Oligodendrocyte precursor cells differentiate into oligodendrocytes to form myelin sheaths. Rab35/ACAP2 and cytohesin-2 antagonistically control oligodendrocyte differentiation and myelination through Arf6 on/off regulation, presenting a unique way of regulating oligodendrocyte differentiation and myelination by a small GTPase network.
Abstract
Oligodendrocyte precursor cells differentiate to produce myelin sheaths that insulate axons to ensure fast propagation of action potentials. Many aspects of differentiation are regulated by multiple extracellular signals. However, their intracellular signalings remain elusive. We show that Rab35 and its effector, ACAP2, a GTPase-activating protein that switches off Arf6 activity, negatively regulate oligodendrocyte morphological differentiation. Knockdown of Rab35 or ACAP2 with their respective small interfering RNAs promotes differentiation. As differentiation initiates, the activities of Rab35 and ACAP2 are down-regulated. The activity of Arf6, in contrast, is up-regulated. Arf6 knockdown inhibits differentiation, indicating that Rab35 and ACAP2 negatively regulate differentiation by down-regulating Arf6. Importantly, as differentiation proceeds, the activity of cytohesin-2, a guanine nucleotide exchange factor that switches on Arf6 activity, is up-regulated. Pharmacological inhibition of cytohesin-2 inhibits differentiation, suggesting that cytohesin-2 promotes differentiation by activating Arf6. Furthermore, using oligodendrocyte-neuronal cocultures, we find that knockdown of Rab35 or ACAP2 promotes myelination, whereas inhibition of cytohesin-2 or knockdown of Arf6 inhibits myelination. Thus Rab35/ACAP2 and cytohesin-2 antagonistically control oligodendrocyte differentiation and myelination through Arf6 regulation, presenting a unique small GTPase on/off switching mechanism.
INTRODUCTION
During development of the CNS, oligodendrocyte precursor cells (OPCs) undergo cell division and migrate along axons to their final destinations, where oligodendrocytes differentiate to wrap axons with myelin sheaths (Miller, 2002; Bradl and Lassman, 2010; Nave, 2010; Simons and Lyons, 2013). The multilayer myelin sheath plays a critical role in insulating axons and thereby markedly increasing their nerve conduction velocity. While the dynamic morphological changes that oligodendrocytes undergo are believed to be achieved by continuous on-and-off reactions of intracellular signals, the whole picture of the relevant mechanisms is still not completely clear. Among a number of signaling components, the signaling network involving small GTPases as guanine nucleotide–dependent molecular switches can regulate continuous morphological changes (Cherfils and Zeghouf, 2013).
It is known that Rac1 and Cdc42, small GTPases of Rho family, act as positive regulators of oligodendrocyte morphological differentiation and are responsible for controlling oligodendrocyte process extension and branching in culture (Liang et al., 2004). Mouse genetic studies indicate that Rac1 and Cdc42 are required for proper axon ensheathment and myelination of axons by oligodendrocytes (Thurnherr et al., 2006); these roles are consistent with their roles as positive myelination regulators. In contrast, RhoA acts as a negative regulator, inhibiting process extension and branching (Liang et al., 2004).
In addition to the Rho GTPases, small GTPase members belonging to the Arf family are also known to contribute to cell morphological changes (D'Souza-Schorey and Chavrier, 2006; Kahn et al., 2006; Casanova, 2007; Donaldson and Jackson, 2011). Arfs can also act as intracellular molecular switches depending on their guanine nucleotide–binding states. Thus the regulators known as guanine nucleotide exchange factors (GEFs, positive regulators) and GTPase-activating proteins (GAPs, negative regulators) participate in oscillating between active GTP-bound states and inactive GDP-bound ones. It is currently unknown whether Arf signals are involved in oligodendrocyte morphological differentiation during the myelination process, although, in the peripheral nervous system (PNS), Arf6-GEF cytohesin-1 (Kolanus et al., 1996; Meacci et al., 1997) and the downstream activation of Arf6 are required for morphological changes in Schwann cells, the myelin-forming glial cells in the PNS, during the myelination process (Yamauchi et al., 2012; Miyamoto et al., 2013a; Torii et al., 2013).
The Rab family proteins are another group of small GTPases that plays an important role in mediating vesicular trafficking (Stenmark, 2009; Pfeffer, 2013). More than 60 Rab proteins have been identified in mammals. Each Rab is specifically localized to a particular membrane transport pathway. Some Rab proteins are well characterized in oligodendrocytes and potentially mediate their morphogenesis through membrane homeostasis. Rab40C, which has been cloned from an oligodendrocyte cDNA library, is localized in the perinuclear recycling compartment (Rodriguez-Gabin et al., 2004). Rab40C participates in endocytic events such as receptor recycling. Rab31, which has also been cloned from an oligodendrocyte cDNA library, is involved in the transport from the trans-Golgi network to the endosomes (Rodriguez-Gabin et al., 2010). In addition, although Rab35 has been cloned from a skeletal muscle cDNA library (Zhu et al., 1994), it also functions as a regulator of the exocytotic pathway in oligodendrocytes (Hsu et al., 2010). Rab35 is known to be a unique Rab protein. In a complete analysis of small GTPases, the major subcellular localization site of Rab35 was identified to be the plasma membrane, in contrast to the other Rab proteins, which are localized in the intracellular vesicles (Heo et al., 2006).
These findings remind us of the possible presence of networks among the small GTPase families in oligodendrocytes and raise important questions concerning how such networks form and whether they act cooperatively or antagonistically in oligodendrocytes. We herein show that Rab35 negatively regulates oligodendrocyte morphological differentiation through ACAP2 (Arf-GAP with coiled-coil, ankyrin repeat, and pleckstrin homology domains 2; Jackson et al., 2000), the GAP that specifically switches off Arf6. In contrast, cytohesin-2 (Chardin et al., 1996; Frank et al., 1998), the GEF protein that specifically switches on Arf6, promotes differentiation. We also find that signaling through Rab35 and ACAP2 and signaling through cytohesin-2 reciprocally regulate myelination by oligodendrocytes, using an oligodendrocyte–dorsal root ganglion (DRG) neuronal coculture system. These results suggest the presence of a previously unknown unique small GTPase network functioning at the stage when oligodendrocytes begin differentiating for myelination.
RESULTS
Rab35 negatively regulates differentiation in FBD-102b cells
FBD-102b cells were established from fetal brains of p53 knockout mice (Horiuchi and Tomooka, 2006). FBD-102b cells retain OPC properties in the presence of 10% fetal bovine serum (FBS). When cultured in a reduced serum medium, FBD-102b cells differentiate into oligodendrocyte phenotypes with highly branched processes that contain myelin marker proteins such as myelin basic protein (MBP). Thus FBD-102b cells are used as a model for analyzing the molecular mechanisms underlying oligodendrocyte differentiation (Horiuchi and Tomooka, 2006; Miyamoto et al., 2007). To quantify differentiation using the method of Sperber and McMorris (2001), we have divided morphological differentiation into three stages based essentially on the complexity of oligodendrocytes’ processes; stage 1 (two or fewer primary processes longer than a cell body), stage 2 (three or four primary processes), and stage 3 (five or more primary processes). Before the induction of differentiation, major morphological cell types exhibit two short processes and thus belong to stage 1. Three days after the induction of differentiation, cells exhibit more highly branched processes and are thus categorized into stage 3 (Miyamoto et al., 2007).
To examine the involvement of Rab35 in oligodendrocyte differentiation, we first knocked down endogenous Rab35 proteins with nonoverlapping small interfering RNAs (siRNA; siRab35#1 and siRab35#2) in FBD-102b cells. Rab35 was specifically knocked down by transfection with each of the respective Rab35 siRNAs but not with the control luciferase siRNA (Figure 1, A and B). In contrast, control β-actin protein was not altered The effect of siRab35#2 on knockdown was slightly stronger than that of siRab35#1. The number of processes was greater in cells transfected with siRab35#1 (44.8 ± 0.805% of stage 3 cells; Figure 1, D and E) than in those transfected with the control siRNA (32.3 ± 0.588% of stage 3 cells; Figure 1, D and E). A similar but slightly stronger effect was observed in transfection with siRab35#2 (53.3 ± 1.44% of stage 3 cells; Figure 1, D and E), consistent reciprocally with the findings concerning the knockdown efficiencies of siRNA#1 and siRNA#2. Transfection with siRab35#1 and siRab35#2 increased the protein levels of MBP by 1.63 ± 0.141–fold and 3.40 ± 0.163–fold, respectively (Figure 1, A and C). Expression of the siRab35#2-resistant form of Rab35 with siRab35#2 reversed the increases in the percentage of stage 3 cells (Figure 1F). These results suggest that Rab35 negatively regulates differentiation in FBD-102b cells.
FIGURE 1:
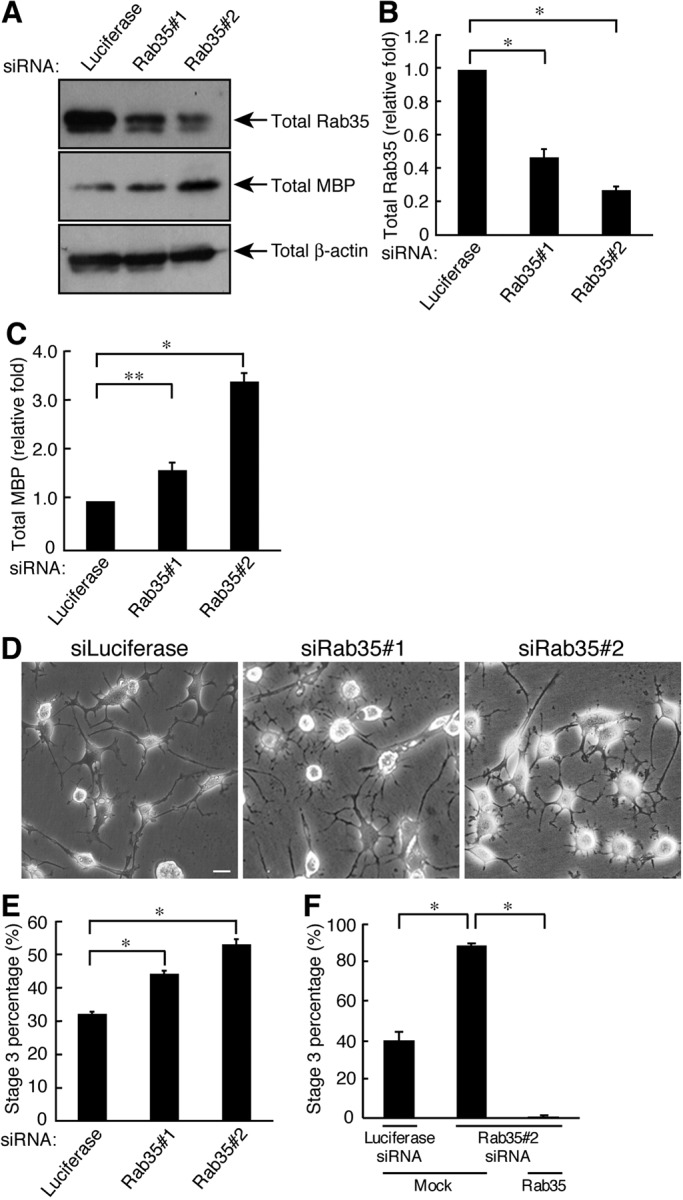
Rab35 is required for FBD-102b cell differentiation. (A) Cells were transfected with an siRNA for control (luciferase), Rab35#1, or Rab35#2 and immunoblotted with an antibody against Rab35, MBP, or β-actin. (B) The bands of each Rab35 protein were semiquantified (n = 5). (C) The bands of each MBP protein were semiquantified (n = 5). (D) Cells transfected with an siRNA for control, Rab35#1, or Rab35#2 were allowed to differentiate for 3 d. Representative images are shown. Scale bar: 20 μm. (E) The percentage of cells in stage 3 was determined (n = 10 fields in three experiments). (F) Cells transfected with an siRNA for control or Rab35#2 as well as control vector or GFP-siRNA–resistant Rab35 were allowed to differentiate for 3 d. The percentage of cells in stage 3 was determined (n = 10 fields in three experiments). Data were evaluated with one-way ANOVA (*, p < 0.01; **, p < 0.05).
Thus we measured the bona fide activity of Rab35 through an affinity-precipitation assay using the Rab35•GTP-binding domain (RBD) of SH3 domain–containing 2 (RUSC2), which specifically binds to active GTP-bound forms of Rab35 proteins (Fukuda et al., 2011). Although expression levels of Rab35 proteins increased following the induction of differentiation (Figure 2, A and C), GTP-bound forms of Rab35 were dramatically decreased, by 27.9 ± 0.0535% and 13.7 ± 0.0261% at 2 and 3 d, respectively (Figure 2, A and B).
FIGURE 2:
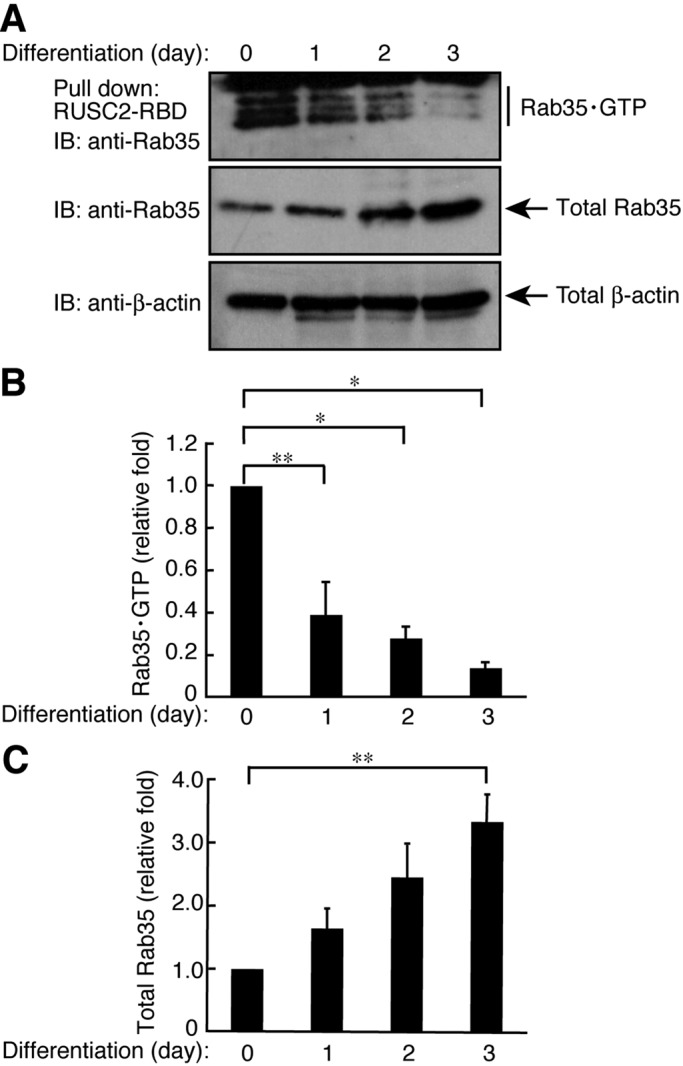
Rab35 is inactivated following differentiation in FBD-102b cells. (A) Cells were allowed to differentiate for 0–3 d and lysed to be used in an affinity precipitation to detect active GTP-bound Rab35 with recombinant GST-RUSC2-RBD. Immunoblots for total Rab35 and β-actin are also shown. (B) Bands corresponding to each GTP-bound Rab35 protein were scanned and semiquantified. (C) Bands corresponding to each total Rab35 protein were scanned and semiquantified. Data were evaluated with one-way ANOVA (*, p < 0.01; **, p < 0.05; n = 4).
The Rab35 effector ACAP2, which switches off Arf6, negatively regulates differentiation in FBD-102b cells
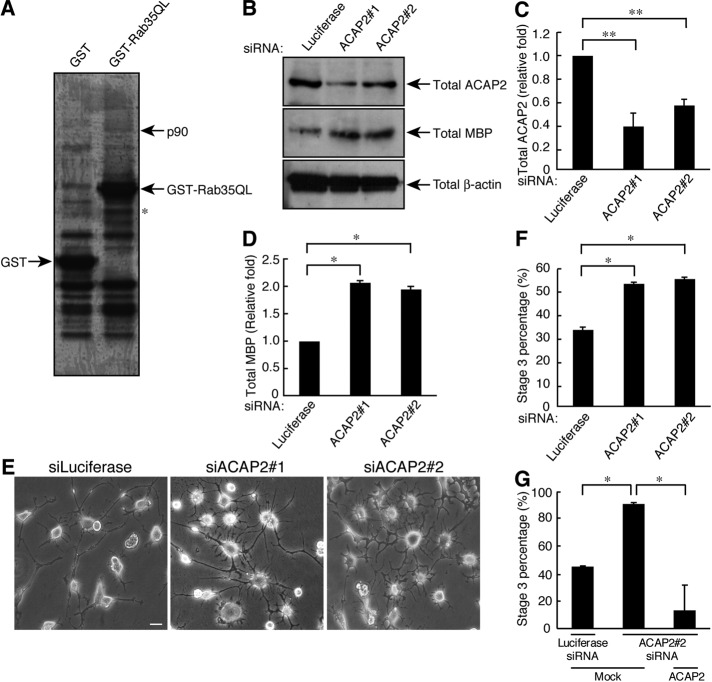
We searched for Rab35-binding protein(s) through biochemical procedures using the lysates of rat brains at postnatal day 1, when oligodendrocyte lineage cells are present as OPCs. Rab35 affinity column chromatography and mass spectrometric analysis identified proteins of ∼90 kDa containing ACAP2 and Hsp90 (Figure 3A). ACAP2 is an Arf6-specific GAP, switching off Arf6 activity. In PC12 cells, it is known as the Rab35 effector that is involved in neurite outgrowth (Kobayashi and Fukuda, 2012); its role in oligodendrocytes, however, is currently unknown. On the other hand, Hsp90 is known to form a complex with guanine nucleotide dissociation inhibitor (GDI), which interacts with Rab1 to modulate its cellular activity (Chen and Balch, 2006). It is possible that Hsp90 may modulate Rab35’s role in oligodendrocytes as a potential GDI for Rab35.
FIGURE 3:
Identification of ACAP2 as a Rab35-interacting protein. (A) Rat brain lysates were loaded onto a glutathione-resin affinity column on which GST or GST-Rab35Q67L was immobilized. The bound proteins were eluted by the addition of glutathione and were evaluated by SDS–PAGE followed by Coomassie staining. The ∼90-kDa stained protein band indicates specific binding to Rab35Q67L. The ∼90 kDa band was identified through proteomic analysis as a mixture of ACAP2 and Hsp90. Asterisk indicates GST protein degradation products. (B) Cells were transfected with an siRNA for ACAP2#1 or ACAP2#2 and immunoblotted with an antibody against ACAP2, MBP, or β-actin. (C) The bands of each ACAP2 protein were semiquantified (n = 6). (D) The bands of each MBP protein were semiquantified (n = 6). (E) Cells transfected with an siRNA for control, ACAP2#1, or ACAP2#2 were allowed to differentiate for 3 d. Representative images are shown. Scale bar: 20 μm. (F) The percentage of cells in stage 3 was determined (n = 10 fields in three experiments). (G) Cells transfected with an siRNA for control or ACAP2#2 as well as control vector or ZsGreen-siRNA–resistant ACAP2 were allowed to differentiate for 3 d. The percentage of cells in stage 3 was determined (n = 10 fields in three experiments). Data were evaluated with one-way ANOVA (*, p < 0.01; **, p < 0.05).
To determine whether ACAP2 is involved in oligodendrocyte differentiation, we transfected the nonoverlapping siRNAs (siACAP2#1 or siACAP2#2) into FBD-102b cells. Transfection with siACAP2#1 and siACAP2#2 knocked down ACAP2 proteins by ∼60 and 40%, respectively (Figure 3, B and C). In contrast, none of the siRNAs had any detectable effect on the control protein. Following ACAP2 knockdown, enhanced differentiation was observed. Differentiation efficiency was assessed based on the percentage of stage 3 cells (Figure 3, E and F; 54.0 ± 0.471% and 56.2 ± 0.620% in siACAP2#1 and siACAP2#2, respectively, compared with 33.8 ±1.02% in the control). In keeping with these findings, MBP protein levels were up-regulated by ACAP2 knockdown (Figure 3, B and D; 2.07 ± 0.0314–fold and 1.95 ± 0.0462–fold in siACAP2#1 and siACAP2#2, respectively). Expression of the siACAP2#2-resistant form of ACAP2 with siACAP2#2 reversed the increases in the percentage of stage 3 cells (Figure 3G). Collectively the FBD-102b cell morphological and biochemical data seen in ACAP2 knockdown are similar to those seen in Rab35 knockdown.
We next tried to measure the Arf6-GAP activity of ACAP2 in FBD-102b cells. We first performed an affinity precipitation of overexpressed myc-tagged ACAP2 with each of several guanine nucleotide–binding Arf6 proteins. Because active GAPs preferentially bind to GTP-bound GTPases, we checked whether ACAP2 forms a complex with GTP-bound Arf6 in vitro (Jackson et al., 2000). As shown in Figure 4A, ACAP2 was specifically coprecipitated with GTPγS-bound Arf6, but not with GDP-bound or nucleotide-free Arf6. This assay revealed that, following the induction of differentiation, levels of precipitated ACAP2 gradually decreased, by 56.9 ± 0.216%, 12.7 ± 0.0839%, and 9.81 ± 0.0493% at 1, 2, and 3 d, respectively (Figure 4, B and C). Expression levels of ACAP2 proteins were comparable during differentiation (Figure 4, B and C).
FIGURE 4:
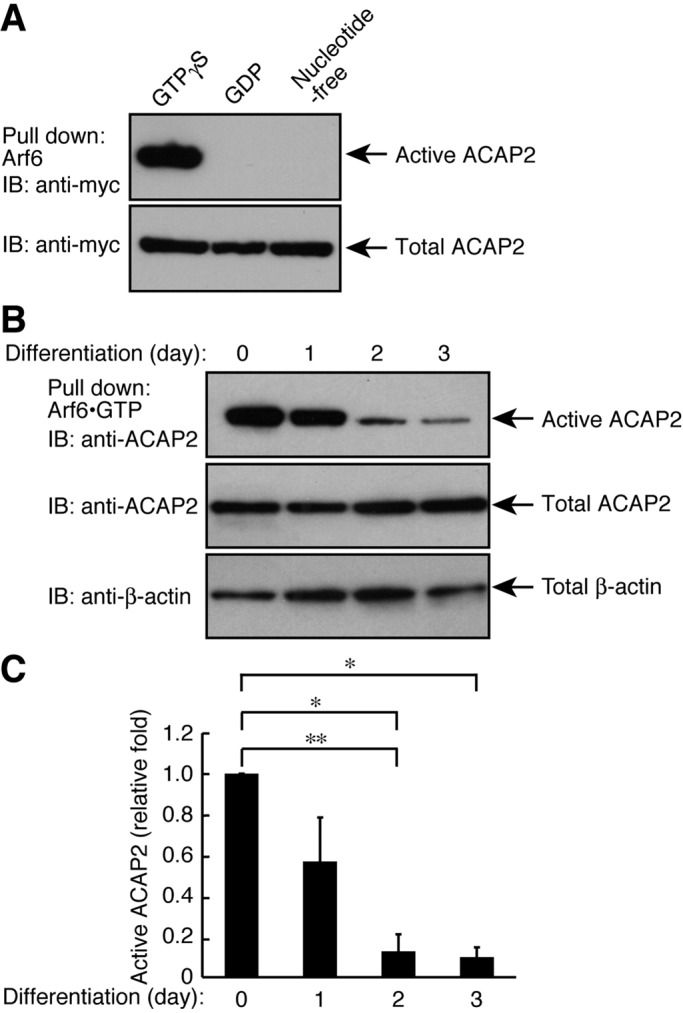
ACAP2 is inactivated following differentiation in FBD-102b cells. (A) The plasmid encoding myc-tagged ACAP2 was transfected into 293T cells and lysed to be used in an affinity precipitation to detect overexpressed, active ACAP2 using various forms of guanine nucleotide–binding recombinant Arf6. The GTP form of Arf6 only binds to ACAP2. Immunoblot for total ACAP2 is also shown. (B) Cells were allowed to differentiate for 0–3 d and lysed to be used in an affinity precipitation of active ACAP2 with recombinant GST-tagged GTPγS-bound Arf6. Immunoblots for total ACAP2 and β-actin are also shown. (C) Bands corresponding to each active ACAP2 protein were scanned and semiquantified (n = 4). Data were evaluated with one-way ANOVA (*, p < 0.01; **, p < 0.05).
Arf6 is required for differentiation in FBD-102b cells
ACAP2 negatively regulates differentiation, and ACAP2 activity decreases as differentiation proceeds. Because ACAP2 is the Arf6-GAP protein and the Arf6 negative regulator, it is thought that Arf6 itself positively regulates differentiation. We previously reported that signaling through Arf6 is also required for Schwann cell morphological changes leading to myelination (Yamauchi et al., 2012). Arf6 primarily mediates intracellular transport and may in part regulate actin cytoskeletal changes (D'Souza-Schorey and Chavrier, 2006; Kahn et al., 2006; Casanova, 2007; Donaldson and Jackson, 2011). Transfection with Arf6 siRNA knocked down Arf6 proteins by ∼60% (Figure 5, A and B). In contrast, Arf6 knockdown had no detectable effect on the control protein (Figure 5A). As expected, Arf6 knockdown decreased the percentages of stage 3 cells (21.5 ± 1.55%) compared with the control (34.3 ± 1.27%) (Figure 5, D and E). Similarly, knockdown decreased MBP protein levels by 0.172 ± 0.0164–fold (Figure 5, A and C). Expression of the siArf6-resistant form of Arf6 with siArf6 reversed the decreases in the percentage of stage 3 cells (Figure 5F). These results suggest that Arf6 is required for oligodendrocyte differentiation, as it is for Schwann cells (Yamauchi et al., 2012).
FIGURE 5:
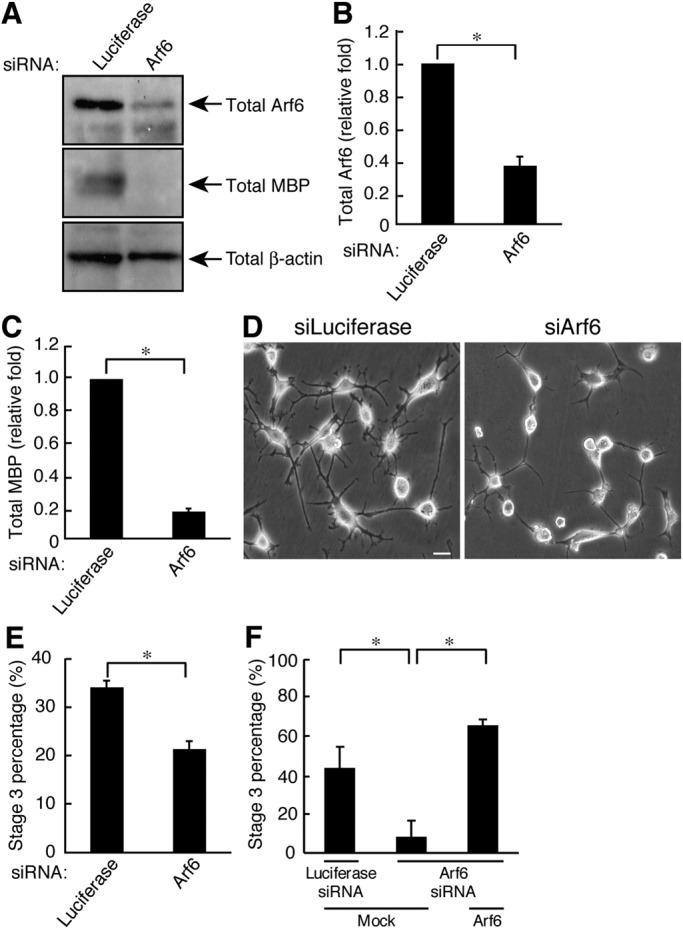
Arf6 is required for FBD-102b cell differentiation. (A) Cells were transfected with either an siRNA for control, or Arf6, and immunoblotted with an antibody against Arf6, MBP, or β-actin. (B) The bands of each Arf6 protein were semiquantified (n = 3). (C) The bands of each MBP protein were semiquantified (n = 3). (D) Cells transfected with an siRNA for control or Arf6 were allowed to differentiate for 3 d. Representative images are shown. Scale bar: 20 μm. (E) The percentage of cells in stage 3 was determined (n = 10 fields in three experiments). (F) Cells transfected with an siRNA for control or Arf6 as well as control vector or ZsGreen-siRNA–resistant Arf6 were allowed to differentiate for 3 d. The percentage of cells in stage 3 was determined (n = 10 fields in three experiments). Data were evaluated with Student's t test (*, p < 0.01).
Accordingly, we next measured the activity of Arf6. We performed an affinity-precipitation assay using glutathione S-transferase (GST)-tagged GGA3-ABD, which specifically binds to active GTP-bound Arf6 (Yamauchi et al., 2009; Torii et al., 2010). The Arf6 activity was up-regulated by 40.0 ± 4.24–fold at 3 d after the induction of differentiation; in contrast, Arf6 expression levels were not altered during differentiation (Figure 6, A and B). These results are consistent with our other results indicating that Arf6 is required for differentiation.
FIGURE 6:
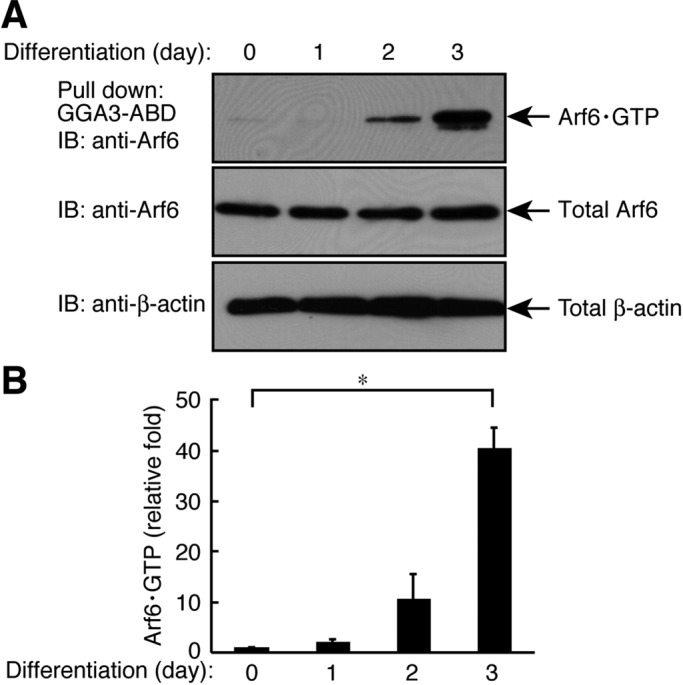
Arf6 is activated following differentiation in FBD-102b cells. (A) Cells were allowed to differentiate for 0–3 d and lysed to be used in an affinity precipitation of active GTP-bound Arf6 with recombinant GST-GGA3-ABD. Immunoblots for total Arf6 and β-actin are also shown. (B) Bands corresponding to each GTP-bound Arf6 protein were scanned and semiquantified (n = 4). Data were evaluated with one-way ANOVA (*, p < 0.01).
We further confirmed that Rab35 and ACAP2 negatively regulate Arf6. Rab35 knockdown increased GTP-bound Arf6 by 2.12 ± 0.177–fold (Supplemental Figure S1, A and B). Similar results were obtained in ACAP2 knockdown. ACAP2 knockdown increased GTP-bound Arf6 by 1.60 ± 0.227–fold (Figure S1, C and D).
Roles of Rab35/ACAP2/Arf6 in oligodendrocyte differentiation
We explored the role of the Rab35/ACAP2/Arf6 signaling unit in primary rat oligodendrocytes. When OPCs were allowed to differentiate for 3 d, their processes became highly branched and abundant with weblike myelin structures that could be detected with an anti-MBP antibody (Figure S2A). We infected primary OPCs with retroviruses harboring short hairpin RNAs (shRNA) for Rab35, ACAP2, or Arf6 and knocked down each of them. Transfection with shRab35 (Figure S2C), shACAP2 (Figure S2D), or shArf6 (Figure S2E) specifically knocked down the respective proteins, whereas expression levels of control β-actin proteins were unaffected. Knockdown of Rab35 and ACAP2 increased differentiation (Figure S2, A and B; 35.8 ± 0.500% and 30.8 ± 0.550% in shRab35 and shACAP2, respectively, compared with 20.9 ± 0.390% in the control). In addition, knockdown of Rab35 or ACAP2 promoted MBP expression (Figure S2, C and D). In contrast, Arf6 knockdown inhibited both differentiation (Figure S2, A and B; 5.60 ± 0.310% in shArf6 compared with 20.9 ± 0.390% in the control) and MBP expression (Figure S2E). These results show that signaling through Rab35 and ACAP2 negatively regulates differentiation in primary oligodendrocytes and support the results obtained from FBD-102b cells.
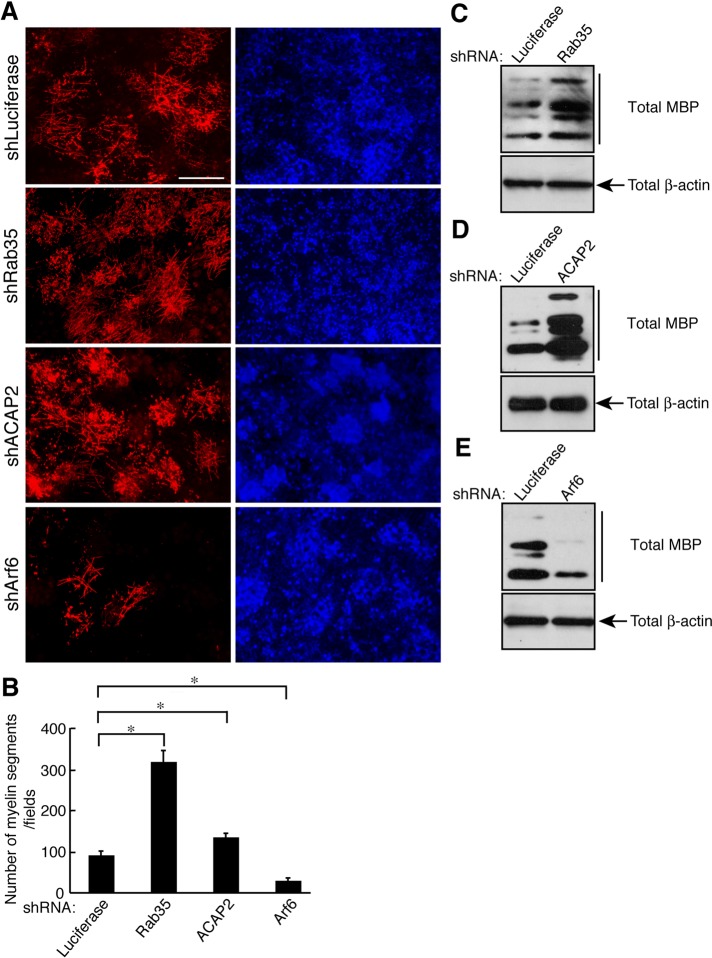
Roles of Rab35/ACAP2/Arf6 in myelin formation in cocultures
To investigate the role of the Rab35/ACAP2/Arf6 signaling unit in myelination by oligodendrocytes, we employed a coculture system using primary oligodendrocytes and primary DRG neurons. Knockdown of Rab35 or ACAP2 in oligodendrocytes promoted formation of MBP-positive myelin segments in cocultures (Figure 7, A and B; 316 ± 29.6 and 133 ± 11.5 in shRab35 and shACAP2, respectively, compared with 89.0 ± 12.6 in the control). On the other hand, Arf6 knockdown inhibited segment formation (Figure 7, A and B; 27.0 ± 8.80 in shArf6 compared with 89.0 ± 12.6 in the control). In concordance, MBP expression was up-regulated by knockdown of Rab35 or ACAP2 and inhibited by Arf6 knockdown (Figure 7, C–E), consistent with our proposal that signaling through Rab35 and ACAP2 negatively regulates oligodendrocyte differentiation and myelination. To further investigate the correlation of Rab35 signaling in the regulation of myelination by oligodendrocytes, we performed affinity-precipitation assays and immunoblotting analyses during mouse spinal cord development. While GTP-bound forms of Rab35 decreased from postnatal days 14–21, the most active phase of myelination, GTP-bound Arf6 gradually increased from postnatal days 7–21 (Figure S3). The levels of ACAP2 and cytohesin-2 proteins remained constant. These results appear to support the in vitro data.
FIGURE 7:
Rab35/ACAP2/Arf6 is required for myelin formation. (A) Primary OPCs infected with an shRNA for control, Rab35, ACAP2, or Arf6 were cocultured with DRG neurons for 4 wk and stained with an anti-MBP antibody (red) and DAPI (blue). Representative images are shown. Scale bar: 50 μm. (B) The percentages of the MBP-positive myelin segments were determined (n = 7 fields in three experiments). Data were evaluated with one-way ANOVA (*, p < 0.01). (C–E) Coculture lysates were immunoblotted with an antibody against MBP or β-actin.
Cytohesin-2 is involved in oligodendrocyte differentiation and myelin formation
While Arf6 is markedly activated as differentiation proceeds in FBD-102 cells, signaling through Rab35 and the effector ACAP2 negatively regulates Arf6. The question presented here is which molecule participates in activating Arf6 as differentiation proceeds. One important group of candidates, the GEF members of the cytohesin family, could activate Arf6, as cytohesin-1 and Arf6 both contribute to myelination by Schwann cells (Yamauchi et al., 2012). To test this possibility, we used SecinH3, the inhibitor specific to cytohesins (Fuss et al., 2006; Hafner et al., 2006), and examined its effect on differentiation. Treatment of FBD-102b cells with SecinH3 resulted in suppressed differentiation (8.36 ± 3.32% of stage 3 cells) compared with the control (57.6 ± 8.12% of stage 3 cells; Figure S4, A and B). Treatment with SecinH3 also decreased MBP expression by 21.4 ± 0.103% (Figure S4, C and D), suggesting that a cytohesin participates in differentiation.
Immunoblotting analysis using an antibody against cytohesin-1, -2, or -3 shows the primary expression of cytohesin-2 in FBD-102b cells, illustrating that cytohesin-2 probably mediates Arf6 activation in differentiation (Figure S4E). Thus we measured the endogenous cytohesin activity (Yamauchi et al., 2012). The affinity-precipitated, active cytoheisn-2 was increased by 3.69 ± 1.95–fold, 8.15 ± 4.19–fold, and 10.3 ± 4.22–fold at 1, 2, and 3 d, respectively. The expression levels of cytohesin-2 were comparable during differentiation (Figure S5, A and B).
We confirmed the role of cytohesin-2 in oligodendrocyte differentiation. Treatment with SecinH3 inhibited both oligodendrocyte differentiation (Figure S6, A and B; 0.341 ± 0.0771% in the presence of SecinH3 compared with 31.9 ± 0.0203% in the control) and MBP expression (Figure S6C). We further examined the effect of SecinH3 on myelin formation in cocultures. We observed that SecinH3 markedly inhibited both the formation of MBP-positive myelin segments (Figure 8, A and B; 3.00 ± 2.66 in the SecinH3 compared with 120 ± 13.8 in the control) and MBP expression (Figure 8C), consistent with the results from FBD-102b cells.
FIGURE 8:
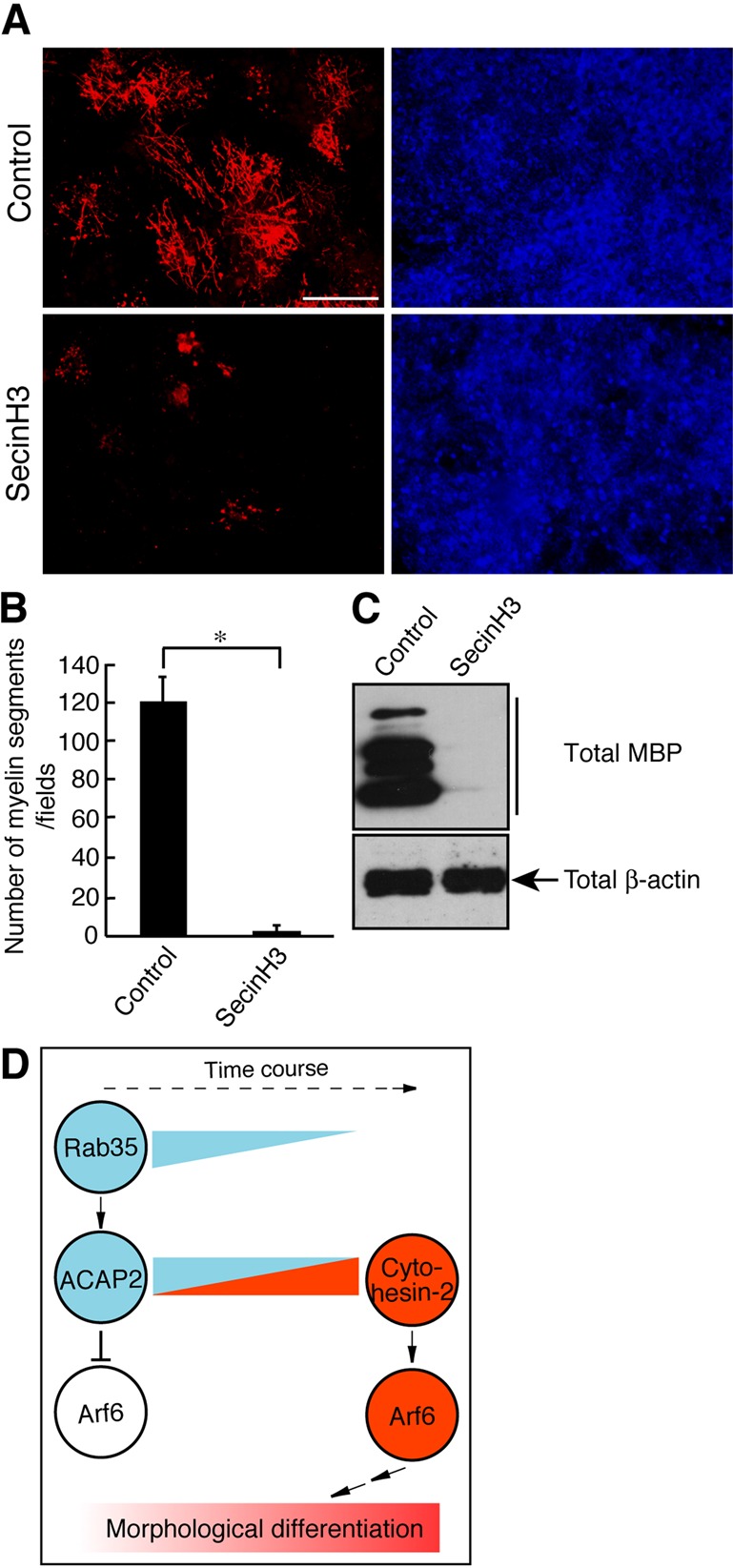
Cytohesin-2 is required for myelin formation. (A) Primary OPCs were cocultured with DRG neurons with or without 10 μM SecinH3 for 3 wk and stained with an anti-MBP antibody (red) and DAPI (blue). Representative images are shown. Scale bar: 50 μm. (B) The percentages of the MBP-positive myelin segments were determined (n = 7 fields in three experiments). (C) Cocultures were lysed and immunoblotted with an antibody against MBP or β-actin. Data were evaluated with the Student's t test (*, p < 0.01). (D) Schematic diagram describing the proposed regulatory mechanism of differentiation.
Finally, we tried to analyze a complex involving signaling molecules described above. We performed a coimmunoprecipitation assay using an anti–cytohesin-2 antibody before and after the induction of differentiation in FBD-102b cells (Figure S7). While Rab35 activity was down-regulated following differentiation (Figure 2A), the levels of coimmunoprecipitated Rab35 proteins were comparable before and after differentiation; however, the levels of Rab35 proteins increased following differentiation. It is likely that Rab35 levels in the complex may have been relatively decreased. The findings that Arf6 levels in the immunoprecipitates increased following differentiation correlate with the data that Arf6 activity is up-regulated following differentiation (Figure 6A). ACAP2 levels in the cytohesin-2 immunoprecipitates were unchanged before and after differentiation. These results again seem to support our idea that Arf6 regulation by Rab35/ACAP2 and cytohesin-2 is involved in oligodendrocyte differentiation and myelination.
DISCUSSION
Myelination of axons by oligodendrocytes is a highly regulated process that requires coordination of dynamic cellular events, including cytoskeletal rearrangements, protein and lipid synthesis, and intracellular trafficking of membranes (Miller, 2002; Bradl and Lassman, 2010; Nave, 2010; Simons and Lyons, 2013). Cytoskeletal changes are essential for the structural and functional organization of myelin sheaths, but the intracellular signaling mechanisms involved in their regulation are largely unknown. In this paper, we show that signaling through Rab35 negatively regulates oligodendrocyte differentiation and myelination. Using lysates of the neonatal rat brain, in which oligodendrocyte lineage cells are present as OPCs before differentiation, we have identified Arf6-specific GAP ACAP2 as a Rab35-binding protein. Indeed, as OPCs differentiate to oligodendrocytes, the activities of ACAP2 and Rab35 become down-regulated. Conversely, following differentiation, the activities of cytohesin-2 and Arf6 are up-regulated (Figure 8D). Because ACAP2 is the GAP protein that switches off Arf6, Arf6 itself has been expected to be the positive regulator in oligodendrocyte differentiation and myelination, consistent with the finding that Arf6 knockdown as well as cytohesin-2 inhibition inhibits these processes. Thus Rab35 and ACAP2 negatively regulate differentiation and myelination by down-regulating Arf6, which is up-regulated by cytohesin-2. The signaling unit presented here is a unique small GTPase network positively or negatively controlling oligodendrocyte differentiation and myelination. In PC12 cells, Rab35 promotes neurite outgrowth (Kobayashi and Fukuda, 2012). The difference in the effects of Rab35 on differentiation may be due to the variation and/or the number of the binding effectors in cell types. This is reminiscent of signaling through RhoA small GTPase. In neurons, RhoA inhibits axonal growth (Nikolic, 2002), whereas RhoA promotes myelination processes in Schwann cells (Melendez-Vasquez et al., 2004). Further studies will allow us to clarify the unique roles of Rab35 in various cell types.
Potential regulatory mechanism of Rab35 in oligodendrocytes
Rab family members, including Rab35, are thought to be regulated by their respective specific GEFs (Stenmark, 2009; Pfeffer, 2013). Increasing evidence shows that proteins containing previously function-unknown domains called DENN (differentially expressed in neoplastic versus normal cells) are GEFs for Rabs (Yoshimura et al., 2010). It was first established that DENND1A (also called connecdenn1) functions as a GEF for Rab35. DENND1A is well characterized to play a key role in the recycling of the major histocompatibility complex class I in lymphocyte early endosomal compartments (Allaire et al., 2010). In recycling, DENND1A, acting together with a clathrin adaptor protein, is recruited to clathrin-coated vesicles, where DENND1A can activate Rab35 (Marat and McPherson, 2010). It is likely that DENND1B (also called connecdenn2) is also a GEF for Rab35 and may play a role in endosomal recycling (Marat and McPherson, 2010). DENND1C (also celled connecdenn3) plays a role in localizing Rab35 to filamentous actin bundles, where DENND1C can activate Rab35 (Marat et al., 2012). DENND1A, DENND1B, and DENND1C are abundantly expressed in the brain (Marat and McPherson, 2010). They may cooperatively or independently act as GEFs to activate oligodendrocyte Rab35 before differentiation.
It is worth noting that disease-associated mutations in the peripheral demyelinating neuropathy Charcot-Marie-Tooth disease type 4B–responsible gene product MTMR13 are often detected in its DENN domain (Azzedine et al., 2003; Senderek et al., 2003). MTMR13’s DENN domain displays GEF activity for Rab21 (Jean et al., 2012). It remains to be precisely determined whether these genes’ mutations result in loss of function or gain of function in humans, but these findings do point out the importance of the regulation of the Rab members’ activities by their GEFs in proper myelination by peripheral myelin-forming Schwann cells. Further studies may allow us to learn whether any DENN domain–containing GEF or Rab member may negatively or positively control myelination processes.
On the other hand, three TBC1D10 family members (TBC1D10A, TBC1D10B, and TBC1D10C) are characterized as Rab35-specific GAPs that switch off Rab35 activity and likely function in oligodendrocytes (Hsu et al., 2010). Proteolipid protein 1 (PLP1), the major protein in myelin in the CNS, is localized to multivesicular bodies, which then fuse with plasma membranes. This fusion participates in PLP1 secretion to the extracellular space as exosome-containing vesicles. Three TBC1D10 family members and Rab35 regulate the release of PLP1-containing exosomes on plasma membranes in the oligodendroglial cell line Oli-neu. In its GTP-binding form, Rab35 can increase the density of vesicles on plasma membranes and control the docking and tethering of the vesicles with plasma membranes. Thus TBC1D10 members serve as Rab35’s negative regulators in releasing PLP1-containing exosomes. Although the role of PLP1-containing exosome formation in oligodendrocytes is not precisely understood, it is possible that these GAPs may act upstream of oligodendrocyte Rab35 to switch off its activity following differentiation.
Potential regulation of oligodendrocyte signaling by Rab35
It is clear that Rab35 and ACAP2 negatively regulate oligodendrocyte differentiation and myelination, but partners other than ACAP2 may interact with Rab35 before differentiation. Rab35 is known to interact with two actin cytoskeleton–associating proteins, molecule interacting with casL 1 (MICAL1) and MICAL-like protein 1 (Fukuda et al., 2008; Rahajeng et al., 2012). MICAL family proteins are thought to provide a link to actin cytoskeletal dynamics and vesicle trafficking (Sharma et al., 2009; Hung et al., 2010). As seen in a detailed immunohistochemical analysis, expression levels of MICAL family proteins are elevated in oligodendrocytes at the injury site after spinal cord injury (Pasterkamp et al., 2006). These findings suggest the role of MICAL family proteins in initiating remyelination. MICAL family proteins may act in parallel with ACAP2, acting downstream of oligodendrocyte Rab35.
Potential role of Arf6 in oligodendrocytes
We find that Arf6 itself positively regulates oligodendrocyte differentiation and myelination. Arf6 regulates actin cytoskeletal protein dynamics through activation of the Arf6 effectors known as the phosphatidylinositol-4-phosphate-5-kinases and phospholipase D isoenzymes (D'Souza-Schorey and Chavrier, 2006; Kahn et al., 2006). In addition, Arf6 has the ability to stimulate indirectly the activity of Rac1, which is directly linked to controlling cell morphological changes through actin cytoskeletal rearrangements (D'Souza-Schorey and Chavrier, 2006; Kahn et al., 2006). Rac1 is known to play a key role in actin cytoskeletal rearrangements required for oligodendrocyte myelination (Liang et al., 2004; Thurnherr et al., 2006; Bacon et al., 2007). The mechanism by which Arf6 activates Rac1 is proposed to occur through the Rac1-GEF Dock180 and the engulfment and cell motility complex (Santy et al. 2005; White et al., 2010). Arf6 also binds to c-Jun N-terminal kinase–interacting protein 3 (JIP3) and JIP4. JIP3 and JIP4 form a complex with kinesin-1 and dynactin to control the microtubule-dependent processes (Montagnac et al., 2009). Because cytoskeletal rearrangements are essential for myelination processes, it is possible that Arf6 effectors act together in oligodendrocyte morphological changes.
We previously reported that cytohesin-1 is a GEF for Arf6 to promote myelination by Schwann cells (Yamauchi et al., 2012; Miyamoto et al., 2013a; Torii et al., 2013). In this study, we find that cytohesin-2 is involved in oligodendrocyte differentiation and myelination, although it is possible that GEFs other than cytohesin family members activate Arf6. We demonstrate that a previously unknown signaling pathway through small GTPases positively or negatively regulates oligodendrocyte differentiation and myelination. These findings may indicate that a unique small GTPase network determines when oligodendrocytes differentiate for myelination. Further studies on the signaling mechanism through Rab35 and Arf6 in this line may promote our understanding of when and how small GTPases regulate myelination processes. They may also shed light on the complicated regulation of the small GTPase network by GEFs and GAPs, which work antagonistically in cells. Detailed studies on signaling networks may help to elucidate a paradigm for remyelination and also nerve regeneration.
MATERIALS AND METHODS
Antibodies
The following antibodies were purchased: anti-Rab35 from the Proteintech Group (Chicago, IL); anti-MBP from Covance (Princeton, NJ) or Merck Millipore (Billerica, MA); anti–β-actin from BD Biosciences PharMingen (Franklin Lakes, NJ); anti-Arf6 and anti–cytohesin-2 from Santa Cruz Biotechnology (Santa Cruz, CA); anti-ACAP2 from Abcam (Cambridge, UK); anti–green fluorescent protein (anti-GFP) from MBL (Nagoya, Japan); anti-ZsGreen from Clontech Takara Bio (Kyoto, Japan); anti-FLAG from Sigma-Aldrich (St. Louis, MO); anti-myc from Nacalai Tesque (Kyoto, Japan) or MBL; horseradish peroxidase–conjugated anti-mouse, anti-rabbit, or anti-goat immunoglobulin G secondary antibodies from GE Healthcare (Little Chalfont, Buckinghamshire, UK); and fluorescence-labeled secondary antibodies from Thermo Fisher Scientific (Waltham, MA).
Plasmids
All sequences were confirmed using automatic sequencers (Life Technologies, Carlsbad, CA). The coding region of human Rab35 protein was amplified by reverse transcription (RT)-PCR from cDNA of human embryonic kidney epithelial-like 293T cells and ligated into the mammalian expression vector pTag-GFPC vector (Clontech Takara Bio). The construct of the full-length Rab35 harboring the Glu-67-to-Leu (Q67L) was created from pTag-GFPC-Rab35 by Pfu-based PCR using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The region encoding mouse full-length ACAP2 was amplified by RT-PCR from cDNA of mouse 3T3-L1 cells and ligated into the pCMV vector containing the myc epitope at the 3′ position. The constructs of the full-length ACAP2 harboring the siACAP2#2- resistant sequence (replacement of GAGGCTGCAAACATTCTCA with GAGGCTGCAAACATCTTGA) was produced from pCMV-ACAP2-myc and then ligated into pIRES2-ZsGreen1 vector (Clontech Takara Bio). The region encoding Arf6 (amino acid residues 13–175) was amplified from total RNA of human brain as previously described (Yamauchi et al., 2012) and was ligated into pIRES2-ZsGreen1. The coding region of the RBD of RUN and RUSC2 was amplified from KIAA0375 (Kazusa DNA Research Institute, Chiba, Japan) and inserted into the Escherichia coli GST-tag expression vector pET42a. The pET42A-Rab35Q67L was created from pTag-GFPC-Rab35Q67L. The pET42a-Arf6 (amino acid residues 13–175) and pET42a-Arf•GTP-binding domain (ABD) of GGA3 plasmids were constructed as previously described (Yamauchi et al., 2009, 2012; Torii et al., 2010).
siRNAs
The siRNA duplexes were synthesized by Eurogentec (Seraing, Belgium). The nucleotide target sequences were 5′-AAGAUUCGGACUGUGGAGAUC-3′ (siRab35#1) and 5′-AAGCUGCAGAUCUGGGACACG-3′ (siRab35#2) for mouse Rab35, 5′-AAGCUUGUGAAACUGUGUAUC-3′ (siACAP2#1) and 5′-AAGAGGCUGCAAACAUUCUCA-3′ (siACAP2#2) for mouse ACAP2, and 5′-AACGUGUGGGAUGUGGGCGGC-3′ (siArf6) for mouse Arf6 (Yamauchi et al., 2009). The target sequence of the control Photinus pyralis luciferase was 5′-AAGCCAUUCUAUCCUCUAGAG-3′. The luciferase siRNA target sequence does not have homology to any other mammalian gene sequences.
Preparation of plasmids encoding sequences for shRNAs
The oligonucleotides (Fasmac, Kanagawa, Japan) used for duplex production were Rab35 (starting from nucleotide 300 of rat Rab35) sense oligonucleotide: 5′-GATCCGCGATGGCTTCATGAAATCTTCAAGAGAGATTTCATGAAGCCATCGCTTTTTTACGCGTG-3′; Rab35 antisense oligonucleotide: 5′-AATTCACGCGTAAAAAAGCGATGGCTTCATGAAATCTCTCTTGAAGATTTCATGAAGCCATCGCG-3′; ACAP2 (starting from nucleotide 459 of rat ACAP2) sense oligonucleotide: 5′-GATCCGAGGCTGCAAACATTCTCATTCAAGAGATGAGAATGTTTGCAGCCTCTTTTTTTACGCGTG-3′; ACAP2 antisense oligonucleotide: 5′-AATTCACGCGTAAAAAAGAGGCTGCAAACATTCTCATCTCTTGAATGAGAATGTTTGCAGCCTCG-3′; Arf6 (starting from nucleotide 174 of rat Arf6) sense oligonucleotide: 5′-GATCCGTTCAACGTGTGGGATGTGTTCAAGAGACACATCCCACACGTTGAACTTTTTTTACGCGTG-3′; and Arf6 antisense oligonucleotide: 5′-AATTCACGCGTAAAAAAGTTCAACGTGTGGGATGTGTCTCTTGAACACATCCCACACGTTGAACG-3′. The control luciferase sense oligonucleotide: 5′-GATCCGGCCATTCTATCCTCTAGAGTTCAAGAGACTCTAGAGGATAGAATGGCCTTTTTTAGATCTC-3′; luciferase antisense oligonucleotide: 5′-AATTCAGATCTAAAAAAGGCCATTCTATCCTCTAGAGTCTCTTGAACTCTAGAGGATAGAATGGCCG-3′. The annealed duplexes were ligated into pSIREN-RetroQ-ZsGreen1 (Clontech Takara Bio), the vector for producing retrovirus.
Cell line culture and transfection
The 293T cells were cultured in DMEM containing 10% heat-inactivated FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Plasmid DNAs were transfected into 293T cells with the Calcium Phosphate Transfection kit (Clontech Takara Bio). Mouse oligodendroglial FBD-102b cells were maintained in a 1:1 mixture of DMEM and F-12 medium containing heat-inactivated FBS at 10%, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1% GlutaMAX (Horiuchi and Tomooka, 2006; Miyamoto et al., 2007). For differentiating FBD-102b cells, cells were plated on poly-l-lysine (PLL)-coated dishes in a 1:1 mixture of DMEM and F-12 medium containing 1% N-2 supplement, 0.1% triodothyronine (T3), and 0.1% thyroxine (T4) in the presence or absence of 10 μM SecinH3 (Merck Millipore, Darmstadt, Germany) for 3 d. Secin-H3 is a specific inhibitor of cytohesins. Plasmid DNAs and/or siRNAs were transfected into FBD-102b cells with the Lipofectamine 2000 reagent (Life Technologies). The culture medium was replaced with the differentiation medium 6 h after transfection. For confirmation of cell viability under these experimental conditions, FBD-102b cells were stained with 0.4% trypan blue. Attached cells incorporating trypan blue made up < 5% of the cells 3 d after transfection. To quantify cell differentiation in detail, we divided FBD-102b cells into three categories (Sperber and McMorris, 2001; Miyamoto et al., 2007): stage 1 (two or fewer primary processes longer than a cell body); stage 2 (three or four primary processes); stage 3 (five or more primary processes).
Retrovirus production
The retroviral expression vectors pVSV-G and pGP were cotransfected into G3Thi cells with the Calcium Phosphate Transfection kit. Two days after transfection, the culture supernatants were centrifuged at 10,000 rpm for 8 h to concentrate the recombinant retroviruses (Miyamoto et al., 2008). The virus pellets were suspended in culture medium and used to infect purified OPCs.
Primary oligodendrocyte culture
OPCs were isolated from embryonic day 16 (E16) Sprague Dawley (SD) rats as previously described (Miyamoto et al., 2008). Briefly, cerebral cortices were dissected, dissociated with 0.25% trypsin, triturated, and passed through mesh with 70-μm pores. Cells were collected; resuspended in MEM containing 10% FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin; and seeded on PLL-coated dishes. After two passages, the cells were cultured on noncoated Petri dishes (Barloworld Scientific, Stone, Staffordshire, UK). On the second day of culture, the medium was changed to DMEM-based serum-free medium with growth factors and N-2 supplement. The cells were cultured for an additional 2 d, then used as OPCs. For differentiating OPCs, the cells were continuously cultured with a differentiation medium with 20 ng/ml T3 and 20 ng/ml T4 in the presence or absence of 10 μM SecinH3 for 3 d. For confirmation of cell viability under these experimental conditions, OPCs were stained with 0.4% trypan blue. Trypan blue–incorporating cells numbered fewer than 5% in each experiment.
Oligodendrocyte-DRG neuronal cocultures
DRG neurons were isolated from E15 SD rat spinal cord regions as previously described (Chan et al., 2004; Yamauchi et al., 2012; Miyamoto et al., 2013b) and then dissociated and plated onto collagen I–coated coverslips. Nonneuronal cells were eliminated by cycling them three times with medium containing 5-fluorodeoxyuridine and uridine. Myelinating cocultures were established by seeding ∼200,000 purified OPCs, each infected with an shRNA, onto purified DRG neurons according to the protocol of Chan et al. (2004). Cocultures were maintained in the presence or absence of 10 μM SecinH3 for 3–4 wk, and the medium was replaced with fresh medium every 3 d. For statistical analysis, the myelin segments (6 fields/coverslip) were counted.
Immunoprecipitation and immunoblotting
Cells or homogenized tissues were lysed in lysis buffer (50 mM HEPES-NaOH, pH 7.5, 20 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethane sulfonylfluoride [PMSF], 1 μg/ml leupeptin, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 0.5% NP-40, 1% 3-((3-cholamidopropyl)dimethylammonium)-1-propanesulfonate [CHAPS], and 0.3% SDS) and centrifuged as previously described (Yamauchi et al., 2012; Miyamoto et al., 2013b). The supernatants were mixed with protein G resin that was preadsorbed with an anti–cytohesin-2 antibody. The immunoprecipitates or proteins in the lysates were denatured and then separated on SDS–polyacrylamide gels. The electrophoretically separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes, blocked with the Blocking One kit (Nacalai Tesque), and immunoblotted first with primary antibodies and then with peroxidase-conjugated secondary antibodies. The bound antibodies were detected using the Chemi-Lumi One L kit (Nacalai Tesque). The band images were captured with a GT-7300U scanner (Epson, Tokyo, Japan) and analyzed with ImageJ software. At least three separate experiments were carried out under each condition, and a representative blot is shown in each of the figures.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde (PFA; Miyamoto et al., 2007). Cocultures were fixed first with 4% PFA and then with 100% cold methanol (Yamauchi et al., 2012). The fixed cultures were permeabilized with phosphate-buffered saline containing 0.1% Tween-20 or 0.3% Triton X-100, blocked with the Blocking One kit, and then incubated first with primary antibodies and then with fluorescence-labeled secondary antibodies. The coverslips or dishes were mounted with the Vectashield reagent containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). The fluorescence images were captured with the fluorescence microscope system (DMI4000B; Leica, Wetzlar, Germany) and analyzed with AF6000 software (Leica).
Recombinant proteins
GST-tagged Arf6, Rab35Q67L, RUSC2-RBD, and GGA3-ABD were purified using E. coli BL21 (DE3) pLysS (Clontech Takara Bio) as previously described (Yamauchi et al., 2005, 2009, 2012; Torii et al., 2010). Briefly, the transformed E. coli cells were treated with 0.4 μM isopropyl-1-thio-β-d-galactopyranoside at 30°C for 2.5 h and harvested by centrifugation. A cell-free extract was made using 500 μg/ml lysozyme and 100 μg/ml DNaseI in extraction buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, 1 μg/ml leupeptin, 1 mM EDTA, and 0.5% Nonidet P-40). All purification steps were performed at 4°C. The lysates were centrifuged, and the supernatants were mixed with glutathione-resin (GE Healthcare). Bound proteins were washed with extraction buffer, eluted with extraction buffer (without 0.5% Nonidet P-40) containing 20 mM glutathione, and dialyzed against extraction buffer without detergent.
Identification of Rab35-binding proteins
Cytoplasmic lysates from a rat brain harvested on postnatal day 1 were loaded onto glutathione-resins preabsorbed with GST alone or GST-fused Rab35Q67L. The column was washed three times with buffer A (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA, 10 mM PMSF, and 10 μg/ml leupeptin), and the bound proteins were eluted three times by the addition of buffer A containing 10 mM glutathione (Arimura et al., 2009). The first elute was subjected to SDS–PAGE, and the proteins were detected with Coomassie brilliant blue solution. Stained protein bands were excised and enzymatically digested in-gel as previously described (Yamauchi et al., 2012). The resultant peptides were analyzed with an Ettan MALDI-TOF Pro mass spectrometer (GE Healthcare) and a Mascot database (Matrix Science, Boston, MA).
Affinity precipitation of active small GTPases
For detection of active GTP-bound Rab35 and Arf6 in the cell lysates, affinity precipitation was performed using 20 μg GST-tagged RUSC2’s RBD (Fukuda et al., 2011) and GST-tagged GGA3’s ABD (Yamauchi et al., 2009; Torii et al., 2010), respectively. Each of the affinity-precipitated small GTPases was detected through immunoblotting with an antibody against Rab35 or Arf6.
Affinity precipitation of active ACAP2
For detection of active ACAP2 in the cell lysates, affinity precipitation was performed using 20 μg GST-tagged, GTPγS-bound Arf6. Active ACAP2 preferentially interacts with GTP-bound Arf6 (Jackson et al., 2000). The affinity-precipitated active ACAP2 was detected through immunoblotting with an antibody against ACAP2.
Detection of active cytohesin-2
For detection of active cytohesin-2 in the cell lysates, we performed affinity-precipitation with 20 μg GST-tagged guanine nucleotide-free Arf6 (Yamauchi et al., 2009, 2012; Torii et al., 2010). Catalytically active GEFs preferentially interact with nucleotide-free GTPases (Cohen et al., 2007). The affinity-precipitated active cytohesin-2 was detected through immunoblotting with an antibody against cytohesin-2.
Statistical analysis
Values shown represent the mean ± SD from separate experiments. Comparisons between two experimental groups were made using Student's t test (*, p < 0.01). A one-way analysis of variance (ANOVA) was followed by a Fisher's protected least significant difference test as a post hoc comparison (*, p < 0.01; **, p < 0.05).
Supplementary Material
Acknowledgments
We thank J. R. Chan (University of California, San Francisco) and H. Saito (National Research Institute for Child Health and Development) for helpful discussions. This work was supported by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and the Japanese Ministry of Health, Labor, and Welfare (MHLW). This work was partially supported by grants from the Kanehara Science Foundation, the Kato Science Foundation, the Kowa Life Science Foundation, the Mochida Science Foundation, the Naito Science Foundation, the Takeda Science Foundation, and the Uehara Science Foundation.
Abbreviations used:
- ANOVA
analysis of variance
- DAPI
4′,6-diamidino-2-phenylindole
- DENN
differentially expressed in neoplastic versus normal cells
- DRG
dorsal root ganglion
- DTT
dithiothreitol
- FBS
fetal bovine serum
- GAP
GTPase-activating protein
- GDI
guanine nucleotide dissociation inhibitor
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- JIP
c-Jun N-terminal kinase–interacting protein
- MBP
myelin basic protein
- MICAL
molecule interacting with casL
- OPC
oligodendrocyte precursor cell
- PFA
paraformaldehyde
- PLL
poly-l-lysine
- PLP1
proteolipid protein 1
- PMSF
phenylmethane sulfonylfluoride
- PNS
peripheral nervous system
- Q67L
Glu-67-to-Leu
- RBD
Rab35•GTP-binding domain
- RUSC2
RUN and SH3 domain containing 2
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- T3
triodothyronine
- T4
thyroxine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0600) on March 5, 2014.
REFERENCES
- Allaire PD, Mart AL, Dall'Armi C, Di Paolo G, McPherson PS, Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell. 2010;37:370–382. doi: 10.1016/j.molcel.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, et al. Anterograde transport of TrkB in axons is mediated by direct interaction with Slp1 and Rab27. Dev Cell. 2009;16:675–686. doi: 10.1016/j.devcel.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Azzedine H, et al. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet. 2003;72:1141–1153. doi: 10.1086/375034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C, Lakics V, Machesky L, Rumsby M. N-WASP regulates extension of filopodia and processes by oligodendrocyte progenitors, oligodendrocytes, and Schwann cells—implications for axon ensheathment at myelination. Glia. 2007;55:844–858. doi: 10.1002/glia.20505. [DOI] [PubMed] [Google Scholar]
- Bradl M, Lassmann H. Oligodendrocytes: biology and pathology. Acta Neuropathol. 2010;119:37–53. doi: 10.1007/s00401-009-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;11:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- Chan JR, Watkins TA, Cosgaya JM, Zhang C, Chen L, Reichardt LF, Shooter EM, Barres BA. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron. 2004;43:183–191. doi: 10.1016/j.neuron.2004.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Béraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec-7 and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Chen CY, Balch WE. The Hsp90 chaperone complex regulates GDI-dependent Rab recycling. Mol Biol Cell. 2006;17:3494–3507. doi: 10.1091/mbc.E05-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnal P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Frank S, Upender S, Hansen SH, Casanova JE. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kobayashi H, Ishibashi K, Ohbayashi N. Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct Funct. 2011;36:155–170. doi: 10.1247/csf.11001. [DOI] [PubMed] [Google Scholar]
- Fuss B, Becker T, Zinke I, Hoch M. The cytohesin Steppke is essential for insulin signaling in Drosophila. Nature. 2006;444:945–948. doi: 10.1038/nature05412. [DOI] [PubMed] [Google Scholar]
- Hafner M, et al. Inhibition of cytohesin by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi M, Tomooka Y. An oligodendroglial progenitor cell line FBD-102b possibly secretes a radial glia-inducing factor. Neurosci Res. 2006;56:213–219. doi: 10.1016/j.neures.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Hsu C, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–727. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, Randazzo PA. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol. 2000;151:627–638. doi: 10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S, Cox S, Schmidt EJ, Robinson FL, Kiger A. Sbf/MTMR13 coordinates PI(3)P and Rab21 regulation in endocytic control of cellular remodeling. Mol Biol Cell. 2012;23:2723–2740. doi: 10.1091/mbc.E12-05-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. Nomenclature of the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Fukuda M. Rab35 regulates Arf6 activity through centaurin-b2 (ACAP2) during neurite outgrowth. J Cell Sci. 2012;125:2235–2243. doi: 10.1242/jcs.098657. [DOI] [PubMed] [Google Scholar]
- Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockingger H, Seed B. αLβ2 integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- Liang X, Draghi NA, Resh MD. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J Neurosci. 2004;24:7140–7149. doi: 10.1523/JNEUROSCI.5319-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, Ioannou MS, McPherson PS. Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol Biol Cell. 2012;23:163–175. doi: 10.1091/mbc.E11-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, McPherson PS. The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J Biol Chem. 2010;285:10627–10637. doi: 10.1074/jbc.M109.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacci E, Tsai SC, Adamik R, Moss J, Vaughan M. Cytohesin-1, a cytosolic guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez-Vasquez CV, Einheber S, Salzer JL. Rho kinase regulates Schwann cell myelination and formation of associated axonal domains. J Neurosci. 2004;24:3953–3963. doi: 10.1523/JNEUROSCI.4920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–467. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Torii T, Nakamura K, Takashima S, Sanbe A, Tanoue A, Yamauchi J. Signaling through Arf6 guanine-nucleotide exchange factor cytohesin-1 regulates migration in Schwann cells. Cell Signal. 2013a;25:1379–1387. doi: 10.1016/j.cellsig.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Torii T, Yamamori N, Ogata T, Tanoue A, Yamauchi J. Akt and PP2A reciprocally regulate the guanine nucleotide exchange factor Dock6 to control axon growth of sensory neurons. Sci Signal. 2013b;6:ra15. doi: 10.1126/scisignal.2003661. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Yamauchi J, Chan JR, Okada A, Tomooka Y, Hisanaga S, Tanoue A. Cdk5 regulates differentiation of oligodendrocyte precursor cells through the direct phosphorylation of paxillin. J Cell Sci. 2007;120:4355–4366. doi: 10.1242/jcs.018218. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Yamauchi J, Tanoue A. Cdk5 phosphorylation of WAVE2 regulates oligodendrocyte precursor cell migration through nonreceptor tyrosine kinase Fyn. J Neurosci. 2008;28:8326–8337. doi: 10.1523/JNEUROSCI.1482-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G, Sibarita JB, Loubéry S, Daviet L, Romao M, Raposo G, Chavrier P. ARF6 interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr Biol. 2009;19:184–195. doi: 10.1016/j.cub.2008.12.043. [DOI] [PubMed] [Google Scholar]
- Nave KA. Myelination and support of axonal integrity by glia. Nature. 2010;468:244–252. doi: 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- Nikolic M. The role of Rho GTPases and associated kinases in regulating neurite outgrowth. Int J Biochem Cell Biol. 2002;34:731–745. doi: 10.1016/s1357-2725(01)00167-4. [DOI] [PubMed] [Google Scholar]
- Pasterkamp RJ, Dai HN, Terman JR, Wahlin KJ, Kim B, Bregman BS, Popovich PG, Kolodokin AL. MICAL flavoprotein monooxygenases: expression during neural development and following spinal cord injuries in the rat. Mol Cell Neurosci. 2006;31:52–69. doi: 10.1016/j.mcn.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahajeng J, Gridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic. 2012;13:82–93. doi: 10.1111/j.1600-0854.2011.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gabin AG, Almazan G, Larocca JN. Vesicle transport in oligodendrocytes: probable role of Rab40c protein. J Neurosci Res. 2004;76:758–770. doi: 10.1002/jnr.20121. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gabin AG, Ortiz E, Demoliner K, Si Q, Almazan G, Larocca JN. Interaction of Rab31 and OCRL-1 in oligodendrocytes: its role in transport of mannose 6-phosphate receptors. J Neurosci Res. 2010;88:589–604. doi: 10.1002/jnr.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Weber S, Ketelsen UP, Schorle H, Rudnik-Schöneborn S, Büttner R, Buchheim E, Zerres K. Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B/11p15. Hum Mol Genet. 2003;12:349–356. doi: 10.1093/hmg/ddg030. [DOI] [PubMed] [Google Scholar]
- Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Lyons DA. Axonal selection and myelin sheath generation in the central nervous system. Curr Opin Cell Biol. 2013;25:512–519. doi: 10.1016/j.ceb.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Sperber BR, McMorris FA. Fyn tyrosine kinase regulates oligodendroglial cell development but is not required for morphological differentiation of oligodendrocytes. J Neurosci Res. 2001;63:303–312. doi: 10.1002/1097-4547(20010215)63:4<303::AID-JNR1024>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Thurnherr T, et al. Cdc42 and Rac1 signaling are both required for and act synergistically in the correct formation of myelin sheaths in the CNS. J Neurosci. 2006;26:10110–10119. doi: 10.1523/JNEUROSCI.2158-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii T, et al. In vivo expression of the Arf6 guanine-nucleotide exchange factor cytohesin-1 in mice exhibits enhanced myelin thickness in nerves. J Mol Neurosci. 2013;51:522–531. doi: 10.1007/s12031-013-0018-4. [DOI] [PubMed] [Google Scholar]
- Torii T, Miyamoto Y, Sanbe A, Nishimura K, Yamauchi J, Tanoue A. Cytohesin-2/ARNO, through its interaction with focal adhesion adaptor protein paxillin, regulates preadipocyte migration via the downstream activation of Arf6. J Biol Chem. 2010;285:24270–24281. doi: 10.1074/jbc.M110.125658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DT, McShea KM, Attar MA, Santy LC. GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180. Mol Biol Cell. 2010;21:562–571. doi: 10.1091/mbc.E09-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, et al. Phosphorylation of cytohesin-1 by Fyn is required for initiation of myelination and the extent of myelination during development. Sci Signal. 2012;5:ra69. doi: 10.1126/scisignal.2002802. [DOI] [PubMed] [Google Scholar]
- Yamauchi J, Miyamoto Y, Tanoue A, Shooter EM, Chan JR. Ras activation of a Rac1 exchange factor, Tiam1, mediates neurotrophin-3-induced Schwann cell migration. Proc Natl Acad Sci USA. 2005;102:14889–14894. doi: 10.1073/pnas.0507125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Miyamoto Y, Torii T, Mizutani R, Nakamura K, Sanbe A, Koide H, Kusakawa S, Tanoue A. Valproic acid-inducible Arl4D and cytohesin-2/ARNO, acting through the downstream Arf6, regulate neurite outgrowth in N1E-115 cells. Exp Cell Res. 2009;315:2043–2052. doi: 10.1016/j.yexcr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AX, Zhao Y, Flier JS. Molecular cloning of two small GTP-binding proteins from human skeletal muscle. Biochem Biophys Res Commun. 1994;205:1875–1882. doi: 10.1006/bbrc.1994.2889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.