the profound erasure of epigenetic marks. Future experiments addressing these redundancies will determine whether the enzymes mediating active demethylation do indeed act in separate, parallel pathways or upstream of or downstream from one another.
With new experimental data, more parallels between PGC and embryo reprogramming have begun to manifest themselves. The protection of imprinted regions in the embryo is demonstrated in PGCs, where imprinted and meiotic genes appear to be specifically targeted for demethylation after being excluded from the wave of passive demethylation. Whether this delay is of functional significance or merely a byproduct of the imprint-specific maintenance mode evolved in embryos yet impacting on PGCs remains to be seen. Importantly, demethylation of imprinted regions following the same kinetics as that of the bulk of the genome has also been observed (Kagiwada et al. 2013), and even in the absence of TET enzymes, meiotic and imprinted genes are eventually reprogrammed (Dawlaty et al. 2013). This is in clear contrast to the case of the early embryo, where it is vital that DNA methylation maintenance of imprints is never overcome.
The novel concept of noncanonical DNA methylation maintenance in the face of reprogramming is now well established during preimplantation development. ZFP57/TRIM28-mediated targeting of scarce nuclear DNMT1 protein to imprinted gene regions ensures their faithful maintenance (Messerschmidt 2012). Are genes such as Trim28 and Zfp57 also expressed in PGCs, and do they influence the demethylation dynamics of these regions? Preliminary findings suggest that ZFP57-binding sites are enriched in “late demethylating” genomic regions (Seisenberger et al. 2012). Also, in preimplantation embryos, DNA methylation maintenance may go beyond imprinting. Many other transiently, differentially methylated regions detected in oocytes are protected through the blastocyst stage (Smallwood et al. 2011; Kobayashi et al. 2012; Smith et al. 2012). Furthermore, ChIP-seq in mESCs has identified numerous additional ZFP57 targets in addition to known imprinted regions. These findings offer an exciting prospect that merits closer scrutiny (Quenneville et al. 2011). Although ZFP57 is the only TRIM28-interacting KRAB zinc finger protein known to display methylation-dependent DNA binding, it is possible that other KRAB family members with similar properties mediate analogous functions at different target sites.
The overall question is: Why do mammals exhibit these complex epigenetic reprogramming processes in the first place? Does the goal of totipotency, which entails such developmental flexibility, offset the lack of guiding, developmental determinants found in frogs, flies, fish, or worms? Did these processes evolve to enable and preserve genomic imprinting as an essential mechanism for mammalian development? Epigenetic reprogramming differs in details among mammalian species, suggesting that demethylation–methylation in PGCs and subsequent demethylation–methylation in the embryo are novel mechanisms and that we are witnessing the evolutionary selection of the optimal one.
Acknowledgments
We thank Heike Wollmann for insightful discussions, valuable input, and proofreading our manuscript.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.234294.113.
References
- Abdalla H, Yoshizawa Y, Hochi S 2009. Active demethylation of paternal genome in mammalian zygotes. J Reprod Dev 55: 356–360 [DOI] [PubMed] [Google Scholar]
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S 2011. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol 30: 267–275 [DOI] [PubMed] [Google Scholar]
- Aran D, Sabato S, Hellman A 2013. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol 14: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, et al. 2012. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet 8: e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS 2009. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 23: 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH 1988. Cloning of a mammalian DNA methyltransferase. Gene 74: 9–12 [DOI] [PubMed] [Google Scholar]
- Bestor TH 2000. The DNA methyltransferases of mammals. Hum Mol Genet 9: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nature Publishing Group 42: 1093–1100 [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève P-O, Clark A, Pradhan S, Jacobsen SE 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539 [DOI] [PubMed] [Google Scholar]
- Branco MR, Oda M, Reik W 2008. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev 22: 1567–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371: 435–438 [DOI] [PubMed] [Google Scholar]
- Carlson LL, Page AW, Bestor TH 1992. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev 6: 2536–2541 [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, Lieber MR, Hsieh C-L 2002. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci 99: 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Li E 2004. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol 60: 55–89 [DOI] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A 2008. X chromosome activity in mouse XX primordial germ cells. PLoS Genet 4: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR 2008. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NM, Kenigsberg E, Tanay A 2011. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell 145: 773–786 [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS 2000. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9: 2885–2894 [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu Y-C, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi S-W, Page DC, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. 2013. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24: 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Chaillet JR 2002. In vivo stabilization of the Dnmt1 (cytosine-5)- methyltransferase protein. Proc Natl Acad Sci 99: 14861–14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia Z-B, Stuart T, Diaz MO, Bushweller JH, Zeleznik-Le NJ 2008. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci 105: 7517–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan C-F, Andrews S, Dean W, Hemberger M, Reik W 2008. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet 4: e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W 2010. Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith AC 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12: 565–575 [DOI] [PubMed] [Google Scholar]
- Fulka H, Mrazek M, Tepla O, Fulka J 2004. DNA methylation pattern in human zygotes and developing embryos. Reproduction 128: 703–708 [DOI] [PubMed] [Google Scholar]
- Gaudet F, Talbot D, Leonhardt H, Jaenisch R 1998. A short DNA methyltransferase isoform restores methylation in vivo. J Biol Chem 273: 32725–32729 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, Golic KG, Jacobsen SE, Bestor TH 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Valle Della G, Robertson KD 2009. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18: 3178–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A 2005. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem 280: 13341–13348 [DOI] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie Z-G, Shi L, He X, Jin S-G, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Guibert S, Weber M 2012. Epigenetics: erase for a new start. Nature 492: 363–364 [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P 2010. Epigenetic reprogramming—taking a lesson from the embryo. Curr Opin Cell Biol 22: 342–350 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X 2012. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40: 4841–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129: 1983–1993 [DOI] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SMC, Surani MA 2007. Germ cell specification in mice. Science 316: 394–396 [DOI] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Chess A 2007. Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P 2009. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet 18: 3375–3383 [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H 2008. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt MM, Wu TL, Cohen G, Linn S 1988. De novo and maintenance DNA methylation by a mouse plasmacytoma cell DNA methyltransferase. J Biol Chem 263: 4392–4399 [PubMed] [Google Scholar]
- Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, et al. 2009. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res 19: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE 1975. DNA modification mechanisms and gene activity during development. Science 187: 226–232 [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104: 829–838 [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y 2011. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y 2011. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21: 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin S-G, Pfeifer GP, Szabó PE 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci 108: 3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ 2011. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449: 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, Fields CR, Delmas AL, Liu X, Qiu J, et al. 2008. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet 17: 690–709 [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M 2013. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32: 340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903 [DOI] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280 [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R 2013. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 368: 20110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa S, Murata T, Ugai H, Yamazaki T, Yokoyama KK 2003. Control elements of Dnmt1 gene are regulated in cell-cycle dependent manner. Nucleic Acids Res 3: 307–308 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8: e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, et al. 2013. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res 23: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Rao A 2013. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr Opin Cell Biol 25: 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota SK, Feil R 2010. Epigenetic transitionsin germ cell development and meiosis. Dev Cell 19: 675–686 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kawamura Y, Uchijima Y, Amamo T, Kobayashi H, Asano T, Kurihara H 2008. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev Biol 313: 335–346 [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M 2008. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 22: 1617–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F 2002. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129: 1807–1817 [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, Li E 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71: 865–873 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R 1993. Role for DNA methylation in genomic imprinting. Nature 366: 362–365 [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai C-L, Wang J, Zhang W, Petersen BE, Yang F-C, Xu M 2011. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097 [DOI] [PubMed] [Google Scholar]
- Liu Y, Toh H, Sasaki H, Zhang X, Cheng X 2012. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev 26: 2374–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM 2013. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341: 1110–1112 [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Kellam LD, Mann MRW, Lei H, Li E, Bartolomei MS, Resnick JL 2006. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 133: 3411–3418 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJG, Boonen SE, Clayton-Smith J, Goodship J, Hahnemann JMD, Kant SG, Njølstad PR, Robin NH, Robinson DO, Siebert R, et al. 2006. A maternal hypomethylation syndrome presenting as transient neonatal diabetes mellitus. Hum Genet 120: 262–269 [DOI] [PubMed] [Google Scholar]
- Mackay DJG, Callaway JLA, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40: 949–951 [DOI] [PubMed] [Google Scholar]
- Maiti A, Drohat AC 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T 2000. Demethylation of the zygotic paternal genome. Nature 403: 501–502 [DOI] [PubMed] [Google Scholar]
- McLaren A 1984. Meiosis and differentiation of mouse germ cells. Symp Soc Exp Biol 38: 7–23 [PubMed] [Google Scholar]
- McLaren A, Lawson KA 2005. How is the mouse germ-cell lineage established? Differentiation 73: 435–437 [DOI] [PubMed] [Google Scholar]
- Meissner A 2011. Guiding DNA methylation. Cell Stem Cell 9: 388–390 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH 1998. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development 125: 889–897 [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM 2012. Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes. Epigenetics 7: 969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB 2012. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335: 1499–1502 [DOI] [PubMed] [Google Scholar]
- Mikl MC, Watt IN, Lu M, Reik W, Davies SL, Neuberger MS, Rada C 2005. Mice deficient in APOBEC2 and APOBEC3. Mol Cell Biol 25: 7270–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem 279: 52353–52360 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2006. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu Y-J, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T 2012. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486: 415–419 [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Rada C 2007. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J Exp Med 204: 7–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213 [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E 1998a. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220 [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E 1998b. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res 26: 2536–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Park JS, Jeong YS, Shin ST, Lee K-K, Kang Y-K 2007. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev Dyn 236: 2523–2533 [DOI] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JPC, Zahn D, Colledge WH, Carlton MBL, Nakano T, Surani MA 2003. Stella is a maternal effect gene required for normal early development in mice. Curr Biol 13: 2110–2117 [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- Polanski Z, Motosugi N, Tsurumi C, Hiiragi T, Hoffmann S 2008. Hypomethylation of paternal DNA in the late mouse zygote is not essential for development. Int J Dev Biol 52: 295–298 [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, Roberts RJ 1999. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem 274: 33002–33010 [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronné L, Valle Della V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern M-H, et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38 [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci 97: 5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam S, Mertineit C, Ding F, Howell CY, Clarke HJ, Bestor TH, Chaillet JR, Trasler JM 2002. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev Biol 245: 304–314 [DOI] [PubMed] [Google Scholar]
- Riggs AD 1975. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14: 9–25 [DOI] [PubMed] [Google Scholar]
- Rouleau J, Tanigawa G, Szyf M 1992. The mouse DNA methyltransferase 5′-region. A unique housekeeping gene promoter. J Biol Chem 267: 7368–7377 [PubMed] [Google Scholar]
- Ruchirawat M, Noshari J, Lapeyre JN 1987. Kinetic mechanisms and interaction of rat liver DNA methyltransferase with defined DNA substrates. Mol Cell Biochem 76: 45–54 [DOI] [PubMed] [Google Scholar]
- Saitou M, Yamaji M 2012. Primordial germ cells in mice. Cold Spring Harb Perspect Biol 4: a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, Jones RH, Marquez VE, Cairns W, Tadayyon M, et al. 2011. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci 108: 5449–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Dean W 2004. Epigenetic reprogramming during early development in mammals. Reproduction 127: 643–651 [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241: 172–182 [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W 2005. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 280: 225–236 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y 2008. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 2008: 129–140 [DOI] [PubMed] [Google Scholar]
- Schiesser SS, Hackner BB, Pfaffeneder TT, Müller MM, Hagemeier CC, Truss MM, Carell TT 2012. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew Chem Int Ed Engl 51: 6516–6520 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458 [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S-I, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa J-PJ 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung H-L, Zhang K, Zhang Y 2013. Genome-wide analysis revealsTET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G 2012. De novo DNA methylation: a germ cell perspective. Trends Genet 28: 33–42 [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S-I, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G 2011. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43: 811–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A 2012. A unique regulatory phase of DNAmethylation in the early mammalianembryo. Nature 484: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H 2009. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol 16: 564–571 [DOI] [PubMed] [Google Scholar]
- Strogantsev R, Ferguson-Smith AC 2012. Proteins involved in establishment and maintenance of imprinted methylation marks. Brief Funct Genomics 11: 227–239 [DOI] [PubMed] [Google Scholar]
- Strome S, Lehmann R 2007. Germ versus soma decisions: lessons from flies and worms. Science 316: 392–393 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Snow MH 1981. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J Embryol Exp Morphol 64: 133–147 [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr ARW, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S-I, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H 2011. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC 2007. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 67: 946–950 [DOI] [PubMed] [Google Scholar]
- Vincent JJ, Huang Y, Chen P-Y, Feng S, Calvopiña JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, et al. 2013. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell 12: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Watanabe D, Suetake I, Tada T, Tajima S 2002. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev 118: 187–190 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, Schöler H, Walter J 2010. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J 29: 1877–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J 2011. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Comms 2: 241. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y 2010. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156: 45–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Péquignot E 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402: 187–191 [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M 2006. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod 75: 705–716 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y 2012. Tet1 controls meiosis by regulating meiotic gene expression. Nature 492: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, Zhang Y 2013. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res 23: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y 2014. Role of Tet1 in erasure of genomic imprinting. Nature 504: 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol 270: 385–395 [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. 2011. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 7: e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau H-T, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. 2012. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem 287: 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]