Abstract
Little is known on how β-barrel proteins are assembled in the outer membrane (OM) of Gram-negative bacteria. SurA has been proposed to be the primary chaperone escorting the bulk mass of OM proteins across the periplasm. However, the impact of SurA deletion on the global OM proteome has not been determined, limiting therefore our understanding of SurA’s function. By using a differential proteomics approach based on 2D-LC-MSn, we compared the relative abundance of 64 OM proteins, including 23 β-barrel proteins, in wild-type and surA strains. Unexpectedly, we found that the loss of SurA affects the abundance of 8 β-barrel proteins. Of all the decreased proteins, FhuA and LptD are the only two for which the decreased protein abundance cannot be attributed, at least in part, to decreased mRNA levels in the surA strain. In the case of LptD, an essential protein involved in OM biogenesis, our data support a role for SurA in the assembly of this protein and suggest that LptD is a true SurA substrate. Based on our results, we propose a revised model in which only a subset of OM proteins depends on SurA for proper folding and insertion in the OM.
Keywords: beta-barrel, periplasm, proteomics, NmpC, Imp
1 Introduction
The outer membrane (OM) of Gram-negative bacteria is a permeability barrier that is essential for their viability and protects them against antimicrobial drugs (reviewed in [1]). The OM is a unique asymmetric lipid bilayer with phospholipids forming the inner leaflet and lipopolysaccharides (LPS) forming the outer leaflet. There are two major groups of proteins present in the OM, lipoproteins that are present in the periplasm but are anchored by a lipid moiety to the inner leaflet of the OM and β-barrel proteins that are integral membrane proteins (OMP). All the components of the OM are synthesized in the cytoplasm or in the inner membrane. After transport across the inner membrane (IM), they have to travel through the periplasm to reach the OM.
Whereas we do not know how phospholipids are transported to the OM, several factors required for LPS transport to the cell surface have recently been identified [2–6]. Regarding the OM proteins, we have a good understanding of the mechanisms of lipoprotein transport and insertion (reviewed in [7]). Although factors necessary for OMP assembly have been identified, the mechanisms by which unfolded β-barrel proteins are transported and finally inserted in the OM are poorly understood.
After their synthesis in the cytoplasm, integral β-barrel proteins bind to the chaperone SecB and are then translocated unfolded across the IM by the Sec machinery [8]. After cleavage of the signal sequence by a leader peptidase, β-barrel proteins are transported across the periplasm and are delivered to an OM assembly complex composed of an essential β-barrel protein, BamA (formerly known as YaeT), and 4 lipoproteins (BamBCDE, formerly known as YfgL, NlpB, YfiO and SmpA, respectively) [9, 10]. Two models have been proposed to explain how unfolded proteins are transported across the hydrophilic environment of the periplasm. The first model is controversial; it suggests that transport takes place at contact sites between the IM and the OM. These contact sites were observed about 40 years ago by Bayer [11] but Kellenberger [12] challenged their existence and proposed that they were an artifact of the fixation technique used by Bayer. The second model, which is more widely accepted, proposes that unfolded proteins are escorted across the periplasm by soluble periplasmic chaperones. In agreement with this model, several proteins that assist in the folding of secreted proteins have been identified in the periplasm [13] [14]. One of them is SurA, a protein whose main function in the periplasm is that of a chaperone [13] [15]. Accordingly, strains lacking SurA exhibit defects that are indicative of OM perturbations [16]. They are hypersensitive to detergents and hydrophobic antibiotics [16, 17] and have a decreased OM density compared to the wild type due to lower levels of several OMPs including OmpA, LamB, OmpF and OmpC [16–18].
The synthesis of many envelope biogenesis factors is controlled by σE, a transcription factor that is induced under envelope stress conditions [19]. It has been shown that the σE stress response is induced in surA strains [16, 20], which leads to the downregulation of the mRNAs coding for several OMPs including OmpF, OmpA and OmpC [21]. For these proteins, it is therefore difficult to discriminate between the effects of SurA’s absence on folding or synthesis. To overcome this problem, Sklar et al. constructed chromosomal depletion strains that allowed them to deplete SurA fast enough to prevent the synthesis defects [18]. This study showed that SurA depletion leads to a marked decrease in OM density, whereas loss of the two other periplasmic chaperones Skp or DegP had no effect. On the basis of these results, these authors proposed that SurA is the primary chaperone responsible for the periplasmic transit of the bulk mass of OMPs.
Our current understanding of SurA’s function is based on studies performed on a few OM proteins for which antibodies are available, such as OmpA, OmpF and LamB. Although these proteins are among the most abundant in the OM, they only represent a small percentage of all the OMPs. In order to determine how global the function of SurA is, it is therefore essential to study the impact of surA deletion on the OM proteome.
We compared the relative abundance of OMPs in wild-type and surA strains by using a label-free differential proteomics approach based on 2D-LC-MS/MS. Our approach allowed us to compare the relative abundance of 64 OM proteins, including 23 β-barrel proteins. We found that 8 of the 23 identified β-barrel proteins were negatively affected by the loss of SurA. Given the dramatic decrease in OM density in surA mutants, this was unexpected. The decreased OMPs include FadL, LptD, FhuA, OmpX, FecA as well as the major OMPs OmpA, OmpF and LamB. The decreased abundance of the three latter proteins in the surA strain has been well documented [16, 18], which validates our proteomics approach. Furthermore, since these three proteins constitute a large portion of the bulk mass of OMPs, we reason that the decrease in OM density reported in cells lacking SurA [16–18] is caused by reduction in these major OMPs. Remarkably, of all the decreased proteins, FhuA and LptD are the only two for which the decrease in protein abundance cannot be due, at least in part, to lower mRNA levels. Moreover, we show that SurA likely plays a direct role in the assembly of LptD. Altogether our data indicate that the biogenesis of LptD, and possibly FhuA, differs from that of most OMPs in that it is highly dependent on SurA, suggesting that they are true SurA substrates.
2 Material and methods
2.1 Bacterial strains and growth conditions
The bacterial strains used in this study are derivatives of MC4100 [22]. All alleles were moved by P1 transduction using standard procedures (Silhavy et al., 1984). Strain CB47 was constructed by introducing the ΔsurA::kan allele from the Keio collection [23] into MC4100. The InCh surA allele from MB292 [18] was introduced into NR754, an ara+ revertant of MC4100 [24]. The resulting strain, NR765, was transduced with the λRS88 lptDsurA’-’lacZ fusion by P1 transduction using MB50 as a donor and selecting for growth on minimal lactose plates supplemented with vitamin B6. An Ara+ transductant, named NR778, was used for β-galactosidase assays. Plasmids pBAD18 [25] and its derivative pBAD18LptD (Silhavy laboratory collection) carrying lptD under the control of the PBAD promoter were introduced into NR756, an ara+ revertant of MC4100 that carries surA::kan allele from AR208 [26]. Unless indicated, all experiments were done by growing cells in LB broth. When appropriate, media was supplemented with kanamycin (25 μg/m), ampicillin at concentrations of 25 μg/ml for strains NR765 and NR778 and 125 μg/ml for plasmid maintenance, or lactose or arabinose [0.2% (w/v)].
2.2 β-Galactosidase assays
Cultures were grown overnight in LB broth at 37°C in the presence or absence of arabinose. β-galactosidase assays were performed using a microtiter plate assay as described previously [24]. The β-galactosidase activities are expressed as Δ(OD420/time)/(OD600 × volume), where volume refers to the amount (in ml) of cell suspension used. For each experiment, every sample was assayed three times and the average activity and standard deviation (SD) are shown. The data presented were derived from a single experiment representative of at least three independent experiments.
2.3 Immunoblotting
1-ml samples from cultures were pelleted (16,000 × g, 2 min). To standardize samples, pellets were resuspended in a volume (ml) of SDS sample buffer equal to OD600/10. Samples were boiled for 10 min and equal volumes were subjected to electrophoresis. We used 10% SDS-PAGE and immunoblotting was performed using LptD (1:7,000 dilution), LamB, and OmpA (1:30,000 dilution) rabbit polyclonal sera as described previously [27]. The intensity of each band in the immunoblot was quantified using ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2008). The band corresponding to the 55 KDa protein, which is recognized by the LptD antiserum, was used as the internal standard for each lane. The relative values for each protein are presented as the intensity value of each band divided by the intensity value of the 55 KDa protein band.
2.4 Preparation of outer membrane fraction and proteolytic digestion
MC4100 and CB47 cells were grown aerobically in LB media, at 37°C to an A600 of 0.8. Envelope proteins were extracted by using the osmotic shock procedure developed by [28] and OM proteins isolated by ultracentrifugation at 100,000 g for 1 h. Protein concentration was determined using the Bradford assay. 150 μg of outer membrane proteins were then precipitated by adding trichloroacetic acid (TCA) to a final concentration of 10% w/v, followed by incubation on ice for 30 min. Samples were then centrifuged at 14,000 rpm for 20 min and the resulting pellets washed with 5% ice cold TCA. The pellets were then resuspended in 100 μl of denaturing buffer (6M urea, 200 mM Tris-HCl pH 8.5, 10 mM EDTA) supplemented with 10 mM DTT. After a 1 h incubation at 25°C, denaturing buffer (100 μl) supplemented with 100 mM iodoacetamide was added to titrate out the remaining DTT and alkylate all reduced cysteines. The reaction was stopped by addition of 10 % TCA and the proteins collected by centrifugation. The resulting pellet was successively washed with 10% TCA and ice cold acetone, dried in a Speedvac and resuspended by sonication in 50 μl 0.1 M NH4HCO3 pH 8.0 containing 0.2 % (w/v) RapiGest (Waters). The samples were then diluted two fold with 50 mM NH4HCO3 pH 8.0 containing 3 μg sequencing grade trypsin, and digested overnight at 30°C. The acido-labile detergent was then removed by acidification with HCl according to the manufacturer’s instructions and the samples were stored at −20°C.
2.5 Differential analysis of periplasmic proteins by label-free 2D-LC-MS/MS
Peptides were loaded onto a strong cation exchange column GROM-SIL 100 SCX (100 × 2 mm, GROM, Rottenburg, Germany) equilibrated with solvent A (5% acetonitrile v/v, 0.05% v/v formic acid pH 2.5 in water) and connected to an Agilent 1100 HPLC system. Peptides were separated using a 50 min elution gradient that consisted of 0%–50% solvent B (5% acetonitrile v/v, 1 M ammonium formate adjusted to pH 3.5 with formic acid in water) at a flow rate of 200 μl/min. Absorbance was monitored at 280 nm to ensure that all samples contained similar amount of material. Fractions were collected at 2 min intervals (20 in total) and dried using a Speedvac. Peptides were resuspended in 10 μl of solvent C (5% acetonitrile v/v, 0.01% v/v TFA in water) and analyzed by LC-MS/MS as described below.
The LC-MS/MS system consisted of an LCQ DECA XP Plus ion trap mass spectrometer (ThermoFisher, San José, CA, USA) equipped with a microflow electrospray ionization source and interfaced to an LCPackings Ultimate Plus Dual gradient pump, Switchos column switching device, and Famos Autosampler (Dionex, Amsterdam, Netherlands). Two reverse phase peptide traps C18 Pepmap 100 Dionex (0.30 mm × 5 mm) were used in parallel with two analytical BioBasic-C18 columns from ThermoFisher (0.18 mm × 150 mm). Samples (6.5 μl) were injected and desalted on the peptide trap equilibrated with solvent C at a flow rate of 30 μl/min. After valve switching, peptides were eluted in backflush mode from the trap onto the analytical column equilibrated in solvent D (5% acetonitrile v/v, 0.05% v/v formic acid in water) and separated using a 100 min gradient from 0% to 70% solvent E (80% acetonitrile v/v, 0.05% formic acid in water) at a flow rate of 1.5 μl/min.
The mass spectrometer in positive mode was set up to acquire one full MS scan in the mass range of 400–2000 m/z, followed by three MS/MS spectra of the three most intense peaks in the mass range 400–1500 m/z. The dynamic exclusion feature was enabled to obtain MS/MS spectra on coeluting peptides, and the exclusion time was set at 2 min.
2.6 Protein identification and quantification
Raw data collection of approximately 55.000 MS/MS spectra per 2D-LC-MS/MS experiment was followed by protein identification using Sequest. In details, peak lists were generated using extract-msn (version 3.0 ThermoFisher) in Bioworks 3.2. From raw files, MS/MS spectra were exported as individuals files in .dta format with the following settings: peptide mass range: 400–3500 Da, minimal total ion intensity 50000, minimal number of fragment ions: 15, precursor mass tolerance: 1.4 Da, group scan: 25, group count: 1. The resulting peak lists were searched against a target-decoy E. coli protein database (release 18.01.2007, 8690 entries comprising forward and reversed sequences) obtained from Swissprot (ftp://ftp.expasy.org/databases/complete_proteomes/fasta/) using TurboSequest (version 27 revision 12 within BioWorks 3.2) by comparison with the theoretical spectra of all possible peptides fragments from the target-decoy database. The following parameters were used: trypsin was selected with proteolytic cleavage only after arginine and lysine but not at Arg/Pro or Lys/Pro, number of internal cleavage sites was set to 1, mass tolerance for precursor and fragment ions was 1.2 Da and 1.1 Da, respectively, considered modifications were +15.99 Da for oxydized methionine and +57.02 Da for carboxyamidomethylcysteine. Peptide matches were filtered using the probability score calculated by TurboSequest and to charge-state versus cross-correlation scores (Xcorr) ensuring an estimated false positive rate below 5% calculated by target-decoy database searching. The filtered Sequest output files for each peptide were grouped according to the protein from which they were derived using the multiconsensus results tool within BioWorks. The analysis was repeated on four biological replicates for each strain. Sampling statistics such as unique peptides, spectral counts and sequence coverage were exported in Microsoft Excel spreadsheets. The spectral counts data were normalized by dividing the protein spectral count in a particular experiment by the average spectral count across all the proteins in that experiment. Relative quantification of protein abundance was estimated by calculating the ratio of normalized spectral counts and statistical significance was tested with the unpaired Student’s t-test and significance was defined as a P < 0.05 (two-tail two-sample equal variance test). Statistical analysis was performed using the Microsoft Excel spreadsheet software.
2.7 RNA extraction and RT-qPCR
For the extraction of total RNA, 20 ml of culture were collected by centrifugation. RNA was isolated using Trizol (Invitrogen) and purified with the RNeasy kit (Qiagen). Following elution, nucleic acid concentrations were determined by spectrophotometry and residual DNA contamination was removed by incubating the samples with TURBO DNase (Ambion) according to the manufacturer’s protocol. After DNase inactivation (phenol-chloroform extraction), the RNA was recovered, quantified, and used as a template for PCR to confirm the degradation of contaminating DNA. RNA was then used for cDNA synthesis by using M-MuLV Reverse Transcriptase (Fermentas). For cDNA synthesis, the manufacturer’s protocol was followed, starting with 0.7 μg total RNA template and 0.2 μg random hexamer. RNA transcripts were quantified on an icycler real-time PCR machine (Biorad, Hercules, CA) by using Brilliant SYBR green QPCR mix (Eurogentec) in 25 μl volumes containing 300 ng cDNA. Real-time PCR oligonucleotide primers were designed using the online Primer3 internet based interface (http://frodo.wi.mit.edu) (Table 4). The final concentration of each primers was 3 μM. The cycling conditions for all amplicons were 2 min at 50°C (UNG activation), 10 min at 95°C (Hot Goldstar DNA polymerase activation), followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Melting curve analysis was performed at the end of the amplification using 0.5°C increasing steps for 70 cycles. Cycle threshold (Ct) values were determined after automatic adjustment of the baseline. For comparative, quantitative analysis, transcript levels were normalized to the level of gapA (glyceraldehyde 3-phosphate dehydrogenase A) and changes were determined. The comparative quantitation method (ΔΔCt) was used to compare the different strains and transformed to absolute values with 2−ΔΔCt for obtaining relative fold changes [29]. All the assays were performed in triplicates.
Table 4.
Oligonucleotides primers for PCR and quantitative real time PCR
| Gene | Sequencea
|
|
|---|---|---|
| Forward | Reverse | |
| insH-nmpC | AGTCATCAACTTACCTTCGC | AGCGGGGAAATTCTTCTCGG |
|
| ||
| gapA | AGGTCTGATGACCACCGTTC | GGAACGCCATACCAGTCAGT |
| lptD | TTAGGCAGCGTAGCCTGAAT | GACGACAAAACGGGTTCACT |
| fecA | ACCAGATTGCCGTAGGTGTC | ACGACTCCAACCAGACCAAC |
| fhuA | AAACGGGCGTTTATGTTCAG | TACCATCCTTCCCAACTTGC |
| fadL | GCCTGAAATGTGGGAAGTGT | CGGTACGGAAGGTCCAGTTA |
Primer sequences are listed from 5′ to 3′.
3 Results
3.1 Proteomic analysis of OM proteins
The E. coli OM contains more than 50 different β-barrel proteins [30], including some, such as BamA (previously YaeT) and LptD (previously Imp), that are essential for OM biogenesis and E. coli’s viability. Until now, due to the limited number of available antibodies and the notoriously difficult quantitative analysis of membrane proteins by 2D-gel electrophoresis, the impact of SurA’s deletion on the global OM proteome has not been clear; the number of OMPs that depend on SurA for their biogenesis was not known. In order to determine if SurA plays a global role in the cell envelope by assisting the folding of many OMPs, we used a differential proteomics approach based on label-free two-dimensional liquid chromatography tandem mass spectrometry analysis (2D-LC-MS/MS) to compare the OM proteome of surA and wild-type strains.
2D-LC-MS/MS allows a global, differential and semi-quantitative analysis of protein expression ratios. Compared to 2D-gels, it offers the advantage of dealing with peptides instead of proteins, which overcomes some of the technical difficulties linked to OMP migration in gels. Importantly, we recently showed that label-free 2D-LC-MS/MS can be reliably used to analyze the protein content of the E. coli cell envelope [31] and of Trypanosoma brucei glycosomes [32].
We prepared extracts containing cell envelope proteins using the protocol developed by Hiniker and Bardwell [28]. The OMPs were then isolated by ultracentrifugation and precipitated. The proteins were solubilized and denatured in the presence of an acid-labile surfactant before digestion with trypsin to improve the efficiency of the proteolytic digestion. Removal of the surfactant by acid hydrolysis eliminates surfactant-caused interference in the analysis of peptides by 2D-LC-MS/MS. The experiments were repeated on four independent cultures for both surA and wild-type strains. In total, 160 raw files containing 360,000 MS/MS spectra were submitted to a database search using TurboSequest against a target-decoy E. coli database obtained from SwissProt. Results were filtered using charge state versus cross-correlation scores (Xcorr) and peptide probabilities to obtain a false positive rate below 5 % calculated according to the formula (FPR= 2× false positives (FP)/(false positives (FP) + true positives (TP)). These results have been collected as supporting information (Supplementary Table 2).
An average of 186 ± 54 and 244 ± 44 proteins were identified for the wild-type and surA strain, respectively. Among them, each run allowed us to identify up to 91 OM proteins (lipoproteins and β-barrel proteins). The remaining proteins were soluble cytoplasmic and periplasmic proteins that contaminated the OM fraction. Of all the identified proteins, only 64 OM proteins that could be identified in at least three runs for either strain were kept for further analysis. These proteins include 23 β-barrel proteins and 41 OM lipoproteins (Supplementary Table 1). This represents ≈ 46 % and ≈ 40 % of all the predicted β-barrel proteins and lipoproteins, respectively [30].
The reproducibility of the 2D-LC-MS/MS approach was good, as attested by the fact that spectral count (SC) values of most proteins are similar in all four independent analyses of the same strain (Supplementary Table 1). The number of SC for a protein is the total number of MS/MS spectra taken on peptides from this protein in a given 2D-LC-MS/MS analysis. The average spectral count across all the proteins was 17.3 ± 2.4 and 16.1 ± 3.6 for wild-type and surA strain, respectively, which denotes a similar sampling rate between samples from the two different strains. We used these values for normalization (see Supplementary Table 1). Moreover, we observed that the total number of spectral counts reported for the OM proteins was similar for the wild-type and surA strains (see Supplementary Table 1), which confirms that the amount of OM material used for the proteomic analysis was similar. We noticed that the SC values from the third analysis of the surA strain (SurA-3, Supplementary Table 1) were lower than those obtained for the other three analyses, likely a result of sample loss during preparation, since the overall amount of peptides that eluted from the SCX column was lower. Nevertheless, we included this sample in our statistical analysis because the effect of the surA deletion on most proteins was comparable to that observed in the other three analyses. We also observed that the SC values of most lipoproteins were higher in the surA strain than in the wild type (Supplementary Table 1), which probably reflects a lower abundance of β-barrel proteins in the total amount of proteins prepared from the OM of that strain.
Analysis of the sequence coverage of the β-barrel proteins showed that the identified peptides are derived from both exposed and OM-embedded segments, as illustrated for OmpA (Supplementary Figure 1). This indicates that the digestion of the OMPs by trypsin was complete and that there is no bias in favor of the soluble protein fragments.
We were surprised to find NmpC among the proteins identified by our proteomics approach. An IS5 insertion has been shown to be present close to the 3′-terminus of the nmpC gene [33]. This insertion deletes 18 residues from the C-terminus of NmpC and generates a hybrid protein by adding 8 residues from an open reading frame extending into the IS5 sequence. The presence of the IS5 insertion was thought to prevent the expression of NmpC in E. coli K12 strains [33]. We confirmed the presence of the IS5 in the nmpC gene of the E. coli strain used in this study (see Supplementary Figure 2). However, the fact that 4 peptides derived from NmpC were unambiguously identified in both wild-type and surA strains (Supplementary Figure 2) indicates that a truncated form of this protein is expressed and targeted to the OM. The fact that the NmpC hybrid protein expressed from a plasmid is capable of being translocated to the OM further supports our finding [33]. Thus, the prevailing view regarding the absence of NmpC in E. coli K12 strains must be revised.
3.2 Loss of SurA only affects a subset of OMPs
To assess the effect on OMP levels caused by the loss of SurA, we compared the abundance of all the identified OMPs in the wild-type and surA strains. For quantification of abundance, we used the number of spectral counts (SC) reported for every protein (Supplementary Table 1), since it is linearly correlated with the protein abundance over a dynamic range of two orders of magnitude [34]. We considered that the abundance of a protein is decreased or increased in the surA strain if the number of SC reported for this protein was changed at least 2 fold compared to the wild type. To test the significance of the data, we used the unpaired Student’s t test and defined significance as a P < 0.05 (2-tail 2-sample equal variance test). It is important to note that the number of SC can be used to compare the abundance of a given protein between two different strains. However, SC values cannot be used to directly compare the abundance of different proteins in the same biological sample because the detection of the tryptic-peptides in the mass spectrometer depends in part on intrinsic properties of the protein sequence that could result in MS/MS under-sampling by the mass spectrometer. For instance, the number of SC reported for the major outer membrane lipoprotein Lpp is rather low (about 10), whereas this protein is the most abundant protein in the OM. Examination of the sequence of Lpp reveals that digestion of this small protein with trypsin only generates 4 peptides that are not easily detected by mass spectrometry analysis.
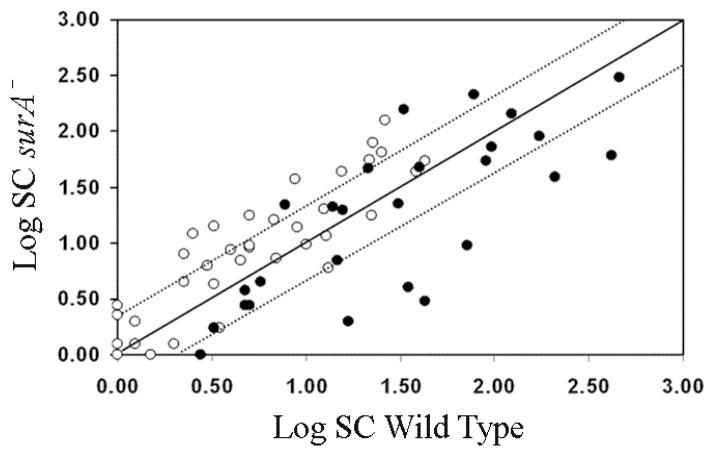
Our proteomics results are summarized in Figure 1. They show that most β-barrels proteins (15 of 23) are not significantly affected by the absence of SurA and that two of them (BamA and TolC) are even more abundant (Tables 1 and 2). The β-barrel proteins YddB and YdiY are also 2 fold less abundant but fail the t test (see Supplementary Table 1). Regarding the lipoproteins, we found that, with the exception of Blc, none of them is significantly decreased in the surA strain and that 14 are even more abundant (Figure 1 and Table 3). These results are consistent with the fact that the assembly of lipoproteins depends on the periplasmic chaperone LolA [7] and not on SurA. The increased abundance of some lipoproteins likely results from σE induction (see below).
Figure 1. A majority of OMPs are not affected by surA deletion.
Two-dimensional logarithmic plot for the mean SC values of the 64 identified proteins. Spots on the diagonal and between the two dotted lines correspond to proteins that are not significantly affected by the loss of SurA (they differ in their SC values by a factor of less than 2). Spots that are above the upper dotted line or below the lower dotted line correspond to proteins whose abundance is increased or decreased by more than 2 fold in the surA strain, respectively. Open circles correspond to lipoproteins and closed circles to β-barrel proteins.
Table 1.
Integral OM proteins whose abundance is significantly changed by at least 2 fold in surA strains
| Proteins ≥ 2 fold less abundant in surA strains (P<0.05) | Proteins ≥ 2 fold more abundant in surA strains (P<0.05) | ||||
|---|---|---|---|---|---|
| Protein | ratio1 | Effect of σE induction on mRNA level | Protein | ratio1 | Effect of σE induction on mRNA level |
| FadL | 0.5 | TolC | 4.9 | ||
| FecA | 0.1 | BamA | 2.8 | ↑ | |
| FhuA | 0.1 | ||||
| LamB | 0.5 | ||||
| LptD | 0.1 | (↑) | |||
| OmpA | 0.2 | (↓) | |||
| OmpF | 0.2 | (↓) | |||
| OmpX | 0.1 | (↓) | |||
Only the proteins whose corresponding genes are induced (↑) or repressed (↓) upon σE activation are shown in grey [21] [36].
. Ratios were calculated by considering that wild-type SC values=1.
Average of four independent experiments.
A complete list of all the SC values for all proteins is available in Supplementary Table 1.
Table 2.
Integral OM proteins identified by 2D-LC-MS/MS that are not significantly affected by the absence of SurA (less than 2 fold change or P > 0.05)
| Protein | ratio1 | Effect of σE induction on mRNA level |
|---|---|---|
| BtuB | 0.7 | |
| CirA | 2.7 | |
| FepA | 0.7 | |
| Fiu | 0.6 | ↓ |
| MipA | 1.2 | |
| NmpC | 0.6 | |
| OmpC | 0.7 | ↓ |
| OmpT | 1.1 | |
| OmpW | 0.8 | |
| PldA | 1.5 | |
| Tsx | 0.8 | ↓ |
| YddB | 0.5 | |
| YdiY | 0.5 |
Table 3.
OM lipoproteins whose abundance is significantly changed by at least 2 fold in surA strains
| Proteins ≥ 2 fold more abundant in in surA strains (P<0.05) | ||
|---|---|---|
| Protein | Ratio1 | Effect of σE induction on mRNA level |
| BamC | 5.0 | ↑ |
| NlpD | 3.8 | |
| RlpA | 4.7 | |
| YbhC | 3.1 | |
| YbjP | 3.7 | |
| YedD | 3.4 | |
| BamB | 4.3 | ↑ |
| BamD | 2.7 | ↑ |
| YggN | Not detected in WT | ↑ |
| YiaD | 2.1 | |
| YraM | 2.3 | |
| CutF | 2.0 | |
| BamE | 2.5 | ↑ |
| Pal | 2.7 | |
Only the proteins whose corresponding genes are induced (↑) or repressed (↓) upon σE activation are shown in grey [21] [36].
. ratios were calculated by considering that wild-type SC values=1.
A complete list of all the identified lipoproteins with the respective SC values is available in Supplementary table 1.
We found 8 of the 23 β-barrel proteins whose levels were decreased in the absence of SurA (Table 1). The decreased proteins include FadL, OmpX, FecA, FhuA and LptD as well as three major OMPs that constitute the bulk mass of the OMPs in E. coli (OmpA, OmpF and LamB). FecA, FhuA and LptD seem to be the most affected by the absence of SurA, the number of SC reported for these proteins in the surA strain being about 10% of the value reported for the wild type. The decrease in abundance that we observe for the major OMPs, as well as the higher abundance of TolC (Table 1), is consistent with previous data [16, 35]. This validates our approach and indicates that our proteomics technique can be reliably used to compare the OM proteome of various E. coli strains.
3.3 For most OMPs, lower protein abundance results from lower mRNA levels
Loss of SurA activates the σE stress response [16, 20], which regulates the expression of many OMPs [21, 36]. While σE upregulates the expression of bamA and lptD, it downregulates the expression of the major OMPs (OmpA, OmpC, OmpF, LamB) and OmpX [21, 36]. Therefore, the decrease in abundance of the major OMPs and OmpX, as well as the increased levels of BamA, that we report in the surA strain (Table 1) partly result from an indirect effect of the surA deletion on the mRNA levels of these proteins [21]. In contrast, the fact that we found the levels of LptD to be greatly reduced despite the fact that lptD expression is expected to increase in the absence of SurA [16, 20, 36] strongly suggests that LptD is a SurA substrate (see our results below).
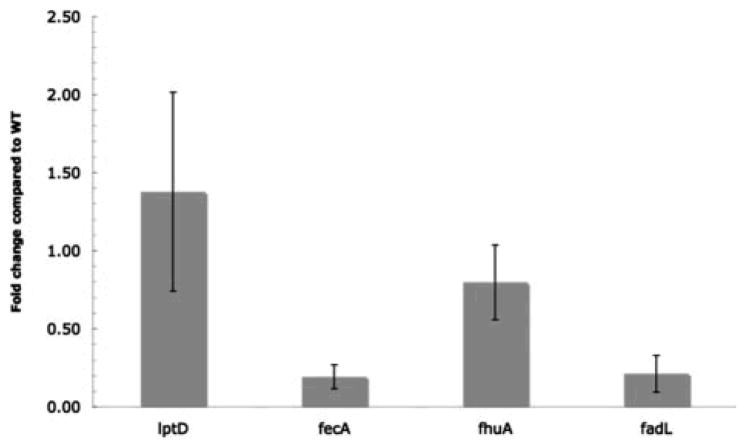
Because of the aforementioned role of σE in the regulation of many OMPs, we determined if the deletion of surA affects the synthesis of fecA, fhuA, and fadL by comparing the mRNA level corresponding to these three proteins by RT-qPCR in both wild-type and surA strains. As shown in Figure 2, fecA and fadL are down-regulated about 5-fold in the surA strain, whereas fhuA is not significantly changed. Therefore, the decrease in protein abundance that we observe for FhuA cannot be attributed to decreased levels of the mRNA. This suggests that FhuA, together with LptD, is a SurA substrate.
Figure 2. The mRNA levels of FadL and FecA, but not FhuA and LptD, are decreased.
The mRNA levels of FadL, FecA, FhuA and LptD were compared in wild-type and surA strains by RT-qPCR. For comparative and quantitative analysis, transcript levels were normalized to the level of gapA (glyceraldehyde 3-phosphate dehydrogenase A).
3.4 LptD is a true SurA substrate
The fact that LptD may be a SurA substrate is especially interesting as LptD is an essential β-barrel protein that is required for the assembly of LPS at the cell surface. In order to further study the dependence of LptD on SurA for folding, we first monitored the levels of LptD in wild-type and surA strains by Western blotting to confirm our findings from the 2D-LC-MS/MS analysis. As shown in Figure 3A, the levels of LptD are dramatically decreased in strains lacking SurA, both in exponential and stationary growth phases. In comparison, LamB and OmpA are also decreased but to a much lower extent. Thus, these data agree with the results of our 2D-LC-MS/MS analysis and confirm that LptD is affected by the absence of SurA.
Figure 3. SurA is required for wild-type levels of LptD.
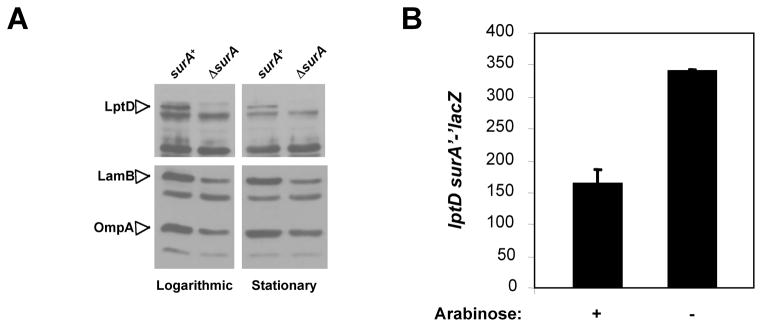
A. Levels of LptD, LamB, and OmpA, three integral β-barrel proteins, were monitored by Western blotting in MC4100 and its ΔsurA::kan derivative CB47 cells in logarithmic (OD600~0.45) and stationary phase (overnight cultures). In both growth phases, all three proteins are decreased in cells lacking SurA, but the decrease in LptD levels is the most dramatic.
B. Lack of SurA increases the synthesis of lptDsurA. Synthesis of a lptDsurA’-’lacZ+ fusion was monitored in NR778, a strain carrying surA under the control of the Para promoter and surA’-’lacZ under the native promoter of the lptDsurA operon. When arabinose is present in the growth medium, this strain is SurA+ but in the absence of arabinose, it lacks SurA and LacZ activity increases.
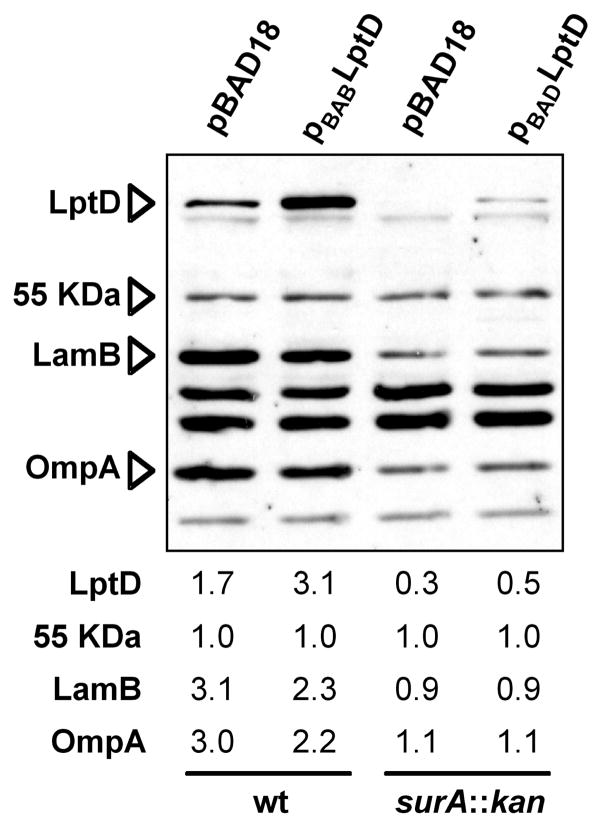
Data reported by Dartigalongue et al. showed that the synthesis of the lptDsurApdxA operon is upregulated by σE [36]. To confirm that this is the case in our surA strain, we monitored the synthesis of a lacZ fusion to the lptDsurApdxA operon [6] in a strain carrying an ectopic copy of surA under the control of the arabinose-inducible Para promoter at the chromosomal att site. In this strain, the native lptDsurA operon has been altered so that the resulting locus has lptD and a surA’-’lacZ gene fusion under the native lptD promoter, as previously described [6]. Therefore, this strain is lptD+ and SurA is only produced from the arabinose promoter. In the presence of arabinose, this strain is effectively surA+ but in the absence of arabinose, it lacks SurA. As shown in Figure 3B, in the absence of arabinose (in the absence of SurA), LacZ activity increases. This indicates that, as expected, lptD expression is upregulated in surA strains. Using RT-qPCR, we confirmed that the levels of the LptD mRNA are unchanged or slightly higher (about 1.4 fold) in the surA strain than in the wild type, which indicates that the decrease in protein abundance is not due to a greater instability of the mRNA (Figure 2). Moreover, we also showed that over-expression of LptD in a strain lacking SurA does not restore wild-type levels (Figure 4), whereas significant amounts of LptD accumulate when the protein is over-expressed in a wild-type strain. Altogether our results indicate that the decreased abundance of LptD results from decreased assembly.
Figure 4. Overexpression of LptD does not restore wild-type levels of LptD in the absence of SurA.
In the presence of inducer arabinose, cells carrying pBAD-LptD accumulate significant amounts of LptD. In a surA::kan mutant, levels of LptD, LamB, OmpA are reduced. Induction of pBAD-LptD increases LptD levels in the surA mutant but they do not restore them to wild-type levels. Values shown under the immunoblot represent the intensity of each band relative to that of the 55 KDa protein that is recognized by the LptD antiserum.
4 Discussion
4.1 A powerful technique to study the OM proteome
By using 2D-LC-MS/MS, we have determined the impact of surA deletion on the abundance of numerous OMPs that had never been analyzed before. This has enabled us to redefine the role of SurA as being a selective periplasmic chaperone and to refine the model for OMPs assembly (see below). Furthermore, the remarkable agreement between our data and those reported previously for the major OMPs validates our results and indicates that our approach based on 2D-LC-MS/MS is reliable. We believe that the use of techniques such as 2D-LC-MS/MS will contribute to a better understanding of the mechanisms that govern OM biogenesis.
Defects in OMP biogenesis lead to both a downregulation of OMP synthesis and an upregulation of degradation of unassembled OMPs in the periplasm [13,21]. Therefore, care must be taken in the selection of strain for proteomic studies like this one. Sklar et al. [18] used a SurA-depletion strain in order to overcome the defect caused by the deletion of surA on the synthesis of the major OMPs mediated by the σE stress response. Unfolded OMPs are degraded by the protease DegP in surA strains [18]; therefore, proteins that do not fold well in the absence of SurA are less abundant in the OM fraction prepared from surA strains. In contrast, during depletion conditions, most unfolded proteins are not degraded during SurA depletion and these unassembled molecules fractionate with the OM [18]. These mistargeted proteins would interfere with the analysis of the properly targeted, folded proteins present in the OM fraction prepared from SurA-depleted strains, limiting therefore our ability to identify the proteins that depend on SurA for folding. Accordingly, since we are interested in finding out the number of different OMPs affected by the absence of SurA, irrespective of the mechanisms leading to the changes, we used a surA null strain in this study.
4.2 Decreased LptD levels likely contribute to the phenotype of surA strains
In contrast to the major OMPs whose decreased abundance likely results from both decreased expression [21] and biogenesis [18], the decreased levels of LptD can be solely attributed to defective assembly in the absence of SurA. Interestingly, lptD and surA are located in an operon, which is conserved among Gram-negative bacteria (see Supplementary Figure 3). Moreover, in γ-proteobacteria, the lptDsurA operon is conserved even though neighboring genes are not. In some instances, the synthesis of both gene products is coupled translationally. These observations reinforce the idea that LptD is a preferred SurA substrate and indicates that the role of SurA in LptD biogenesis might be widely conserved among Gram-negative bacteria.
The dependence of LptD on SurA for folding suggests that some phenotypes previously attributed to the loss of the SurA periplasmic chaperone may be caused instead by the decreased levels of LptD in surA strains. For instance, one of the phenotypes characteristic of surA strains is that they exhibit increased sensitivity to toxic small molecules, such as detergents and hydrophobic antibiotics [16]. However, strains expressing low levels of LptD are also sensitive to such compounds [37]. We suggest that most, if not all, of the OM defects in surA strains is caused by the low levels of LptD.
4.3 A proteomic snapshot of σE induction upon surA deletion
Our analysis revealed that the levels of 14 lipoproteins and 2 β-barrel proteins are increased in the surA strain. It is likely that this higher protein abundance results from an increased transcription of the genes coding for these proteins mediated by σE. When assembly of β-barrel proteins is compromised, unfolded intermediates that accumulate in the periplasm are detected by the σE stress response [19]. Activation of this response leads to the up-regulation of the synthesis of many envelope biogenesis factors, such as periplasmic chaperones and the components of the BamABCDE complex, which is required for the assembly of β-barrel proteins at the OM [9, 10]. Indeed, we detected increased levels of the β-barrel protein BamA and each of its four lipoprotein partners BamBCDE (Tables 1 and 3). Thus, we believe that in the absence of SurA, unfolded OMPs trigger the σE stress response and as a result, the levels of the machinery required for OMP biogenesis are increased. Whether the other OM lipoproteins that are more abundant in the surA strain are also regulated by σE, or some other envelope stress response, remains to be determined.
The levels of another β-barrel protein, TolC, were also increased in the absence of SurA. TolC is an unusual OMP that has a novel multimeric β-barrel conformation and previous studies have shown that the structure of this protein reflects a unique assembly pathway that does not involve the known periplasmic chaperones, including SurA [38]. The gene coding for TolC does not belong to the σE regulon and we propose that the increase in TolC levels results from a decreased competition of the major OMPs for the Bam complex. This hypothesis was first proposed by Charslon et al. [35] to explain the increased abundance of TolC that they observed in strains lacking BamB, a non-essential component of the Bam complex, that, when absent, leads to the reduction of the levels of major OMPs. SurA and BamB have been proposed to function in the same pathway because surA and bamB strains are quantitatively indistinguishable in term of LamB biogenesis [39]. Our data confirm that the absence of SurA confers phenotypes similar to those recently reported for bamB strains. Both strains have increased levels of TolC but decreased levels of OmpA, OmpF, LamB and, to a lower extent, OmpC [9, 27, 35]. Absence of either SurA or BamB induces the σE response [40], which leads to a decrease in the synthesis of major OMPs [21].
However, both surA and bamB mutants differ with respect to LptD. While we report here that the absence of SurA causes a dramatic reduction in the levels of LptD, LptD levels do not decrease in bamB cells [27]. This cannot be explained by differences in lptD synthesis, since the loss of SurA induces the σE response to a greater extent than the loss of BamB does [40]. Instead, we believe that this reflects the fact that LptD is a substrate of SurA but that its assembly by the Bam complex is not strongly dependent on BamB. This likely explains why OM defects are more pronounced in mutants lacking SurA than in mutants lacking BamB.
4.4 A revised model for OMP transport and assembly
Sklar et al. have proposed that SurA is the primary chaperone in the periplasm responsible for the transit of the bulk mass of OMPs and that a function of the other folding factors DegP and Skp is to rescue OMPs that fall off the SurA pathway [18]. However, our data are consistent with a model in which only a subset of OMPs, including the most abundant OMPs, depends on SurA for proper folding and insertion in the OM. Additional periplasmic chaperones must participate in the biogenesis of those OMPs whose levels are not decreased in the absence of SurA. It is possible that DegP (which also has a protease activity) and Skp are the primary chaperones for these OMPs and that because they do not contribute much to the bulk mass of proteins of the OM, no change in OM density was detected by Sklar et al in degP and skp strains [18].
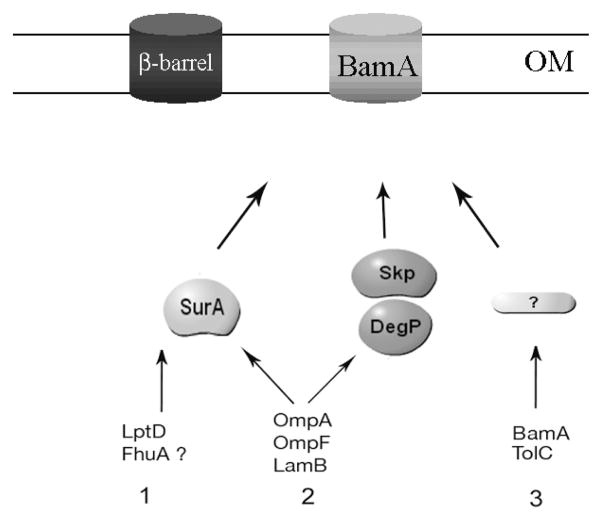
On the basis of our results, we propose a refined model for SurA’s function in the periplasm (Figure 5). First, there are the true SurA substrates, such as LptD and probably FhuA. These proteins are strongly affected by the absence of SurA and the other periplasmic folding factors do a poor job in assisting them to fold. Second, there are proteins such as the major OMPs, OmpA and OmpF. These proteins constitute a large fraction of the bulk mass of OMPs. They seem to preferentially depend on SurA for transport across the periplasm [18] and their synthesis is greatly reduced in the absence of this protein [21]. When SurA is absent, the lowered levels of these proteins can be assembled, perhaps inefficiently, by other chaperones, likely DegP and Skp [18]. The third group of OMPs corresponds to proteins, such as BamA and TolC, whose assembly is SurA-independent.
Figure 5. A refined model for SurA’s function in the E. coli periplasm.
Unfolded β-barrel proteins are transported across the IM by the Sec translocon. After cleavage of the signal sequence, they are escorted across the periplasm by periplasmic chaperones before reaching the OM where they are inserted by the Bam complex [1]. There are 3 known periplasmic chaperones that interact with OMPs: SurA, Skp and DegP. Our results indicate that only a subset of OMPs strongly depend on SurA for biogenesis. We propose to divide the OMPs in three groups. Group 1 includes β-barrel proteins, such as LptD and possibly FhuA, that greatly depend on the presence of SurA for biogenesis. Group 2 includes β-barrel proteins such as the major OMPs OmpA, OmpF and LamB that preferentially interact with SurA for biogenesis but are also able to interact with the other periplasmic chaperones. Group 3 includes β-barrel proteins such as BamA and TolC that do not seem to require the presence of SurA for biogenesis. Werner et al. have shown that the assembly of TolC does not depend on Skp and DegP [37] and the fact that a skPdegP double mutant is viable suggest that these two chaperones are not required for the assembly of the essential protein BamA. BamA and TolC are therefore likely to depend on other periplasmic folding factors that are yet to be identified.
Acknowledgments
We thank Geneviève Connerotte and Martine De Cloedt for technical help and Emile Van Schaftingen for helpful criticism of the manuscript. We also thank Emeline de Viron and Frédéric Sorgeloos for helping us with the RT-qPCR experiments. JFC is Chercheur Qualifié, PL is Collaborateur Scientifique, and DV is Collaborateur logistique of the Belgian FRS-FNRS. This work was supported by the Interuniversity Attraction Pole Programme - Belgian Science Policy to JFC and PL (network P6/05) and to DV (network P6/28). This research was supported in part by grants from the FRS-FNRS to JFC. T.J.S is supported by National Institute of General Medical Sciences Grant GM34821.
Footnotes
The authors have declared no conflict of interest.
References
- 1.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 2.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperandeo P, Lau FK, Carpentieri A, De Castro C, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol. 2008;190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, McCandlish AC, Gronenberg LS, Chng SS, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 7.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1694:IN1–9. [PubMed] [Google Scholar]
- 8.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 9.Wu T, Malinverni J, Ruiz N, Kim S, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayer ME. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- 12.Kellenberger E. The ‘Bayer bridges’ confronted with results from improved electron microscopy methods. Mol Microbiol. 1990;4:697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 13.Duguay AR, Silhavy TJ. Quality control in the bacterial periplasm. Biochim Biophys Acta. 2004;1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Messens J, Collet JF. Pathways of disulfide bond formation in Escherichia coli. Int J Biochem Cell Biol. 2006;38:1050–1062. doi: 10.1016/j.biocel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Behrens S, Maier R, de Cock H, Schmid FX, Gross CA. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 2001;20:285–294. doi: 10.1093/emboj/20.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouviere PE, Gross CA. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 17.Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 20.Missiakas D, Betton JM, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 21.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Button JE, Silhavy TJ, Ruiz N. A suppressor of cell death caused by the loss of sigmaE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J Bacteriol. 2007;189:1523–1530. doi: 10.1128/JB.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Hiniker A, Bardwell JC. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J Biol Chem. 2004;279:12967–12973. doi: 10.1074/jbc.M311391200. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: an integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33:D329–333. doi: 10.1093/nar/gki028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vertommen D, Depuydt M, Pan J, Leverrier P, et al. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol. 2008;67:336–349. doi: 10.1111/j.1365-2958.2007.06030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vertommen D, Van Roy J, Szikora JP, Rider MH, et al. Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol Biochem Parasitol. 2007 doi: 10.1016/j.molbiopara.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Blasband AJ, Marcotte WR, Jr, Schnaitman CA. Structure of the lc nComputer membrane porin protein genes of lambdoid bacteriophage. J Biol Chem. 1986;261:12723–12732. [PubMed] [Google Scholar]
- 34.Liu H, Sadygov RG, Yates JR. 3rd, A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 35.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli sigma E regulon. J Biol Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 37.Abe S, Okutsu T, Nakajima H, Kakuda N, et al. n-Hexane sensitivity of Escherichia coli due to low expression of imp/ostA encoding an 87 kDa minor protein associated with the outer membrane. Microbiology. 2003;149:1265–1273. doi: 10.1099/mic.0.25927-0. [DOI] [PubMed] [Google Scholar]
- 38.Werner J, Augustus AM, Misra R. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J Bacteriol. 2003;185:6540–6547. doi: 10.1128/JB.185.22.6540-6547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]