Abstract
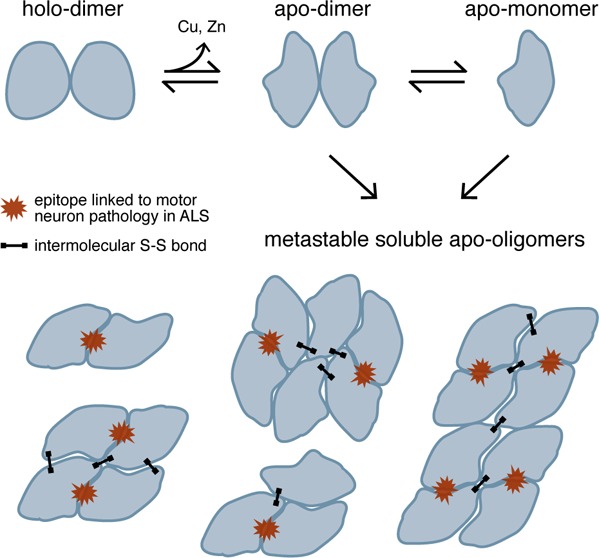
Soluble misfolded Cu/Zn superoxide dismutase (SOD1) is implicated in motor neuron death in amyotrophic lateral sclerosis (ALS); however, the relative toxicities of the various non-native species formed by SOD1 as it misfolds and aggregates are unknown. Here, we demonstrate that early stages of SOD1 aggregation involve the formation of soluble oligomers that contain an epitope specific to disease-relevant misfolded SOD1; this epitope, recognized by the C4F6 antibody, has been proposed as a marker of toxic species. Formation of potentially toxic oligomers is likely to be exacerbated by an oxidizing cellular environment, as evidenced by increased oligomerization propensity and C4F6 reactivity when oxidative modification by glutathione is present at Cys-111. These findings suggest that soluble non-native SOD1 oligomers, rather than native-like dimers or monomers, share structural similarity to pathogenic misfolded species found in ALS patients and therefore represent potential cytotoxic agents and therapeutic targets in ALS.
Accumulating evidence supports a prominent contribution of misfolding and aggregation of SOD1 to the dysfunction and progressive death of motor neurons in ALS. Over 140 mutations (mostly missense) in the SOD1 gene have been identified in patients with familial ALS (FALS), most of which destabilize the native SOD1 homodimer and/or increase aggregation propensity.1,2 Current evidence supports the pathogenic capacity of soluble misfolded SOD1, rather than the large insoluble aggregates that appear only near the onset of paralysis in ALS mouse models.3−7 However, little is known about the structural features of soluble non-native SOD1 conformers or the factors in the cellular environment that influence misfolding and aggregation. Soluble misfolded WT SOD1 has been found in the spinal cord from sporadic ALS patients that do not carry mutations in SOD1,8,9 demonstrating the sufficiency of nongenetic factors to induce the formation of potentially toxic oligomers by SOD1.
To identify misfolded SOD1 conformers with greatest relevance to ALS pathology, we probed isolated oligomeric species with a conformation-specific antibody (C4F6) to identify those with potential cytotoxicity. In FALS patients and mouse models, C4F6 specifically recognizes soluble SOD1 found only in disease-affected tissue, revealing a connection between FALS pathology and the as-yet unidentified epitope bound by C4F6.7 Here, we show that higher-order non-native oligomers of mutant SOD1, but not dimers or monomers, contain the epitope recognized by the C4F6 antibody. To assess the impact of the cellular redox environment on the formation of potentially toxic soluble oligomers, we determine the effect of a physiologically prevalent oxidative modification (glutathionylation at Cys-111) on oligomerization. Cys-111 glutathionylation increases both the abundance of soluble oligomers and exposure of the disease-specific epitope recognized by C4F6, revealing a novel mechanism by which oxidative stress modulates potentially toxic SOD1 aggregation. Our results suggest that SOD1 acquires pathogenic features upon the formation of soluble non-native oligomeric assemblies, indicating a particular relevance of these species to neuronal dysfunction in ALS.
Experimental Procedures
Cloning, Expression, and Purification of Recombinant SOD1 from S. cerevisiae
Mutagenesis of constructs for the expression of human SOD1, expression in S. cerevisiae, and SOD1 purification were performed according to previously published methods.10,11 The final step of purification of recombinant SOD1 is anion-exchange chromatographic separation using a MonoQ HR 10/10 column connected to an AKTA purifier system (GE Healthcare), which separates a population of predominantly unmodified SOD1 from one enriched in SOD1 that is glutathionylated at Cys-111.10 Samples were stored at −80 °C in 20 mM Tris and 150 mM NaCl at pH 7.4 until use.
High-Resolution Mass Determination of Intact Recombinant SOD1
SOD1 sample buffer was exchanged with 10 mM ammonium acetate using 2 kDa VIVACON 500 filtration devices (Sartorius Stedim Biotech GmbH), after which samples were collected by centrifugation of the inverted concentrator body within a fresh tube. These samples were then diluted 1:10 in a 50% acetonitrile/49% water/1% formic acid mixture and directly infused into the LTQ Orbitrap Velos (Thermo Fisher Scientific) using a Picoview nanoelectrospray source (New Objective). Spectra were collected with the Orbitrap analyzer in positive ion mode at a resolution of 30,000 (at 400 m/z), with a maximum ion injection time of 200 ms, a spray voltage of 5 kV, and the automatic gain control (AGC) set to 2 × 105. Spectra were deconvoluted using Promass for Xcalibur, version 2.5 SR-1 (Thermo Fisher Scientific).
Time-Resolved Analytical Size Exclusion Chromatography (SEC)
Cu2+ and Zn2+ were removed from as-isolated remetalated SOD1 by dialysis against 50 mM sodium acetate and 10 mM EDTA at pH 3.8 for 1.5 h in the case of mutant SOD1 and 2 h in the case of the WT enzyme. Removal of EDTA and return to physiological pH were achieved by overnight dialysis against 20 mM Tris and 150 mM NaCl at pH 7.4. All dialysis was performed at 4 °C. Demetalated (“apo”) SOD1 was brought to a concentration of 100 μM in 20 mM Tris and 150 mM NaCl at pH 7.4 and incubated in a 37 °C water bath. At each indicated time point, an aliquot containing 64 μg of apo-SOD1 was removed, filtered using a 0.22 μm centrifugal filter, and injected onto a Superdex 200 10/300 GL column (GE Healthcare) at 4 °C equilibrated in 20 mM Tris and 150 mM NaCl at pH 7.4.
Estimation of Molecular Weight of Oligomers Using Size Exclusion Chromatography Combined with Multiangle Light Scattering (SEC-MALS)
Apo-SOD1 incubated for 1 week under the pH, temperature, concentration, and ionic strength conditions listed above was analyzed using a DAWN HELEOS II light scattering instrument (Wyatt Technology), which detects scattered light at 18 angles with respect to the incident beam. The light scattering instrument is interfaced to an Agilent FPLC System with a connected Superdex 200 10/300 GL column (GE Healthcare), a T-rEX refractometer, and a dynamic light scattering module (Wyatt Technology). SEC separation and detection of scattered light, absorbance at 280 nm, and differential refractive index were performed at room temperature. Data were analyzed, and weight average molar masses as a function of elution volume were determined using ASTRA software (Wyatt Technology) with the Zimm fit method, which assumes weak protein–solvent interactions.12
Measurement of C4F6 Epitope Exposure of Isolated Apo-SOD1 Oligomer Populations
Apo-SOD1 oligomers were prepared by incubation at 100 μM in 20 mM Tris and 150 mM NaCl at pH 7.4 at 37 °C for 1 week. Samples containing 640 μg of apo-SOD1 were filtered and separated by SEC as described above. Immediately following elution, individual oligomeric populations were collected and individually loaded onto PVDF membranes equilibrated in 20 mM Tris at pH 7.4 using a chilled Minifold I dot-blot system (S&S). Samples were blotted in duplicate simultaneously; one blot was immediately incubated with monoclonal antibody to misfolded SOD1 (C4F6, MediMabs) diluted 1:250 in blocking buffer (TBS-T with 5% (w/v) nonfat dry milk); the duplicate blot was stained with Ponceau S in 5% acetic acid to visualize total protein loaded onto the membrane. Duplicate blotting was carried out in lieu of fixation with Ponceau S prior to incubation with primary antibody due to our observation of increased C4F6 reactivity following Ponceau S staining and destaining, as well as to minimize the time elapsed between isolation of oligomers by SEC and probing with C4F6. Blots were incubated with C4F6 overnight at 4 °C, and C4F6 binding was visualized using HRP-conjugated antimouse antibodies (GE Healthcare, Pierce, Millipore). To quantify abundance of individual oligomeric populations represented in SEC chromatograms, A280 data from Ve = 7.5–19 mL were deconvoluted into multiple single Gaussian distributions using Matlab (Mathworks), and the area under each Gaussian curve was calculated as a percentage of the total area under all Gaussian curves in the deconvoluted chromatogram. For comparison of oligomeric populations in glutathionylated and unmodified apo-SOD1, oligomers were grouped based on Ve at the center of the Gaussian curve obtained by deconvolution: O1 consists of oligomers eluting between 14 and 15 mL, O2 consists of oligomers eluting between 11.3 and 13.5 mL, O3 consists of oligomers eluting between 9.2 and 11.2 mL, and Vo consists of oligomers eluting between 8.0 and 9.0 mL (corresponding to the approximate void volume of the column).
In Vitro Glutathionylation of SOD1
SOD1 was glutathionylated in vitro by incubating at 37 °C for 30 min with 1000-fold molar excess oxidized glutathione (GSSG) in 50 mM CAPS buffer at pH 9.7. Untreated SOD1 was subjected to the same incubation in 50 mM CAPS buffer at pH 9.7, containing no GSSG. Following this incubation, untreated and GSSG-enriched SOD1 samples were demetalated as described above, then brought to 100 μM apo-SOD1. A 64 μg aliquot was removed, filtered using a 0.22 μm centrifugal filter, and injected onto a Superdex 200 10/300 GL column (GE Healthcare) at 4 °C equilibrated in 20 mM Tris and 150 mM NaCl at pH 7.4.
Effect of Reducing Agent Treatment on Apo-SOD1 Oligomer Stability
Oligomers of apo-SOD1 were prepared as described above, and DTT was added to a final concentration of 1 mM to the sample and SEC running buffer. Aliquots from the mixture of oligomers were separated by SEC as described above immediately following the addition of DTT and after 2 h and overnight incubation at room temperature.
Results
Formation of Metastable Soluble Oligomers by Apo-SOD1 with FALS-Linked Substitutions
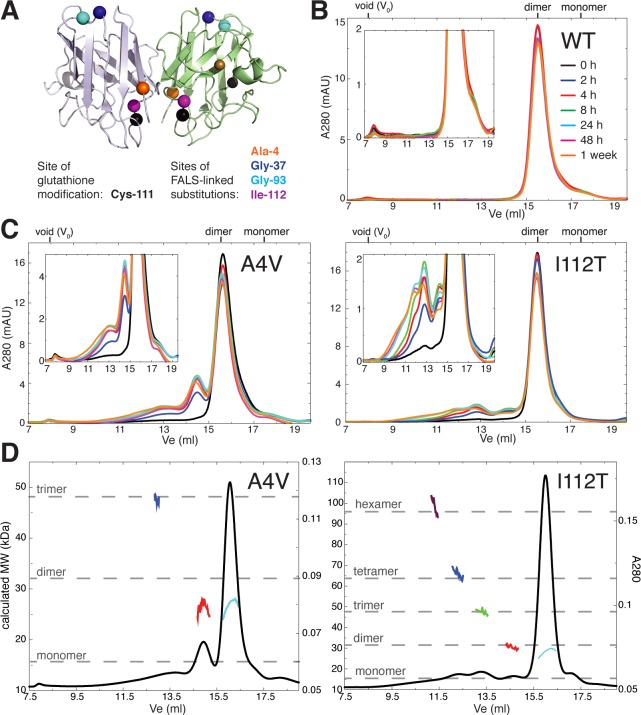
To identify potentially disease-relevant metastable SOD1 oligomers, we incubated apo-SOD1 at physiological pH, temperature, ionic strength, and SOD1 concentration for up to one week, separating the reaction mixture by size exclusion chromatography (SEC) at multiple time points. We use recombinant protein in which SOD1’s native free cysteines (Cys-6 and Cys-111) are retained, as they have been demonstrated to play crucial roles in oligomerization.13,14 Metal-free (“apo”) SOD1 is utilized in all experiments since it is widely considered to be the common precursor to misfolded and aggregated species.4,15,16 We analyze soluble oligomers because of their particular relevance to ALS pathology; apo-SOD1 remains soluble throughout the 1-week incubation period, as evidenced by the minimal changes in total A280 from SEC chromatograms (Figure 1B,C). WT SOD1 (Figures 1B and 2B) and SOD1 containing the FALS-linked G93A and G37R substitutions (Figure 2B) have low propensities to form soluble oligomers under these conditions, whereas SOD1 with the A4V or I112T substitutions shows substantial oligomerization (Figures 1C and 2B). Analysis of SEC-separated oligomers with multiangle light scattering is consistent with the presence of native-like dimers, non-native-like expanded dimers, trimers, tetramers, and hexamers (Figure 1D). The presence of an expanded dimer is inferred from the SEC-MALS data based on the presence of a peak eluting before the native-like dimer (indicating its larger hydrodynamic radius), yet having a calculated molecular weight equivalent to that of the native-like dimer (red vs cyan curves, Figure 1D). In the case of the aggregation-prone A4V and I112T variants, small soluble oligomers are apparent by 2 h of incubation at 37 °C (Figure 1C) and remain detectable throughout the 1-week incubation period. The smallest non-native oligomers (those eluting near 13 and 14.5 mL following injection onto the gel filtration column) increase in abundance for the first 8–24 h, after which their populations decline concomitant with the appearance of higher-order species (Figure 1C).
Figure 1.
Formation of metastable soluble non-native oligomers of metal-free SOD1. (A) Positions of the glutathione modification and of the FALS-linked amino acid substitutions included in the current study; residue positions are indicated by colored spheres on the background of the WT SOD1 crystal structure (PDB ID: 1spd). (B) SEC chromatograms showing aggregation of 100 μM apo-WT SOD1 at 37 °C in 20 mM Tris and 150 mM NaCl at pH 7.4 for up to 1 week. (C) Aggregation of 100 μM apo-SOD1 with the indicated FALS-linked amino acid substitutions under identical conditions as those described for B. Insets show chromatograms with expanded y-axes. (D) Weight average molar masses of metastable soluble SOD1 oligomers separated by SEC, as determined by multiangle light scattering (SEC-MALS). Black curves, absorbance at 280 nm (A280) vs elution volume (Ve); colored curves, molecular weight (MW) calculated at each Ve from the intensities of scattered light at multiple fixed detectors. Dashed gray lines indicate approximate theoretical molecular masses for SOD1 oligomers.
Figure 2.
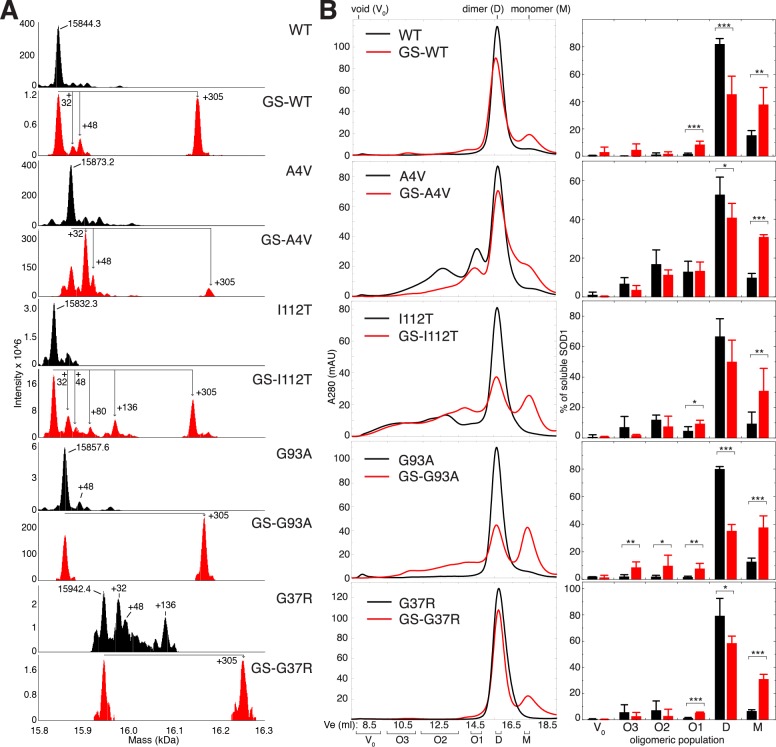
Cys-111 glutathionylation that occurs endogenously promotes the formation of non-native apo-SOD1 oligomers. (A) Analysis of full-length SOD1 by mass spectrometry. For each SOD1 variant studied, deconvoluted mass spectra are shown for two populations separated by ion exchange chromatography: one in which unmodified SOD1 is the predominant species (black spectra) and one enriched in post-translationally modified SOD1 (red spectra). Labeled masses correspond to the average masses obtained by the deconvolution of spectra using ProMass for Xcalibur software. Mass shifts consistent with glutathionylation (+305), irreversible oxidation (+32, +48), and phosphorylation (+80) are labeled. Occasionally, a mass shift of +136 was observed and is consistent with SOD1 containing phosphorylation (+80), oxidation (+32), and a nonspecific sodium adduct originating from the sample buffer (+22), all of which have been previously observed on SOD1.8,10 (B) Left, SEC chromatograms showing populations of soluble oligomers of unmodified (black) and glutathionylated (red) apo-SOD1 incubated at 100 μM (initial dimeric concentration) for 1 week at 37 °C in 20 mM Tris and 150 mM NaCl at pH 7.4. Right, oligomeric populations quantified by deconvolution of SEC data and integration of Gaussian curves corresponding to individual oligomeric populations. Bar heights represent average values, and error bars represent SD from at least 3 independent experiments. Student’s t test was used to compare the abundance of oligomers in the presence and absence of Cys-111 glutathionylation. * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001.
Glutathionylation at Cys-111 Induces Monomerization of Apo-SOD1 and Increases Propensity to Form Non-Native Oligomers
Protein S-glutathionylation is a reversible post-translational modification that serves, in addition to regulatory and signaling functions, as a protective measure against irreversible oxidation of cysteines.17 SOD1 glutathionylated at Cys-111 is abundant in SOD1 isolated from human tissue or expressed in S. cerevisiae can be partially resolved from the unmodified enzyme by ion-exchange chromatography and destabilizes the holo-SOD1 dimer.10,18−21 To assess the effects of Cys-111 glutathionylation on the assembly of soluble SOD1 oligomers, we analyzed the impact of this modification on the oligomeric distributions of soluble WT and mutant apo-SOD1. For each SOD1 variant studied, a predominantly unmodified SOD1 population and one enriched in glutathionylated SOD1 (GS-SOD1) (Figure 2A) were incubated at physiological pH, temperature, ionic strength, and SOD1 concentration. We assess the effect of Cys-111 glutathionylation on oligomer formation by comparing these two populations of recombinant human SOD1, which are isolated by ion-exchange chromatography.10 The GS-SOD1-enriched population contains SOD1 modified endogenously during expression in S. cerevisiae, and only singly modified (one glutathione moiety per monomer) SOD1 is detected. We first analyzed this GS-SOD1-enriched fraction without further in vitro glutathionylation to determine the effect of Cys-111 glutathionylation alone, without modification of Cys-6, Cys-57, or Cys-146 (which are not glutathionylated in SOD1 isolated from human tissue due to the intermolecular Cys-57-Cys-146 disulfide bond and the low solvent accessibility of Cys-610).
For the wild type as well as all FALS mutants studied, glutathionylation of apo-SOD1 results in a significant increase in the proportion of soluble protein present as monomers (Figure 2B), in accordance with previous findings.10,18,21 Glutathionylation also significantly increases the abundance of several non-native higher-order species, especially in G93A SOD1 (Figure 2B). In all variants except A4V SOD1, glutathionylation significantly increases the formation of the oligomeric population eluting just prior to the native-like dimer: O1, the putative expanded dimer (Figures 2B and 1D). In vitro enrichment of SOD1 with oxidized glutathione (GSSG) recapitulates the increased abundance of apo-SOD1 monomers and small oligomers (Figure 3); this effect was also observed for the A4V variant, which contains a relatively low amount of endogenously modified GS-SOD1 (Figure 2A).
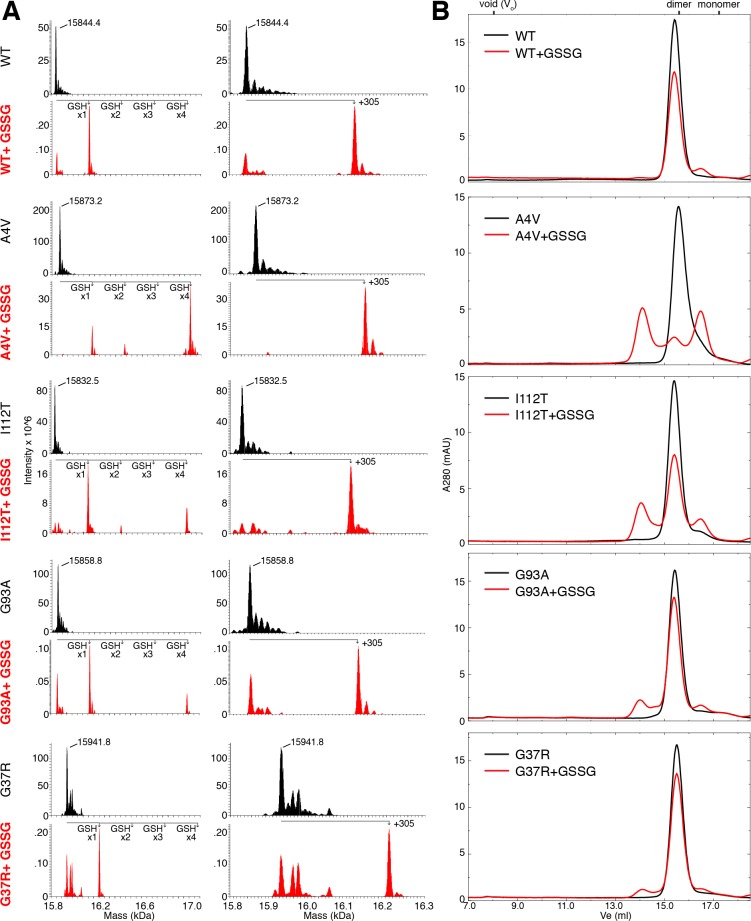
Figure 3.
In vitro glutathionylation of SOD1 also promotes dimer dissociation and formation of soluble non-native oligomers. (A) Deconvoluted mass spectra for full-length SOD1 (unenriched and treated with GSSG). Labeled masses correspond to the average masses obtained by deconvolution of spectra using ProMass for Xcalibur software. Mass shifts corresponding to the addition of one, two, three, and four glutathione adducts are shown at left. At right, spectra are expanded to show only the mass range corresponding to unmodified SOD1 and SOD1 containing a single glutathione modification. (B) SEC chromatograms showing oligomeric populations of apo-SOD1 incubated in the presence or absence of GSSG prior to demetalation, then immediately separated by SEC at 4 °C.
Glutathionylation of SOD1 in vitro generates protein with a single glutathione adduct, as well as species containing 2 (most likely corresponding to the modification of both free cysteines, at positions 6 and 111) or 4 adducts (corresponding to the modification of all cysteines in SOD1, including those at positions 57 and 146 that participate in the native intramonomer disulfide bond); species with multiple glutathione adducts were only observed for the A4V, I112T, and G93A variants (Figure 3). In the latter case, disruption of the native disulfide bond likely contributes to the observed misfolding and aggregation. The increased abundance of monomeric and oligomeric SOD1 in endogenously modified samples and in vitro glutathionylated WT and G37R SOD1, which contain only a single glutathione modification (Figures 2 and 3), supports the conclusion that Cys-111 glutathionylation is predominantly responsible for the observed promotion of dimer dissociation and oligomerization.
Metastable Oligomers Show Enhanced Exposure of an Epitope Common to SOD1 Found in ALS Patients
Though soluble misfolded SOD1 (as opposed to that which is present in insoluble aggregates) is increasingly implicated in motor neuron dysfunction,4,5,22,23 the potential cytotoxicities of individual oligomeric species have not been evaluated. Direct determination of the effects of specific oligomers on motor neuron viability is complicated by the difficulty of delivering metastable protein assemblies to the cytoplasm of living cells. To begin to evaluate the cytotoxic potential of the apo-SOD1 oligomers isolated by SEC, we probed for exposure of an epitope known to be exposed on misfolded SOD1 in disease-affected cell populations of ALS patients.7,8 The various apo-SOD1 oligomeric populations isolated by SEC and dimeric holo-SOD1 were bound to PVDF membranes and probed with the C4F6 conformation-specific antibody (Figure 4). The species with greatest reactivity to C4F6 are higher-order non-native oligomers, those eluting at postinjection volumes ranging from the column void to ∼14.5 mL, just prior to the elution of native-like SOD1 dimer (Figures 2B and 4). Monomeric apo-SOD1 is not C4F6-reactive in any of the SOD1 variants studied, while dimeric holo- and apo-SOD1 is faintly reactive in some cases. Oligomers of glutathionylated apo-SOD1 were also probed with C4F6 to determine whether this modification induces structural rearrangements that enhance exposure of the disease-specific epitope. In the case of SOD1 with the FALS-linked A4V or I112T substitutions, glutathionylation enhances exposure of the C4F6-recognized epitope in higher-order soluble oligomers (Figure 4), suggesting that glutathionylation substantially alters the conformations of oligomers formed by these disease-associated mutant proteins.
Figure 4.
Non-native oligomers of SOD1 are potentially toxic in ALS. Dot blots of apo-SOD1 oligomers isolated by SEC and probed with the C4F6 monoclonal conformation-specific antibody, which has been proposed to recognize a toxic subset of misfolded SOD1.7 Dimeric SOD1 isolated from S. cerevisiae and remetalated (“holo”) was also probed to determine initial C4F6 reactivity prior to the removal of metals and oligomerization. The amounts of each oligomeric population bound to the membrane were visualized by Ponceau S staining.
Cys-111 Modulates Soluble Oligomer Formation through Mechanism(s) Independent of Intermolecular Disulfide Bonding
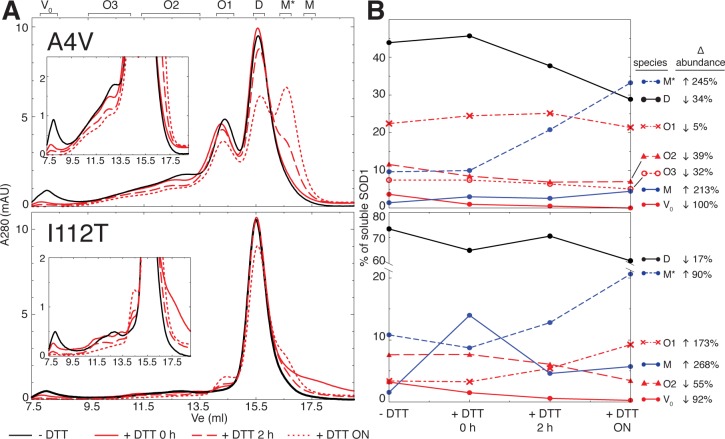
The two free cysteines of SOD1, especially Cys-111, have been recognized to modulate its aggregation propensity.13,14 The most commonly assumed mechanism by which free cysteines affect aggregation is their participation in aberrant disulfide bonds, including “scrambled” non-native intramolecular disulfide bonds and intermolecular disulfide bonds that stabilize oligomers.24,25 However, our findings of decreased dimer stability18 (Figure 2B) and increased aggregation propensity (Figure 2B) in GS-SOD1 suggest that the propensity of Cys-111 to be glutathionylated contributes to its role in SOD1 aggregation. We therefore sought to determine whether intermolecular disulfide bonds are required for the stability of the apo-SOD1 oligomers we observed in vitro. Oligomers of A4V and I112T SOD1 generated by incubation at physiological pH, temperature, ionic strength, and SOD1 concentration for one week (WT, G93A, and G37R SOD1 form few oligomers under these conditions (Figures 1 and 2B)) were incubated at room temperature with the reducing agent DTT, and aliquots were removed at various time points for SEC analysis. The largest oligomers, those eluting from the void volume to ∼13.5 mL postinjection on the Superdex 200 10/300 GL column, are most sensitive to dissociation by treatment with reducing agent (Figure 5). While the abundance of these oligomers decreases steadily following the addition of DTT, the oligomeric population eluting at ∼14.5 mL (O1) is relatively stable in the presence of the reducing agent (in A4V, O1 abundance remains largely unchanged for at least 2 h following the introduction of DTT; in I112T, O1 abundance increases in the presence of DTT (Figure 5)). The relative resistance of the O1 population to dissociation by DTT indicates that the stability of these oligomers is less dependent on the presence of intermolecular disulfide bonds or that any intermolecular disulfide bonds present are not sufficiently solvent-exposed to be reduced by DTT added to the buffer. The increase in O1 abundance in I112T may be attributed to both continual accumulation of this oligomer throughout the period of incubation with DTT or the generation of oligomers of this size upon dissociation of larger oligomers.
Figure 5.
Intermolecular disulfide bonding is not universally required for the persistence of metastable non-native oligomers in vitro. (A) Apo-SOD1 oligomers generated by incubation for 1 week at 37 °C in 20 mM Tris and 150 mM NaCl at pH 7.4 were separated by SEC in the absence (black curves) of DTT and in the presence of 1 mM DTT following incubation at room temperature (red curves). Designations of oligomeric populations (D, M, O1, O2, O3, and V0) correspond to those in Figure 2A, while M* denotes a species that that appears subsequent to DTT treatment and whose elution volume is consistent with an expanded monomer. (B) Quantification of oligomeric populations prior to DTT treatment and after room temperature incubation with 1 mM DTT for the indicated time periods. ON = overnight incubation.
Discussion
Relevance of the in Vitro System to Pathological SOD1 Aggregation in ALS
Although misfolding and aggregation of SOD1 is believed to be a major contributor to ALS pathology, little is known about the potential toxicities of individual aggregate species or the cellular determinants of their formation. Here, we examine the propensities of WT and FALS mutant SOD1 to form metastable soluble oligomers with an epitope linked to toxicity in ALS and explore the effects of oxidative modification of Cys-111, a residue known to modulate SOD1 aggregation. We assess oligomerization of SOD1 under conditions approximating physiological pH (7.4), temperature (37 °C), and SOD1 concentration (100 μM,24) without agitation. The degrees to which the various FALS-linked mutations increase SOD1 destabilization and aggregation propensity have been shown to be correlated with disease severity.2 Although this correlation was recently challenged,26 this latter study employed the use of SOD1 in which both free cysteines (at positions 6 and 111) are mutated to alanine and serine, respectively, potentially restricting the applicability to physiological SOD1 aggregation. We find that the A4V and I112T substitutions, which are found in patients with rapidly progressing FALS,2 exhibit the highest propensities to form soluble oligomers (Figures 1C and 2B). These results would be predicted by the correlation of aggregation propensity with disease severity, as would the minimal aggregation of SOD1 containing the G37R substitution (Figure 2B), which causes relatively slowly progressing paralysis in ALS patients and mouse models.2,27 The higher oligomerization propensities of the A4V and I112T variants may stem from the proximity of these substitutions to the native homodimeric interface (Figure 1A).
Identification of Species with Potential Toxicity in ALS
While we observe differences in aggregation propensities among the SOD1 variants studied, the formation of certain metastable non-native oligomers (such as those eluting at 14.5 mL) by both wild type and FALS mutant SOD1 suggests that some common mechanism(s) underlie SOD1 oligomerization. To explore potentially unifying conformational changes that occur as apo-SOD1 transitions from dimeric to monomeric and higher-order oligomeric species, we analyzed the exposure of a putatively disease-relevant epitope. The epitope recognized by C4F6 is not known, but this antibody binds to soluble misfolded SOD1 in disease-affected motor neuron populations in ALS patient spinal cord.7,8 For this reason, the C4F6 monoclonal antibody has been proposed to recognize an epitope present specifically in toxic conformers of SOD1.7 We find that the C4F6 antibody binds to several higher-order oligomers of apo-SOD1 but not to monomers and rarely to native-like dimers (Figure 4). The C4F6 antibody was raised against apo-G93A and has been shown to have specific reactivity to this sequence element (WT SOD1, FALS mutants other than G93A, and SOD1 with other substitutions other than alanine at position 93 show little reactivity to C4F6 when denatured).8,27 However, C4F6 also exhibits conformation-specific reactivity, which is not restricted to G93A under nondenaturing conditions.8 We probe apo-SOD1 oligomers under nondenaturing conditions in order to detect species with the conformation-specific epitope recognized by C4F6 in ALS-affected motor neurons. The strong C4F6 reactivity of unmodified apo-G93A oligomers may partially be due to the recognition of the G93A sequence epitope. Previous work has implicated soluble misfolded SOD1 in a range of oligomeric states in specific cytotoxic phenomena;5,22,23 our findings suggest that, of the pool of soluble species formed by apo-SOD1 in vitro, metastable oligomers larger than the native dimer are the most likely toxic culprits.
Oxidative Modification of Cys-111 Induces Conformational Changes That Promote Oligomer Assembly and Exposure of the Disease-Linked C4F6 Epitope
Oxidation of SOD1 has been shown to induce its misfolding and aggregation,8,28,29 and various oxidized forms of SOD1 have been linked to toxicity in cultured neurons,30 a mouse model of FALS,31 and a subset of sporadic ALS cases.32 In particular, the presence of an oxidizable cysteine at position 111 has been shown to promote SOD1 aggregation,13,29 an effect that has been widely attributed to the stabilization of insoluble aggregates and soluble oligomers by intermolecular disulfide bonds involving Cys-111.14,24,31,33 However, others have suggested that intermolecular disulfide cross-linking is a secondary event to non-native oligomer assembly and is not universally present in SOD1 oligomers.34,35
We find that glutathionylation of Cys-111, a reversible oxidative modification present extensively on SOD1 from human tissue,10,20,36,37 increases the proportion of monomeric apo-SOD1 in all studied variants and enhances the formation of several soluble non-native oligomers (Figure 2B). This observation, as well as the increased C4F6 reactivity of GS-I112T-SOD1 and GS-A4V-SOD1 oligomers (Figure 4), suggests that conformational changes in SOD1 induced by Cys-111 glutathionylation18,19 have significant effects on the abundance and morphologies of SOD1 oligomers. In particular, the altered morphologies of GS-SOD1 oligomers suggest that glutathionylation induces substantial conformational change(s) not limited to those which result in a weakened dimer interface. That is, rather than simply increasing the population of monomers available to oligomerize, glutathionylation alters the structural features of oligomers themselves. The alteration of oligomer structure induced by this modification is particularly notable since it increases exposure of an epitope linked to toxicity in ALS. We also find that while intermolecular disulfide bonds stabilize higher-order soluble SOD1 oligomers, these bonds are absent or not essential for stability in the smallest and earliest-appearing non-native oligomers (the O1 population, Figure 5).
Taken together, these results suggest that intermolecular disulfide cross-linking represents just one mechanism by which Cys-111 facilitates oligomerization. At the earliest stages of SOD1 misfolding and aggregation, oxidative modification of Cys-111 induces conformational changes that destabilize the dimer18,19 and favor assembly into potentially toxic non-native oligomers (Figures 2B and 4). Given the central role of Cys-111 in SOD1 aggregation (13) and the abundance of glutathionylated SOD1 in human tissue,10 we hypothesize that Cys-111 glutathionylation is a physiologically relevant mechanism by which oxidative stress induces aberrant oligomerization of SOD1.
Overall, our results highlight the toxic potential of soluble oligomers of apo-SOD1 and demonstrate the ability of Cys-111 oxidation to promote the formation of oligomers with the disease-linked epitope. The latter finding implicates oxidative stress as a factor in the cellular environment that can induce the formation of potentially toxic SOD1 oligomers. Our use of C4F6 binding as a proxy for disease relevance is, to the best of our knowledge, the first evaluation of the potential toxicities of SOD1 oligomers isolated in vitro. Enhanced exposure of the disease-linked epitope in non-native SOD1 oligomers supports a cytotoxic role for these assemblies, in parallel with previous findings directly demonstrating the toxicity of small oligomers of Aβ and α-synuclein in models of Alzheimer’s disease and Parkinson’s disease, respectively.38,39 A pattern is thus emerging among numerous neurodegenerative disorders in which small oligomers exert neurotoxic effects that are mitigated by assembly into large, insoluble species such as amyloid fibrils.40,41 Inhibition of small oligomer formation of disease-linked proteins therefore represents a therapeutic approach with potentially broad applicability to many neurodegenerative disorders. Knowledge of atomic-level structural features of putatively toxic soluble SOD1 oligomers and identification of factors modulating their formation would facilitate the direct determination of their contribution(s) to cellular pathology, as well as provide an avenue for the development of antioligomerization therapeutics for ALS.
Acknowledgments
We thank Dr. Joan S. Valentine for graciously providing the EG118 yeast strain and yEP351:hwtSOD1 vector and Drs. Ashutosh Tripathy, Xian Chen, and David Smalley for instrument training and helpful discussions.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FALS
familial amyotrophic lateral sclerosis
- GSSG
oxidized glutathione
- MALS
multiangle light scattering
- SEC
size exclusion chromatography
- SOD1
Cu/Zn superoxide dismutase
This work was supported by the National Institutes of Health grant R01GM080742 to N.V.D. R.L.R. was supported by the National Institutes of Health Predoctoral Fellowship F31NS073435 from the National Institute of Neurological Disorders and Stroke.
This research is based in part upon work conducted using the UNC Michael Hooker Proteomics Center, which is supported in part by the NIH-NCI Grant No. CA016086 to the Lineberger Comprehensive Cancer Center.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Khare S. D.; Caplow M.; Dokholyan N. V. (2006) FALS mutations in Cu, Zn superoxide dismutase destabilize the dimer and increase dimer dissociation propensity: a large-scale thermodynamic analysis. Amyloid 13, 226–235. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Johnson J. L.; Agar N. Y. .; Agar J. N. (2008) Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 6, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Xu G.; Borchelt D. R. (2002) High molecular weight complexes of mutant superoxide dismutase 1: age-dependent and tissue-specific accumulation. Neurobiol. Dis. 9, 139–148. [DOI] [PubMed] [Google Scholar]
- Zetterström P.; Stewart H. G.; Bergemalm D.; Jonsson P. A.; Graffmo K. S.; Andersen P. M.; Brännström T.; Oliveberg M.; Marklund S. L. (2007) Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl. Acad. Sci. U.S.A. 104, 14157–14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H.; Kadowaki H.; Nagai A.; Maruyama T.; Yokota T.; Fukutomi H.; Noguchi T.; Matsuzawa A.; Takeda K.; Ichijo H. (2008) ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 22, 1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M.; Durazo A.; Whitelegge J. P.; Borchelt D. R. (2010) An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Hum. Mol. Genet. 19, 4774–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton T. E.; Li Y.; Cooper D.; Gearing M.; Julien J.-P.; Rothstein J. D.; Boylan K.; Glass J. D. (2012) Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proc. Natl. Acad. Sci. U.S.A. 109, 5505–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco D. A.; Morfini G.; Karabacak N. M.; Song Y.; Gros-Louis F.; Pasinelli P.; Goolsby H.; Fontaine B. A.; Lemay N.; McKenna-Yasek D.; Frosch M. P.; Agar J. N.; Julien J.-P.; Brady S. T.; Brown R. H. Jr. (2010) Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 13, 1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K.; Andersen P. M.; Marklund S. L.; Brännström T. (2011) Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 121, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox K. C.; Zhou L.; Jordon J. K.; Huang Y.; Yu Y.; Redler R. L.; Chen X.; Caplow M.; Dokholyan N. V. (2009) Modifications of superoxide dismutase (SOD1) in human erythrocytes: a possible role in amyotrophic lateral sclerosis. J. Biol. Chem. 284, 13940–13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goscin S. A.; Fridovich I. (1972) The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochim. Biophys. Acta 289, 276–283. [DOI] [PubMed] [Google Scholar]
- Sahin E., and Roberts C. (2012) Size-Exclusion Chromatography with Multi-angle Light Scattering for Elucidating Protein Aggregation Mechanisms, in Therapeutic Proteins (Voynov V., and Caravella J. A., Eds.), pp 403–423, Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Cozzolino M.; Amori I.; Grazia Pesaresi M.; Ferri A.; Nencini M.; Teresa Carrì M. (2008) Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 283, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri A.; Fiorenzo P.; Nencini M.; Cozzolino M.; Pesaresi M. G.; Valle C.; Sepe S.; Moreno S.; Carrì M. T. (2010) Glutaredoxin 2 prevents aggregation of mutant SOD1 in mitochondria and abolishes its toxicity. Hum. Mol. Genet. 19, 4529–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S. D.; Caplow M.; Dokholyan N. V. (2004) The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 101, 15094–15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F.; Dokholyan N. V. (2008) Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc. Natl. Acad. Sci. U.S.A. 105, 19696–19701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I.; Rossi R.; Colombo G.; Giustarini D.; Milzani A. (2009) Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96. [DOI] [PubMed] [Google Scholar]
- Redler R. L.; Wilcox K. C.; Proctor E. A.; Fee L.; Caplow M.; Dokholyan N. V. (2011) Glutathionylation at Cys-111 induces dissociation of wild type and FALS mutant SOD1 dimers. Biochemistry 50, 7057–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor E. A.; Ding F.; Dokholyan N. V. (2011) Structural and thermodynamic effects of post-translational modifications in mutant and wild type Cu, Zn superoxide dismutase. J. Mol. Biol. 408, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund S. L.; Andersen P. M.; Forsgren L.; Nilsson P.; Ohlsson P.-I.; Wikander G.; Öberg A. (1997) Normal binding and reactivity of copper in mutant superoxide dismutase isolated from amyotrophic lateral sclerosis patients. J. Neurochem. 69, 675–681. [DOI] [PubMed] [Google Scholar]
- McAlary L.; Yerbury J. J.; Aquilina J. A. (2013) Glutathionylation potentiates benign superoxide dismutase 1 variants to the toxic forms associated with amyotrophic lateral sclerosis. Sci. Rep. 3, 3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi H.; Almer G.; Yamashita S.; Guégan C.; Nagai M.; Xu Z.; Sosunov A. A.; McKhann G. M. II; Przedborski S. (2006) Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl. Acad. Sci. U.S.A. 103, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushitani M.; Kurisu J.; Tsukita K.; Takahashi R. (2002) Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. J. Neurochem. 83, 1030–1042. [DOI] [PubMed] [Google Scholar]
- Banci L.; Bertini I.; Durazo A.; Girotto S.; Gralla E. B.; Martinelli M.; Valentine J. S.; Vieru M.; Whitelegge J. P. (2007) Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc. Natl. Acad. Sci. U.S.A. 104, 11263–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toichi K.; Yamanaka K.; Furukawa Y. (2013) Disulfide scrambling describes the oligomer formation of superoxide dismutase (SOD1) proteins in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 288, 4970–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassall K. A.; Stubbs H. R.; Primmer H. A.; Tong M. S.; Sullivan S. M.; Sobering R.; Srinivasan S.; Briere L.-A. K.; Dunn S. D.; Colon W.; Meiering E. M. (2011) Decreased stability and increased formation of soluble aggregates by immature superoxide dismutase do not account for disease severity in ALS. Proc. Natl. Acad. Sci. U.S.A. 108, 2210–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushitani M.; Ezzi S. A.; Julien J.-P. (2007) Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 104, 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhit R.; Cunningham P.; Furtos-Matei A.; Dahan S.; Qi X.-F.; Crow J. P.; Cashman N. R.; Kondejewski L. H.; Chakrabartty A. (2002) Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 277, 47551–47556. [DOI] [PubMed] [Google Scholar]
- Fujiwara N.; Nakano M.; Kato S.; Yoshihara D.; Ookawara T.; Eguchi H.; Taniguchi N.; Suzuki K. (2007) Oxidative modification to cysteine sulfonic acid of Cys111 in human copper-zinc superoxide dismutase. J. Biol. Chem. 282, 35933–35944. [DOI] [PubMed] [Google Scholar]
- Ezzi S. A.; Urushitani M.; Julien J.-P. (2007) Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J. Neurochem. 102, 170–178. [DOI] [PubMed] [Google Scholar]
- Furukawa Y.; Fu R.; Deng H.-X.; Siddique T.; O’Halloran T. V. (2006) Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc. Natl. Acad. Sci. U.S.A. 103, 7148–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guareschi S.; Cova E.; Cereda C.; Ceroni M.; Donetti E.; Bosco D. A.; Trotti D.; Pasinelli P. (2012) An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc. Natl. Acad. Sci. U.S.A. 109, 5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y.; O’Halloran T. V. (2005) Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J. Biol. Chem. 280, 17266–17274. [DOI] [PubMed] [Google Scholar]
- Karch C. M.; Borchelt D. R. (2008) A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 283, 13528–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Shang H.; Qiu X.; Fujiwara N.; Cui L.; Li X.-M.; Gao T.-M.; Kong J. (2012) Oxidative modification of cysteine 111 promotes disulfide bond-independent aggregation of SOD1. Neurochem. Res. 37, 835–845. [DOI] [PubMed] [Google Scholar]
- Nakanishi T.; Kishikawa M.; Miyazaki A.; Shimizu A.; Ogawa Y.; Sakoda S.; Ohi T.; Shoji H. (1998) Simple and defined method to detect the SOD-1 mutants from patients with familial amyotrophic lateral sclerosis by mass spectrometry. J. Neurosci. Methods 81, 41–44. [DOI] [PubMed] [Google Scholar]
- Schininà M. E.; Carlini P.; Polticelli F.; Zappacosta F.; Bossa F.; Calabrese L. (1996) Amino acid sequence of chicken Cu, Zn-containing superoxide dismutase and identification of glutathionyl adducts at exposed cysteine residues. Eur. J. Biochem. 237, 433–439. [DOI] [PubMed] [Google Scholar]
- Walsh D. M.; Klyubin I.; Fadeeva J. V.; Cullen W. K.; Anwyl R.; Wolfe M. S.; Rowan M. J.; Selkoe D. J. (2002) Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539. [DOI] [PubMed] [Google Scholar]
- Winner B.; Jappelli R.; Maji S. K.; Desplats P. A.; Boyer L.; Aigner S.; Hetzer C.; Loher T.; Vilar M.; Campioni S.; Tzitzilonis C.; Soragni A.; Jessberger S.; Mira H.; Consiglio A.; Pham E.; Masliah E.; Gage F. H.; Riek R. (2011) In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U.S.A. 108, 4194–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze M. D.; Bitan G.; Teplow D. B. (2002) Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 69, 567–577. [DOI] [PubMed] [Google Scholar]
- Haass C.; Selkoe D. J. (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112. [DOI] [PubMed] [Google Scholar]