Abstract
DNA damage checkpoint pathways sense DNA lesions and transduce the signals into appropriate biological responses, including cell cycle arrest, induction of transcriptional programs, and modification or activation of repair factors. Here we show that the Saccharomyces cerevisiae Sae2 protein, known to be involved in processing meiotic and mitotic double-strand breaks, is required for proper recovery from checkpoint-mediated cell cycle arrest after DNA damage and is phosphorylated periodically during the unperturbed cell cycle and in response to DNA damage. Both cell cycle- and DNA damage-dependent Sae2 phosphorylation requires the main checkpoint kinase, Mec1, and the upstream components of its pathway, Ddc1, Rad17, Rad24, and Mec3. Another pathway, involving Tel1 and the MRX complex, is also required for full DNA damage-induced Sae2 phosphorylation, that is instead independent of the downstream checkpoint transducers Rad53 and Chk1, as well as of their mediators Rad9 and Mrc1. Mutations altering all the favored ATM/ATR phosphorylation sites of Sae2 not only abolish its in vivo phosphorylation after DNA damage but also cause hypersensitivity to methyl methanesulfonate treatment, synthetic lethality with RAD27 deletion, and decreased rates of mitotic recombination between inverted Alu repeats, suggesting that checkpoint-mediated phosphorylation of Sae2 is important to support its repair and recombination functions.
Genetic inheritance requires exceptional genetic stability over many generations of cells and organisms. To ensure that cells pass accurate copies of their genomes on to the next generation, evolution has overlaid the core cell cycle machinery with a series of surveillance pathways, termed checkpoints, that provide the cells with the capacity to survive genotoxic insults. These protective mechanisms are signal transduction pathways specialized in detecting abnormal DNA structures and in coordinating cell cycle progression with DNA repair. Their activation leads to cell cycle progression delay and concomitant activation of DNA repair pathways, thus preventing replication or segregation of damaged DNA molecules.
The keystone of the DNA damage checkpoint is a protein kinase family related to phosphoinositide 3-kinase, among which are Saccharomyces cerevisiae Mec1 (48, 61, 90) and Tel1 (26, 52), Schizosaccharomyces pombe Rad3 (5), Drosophila melanogaster Mei-41 (29), and mammalian ATR (5) and ATM (72). These protein kinases respond to various stresses by phosphorylating key proteins, thus regulating numerous processes, depending on the spectra of their substrates (for reviews, see references 1 and 75). In particular, S. cerevisiae Mec1 and S. pombe Rad3, more closely related to human ATR, are the prototype transducers of the DNA damage and replication stress signals; they respond to UV damage, double-strand breaks (DSBs), and stalled replication forks. Conversely, yeast Tel1, similar to human ATM, is likely involved only in the response to DSBs (for reviews, see references 57 and 75).
Tel1 and Mec1 also contribute to telomere length maintenance. In fact, TEL1 deletion causes marked telomere shortening in yeast (26, 52), and mec1 tel1 cells show more-dramatic telomere shortening than each single mutant, similar to that seen in cells lacking active telomerase (67). The absence of Tel1 does not affect either telomerase catalytic activity or binding to telomeric DNA of the Cdc13 protein, which mediates telomerase action (9, 83). However, when telomerase is artificially targeted to telomeres, their lengthening is at least as effective in mec1 tel1 as in wild-type cells, suggesting that Mec1 and Tel1 may act by recruiting to telomeres either telomerase or telomerase-activating factors (83).
One characteristic of ATR-related proteins is their need for an accessory protein. Mec1 physically interacts with the checkpoint protein Ddc2 (also called Lcd1 or Pie1) (60, 69, 89), functionally related to Rad26 and ATRIP, which bind S. pombe Rad3 and human ATR, respectively (12, 20). Although the Mec1-Ddc2 complex plays a key role in the DNA damage checkpoint, full Mec1-dependent activation of downstream targets requires other factors, such as the Ddc1/Rad17/Mec3 and Rad24/Rfc2 to -5 complexes (for a review, see reference 42), presumed to be structure-specific DNA damage sensors based on their similarities to the proliferating cell nuclear antigen (PCNA) and its accessory factor replication factor C (RFC), respectively (7, 25, 46, 79).
Once DNA perturbations are sensed, checkpoint signals are propagated through the protein kinases Chk1 and Rad53, which also undergo phosphorylation in response to DNA damage in a Mec1-dependent manner (70, 71). While Rad53 is required for proper response to DNA damage in all the cell cycle phases, Chk1 contributes only to the activation of the G2/M checkpoint in a Rad53-independent way (71). The DNA damage-sensing functions are then linked with the downstream effectors through DNA damage-specific or S-phase-specific mediators. In particular, while Rad9 is required to activate Rad53 in response to DNA damage by acting as a scaffold protein upon which Rad53 may autophosphorylate and self-activate (24, 73), Mrc1 seems to act as the Rad9 counterpart in activating Rad53 in response to DNA replication blocks (2, 58).
Whereas human ATM plays a key role in responding to DSBs in human cells, S. cerevisiae Tel1 has only a secondary role in the DNA damage response. In fact, Tel1, which is primarily involved in telomere metabolism, seems to respond to unprocessed DSBs by controlling a checkpoint that becomes apparent only in the absence of Mec1, and converges with the canonical Mec1 pathway on the effector kinase Rad53 (84). A trimeric complex, known as MRN (Mre11-Rad50-Nbs1) in mammals and MRX (Mre11-Rad50-Xrs2) in S. cerevisiae, is a substrate for the ATM/Tel1 kinase. In fact, mammalian Mre11 and Nbs1 are phosphorylated in response to DNA damage, and their phosphorylation depends on ATM (21, 22, 39, 91, 93). This response is evolutionarily conserved, as DNA damage also stimulates Tel1-dependent phosphorylation of the S. cerevisiae Mre11 and Xrs2 proteins (15, 27). In both yeast and humans, this complex plays multiple roles in chromosome metabolism and DNA repair. It has been implicated in telomere maintenance, regulation of DNA replication, nonhomologous end joining, and meiotic DSB formation or resection (for reviews, see references 16, 62, and 86). Moreover, it may control DSB processing during mitotic homologous recombination, thus amplifying the checkpoint signal and stimulating the Mec1/Tel1 kinases (84). Finally, the MRX complex is involved in the S. cerevisiae checkpoint response as part of the DNA damage-sensing apparatus, since mre11Δ, rad50Δ, and xrs2Δ yeast cells are defective in S-phase checkpoint activation in response to DSB-inducing agents and hydroxyurea (HU) (15, 27).
Some processes involving the MRX complex also require the SAE2/COM1 gene, whose loss-of-function alleles were originally identified together with the rad50s and mre11s separation-of-function alleles in a search for mutants whose meiotic products died if DSBs were made but could survive when meiotic recombination and reductional chromosome segregation were prevented by mutations inactivating the SPO11 gene (49, 65). The topoisomerase II-like Spo11 protein is necessary for the creation of meiotic DSBs, which are subjected to rapid resection of their 5′ strand termini, yielding molecules with 3′ single-stranded tails, presumably used to form strand exchange products during recombination (62). While the MRX complex participates in both the formation and the processing of DSBs at meiotic recombination hot spots, the Sae2 protein seems to participate only in DSB processing. In fact, in contrast to mrx-null alleles, which are unable to initiate meiotic DSBs, both sae2Δ and the rad50s and mre11s separation-of-function alleles were shown to allow SPO11-mediated DSB formation, but not single-strand endonucleolytic removal of Spo11 from the DNA ends, thus uncoupling cleavage of DNA strands from subsequent exonucleolytic resection (34). Since the MRX complex has nuclease activity, the lack of Sae2 might directly modify its in vivo nuclease function, and the rad50s allele might affect its ability to interact with Sae2, as previously suggested (66).
Although Sae2, as well as the MRX complex, is dispensable for some mitotic homologous recombination processes requiring Rad52 and Rad51 (6, 32, 47, 82), several observations indicate that Sae2 may also have mitotic functions. (i) sae2Δ cells have been shown to be hypersensitive to the alkylating agent methyl methanesulfonate (MMS), and the fidelity of mitotic DSB repair in these cells has been shown to be affected (49, 66). (ii) Sae2, along with MRX, is also required for repair of mitotic hairpin-capped DSBs induced by inverted Alu sequences, indicating that Sae2 may participate in making protected DNA ends accessible to resection in mitotic cells also (40). (iii) A lack of Sae2 enhances Tel1-mediated Rad53 phosphorylation after DNA damage, and this enhancement requires the MRX complex, suggesting that sae2Δ cells may accumulate DNA lesions specifically sensed by the Tel1/MRX-dependent checkpoint (84). (iv) High levels of Sae2 not only cause telomere lengthening in a Tel1-dependent manner, but also accelerate the rebalancing of telomere length when sudden telomere elongation is induced by TEL1 overexpression, suggesting a role for Sae2 in unprotected telomeric end processing (87).
We now show that the functions of Sae2 in DNA repair and recombination require the checkpoint pathway that responds to DNA damage during the mitotic cell cycle. Sae2, whose lack delays the recovery from checkpoint-mediated cell cycle arrest, undergoes Mec1- and Tel1-dependent phosphorylation both periodically during the unperturbed cell cycle and in response to DNA damage independently of cell cycle progression. Site-directed mutagenesis of the favored ATM/ATR phosphorylation sites indicates that they are essential not only for DNA damage-induced Sae2 phosphorylation but also for Sae2 functions in vivo.
MATERIALS AND METHODS
Plasmids.
To construct plasmid pML469.14, containing the 1,037-bp SAE2 open reading frame flanked by 329 bp upstream and 226 bp downstream, the 1,592-bp EcoRI-EcoRI fragment from a yeast genomic DNA library was cloned into the EcoRI-EcoRI sites of plasmid YIplac128 (23). This plasmid was used as a template to create alanine substitution mutants by QuikChange site-directed mutagenesis (Stratagene), thus generating plasmids pML473.15, pML474.35, pML488.15, pML475.5, and pML468.6, carrying, respectively, the sae21-5 (alanine substitutions at S72, S73, T75, S76, and T90), sae26-9 (alanine substitutions at S249, S278, T279, and S289), sae22,5,6,8,9 (alanine substitutions at S73, T90, S249, T279, and S289), sae21-2,5-9 (alanine substitutions at S72, S73, T90, S249, S278, T279, and S289), and sae21-9 (alanine substitutions at S72, S73, T75, S76, T90, S249, S278, T279, and S289) alleles, as confirmed by subsequent nucleotide sequence determination. The 1,592-bp EcoRI-EcoRI fragments from plasmids pML473.15, pML474.35, pML488.15, pML475.5, and pML468.6, carrying, respectively, the sae21-5, sae26-9, sae22,5,6,8,9, sae21-2,5-9, and sae21-9 alleles, were cloned into the EcoRI site of plasmid YIplac204 (23) to produce plasmids pML484, pML482, pML487, pML478, and pML476.
Yeast strains and media.
The relevant genotypes of all the yeast strains used in this study are listed in Table 1. All the strains were constructed during this study, and all were derivatives of W303 (K699) (MATa or MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad5), with the exception of strains YLL1357, YLL1368.21, YLL1375.39, YLL1406, YLL1407, and YLL1408, which were generated from strain HS21 (MATα ade5-1 his7-2 ura3Δ trp1-289 leu2-3,112::p305L3 LEU2 lys2::AluIR), kindly provided by M. A. Resnick (Research Triangle Park, N.C.) (see below).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype |
|---|---|
| YLL254 | MATarad27Δ::KANMX4 |
| YLL1069.3 | MATasae2Δ::KANMX4 |
| YLL1072.1 | MATaMRE11-HA3::URA3 |
| YLL1103 | MATaSAE2-HA3::URA3 |
| YLL1345.6 | MATabar1Δ::KANMX4 ddc1Δ::HIS3 SAE2-HA3::URA3 |
| YLL1346.2 | MATabar1Δ::KANMX4 ddc1Δ::HIS3 rad9Δ::URA3 SAE2-HA3::URA3 |
| YLL1347.1 | MATasae2Δ::KANMX4::sae21-9::LEU2 |
| YLL1348.1 | MATasae2Δ::KANMX4::SAE2::LEU2 |
| YLL1351.8 | MATasae2Δ::KANMX4::SAE2-HA3::URA3::LEU2 |
| YLL1352.2 | MATasae2Δ::KANMX4::sae21-9-HA3::URA3::LEU2 |
| YLL1353.1 | MATasae2Δ::KANMX4::sae21-5::LEU2 |
| YLL1354.1 | MATasae2Δ::KANMX4::sae26-9::LEU2 |
| YLL1355.1 | MATasae2Δ::KANMX4::sae21-2,5-9::LEU2 |
| YLL1359.4 | MATasae2Δ::KANMX4::sae21-5-HA3::URA3::LEU2 |
| YLL1360.31 | MATasae2Δ::KANMX4::sae26-9-HA3::URA3::LEU2 |
| YLL1361.44 | MATasae2Δ::KANMX4::sae21-2,5-9-HA3::URA3::LEU2 |
| YLL1395.1 | MATasae2Δ::KANMX4::sae22,5,6,8,9::LEU2 |
| YLL1405 | MATasae2Δ::KANMX4::sae22,5,6,8,9-HA3::URA3::LEU2 |
| YLL1357 | MATα leu2-3,112::p305L3 LEU2 lys2::AluIR sae2Δ::KANMX4 |
| YLL1368.21 | MATα leu2-3,112::p305L3 LEU2 lys2::AluIR sae2Δ::KANMX4::sae21-9::TRP1 |
| YLL1375.39 | MATα leu2-3,112::p305L3 LEU2 lys2::AluIR sae2Δ::KANMX4::sae21-2,5-9::TRP1 |
| YLL1406 | MATα leu2-3,112::p305L3 LEU2 lys2::AluIR sae2Δ::KANMX4::sae26-9::TRP1 |
| YLL1407 | MATα leu2-3,112::p305L3 LEU2 lys2::AluIR sae2Δ::KANMX4::sae21-5::TRP1 |
| YLL1408 | MATα leu2-3,112::p305L3 LEU2 lys2::AluIR sae2Δ::KANMX4::sae22,5,6,8,9::TRP1 |
| DMP3806/2A | MATamec1Δ::HIS3 sml1Δ::KANMX4 SAE2-HA3::URA3 |
| DMP3807/2D | MATatel1Δ::HIS3 SAE2-HA3::URA3 |
| DMP3909/7D | MATabar1Δ::KANMX4 SAE2-HA3::URA3 |
| DMP3916/5B | MATamec1Δ::HIS3 sml1Δ::KANMX4 tel1Δ::HIS3 SAE2-HA3::URA3 |
| DMP3919/3B | MATabar1Δ::KANMX4 mec1Δ::HIS3 sml1Δ::KANMX4 SAE2-HA3::URA3 |
| DMP3919/11A | MATabar1Δ::KANMX4 mec1Δ::HIS3 sml1Δ::KANMX4 tel1Δ::HIS3 SAE2-HA3::URA3 |
| DMP3920/16D | MATabar1Δ::KANMX4 tel1Δ::HIS3 SAE2-HA3::URA3 |
| DMP4048/7D | MATabar1Δ::KANMX4 rad50Δ::HIS3 SAE2-HA3::URA3 |
| DMP4049/5C | MATα bar1Δ::KANMX4 rad9Δ::URA3 SAE2-HA3::URA3 |
| DMP4050/17A | MATabar1Δ::KANMX4 rad50Δ::HIS3 telIΔ::HIS3 SAE2-HA3::URA3 |
| DMP4052/18A | MATabar1Δ::KANMX4 rad50Δ::HIS3 mec1Δ::HIS3 sml1Δ::KANMX4 SAE2-HA3::URA3 |
| DMP4105/3B | MATabar1Δ::KANMX4 mrc1Δ::HIS3 SAE2-HA3::URA3 |
| DMP4106/27A | MATabar1Δ::KANMX4 rad53Δ::HIS3 sml1Δ::KANMX4 SAE2-HA3::URA3 |
| DMP4137/1D | MATabar1Δ::KANMX4 rad17Δ::LEU2 SAE2-HA3::URA3 |
| DMP4137/17A | MATabar1Δ::KANMX4 rad24Δ::TRP1 SAE2-HA3::URA3 |
| DMP4138/7C | MATabar1Δ::KANMX4 mec3Δ::TRP1 SAE2-HA3::URA3 |
| DMP4141/12C | MATabar1Δ::KANMX4 chk1Δ::HIS3 SAE2-HA3::URA3 |
| DMP4224/5B | MATasae2Δ::KANMX4 MRE11-HA3::URA3 |
| DMP4295/10A | MATabar1Δ::KANMX4 mec1Δ::HIS3 sml1Δ::KANMX4 rad53Δ::HIS3 SAE2-HA3::URA3 |
| DMP4296/1D | MATabar1Δ::KANMX4 tel1Δ::HIS3 chk1Δ::HIS3 SAE2-HA3::URA3 |
| DMP4297/5A | MATabar1Δ::KANMX4 mec1Δ::HIS3 sml1Δ::KANMX4 chk1Δ::HIS3 SAE2-HA3::URA3 |
| DMP4298/2B | MATabar1Δ::KANMX4 rad53Δ::HIS3 sml1Δ::KANMX4 tel1Δ::HIS3 SAE2-HA3::URA3 |
Strains YLL1069.3 and YLL1357, where 968 bp of the SAE2 coding region was replaced by the KANMX4 gene, were generated from strains K699 and HS21 by a one-step PCR disruption method (88). Strains DMP4048/7D and DMP4105/3B, where 3,890 bp of the RAD50 coding region and 2,880 bp of the MRC1 coding region, respectively, were replaced by the Kluyveromyces lactis HIS3 gene, were generated from strain DMP3909/7D by a one-step PCR disruption method (88). Deletions of the MEC1, TEL1, SML1, BAR1, RAD53, CHK1, RAD9, DDC1, MEC3, RAD17, and RAD24 genes were constructed as previously described (43, 44, 59, 60).
Strains carrying the SAE2-HA3 allele at the SAE2 chromosomal locus were generated by a PCR one-step tagging method (35) using plasmid 3748, kindly provided by K. Nasmyth (Institute for Molecular Pathology, Vienna, Austria), as a template and oligonucleotides PRP326 (5′-TTA AAT CAA ATT GTT GAC GAT GGA TGT TTC TTC TGG AGT GAT AAA TTA TTG CAG ATA TAT GCT AGA TGT TCC GGT TCT GCT GCT AG-3′) and PRP327 (5′-AAA GCC CTT TCA ACC ATA CCA AAA AAA ATG TAT TTG AAG TAA TGA ATA AAG AAT GAT GAT CGC TGG CGT CCT CGA GGC CAG AAG AC-3′) as primers.
Strains carrying the MRE11-HA3 allele at the MRE11 chromosomal locus were generated by a PCR one-step tagging method (35) using plasmid 3748 as the template and oligonucleotides PRP333 (5′-GGA AAA GGA AGA GCA TCA AGG ACG CCA AAG ACG GAT ATT CTT GGA AGT CTC CTT GCT AAG AAA AGA AAA TCC GGT TCT GCT GCT AG-3′) and PRP334 (5′-CCT TGT TGT TCG CGA AGG CAA GCC CTT GGT TAT AAA TAG GAT ATA ATA TAA TAT AGG GAT CAA GTA CAA CCT CGA GGC CAG AAG AC-3′) as primers. The SAE2-HA3 and MRE11-HA3 alleles were shown to be fully functional, since all the strains carrying the tagged alleles were indistinguishable from the isogenic untagged strains with respect to viability, growth rates at any temperature, and sensitivity to UV radiation, MMS, and HU.
ApaI digestion of the integrative plasmids pML469.14, pML473.15, pML474.35, pML488.15, pML475.5, and pML468.6 was used to direct the integration of these plasmids to the SAE2 promoter region of a W303-derivative sae2Δ strain (YLL1069.3), giving rise to strains YLL1348.1, YLL1353.1, YLL1354.1, YLL1395.1, YLL1355.1, and YLL1347.1, carrying, respectively, single copies of the SAE2, sae21-5, sae26-9, sae22,5,6,8,9, sae21-2,5-9, and sae21-9 alleles at the SAE2 chromosomal locus. ApaI-directed integration of plasmids pML484, pML482, pML487, pML478, and pML476 into the SAE2 promoter region of an HS21-derivative sae2Δ strain (YLL1357) was also used to construct strains YLL1407, YLL1406, YLL1408, YLL1375.39, and YLL1368.21, carrying, respectively, single copies of the sae21-5, sae26-9, sae22,5,6,8,9, sae21-2,5-9, and sae21-9 alleles at the SAE2 chromosomal locus.
The SAE2, sae21-5, sae26-9, sae22,5,6,8,9, sae21-2,5-9, and sae21-9 alleles of strains YLL1348.1, YLL1353.1, YLL1354.1, YLL1395.1, YLL1355.1, and YLL1347.1 were then hemagglutinin (HA) epitope tagged by PCR one-step tagging as described above to produce strains YLL1351.8, YLL1359.4, YLL1360.31, YLL1405, YLL1361.44, and YLL1352.2, respectively.
The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. Standard yeast genetic techniques and media were used according to reference 68. Cells were grown in YEP medium (1% yeast extract, 2% Bacto Peptone, 50 mg of adenine/liter) supplemented with 2% glucose (YEPD). Transformants carrying the KANMX4 cassette were selected on YEPD plates containing 400 μg of G418 (U.S. Biological)/ml.
Recombination rates.
Rates of homologous recombination induced by inverted Alu repeats were determined in fluctuation tests (36) by using at least 20 independent cultures from wild-type (HS21), sae2Δ (YLL1357), sae21-5 (YLL1407), sae26-9 (YLL1406), sae22,5,6,8,9 (YLL1408), sae21-2,5-9 (YLL1375.39), and sae21-9 (YLL1368.21) cell cultures, as previously described (40).
Western blot analysis, immunoprecipitation, and phosphatase treatment.
For Western blot analysis, protein extracts were prepared by trichloroacetic acid precipitation as previously described (43). Immunoprecipitation and phosphatase treatments were performed as previously described (43). Phosphorylated forms of Sae2 were detected by using rabbit polyclonal antibodies that recognize phosphorylated (S/T)Q motifs (Cell Signaling Technology). HA-tagged proteins were detected with the 12CA5 monoclonal antibody. Rad53 was detected by using anti-Rad53 polyclonal antibodies, kindly provided by J. Diffley (Clare Hall Laboratories, South Mimms, United Kingdom). Secondary antibodies were purchased from Amersham, and proteins were visualized by an enhanced chemiluminescence system according to the manufacturer's instructions.
Synthetic-effect assays.
Strains YLL1353.1, YLL1354.1, YLL1395.1, YLL1355.1, and YLL1347.1, carrying, respectively, single copies of the sae21-5, sae26-9, sae22,5,6,8,9, sae21-2,5-9, and sae21-9 alleles at the SAE2 chromosomal locus (see under “Yeast strains and media” above), were crossed to W303-derivative strains carrying the rad27Δ allele (YLL254). For each sporulated diploid, 24 tetrads were dissected, and spores were scored for the ability to form colonies on YEPD plates at 25°C. Segregation of the sae2 and rad27Δ mutations in the viable spores was assayed by monitoring the LEU2 marker linked to the sae2 alleles and temperature sensitivity due to the rad27Δ allele. Synthetic lethality was inferred when no viable double-mutant spores were observed from a diploid, and if there was no significant deviation from the 1 parental ditype:4 tetratype:1 nonparental ditype ratio of tetrad types predicted from the segregation of two unlinked genes. When segregants containing both mutations were viable, serial dilutions of three independent cultures each of two single- and two double-mutant segregants for each cross were spotted onto YEPD plates with or without MMS (0.005%). YEPD plates without MMS were prepared in triplicate, and two of these plates were incubated at 30 and 37°C, respectively, for 3 days, while the other plates were incubated at 25°C for 4 days before determination of the number of CFU for each strain under the different conditions. No significant differences were found among strains with the same genotypes.
Other techniques.
Synchronization experiments were performed as described previously (60). Flow cytometric DNA quantitation was determined on a Becton-Dickinson FACScan instrument. All experiments were performed at 26°C.
RESULTS
Sae2 is phosphorylated periodically during the unperturbed cell cycle and in response to DNA damage.
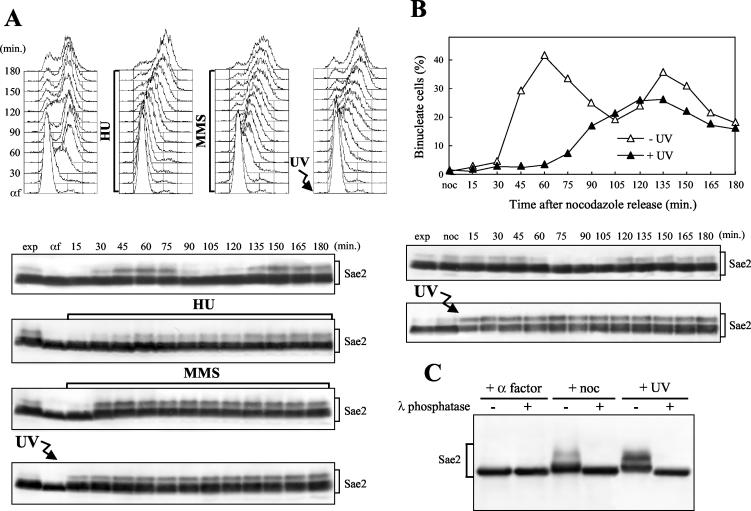
To gain insights into Sae2 function(s) and regulation, we generated a strain expressing fully functional 3HA-tagged Sae2 (see Materials and Methods). As shown in Fig. 1, when anti-HA antibodies were used in Western blot analysis of cell extracts from exponentially growing SAE2-HA3 cells, they specifically detected Sae2-HA3 protein species with different electrophoretic mobilities that did not appear in extracts from cells carrying the untagged SAE2 allele (data not shown). When SAE2-HA3 cells were arrested in G1 with α-factor and then released into the cell cycle under unperturbed conditions, the slowest-migrating Sae2-HA3 species, which were absent in α-factor-arrested cells, accumulated periodically during the cell cycle (Fig. 1A), increasing in level when cells progressed through the S, G2, and M phases (30 to 75 min), disappearing when almost all cells were ready to divide (90 min), and then reaccumulating as cells entered the next S phase. The events leading to the disappearance of the slowest-migrating Sae2 species took place during the exit from mitosis, as evidenced by the fact that these species were detectable in nocodazole-arrested cells and disappeared concomitantly with cell division after release into the cell cycle (Fig. 1B).
FIG. 1.
Sae2 phosphorylation during the unperturbed cell cycle and in response to genotoxic treatments. (A) Exponentially growing YLL1103 cells, expressing Sae2-HA3 from the SAE2 promoter, were synchronized with α-factor (αf) and released from the pheromone block in YEPD, YEPD containing 50 mM HU, or 0.02% MMS, or were UV irradiated (40 J/m2) prior to the release in YEPD. Cell samples collected at the indicated times after α-factor release were analyzed by fluorescence-activated cell sorting (top), and protein extracts were prepared and analyzed by Western blotting with anti-HA antibodies (bottom). (B) Exponentially growing YLL1103 cells were synchronized with nocodazole (noc) and released from the G2 block in YEPD, or were UV irradiated (45 J/m2) prior to the release in YEPD. Cell samples were collected at the indicated times after release from nocodazole to analyze the percentage of binucleate cells by propidium iodide staining (top) and the Sae2-HA3 protein as in panel A (bottom). (C) Protein extracts from α-factor-arrested (+ α factor), nocodazole-arrested (+ noc), or UV-treated exponentially growing (+ UV) YLL1103 cells were immunoprecipitated with anti-HA antibodies. Immunoprecipitates were then incubated at 30°C with (+) or without (−) λ phosphatase before electrophoresis and Western blot analysis using anti-HA antibodies. exp, exponentially growing cells.
Treatment of wild type cells with UV radiation or MMS led to accumulation of Sae2 species whose electrophoretic mobilities were indistinguishable from that of the slowly migrating forms observed during unperturbed S phase (Fig. 1). When G1-arrested SAE2-HA3 cell cultures were released from α-factor after UV irradiation or in the presence of MMS, these Sae2 species became detectable immediately after UV treatment and about 30 min after MMS addition, and they persisted until the end of the experiments (Fig. 1A, bottom).
Release into the cell cycle of G1-arrested cells in the presence of the DNA synthesis inhibitor HU also led to accumulation of Sae2 modified forms about 30 min after release, concomitantly with S-phase entry (Fig. 1A), but neither the amount nor the kinetics of Sae2 modification after HU treatment seemed to significantly differ from that observed in S/G2 untreated cell cultures, although modified Sae2 persisted in the HU-treated cell culture until the end of the experiment, likely due to the prolonged S phase.
As was observed after DNA damage in G1, modified Sae2 also accumulated in response to DNA damage in G2 (Fig. 1B). In fact, the amount of slowly migrating Sae2 species, which were already present in nocodazole-arrested, unirradiated cells, increased immediately after these cells were UV irradiated and then released into the cell cycle.
The observed changes in Sae2 electrophoretic mobility after DNA damage and during the cell cycle were due to phosphorylation events. In fact, Western blot analysis of anti-HA immunoprecipitates from UV-treated and nocodazole-arrested SAE2-HA3 cells after bacteriophage λ phosphatase treatment revealed a single Sae2 band whose electrophoretic mobility was faster than that of any Sae2 species detectable in the same immunoprecipitates in the absence of phosphatase (Fig. 1C). A similar single Sae2 band was detectable also in anti-HA immunoprecipitates from α-factor-arrested SAE2-HA3 cells, and its electrophoretic mobility was not affected by λ phosphatase treatment (Fig. 1C). Thus, if Sae2 phosphorylation events take place in α-factor-arrested cells, they do not cause detectable changes in Sae2 electrophoretic mobility, while all the Sae2 species detectable by this analysis in UV- and nocodazole-treated cell extracts represent phosphorylated Sae2 forms.
UV- and bleomycin-induced Sae2 phosphorylation does not require cell cycle progression.
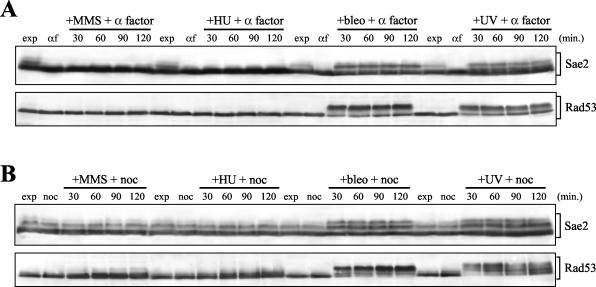
In order to distinguish between cell cycle- and DNA damage-dependent Sae2 phosphorylation, we performed Western blot analysis with anti-HA antibodies on protein extracts of SAE2-HA3 cells, which were kept blocked in G1 or in G2 after UV irradiation or MMS, HU, or bleomycin addition. As a control for the phosphorylation response to genotoxic treatment, we probed the same extracts with antibodies raised against the Rad53 DNA damage checkpoint kinase, whose phosphorylation is also detectable as changes in electrophoretic mobility (70). As shown in Fig. 2A, neither Sae2 nor Rad53 phosphorylated forms were detectable during the unperturbed G1 phase, but they both appeared immediately after bleomycin addition or UV irradiation in G1-arrested cells kept in G1 by α-factor addition, and they persisted until the end of the experiment. The same genotoxic treatments also increased the amounts of phosphorylated Sae2 and Rad53 in G2-arrested cells that were kept in G2 by nocodazole addition (Fig. 2B). As was observed for Rad53, whose phosphorylation can be induced by MMS and HU treatments only when cells are allowed to proceed further into S phase (77), neither MMS nor HU addition caused significant changes in Sae2 electrophoretic mobility in G1- or G2-arrested cells (Fig. 2).
FIG. 2.
DNA damage-induced Sae2 phosphorylation in G1- and G2-arrested cells. (A) Exponentially growing cell cultures of strain DMP3909/7D, expressing Sae2-HA3 from the SAE2 promoter in a bar1Δ background, were synchronized with α factor (αf) and resuspended in YEPD containing 1 μg of α factor/ml in the presence of either 0.02% MMS (+MMS + α factor), 150 mM HU (+HU + α factor), or 20 mU of bleomycin/ml (+bleo + α factor), or after UV-irradiation (40 J/m2) (+UV + α factor). (B) Exponentially growing cell cultures of strain DMP3909/7D were synchronized with nocodazole (noc) and resuspended in YEPD containing nocodazole in the presence of either 0.02% MMS (+MMS + noc), 150 mM HU (+HU + noc), or 20 mU of bleomycin/ml (+bleo + noc), or after UV irradiation (40 J/m2) (+UV + noc). Protein extracts prepared from cell samples collected at the indicated times were subjected to Western blot analysis with anti-HA (Sae2) and anti-Rad53 (Rad53) antibodies. exp, exponentially growing cells.
Checkpoint proteins involved in DNA damage-induced Sae2 phosphorylation.
To identify the pathway(s) leading to DNA damage-induced Sae2 phosphorylation, we used Western blot analysis to verify whether this modification was affected in DNA damage checkpoint mutants. It is worth pointing out that all the SAE2-HA3 strains carrying the mec1Δ or rad53Δ allele also carried the sml1Δ allele (Table 1), which suppresses the lethality of MEC1 and RAD53 deletions but not their effects on the DNA damage response (19, 94), and that the sml1Δ allele did not per se cause any detectable effect on Sae2 electrophoretic mobility (data not shown).
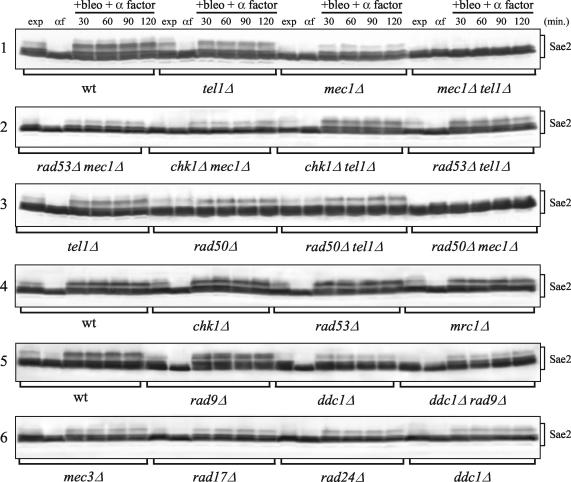
Bleomycin-induced Sae2 phosphorylation in G1-arrested cells was severely reduced, compared to that for the wild type, for the mec1Δ, ddc1Δ, rad17Δ, mec3Δ, and rad24Δ mutants (Fig. 3, gels 1 and 6), all of which are defective in the canonical Mec1-dependent DNA damage checkpoint pathway. The residual Sae2 phosphorylation detectable in bleomycin-treated mec1Δ cells was dependent on the Tel1 and MRX proteins (Fig. 3, gels 1 and 3). In fact, both the tel1Δ and rad50Δ single deletions decreased the amount of the bleomycin-induced Sae2 mobility shift compared to that of the wild type, although to a lesser extent than the mec1Δ allele under the same conditions. Moreover, the same genotoxic treatment could not induce detectable Sae2 phosphorylation in either mec1Δ tel1Δ or mec1Δ rad50Δ G1-arrested double mutants. Finally, bleomycin-induced Sae2 phosphorylation was reduced in the rad50Δ tel1Δ double mutant and the rad50Δ single mutant to the same extent as in the wild type (Fig. 3, gel 3), thus indicating that Tel1 and MRX likely act in the same Sae2 phosphorylation pathway, while Mec1 acts in a different pathway.
FIG. 3.
DNA damage-induced Sae2 phosphorylation in checkpoint mutants. Strains expressing Sae2-HA3 from the SAE2 promoter in a bar1Δ background were as follows: wild type (wt) (DMP3909/7D), tel1Δ (DMP3920/16D), mec1Δ sml1Δ (DMP3919/3B), mec1Δ tel1Δ sml1Δ (DMP3919/11A), rad53Δ mec1Δ sml1Δ (DMP4295/10A), chk1Δ mec1Δ sml1Δ (DMP4297/5A), chk1Δ tel1Δ (DMP4296/1D), tel1Δ rad53Δ sml1Δ (DMP4298/2B), rad50Δ (DMP4048/7D), rad50Δ tel1Δ (DMP4050/17A), rad50Δ mec1Δ sml1Δ (DMP4052/18A), chk1Δ (DMP4141/12C), rad53Δ sml1Δ (DMP4106/27A), mrc1Δ (DMP4105/3B), rad9Δ (DMP4049/5C), ddc1Δ (YLL1345.6), ddc1Δ rad9Δ (YLL1346.2), mec3Δ (DMP4138/7C), rad17Δ (DMP4137/1D), and rad24Δ (DMP4137/17A). Cell cultures were arrested in G1 with α-factor and then transferred in YEPD containing 20 mU of bleomycin/ml and 1 μg of α-factor/ml (+bleo + α factor). Time zero (αf) corresponds to cell samples taken immediately before bleomycin addition. Protein extracts prepared from cell samples collected at the indicated times were subjected to Western blot analysis with anti-HA antibodies. exp, exponentially growing cells.
The Rad53, Chk1, Mrc1, and Rad9 proteins, acting downstream of both Mec1 and Tel1 in the checkpoint pathway, did not appear to be required for DNA damage-induced Sae2 phosphorylation. In fact, similar amounts of slowly migrating Sae2 species were observed in G1-arrested, bleomycin-treated rad53Δ, chk1Δ, mrc1Δ, rad9Δ, and wild-type cells (Fig. 3, gels 4 and 5). Moreover, deletion of RAD53 and CHK1 in mec1Δ and tel1Δ cells or of RAD9 in ddc1Δ cells did not reduce the extent of detectable bleomycin-induced Sae2 phoshorylation compared to that for the single mec1Δ, tel1Δ, and ddc1Δ single mutants, respectively (Fig. 3, gels 1, 2, and 5).
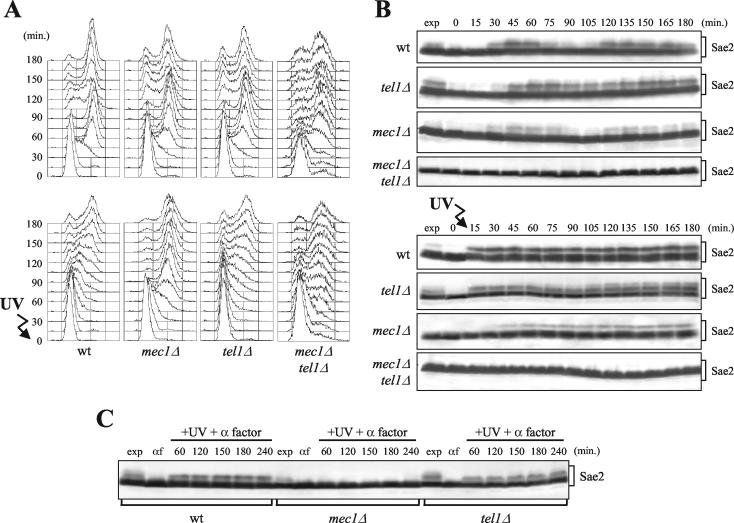
To better understand the requirements of Mec1 and Tel1 for Sae2 phosphorylation, we analyzed the kinetics of the Sae2 mobility shift under unperturbed conditions and in response to DNA damage in the absence of Mec1 and/or Tel1. When G1-arrested mec1Δ, tel1Δ, or mec1Δ tel1Δ cell cultures were released into the cell cycle without any genotoxic treatment, Sae2 phosphorylation was not detectable in mec1Δ tel1Δ cells throughout the experiment, and its amount was reduced in mec1Δ cells compared to that for the wild type, indicating that Mec1 and Tel1 promote Sae2 phosphorylation during the unperturbed S phase (Fig. 4A and B, top). Under these conditions, the amount of slowly migrating Sae2 species fluctuated during the cell cycle with similar kinetics in wild-type and mec1Δ cells, reaching different maximal levels during S phase and decreasing at the end of the first cell cycle, while the same Sae2 species did not appear to decrease significantly at the end of the first cell cycle in tel1Δ cells (Fig. 4B, top). This might suggest that the lack of Tel1 may impair DNA intermediate processing during the unperturbed S phase, thus triggering Sae2 phosphorylation also in the absence of exogenous DNA damage.
FIG. 4.
Cell cycle- and DNA damage-dependent Sae2 phosphorylation in the absence of Mec1 and/or Tel1. Strains expressing SAE2-HA3 from the SAE2 promoter were as follows: wild type (YLL1103), tel1Δ (DMP3807/2D), mec1Δ sml1Δ (DMP3806/2A), and mec1Δ tel1Δ sml1Δ (DMP3916/5B). (A and B) Cell cultures growing logarithmically in YEPD were arrested in G1 with α-factor and released from the pheromone block at time zero in YEPD, or were UV-irradiated (40 J/m2) prior to the release in YEPD. Samples of untreated and UV-treated cell cultures were withdrawn at the indicated times after α-factor release in order to analyze the DNA content by fluorescence-activated cell sorting in nonirradiated (A, top) and UV-irradiated (A, bottom) cell cultures and to analyze Sae2 phosphorylation by Western blot analysis with anti-HA antibodies of extracts from nonirradiated (B, top) and UV-irradiated (B, bottom) cell cultures. (C) Cell cultures were synchronized with α-factor and transferred in YEPD containing α factor after UV irradiation (40 J/m2) (+UV + α factor). Time zero (αf) corresponds to cell samples taken immediately before UV treatment. Protein extracts from samples withdrawn at the indicated times were treated as described for panel B. exp, exponentially growing cells.
Interestingly, when G1-arrested cells were UV irradiated before release into the cell cycle, phosphorylated Sae2 appeared immediately in wild-type and tel1Δ cells, while similar Sae2 species became detectable in mec1Δ cells only 45 min after release, concomitantly with entry into S phase, and in smaller amounts than in the wild type (Fig. 4A and B, bottom). Moreover, phosphorylated Sae2 was detectable as a mobility shift after UV irradiation in G1 even in wild-type and tel1Δ cells kept in G1 by α-factor addition, while it was below the detection level in mec1Δ cells under the same conditions (Fig. 4C). Conversely, Sae2 phosphorylation was triggered by bleomycin treatment, which is known to generate DNA DSBs, in both mec1Δ and tel1Δ cells either released into the cell cycle (data not shown) or kept in G1 by α-factor addition (Fig. 3, gel 1), although to a lesser extent than in the wild type. Since, in the absence of Mec1, DNA damage-induced Sae2 phosphorylation is dependent on Tel1, this suggests that UV-induced DNA damage might need to undergo some kind of processing during S phase in order to trigger Tel1-dependent Sae2 phosphorylation and/or that the Tel1 kinase may be less efficiently activated in G1 than Mec1.
No Sae2 mobility shift was detectable in either UV-irradiated (Fig. 4B, bottom) or bleomycin-treated (Fig. 3) mec1Δ tel1Δ double-mutant cells, further confirming that Sae2 phosphorylation likely depends totally on Mec1 and Tel1.
DNA damage response in sae2Δ cells.
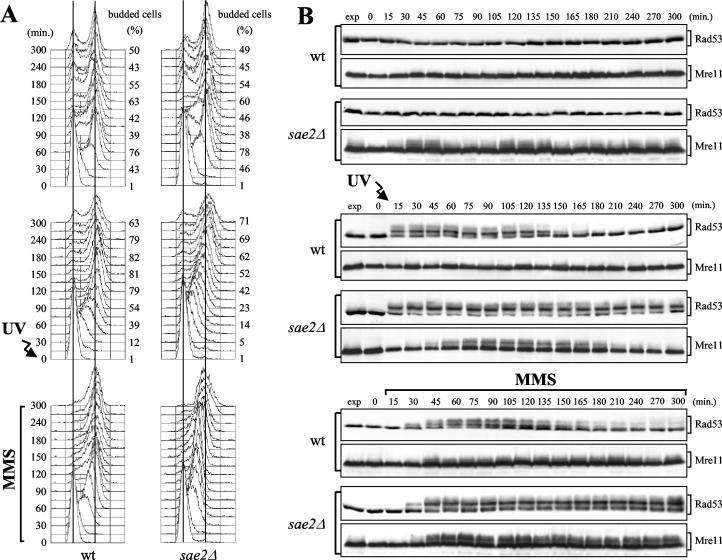
The results described above prompted us to study in more detail a possible role of Sae2 in the checkpoint signal transduction cascade. As shown in Fig. 5 (top), when undamaged G1-arrested wild-type and sae2Δ cells were released from the block, both cell types underwent budding and DNA replication with similar kinetics, and Rad53 remained unphosphorylated throughout the experiment. However, phosphorylated Mre11 became detectable as a mobility shift in unperturbed sae2Δ cells entering and progressing through S phase, but not in wild-type cells under the same conditions (Fig. 5B). Thus, the lack of Sae2 during DNA replication might cause accumulation of lesions that specifically trigger MRX phosphorylation in the absence of exogenous DNA damage.
FIG. 5.
Checkpoint response to DNA damage in sae2Δ cells. Exponentially growing wild-type (wt) (YLL1072.1) and sae2Δ (DMP4224/5B) cell cultures were synchronized with α factor and released from the pheromone block either in YEPD without any treatment (top), in YEPD after UV-irradiation (30 J/m2) (center), or in YEPD containing 0.01% MMS (bottom). Samples were withdrawn at the indicated times after α-factor release (time zero) in order to analyze the DNA content by fluorescence-activated cell sorting and the percentage of budded cells (A) and to monitor Rad53 and Mre11 phosphorylation by Western blot analysis with anti-Rad53 and anti-HA antibodies, respectively (B). exp, exponentially growing cells.
When cells were UV irradiated in G1 before release into the cell cycle, both wild type and sae2Δ cells delayed budding and S-phase progression compared to untreated cultures (Fig. 5A, center). Interestingly, wild-type cells completed DNA replication within 75 min and reached the highest percentage of budding within 120 min after release, while sae2Δ cultures contained mostly S-phase cells after 105 min and the percentage of budded cells was still increasing after 150 min (Fig. 5A, center). Furthermore, when G1-arrested cells were released into the cell cycle in the presence of MMS, both wild-type and sae2Δ cells slowed down S-phase progression compared to untreated cultures, but most wild-type cells reached 2C DNA content within 75 min after release, while sae2Δ cells progressed through S phase much more slowly and completed DNA replication only after 180 min (Fig. 5A, bottom).
The reduced ability of sae2Δ cells to complete DNA replication in the presence of DNA damage correlated with the persistence of DNA damage-induced Rad53 phosphorylated forms. These became detectable concomitantly in wild-type and sae2Δ cells after UV irradiation or MMS addition, thus indicating that SAE2 deletion was not affecting the timing of checkpoint activation. However, the amount of phosphorylated Rad53 started to decrease 120 to 135 min after UV irradiation and 135 to 150 min after MMS addition in wild-type cells, as expected when the checkpoint is turned off, while it remained constant until the end of the experiment in sae2Δ cells (Fig. 5B, center and bottom).
As was observed after DNA damage in G1, when cell cultures were released from G2 nocodazole arrest after UV irradiation, sae2Δ cells divided their nuclei more much slowly than wild-type cells and accumulated UV-induced Rad53 phosphorylated forms (data not shown).
Thus, sae2Δ cells undergo a checkpoint-mediated cell cycle arrest that lasts longer than that in wild type cells, indicating that Sae2 is not required for checkpoint activation but may be required to properly turn off the checkpoint signal by contributing to the repair of DNA lesions.
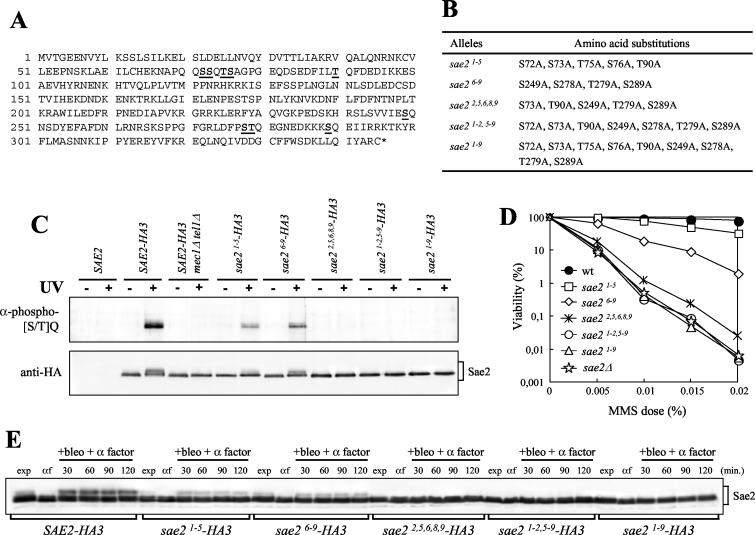
Mutagenesis of putative Sae2 phosphorylation sites.
Analysis of the Sae2 amino acid sequence revealed five serine or threonine residues (S73, T90, S249, T279, and S289) (Fig. 6A), located in canonical (S/T)Q motifs, which are favored for phosphorylation by ATM/ATR kinases (1, 75). As shown in Fig. 6C, antibodies specifically recognizing phosphorylated (S/T)Q sequences detected Sae2 in anti-HA immunoprecipitates from G1-arrested, UV-treated SAE2-HA3 cells, indicating that serine or threonine residues in Sae2 (S/T)Q motifs can be phosphorylated after DNA damage in vivo. Sae2 phosphorylation at these sites is induced by DNA damage and depends on Mec1/Tel1 activities. In fact, anti-phospho-(S/T)Q antibodies failed to detect Sae2 in Sae2-HA3 immunoprecipitates from cell extracts of α-factor-arrested, nonirradiated wild-type cells or UV-irradiated mec1Δ tel1Δ cells (Fig. 6C).
FIG. 6.
Substitutions of S or T to A at putative Sae2 phosphorylation sites affect Sae2 phosphorylation and cell survival after MMS treatment. (A) Sae2 amino acid sequence. Numbers on the left indicate the position of the first amino acid residue in each row. The S or T putative phospho-acceptor residues are boldfaced and underlined. (B) Mutant sae2 alleles obtained by site-directed mutagenesis are listed, together with the resulting serine- or threonine-to-alanine changes. (C) Strains were as follows: SAE2 (YLL1348.1), SAE2-HA3 (YLL1351.8), SAE2-HA3 mec1Δ tel1Δ sml1Δ (DMP3916/5B), sae21-5-HA3 (YLL1359.4), sae26-9-HA3 (YLL1360.31), sae22,5,6,8,9-HA3 (YLL1405), sae21-2,5-9-HA3 (YLL1361.44), and sae21-9-HA3 (YLL1352.2). Immunoprecipitations with anti-HA antibodies were performed on protein extracts prepared from nonirradiated (−) or UV-irradiated (+) α-factor-arrested cells of the indicated genotypes. Immunoprecipitates were subjected to Western blot analysis using polyclonal rabbit anti-phosphorylated-(S/T)Q antibodies (α-phospho-[S/T]Q) and anti-HA antibodies. (D) Dose-response killing curves were determined by plating serial dilutions of wild-type (YLL1348.1), sae2Δ (YLL1069.3), sae21-5 (YLL1353.1), sae26-9 (YLL1354.1), sae22,5,6,8,9 (YLL1395.1), sae21-2,5-9 (YLL1355.1), and sae21-9 (YLL1347.1) cell cultures, growing exponentially in YEPD, onto YEPD plates with or without MMS at the indicated concentrations. Plates were incubated at 26°C, and CFU were counted after 3 days. (E) α-factor-arrested cultures of the strains listed in the legend to panel C were transferred to YEPD medium containing 20 mU of bleomycin/ml and 1 μg of α factor/ml (+bleo + α factor). Time zero (αf) corresponds to cell samples taken immediately before bleomycin addition. Protein extracts prepared from cell samples collected at the indicated times were subjected to Western blot analysis with anti-HA antibodies. exp, exponentially growing cells.
We then investigated whether these residues were important for supporting Sae2 functions in vivo by constructing several sae2 mutants with multiple changes to alanine of the serine or threonine residues located in the (S/T)Q motifs (Fig. 6B). We also mutagenized to alanine the S72, T75, and S76 residues, belonging to two additional QS and QTS sequences close to the first SQ motif in the N-terminal part of the protein, and the S278 residue, close to the TQ motif in the C-terminal part of the protein (Fig. 6A). All these residues were numbered progressively from 1 to 9, starting from the N-terminal-most residue (Fig. 6B). As shown in Fig. 6C, when Sae2-HA3 immunoprecipitates from G1-arrested, UV-treated cells were subjected to Western blot analysis with the anti-phospho-(S/T)Q antibodies, Sae2-HA3 was detectable, although to a lesser extent than for the wild type, in immunoprecipitates from strains carrying either the sae21-5 (S72, S73, T75, S76, and T90 mutated to alanine) or the sae26-9 (S249, S278, T279, and S289 mutated to alanine) mutant allele. The decrease in the amount of anti-phospho-(S/T)Q-responsive Sae2 paralleled that of the slowest-migrating Sae2 protein species detected in the same immunoprecipitates by anti-HA antibodies (Fig. 6C). Thus, within the limits of this analysis, multiple (S/T)Q sites appear to contribute to in vivo DNA damage-induced Sae2 phosphorylation, which was indeed abolished in strains expressing the sae22,5,6,8,9 (S73, T90, S249, T279, and S289 mutated to alanine), sae21-2,5-9 (S72, S73, T90, S249, S278, T279, and S289 mutated to alanine), or sae21-9 (S72, S73, T75, S76, T90, S249, S278, T279, and S289 mutated to alanine) allele. In fact, the anti-phospho-(S/T)Q antibodies failed to detect Sae2-HA3, and the anti-HA antibodies failed to detect the slowest-migrating Sae2-HA3 protein species, in Western blot analyses of immunoprecipitates prepared from sae22,5,6,8,9, sae21-2,5-9 or sae21-9 G1-arrested, UV-irradiated cells (Fig. 6C). Altogether, these data indicate that replacement of all the serine or threonine residues located in the canonical (S/T)Q motifs with alanine is sufficient to reduce UV-induced Sae2 phosphorylation below the level of detection by both methods.
The effects of the sae2 alleles described above on Sae2 phosphorylation were not specific for the response to UV-induced DNA damage. In fact, Western blot analysis with anti-HA antibodies of G1-arrested, bleomycin-treated sae21-5 and sae26-9 cells, which expressed UV-induced phosphorylated Sae2 forms still detectable by the anti-phospho-(S/T)Q antibodies (Fig. 6C), showed only a partial decrease in the amount of the bleomycin-induced slowly migrating Sae2 protein species compared to that for the wild type (Fig. 6E). Conversely, the sae22,5,6,8,9, sae21-2,5-9, and sae21-9 alleles, which completely abolished UV-induced Sae2 (S/T)Q phosphorylation (Fig. 6C), did not allow any Sae2 mobility shift under the same conditions (Fig. 6E), indicating that the same amino acid changes affect Sae2 phosphorylation in response to different kinds of DNA damage.
Sae2 phosphorylation-defective alleles impair Sae2 functions.
Since SAE2 deletion causes hypersensitivity to MMS (49) (Fig. 6D), we first analyzed the MMS sensitivity of the sae2 mutants described above. As shown in Fig. 6D, the sae21-5 and sae26-9 mutants, in which DNA damage-induced Sae2 phosphorylation was only partially affected, were slightly more MMS sensitive than the wild type, although to different extents. Strikingly, the sae22,5,6,8,9, sae21-2,5-9, and sae21-9 mutants were dramatically more sensitive than the wild type to the same drug, either as sensitive as sae2Δ cells (sae21-2,5-9 and sae21-9) or slightly more resistant (sae22,5,6,8,9). Altogether, these data implicate the S and T residues of Sae2 (S/T)Q motifs both in Sae2 DNA damage-induced phosphorylation and in the DNA damage response functions of Sae2.
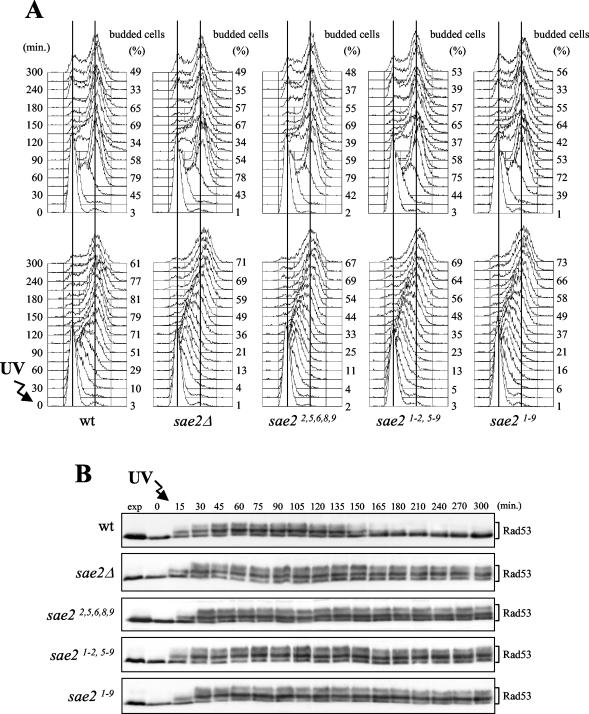
As shown in Fig. 7, the sae22,5,6,8,9, sae21-2,5-9, and sae21-9 mutants were also as defective as sae2Δ cells in resuming cell cycle progression after checkpoint activation. In fact, when cell cultures were UV irradiated in G1 before release into the cell cycle, the sae2 mutant cells were still mostly in S phase 150 min after release, and the percentage of budded cells was still increasing after 180 min, while most wild-type cells under the same conditions completed DNA replication and budding within 120 and 150 min, respectively (Fig. 7A). As observed for sae2Δ cells (Fig. 5 and 7), the reduced ability of sae22,5,6,8,9, sae21-2,5-9, and sae21-9 mutants to complete DNA replication after UV irradiation correlated with the persistence of DNA damage-induced phosphorylated Rad53 forms. In fact, the amount of phosphorylated Rad53, which started to decrease 150 min after UV irradiation in wild-type cells, remained constant until the end of the experiment in all the UV-treated sae2 mutant cultures (Fig. 7B). It is worth pointing out that the same mutants were also similar to sae2Δ cells in the accumulation of phosphorylated Mre11 forms during the unperturbed S phase (data not shown).
FIG. 7.
DNA damage checkpoint response in sae2 phosphorylation-defective mutants. Exponentially growing wild-type (K699), sae2Δ (YLL1069.3), sae22,5,6,8,9 (YLL1395.1), sae21-2,5-9 (YLL1355.1), and sae21-9 (YLL1347.1) cell cultures, growing exponentially in YEPD, were synchronized with α factor and released from the pheromone block in YEPD (A, top), or were UV irradiated (30 J/m2) (A, bottom) prior to the release in YEPD. Samples were withdrawn at the indicated times after α-factor release (time zero) in order to analyze the DNA content by fluorescence-activated cell sorting and determine the percentage of budded cells (A) and to detect Rad53 by Western blot analysis with anti-Rad53 antibodies in UV-irradiated cell extracts (B). exp, exponentially growing cells.
Since SAE2 deletion was shown to be synthetic lethal with deletion of the RAD27 gene, encoding a nuclease implicated in 5′-end processing of the Okazaki fragments (17), we also looked for possible synthetic effects of rad27Δ and sae2 phosphorylation-defective mutations. As described in detail in Materials and Methods, dissection of meiotic tetrads from heterozygous rad27Δ/RAD27 sae2/SAE2 diploid strains did not allow recovery of any viable sae21-2,5-9 rad27Δ or sae21-9 rad27Δ double-mutant segregants, while both sae26-9 rad27Δ and sae22,5,6,8,9 rad27Δ segregants gave rise to microcolonies that either did not grow when streaked onto YEPD plates or showed very severe growth defects at all temperatures. Conversely, the sae21-5 rad27Δ combination did not cause any synthetic effects on cell growth or MMS sensitivity compared to the single mutants.
Finally, as a further sensitive test for Sae2 functions, we analyzed the effects of all the sae2 phosphorylation-defective mutations described above on hairpin-capped DSB repair, which was known to be defective in sae2Δ cells (40). We therefore measured the rate of mitotic recombination induced by inverted Alu repeats between two lys2 alleles (40), after the SAE2 chromosomal allele had been replaced with the sae21-5, sae26-9, sae22,5,6,8,9, sae21-2,5-9, or sae21-9 allele in strain HS21 (kindly provided by M. A. Resnick), which is suitable for this assay (see Materials and Methods). As shown in Table 2, the rate of recombination between inverted Alu repeats was much lower in the sae2Δ, sae21-2,5-9, and sae21-9 mutants (about 40-fold), as well as in the sae26-9 and sae22,5,6,8,9 mutants (about 25-fold), than in the wild type. Conversely, the sae21-5 allele affected this process only weakly, causing a ∼4-fold decrease in the recombination rate compared to that for the wild type.
TABLE 2.
Effects of sae2 phosphorylation-defective alleles on recombination stimulated by inverted Alus
| Strain | Recombination rate (10−7)a |
|---|---|
| Wild type | 1,220 (±198) |
| sae2Δ | 16 (±4) |
| sae21-5 | 299 (±12) |
| sae26-9 | 49 (±13) |
| sae22,5,6,8,9 | 45 (±11) |
| sae21-2,5-9 | 29 (±10) |
| sae21-9 | 31 (±9) |
Recombination rates induced by inverted Alu repeats in wild-type (HS21), sae2Δ (YLL1357), sae21-5 (YLL1407), sae26-9 (YLL1406), sae22,5,6,8,9 (YLL1408), sae21-2,5-9 (YLL1375.39), and sae21-9 (YLL1368.21) strains were determined as described previously (40). Standard deviations observed for the different cell cultures used for the fluctuation tests (see Materials and Methods) are given in parentheses.
DISCUSSION
Sae2 as a target of the DNA damage checkpoint.
It is now clear that DNA damage checkpoints regulate a multifaceted and vast DNA damage response network and that defects in these checkpoints sensitize cells to DNA damage and replication blocks by failing not only to arrest the cell cycle but also to activate and/or optimally recruit the DNA damage repair machinery to the sites of damage. In agreement with a role of the ATM/ATR family kinases in modulating the DNA recombination and repair responses, the original S. cerevisiae mec1/esr1 mutants showed reduced levels of meiotic recombination compared to that of the wild type (33). Moreover, rad17, rad24, and mec1 mutants show defects in promoting proper recombination partner choice and delays in processing and repairing HO-induced broken ends (3, 28). Finally, ATM is required for homologous-recombination-mediated repair of DSBs and is a member of the recombinational repair epistasis group (11, 13, 37, 50, 51, 74).
Among the several possible roles envisaged for ATM/ATR kinases in recombination and repair processes, it has been proposed that, once recruited to a DSB, they may orchestrate the response to the damage by phosphorylating substrates required for cell cycle arrest and DSB transcriptional and repair responses. In agreement with this hypothesis, DNA damage-induced phosphorylation of the Rad55 and Srs2 proteins, both involved in DNA repair and recombination (for a review, see reference 62), was found to be checkpoint dependent (4, 38). In this view, the identification of other recombination or repair factors under DNA damage checkpoint control may help to elucidate these complex interactions.
We have shown that the Sae2 protein, which is known to act in concert with the MRX complex in processing meiotic and mitotic DSBs (34, 40, 55, 66), is phosphorylated in response to different genotoxic treatments in a checkpoint-dependent manner, suggesting that it may connect the recombination and repair processes to the DNA damage checkpoint pathways. Among the different genotoxic agents, UV irradiation and bleomycin treatment, but not MMS or HU, can induce Sae2 phosphorylation in G1-arrested cells. Since bleomycin induces DSBs (30), this finding suggests that these Sae2 modifications are likely induced by DSB formation and/or processing of the UV-induced lesions to reveal single-stranded DNA.
DNA damage-induced phosphorylation of Sae2 correlates with the activation of DNA damage checkpoint pathways. In fact, Sae2 phosphorylation occurs immediately after DNA damage, and its kinetics are similar to those of the DNA damage checkpoint kinase Rad53. Moreover, Mec1 and its accessory proteins Ddc1, Rad17, Mec3, and Rad24 are necessary for DNA damage-induced Sae2 phosphorylation. In the absence of Mec1 or of any of its accessory proteins, the amount of detectable DNA damage-induced phosphorylated Sae2 appears to be consistently lower than that for the wild type, and residual Sae2 phosphorylation in mec1Δ cells requires functional Tel1 and MRX complex. Based on epistasis analysis, Tel1 and MRX act in the same pathway to induce Sae2 phosphorylation after DNA damage, in agreement with the observations that Tel1 physically interacts with Xrs2 and that its association with DSBs is dependent on the C terminus of Xrs2 (56). In tel1Δ and mrxΔ cells, where the Mec1-dependent checkpoint is active, the amount of DNA damage-induced phosphorylated Sae2 is only slightly lower than that in wild-type cells, indicating that Mec1 plays a major role in Sae2 phosphorylation. Conversely, neither the Rad9 and Mrc1 mediators, which are involved in transducing the Mec1- and Tel1-dependent checkpoint signals to Rad53 and Chk1, nor the same downstream kinases (2, 24, 58, 73) appear to be required for Sae2 phosphorylation.
Phosphorylated Sae2 is detected by polyclonal antibodies specifically recognizing phospho-(S/T)Q motifs, which are favored for phosphorylation by Mec1/Tel1 kinases, suggesting that Sae2 can be phosphorylated at these sites in vivo. Indeed, both Sae2 recognition by the anti-phospho-(S/T)Q antibodies and the Sae2 electrophoretic mobility shift after DNA damage are abolished in tel1Δ mec1Δ strains. Moreover, alanine replacement of serine or threonine residues altering all the Sae2 (S/T)Q motifs, either alone or together with two closely related motifs, reduces DNA damage-induced Sae2 phosphorylation, if any, below the level of detection both by anti-phospho-(S/T)Q antibodies and by the Sae2 electrophoretic mobility shift.
Last but not least, Mec1-/Tel1-dependent Sae2 phosphorylation appears to be important for supporting Sae2 functions in vivo. In fact, the sae22,5,6,8,9 and sae21-2,5-9 alleles cause defects very similar to those caused by SAE2 deletion with respect to sensitivity to MMS, mitotic recombination induced by inverted Alu repeats, and resumption of cell cycle progression once the checkpoint is activated. Moreover, combination of the sae22,5,6,8,9 or the sae21-2,5-9 allele with RAD27 deletion, which impairs Okazaki fragment maturation and is synthetic lethal with sae2Δ (17, 80), results in synthetic lethality or dramatic growth defects, respectively.
Although we could not rule out the possibility that the primary effect of these mutations on Sae2 functions might be due to protein-folding alterations, we believe that these data, taken together, strongly support the hypothesis that Sae2 is likely a target of the Mec1- and Tel1-dependent checkpoints.
DNA alterations triggering Sae2 phosphorylation.
Lack of Mec1 appears not only to cause a significant decrease in the overall amount of phosphorylated Sae2 but also to impair the ability of specific DNA lesions to induce Sae2 phosphorylation. In fact, UV irradiation in G1, which results in an immediate Sae2 mobility shift in both wild-type and tel1Δ cells, seems to trigger this modification in mec1Δ mutants only when they undergo DNA replication. In contrast, slowly migrating Sae2 species appear immediately in G1-arrested mec1Δ cells treated with bleomycin, which induces DSB formation (30). Since Tel1, which is known to be specifically activated by DSBs (84), is responsible for Sae2 phosphorylation in mec1Δ cells, this suggests that either Mec1 can be activated more efficiently than Tel1 when DNA is UV irradiated in G1 or the Tel1-dependent pathway is not able to sense UV-induced lesions until their DNA replication in the absence of Mec1 generates DSBs. In agreement with the last hypothesis, it has been shown that mec1Δ mutants, which are unable to complete DNA replication under stress conditions (19, 78), accumulate aberrant DNA structures and cause replication fork collapse when DNA replication is blocked (45, 76, 78).
Interestingly, Sae2 undergoes Mec1- and Tel1-dependent phosphorylation during the unperturbed S phase, suggesting that the DNA replication process by itself may generate signals that can be sensed by the checkpoint apparatus. Consistently, also the checkpoint proteins Ddc1 and Ddc2 undergo Mec1-dependent phosphorylation during the unperturbed S phase (43, 60). Moreover, the DNA damage checkpoint pathway is necessary to suppress genome instability resulting from the aberrant repair of DNA damage, which normally occurs during DNA replication. In fact, like mutations altering DNA replication and repair genes, mutations in S-phase checkpoint genes cause increased rates of gross chromosomal rearrangements in the absence of exogenous sources of DNA damage (10, 54). In turn, Sae2 may be involved in processing aberrant replication and recombination intermediates that may occur during S phase. In fact, Sae2 loss of function is lethal in the absence of either the RAD27 gene, encoding the yeast homolog of the mammalian FEN1 flap endonuclease (17), or the RecQ-like DNA helicase, encoded by the SGS1 gene (81); both these genes are necessary for the suppression of rearrangements underlying genome instability (10, 53).
Modulation of the DSB processing machinery by the DNA damage checkpoint.
The Mre11 protein possesses both single- and double-stranded DNA endonuclease and 3′-5′ double-stranded DNA exonuclease activities (6, 18, 31, 63, 64, 85), which are important for the initial DSB processing steps. Although Sae2 is not known to be a component of the MRX complex, sae2Δ strains accumulate unprocessed DSBs and show meiotic recombination defects undistinguishable from those of nuclease-deficient mre11 mutants (32, 40, 47, 55, 66, 82). This suggests that Sae2 may be part of the MRX complex or somehow play a role in regulating its nuclease activity and/or its recruitment to broken DNA ends. In this view, Mec1- and Tel1-dependent phosphorylation of Sae2 may regulate its biochemical properties and may therefore be required to commit the resection complex to specific repair activities, once the checkpoint has been triggered.
The checkpoint cascade may also regulate Sae2 and MRX functions in order to modulate the accessibility of broken ends to DNA repair activities. Previous work has shown that two closely spaced inverted Alu sequences (Alu-IR) are highly unstable in eukaryotes and are thought to cause genomic instability (41). These sequences are found to be recombination hot spots, inducing mitotic DSBs with terminal hairpin structures (40, 41). Recombination stimulated by hairpin-capped DSBs, which can be induced by Alu inverted repeats, depends on Sae2 and MRX (40), suggesting that Sae2, along with MRX, may allow protected DNA ends to become accessible to resection. Since hairpin structures are substrates for the nuclease activity of Mre11 in vitro (63), it is tempting to speculate that the MRX complex, along with Sae2, can cleave and process hairpin structures arising during repair-associated DNA synthesis, thus preventing inverted chromosome duplication and genome instability. Similarly, the inability of sae2Δ cells to remove Spo11 in meiosis results in accumulation of unprocessed DSBs, leading to a checkpoint-dependent delay of the meiotic cell cycle, which requires the meiosis-specific Rad53 paralog Mre4/Mek1 (92). Since sae2Δ and mrx mutants show sensitivity to MMS and HU during vegetative growth, Sae2 and MRX may be required to process DSBs arising from failure to repair DNA adducts induced by DNA-modifying agents, as well as to cleave hairpin structures that might accumulate in stalled replication forks. Finally, it has been suggested that Sae2 may contribute to ensuring that both ends of a DSB participate in a recombination event, thus avoiding break-induced replication (66). Thus, DNA damage checkpoint-mediated phosphorylation of Sae2 might also influence repair partner choice.
Also the MRX complex, which is required for prevention of chromosome rearrangements in mitotic cells (10), appears to be under checkpoint control. In fact, the checkpoint kinase Tel1 is required for the DNA damage-induced phosphorylation of Mre11 and Xrs2 in vivo, and human ATM phosphorylates NBS1 in response to DNA damage, an event that is required for activation of the S-phase checkpoint (8, 15, 22, 27, 39, 91, 93). However, while mrx-null mutants are defective in the initiation of the S-phase DNA damage checkpoint, indicating that the MRX complex is part of the DNA damage checkpoint-sensing apparatus (8, 15, 21, 27), deletion of SAE2 does not cause any defect in checkpoint activation. On the contrary, once the DNA damage checkpoint is activated by exogenous DNA damage, the lack of Sae2 delays the recovery from checkpoint-mediated cell cycle arrest. These data indicate that Sae2 is not involved in DNA damage sensing but is instead likely required for processing checkpoint-activating lesions, further supporting a role for Sae2 as a target of the DNA damage checkpoint cascade. In agreement with a function of Sae2 in processing DNA lesions sensed by the checkpoints, it has been shown that the block of MMS-damaged DNA processing in sae2Δ mutants activates the Tel1-dependent checkpoint, leading to Mre11 and Xrs2 phosphorylation (84). The Sae2-dependent resection machinery is necessary also during unperturbed DNA replication. In fact, sae2Δ cells progressing through S phase in the absence of exogenous DNA damage contain phosphorylated Mre11, which is not detectable in wild-type cells under the same conditions, suggesting that DNA replication in the absence of Sae2 may have an active role in generating structures, like DSBs, that are able to activate Tel1 with subsequent Mre11 phosphorylation. Defects in the MRX complex may have the same effects, since removal of Mre11 from frog replicating extracts results in the formation of DSBs in the newly replicated DNA (14).
Although Mec1 and Tel1 have some overlapping functions, it is interesting that Mec1 appears to play a major role in Sae2 phosphorylation after genotoxic treatment, while DNA damage-induced Mre11 and Xrs2 phosphorylation depends primarily on Tel1. Although Sae2 and MRX seem to be related to each other, the differences underlying their phosphorylation may reflect a different regulation of their activity.
Acknowledgments
We thank M. Romano for some preliminary experiments, M. A. Resnick for strain HS21, S. Piatti for critical reading of the manuscript, and all the members of our laboratory for useful discussions and criticism.
This work was supported by grants from Telethon-Italy (E.1247), Associazione Italiana per la Ricerca sul Cancro, and Cofinanziamento 2003 MIUR/Università di Milano-Bicocca (to M.P.L.) and from Fondo per gli Investimenti della Ricerca di Base (FIRB) and Progetto Strategico MIUR-Legge 449/97 (to G.L.). H.C.-L. was supported by an EC Research Training Network grant (HPRN-CT-2002-00238).
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 3.Aylon, Y., and M. Kupiec. 2003. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol. Cell. Biol. 23:6585-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashkirov, V. I., J. S. King, E. V. Bashkirova, J. Schmuckli-Maurer, and W. D. Heyer. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20:4393-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley, N. J., D. A. Holtzman, G. Flaggs, K. S. Keegan, A. DeMaggio, J. C. Ford, M. Hoekstra, and A. M. Carr. 1996. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 15:6641-6651. [PMC free article] [PubMed] [Google Scholar]
- 6.Bressan, D. A., B. K. Baxter, and J. H. J. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspari, T., M. Dahlen, G. Kanter-Smole, H. D. Lindsay, K. Hofmann, K. Papadimitriou, P. Sunnerhagen, and A. M. Carr. 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20:1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell, and N. Rhind. 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 23:6564-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, S. W. L., J. Chang, J. Prescott, and E. H. Blackburn. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11:1240-1250. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 11.Chen, G., S. S. Yuan, W. Liu, Y. Xu, K. Trujillo, B. Song, F. Cong, S. P. Goff, Y. Wu, R. Arlinghaus, D. Baltimore, P. J. Gasser, M. S. Park, P. Sung, and E. Y. Lee. 1999. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J. Biol. Chem. 274:12748-12752. [DOI] [PubMed] [Google Scholar]
- 12.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 13.Cortez, D., Y. Wang, J. Quin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8:137-147. [DOI] [PubMed] [Google Scholar]
- 15.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Amours, D., and S. P. Jackson. 2002. The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 17.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 5:1129-1135. [DOI] [PubMed] [Google Scholar]
- 19.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 21.Falck, J., J. H. Petrini, B. R. Williams, J. Lukas, and J. Bartek. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 3:290-294. [DOI] [PubMed] [Google Scholar]
- 22.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 23.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 25.Green, C. M., H. Erdjument-Bromage, P. Tempst, and N. F. Lowndes. 2000. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10:39-42. [DOI] [PubMed] [Google Scholar]
- 26.Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier, and T. D. Petes. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82:823-829. [DOI] [PubMed]
- 27.Grenon, M., C. S. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 28.Grushcow, J. M., T. M. Holzen, K. J. Park, T. Weinert, M. Lichten, and D. K. Bishop. 1999. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hari, K. L., A. Santerre, J. J. Sekelsky, K. S. McKim, J. B. Boyd, and R. S. Hawley. 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82:815-821. [DOI] [PubMed] [Google Scholar]
- 30.Hoehn, S. T., H. D. Junker, R. C. Bunt, C. J. Turner, and J. Stubbe. 2001. Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage. Biochemistry 40:5894-5905. [DOI] [PubMed] [Google Scholar]
- 31.Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 4:473-485. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato, R., and H. Ogawa. 1994. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 22:3104-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keeney, S., and N. Kleckner. 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA 92:11274-11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 36.Lea, D. E., and C. A. Coulson. 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 37.Li, S., N. S. Y. Ting, L. Zheng, P. L. Chen, Y. Ziv, Y. Shiloh, E. Y. Lee, and W. H. Lee. 2000. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406:210-215. [DOI] [PubMed] [Google Scholar]
- 38.Liberi, G., I. Chiolo, A. Pellicioli, M. Lopes, P. Plevani, M. Muzi-Falconi, and M. Foiani. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19:5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. J. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 40.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 41.Lobachev, K. S., J. E. Stenger, O. G. Kozyreva, J. Jurka, D. A. Gordenin, and M. A. Resnick. 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19:3822-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longhese, M. P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532:41-58. [DOI] [PubMed] [Google Scholar]
- 43.Longhese, M. P., V. Paciotti, R. Fraschini, R. Zaccarini, P. Plevani, and G. Lucchini. 1997. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16:5216-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longhese, M. P., R. Fraschini, P. Plevani, and G. Lucchini. 1996. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol. Cell. Biol. 16:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 46.Majka, J., and P. M. Burgers. 2003. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 100:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malkova, A., L. Ross, D. Dawson, M. F. Hoekstra, and J. E. Haber. 1996. Meiotic recombination initiated by a double-strand break in rad50Δ yeast cells otherwise unable to initiate meiotic recombination. Genetics 143:741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mallory, J. C., and T. D. Petes. 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97:13749-13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKee, A. H. Z., and N. Kleckner. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene, SAE2. Genetics 146:797-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyn, M. S. 1993. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science 260:1327-1330. [DOI] [PubMed] [Google Scholar]
- 51.Morrison, C., E. Sonoda, N. Takao, A. Shinohara, K. Yamamoto, and S. Takeda. 2000. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 19:463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins, and P. Hieter.1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82:831-840. [DOI] [PubMed] [Google Scholar]
- 53.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 54.Myung, K., A. Dutta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 55.Nairz, K., and F. Klein. 1997. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11:2272-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakada, D., K. Matsumoto, and K. Sugimoto. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 16:1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 58.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paciotti, V., G. Lucchini, P. Plevani, and M. P. Longhese. 1998. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 17:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 61.Paciotti, V., M. Clerici, M. Scotti, G. Lucchini, and M. P. Longhese. 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21:3913-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 7:969-979. [DOI] [PubMed] [Google Scholar]
- 64.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 10:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prinz, S., A. Amon, and F. Klein. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146:781-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rattray, A. J., C. B. McGill, B. K. Shafer, and J. N. Strathern. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J. 19:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 71.Sanchez, Y., J. Bachant, H. Wang, F. H. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 72.Savitsky, K., et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268:1749-1753. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 74.Shafman, T., K. K. Khanna, P. Kedar, K. Spring, S. Kozlov, T. Yen, K. Hobson, M. Gatei, N. Zhang, D. Watters, M. Egerton, Y. Shiloh, S. Kharbanda, D. Kufe, and M. F. Lavin. 1997. Interaction between ATM protein and c-Abl in response to DNA damage. Nature 387:520-523. [DOI] [PubMed] [Google Scholar]
- 75.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 76.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 77.Tercero, J. A., M. P. Longhese, and J. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 78.Tercero, J. A., and J. F. X. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 79.Thelen, M. P., C. Venclovas, and K. Fidelis. 1999. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell 96:769-770. [DOI] [PubMed] [Google Scholar]
- 80.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 81.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 82.Tsubouchi, H., and H. Ogawa. 1998. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 18:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsukamoto, Y., A. K. P. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 84.Usui, T., H. Ogawa, and J. H. J. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 85.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 86.Van Den Bosch, M., R. T. Bree, and N. F. Lowndes. 2003. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 9:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Viscardi, V., E. Baroni, M. Romano, G. Lucchini, and M. P. Longhese. 2003. Sudden telomere lengthening triggers a Rad53-dependent checkpoint response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:3126-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 13:1793-1808. [DOI] [PubMed] [Google Scholar]
- 89.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 91.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O'Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbun, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477-482. [DOI] [PubMed] [Google Scholar]
- 92.Xu, L., B. M. Weiner, and N. Kleckner. 1997. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11:106-118. [DOI] [PubMed] [Google Scholar]
- 93.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]
- 94.Zhao, X., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]