Abstract
E2F proteins can either activate or repress transcription. Following mitogenic stimulation, repressive E2F4-p130-histone deacetylase complexes dissociate from, while activating species (E2F1, -2, and -3) associate with, target promoters. Histones H3 and H4 simultaneously become hyperacetylated, but it remains unclear whether this is a prerequisite or a consequence of E2F binding. Here, we show that activating E2F species are required for hyperacetylation of target chromatin in human cells. Overexpression of a dominant-negative (DN) E2F1 mutant in serum-stimulated T98G cells blocked all E2F binding, H4 acetylation, and, albeit partially, H3 acetylation. Target gene activation and S-phase entry were also blocked by DN E2F1. Conversely, ectopic activation of E2F1 rapidly induced H3 and H4 acetylation, demonstrating a direct role for E2F in these events. E2F1 was previously shown to bind the histone acetyltransferases (HATs) p300/CBP and PCAF/GCN5. In our hands, ectopically expressed E2F1 also bound the unrelated HAT Tip60 and induced recruitment of five subunits of the Tip60 complex (Tip60, TRRAP, p400, Tip48, and Tip49) to target promoters in vivo. Moreover, E2F-dependent recruitment of Tip60 to chromatin occurred in late G1 following serum stimulation. We speculate that the activities of multiple HAT complexes account for E2F-dependent acetylation, transcription, and S-phase entry.
The fundamental unit of eukaryotic chromatin is the nucleosome, in which genomic DNA is tightly wrapped around a histone octamer comprising two copies of each core histone (H2A, H2B, H3, and H4) (25). Chromatin structure and gene transcription are regulated by covalent modifications in the N-terminal tails of histones, of which the best characterized is lysine acetylation (39, 42, 46). Generally, hyperacetylation correlates with transcriptional activation, while deacetylation is associated with repression. The enzymes that catalyze these transitions are histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. These enzymes are recruited to target genes by sequence-specific transcription factors, allowing both selective and diversified use of their coactivator and corepressor functions.
Transcription factors of the E2F family regulate cell cycle progression in higher eukaryotes (reviewed in references 7, 44, and 48). In mammals, six E2F species have been described: E2F1, -2, and -3 are believed to act primarily as transactivators and are thus commonly referred to as “activating” E2Fs, while E2F4 and -5 appear to be critical for repression of target genes. All of these proteins, however, possess a C-terminal transactivation domain (TAD) and thus have the potential to activate transcription. Through a subdomain within the TAD, E2Fs are bound by retinoblastoma (Rb) family proteins, or “pocket proteins”: E2F1, -2, and -3 interact with pRb, while E2F4 and -5 bind p107 and p130. These interactions are thought to counteract E2F-dependent gene activation by two mechanisms: (i) interference, i.e., preventing association of the TAD with basal transcription factors, and (ii) active repression, i.e., assembly of transcriptional repressor complexes. These include HDACs (2, 10, 28), histone methyltransferases (34), and the chromatin remodeling complex SWI/SNF (reviewed in reference 18). As a result, in quiescent cells, E2F target genes exhibit a repressed “heterochromatic” state, in which histones H3 and H4 are hypoacetylated and lysine 9 of histone H3 is methylated (11, 34, 38, 47).
In contrast to repression, the mechanisms of transactivation by E2F have not yet been elucidated in molecular detail. While E2F target genes are bound by E2F1 to -3 and exhibit hyperacetylated histones in the late-G1 and S phases (47), the causal relationship between these events remains unclear. In favor of a direct role of E2Fs in histone acetylation, they interact with HATs, including PCAF/GCN5 and p300/CBP. In reporter gene assays, these cofactors enhance E2F-dependent transcription, possibly through acetylation of E2F itself (1, 14, 26, 29, 30, 37, 49). E2F1 also binds TRRAP (26, 31), a subunit of the HAT complexes PCAF/GCN5 (51) and Tip60 (20). In spite of these observations, there is no direct proof that E2Fs act upstream, rather than downstream, of histone acetylation. Under certain circumstances, the simple removal of repressive E2F-HDAC complexes can induce downstream responses (19, 40, 56), suggesting that promotion of acetylation by E2F may not be required. Instead, like other transcription factors such as yeast SBF (4), E2F1 to -3 may bind chromatin only following histone acetylation and may activate transcription by recruiting basal factors. Consistent with this hypothesis, E2Fs have been shown to bind TBP, TFIIH (8, 17, 36, 50), and the Mediator subunit RING3 (6). Here, we show that chromatin association of activating E2F complexes is required to promote histone acetylation in late G1. We also provide evidence that five subunits of the Tip60 HAT complex (15, 20) are recruited to chromatin in an E2F-dependent manner.
MATERIALS AND METHODS
Cells, ChIP, and mRNA analysis.
T98G cells (American Type Culture Collection) and U2OS cells expressing estrogen receptor (ER)-E2F chimeras (32) were cultured in Dulbecco's minimal essential medium with 10% fetal calf serum and were rendered quiescent by contact inhibition followed by serum removal for 2 or 4 days, respectively. For cell cycle reentry, T98G cells were harvested by trypsinization and reseeded onto plates containing Dulbecco's minimal essential medium-10% fetal calf serum at a density of 1:3. U2OS ER-E2F1 cells were directly induced by stimulation with 4-hydroxytamoxifen (OHT). Recombinant AdEasy adenoviruses expressing human wild-type (WT) E2F1 or E2F1-Eco132 were used as described previously (13). RNA extraction, cDNA synthesis, quantitative chromatin immunoprecipitation (ChIP), real-time PCR, and calculations of DNA recovery in ChIP were exactly as described in previous work (12, 13), with no further modifications. The following antibodies were used: E2F1 C-20 (sc-193; Santa Cruz), KH20 and KH95 (05-379; Upstate Biotech), E2F2 C-20 (sc-633), E2F3 C-18 (sc-878), E2F4 A-20 (sc-1082), p130 C-20 (sc-317), pan-acetyl-histone H3 (06-599; Upstate Biotech), and pan-acetyl-histone H4 (06-866; Upstate Biotech). The antisera against TRRAP (13), Tip60, Tip48, Tip49 (12), and p400 (15) were previously described. For ChIP, 2 μg of each antibody was used to precipitate chromatin from 1 × 107 to 4 × 107 cells. The PCR primers used for ChIP and mRNA analysis, performed by reverse transcription (RT)-PCR as previously described (13), are listed in Table 1.
TABLE 1.
Nucleotide sequences of the PCR primers used in this study
| Gene | Positiona | Accession no. | Forward primer | Reverse primer |
|---|---|---|---|---|
| AchR | In promoter | Y00508 | CCTTCATTGGGATCACCACG | GGAGATGAGTACCAGCAGGTTG |
| MCM4 | −2355 | NT_023806 | TTGAAACCCTGAGAAGAGAAAGTAATC | TCAAACAAAAAACTAACATTGCGTG |
| −1665 | TTCTCAAAGTAGGAACGGCAAAT | GCCCGGCCAAGAGAAAGTAT | ||
| −966 | CTGCTGCTCAGGACGCATT | CCTGGCCGGTCATCAACT | ||
| E2F site (−208) | CCGAGCGAGGCCTACTTCT | GGACAGTGCCGCTTCTTTCA | ||
| +580 | ACGTAGAGGCGAGGATTCCAC | CAGGTCCACTCCAGGCGA | ||
| +1325 | TGTGTAGGCAGCAGCAGAAGA | TCTGTTCCCCAGATCACAAGTTT | ||
| +9727 | GGCCTCACTGCGTACGTAATG | CAGGACCAGCTGCCTTGTCT | ||
| MCM3 | −2013 | AL034343 | GCAACAACCTGATTGATTTTGG | CATGAAATCTTTAAGCAGAGGAAATG |
| −1347 | TTGGCTAAGGATCGATTTATCTGC | ACTGTATGTTTTGTGCCATCCTAGTG | ||
| −677 | TTTCCAAAATTTGTATAACACACGATG | GTTTACCCTGCCCCCTAGAGA | ||
| E2F site (+95) | TCTTTGGCAGCGGGCAT | CGCAGCTCCACATCGTCC | ||
| +683 | GGATTTTATGTGCAACCCAAGAC | GATGGTTTTTTCAACATTCTATGGC | ||
| +1365 | GGCTGTTTTGCAGGAAGACC | GATCAGCTCCCGAACTTTGC | ||
| +2247 | TACGTGGTAAAACTTGGAGGGC | TTCATCATTCTACTAATACATGTGATTACTCAA | ||
| PCNA | −711 | J04718 | TGTGATGCTATGTTTTAAAGAGGTACTGA | AATTTTATGCCACGTACATCTTTTTATC |
| −240 | CTCCACATATGCCCGGACTT | CCTCTTTGACTCCTGAACCCG | ||
| E2F site (+547) | CTGGCTGCTGCGCGA | CACCACCCGCTTTGTGACT | ||
| +1506 | CTCAAGCAGTCCTCTTGCCTTAGT | GGCCCGTAGTCCTTCCTAGC | ||
| +2132 | TTTGAGTGGAGTTTGTTAAGAAATTACG | TTCTGATTACTTTAAAATATGACTTGCGT | ||
| +2716 | ATCAAGTCCACACCGGCTCT | TTATACGCCTAACTTCCCAATCCT | ||
| p107 | −3421 | AL136172 | GGAGCTTGGCTTAACCCTGA | TTTGAAAATGTTCTAGTGACTCCCC |
| −2057 | CACACAATGGGATCAAAATAGAAACA | TTTTGAGCCAGATACACATGAAGG | ||
| −1682 | TACAGGTGTGAGCCACTGCAAC | CTGAATAGAAAATGAAGTATGGAAGGC | ||
| −1228 | ACTGGGATTACAGGCAAAGCA | CTGTCTTCGTTCTTTTGTTGTTCC | ||
| −559 | CCACTCACTGGGCACTAGGAC | AAAGTAGCCCCTGGACGAAGA | ||
| E2F site (−64) | TGGATGACAACACGTCCCG | CACGGCCCCCGACTTC | ||
| +970 | AATGAGTATTAGACTGCTTTCCAGGTG | AAGACAACCTTCTGAGCCTTGC | ||
| +1357 | AGCATAAGCCCAGTGTAGACAGAA | TGCTACATGAAATACACTCCATCACA | ||
| +1540 | ATGTGTGCCTATATATGAAGGAGTAGTACCT | CCATTGTCCTTTACCCAGTCTATCC | ||
| +2436 | GATGTTCCTGCTTCAGCCTCC | TGGGCAACATAGTCAGATACTATCTCC | ||
| Ubiquitin | mRNA | M26880 | CACTTGGTCCTGCGCTTGA | CAATTGGGAATGCAACAACTTTAT |
| MCM4 | mRNA | XM_030274 | TGTGGCAGCATGCAAAGAA | GGGTCAATAAAACGCTGAAGAAA |
| MCM3 | mRNA | NM_002388 | GCTCCTCTGGAGTGGGTCTG | TCCTGTTTCCTGGTCTGTGGT |
| PCNA | mRNA | NM_002592 | CCTGGTCCAGGGCTCCAT | ATATCCCAGCAGGCCTCGT |
| p107 | mRNA | NM_002895 | CTTTTTGGGAGCAGACGCAG | GTGAACTTTCGAGGTGTTCCAAT |
The primer pairs used for ChIP and the corresponding amplicons (shown in Fig. 1) are numbered according to the position of the first base of the forward primer relative to the transcription start site. The primers used for mRNA analysis are indicated as “mRNA” in the same column.
Coimmunoprecipitation assay.
Human Tip60 and hemagglutinin (HA)-tagged E2F1 proteins were transiently expressed in U2OS cells by calcium phosphate-mediated transfection. Cells were resuspended in 0.5 ml of lysis buffer (33) (50 mM Tris, pH 8, 0.4% Nonidet P-40, 300 mM NaCl, 10 mM MgCl2, 2.5 mM CaCl2) supplemented with protease inhibitors (Complete EDTA free; Roche Diagnostics) and DNase (10 U/μl; Roche) and incubated for 10 min on ice. One-tenth of the lysate was saved for immunoblotting, and the rest was used for two parallel immunoprecipitations followed by immunoblotting (see Fig. 9). We used the anti-Tip60 antibody CLHF (12), mouse anti-HA (Covance), and the anti-E2F1 monoclonal KH20. Immunoprecipitation was performed with protein A (for anti-Tip60) or protein G beads (for anti-HA) for 2 h at 4°C, followed by three washes with wash buffer (50 mM Tris, pH 8, 150 mM NaCl, 5 mM MgCl2, 0.4% Nonidet P-40). Bound proteins were loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Western blotting was performed by standard procedures and revealed by chemiluminescence with an ECL detection system (Amersham).
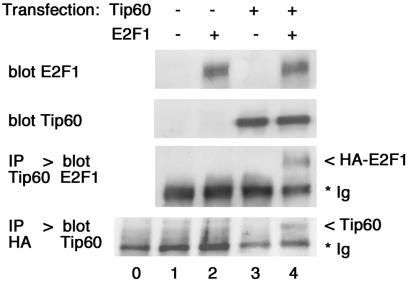
FIG. 9.
E2F1 and Tip60 interact in cells. U2OS cells were transfected with vectors expressing human Tip60 and HA-E2F1 as indicated (lanes 1 to 4). Whole-cell lysates were immunoprecipitated (IP) with antibodies against Tip60 or HA, followed by immunoblotting (blot). Lane 0, the anti-HA antibody was loaded without cell lysate. The positions of the immunoglobulin (Ig), HA-E2F1, and Tip60 bands are indicated to the right.
RESULTS
Kinetics of E2F binding and histone acetylation at E2F target sites.
In order to study the binding of E2F to chromatin and associated events in live cells, we used a quantitative ChIP assay previously developed by our group (13). Chromatin was cross-linked in vivo by formaldehyde fixation of cultured cells. After shearing by sonication in a sodium dodecyl sulfate-containing buffer, cross-linked protein-DNA complexes were immunoprecipitated (always in triplicate) with antibodies against E2F family proteins or against acetylated forms of histones H3 and H4. Cross-links were reversed and DNA was purified from the precipitates. Real-time PCR was used to amplify known E2F-binding sites in the mcm4, mcm3, PCNA, and p107 promoters (5, 24, 27, 47) (Fig. 1). For each sample (e.g., time point), parallel PCRs were run on triplicate isolates of DNA from each of the following: immune precipitates, control precipitates, and total input chromatin. The recovery of a given site (or amplicon, as shown in Fig. 1) in each precipitate was expressed as a percentage of its abundance in input chromatin, as previously described (13).
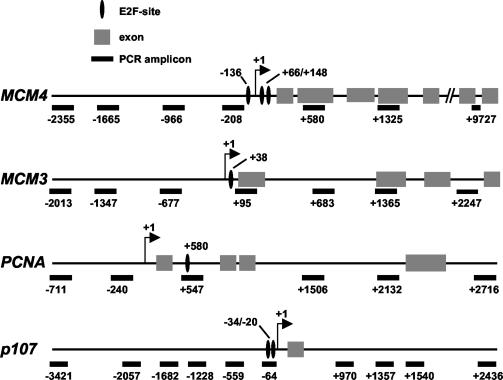
FIG. 1.
Maps of the E2F target genes studied in this work.
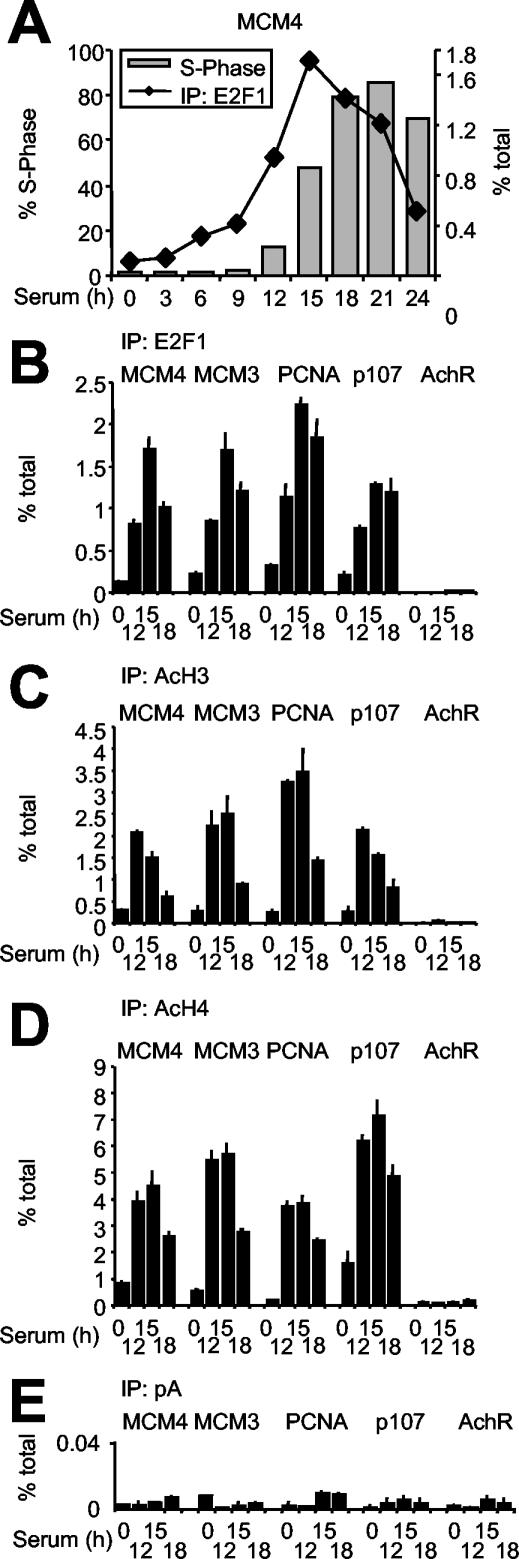
We used T98G cells, which are known to progress synchronously through G1 and S phase following serum stimulation (Fig. 2A). As previously reported (47), the association of E2F1 with target promoters peaked in late G1 (15 h) (Fig. 2A and B). As negative controls, the AchR promoter did not bind E2F (Fig. 2B), and none of the tested sequences was enriched in nonimmune precipitates (Fig. 2E). Amplification of sequences on either side of the E2F consensus sites in mcm4, mcm3, PCNA, and p107 showed that E2F was localized over the predicted binding sites (Fig. 3A). This was true in quiescent cells (0 h), with minimal levels of E2F1 binding, as well as in late G1 (15 h). The binding kinetics for E2F2 and E2F3 were similar to that of E2F1. E2F4, in association with p130, was the main species bound to chromatin in quiescent cells and decreased upon cell cycle entry (data not shown; see Fig. 4F). Concomitant with the dissociation of E2F4/p130 and association of E2F1 to -3, acetylation of histones H3 and H4 peaked in late G1 (Fig. 2C and D). This modification was localized to a region of 2 to 3 kb centered to the E2F-binding sites (Fig. 3B and C). Although differences in the detailed kinetics of E2F-chromatin interactions may be seen with different target genes and/or cell lines (53), our observations reproduce previous data with T98G cells (47) as well as mouse fibroblasts (38).
FIG. 2.
E2F1 binding and histone acetylation on E2F target genes. (A) E2F binding to the MCM4 promoter in serum-stimulated T98G cells as determined by ChIP (% total) overlapped with the percentage of cells in S phase as determined by BrdU incorporation and fluorescence-activated cell sorting. (B) ChIP analysis with E2F1 antibodies on quiescent T98G cells (0 h) and cells serum stimulated for 12, 15, or 18 h, as indicated. The amplified E2F-binding sites (MCM4, MCM3, PCNA, and p107) and control site (AchR) are indicated at the top. (C to E) ChIP as for panel B, but with acetyl-H3 (C), acetyl-H4 (D), or no specific antibodies (E).
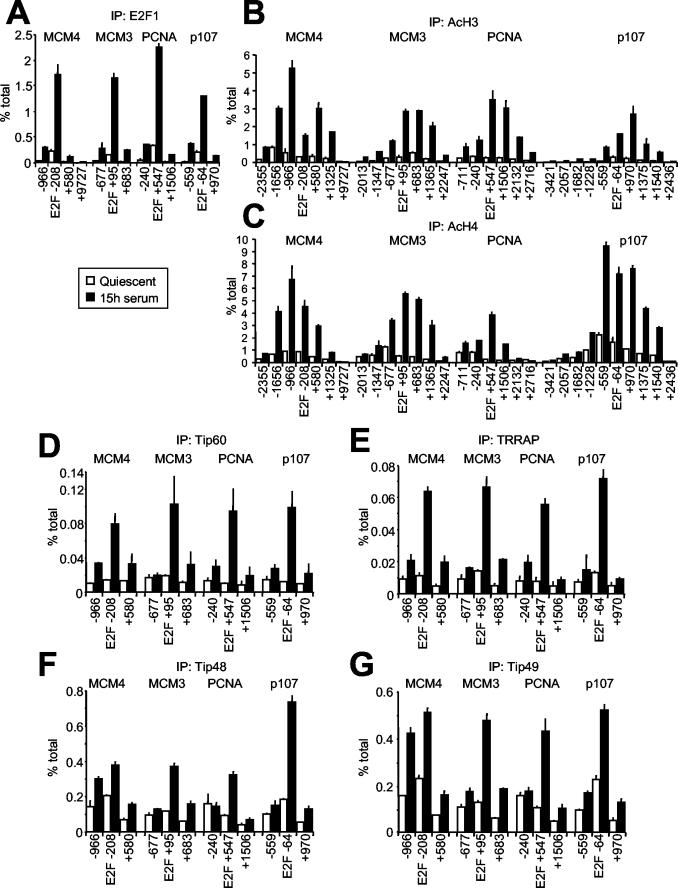
FIG. 3.
E2F1 binding, acetylation, and cofactor recruitment colocalize on chromatin. Chromatin recovered from T98G cells at quiescence or in late G1 was analyzed by ChIP with antibodies for E2F1 (A), acetyl-H3 (B), acetyl-H4 (C), Tip60 (D), TRRAP (E), Tip48 (F), or Tip49 (G). DNA was amplified with a series of primer pairs yielding amplicons located at increasing distances from the E2F-binding site. Numerical positions are given relative to the E2F site and correspond to the 5′ end of each amplicon (Table 1 and Fig. 1).
FIG. 4.
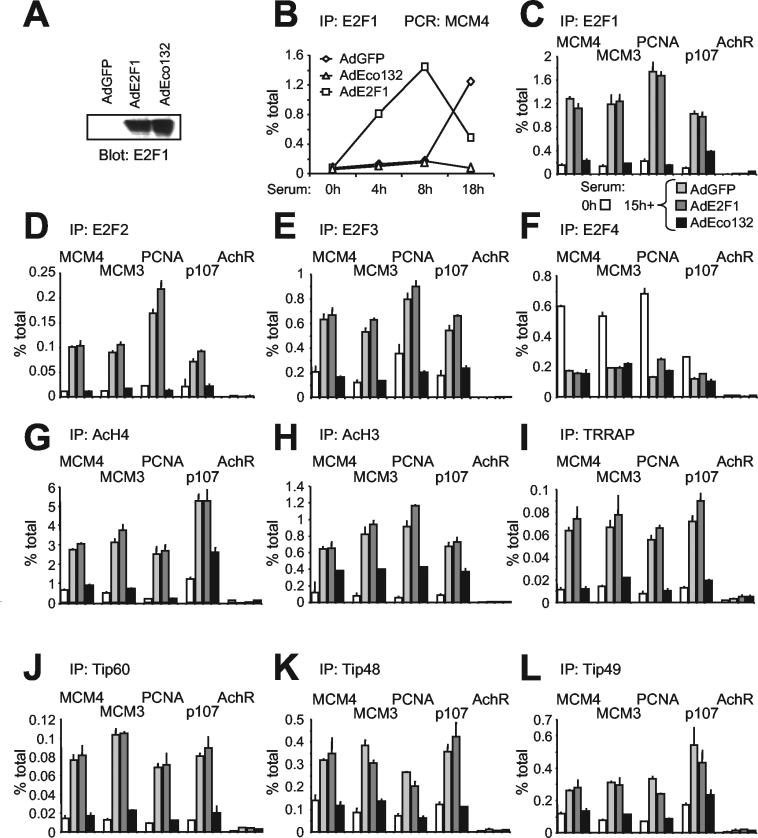
Activating E2F is essential for induction of H4 acetylation. Quiescent T98G cells were serum stimulated and simultaneously infected with adenoviruses expressing green fluorescent protein alone (AdGFP) or green fluorescent protein plus WT E2F1 (AdE2F1) or mutant E2F1-Eco132 (AdEco132). (A) Expression of E2F1 proteins in infected cells, determined by immunoblotting after 15 h. (B) ChIP analysis demonstrating that expression of WT E2F1 induced premature accumulation of E2F1 on the MCM4 promoter, while expression of E2F1-Eco132 prevented binding of endogenous E2F1. AdGFP did not modify the binding of endogenous E2F1 relative to that of noninfected controls (data not shown). (C) Binding of E2F1 to E2F targets (MCM4, MCM3, PCNA, and p107) and control promoter (AchR). ChIP was from quiescent T98G cells and cells stimulated for 15 h with serum alone (not shown) or serum plus AdGFP, AdE2F1, or AdEco132 as indicated. (D to L) Data represented as in panel C but for ChIP with antibodies specific to E2F2 (D), E2F3 (E), E2F4 (F), acetyl-H4 (G), acetyl-H3 (H), TRRAP (I), Tip60 (J), Tip48 (K), or Tip49 (L).
Activating E2F complexes are required for H3 and H4 acetylation in late G1.
The timing and localization of E2F1-3 binding and histone acetylation are compatible with two alternative scenarios: (i) E2F proteins might directly induce, and hence be required for, acetylation, and (ii) acetylation induced by other factors might be a prerequisite for E2F binding. In order to address the role of E2F, we chose to block the binding of all E2F species to chromatin by overexpressing a dominant-negative (DN) mutant of E2F1. To this aim, we subcloned the mutant E2F1-Eco132 (21) in an adenoviral vector. This mutant bears a point mutation that prevents DNA binding but retains all protein-protein interaction surfaces, including the dimerization domain. Thus, overexpression of E2F1-Eco132 would be expected to block the binding of endogenous E2F to promoters by titrating dimerization partner proteins. In order to test this prediction, quiescent T98G cells were seeded in fresh medium and simultaneously infected with one of three adenoviruses: AdGFP (control), AdE2F1 (expressing WT E2F1), and AdEco132 (mutant E2F1). Both forms of E2F1 accumulated to similar levels in the cells (Fig. 4A). We then monitored E2F1 binding to mcm4 by ChIP (Fig. 4B). AdGFP had no incidence upon the association of endogenous E2F1 to chromatin in late G1 (18 h). AdEco132, in contrast, eliminated E2F1 binding. As expected, interfering activity depended upon the E2F-Eco132 mutation, since AdE2F1 led to premature binding of E2F1 in early G1 (4 to 8 h) (Fig. 4B). The disruption of E2F1 binding by Eco132 was observed on all target genes at the 15-h time point (Fig. 4C). Note that at this time point, binding by exogenous E2F1 in AdE2F1-infected cells was already past its peak level and was comparable to the level of endogenous E2F1 binding seen in control cells (Fig. 4B and C). Furthermore, E2F-Eco132 also eliminated chromatin association of E2F2 and E2F3 (Fig. 4D and E). Consistent with the observation that E2F4-p130 repressive complexes dissociate from chromatin upon cell cycle entry (38, 47), late-G1 cells showed reduced binding of E2F4 (Fig. 4F) and p130 (data not shown) relative to quiescent cells, which was not further reduced by infection with either of the adenoviruses.
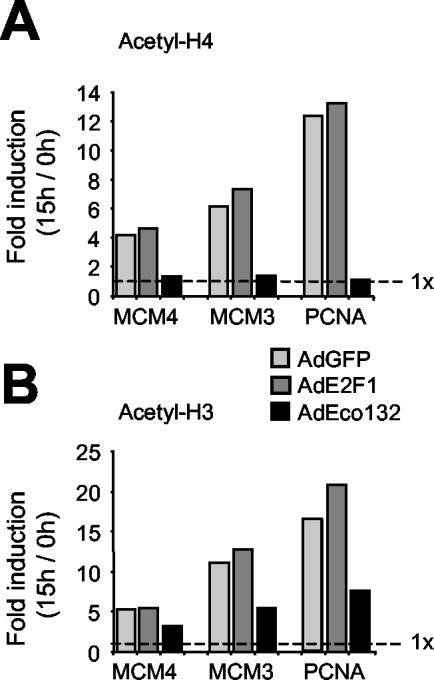
We next investigated whether the Eco132 mutant had an effect on histone acetylation. AdEco132 caused a complete block on histone H4, with acetylation levels on mcm4, mcm3, and PCNA remaining at the basal levels observed in quiescent cells (Fig. 4G; note that on p107 interference by AdEco132 was incomplete, correlating with residual binding of E2F1 [Fig. 4C]). This blocking effect of AdEco132 is better illustrated in Fig. 5A, showing the extent of serum-induced H4 acetylation (n-fold induction) relative to quiescent cells (0 h). In summary, removal of E2F complexes from target promoters eliminates any effect of serum on H4 acetylation. These data have two major implications: (i) binding of activating E2F species in late G1 is required to induce and/or maintain H4 acetylation, and (ii) the removal of repressor complexes (i.e., E2F4-p130) is insufficient for H4 acetylation.
FIG. 5.
Differential effect of DN E2F1 (AdEco132) on histones H4 and H3. The data from Fig. 4G for histone H4 (A) and 4H for histone H3 (B) are expressed as n-fold induction of acetylation at 15 h relative to that of quiescent cells (0 h). The elimination of serum-induced H4 acetylation by AdEco132 and its partial effect on H3 are evident from this display.
An important difference was revealed when studying H3. In this case, AdEco132 reduced, but did not eliminate, acetylation (Fig. 4H and 5B). Thus, although maximal H3 hyperacetylation in late G1 also requires E2F, a significant fraction of it is retained upon removal of all E2F species from target promoters. This implies that H3 is modified in part by HATs that do not require recruitment or that are recruited by a transcription factor(s) distinct from E2F.
E2F binding is required for target gene activation and S-phase entry.
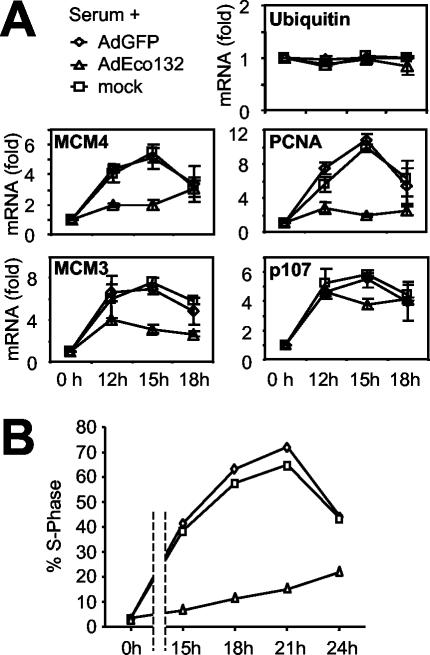
Since E2F1-Eco132 negates binding of E2F family proteins after serum stimulation and prevents hyperacetylation, we investigated whether it also affected expression of E2F target genes. Total mRNA was prepared from serum-stimulated T98G cells and analyzed by quantitative RT-PCR. While mRNA levels of ubiquitin (not an E2F target) did not fluctuate, mRNAs of all four E2F target genes were induced (Fig. 6A). This induction was not affected by infection with AdGFP but was strongly reduced by AdEco132. Interestingly, this did not hold true for the p107 gene, whose mRNA induction was not affected, correlating with residual E2F1 binding and H4 acetylation. Eco132-expressing cells also failed to enter S phase (Fig. 6B). A similar phenotype has recently been described in mouse embryo fibroblasts expressing another E2F1 mutant, E2F-DB, which lacks the TAD (40). Altogether, these data suggest that the transactivation function of E2F proteins is critical for gene activation and cell cycle entry upon exit from quiescence.
FIG. 6.
E2F is essential for target gene activation and S-phase entry. (A) T98G cells were serum stimulated and either mock infected or infected with AdGFP or AdEco132, as indicated. mRNA levels were quantified by RT-PCR and are shown as n-fold induction relative to that of quiescent cells (0 h). We monitored the mRNAs for ubiquitin (control), MCM4, MCM3, PCNA, and p107 as indicated within each panel. (B) Percentage of cells in S phase in the same populations as in panel A, as determined by BrdU incorporation and analysis in a fluorescence-activated cell sorter.
E2F1 induces H3 and H4 acetylation.
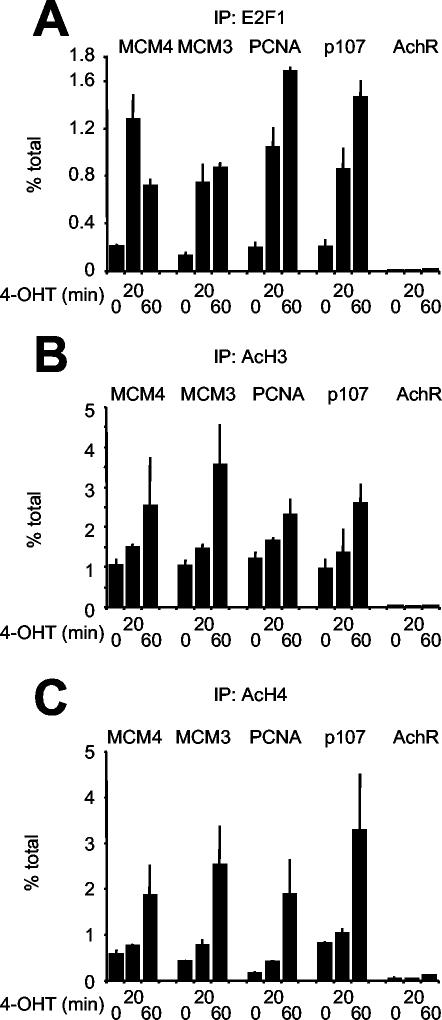
The above data show that activating E2F species (E2F1, -2, or -3) are required for histone acetylation (only in part for H3), activation of gene expression, and cell cycle entry. In order to address whether E2F can directly induce those changes, U2OS cells expressing a conditional ER-E2F1 chimera were treated with 4-OHT, which activates ER-E2F1 and induces multiple E2F target genes (32). ChIP with anti-E2F1 antibodies demonstrated rapid association of ER-E2F1 with target promoters, while AchR remained unbound (Fig. 7A). As for endogenous E2F, binding of ER-E2F1 was localized over the predicted sites on all the promoters, and the control promoter AchR was not bound. Control nonimmune ChIPs did not enrich any of the promoters (data not shown). Concomitant with ER-E2F1 binding, OHT treatment induced rapid hyperacetylation of histones H3 and H4 (Fig. 7B and C). We conclude that E2F1 is not only required for but can directly induce hyperacetylation of histones H3 and H4. While ER-E2F1 also induced mRNA accumulation in both U2OS and T98G cells (data not shown), this response was generally weaker than that seen in serum-stimulated cells (Fig. 6A). Thus, other serum-dependent signals must contribute to the full activation of E2F target genes.
FIG. 7.
ER-E2F1 rapidly binds chromatin and induces histone acetylation. Chromatin from quiescent U2OS cells expressing an ER-E2F1 chimera was prepared before (0 min) or after (20 and 60 min) OHT treatment and analyzed by ChIP with antibodies against E2F1 (A), acetyl-H3 (B), or acetyl-H4 (C).
Notably, the domains of hyperacetylation induced by ER-E2F1 in U2OS cells (data not shown) were superimposable with those observed in late G1 in T98G cells (Fig. 3B and C), covering a region of 2 to 3 kb (≤20 nucleosomes) roughly centered on the E2F-binding sites and the transcription start sites (indeed, E2F-binding sites are generally proximal to—but can also occur on either side of—the start sites [Fig. 1]). The same phenomenon has been seen with other transcription factors in vivo, such as Myc (13). In vitro experiments also showed that HATs recruited to a specific site by a given transcription factor can acetylate multiple nucleosomes in target chromatin (9, 52).
E2F-dependent recruitment of the Tip60 HAT complex to chromatin.
Since activating E2F species induce histone acetylation, we reasoned that they must recruit one or more HATs to target loci. We initially tested the HAT-associated protein TRRAP, which was known to bind E2F1 (31). OHT treatment of ER-E2F1 cells followed by ChIP with a TRRAP-specific antibody showed that TRRAP was recruited to E2F target sites but not to the AchR promoter (Fig. 8A). This occurred with similar kinetics as ER-E2F1 binding (Fig. 7A). As previously observed at Myc target loci (12, 13), the extent of chromatin recovery with TRRAP antibodies was modest but was consistently elevated above the background observed at the nontarget AchR promoter or in nonimmune precipitates (Fig. 8A; data not shown). Binding of TRRAP is likely to reflect recruitment of the HAT complexes GCN5/PCAF (51) and/or Tip60 (20). So far, we failed to observe recruitment of GCN5 or the unrelated HAT p300 to E2F target genes (data not shown). It should be considered, however, that not all antibodies work in ChIPs and that our negative results should not be interpreted as evidence against a role of these HATs in E2F activity. On the other hand, we obtained clear evidence for recruitment of Tip60 and will henceforth concentrate on this HAT complex.
FIG. 8.
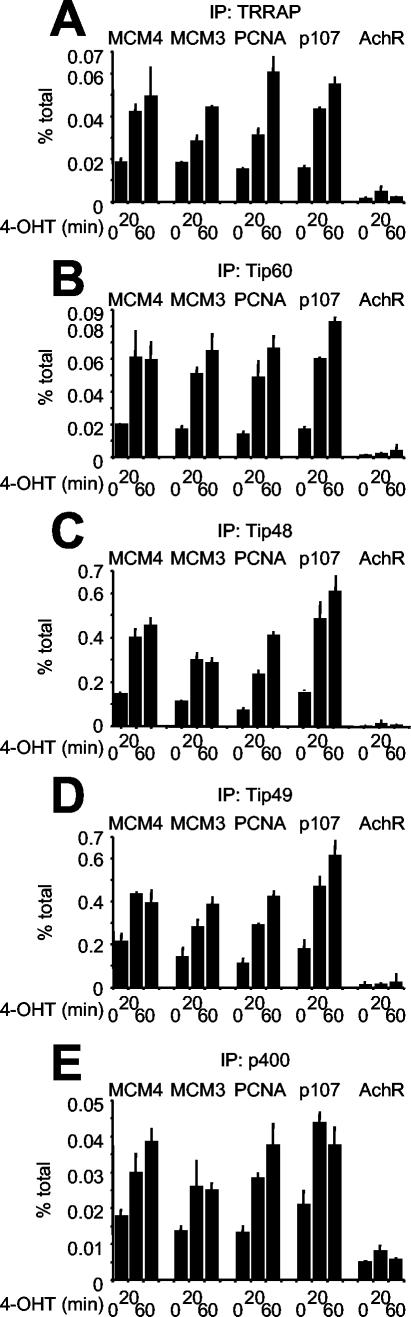
ER-E2F1 induces recruitment of the Tip60 complex. U2OS cells expressing an ER-E2F1 chimera were analyzed by ChIP following 4-OHT stimulation as for Fig. 7, but with antibodies against TRRAP (A), Tip60 (B), Tip48 (C), Tip49 (D), and p400 (E).
Besides TRRAP, we studied four known subunits of the Tip60 complex: Tip60 itself, Tip48, Tip49, and p400 (15, 20). Like TRRAP, these proteins selectively associated with E2F target promoters following ER-E2F1 activation (Fig. 8). Furthermore, maximal enrichment of ER-E2F1 and each Tip60 subunit was colocalized on the E2F target sites (data not shown; see similar data in Fig. 3D to G for serum-induced recruitment). We conclude that ER-E2F1 induces recruitment of TRRAP, Tip60, Tip48, Tip49, and p400 to chromatin. TRRAP and Tip60 were also recruited upon activation of ER-E2F2 and ER-E2F3 (data not shown). Although independent interactions with each subunit cannot be ruled out, our data are most compatible with recruitment of the whole Tip60 complex by activating E2Fs.
We used coimmunoprecipitation to confirm that E2F1 and Tip60 can interact physically. Tip60 and HA-tagged E2F1 were overexpressed by cotransfection in U2OS cells and analyzed by coimmunoprecipitation. Anti-Tip60 precipitation followed by E2F1 immunoblotting revealed that HA-E2F1 specifically coprecipitated with Tip60 (Fig. 9, lane 4). Conversely, Tip60 was also detected in anti-HA immunoprecipitates. Coprecipitation was selective, since it was not detected in cells transfected with control vectors or with either HA-E2F1 or Tip60 alone (lanes 1 to 3). It remains to be determined whether this interaction is direct or occurs via association of either protein with cellular TRRAP (20, 31). So far, we have not been able to detect coprecipitation between endogenous E2F1 and Tip60, possibly due to low expression levels and the small fraction of either protein involved. This notwithstanding, our data show that E2F1 and Tip60 can bind each other in cells. Furthermore, the following ChIP experiments strongly indicate an interaction—whether direct or indirect—between endogenous E2F and Tip60 on chromatin.
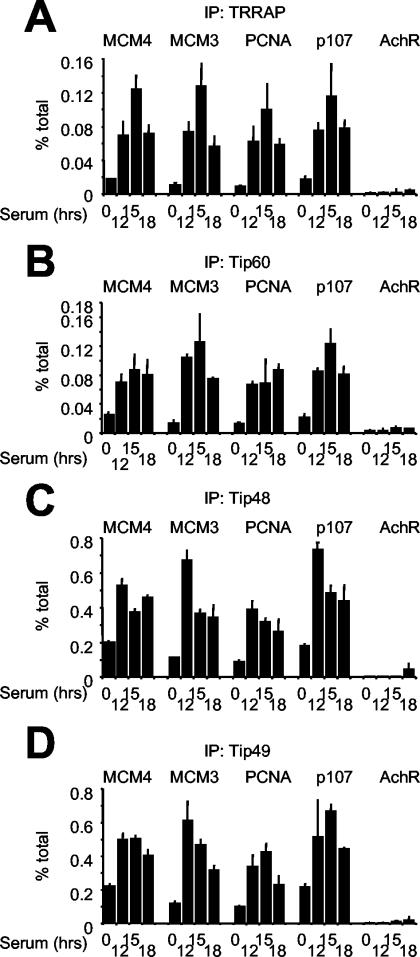
We asked whether the Tip60 complex was recruited by endogenous E2F following serum stimulation of T98G cells. By ChIP, we observed loading of TRRAP, Tip60, Tip48, and Tip49 onto E2F target sites but not onto AchR in late G1 (Fig. 10A to D). Immunoprecipitation of these subunits yielded recoveries that were significantly above the background seen in nonimmune immunoprecipitations (Fig. 2E), while p400 antibodies did not consistently do so (data not shown). Compared with the binding of E2F1 to -3, the cofactors showed consistent kinetics (Fig. 2B and 10A to D) and colocalization on E2F target sites (Fig. 3A and D to G). In T98G cells, the TRRAP and Tip60 proteins were expressed at constant levels throughout the serum time courses (data not shown), further indicating that their binding to chromatin was not simply due to enhanced expression but to localized recruitment by E2F. Finally, to directly address the role of E2F, we overexpressed the DN E2F1 mutant Eco132. As seen above for E2F1-3 (Fig. 4C to E), serum-induced association of TRRAP, Tip60, Tip48, and Tip49 with chromatin was blocked by AdEco123 but not by AdGFP or AdE2F1 (Fig. 4I to L). Together with the rapid recruitment seen upon ER-E2F activation, these data show that activating E2F species are responsible for recruitment of the Tip60 complex in late G1.
FIG. 10.
The Tip60 complex is recruited to E2F target genes in late G1. Serum-stimulated T98G cells were analyzed as for Fig. 2B, but with antibodies against TRRAP (A), Tip60 (B), Tip48 (C), and Tip49 (D).
Of note, Tip48 and Tip49 were detectable on promoters prior to serum stimulation (Fig. 10C and D) and on wider domains (Fig. 3D to G) relative to E2F, Tip60, and TRRAP. Unlike the increased binding seen in late G1, basal association of Tip48/49 was E2F independent, as it was not affected by DN E2F1 (Fig. 4K and L, compare white and black bars). In no circumstance did Tip48 or Tip49 bind the negative control AchR (Fig. 10). In an analogous manner, we observed basal Myc-independent association of Tip48/49 with the nucleolin promoter, while serum- and Myc-dependent recruitment, in this case, were maximal in intron 1 (12). These observations might reflect the fact that Tip48/49 heteromers (22, 54), besides associating with Tip60/p400 (15, 20), participate in other transcriptional regulatory complexes (16, 23, 43).
DISCUSSION
Genetic analysis of E2F1, -2, and -3 has revealed nonidentical functions (3, 41, 45, 48). Most importantly, however, these activating E2F proteins have a redundant and essential role in cellular proliferation (55). These biological similarities are likely to reflect a common function at the molecular level, and in particular in the transactivation of target genes. Following mitogenic stimulation of quiescent cells, a number of dynamic changes occur on the chromatin of E2F target genes (see the introduction). In particular, activating E2F species associate with target promoters in late G1, concomitant with hyperacetylation of histones H3 and H4 (47). It remained unknown, however, whether E2F promotes histone acetylation or acts on a downstream event in the transcriptional activation cascade. Our data establish that E2F directly promotes acetylation of target chromatin in vivo and is required for hyperacetylation of histones H3 and H4 in late G1 after mitogenic stimulation.
We used T98G cells, in which the dynamics of chromatin modifications were previously characterized (47). Adenovirus-mediated expression of a DN E2F species upon serum stimulation blocked binding of all E2F species to target promoters, as assayed by quantitative ChIP. The hyperacetylation of chromatin on those promoters was also impaired, implying that E2F is required for this event. In support of this conclusion, acute activation of a recombinant ER-E2F1 protein induced rapid acetylation of histones H3 and H4. DN E2F blocked not only acetylation but also the activation of E2F target genes and, as previously reported (40), entry into S phase. Hence, the hyperacetylation of target promoters mediated by E2F is likely to be an essential step in the transcriptional activation cascade that follows serum stimulation.
The HATs p300/CBP and GCN5/PCAF have been shown to interact with E2F (see the introduction). We demonstrate that E2F can bind an additional HAT, Tip60. Activation of ER-E2F1 induced recruitment of Tip60 to target promoters, alongside four additional subunits of the Tip60 complex: TRRAP, p400, Tip48, and Tip49 (15, 20). Four of these proteins (Tip60, TRRAP, Tip48, and Tip49) were detectably enriched on E2F target promoters in late G1 in a manner sensitive to blockade by DN E2F. Hence, E2F recruits the Tip60 complex to target chromatin in vivo. It remains to be addressed whether this occurs through direct interaction of E2F with TRRAP, Tip60, and/or other subunits of the complex.
So far, our attempts to interfere with Tip60 function, either by RNA interference or with a DN mutant, have failed to reveal an essential role of Tip60 in E2F-regulated transcription (data not shown). We do not know whether this is due to technical limitations (e.g., incomplete Tip60 knockdown) or to functional redundancy of Tip60 with different HATs. In favor of the latter possibility, a HAT activity distinct from Tip60 but of similar enzymatic specificity can be detected in p400- and TRRAP-containing complexes (35). Other E2F-interacting HATs, including GCN5/PCAF and p300/CBP (see the introduction) may also account for functional redundancy. In spite of consistent efforts, these HATs have not yet been detected by ChIP on E2F target genes (see Results) (47). Nonetheless, their interaction with E2F strongly suggests that they are also recruited to chromatin.
Altogether, the available data suggest that a complex web of histone modifications occurs on E2F target genes. In particular, hyperacetylation of H3 and H4 is likely to reflect modification of various lysines in their N-terminal tails, catalyzed by multiple HATs, which include p300/CBP, GCN5/PCAF, Tip60, and possibly other, yet unidentified species. In this context, it is worth reminding that while H4 acetylation in late G1 was fully dependent upon activating E2Fs, a substantial fraction of H3 acetylation was not and occurred upon mere dissociation of repressive E2F4-p130-HDAC complexes. It is tempting to speculate that, in the order of events leading to transcriptional activation of target genes, E2F-independent H3 acetylation may be a prerequisite for chromatin association of E2F1 to -3 and subsequent acetylation of both H3 and H4. Current models predict that different histone modifications determine the specificity of downstream responses (42, 46). Understanding E2F-regulated transcription will require systematic analysis of all HATs and acetylated lysines that occur on target promoters.
Acknowledgments
We thank Kristian Helin for reagents and for critical reading of the manuscript, Sandra Zurawski for the purification of adenoviruses, and Emma Lees for her support.
DNAX is supported by Schering-Plough Corp. Work in Bruno Amati's lab at the European Institute of Oncology is supported by the Italian Association for Cancer Research (AIRC) and the Italian Health Ministry.
REFERENCES
- 1.Ait-Si-Ali, S., A. Polesskaya, S. Filleur, R. Ferreira, A. Duquet, P. Robin, A. Vervish, D. Trouche, F. Cabon, and A. Harel-Bellan. 2000. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene 19:2430-2437. [DOI] [PubMed] [Google Scholar]
- 2.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 3.Cloud, J. E., C. Rogers, T. L. Reza, U. Ziebold, J. R. Stone, M. H. Picard, A. M. Caron, R. T. Bronson, and J. A. Lees. 2002. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol. Cell. Biol. 22:2663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 5.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis, G. V., C. Vaziri, N. Guo, and D. V. Faller. 2000. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 11:417-424. [PMC free article] [PubMed] [Google Scholar]
- 7.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 8.Emili, A., and C. J. Ingles. 1995. Promoter-dependent photocross-linking of the acidic transcriptional activator E2F-1 to the TATA-binding protein. J. Biol. Chem. 270:13674-13680. [DOI] [PubMed] [Google Scholar]
- 9.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira, R., L. Magnaghi-Jaulin, P. Robin, A. Harel-Bellan, and D. Trouche. 1998. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. USA 95:10493-10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira, R., I. Naguibneva, M. Mathieu, S. Ait-Si-Ali, P. Robin, L. L. Pritchard, and A. Harel-Bellan. 2001. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H.-M. Chan, D. M. Livingston, and B. Amati. 2003. Myc recruits the Tip60 histone acetyl-transferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry, C. J., A. Pearson, E. Malinowski, S. M. Bartley, J. Greenblatt, and P. J. Farnham. 1999. Activation of the murine dihydrofolate reductase promoter by E2F1. A requirement for CBP recruitment. J. Biol. Chem. 274:15883-15891. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 16.Gstaiger, M., B. Luke, D. Hess, E. J. Oakeley, C. Wirbelauer, M. Blondel, M. Vigneron, M. Peter, and W. Krek. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302:1208-1212. [DOI] [PubMed] [Google Scholar]
- 17.Hagemeier, C., A. Cook, and T. Kouzarides. 1993. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 21:4998-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J. K., S. Fredersdorf, T. Kouzarides, K. Martin, and X. Lu. 1997. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 11:1840-1852. [DOI] [PubMed] [Google Scholar]
- 20.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, D. G., J. K. Schwartz, W. D. Cress, and J. R. Nevins. 1993. Expression of transcription factor E2F-1 induces quiescent cells to enter S phase. Nature 365:349-352. [DOI] [PubMed] [Google Scholar]
- 22.Kanemaki, M., Y. Kurokawa, T. Matsu-ura, Y. Makino, A. Masani, K. Okazaki, T. Morishita, and T. A. Tamura. 1999. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 274:22437-22444. [DOI] [PubMed] [Google Scholar]
- 23.Kanemaki, M., Y. Makino, T. Yoshida, T. Kishimoto, A. Koga, K. Yamamoto, M. Yamamoto, V. Moncollin, J. M. Egly, M. Muramatsu, and T. Tamura. 1997. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 235:64-68. [DOI] [PubMed] [Google Scholar]
- 24.Kel, A. E., O. V. Kel-Margoulis, P. J. Farnham, S. M. Bartley, E. Wingender, and M. Q. Zhang. 2001. Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J. Mol. Biol. 309:99-120. [DOI] [PubMed] [Google Scholar]
- 25.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 26.Lang, S. E., S. B. McMahon, M. D. Cole, and P. Hearing. 2001. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 276:32627-32634. [DOI] [PubMed] [Google Scholar]
- 27.Leone, G., J. DeGregori, Z. Yan, L. Jakoi, S. Ishida, R. S. Williams, and J. R. Nevins. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 12:2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzio, G., C. Wagener, M. I. Gutierrez, P. Cartwright, K. Helin, and M. Giacca. 2000. E2F family members are differentially regulated by reversible acetylation. J. Biol. Chem. 275:10887-10892. [DOI] [PubMed] [Google Scholar]
- 31.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 32.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolas, E., V. Morales, L. Magnaghi-Jaulin, A. Harel-Bellan, H. Richard-Foy, and D. Trouche. 2000. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem. 275:9797-9804. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas, E., C. Roumillac, and D. Trouche. 2003. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol. Cell. Biol. 23:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, J., M. A. Wood, and M. D. Cole. 2002. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol. 22:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, A., and J. Greenblatt. 1997. Modular organization of the E2F1 activation domain and its interaction with general transcription factors TBP and TFIIH. Oncogene 15:2643-2658. [DOI] [PubMed] [Google Scholar]
- 37.Pediconi, N., A. Ianari, A. Costanzo, L. Belloni, R. Gallo, L. Cimino, A. Porcellini, I. Screpanti, C. Balsano, E. Alesse, A. Gulino, and M. Levrero. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5:552-558. [DOI] [PubMed] [Google Scholar]
- 38.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 40.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 41.Saavedra, H. I., B. Maiti, C. Timmers, R. Altura, Y. Tokuyama, K. Fukasawa, and G. Leone. 2003. Inactivation of E2F3 results in centrosome amplification. Cancer Cell 3:333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiber, S. L., and B. E. Bernstein. 2002. Signaling network model of chromatin. Cell 111:771-778. [DOI] [PubMed] [Google Scholar]
- 43.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 44.Stevaux, O., and N. J. Dyson. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14:684-691. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, C., and N. B. La Thangue. 2003. E2F and cell cycle control: a double-edged sword. Arch. Biochem. Biophys. 412:157-169. [DOI] [PubMed] [Google Scholar]
- 46.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 48.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 49.Trouche, D., A. Cook, and T. Kouzarides. 1996. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 24:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandel, L., and T. Kouzarides. 1999. Residues phosphorylated by TFIIH are required for E2F-1 degradation during S-phase. EMBO J. 18:4280-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 52.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321-330. [DOI] [PubMed] [Google Scholar]
- 55.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]