Abstract
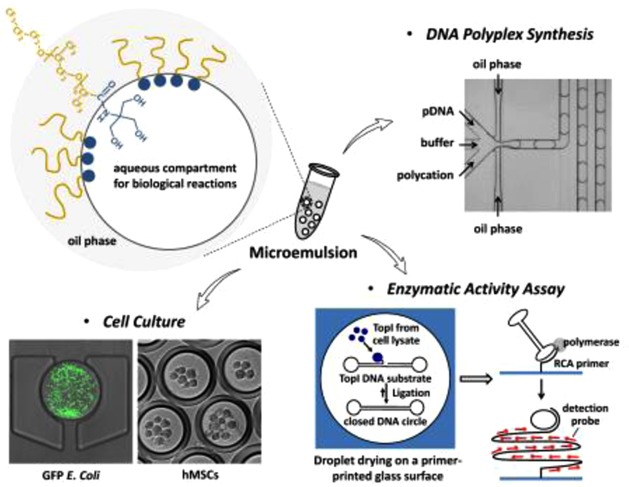
Microemulsion represents an attractive platform for fundamental and applied biomedical research because the emulsified droplets can serve as millions of compartmentalized micrometer-sized reactors amenable to high-throughput screening or online monitoring. However, establishing stable emulsions with surfactants that are compatible with biological applications remains a significant challenge. Motivated by the lack of commercially available surfactants suitable for microemulsion-based biological assays, this study describes the facile synthesis of a biocompatible fluorosurfactant with nonionic tris(hydroxymethyl)methyl (Tris) polar head groups. We have further demonstrated compatibility of the developed surfactant with diverse emulsion-based applications, including DNA polymeric nanoparticle synthesis, enzymatic activity assay, and bacterial or mammalian cell culture, in the setup of both double- and multiphases of emulsions.
Keywords: biomaterials, emulsions, fluorosurfactant, microdroplets, microfluidics
Microemulsion is an attractive platform for fundamental and applied biomedical research because the emulsified droplets can serve as millions of compartmentalized micrometer-sized reactors in a single vessel.1,2 For instance, water-in-oil (w/o) droplets are one of the commonly used miniaturized incubators for manipulation of a single gene, cell, or organism.1,3−5 The mini-incubators can maintain a local concentration of reagents at an effective level, allowing reduced sample consumption.3 Recent development in active droplet sorting and manipulation techniques, such as those activated by dielectrophoresis,6 fluorescence,7,8 and standing surface acoustic waves,9 has further promoted the droplet-based microfluidics for high-throughput in vitro screening. In addition, confined diffusion in the small volume of emulsified droplets benefits from efficient molecular transport and uniform mixing, thus resulting in reduction of reaction time.10 Although microemulsion is an attractive platform for diverse biological applications, establishing a suitable emulsification system remains a significant challenge.11 Formulation of stable w/o emulsions requires a proper surfactant for a specific water/oil pair. Surfactant is essential for maintaining the interfacial force between the two phases, thereby preventing the emulsion from collapsing or coalescing. Since any reagent entrapped in the w/o emulsion can come into contact with the polar portion of the amphiphilic surfactant, the composition of the surfactant is critical. The efficiency and performance of biological reactions in the microemulsion would be affected if there is any adverse interaction between the surfactant and the biomolecules or ionic components dissolved in the aqueous phase.11
Microfluidics has recently emerged as an attractive strategy for the generation of well-controlled microemulsions.2,12,13 Within the reported designs, polydimethylsiloxane (PDMS) is one of the most widely used polymers in soft lithography for the fabrication of microfluidic devices. However, when PDMS is in contact with conventional silicon and hydrocarbon oils, the polymer may swell or shrink, leading to device delamination or channel blockage, and thus unstable emulsion generation. Therefore, there is a growing trend in the use of fluorinated oil as an alternative in the generation of microemulsions, due to the minimized deformation of PDMS polymers.11 Moreover, fluorinated oil has minimal interaction with biomolecules and higher solubility of respiratory gases than water, making it particularly appealing for long-term cell culture and tissue engineering applications.4,14 Fluorosurfactants, or fluorinated surfactants, soluble in both water and fluorinated oil, are also less volatile and more stable at high temperature than hydrocarbon-based surfactants due to the stability of the carbon–fluorine bond.11 Fluorinated emulsion is thus appealing, but the availability of fluorosurfactants is limited.4,11,15−18
Perfluoropolyethers (PFPE), a unique class of commercially available fluorosurfactants from DuPont, such as Krytox 157FSH, can generate emulsions in PDMS microchannels, but the emulsions are progressively unstable.15 Moreover, the carboxylic hydrophilic head groups of PFPE would interact with many biomolecules or polyelectrolytes, limiting its appeal for biological applications. Commercially available surfactants with short fluorinated tails, such as 1H,1H,2H,2H-perfluoro-1-octanol (PFO), have been reported for the study of chemical reactions and screening of protein crystallization conditions.19,20 However, this category of surfactants still suffers from the problem of nonspecific binding with proteins, such as fibrinogen.21 Chemistry innovation has subsequently been demonstrated using amphiphilic molecules, such as perfluoropolyether and perfluoroalkyl chains, for the modification of the PFPE head groups.4,17 Among these, the polyethylene glycol (PEG) based block copolymer has shown the most promising results in preventing nonspecific adsorption of biological materials, as well as enabling effective encapsulation of mammalian cells.22,23 However, the modification chemistry is relatively complex and the optimized PEG reactant is not commercially available.15,24 In this study, we present a novel biocompatible PFPE-based fluorinated surfactant by a facile synthesis of converting the ionic carboxyl head groups of PFPE into nonionic tris(hydroxymethyl)methyl groups (Tris) (Figure 1a). We further demonstrate the biocompatibility and versatility of PFPE–Tris by conducting three different biological reactions, namely, DNA polymeric nanoparticle production, enzyme detection with clinical samples, and cell cultures, in the PFPE–Tris-based w/o microemulsions.
Figure 1.
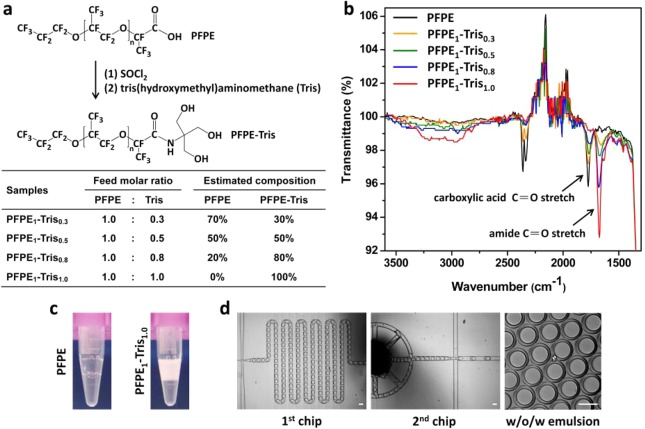
(a) Schematic illustration of the synthesis of PFPE–Tris surfactants with estimated composition and (b) the corresponding FT-IR spectra. The absorbance peak at 1775 cm–1 corresponds to the carboxylic acid C=O stretch of unreacted PFPE, whereas the absorption peak at 1675 cm–1 represents the converted amide C=O stretch. The maximal conversion occurred when the absorbance peak of 1775 cm–1, representing the amount of the unreacted PFPE, completely disappeared, at the feed molar ratio of 1.0. The percentage of conversion, shown as a table in (a), was estimated by the relative peak height on the FT-IR spectra. (c) Photographs of off-chip collected w/o emulsion generated by a flow-focusing droplet generator with 2 wt % unmodified PFPE or PFPE1–Tris1.0 dissolved in HFE-7500 as the oil phase. (d) Formation of water-in-oil-in-water (w/o/w) double emulsion by a two-step emulsification process using two integrated microfluidic chips (scale bar: 100 μm) and 2 wt % PFPE1–Tris1.0 in HFE-7500 as the oil phase.
Results and Discussion
Synthesis and Characterization of the PFPE–Tris-Based Surfactant
The amphiphilic characteristics of PFPE were modulated by the feed molar ratios of Tris to PFPE of 0.3, 0.5, 0.8, and 1.0. The viscosity of the final product increased with the increased molar ratio of Tris. For example, PFPE1–Tris1.0 became wax-like at room temperature (see Figure S1 in the Supporting Information). The conversion of the COOH group into the nonionic Tris was verified by Fourier transform infrared spectroscopy (FT-IR) (Figure 1b). As the molar ratio of Tris increased, the absorbance peak at 1775 cm–1 (corresponding to a carboxylic acid C=O stretch of unreacted PFPE) decreased and was accompanied by the appearance of an absorption peak at 1675 cm–1 (corresponding to an amide C=O stretch). The maximal conversion plateaued at PFPE1–Tris1.0, when the absorbance peak of 1775 cm–1, representing the amount of the unreacted PFPE, completely disappeared. Additional increase of the Tris reactant did not result in significant change of the FT-IR spectrum (see Figure S1 in the Supporting Information). To evaluate the performance of the modified PFPEs, we compared PFPE1–Tris1.0 with the unmodified PFPE in generating emulsions in a flow-focusing-based droplet generator,10 using 2 wt % of surfactant dissolved in Novec HFE-7500 fluorinated oil as the continuous phase. As shown in Figure 1c, when the unmodified PFPE was used, the w/o emulsions tended to coalesce and phase-separate immediately after collection in the tube. In contrast, PFPE1–Tris1.0-based emulsions exhibited superior stability, with droplets remaining a uniform micrometer-size during collection, with no obvious coalescence or phase separation observed even after one week of storage. We further demonstrated the applicability of PFPE–Tris surfactants for generation of multiphase emulsions. As shown in Figure 1d, water-in-oil-in-water (w/o/w) emulsions were prepared by a two-step emulsification process using two serially connected microfluidic chips, as demonstrated previously.25 Briefly, the w/o emulsion formed in the first chip was flowed into the second chip, forming a highly stable w/o/w emulsion. This highlights the potential of PFPE–Tris in generating highly stable emulsions for multistep processing in biological studies.
Preparation of DNA Nanocomplexes in PFPE–Tris-Based Microemulsions
We then investigated the compatibility of PFPE–Tris in diverse droplet-based applications, including DNA nanocomplex synthesis, an enzymatic activity assay, bacterial growth analysis, and mammalian cell culture. We have previously shown that the self-assembly of polycation and plasmid DNA in a confined picoliter-sized volume, as in microfluidics-generated droplets, would lead to nanocomplexes (or polyplexes) that are more uniform in size, more compact, and higher in transfection efficiency.10,26,27 The choice of a surfactant is critical because the polycation or DNA might interact with the polar end of the surfactant. In this study, we first addressed the issue of interaction by mixing the polycation (jetPEI) and DNA in w/o emulsion droplets prepared by bulk-mixing. This configuration would not generate uniform droplets but would eliminate any possible complications coming from interaction of the reactants with the PDMS microchannels. As shown in Figure 2, the transfection efficiency of the polyplexes synthesized in the fluorinated microemulsions was correlated with the composition of the surfactants. At N/P ratio = 5, defined as the ratio of nitrogen (N) in the polymer to phosphate (P) in the nucleic acid, the percentage of HEK cells transfected (GFP+ cells; GFP = green fluorescent protein) was only 4% when the unmodified PFPE was used, compared to 60% in the positive control obtained from a standard transfection protocol using commercially available jetPEI without any involvement of surfactant (Figure 2a). As the Tris component in PFPE increased, the transfection efficiency increased toward the 60% transfection level. In addition to comparing the percent of cells transfected, we also assessed the transfection efficiency by measuring the total fluorescence intensity. The data normalized to the control are shown in Figure 2b. Again, at a N/P ratio of 5 or 6, the transfection level was below the control with the unmodified or lightly modified PFPE (PFPE1–Tris0.3), but equaled or exceeded the control at higher N/P ratios.
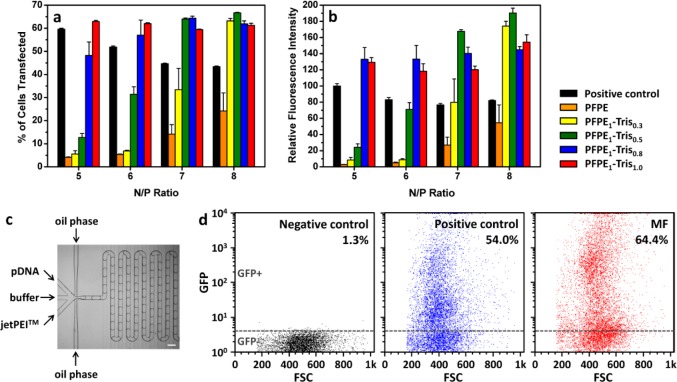
Figure 2.
Transfection efficiency of jetPEI polyplexes synthesized in w/o emulsion droplets stabilized with different synthesized surfactants. (a) Percent of cells transfected and (b) relative fluorescence intensity of transfected GFP+ cells (n = 3). The w/o emulsion in (a) and (b) was formed by the vortexing method. (c) Setup of microfluidic-assisted synthesis of jetPEI polyplexes using PFPE1–Tris1.0 and at a N/P ratio of 5 (scale bar: 200 μm) and (d) the corresponding transfection efficiency measured by GFP+ and GFP– cell population in the GFP/FSC plot. The cells without any treatment were used as a negative control to identify GFP– cells. Standard jetPEI polyplex formulation, without emulsion treatment, was used as a positive control.
We hypothesized that the poor transfection observed at the low N/P ratios was caused by interactions between the negatively charged carboxyl group of PFPE and the positively charged jetPEI molecules. This was inferred in an experiment where 100% of the DNA could be recovered from w/o emulsions stabilized with all the surfactants tested (Figure S2 in Supporting Information) and corroborated by the transfection results using a luciferase reporter gene at N/P = 8, when the excess polycation would compensate for any loss in the interaction with the unmodified PFPE and PFPE1–Tris0.3 (Figure S3).
The compatibility of PFPE1–Tris1.0 with microfluidic usage was then established in the synthesis of polyplexes in a w/o emulsion generated in a PDMS microchannel (Figure 2c). Figure 2d shows that, even at the low N/P ratio of 5, the percentage of cells transfected was higher than the positive control, as evidenced by the shift toward higher GFP intensities. Collectively, the results of Figures 2b,d and S3 confirmed that the polyplexes synthesized in the confined volume of a w/o droplet could perform as well as, or better than the polyplexes prepared in bulk. This is consistent with our previous observations that self-assembly of these nanocomplexes in a small confined volume is advantageous and that the choice of a suitable surfactant is important.10,26
Enzymatic Activities at the Single-Cell Level
To further investigate the biocompatibility of PFPE–Tris, we conducted an enzymatic activity assay in the droplets based on the previously developed REEAD (rolling-circle enhanced enzyme activity detection) on a chip setup.28,29 Briefly, cell suspension, a low salt buffer, and DNA REEAD substrate(s) designed to specifically monitor the activity of the enzyme topoisomerase I (TopI) were entrapped in w/o emulsion droplets generated in PDMS microchannels using PFPE1–Tris1.0 (Figure 3a). TopI released by cell lysis interacted with the REEAD substrates, forming closed DNA circles. Subsequently, droplets were collected on a drop-trap placed on top of a primer-activated glass slide (Figure 3b). Here the enzymatic products, i.e., the closed DNA circles, would hybridize to a specific primer printed on the glass surface and were subjected to isothermal rolling circle amplification (RCA, Figure 3c). Hybridization of appropriate fluorescent probes to the RCA amplified product allowed the enzymatic activity to be quantified through the detected fluorescent signals observed in a fluorescence microscope. We demonstrated the utility of the REEAD assay in detecting Plasmodium parasites that are present in saliva of malaria-infected patients30via the detection of Plasmodium topoisomerase I (pTopI). The saliva sample was introduced into the droplets along with the REEAD substrates recognized by Plasmodium topoisomerase I (pTopI) and human topoisomerase I (hTopI), respectively.29Figure 3d indicates that enzymatic activities of pTopI (red) and hTopI (green) originated from the parasites and the human cells, respectively, were detectable in the saliva of the patient. These experiments showed that PFPE1–Tris1.0 did not interfere with any of the steps in the assay and that it would be suitable for cell-based lab-on-chip diagnostics.
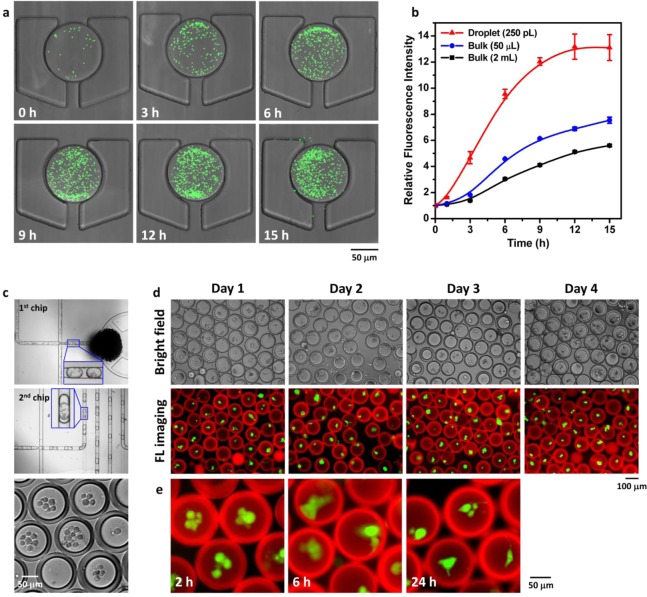
Figure 3.
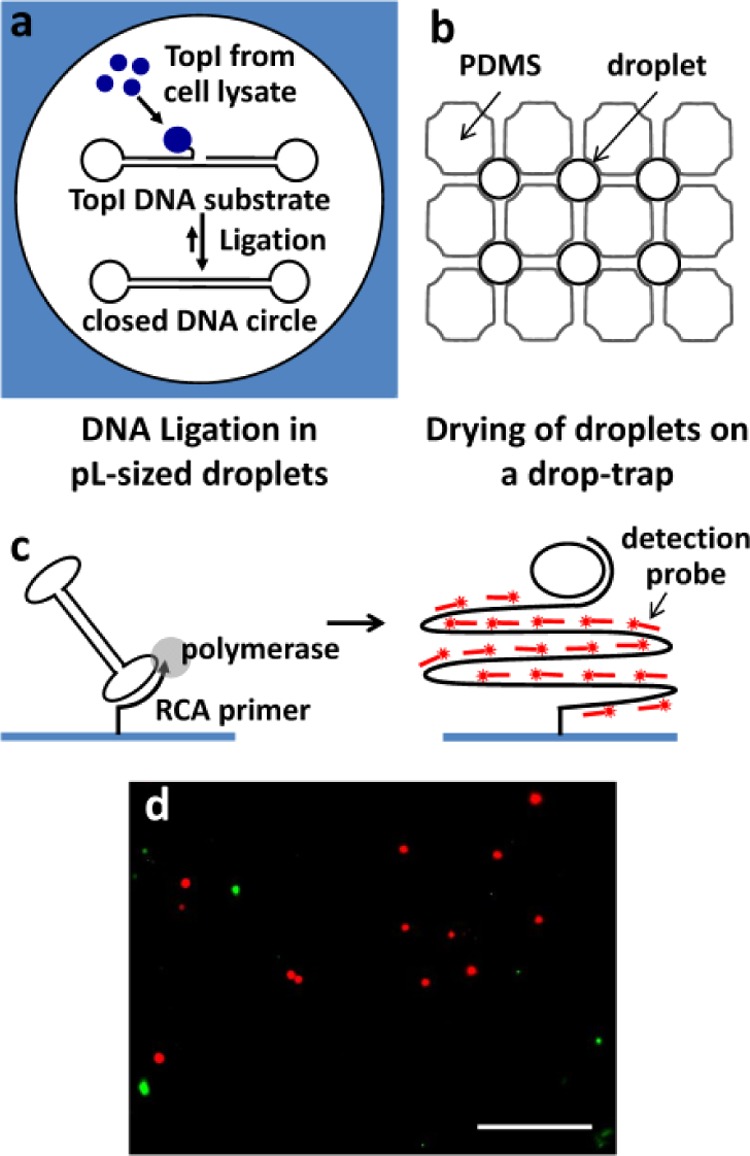
Detection of enzymatic activities for diagnostics by (a) confining the topoisomerase I (TopI) reaction in w/o emulsion droplets, (b) collection and air drying of droplets in a drop-trap, and then (c) performing isothermal rolling circle amplification (RCA) reaction initiated by hybridization of the closed DNA circle with a specific primer printed on the glass surface. (d) Detection of malaria-causing Plasmodium parasites in saliva sample. Plasmodium TopI (red) activity is clearly differentiated from human TopI (green) activity. Scale bar: 25 μm.
Cell Culture and Long-Term Cultivation in Microemulsions
Finally we examined if the w/o emulsion and w/o/w double emulsion stabilized by PFPE1–Tris1.0 (Figure 1d) would be suitable for cell culture and cell-cluster cultivation. Figure 4a shows GFP-expressing Escherichia coli (E. coli) entrapped in an individual w/o emulsion droplet collected on the drop-trap. The rapid bacterial growth was monitored by the increase in GFP signal. The bacterial growth curve (Figure 4b) displayed the typical three phases: lag phase, exponential growth phase, and stationary phase. Interestingly, the bacterial growth rate appeared to inversely correlate with the culture volume given the same starting cell density. This may be attributed to the more efficient gas transport that benefits bacterial growth in the smaller culture volume. The culture in 50 μL had a much shallower depth of medium in the culture dish than the 2 mL culture. The 250 pL droplet in turn had a much higher surface-to-volume (S/V) ratio than the culture dish format. This is consistent with literature reports that bacterial growth rates are highly dependent on the S/V ratio of liquid cultures.31 The E. coli observed outside the intact droplet at 15 h came from bacteria bursting out of other droplets in the drop-trap collection.
Figure 4.
Use of PFPE1–Tris1.0-stabilized emulsions for bacterial or mammalian cell culture. (a) Confocal images of GFP-expressing E. coli cultured in w/o emulsion droplets (250 pL) collected in a drop-trap; (b) their corresponding proliferation rate and comparison with conventional batch/bulk culture (50 μL or 2 mL) (n = 5). (c) Photographs of two-step microfluidic encapsulation of bone marrow-derived human mesenchymal stem cells (hMSCs) in w/o/w double emulsion; fluorescence images of the encapsulated hMSCs for (d) cell viability assay and (e) cell attachment and spreading observation at the indicated time points taken by a fluorescence microscope. The live cells were stained with calcein-AM (green), and the oil layer composed of 2 wt % PFPE1–Tris1.0 in HFE-7500 was stained with DAPI (red).
Compared to bacteria, mammalian cells are more sensitive to their cultivation conditions. Mammalian cells cultured in w/o emulsion droplets often suffer from local acidification of culture medium and high osmolarity caused by accumulation of two major cellular metabolic byproducts, lactate and ammonium ions.32Figure 4c shows the encapsulation of bone marrow-derived human mesenchymal stem cells (hMSCs) in w/o/w double emulsion, where the thin (∼15–20 μm) oil layer (HFE-7500) allowed nutrient and waste product exchange between the interior and exterior aqueous environment of the droplet.25Figure 4d shows that hMSCs cultured in these w/o/w droplets maintained their viability (>85%) for at least 4 days (Figure S4 in the Supporting Information). There was a good correlation between cells in the bright field and those under fluorescence imaging, where the green signal indicated live cells stained by calcein-AM. Another indication of the compatibility of the surfactant is the morphology of the hMSCs in the droplet. hMSCs are anchorage-dependent cells,33 which adhere to a culture dish and spread in standard culture conditions. Shown as a comparison in Figure 4e, the hMSCs cultured in the emulsion droplets initially showed a rounded morphology at 2 h, but spread at later time points, suggesting that the PFPE1–Tris1.0-stabilized interface could serve as a support for hMSC attachment. It must be noted that formation of hMSC aggregates or clusters, which also happened in these droplets, could also be valuable for differentiation purposes.25 The important point is that the hMSCs can retain their viability in these droplets.
Conclusions
Surfactant development is underappreciated. Yet it is central to the development of any microemulsion-based applications. The limited choice of commercially available surfactants motivated us to come up with an alternative. In this study, we describe a series of PFPE–Tris-based surfactants that can be readily synthesized in any lab and demonstrate their suitability in the production of stable single and double emulsions. We further establish their compatibility with diverse emulsion-based biological applications, including synthesis of DNA-based polymeric nanoparticles, investigation of enzymatic activities in saliva for diagnosis of infectious disease, and culture of bacterial and mammalian cells. This study is expected to accelerate the development in microemulsion-based biological assays, particular those associated with microfluidics, by providing an easy-to-synthesize fluorosurfactant.
Materials and Methods
Materials
DuPont Krytox 157-FSH (a perfluoropolyether, MW 7000–7500) and 3M Novec HFE-7100 and HFE-7500 were purchased from Miller-Stephenson Chemical Co. Inc. (Danbury, CT, USA). All other analytical grade chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Synthesis of PFPE–Tris
The PFPE–Tris was synthesized via a two-step process: (1) PFPE–COOH to PFPE–COCl and (2) PFPE–COCl to PFPE–Tris. First, the PFPE was treated with thionyl chloride (SOCl2), a commonly used compound in organic synthesis, to convert the carboxyl end group to an acyl chloride group, due to the ease of purification.34 Briefly, SOCl2 (0.81 g, 6.897 mmol) in a 10:1 molar excess relative to the PFPE was added to an HFE-7100 solution of PFPE (5 g, 0.6879 mmol) in a three-neck flask purged with argon gas. The reaction was refluxed with a condenser and stirred for 24 h at 50 °C. The resulting mixture was concentrated using a rotary evaporator while excess SOCl2, hydrochloric acid, and sulfur dioxide byproducts, all being gaseous compounds, were removed under vacuum.34 The obtained PFPE-COCl was then used to prepare PFPE–Tris. In the second step, a mixture of HFE-7100 (10 mL) and benzotrifluoride (6 mL) was added to dissolve PFPE–COCl, followed by the addition of respective amounts of tris(hydroxymethylaminomethane) (0.025, 0.042, 0.067, or 0.084 g) under a slow argon purge. The refluxing reaction was conducted at 60 °C for 24 h. The purification of unreacted Tris, if there was any, employed the water solubility of Tris, as elaborated in the following. The crude product was first obtained by evaporation of the solvents, HFE-7100 (boiling point: 61 °C) and benzotrifluoride (boiling point: 103.46 °C), using a rotary evaporator. The obtained product was then redissolved in a small amount of HFE-7100, along with excess water for the extraction of unreacted Tris (note that Tris is soluble in water but not in fluorocarbon oil). Due to the higher density of HFE-7100 compared to water, the top layer would be the water phase containing the unreacted Tris. This phase could then be easily discarded subsequently. The same purification procedure was repeated five times. Finally, the purified surfactant was then dried under vacuum (in a yield of ca. 68–80%). The synthesized PFPE–Tris was analyzed by Fourier transformed infrared spectroscopy (Thermo Scientific Nicolet 6700 FT-IR spectrometer, Madison, WI, USA) to gauge the fraction of PFPE being converted to PFPE–Tris.
Fabrication of PDMS Chips
All microfluidic chips were fabricated using conventional soft lithography techniques.35 PDMS prepolymer was cured on an SU-8 Master (MicroChem, Newton, MA, USA) followed by bonding of fabricated PDMS onto a glass slide. PDMS prepolymer (Sylgard 184 silicone elastomer kit, Dow Corning, Midland, MI, USA) was prepared by mixing PDMS monomer base and curing agent in a 10:1 mass ratio, then applied to the SU-8 Master layered on a silicon wafer placed in a Petri dish and cured at 70 °C for 1 h. Solidified PDMS replica was peeled off from the wafer, cut out, and hole punched (hole puncher, Technical Innovations, Brazoria, TX, USA) as tubing inlets and outlets. To close the channels, the obtained PDMS chip was bonded to a cover slide coated with or without a thin layer of PDMS through thermal curing or oxygen plasma treatment for 30 s at 20 W (Plasma Asher, Quorum Technologies, West Sussex, RH, USA), respectively. The bonded PDMS chip was then left in an oven at 95 °C overnight to enhance the bonding strength. To increase the hydrophilicity of the PDMS microchannels for the formation of the w/o/w double emulsion, the oxygen plasma-treated chip was coated with poly(acrylic acid) following a reported protocol.36 The channel depth of the produced chips is around 50–70 μm.
Synthesis of jetPEI/DNA Nanocomplexes
To determine if the synthesized PFPE–Tris surfactants could offer a compatible interface for emulsion droplets as biocompatible microreactors, 50 μL of jetPEI (Polyplus-Transfection Inc., New York, NY, USA) was added to 50 μL of DNA solution under gentle vortex (following the manufacturer’s protocol), and this 100 μL mixture was immediately added into an equal volume of HFE-7500 oil containing 0.5 wt % PFPE surfactant and vortexed for an additional 1 to 2 s.
Alternatively, jetPEI/DNA complexes at a N/P ratio of 5 were synthesized in picoliter droplets (∼1250 pL) generated by a flow-focusing microfluidic device. The HFE-7500 carrier oil with low surfactant concentration (0.05 wt % PFPE1–Tris1.0) was used to provide enough emulsion stability and but could be easily broken down using a droplet-breaking agent (Droplet Destabilizer, RainDance Technologies, in 1:4 volume ratio). The flow rate of three streams of aqueous reagents (i.e., jetPEI, 0.15 M NaCl, pDNA) and carrier oil was controlled at 15 and 45 μL/min, respectively. W/O emulsions containing microfluidic-generated DNA nanoparticles were incubated for 15 min in a microcentrifuge tube, broken with droplet-releasing agent, and an aqueous phase containing the DNA nanoparticles was taken for cell transfection.
In Vitro Transfection
At 24 h prior to transfection, human embryonic kidney (HEK) 293 cells (ATCC CRL-1573) were seeded (1 × 105 cells/well) in 24-well plates and grown in complete media (MEM supplemented with 10% FBS, 2 mM l-glutamine, 50 U/mL penicillin, and 50 U/mL streptomycin, Invitrogen, Carlsbad, CA, USA). Prior to transfection, the media were removed and replenished with reduced-serum media (Opti-MEM) containing jetPEI/DNA complexes at a concentration of 1 μg DNA/well. Each preparation was performed in triplicate. At 4 h post-transfection, the transfection media containing nanocomplexes were replaced with FBS-containing media. To quantify GFP transfection efficiency, cells were incubated for 24 h post-transfection, detached by 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA), and fixed using 4% paraformaldehyde. Cells were analyzed by a flow cytometer equipped with a 488 nm argon laser (FACS Calibur, BD Biosciences, Franklin Lakes, NJ, USA). To quantify luciferase expression, cells were incubated for 48 h post-transfection and lysed with 160 μL of Glo lysis buffer (Promega, Madison, WI, USA), and cell debris was removed by centrifugation (15 000 rpm, 5 min). Subsequently, 50 μL of Steady-Glo luciferferase assay reagent (Promega, Madison, WI, USA) was added to 50 μL of the supernatant lysate. After 5 min, relative luminescence of the sample was determined by a microplate reader (FLUOstar Optima, BMG Labtech GmbH, Germany) and normalized to the total cell protein concentration by the bicinchoninic acid (BCA) protein assay.
Enzymatic Activity Assay
The enzymatic activity assay was conducted by using a previously developed microfluidic setup consisting of a flow-focusing droplet generator and a drop-trap.28,29 All oligonucleotides for construction of TopI DNA substrates, PCA primer, and fluorescently labeled detection probes were purchased from DNA Technology A/S (Risskov, Denmark). The sequences of the used oligonucleotides have been published previously.29,37
The saliva sample29 from the patient was introduced into the droplet generator, along with the lysis buffer (10 mM Tris-HCL pH 7.5, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.2% Tween 20) and hTopI and pTopI substrates (final concentration were 67 and 167 nM, respectively in the droplets). The generated droplets were harvested in Eppendorf tubes and placed on a primer-printed glass slide (CodeLink activated slides from SurModics) prepared as in the previous report.29 The PDMS drop-trap was gently placed on top of the glass slide. The geometry of the drop-trap was designed according to the size of generated droplets. The droplets were left to exsiccate for 16 h. Washing, RCA, and hybridization of probes were performed as previously described.37 Epifluorescent and bright field images were captured with an inverted fluorescence microscope (Axio Observer, Zeiss, Germany). Monocolor emission from each fluorophore was collected and filtered through appropriate filters and dichroics. Image processing and analysis was performed with MetaMorph (v.7.6.5).
Bacterial Growth in Emulsion Droplets
GFP-expressing Escherichia coli (E. coli) were grown at 37 °C under shaking at 250 rpm for 24 h in M9 growth medium with 0.1% kanamycin to allow saturation. On the following day, a 1/10 dilution of saturated E. coli culture in fresh medium was made and used for bacterial encapsulation in w/o emulsion, which was performed in a flow-focusing droplet generator at a control flow rate of 5 μL/min for both disperse phase (diluted bacterial culture) and continuous phase (2 wt % PFPE1–Tris1.0 in HFE-7500 oil). Generated w/o emulsion droplets were collected off-chip, reinjected into a trapping array, and analyzed for GFP signal (500–600 nm) under illumination of 488 nm using confocal laser scanning microscopy (Leica TCS SP5, Germany). The obtained images were further analyzed using ImageJ software to quantify GFP fluorescence signal intensity per droplet.25 Five individual droplets were defined as region of interest (ROI) for analysis to obtain a data point. In addition, the proliferation rate of GFP-E. coli in conventional batch/bulk culture (50 μL or 2 mL) was assayed using a 96-well microplate reader (FLUOstar OPTIMA, BMG Labtech) with an excitation filter of 485 nm and an emission filter of 520 nm to quantify GFP signal. The obtained signal or fluorescence intensities were normalized to the initial value (time = 0 h).
Encapsulation of hMSCs in W/O/W Emulsion
Double emulsion droplets were generated using a two-microfluidic-chip setup where the w/o emulsions were first formed in an untreated PDMS chip and then fed to a hydrophilic (poly(acrylic acid)-coated) chip to generate w/o/w double emulsions.
Bone marrow-derived hMSCs were kindly provided by Tulane University Health Sciences Center. Cells were cultured in α-minimum essential medium with 20% fetal bovine serum and 1% penicillin/streptomycin at 37 °C and 5% CO2. The 5–10th passages of the obtained hMSCs were used in this study. For cell encapsulation experiments, cells were trypsinized and suspended at (0.5–1) × 107 cell/mL in culture medium supplemented with 0.1 wt % Pluronic F-127. The oil phase used was HFE-7500 supplemented with 2 wt % PFPE1–Tris1.0 surfactant. The outer water phase used was culture medium supplemented with 5 wt % Pluronic F-127. The flow rates of three phases in emulsion generation (inner medium:middle oil:outer medium) were chosen to be 5:5:15 μL/min respectively.25,38 The formed double emulsion geometry of ∼100 μm in diameter with a thickness of the oil layer around 15–20 μm was chosen, yet not optimized, for both stable double emulsions and high cell viability, given the experimental conditions, such as culture conditions and cell density, defined as mentioned above. However, a range of oil layer thickness (6.4–20 μm) was tested, and no major difference was observed in terms of cell viability, most likely due to the (1) good oxygen solubility and (2) comparable diffusion coefficient in fluorocarbon and in water.39 The droplets were collected and transferred to 96-well plates for subsequent culture and analysis. Cell viability was assessed by adding calcein-AM (Molecular Probes, Eugene, OR, USA),4,23,40 a nonfluorescent and cell-membrane-permeable compound, into the external culture medium, and the mixture was incubated for 20–40 min before imaging, according to the manufacturer’s protocol. The calcein-AM was able to pass through the oil layer of the w/o/w double emulsions and be converted to green fluorescent calcein when hydrolyzed by intracellular esterases in live cells. Survival rate was evaluated by the ratio of emulsions containing live cells (green, calcein-AM+) in all the emulsions examined (Figure S4 in the Supporting Information).
Acknowledgments
This work is supported by NIH (EB015300, AI096305), NSF (EEC-0425626), National Science Council in Taiwan (NSC 100-2917-I-564-035), Danish Research Council (116325/FTP), Lundbeck Foundation in Denmark (R95-A10275), and National University of Singapore (NUS-OGS). We also thank RainDance Technologies for providing the droplet destabilizer.
Supporting Information Available
(1) Photograph of the synthesized PFPE–Tris surfactants with increasing the feed molar ratio of Tris to PFPE from 0.3 to 0.5, 0.8, and 1.0 and their full-scale FT-IR spectra. (2) Comparison of FT-IR spectra of the synthesized PFPE–Tris surfactants at Tris/PFPE feed molar ratios of 1.0 (PFPE1–Tris1.0) and 1.2 (PFPE1–Tris1.2). (3) Amount of DNA recovered from the w/o emulsion after disrupting the droplets by a droplet-breaking agent. (4) Luciferase activities of the transfected cells. (5) Survival rate of hMSCs cultured in the emulsion droplets. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
∥ Department of Chemical Engineering, National Tsing Hua University, Hsinchu 30013, Taiwan.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Griffiths A. D.; Tawfik D. S. Miniaturising the Laboratory in Emulsion Droplets. Trends Biotechnol. 2006, 24, 395–402. [DOI] [PubMed] [Google Scholar]
- Song H.; Chen D. L.; Ismagilov R. F. Reactions in Droplets in Microflulidic Channels. Angew. Chem., Int. Ed. 2006, 45, 7336–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik D. S.; Griffiths A. D. Man-Made Cell-Like Compartments for Molecular Evolution. Nat. Biotechnol. 1998, 16, 652–656. [DOI] [PubMed] [Google Scholar]
- Clausell-Tormos J.; Lieber D.; Baret J. C.; El-Harrak A.; Miller O. J.; Frenz L.; Blouwolff J.; Humphry K. J.; Koster S.; Duan H.; etal. Droplet-Based Microfluidic Platforms for the Encapsulation and Screening of Mammalian Cells and Multicellular Organisms. Chem. Biol. 2008, 15, 427–437. [DOI] [PubMed] [Google Scholar]
- Eisenstein M. Tiny Droplets Make a Big Splash. Nat. Methods 2006, 3, 71–71. [DOI] [PubMed] [Google Scholar]
- Ahn K.; Kerbage C.; Hunt T. P.; Westervelt R. M.; Link D. R.; Weitz D. A. Dielectrophoretic Manipulation of Drops for High-Speed Microfluidic Sorting Devices. Appl. Phys. Lett. 2006, 88, 024104. [Google Scholar]
- Guo F.; Lapsley M. I.; Nawaz A. A.; Zhao Y.; Lin S. C. S.; Chen Y.; Yang S.; Zhao X. Z.; Huang T. J. A Droplet-Based, Optofluidic Device for High-Throughput, Quantitative Bioanalysis. Anal. Chem. 2012, 84, 10745–10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J. C.; Miller O. J.; Taly V.; Ryckelynck M.; El-Harrak A.; Frenz L.; Rick C.; Samuels M. L.; Hutchison J. B.; Agresti J. J.; etal. Fluorescence-Activated Droplet Sorting (FADS): Efficient Microfluidic Cell Sorting Based on Enzymatic Activity. Lab Chip 2009, 9, 1850–1858. [DOI] [PubMed] [Google Scholar]
- Li S.; Ding X.; Guo F.; Chen Y.; Lapsley M. I.; Lin S. C. S.; Wang L.; McCoy J. P.; Cameron C. E.; Huang T. J. An On-Chip, Multichannel Droplet Sorter Using Standing Surface Acoustic Waves. Anal. Chem. 2013, 85, 5468–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. P.; Grigsby C. L.; Zhao F.; Leong K. W. Tuning Physical Properties of Nanocomplexes through Microfluidics-Assisted Confinement. Nano Lett. 2011, 11, 2178–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J. C. Surfactants in Droplet-Based Microfluidics. Lab Chip 2012, 12, 422–433. [DOI] [PubMed] [Google Scholar]
- Duraiswamy S.; Khan S. A. Droplet-Based Microfluidic Synthesis of Anisotropic Metal Nanocrystals. Small 2009, 5, 2828–2834. [DOI] [PubMed] [Google Scholar]
- Teh S. Y.; Lin R.; Hung L. H.; Lee A. P. Droplet Microfluidics. Lab Chip 2008, 8, 198–220. [DOI] [PubMed] [Google Scholar]
- Riess J. G.; Krafft M. P. Fluorinated Materials for in Vivo Oxygen Transport (Blood Substitutes), Diagnosis and Drug Delivery. Biomaterials 1998, 19, 1529–1539. [DOI] [PubMed] [Google Scholar]
- Holtze C.; Rowat A. C.; Agresti J. J.; Hutchison J. B.; Angile F. E.; Schmitz C. H. J.; Koster S.; Duan H.; Humphry K. J.; Scanga R. A.; et al. Biocompatible Surfactants for Water-in-Fluorocarbon Emulsions. Lab Chip 2008, 8, 1632–1639. [DOI] [PubMed] [Google Scholar]
- Meier M.; Kennedy-Darling J.; Choi S. H.; Norstrom E. M.; Sisodia S. S.; Ismagilov R. F. Plug-Based Microfluidics with Defined Surface Chemistry to Miniaturize and Control Aggregation of Amyloidogenic Peptides. Angew. Chem., Int. Ed. 2009, 48, 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt D. J.; Payne R. J.; Chow W. Y.; Abell C. Fluorosurfactants for Microdroplets: Interfacial Tension Analysis. J. Colloid Interface Sci. 2010, 350, 205–211. [DOI] [PubMed] [Google Scholar]
- Holt D. J.; Payne R. J.; Abell C. Synthesis of Novel Fluorous Surfactants for Microdroplet Stabilisation in Fluorous Oil Streams. J. Fluor. Chem. 2010, 131, 398–407. [Google Scholar]
- Song H.; Ismagilov R. F. Millisecond Kinetics on a Microfluidic Chip Using Nanoliters of Reagents. J. Am. Chem. Soc. 2003, 125, 14613–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.; Tice J. D.; Ismagilov R. F. Formation of Droplets of Alternating Composition in Microfluidic Channels and Applications to Indexing of Concentrations in Droplet-Based Assays. Anal. Chem. 2004, 76, 4977–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach L. S.; Song H.; Ismagilov R. F. Controlling Nonspecific Protein Adsorption in a Plug-Based Microfluidic System by Controlling Interfacial Chemistry Using Fluorous-Phase Surfactants. Anal. Chem. 2005, 77, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti J. J.; Antipov E.; Abate A. R.; Ahn K.; Rowat A. C.; Baret J. C.; Marquez M.; Klibanov A. M.; Griffiths A. D.; Weitz D. A. Ultrahigh-Throughput Screening in Drop-Based Microfluidics for Directed Evolution. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouzes E.; Medkova M.; Savenelli N.; Marran D.; Twardowski M.; Hutchison J. B.; Rothberg J. M.; Link D. R.; Perrimon N.; Samuels M. L. Droplet Microfluidic Technology for Single-Cell High-Throughput Screening. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 14195–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtze C.; Agresti J. J.; Weitz D. A.; Ahn K.; Hutchison J. B.; Gifiths A.; Harrak A. E.; Miller O. J.; Barat J. C.; Taly V.; et al. Fluorocarbon Emulsion Stabilizing Surfactants. U.S. Pat. Appl. 12/310,048, 2010.
- Zhang Y.; Ho Y. P.; Chiu Y. L.; Chan H. F.; Chlebina B.; Schuhmann T.; You L.; Leong K. W. A Programmable Microenvironment for Cellular Studies via Microfluidics-Generated Double Emulsions. Biomaterials 2013, 34, 4564–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigsby C. L.; Ho Y. P.; Lin C.; Engbersen J. F. J.; Leong K. W. Microfluidic Preparation of Polymer-Nucleic Acid Nanocomplexes Improves Nonviral Gene Transfer. Sci. Rep. 2013, 3, 3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua K. K.; Leong K. W.; Nair S. K. Transfection Efficiency and Transgene Expression Kinetics of mRNA Delivered in Naked and Nanoparticle Format. J. Controlled Release 2013, 166, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul S.; Ho Y. P.; Koch J.; Andersen F. F.; Stougaard M.; Leong K. W.; Knudsen B. R. Detection of Single Enzymatic Events in Rare or Single Cells Using Microfluidics. ACS Nano 2011, 5, 8305–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul S.; Nielsen C. J. F.; Labouriau R.; Roy A.; Tesauro C.; Jensen P. W.; Harmsen C.; Kristoffersen E. L.; Chiu Y. L.; Frohlich R.; et al. Droplet Microfluidics Platform for Highly Sensitive and Quantitative Detection of Malaria-Causing Plasmodium Parasites Based on Enzyme Activity Measurement. ACS Nano 2012, 6, 10676–10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C.; Buppan P.; Jongwutiwes S. Improved Performance with Saliva and Urine as Alternative DNA Sources for Malaria Diagnosis by Mitochondrial DNA-Based PCR Assays. Clin. Microbiol. Infect. 2011, 17, 1484–1491. [DOI] [PubMed] [Google Scholar]
- Martinez H.; Buhse T.; Rivera M.; Ayala G.; Parmananda P.; Sanchez J. Effect of the Volume-to-Surface Ratio of Cultures on Escherichia coli Growth: An Experimental and Theoretical Analysis. Curr. Microbiol. 2012, 65, 60–65. [DOI] [PubMed] [Google Scholar]
- Glacken M. W.; Fleischaker R. J.; Sinskey A. J. Reduction of Waste Product Excretion via Nutrient Control: Possible Strategies for Maximizing Product and Cell Yields on Serum in Cultures of Mammalian Cells. Biotechnol. Bioeng. 1986, 28, 1376–1389. [DOI] [PubMed] [Google Scholar]
- Singh A.; Elisseeff J. Biomaterials for Stem Cell Differentiation. J. Mater. Chem. 2010, 20, 8832–8847. [Google Scholar]
- Wade L. G.Organic Chemistry, 6th ed.; Pearson Hall: Upper Saddle River, NJ, 2006. [Google Scholar]
- Qin D.; Xia Y.; Whitesides G. M. Soft Lithography for Micro- and Nanoscale Patterning. Nat. Protoc. 2010, 5, 491–502. [DOI] [PubMed] [Google Scholar]
- Abate A. R.; Thiele J.; Weinhart M.; Weitz D. A. Patterning Microfluidic Device Wettability Using Flow Confinement. Lab Chip 2010, 10, 1774–1776. [DOI] [PubMed] [Google Scholar]
- Andersen F. F.; Stougaard M.; Jorgensen H. L.; Bendsen S.; Juul S.; Hald K.; Andersen A. H.; Koch J.; Knudsen B. R. Multiplexed Detection of Site Specific Recombinase and DNA Topoisomerase Activities at the Single Molecule Level. ACS Nano 2009, 3, 4043–4054. [DOI] [PubMed] [Google Scholar]
- Chan H. F.; Zhang Y.; Ho Y. P.; Chiu Y. L.; Jung Y.; Leong K. W. Rapid Formation of Multicellular Spheroids in Double-Emulsion Droplets with Controllable Microenvironment. Sci. Rep. 2013, 3, 3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navari R. M.; Rosenblum W. I.; Kontos H. A.; Patterson J. L. Jr. Mass Transfer Properties of Gases in Fluorocarbons. Res. Exp. Med. 1977, 170, 169–180. [DOI] [PubMed] [Google Scholar]
- Chiu Y. L.; Chen S. C.; Su C. J.; Hsiao C. W.; Chen Y. M.; Chen H. L.; Sung H. W. pH-Triggered Injectable Hydrogels Prepared from Aqueous N-Palmitoyl Chitosan: In Vitro Characteristics and in Vivo Biocompatibility. Biomaterials 2009, 30, 4877–4888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.