Abstract
There is a discrepancy in telomere length as measured by signal intensity of telomere restriction fragments on gels and fluorescence in situ hybridization analysis. This difference has been ascribed to the X-region, a segment of subtelomeric DNA that is resistant to being cut by restriction enzymes. To explore the nature of this region, we analyzed the digestibility of an artificial seeded telomere in HeLa cells as well as the Xp/Yp autosomal telomere in human BJ fibroblasts. We found that there is a substantial fraction of subtelomeric DNA containing restriction sites that is not digested with enzymes such as EcoRI, NlaIII, and SphI. Comparison of methylation-sensitive and -resistant enzymes excluded the possibility of the X-region being maintained by DNA methylation. We show that the X-region represents a variable domain whose size changes with telomere length, and neither non-TTAGGG sequences nor cytidine methylation can adequately explain the size of the X-region.
Telomeres play an important role in human replicative aging and cancer (reviewed in reference 31). The repression of telomerase (the enzyme that maintains telomere length) in most tissues during development results in telomere shortening with ongoing cell divisions (15). Replicative senescence occurs after enough divisions (typically 50 to 70 for fibroblasts in culture) have occurred to make telomeres sufficiently short. Most cancer cells escape from the limits of replicative senescence by upregulating telomerase to levels that can maintain telomeres (26).
Human germ line telomeres contain 15- to 20-kb-long tracks of repetitive TTAGGG hexamers (21), ending with the G-rich strand forming a 3′ single-stranded overhang (19). This overhang can invade the more-proximal duplex telomeric repeats to form a lariat-like structure called a t-loop (telomere loop) that may be involved in preventing the end of the chromosome from being recognized as a double-strand break needing repair (13).
In addition to providing a solution to the inability of lagging-strand DNA synthesis to replicate the very 3′ end of linear molecules and hiding the ends from the double-strand break recognition system, telomeres also anchor the chromosomal ends to the nuclear matrix and serve as sites of chromosome alignment during meiosis (7, 27). This latter function, facilitating alignment at the ends of the chromosomes, may provide a partial explanation for the increased density of genes near chromosome ends and the increased frequency of meiotic recombination in that region (reviewed in reference 20). In contrast to the increased recombination on a megabase scale near telomeres that is observed in this gene-rich region, there is a more-proximate inhibition of recombination manifested by a dramatic linkage disequilibrium that can, in some cases, extend 100 kb beyond the base of the telomere (reviewed in reference 20). For example, there are three predominant haplotypes for the subtelomeric region of chromosome 16p that can differ in size by 260 kb (5, 30). This inhibition of recombination has been analyzed at the level of single-nucleotide polymorphisms for chromosome 12q. Over 20 single-nucleotide polymorphisms within the largely identical 2-kb region next to the telomere segregate into only three predominant haplotypes in Caucasians (2). This linkage disequilibrium within the nonrepetitive subtelomeric sequences extends into the repetitive region as well. Variants of the sequence TTAGGG (such as TGAGGG) are present at the very base of the telomeres, presumably because this region is so far from the ends that it is not regenerated by telomerase in the germ line (1). Remarkably specific patterns of sequence variants are maintained within this repetitive region among the 12q haplotypes (2). Specific repetitive variant haplotypes have also been demonstrated on chromosome 16p and the Xp/Yp autosomal region as well (5). The barrier to recombination at telomeres prevents recombination from being used to maintain telomere length under normal circumstances, and it is only found as an alternative to telomerase in a small fraction of immortal cells (4).
DNA base modifications such as methylation are used by many organisms to protect themselves from viral parasites, not only to repress transcription but also to inhibit illegitimate recombination between sequences integrated at different locations. This is thought to be the evolutionary reason why various repetitive elements are methylated in the mammalian genome (reviewed in reference 33). The extremely repetitive nature of telomeres, the normal lack of recombination as a length maintenance mechanism, and the linkage disequilibrium including subtelomeric regions suggest the possibility of extensive base modifications at telomeres. Although unusual base modifications have been described at trypanosome telomeres (29), no such modifications have yet been reported in human telomeres. The best evidence to date suggesting the presence of extensive subtelomeric DNA modifications is the proposed existence of an approximately 2- to 4-kb region of subtelomeric DNA that is resistant to enzymatic digestion (18) and thus contributes to the apparent size of telomeres on gels. This prediction is based on the rate of decrease of telomeric probe signals with size that results in a disappearance of signal when telomeres are still 2 to 4 kb in size (6). This calculation has been confirmed by a comparison of in situ hybridization signals to telomeres in metaphase chromosomes and telomere sizes as determined on gels (16). The published distribution of variant sequences at the base of chromosomes 12p, 16p, and Xp/Yp (that would not hybridize to the canonical TTAGGG probes) suggests that they might contribute less than 500 bp to the X-region, leaving the cause of the remainder undetermined.
Here we show that known restriction sites within the subtelomeric DNA are resistant to digestion with different types of restriction enzymes, and the extent of this resistance varies with telomere length. Comparing the digestion using methylation-resistant versus -sensitive restriction enzymes excludes a major or exclusive role for 5-methylcytidine and suggests that other nucleotide modifications are present near human telomeres.
MATERIALS AND METHODS
Quantitative florescence in situ hybridization (Q-FISH).
Metaphases were arrested with Colcemid (0.01 μg/ml) for 18 h. Chromosome preparations were made by exposing the cells to 0.075 M KCl for 30 min and fixing in fresh fixative (methanol-glacial acetic acid at 3:1). Cells were first mixed with an internal standard (a clone of mouse 3T3 cells [ATCC CCL-92]; telomere restriction fragment [TRF], 17 kb) and then dropped onto glass slides, air dried, and dehydrated in an ethanol series (25, 50, and 100%). The spreads were fixed with 4% formaldehyde for 2 min at room temperature, washed with phosphate-buffered saline (three times for 5 min), and treated with pepsin (Sigma) at 1 μg/ml at pH 2 for 15 min at 37°C. The slides were dehydrated again and air dried. The hybridization mixture [300 ng of Cy3-labeled (CCCTAA)3-N3′-P5′phospho-amidate probe/ml (kindly provided by S. Gryaznov, Geron, Inc.) in 70% formamide, 0.25% blocking reagent (Roche), and 5 mM MgCl2 in 10 mM Tris-HCl (pH 7.2)] was added and overlaid with a coverslip. The DNA was denatured (10 min at 80°C) and incubated for at least 3 h at room temperature. Unbound probe was removed with 70% formamide, 0.1% bovine serum albumin in 10 mM Tris-HCl (pH 7.2) four times for 15 min with shaking, followed by 150 mM NaCl-0.05% Tween 20 in 50 mM Tris-HCl (pH 7.2) four times for 5 min. Slides were dehydrated with ethanol, air dried, and counterstained in the dark with 4′-6-diamidino-2-phenylindole (DAPI; Vector Laboratories). The slides were examined and digitally imaged on a Zeiss Axioscope (100×, 1.4 numerical aperture, Plan Neoflour oil-immersion objective) that was equipped with precision Cy3-DAPI band-pass filters. Photographed and unmodified pictures were analyzed with Image J software (freely available at http://rsb.info.nih.gov/ij/). After recording the range of pixels in the image, it was converted to an eight-bit format and background was subtracted using a rolling ball algorithm with the radius set to a value of six. After adjusting the threshold to eliminate nontelomeric signals, the image was quantitated and normalized for the initial pixel range. The intensity of human telomeres was converted to kilobases by comparison to the telomere signals from the mouse cells on the very same slide. This analysis assumes that the X-region forms a sufficiently small fraction of the length of 17-kb mouse telomeres that it can be ignored.
TRF analysis.
Mean telomere length was evaluated by using TRF analysis, a variation of standard Southern analysis. DNAs isolated from different cell clones were digested with either NlaIII, MboI, Sau3A, or a mixture of six enzymes (AluI, CfoI, HaeIII, HinfI, MspI, RsaI) and resolved on a 0.7% agarose gel. The denatured and dried gel was hybridized with 32P-labeled oligonucleotides [(TTAGGG)4] and exposed to a PhosphorImager screen, and the weighted mean telomere length was calculated as described previously (23).
Southern blotting.
Genomic DNA (20 μg) was digested with restriction enzymes and separated on a 0.8% agarose gel. After denaturing the gel, the DNA was transferred onto a positively charges nylon membrane (Zetaprobe; Bio-Rad) using capillary transfer followed by UV fixation. The membrane was hybridized with a specific probe labeled with a random primed labeling kit (Invitrogen) using the manufacturer's protocol.
Telomere preparation and ligation to primers.
Telomeres were purified from 100 μg of genomic DNA digested with NlaIII or SphI as described previously (32). In brief, a biotinylated oligonucleotide, (CCCTAA)6, was annealed to the G-rich overhang of telomeres in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) and recovered with streptavidin-coated micro beads (Miltenyi Biotec) using MiniMACS separation columns (Miltenyi Biotec). The telomeres were released into 200 μl of Tris-EDTA at 65°C and redigested to ensure complete digestion of all available recognition sites (using approximately 10 U of enzyme for a few nanograms of purified telomeres). The last digested restriction site remaining attached to the telomere was tagged by ligation to a double-stranded biotinylated linker, SA (5′-biotinyl-TTTGGATTTGCTGGTGCAGTACAACTAGGCTTAATAGGGACATG-3′ annealed to 5′-TCCCTATTAAGCCTAGTTGTACTGCACCAGCAAATCC-3′), having an overhang compatible with NlaIII and SphI sites. A 0.5-pmol aliquot was ligated to 100 μl of the telomere preparation at 16°C overnight using 5 U of T4 ligase (Roche). The ligated telomeres were then repurified using 0.8% agarose gels. After the 2-kb size marker had migrated about 4 cm, the gel was cut at 2 kb and everything below that size marker was discarded. The electrodes were then reversed, and the DNA was back-migrated until the ethidium bromide-stained molecular weight standard had returned to the origin. To ensure a constant and reproducible rate of back-migration, the lower part of the gel that was removed was replaced with fresh 0.8% agarose and the slots at the origin were replaced with 1% low-melting-point agarose. Streptavidin-coated beads (Dynal) were used to recover biotinylated linker-telomere complexes out of the gelase-treated (Epigenetics) low-melting-point agarose blocks. After washing with Tris-EDTA-0.05% Triton, aliquots of the telomere-bead complexes were added directly to PCRs.
PCR analysis of subtelomeres.
The subtelomeric fraction that stays attached to the telomere after digestion with restriction enzymes was determined using PCR with the following primers (see Fig. 4 and 5, below, for primer locations): XpYpE2F, 5′-TTGTCTCAGGGTCCTAGTG-3′; XpYpB2, 5′-TCTGAAAGTGGACCTATCAG-3′; XpYpE2R, 5′-CACTAGGACCCTGAGACAA-3′; XpYp2NR, 5′-TCTCGGGAGTCCCCGTCTAT-3′; XpYp4NR, 5′-GATATGGCCCACTCAGGCAC-3′; XpYp8NR, 5′-GAACATAGAATTGGAAAACGCG-3′; SA, 5′-GGATTTGCTGGTGCAGTACA-3′. Telomeres from HeLa cells containing a luciferase gene immediately adjacent to a newly seeded telomere were amplified with LucF (5′-AGATGCACATATCGAGGTGAAC-3′), LucR (5′-CATACTGTTGAGCAATTCAC-3′) (annealing temperature, 50°C), PolyAF (5′-GATCGTGGATTACGTCGCC-3′), and PolyAR (5′-CTTGTTTATTGCAGCTTATAATGG-3′) (annealing temperature, 50°C). Sequence analysis revealed a nucleotide polymorphism generating an additional NlaIII site at position bp 1712 (in HUMTARS7AL; gi338679) in our HeLa cells compared to the published sequence. The following primers telomere proximal from this site were used to amplify the Xp/Yp region from this HeLa-derived cell clone to measure the abundance of total telomeric DNA in the different preparations, XpYpHF (5′-TGAAGCTGCAGACCTTTGCG-3′) and XpYpB2 (5′-TCTGAAAGTGGACCTATCAG-3′). All amplifications were performed using a GeneAmp PCR System 9700 (PE Applied Biosystems) running a profile of 35 cycles of 95°C for 20 s, 55°C (if not otherwise noted) for 20 s, and 72°C for 40 s. We used 1 U of HotStar Taq polymerase (Qiagen), 200 μM deoxynucleoside triphosphates, and 0.1 μM primer in PCR buffer (provided by the manufacturer; containing 1.5 mM MgCl2) per 50-μl reaction mix.
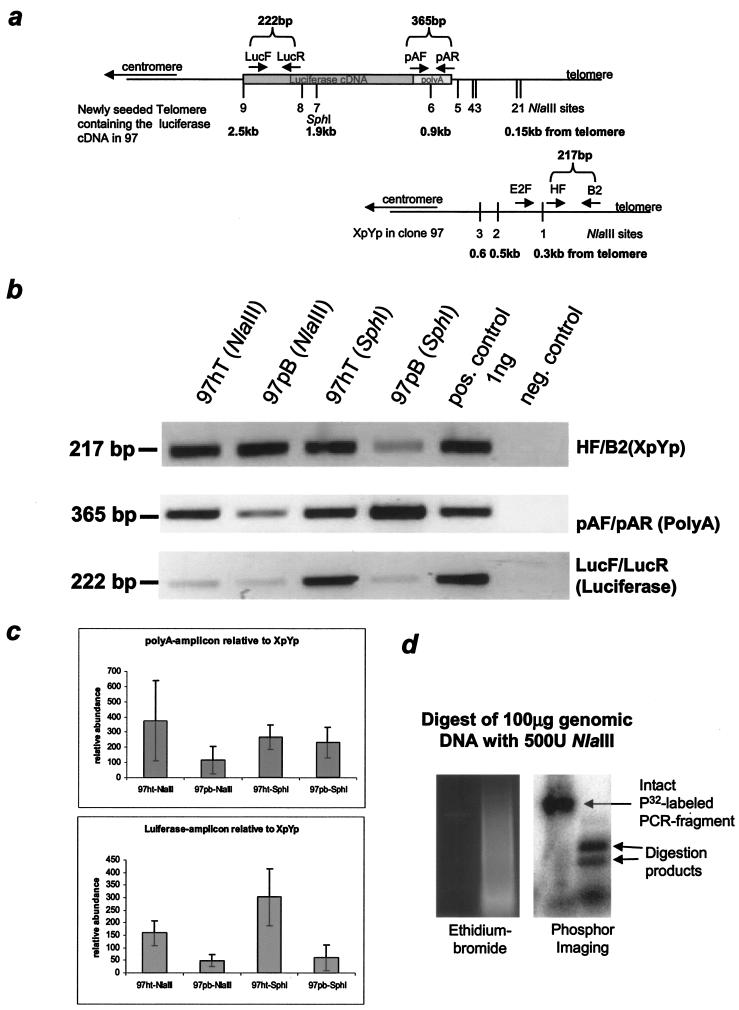
FIG.4.
PCR analysis of the X-region at a newly seeded telomere in 97hT and 97pB. (a) Map of the first nine NlaIII sites and one SphI site and their distance from the telomere, which also has been confirmed with sequence analysis of a newly seeded telomere and the Xp/Yp telomere in 97hT and 97pB. (b) Representative PCR amplification from telomere preparations after NlaIII or SphI digestion using primer pairs that are located in the subtelomeric region. (c) Semiquantitative analysis using spot densitometry (AlphaImager software) from three independent telomere preparations. The results of each amplification have been normalized to a standardized positive control (1 ng of genomic DNA of 97), and the averages from eight different PCRs have been normalized to total telomere input (XpYp PCR). (d) Control showing the complete digestion by NlaIII of a radioactively labeled PCR fragment mixed with genomic DNA (before telomere preparation), shown on an ethidium bromide-containing gel, and the subsequent PhosphorImager analysis.
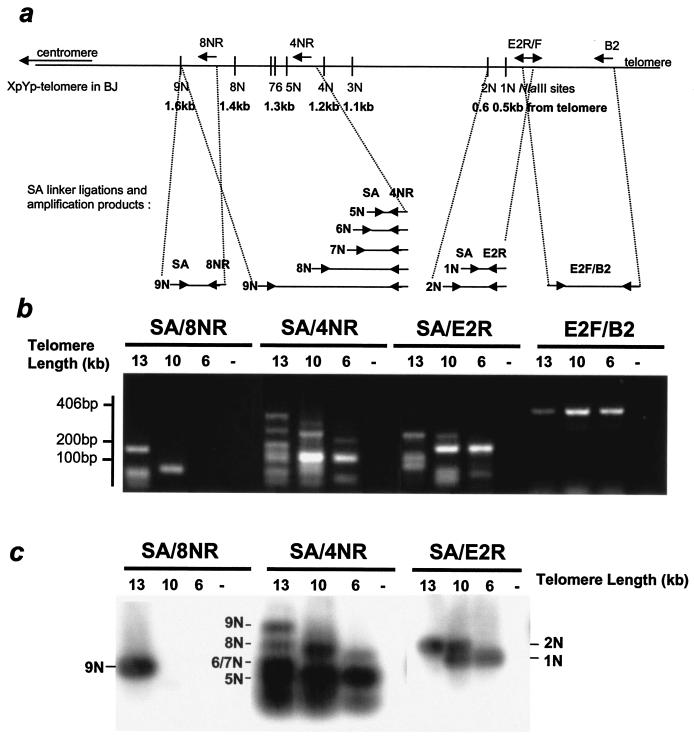
FIG. 5.
PCR analysis of the X-region at the Xp/Yp telomere in different-aged BJ fibroblasts. (a) The NlaIII sites mapped according to the published sequence of the Xp/Yp telomere (HUMTARS7AL, gi338679) have been confirmed with sequence analysis (data not shown). Due to sequence polymorphism, the first NlaIII site in the HeLa clone 97 is only about 300 bp away from the telomere. Arrows indicate possible ligations of a linker (SA) and their corresponding amplification products. (b) PCR amplification from telomere preparations after NlaIII digestion of BJs with different telomere lengths using an SA linker-specific primer and reverse primer that are located in the Xp/Yp subtelomeric region. The telomere lengths are slightly different from those shown in Fig. 2 because they represent averages from multiple TRF analyses using NlaIII. (c) Amplification products transferred to nylon membrane and hybridized to an Xp/Yp-specific probe amplified from genomic DNA from BJ cells.
RESULTS
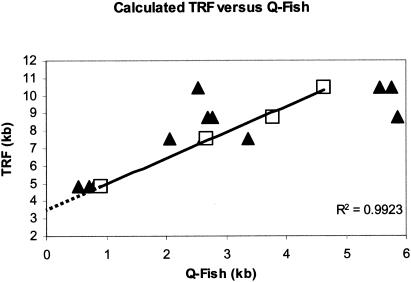
Previous studies have found a 2- to 4-kb X-region by comparing telomere sizes by TRF and Q-FISH. We confirmed these observations using BJ foreskin fibroblasts. Figure 1 shows telomere length measurements of BJ cells (population doubling [PD], 17 to 94) determined by TRF using a mixture of six restriction enzymes (see Materials and Methods). The average telomere fluorescence per chromosome (n = 40 to 60 spreads) was converted to kilobases by normalization to the intensities from intermixed mouse cells (telomere length of 17 kb) present on the same slides as an internal standard. TRF analysis generated telomere lengths that were longer than those measured by Q-FISH. Extrapolation of the data to zero suggested the X-region to be approximately 3.5 kb. The published subtelomeric regions of chromosomes 4p (gi 2121307), 7q (gi 3004858), 16p (gi 1845164), Xq (gi 8979791), and Xp/Yp (gi 338679) indicated an average distance to the most telomeric restriction site for these six enzymes of only 139 bp, considerably less than the observed X-region.
FIG. 1.
Telomere length analysis of different-aged normal human fibroblasts (BJ) using Q-FISH versus TRF analysis. The best value for the TRF at each PD has been plotted versus telomere length determined by Q-FISH. Each point (solid triangle) represents the average of 15 to 20 spreads on a single slide. The open symbols represent the average Q-FISH value for each TRF length. An additional set of values at a TRF length of 10 kb was determined from BJ cells at PD 166 that had elongated telomeres due to the expression of hTERT. The regression line was determined from the average Q-FISH values at each TRF length. The dotted line indicates the regression to zero.
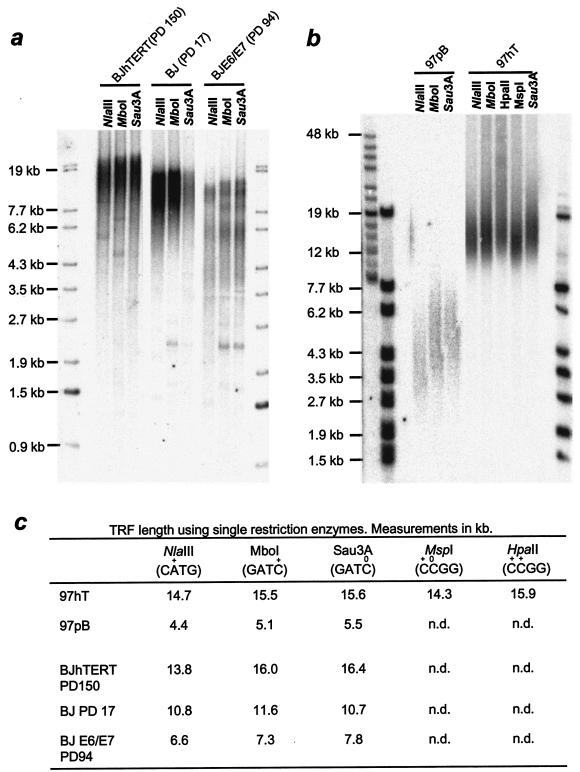
An earlier report suggested the X-region results from 5-methylcytidine modifications (8). We compared methylation-sensitive versus -resistant restriction enzymes to analyze telomere restriction fragment sizes in BJ fibroblasts or HeLa cells harboring a newly seeded telomere (clone 97, as described in reference 3). For both cell lines, we analyzed cells with different telomere lengths. BJ fibroblasts with long telomeres (elongated by introducing the catalytic subunit of telomerase hTERT) were compared to young BJ cells at PD 17 and to BJ cells expressing E6/E7 to drive the cells beyond mortality stage 1 and thus having very short telomeres. HeLa cells overexpressing an exogenous telomerase (97hT) with 12-kb telomeres were compared to vector-only controls (97pB) with approximately 4-kb telomeres. Samples were digested with NlaIII (which digests CATG but not Cm6ATG), MboI (which digests either GATC or GATm5C), and Sau3A (which cuts only the unmethylated sequence). Figure 2 shows that the difference in telomere length after digestion with either MboI or Sau3A was very small for all analyzed samples. The average difference in telomere length was only 0.2 kb, and only minimal differences in the smear to smaller sizes could be seen on the gels. The methylation-sensitive enzymes MspI (digests at Cm5CGG) and HpaII (does not digest Cm5CGG) both contain a CpG sequence. A 1.7-kb difference in telomere size was observed when these enzymes were used to digest 97hT DNA, confirming that many telomeric CpG sites are methylated. However, MspI is one of the six enzymes used to digest the DNA in Fig. 1. The approximately 3.5-kb X-region that was observed lacked digestible MspI sites and thus could not be explained solely by cytidine methylation. NlaIII yielded telomere lengths approximately the same size as those with MspI and 1 kb shorter than those with either MboI or Sau3A. Given the sequence diversity in the population of telomeres, it is improbable that the frequency of NlaIII, MspI, and MboI sites differ. Overall, these results suggest that the X-region is not maintained primarily by 5-methylcytidine. Additional treatments of the DNA with RNase A, RNase H, or another round of phenol-chloroform extraction did not influence the TRF length (data not shown).
FIG. 2.
TRF analysis using methylation-specific enzymes. DNA of human BJ fibroblasts with different telomere lengths (a) and HeLa cells with (97hT) and without (97pB) overexpressed telomerase (b) were digested with NlaIII, MboI, or its methylation-sensitive isochizomere, Sau3A, separated on a 0.8% agarose gel, and hybridized to a telomere (CCCTAA)4-specific probe. Additional digests using an additional pair of methylation-sensitive enzymes, MspI and HpaII, were performed with 97hT DNA. Samples in panel b have been separated on a 0.8% pulse-field gel. (c) Results from quantitating these gels.
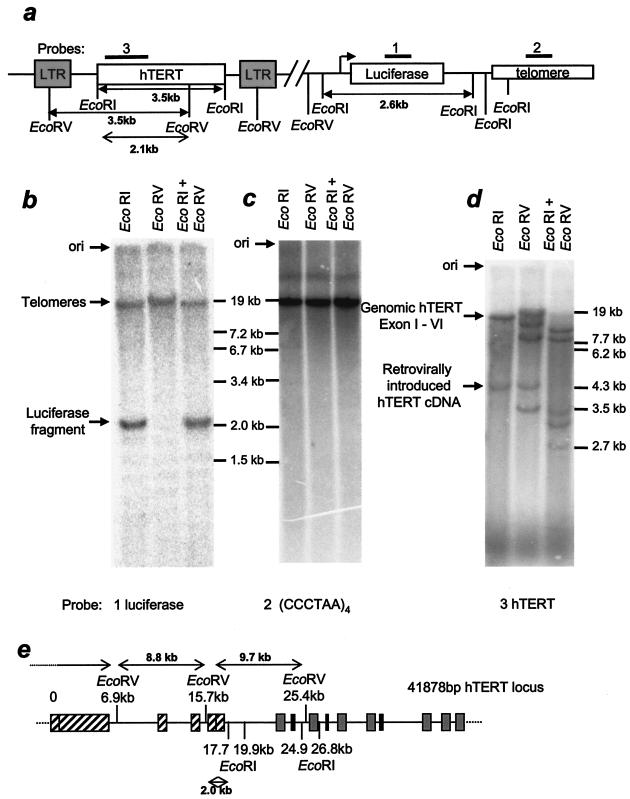
Because of sequence polymorphism and shared sequences in subtelomeric regions (reviewed in reference 20), the presence of up to 92 different telomeres in a normal diploid cell makes the analysis of individual restriction sites difficult. We thus first analyzed specific sites in a luciferase gene placed adjacent to a healed telomere in HeLa cells (3). Based on the original plasmid sequence, we mapped a variety of restriction enzyme sites, such as EcoRI or EcoRV sites (Fig. 3a). EcoRV digestion should leave the luciferase cDNA attached to telomeres, while EcoRI digestion should release a 2.6-kb-long luciferase fragment from the telomere. When probed in a Southern blot assay, we found the corresponding band at 2.6 kb after EcoRI digestion but also that a large fraction of the signal was cohybridizing to telomeres (Fig. 3b). The telomeres of this particular clone are very long, and under the conditions used in this Southern blot assay (digested with restriction enzymes having six-base rather than four-base recognition sites) ran near the mobility limit as a tight band (Fig. 3c). This indicates that in many cases all three of the EcoRI sites telomere proximal to the luciferase cDNA are resistant to digestion. Since the interpretation of these results requires a complete digestion of genomic DNA, we verified the digestion of a 3.5-kb-long fragment from retrovirally inserted hTERT cDNA using EcoRI and EcoRV (Fig. 3d, lanes 1 and 2). Single digestion with EcoRI showed also the expected band (>17.7 kb according to the published sequence for human telomerase [gi 37651900]) corresponding to hybridization to the first six exons of the endogenous hTERT gene. A schematic of the hTERT genomic locus is shown in Fig. 3e. Since this clone of HeLa cells contains multiple copies of hTERT (Y. Zou, unpublished data), an additional band not explained by the database sequence in the EcoRV digest is probably due to gene duplication but is not relevant for our analysis. The change in mobility of all of the bands that had both sites in the double digestion confirmed that the DNA was completely digested.
FIG. 3.
Southern blot of a subtelomeric-located luciferase cDNA fragment. (a) Schematic map of restriction enzyme sites of an integrated retroviral vector containing the hTERT cDNA and a newly seeded telomere containing the luciferase cDNA in a HeLa cell clone (97hT). Three identical blots of 97hT DNA that had been digested with EcoRI, EcoRV, or a combination of both have been hybridized to a luciferase-specific (b), a telomere-specific (c), or an hTERT-specific (d) probe. (e) Schematic map of restriction enzymes in the hTERT locus at chromosome 5p15.33. Striped exons are recognized by the hTERT probe that we used.
We used PCR to confirm that other restriction sites within the telomeric luciferase gene were resistant to digestion. DNA from HeLa cells with long (97hT; 12 kb) and short (97pB; 4 kb) telomeres was digested with NlaIII or SphI and then telomeres were purified based on the presence of a 3′ G-rich overhang. The purified telomeres were then redigested with the same enzyme to absolutely ensure that all available sites were digested before being gel purified based on size, so that any fragments smaller than 2 kb were eliminated (see Materials and Methods) and amplified with three different sets of primers (an outline of the used primer sets is shown in Fig. 4a). The purification step reduces the input DNA by at least 3,000-fold (giving at least a 1,000-fold purification) (data not shown); thus, the second digestion used at least a 1,000-fold excess of enzyme to ensure that all digestible sites were actually cut. The 217-bp PCR amplicon from the Xp/Yp chromosome was internal to any of the NlaIII and SphI sites and thus served as an internal control for the amount of input telomeres (Fig. 4b, top panel). The 365-bp amplicon spanning the sixth NlaIII site, in the luciferase poly(A) sequence 0.9 kb from the telomere, was internal to the SphI site and thus measured the specific input of the luciferase-carrying telomere for SphI-digested DNA (Fig. 4b, middle panel). This fragment would not be present in equal abundance in NlaIII-digested purified telomeres unless the sixth NlaIII site (and the five more-telomere-proximal NlaIII sites) was resistant to digestion. The relative abundance of the 222-bp amplicon located approximately 2.2 kb from the telomere would reflect resistance of one SphI site or eight telomere-proximal NlaIII sites (Fig. 4b, lower panel). In comparison to the 217-bp Xp/Yp general telomeric control and the 365-bp luciferase-specific SphI control [within the poly(A) sequence], all of the bands in both cell types were present in proportions that indicated that most of the tested sites were resistant to digestion. The shown gels are representative data from three independent telomere preparations and at least eight PCRs. A semiquantitative analysis of the band intensities using spot densitometry (AlphaImager software) is shown in Fig. 4c. To compare independent PCRs, we normalized each PCR to a 1-ng input control using genomic DNA from 97 cells. The amplification amounts for the poly(A) and luciferase amplicon have been normalized to the Xp/Yp (total telomere) control. Interestingly, we find in all cases there is at least twice as much product from each amplified region of this particular telomere in samples generated from cells with longer telomeres. These data rely again on the complete digestion of the input DNA. Figure 4d documents an experiment in which we added 50 ng of a radioactively labeled DNA fragment of about 800 bp that has one internal NlaIII site (lane 1) to 100 μg of genomic DNA. After digestion in the presence of 500 U of NlaIII we found the fragment entirely digested (lane 2), indicating that NlaIII digestion was complete after the first round. Nonetheless, we continued treating the telomere-enriched DNA samples a second time to avoid any question of the completeness of the digestion.
These results with an artificial healed telomere in a tumor cell line were confirmed for the Xp/Yp telomeres in normal diploid BJ fibroblasts. DNA from three BJ derivatives with different telomere lengths (the same as shown in Fig. 2) was digested with NlaIII and purified based upon the presence of G-rich overhangs. After redigesting the DNA, the PCR linker SA was ligated to the exposed NlaIII sites. The telomeres were then repurified as mentioned above. The ligated SA linker marked the most-telomere-proximal NlaIII site that could be digested while still remaining attached to the telomere. It served as the forward primer for a series of PCRs. Under the conditions of PCR used in this study, amplicons longer than 600 bp failed to amplify in the presence of coamplified shorter fragments 100 to 200 bp long. In order to avoid bias due to the preferential amplification of small PCR products, several different reverse primers were used, as illustrated in the map in Fig. 5a.
The E2F/B2 amplicon is internal to any of the NlaIII sites on chromosome Xp/Yp and documented an approximately equal input for the different DNAs used in this analysis (Fig. 5b). All of the expected PCR fragments could be identified in the different samples. In order to confirm the specificity of the reaction, the PCR fragments were transferred to a nylon membrane and hybridized to a radioactive Xp/Yp probe. The results in Fig. 5c demonstrate that not only were all of the predicted fragments present, but also their abundance varied in proportion to telomere length. For example, the fragment representing the ligation of the SA primer to the ninth NlaIII (9N) site 1.6 kb from the telomere requires that all eight telomere-proximal sites are resistant to NlaIII digestion. The 9N amplicon corresponding to the ninth site was present in BJ cells with 13-kb telomeres but could not be detected in BJ cells with 10- or 6-kb telomeres.
Taken together, these data indicate that the X-region varies with telomere length on one specific telomere analyzed. The generalization of this conclusion to the majority of telomeres was confirmed analyzing the X-region as a function of TRF length for both six-enzyme- and NlaIII-digested DNA. At long telomere lengths, NlaIII yielded slightly larger TRF sizes than the six-enzyme digestion, but this difference disappeared as telomeres became shorter (Fig. 6c and d). The difference between the telomere length determined by TRF and Q-FISH analyses represents the X-region for any given sample. This difference of TRF (kilobases) minus Q-FISH (kilobases) for these enzymes was plotted versus the average Q-FISH values from Fig. 1 (Fig. 6a and b). The slope of the regression lines indicates that the X-region increases by about 0.4 to 0.7 kb per kb of telomere length. In other words, approximately one-third of the rate of TRF shortening in these cells (about 500 bp per 1.5 kb of shortening) is due to a decrease in the X-region rather than a decrease in the actual size of the telomere.
FIG. 6.
X-region at different telomere lengths in BJ fibroblasts. Regression of the X-region obtained from TRF measurements using NlaIII (a) or six enzymes (AluI, CfoI, HaeIII, HinfI, MspI, and RsaI) (b) versus telomere length analyzed with Q-FISH. The Q-FISH values are the average value shown in Fig. 1. The X-region values were determined from the regression lines for TRF values from multiple samples at multiple population doubling levels, as shown in panel d. (c) Representative TRF gel of selected samples digested with either NlaIII or six enzymes. (d) TRF analysis using NlaIII or six enzymes from independent samples of BJ fibroblasts run on six to eight different gels, each plotted versus population doublings.
DISCUSSION
Previous studies have provided indirect evidence that a significant amount of non-TTAGGG sequences contribute to the size of telomeres determined on TRF gels (6, 18), but the nature of the X-region has remained undefined. The observations that telomeres are late replicating in yeast (9), attach to the nuclear matrix (7), and repress genes placed next to them (telomere position effects [3, 12, 17, 34]) have contributed to the concept that telomeres are heterochromatic. The observation that at least some subtelomeric sites in white blood cells are resistant to digestion by methylcytidine-sensitive enzymes (8) reinforced the notion that the X-region simply represented the usual association of CpG modification with gene silencing and heterochromatin. The present results comparing methylation-sensitive and -insensitive restriction enzymes and looking at specific sites in both endogenous and healed telomeres demonstrate a much more interesting and dynamic picture of the subtelomeric region. Not only are sites resistant to digestion in a pattern that cannot be adequately explained by cytidine methylation, but also the X-region represents a variable domain whose size changes significantly as a function of telomere length.
Cytidine methylation is the only known base modification to occur in mammalian DNA (14). It is primarily present in CpG dinucleotides, although some modifications of CpA and CpT have been reported (24). Neither MboI (GATC) nor NlaIII (CATG) are sensitive to cytidine methylation. NlaIII sites are sensitive to adenine methylation in bacterial cells (22, 25), but such modifications have not been observed in mammalian DNA. The thymidine modification beta-d-glucosylhydroxymethyluracil (base J) is present in repetitive sequences, primarily telomeric DNA, of kinetoplastid protozoans such as trypanosomes (10, 11, 29). It is involved in vesicular stomatitis virus gene silencing and produces resistance to enzymatic digestion. Antibodies that detect base J do not react with mammalian DNA (28), so it is unlikely to represent the modification that produces the X-region in human cells. The specific structure preventing enzyme digestion in this region remains to be determined.
The X-region represents the amount of TRF length that is due to telomeric sequence variants at the base of the telomere plus the average distance over which no restriction sites can be digested. The total number of sites that must be modified increases as the distance from the telomere increases, and to be seen as an X-region every single site needs to be modified (to a first approximation, any single unmodified site will be cut). The present data thus address only the distance over which the efficiency of modification is so high that every single telomere-proximal site has been modified, not how far into the subtelomeric DNA potential base modifications can spread.
Telomeres are unique structures in the genome. Blocks of 10 to 15 kb of highly repetitive TTAGGG sequences present in each of 92 different telomeres of a typical young normal human diploid cell would be expected to be enormously recombinogenic in the absence of mechanisms to prevent it. The total absence of both CpG and CpA sequences in the telomeric DNA sequence prevents the use of most of the known mammalian modifications thought to contribute to reduced recombination between repetitive elements, although the presence of CpT sequences makes some telomeric methylation possible. It is possible that the DNA base modifications producing the X-region are a substitute for cytidine methylation and are involved in preventing recombination over telomeres. This would also produce the linkage disequilibrium seen in the immediately subtelomeric region and the unusual pattern of exchanges between telomeres of up to 50 kb into the subtelomeric region (20), and they could contribute to the presence or extent of telomere position effects in mammalian cells. Changes in telomeric DNA modifications might also contribute to the increased recombination seen in alternative lengthening of telomere (ALT) pathway cells.
Acknowledgments
We thank William Walker and Patrice Evans for technical assistance.
This work was supported by the National Institutes of Health (AG01228). W.E.W. is an Ellison Medical Foundation Senior Scholar.
REFERENCES
- 1.Allshire, R. C., M. Dempster, and N. D. Hastie. 1989. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 17:4611-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird, D. M., J. Coleman, Z. H. Rosser, and N. J. Royle. 2000. High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum. Genet. 66:235-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baur, J. A., Y. Zou, J. W. Shay, and W. E. Wright. 2001. Telomere position effect in human cells. Science 292:2075-2077. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, T. M., L. Marusic, S. Bacchetti, M. Namba, and R. R. Reddel. 1997. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum. Mol. Genet. 6:921-926. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, J., D. M. Baird, and N. J. Royle. 1999. The plasticity of human telomeres demonstrated by a hypervariable telomere repeat array that is located on some copies of 16p and 16q. Hum. Mol. Genet. 8:1637-1646. [DOI] [PubMed] [Google Scholar]
- 6.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lange, T. 1992. Human telomeres are attached to the nuclear matrix. EMBO J. 11:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lange, T., L. Shiue, R. M. Myers, D. R. Cox, S. L. Naylor, A. M. Killery, and H. E. Varmus. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson, B. M., and W. L. Fangman. 1992. A position effect on the time of replication origin activation in yeast. Cell 68:333-339. [DOI] [PubMed] [Google Scholar]
- 10.Gommers-Ampt, J., J. Lutgerink, and P. Borst. 1991. A novel DNA nucleotide in Trypanosoma brucei only present in the mammalian phase of the life-cycle. Nucleic Acids Res. 19:1745-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gommers-Ampt, J. H., F. Van Leeuwen, A. L. de Beer, J. F. Vliegenthart, M. Dizdaroglu, J. A. Kowalak, P. F. Crain, and P. Borst. 1993. β-d-Glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell 75:1129-1136. [DOI] [PubMed] [Google Scholar]
- 12.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 13.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 14.Gruenbaum, Y., R. Stein, H. Cedar, and A. Razin. 1981. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 124:67-71. [DOI] [PubMed] [Google Scholar]
- 15.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 16.Hultdin, M., E. Gronlund, K. Norrback, E. Eriksson-Lindstrom, T. Just, and G. Roos. 1998. Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res. 26:3651-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koering, C. E., A. Pollice, M. P. Zibella, S. Bauwens, A. Puisieux, M. Brunori, C. Brun, L. Martins, L. Sabatier, J. F. Pulitzer, and E. Gilson. 2002. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 3:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy, M. Z., R. C. Allsopp, A. B. Futcher, C. W. Greider, and C. B. Harley. 1992. Telomere end-replication problem and cell aging. J. Mol. Biol. 225:951-960. [DOI] [PubMed] [Google Scholar]
- 19.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 20.Mefford, H. C., and B. J. Trask. 2002. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3:91-102. [DOI] [PubMed] [Google Scholar]
- 21.Moyzis, R. K., J. M. Buckingham, L. S. Cram, M. Dani, L. L. Deaven, M. D. Jones, J. Meyne, R. L. Ratliff, and J. R. Wu. 1988. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 85:6622-6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, M., E. Raschke, and M. McClelland. 1993. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 21:3139-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouellette, M. M., M. Liao, B. S. Herbert, M. Johnson, S. E. Holt, H. S. Liss, J. W. Shay, and W. E. Wright. 2000. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 275:10072-10076. [DOI] [PubMed] [Google Scholar]
- 24.Ramsahoye, B. H., D. Biniszkiewicz, F. Lyko, V. Clark, A. P. Bird, and R. Jaenisch. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 97:5237-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, R. J., and D. Macelis. 1993. REBASE—restriction enzymes and methylases. Nucleic Acids Res. 21:3125-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 27.Smilenov, L. B., S. Dhar, and T. K. Pandita. 1999. Altered telomere nuclear matrix interactions and nucleosomal periodicity in ataxia telangiectasia cells before and after ionizing radiation treatment. Mol. Cell. Biol. 19:6963-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Leeuwen, F., M. C. Taylor, A. Mondragon, H. Moreau, W. Gibson, R. Kieft, and P. Borst. 1998. β-d-Glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. USA 95:2366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Leeuwen, F., E. R. Wijsman, E. Kuyl-Yeheskiely, G. A. van der Marel, J. H. van Boom, and P. Borst. 1996. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 24:2476-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkie, A. O., D. R. Higgs, K. A. Rack, V. J. Buckle, N. K. Spurr, N. Fischel-Ghodsian, I. Ceccherini, W. R. Brown, and P. C. Harris. 1991. Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64:595-606. [DOI] [PubMed] [Google Scholar]
- 31.Wright, W. E., and J. W. Shay. 2002. Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 20:682-688. [DOI] [PubMed] [Google Scholar]
- 32.Wright, W. E., V. M. Tesmer, K. E. Huffman, S. D. Levene, and J. W. Shay. 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 34.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]