Abstract
In mice and humans, there are two known members of the Huntingtin interacting protein 1 (HIP1) family, HIP1 and HIP1-related (HIP1r). Based on structural and functional data, these proteins participate in the clathrin trafficking network. The inactivation of Hip1 in mice leads to spinal, hematopoietic, and testicular defects. To investigate the biological function of HIP1r, we generated a Hip1r mutant allele in mice. Hip1r homozygous mutant mice are viable and fertile without obvious morphological abnormalities. In addition, embryonic fibroblasts derived from these mice do not have gross abnormalities in survival, proliferation, or clathrin trafficking pathways. Altogether, this demonstrates that HIP1r is not necessary for normal development of the embryo or for normal adulthood and suggests that HIP1 or other functionally related members of the clathrin trafficking network can compensate for HIP1r absence. To test the latter, we generated mice deficient in both HIP1 and HIP1r. These mice have accelerated development of abnormalities seen in Hip1 -deficient mice, including kypholordosis and growth defects. The severity of the Hip1r/Hip1 double-knockout phenotype compared to the Hip1 knockout indicates that HIP1r partially compensates for HIP1 function in the absence of HIP1 expression, providing strong evidence that HIP1 and HIP1r have overlapping roles in vivo.
Huntingtin interacting protein 1-related (HIP1r) was originally identified in 1998 due to its homology to Huntingtin interacting protein 1 (HIP1) (24) . The yeast orthologue of HIP1 and HIP1r, Sla2p, is necessary for endocytosis, proper cytoskeletal function, and growth at high temperatures (9, 30). Both HIP1 and HIP1r have been implicated in endocytosis or trafficking of clathrin-coated vesicles. Domains shared between HIP1 and HIP1r include the epsin N-terminal homology (ENTH) domain, a central coiled-coil region containing a leucine zipper, and a carboxyl-terminal TALIN homology domain. TALIN is an actin-binding protein implicated in both cell-substratum and cell-cell interactions (23).
The ENTH domains bind inositol lipids and have thus far only been found in endocytic proteins. The founding mammalian members of the group of proteins with ENTH domains are epsin, AP180, and CALM. ENTH domains bind the plasma membrane lipid, phosphatidylinositol-4,5-bisphosphate (PtdIns-4,5-P2), and have well-established roles in clathrin-mediated endocytosis (7, 11). In contrast, the ENTH domains of HIP1 and HIP1r preferentially bind the intracellular membrane lipids, phosphatidylinositol-3,4-bisphosphate (PtdIns-3,4-P2) and phosphatidylinositol-3,5-bisphosphate (PtdIns-3,4-P2) (10). This suggests that the HIP1 family may have distinct functions associated with intracellular trafficking in addition to their roles in clathrin-mediated receptor internalization. In fact, recent evidence points toward different functions at a molecular level for the ENTH domain of epsin 1 versus that of AP180 (more recently referred to as the ANTH domain). In the case of epsin, the ENTH domain has been shown to promote tubulation of lipid micelles, implying that this domain facilitates curvature of membranes (6, 26). Thus, its function is consistent with a role in the internalization phase of endocytosis. On the other hand, the ENTH/ANTH domain of AP180 does not promote the curvature of the membrane. The HIP1 and HIP1r ENTH domains are also referred to as ANTH domains due to their greater homology to the ANTH domain of AP180 compared to the ENTH domain of epsin. Finally, the recently discovered ENTH domain-containing protein, enthoprotin (also referred to as Clint and EpsinR), has been variably reported to bind PtdIns-4-P, PtdIns-5-P, or PtdIns-3,4-P2 , as well as the clathrin-Golgi adapter protein, AP1 (γ -adaptin) (8, 13, 18, 29). Enthoprotin is therefore thought to function in vesicle trafficking from the Golgi complex to the plasma membrane via its binding to γ -adaptin and intracellular membrane lipids (8, 13, 18, 29). HIP1, although sharing the affinity for PtdIns-3,4-P2 with enthoprotin, binds to the clathrin adapter protein, α-adaptin (AP2), which in contrast to its interacting lipids localizes to the plasma membrane clathrin trafficking network. Thus, the ENTH/ANTH family of proteins comprises a diverse family of proteins with distinct functions in trafficking that may be based partly on their subcellular localization as mediated by differential lipid binding.
HIP1 and HIP1r have been shown to colocalize partially with clathrin, AP-2, and endocytosed transferrin, and both proteins biochemically fractionate with clathrin-coated vesicles (4, 5, 16, 19, 21, 28). Although HIP1 and HIP1r belong to the same family and share these common domains and properties, HIP1r differs from HIP1 in several important ways (10). For example, HIP1r is expressed more ubiquitously than HIP1, and only HIP1r binds F-actin via the TALIN homology domain in vitro (4, 15). Unlike HIP1, HIP1r does not interact with Huntingtin in the yeast two-hybrid system (2), does not bind AP2 directly, and has a lower affinity for clathrin compared to HIP1 (15). These findings suggest that HIP1 and HIP1r may play both distinct and overlapping roles in clathrin-mediated vesicle trafficking. Specifically, the ability of HIP1r to bind both actin and clathrin has raised the possibility that HIP1r but not HIP1 links actin cytoskeletal functions and receptor-mediated endocytosis (4).
Deletion of Hip1 in the mouse results in varied effects on several different organs. We have found that Hip1−/− mice have increased apoptosis of postmeiotic spermatids and testicular degeneration (21), which results in reduced fertility of male Hip1−/− mice (19a). An independently derived Hip1 knockout developed tremor, gait ataxia, and thoracolumbar kyphosis, in addition to testicular degeneration (17). In addition to spinal deformities, one mutation of Hip1 has resulted in microophthalmia and cataracts (19a). In primary hippocampal neurons, the absence of HIP1 results in a possible defect in clathrin-mediated trafficking of GluR1-containing AMPA receptors in which intracellular/surface glutamate receptor ratios are diminished in the Hip1 knockout cells after ligand stimulation (17).
To investigate the role of HIP1r in vivo, we generated mice with a targeted deletion of the Hip1r exons 2 to 8. Mice deficient in HIP1r are viable and fertile and have no gross morphological abnormalities by 1 year of age. Analysis of Hip1r−/− mouse embryonic fibroblasts (MEFs) also showed no abnormalities in constitutive or stimulated clathrin-mediated endocytosis in the Hip1r knockout background. Hip1r mutant mice also were crossed with Hip1 mutant mice to produce double-knockout mice. Hip1r/Hip1 double-mutant mice showed accelerated development and penetrance of abnormal traits, including spinal defects and growth arrest, that occur in adult Hip1 mutant mice (17). Our findings demonstrate that HIP1r is not necessary for normal development and growth of the mouse embryo or adult. We suggest that HIP1 or other functionally related members of the clathrin trafficking network may compensate for the loss of HIP1r expression. In support of this hypothesis, HIP1r acts to partly compensate for the loss of HIP1 function in the mutant Hip1 background.
MATERIALS AND METHODS
Immunohistochemistry.
A normal human tissue microarray was stained with the anti-HIP1r monoclonal antibody 1E1 by standard methods (10) (VastArrays; ResGen).
Construction of Hip1r targeting vector.
The 5′ end of the mouse Hip1r gene was isolated by PCR screening of a bacterial artificial chromosome (BAC) library and confirmed by hybridization with a cDNA probe spanning nucleotides 300 to 433 of murine Hip1r coding sequence (as numbered in the NCI database; accession no. AF221713). The BAC clone was digested with a panel of 10 restriction enzymes and the fragments subcloned into the pZERO-2 vector. Three subclones (XhoI-3, HindIII-2, and BamHI-3) were analyzed by restriction digests and Southern blotting to create a plasmid contig of 22.7 kb (see Fig. S1 in the supplementary material). The exon-intron junctions were confirmed by comparing the cDNA coding sequence of the human Hip1r with mouse cDNA sequence (accession no. AF221713 [mouse] and AB013384 [human]) and by sequencing several genomic subclones with exon-specific primers. Subclone HindIII-2 was digested with KpnI, and the released 2.6-kb fragment was inserted into the KpnI site of the 38loxpNeo targeting vector to create plasmid #1 containing the 5′ arm. Subclone BamHI-3 was digested with SpeI, blunt ended, and ligated to the XhoI-digested and blunt-ended plasmid #1 to create the final vector, 38loxpNeoHIP1r. This vector has the reverse-oriented floxed neomycin resistance cassette in the place of exons 2 to 8 of the Hip1r gene.
Southern blot analysis.
Standard genotypic analysis from tail biopsies of 3-week-old mice for the Hip1 knockout were performed as previously described (19a, 21). For the Hip1r mutant, the 5′ probe distinguished 10.5-kb (wild-type) and 9.2-kb (recombinant) fragments from BamHI-digested genomic DNA. The 3′ probe recognized 11.1-kb (wild-type) and 7.8-kb (recombinant) bands from BamHI-digested genomic DNA (see Fig. 2B).
FIG. 2.
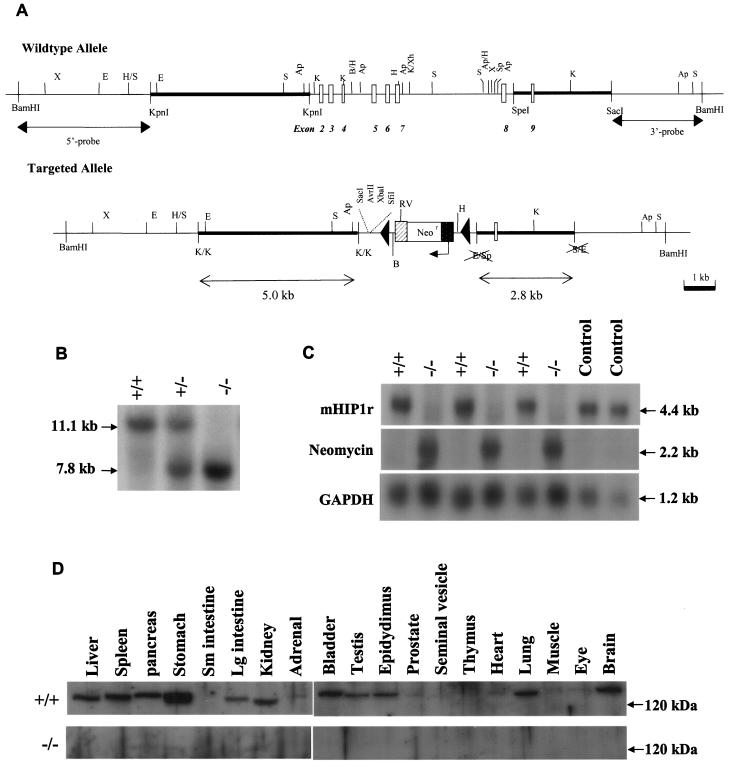
Disruption of the murine Hip1r gene by homologous recombination. (A) Hip1r targeting strategy. Hip1r exons 2 to 8 were replaced by the neomycin resistance gene in the opposite orientation as the Hip1r gene in the targeted allele. The genomic sequences recognized by the 5′ and 3′ Southern blot probes are shown (5′ probe and 3′ probe). K, KpnI; Ap, ApaI; B, BamHI; E, EcoRI; Rv, EcoRV; H, HindIII; S, SacI; Sp, SpeI; X, XbaI; Xh, XhoI. (B) Genotype analysis of the F2 generation. Representative BamHI-digested genomic DNA fragments detected by hybridization with the 3′ Southern blot probe are shown. (C) Northern blot analysis of mouse brain. Hybridization of poly(A) mRNA with a mouse Hip1r cDNA probe identified the 4.4-kb Hip1r mRNA transcript (top panel). The neomycin probe recognized a 2.2-kb transcript corresponding to the neomycin-resistant mRNA transcript (middle panel). GAPDH was used as a loading control (bottom panel). (D) Western blot analysis. Tissue extracts of organs from wild-type (top panel) and Hip1r−/− (bottom panel) mice were blotted with anti-HIP1r polyclonal antibody UM359.
RNA isolation and Northern blot analysis.
Total RNA was isolated from mouse brain by using TRIzol reagent (Invitrogen). Poly(A) RNA was purified by using the Poly(A)Purist MAG protocol according to manufacturer's directions (Ambion). Then, 5 μ g of poly(A) RNA was separated on a 1% agarose gel with 6% formaldehyde, stained with ethidium bromide, transferred to Nytran membrane, and cross-linked. The membrane was prehybridized in buffer containing 5× SSC, 5 × Denhardt solution, 1% sodium dodecyl sulfate (SDS) (wt/vol), and 100 μ g of denatured salmon sperm DNA/ml for 3 h at 65°C. For the mouse HIP1r Northern blot probe, a 700-bp HindIII- and BamHI-digested fragment from murine Hip1r cDNA was used as the probe. The neomycin probe DNA fragment was amplified by PCR with the targeting vector as the template with the following primers: Neo F 1440K 5′-AGGATCTCCTGTCATCTCA-CCTTGCTCCTG-3′ and Neo R 1441K 5′-AAGAACTCGTCAAGAAGGCGATAGAAGGCG-3′. The thermocycling conditions were 94°C for 11.5 min; 30 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min; and 72°C for 10 min. The 0.5-kb PCR product was detected on a 1% agarose-Tris-borate-EDTA gel and purified by using the Qiaex II agarose gel extraction protocol (Qiagen). Generation of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe has been described previously (31). The probes were 32 P labeled with a random primed labeling kit according to manufacturer's directions (Roche). The blot was hybridized overnight at 65°C, washed twice in 2× SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 20 min, once in 1× SSC for 10 min, and twice in 0.1× SSC for 10 min. The blot was exposed for 3 to 4 days on Kodak Biomax film.
Isolation and maintenance of MEFs.
Hip1r heterozygous F1 mice were intercrossed, and embryos were collected at day 14.5 of gestation. Embryo lengths were measured to confirm the predicted gestational age (11.5 to 12.5 mm = 14.5 days) (14). After removal of the head and liver, embryos were minced and incubated in 2 ml of 0.05% trypsin-EDTA (Invitrogen) at 37°C for 10 min. After trituration to generate single cell suspensions, cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum, gentamicin, glutamine, minimal essential medium nonessential amino acids, and β -mercaptoethanol in a 37°C incubator with 5% CO2 . MEF cell lines were immortalized after 30 serial passages by using the NIH 3T3 protocol (27). Genotypes were determined by both Southern and Western blot analysis for all cell lines.
Isolation of keratinocytes.
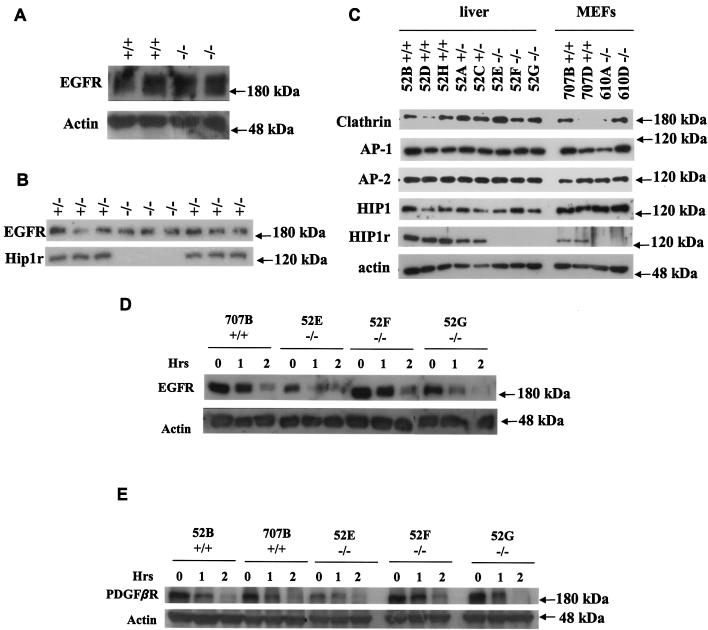
Epidermal keratinocytes were isolated as described by Dlugosz et al. (3). Briefly, 1- to 2-day-old mice from a Hip1r−/− and Hip1r+/− intercross were sacrificed, soaked in 5% iodine, and then washed in 70% ethanol. After removal of the tail and limbs, the skin of each animal was removed in one piece and floated dermal side down in 0.25% trypsin overnight at 4°C. The epidermal layer was then separated from the dermis and vortexed in high-calcium medium, inducing the keratinocytes to disperse into the medium. The cells were then pelleted and plated in calcium-containing medium. After 24 h, the cells were changed to a low-calcium medium. Finally, after 2 days of growth, cells were lysed and analyzed for total epidemal growth factor receptor (EGFR) levels by Western analysis with anti-EGFR sheep polyclonal antibody (Upstate Biotechnology).
Western blot of mouse tissues.
For analysis of the epidermis, skin from 1.5 cm of tail was scraped by using a no. 10 blade to isolate the epidermis. Samples were homogenized in 100 μ l of lysis buffer containing 1% Triton X-100 and protease inhibitors. For all other tissues, 10 to 20 mg of fresh or frozen tissue was added to 200 to 300 μ l of lysis buffer, homogenized for 10 s by using a small homogenizer, and incubated on ice for 15 min. Samples were centrifuged at 14,000 rpm in a 4°C microcentrifuge for 15 min, and supernatants were transferred to a new tube. Protein concentrations were determined by Bradford assay (Bio-Rad). Then, 50 μ g of protein was subjected to SDS-8% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose. HIP1r was identified by using the polyclonal anti-HIP1r antibody UM359 (10) and horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody. EGFR was detected by using an anti-EGFR sheep polyclonal antibody (1:500; Upstate Biotech). Other antibodies used were monoclonal anticlathrin (1:1,000; TD.1), monoclonal anti-adaptin γ (1:1,000; Transduction Laboratories), polyclonal anti-adaptin α (1:1,000; Santa Cruz), polyclonal anti-HIP1 (1:5,000, UM354; Assay Designs, Inc.), polyclonal anti-HIP1r (1:10,000, UM365), and polyclonal antiactin (1:500, Sigma). Signal was detected by reaction with enhanced chemiluminescence reagent for HIP1r and HIP1 or SuperSignal West Pico Chemiluminescent substrate (Pierce) for other proteins according to the manufacturer's directions.
Cell proliferation assay.
Cell proliferation was measured by MTT assay according to the manufacturer's directions (Roche). A total of 2 × 103 or 2 × 104 cells suspended in 100 μ l of medium from each MEF line were plated in quadruplicate in 96-well plates. Measurements of absorbance at 600 nm were taken at 1, 2, 4, 8, and 11 days of growth by using an enzyme-linked immunosorbent assay plate reader.
Growth factor stimulation.
MEF cell lines were plated onto 100-mm dishes and grown to 70 to 80% confluence. Cells were serum starved for 18 h, pretreated with cycloheximide to inhibit protein synthesis (100 μ g/ml for 30 min), and stimulated with EGF (100 ng/ml) or platelet-derived growth factor ββ (PDGF ββ ; 50 ng/ml). Cells were harvested 0, 1, and 2 h poststimulation and lysed in buffer containing 1% Triton X-100 and protease inhibitors. Protein concentrations were determined by using Bradford reagent (Bio-Rad). Then, 50 μ g of protein was subjected to SDS-7% PAGE and transferred to nitrocellulose. To detect the EGFR, a sheep polyclonal anti-EGFR antibody was utilized (2 μ g/ml) according to the manufacturer's directions (Upstate Biotechnology). HRP-conjugated anti-sheep secondary antibody (1:2,000) and SuperSignal West Pico chemiluminescent substrate (Pierce) were used to detect the bound primary antibody. To detect the PDGF β R, a polyclonal anti-PDGF β R antibody (1:1,000; BD Pharmingen) and secondary HRP-conjugated anti-rabbit antibody (1:5,000) were used. Antiactin (1:500; Sigma) staining was used as a loading control.
EGF and transferrin uptake and immunofluorescence.
For EGF internalization, cells were grown on coverslips until 70 to 80% confluent and starved in serum-free medium for 16 h. Samples were washed in ice-cold DMEM, incubated for 1 h at 4°C with 500 ng of Alexa Fluor 488-EGF (Molecular Probes)/ml, and then shifted to 37°C for 30 min to allow internalization. For transferrin internalization, cells were starved for 3 h in serum-free medium, washed in ice-cold DMEM, incubated for 1 h at 4° with 50 μ g of Texas red-transferrin (Molecular Probes)/ml, and then shifted to 37°C for 30 min. Cells were fixed with 4% paraformaldehyde and mounted onto slides with Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole). Samples were visualized by using an Olympus fluorescence microscope, and images were captured digitally.
RESULTS
HIP1r expression in human tissue.
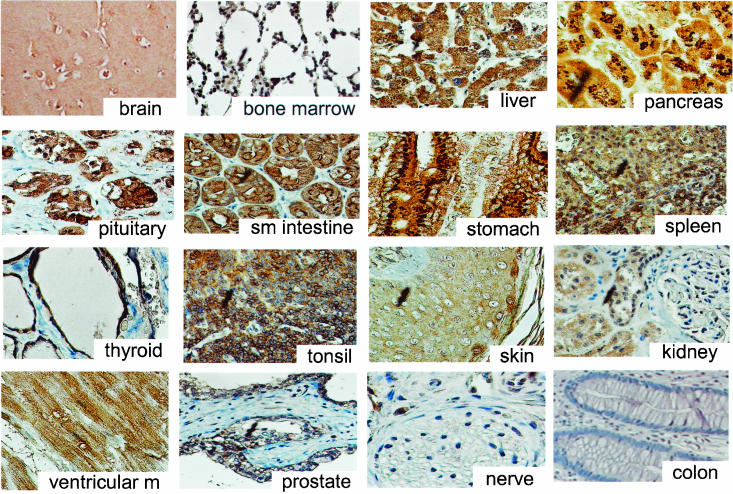
By further defining the expression pattern of the HIP1r protein, we hoped to predict where the effects of HIP1r loss in the Hip1r knockout mouse may be most obvious phenotypically. To do this, a normal human tissue microarray was stained by immunohistochemistry for HIP1r with the anti-HIP1r monoclonal antibodies 1E1 (Fig. 1) and 1C5 (data not shown) (10). Strong HIP1r expression was observed in several tissues, including the brain, bone marrow cells, liver, pancreas, pituitary, small intestine, stomach, spleen, thyroid, and tonsils. Moderate HIP1r staining was present in the epidermis of the skin, proximal tubules of the kidney, and ventricular muscle of the heart. In contrast to HIP1 expression (22), there was little HIP1r staining in peripheral nerves and blood vessel endothelium, whereas expression of HIP1r was observed in the HIP1-negative prostate epithelium. Like HIP1, the colonic epithelium also showed low levels of HIP1r.
FIG. 1.
HIP1r expression in normal human tissues. Photomicrographs of a normal human tissue microarray stained with anti-HIP1r MAb 1E1. Examples of tissues with high (brain, bone marrow, pancreas, pituitary, small intestine, stomach, spleen, thyroid, and tonsil), moderate (skin, kidney, ventricular muscle, and prostate), and low (peripheral nerve and colon) HIP1r staining are shown.
Mapping and targeted deletion of Hip1r .
In order to understand the function of HIP1r in vivo, we generated a mouse with a targeted deletion of exons 2 to 8 of the Hip1r gene. Exons 2 to 9 of the mouse Hip1r gene were first isolated in a single BAC clone and used to construct the targeting vector (see supplemental Fig. S1). Several primers covering a region 5′ of exon 2 failed to document the presence of exon 1. Hence, the exact genomic location of mouse exon 1 was not defined. Exons 2 to 8 of the Hip1r gene (encompassing nucleotides 92 to 719 of the murine Hip1r cDNA [accession no. AF221713]) were replaced with a neomycin resistance cassette in reverse orientation flanked by loxp sites. Portions (5.0 and 2.8 kb) of genomic sequence at the 5′ and 3′ ends, respectively, were used as the homologous arms (Fig. 2A). The Hip1r targeting construct was electroporated into RW1 129Sv mouse embryonic stem (ES) cells (obtained from Incyte Genomics, St. Louis, Mo.). Selection for neomycin resistance produced multiple clones. Each ES clone was subjected to Southern blotting with both 5′ and 3′ probes for Hip1r. Of 384 clones tested, 6 ES clones were found to be correctly targeted at both the 5′ and the 3′ ends (see Fig. S2 in the supplemental material). ES cells from two of these clones, 4C9 and 2F2, were injected into C57BL/6 blastocysts. The blastocysts were then implanted successfully into pseudopregnant females. For each clone, Southern blot analysis of tails from the resulting chimeras indicated that all animals had significant contributions from the 129 Sv ES cell lineage. Male chimeras were then mated with C57BL/6 female mice to produce the F1 generation. Southern blot analysis of the genomic DNA from F1 agouti mice indicated germ line transmission of the recombinant Hip1r allele for both the 4C9 and 2F2 lines. Mice were then expanded for subsequent analysis.
HIP1r -deficient mice are viable and fertile.
F1 heterozygous mice were intercrossed, and the resultant F2 offspring were genotyped for Hip1r by Southern blot (Fig. 2B). Mating of the F1 generation produced Hip1r+/+, Hip1r+/−, and Hip1r−/− mice in normal Mendelian ratios (24, 53, and 23%, respectively [316 total mice]). Northern blot analysis of brain with a mouse Hip1r probe showed the absence of Hip1r mRNA in Hip1r−/− mice (Fig. 2C, top panel). Overblotting with a neomycin probe showed the expected expression of the neomycin mRNA only in the Hip1r−/− mice (Fig. 2C, middle panel).
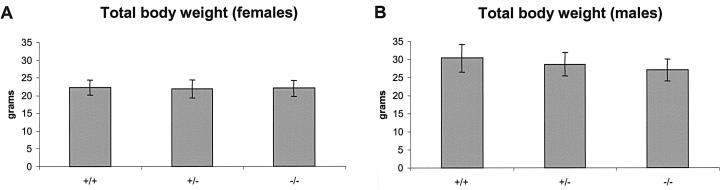
In the mixed 129 Svj/C57BL/6 background, Hip1r−/− mice were completely viable. Total necropsy and evaluation of individual tissues by hematoxylin and eosin staining of paraffin sections showed no pathological changes in Hip1r−/− mice; these tissues included the liver, spleen, pancreas, stomach, small and large intestines, kidney, adrenal glands, bladder, testis, epididymis, prostate, seminal vesicle, ovary, thymus, heart, lung, muscle, eye, and brain. Hip1r−/− mice exhibited no gross phenotype by 12 months of age. Peripheral blood counts of Hip1r−/− mice were also normal (data not shown). A survey of organs by Western blot analysis with the polyclonal anti-HIP1r antibody confirmed the absence of HIP1r expression in all of the knockout tissues (Fig. 2D, bottom panel). Finally, Hip1r−/− male and female mice were fertile, produced normal-sized litters, and were of normal weight. To document the latter, Hip1r+/+, Hip1r+/−, and Hip1r−/− mice in the colony that were 8 weeks of age or older were weighed. Comparison of homozygous wild type, heterozygous, and homozygous null mice showed no significant differences in body weights for either females (Fig. 3A, n = 75) or males (Fig. 3B, n = 55).
FIG. 3.
Growth analysis of Hip1r−/− mice. (A) Average weights of adult female (>8 weeks old) Hip1r+/+ (n = 12), Hip1r+/− (n = 32) and Hip1r−/− (n = 31) mice. (B) Average weights of adult male (>8 weeks old) Hip1r+/+ (n = 15), Hip1r+/− (n = 31), and Hip1r−/− (n = 9). Error bars represent standard deviations.
Growth analysis of Hip1r−/− MEFs.
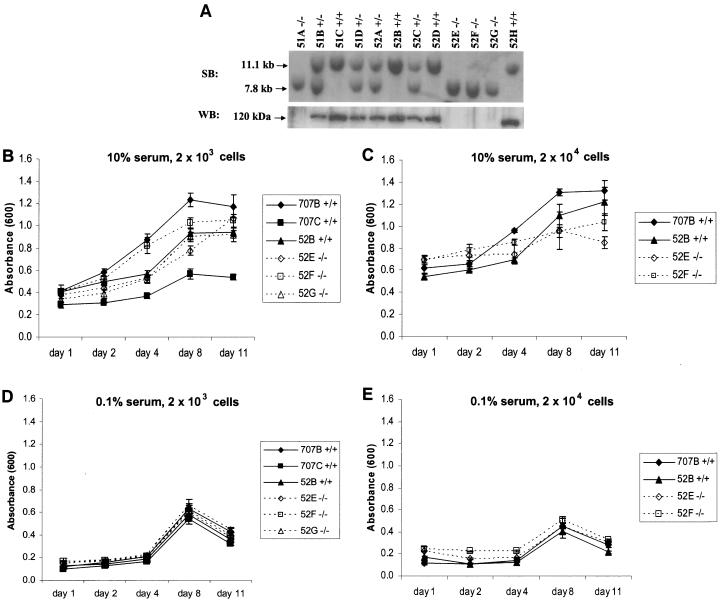
To further investigate the effects of HIP1r absence on cellular function, Hip1r+/+, Hip1r+/−, and Hip1r−/− MEFs from 14.5-day-old embryos were isolated and immortalized (see Materials and Methods). Southern and Western blot analyses of the resulting MEF cell lines confirmed the mutation of the Hip1r gene and the absence of HIP1r protein, respectively (Fig. 4A). Since HIP1 is overexpressed in tumors (22) and is able to transform fibroblasts (20), the HIP1 family may be involved in cellular proliferation. Therefore, we tested the effects of HIP1r deficiency on cellular proliferation of MEFs by using the MTT assay. This colorimetric assay measures the amount of purple formazan crystals metabolized from the yellow tetrazolium salt MTT by viable cells. Formazan is then solubilized and quantified by spectrophotometry at a wavelength of 600 nm. The amount of formazan crystals formed directly correlates with the increase in total metabolic activity associated with an increase in the number of living cells. This assay was used to compare the proliferation rates of Hip1r−/− MEF cell lines compared to wild-type MEFs from the same litter. High and low numbers of cells from each cell line were plated in quadruplicate and then measurements taken at 1, 2, 4, 8, and 11 days of growth. We found no differences in cellular proliferation rates in the Hip1r−/− MEFs compared to wild-type MEFs when cells were grown under normal conditions, at both low and high density plating (Fig. 4B and C). Hip1r−/− MEFs also did not have altered growth in limiting serum (0.1%) at either low or high initial plating densities (Fig. 4D and E). This suggests that whereas the overexpression of HIP family members can promote higher growth rates, the absence of expression of one family member, HIP1r, does not affect normal cellular proliferation.
FIG. 4.
Growth analysis of Hip1r−/− MEFs. (A) Southern blot of MEF lines established from the F2 generation hybridized with the 3′ HIP1r probe (top panel). The same MEF lines were analyzed by Western blot with the anti-HIP1r antibody UM359 (bottom panel). Note the intermediate expression levels of HIP1r in the heterozygous cell lines. (B) Growth analysis of MEFs in 10% serum by MTT assay. Cells were plated at 2 × 103 per well in quadruplicate and analyzed for 11 days. (C) Growth analysis after plating 2 × 104 cells per well in 10% serum. (D to E) Growth analysis of MEFs in 0.1% serum. Cells were plated at 2 × 103 (D) or 2 × 104 (E) cells per well.
Steady-state levels of endocytic proteins in HIP1r-deficient cells.
To begin to test whether Hip1r−/− mice had defects in receptor-mediated endocytosis, we first looked at EGFR levels in the skin. We focused on this tissue since it contains high levels of HIP1r (Fig. 1), and EGFR proteins and EGF-mediated signaling pathways are important for normal skin properties (25) . Since we have observed previously that overexpression of HIP1r stabilizes the EGFR (10), EGFR levels in the absence of HIP1r might be predicted to be diminished compared to wild-type skin. We have not yet seen, by Western blot analysis, a difference in the steady-state levels of EGFR in the epidermis (Fig. 5A) or isolated keratinocytes (Fig. 5B) between Hip1r+/+ and Hip1r+/− mice and Hip1r−/− mice.
FIG. 5.
Growth factor receptor levels and endocytosis in Hip1r−/− cells. (A) EGFR levels in the epidermis of Hip1r−/− mice. A total of 100 μ g of protein was separated by SDS-PAGE, and EGFR levels were detected by using anti-EGFR sheep polyclonal antibody. (B) Keratinocytes were isolated from newborn Hip1r homozygous or heterozygous mutant mice and lysed 2 days after plating, and 50 μ g of protein was separated by SDS-PAGE. EGFR levels were detected with anti-EGFR sheep polyclonal antibody. (C) Expression of endocytic proteins in livers from Hip1r−/− embryos (isolated when MEF lines were created) and Hip1r−/− MEFs. A total of 50 μ g of protein was run on 8% gels and then blotted for the clathrin heavy chain, the γ subunit of AP-1, the α subunit of AP-2, HIP1, and HIP1r. (D) EGFR half-life in Hip1r−/− MEFs after starvation and stimulation with 100 ng of EGF/ml. Lysates were separated by SDS-PAGE, transferred to nitrocellulose, and blotted with anti-EGFR sheep polyclonal antibody. (E) PDGF β R levels in Hip1r−/− MEFs after starvation and stimulation with 50 ng of PDGF ββ /ml. PDGF β R was detected with an anti-PDGF β R polyclonal antibody. Actin levels were used as a loading control.
To further test whether cells from Hip1r−/− mice exhibited defects in endocytosis, expression of various endocytic proteins were measured in Hip1r−/− embryo livers and MEFs derived from Hip1r−/− mice. We found that levels of adaptin γ (a subunit of AP-1) and adaptin α (a subunit of AP-2) from Hip1r−/− cells were similar to the expression found in wild-type cells (Fig. 5C). Although clathrin levels were quite variable among the tested samples, there was no consistent change in the level of clathrin expression in the absence of HIP1r expression (Fig. 5C). We also observed that levels of HIP1 were not altered in cells that lack HIP1r expression (Fig. 5C).
Growth factor receptor stability.
Since HIP1r did not affect steady-state levels of EGFR or levels of endocytic proteins, we tested whether absence of HIP1r alters the growth factor receptor half-life after ligand stimulation in the immortalized MEFs. We have previously shown that growth factor receptor levels and their half-life after ligand simulation is prolonged in cells with overexpression of HIP1 or HIP1r (10, 20). First, we analyzed the EGFR levels in Hip1r−/− MEFs after starvation and stimulation with EGF. Briefly, cells were starved for 18 h in serum-free media, treated with cycloheximide for 30 min to inhibit protein synthesis, and stimulated with 100 ng of EGF/ml for 0, 1, or 2 h. Western blot analysis of EGFR levels in three independent Hip1r−/− MEF lines after EGF stimulation showed that the EGFR half-life and steady-state levels were similar to wild-type MEFs (Fig. 5D). The half-life of the EGFR was approximately 1 h for both wild-type and Hip1r−/− MEF lines. Although we have found that overexpression of HIP1 or HIP1r prolongs the half-life of ligand-stimulated EGFR (10), these data suggest that HIP1r expression is not necessary for ligand-stimulated EGFR degradation.
Like EGFR levels after ligand stimulation, we previously observed that levels of the PDGF β R are stabilized after stimulation with PDGF ββ in cells by overexpression of full-length HIP1r or HIP1 (10). In addition, the levels of the PDGF β R diminish more quickly after ligand stimulation when cells are transfected with the HIP1 or HIP1r mutants lacking the ANTH domain (10). Consequently, we tested the response of the PDGF β R to ligand stimulation in the Hip1r−/− MEF lines. A result similar to the results for the EGFR was found for the PDGF β R, where there was little difference in the kinetics of PDGF β R degradation after ligand stimulation (Fig. 5E). The PDGF β R half-life after stimulation was roughly 1 h in both the wild-type and the homozygous Hip1r -null MEFs. Here again, the absence of HIP1r does not appear to affect the degradation of the PDGF β R after stimulation with its ligand.
Endocytic uptake in Hip1r -deficient MEFs.
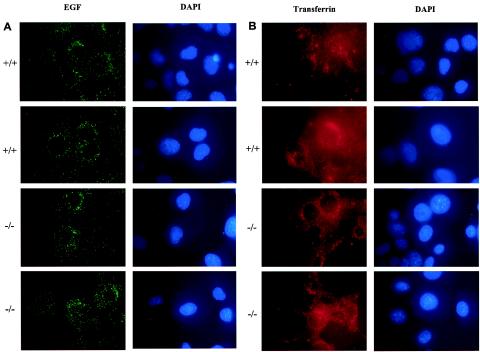
Since HIP1 proteins interact with clathrin and affect the stability of growth factor receptors when overexpressed, we tested whether loss of HIP1r affected uptake of EGF or transferrin in the Hip1r−/− MEF lines. We utilized an immunofluorescence-based internalization assay wherein EGF coupled to the fluorophore Alexa Fluor 488 is used to assay receptor-mediated endocytosis. Briefly, cells were starved overnight before stimulation with 100 ng of Alexa Fluor 488-EGF/ml. Samples were incubated at 4°C for 1 h to allow binding to receptors, shifted to 37°C for 30 min to initiate internalization, and fixed for immunofluorescence analysis. Internalized EGF forms a punctate pattern of vesicles when visualized by immunofluorescence (Fig. 6A, left column). There was no change in the ability to internalize EGF in the Hip1r−/− MEFs (Fig. 6A, third and fourth rows) compared to wild-type MEFs (Fig. 6A, first and second rows) after 30 min. Three independent cell lines of each genotype were tested, and representative figures are shown.
FIG. 6.
Endocytosis in Hip1r−/− MEFs. (A) Immunofluorescence of EGF internalization in Hip1r+/+ and Hip1r−/− MEFs. Cells were starved overnight, incubated with Alexa Fluor 488-EGF (green) at 4°C for 1 h, and then shifted to 37°C for 30 min. Nuclei were visualized by DAPI staining. Two representative fields from wild-type MEFs (first and second rows) and homozygous Hip1r -null MEFs (third and fourth rows) are shown. (B) Immunofluorescence of transferrin internalization in Hip1r+/+ and Hip1r−/− MEFs. Cells were starved for 3 h, incubated with Texas red-transferrin (red) at 4°C for 1 h, and then shifted to 37°C for 30 min. Two representative fields from Hip1r+/+ MEFs (first and second rows) and Hip1r−/− MEFs (third and fourth rows) are shown.
Next, constitutive endocytosis of transferrin was examined. MEFs were starved for 3 h, incubated with 50 μ g of Texas red-conjugated transferrin/ml at 4°C for 1 h, and then shifted to 37°C for 30 min to allow internalization. Immunofluorescence analysis of Hip1r+/+ MEFs showed a punctate pattern of staining in the cytoplasm (Fig. 6B, first and second rows). As in the case of EGF internalization, transferrin-stimulated Hip1r−/− MEFs had a similar pattern and frequency of transferrin internalization compared to Hip1r+/+ MEFs after 30 min (Fig. 6B, third and fourth rows).
Generation of Hip1r/Hip1 double-knockout mice.
One explanation for the lack of an altered phenotype in Hip1r−/− mice is that HIP1 is able to compensate for the lack of HIP1r function. In order to test this hypothesis, Hip1r−/− mice were crossed with previously generated Hip1null/null mice (19a) to produce double heterozygous mice. We then attempted to generate double-knockout mice by intercrossing the double heterozygous mice, but did not observe any Hip1r−/−; Hip1null/null mice from a total of 196 offspring (Table 1). During this experiment, it was reported that both mouse Hip1r and mouse Hip1 are located on chromosome 5 (1), providing an explanation for why Hip1r and Hip1 genes did not segregate independently. Fortunately, due to the occurrence of single crossover events between the Hip1r and Hip1 genes on chromosome 5, Hip1r+/−; Hip1null/null and Hip1r−/−; Hip1+/null mice were successfully produced. The single Hip1r+/+; Hip1+/+ offspring produced from the double heterozygous matings was the result of simultaneous crossover events occurring in the germ cells of both parents (Table 1). Intercrossing Hip1r+/−; Hip1null/null and Hip1r−/−; Hip1+/null mice or Hip1r−/−; Hip1+/null and Hip1r−/−; Hip1+/null mice, as expected, successfully generated double-knockout offspring.
TABLE 1.
Genotypes of 196 offspring from Hip1r/Hip1 double-heterozygous matingsa
| Genotype
|
Observed
|
Expected
|
|||
|---|---|---|---|---|---|
| HIP1r | HIP1 | nb | Rate (%) | n | Rate (%) |
| +/+ | +/+ | 1 | 0.5 | 12.3 | 6.25 |
| +/+ | −/− | 37 | 18.9 | 12.3 | 6.25 |
| +/+ | +/− | 10 | 5.1 | 24.5 | 12.5 |
| +/− | +/+ | 13 | 6.6 | 24.5 | 12.5 |
| +/− | +/− | 72 | 36.7 | 49 | 2.5 |
| +/− | −/− | 7 | 3.6 | 24.5 | 12.5 |
| −/− | +/− | 9 | 4.6 | 24.5 | 12.5 |
| −/− | +/+ | 47 | 24.0 | 12.3 | 6.25 |
| −/− | −/− | 0 | 0 | 12.3 | 6.25 |
As determined by chi-square goodness-of-fit analysis, χ2 = 8.54 and P < 0.001.
n, number of offspring.
The Hip1 knockout phenotype is accelerated in the absence of HIP1r expression.
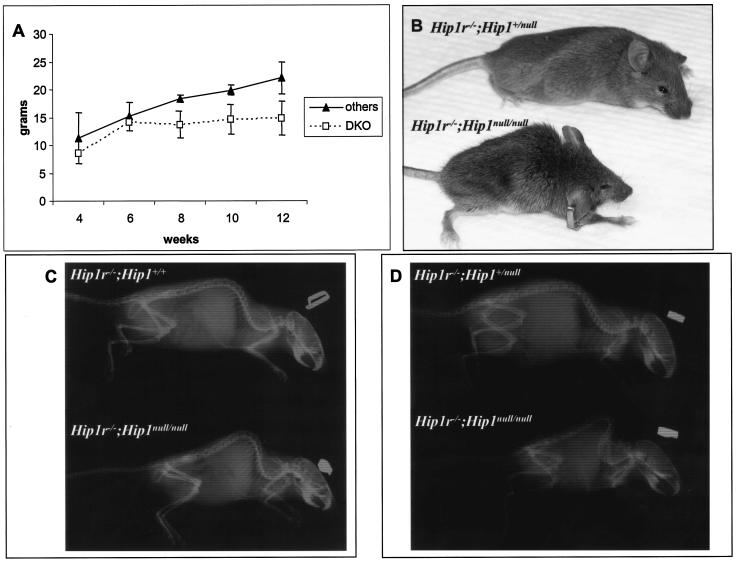
Deletion of Hip1 results in a complex phenotype whose mechanism is currently unclear. We and others have observed the development of adult onset kypholordosis and reduced body mass with resulting abnormal gait in HIP1-deficient mice (17, 19a). In addition, testicular degeneration, abnormalities in hematopoietic progenitor frequency, microophthalmia, and cataracts have also been observed in another more recently characterized Hip1null/null line (19a).In the HIP1r-deficient background, we observed accelerated development of the two abnormal adult onset traits associated with Hip1 mutations. Hip1r−/−; Hip1null/null mice had reduced body mass (Fig. 7A) and a hunched posture (Fig. 7B) resulting from severe spinal deformities that could easily be observed on X-ray images (Fig. 7C, 14-week-old female, and Fig. 7D, 6-week-old male) compared to their littermates. The dwarfism was less apparent at weaning but was obvious in all of the double-mutant mice that survived into early adulthood (Fig. 7A). Histologic analysis of brain, lungs, liver, intestine, pancreas, kidneys, and spleen displayed no morphological abnormalities despite spontaneous death of two of the generated knockout mice at 3 months of age.
FIG. 7.
Dwarfism and kypholordosis in Hip1r−/−; Hip1null/null mice. (A) Growth curve of female Hip1r/Hip1 mutant mice. Hip1r−/−; Hip1null/null mice are shown (n = 4) in comparison to all other female littermates that had at least one normal Hip1r or Hip1 allele (Hip1r+/−; Hip1+/null, Hip1r+/−; Hip1null/null, Hip1r−/−; Hip1+/null, and Hip1r−/−; Hip1+/+) as indicated (n = 11). The male double-mutant mice (n = 3) were also obviously dwarfed compared to their littermate controls (n = 8) (data not shown). (B) Representative photograph of two 13-week-old female littermates. Note the reduced body mass and kypholordosis of the Hip1r−/−; Hip1null/null mouse (bottom) compared to its Hip1r−/−; Hip1+/null littermate (top). (C) Radiograph of 14-week-old female littermates. The Hip1r−/−; Hip1null/null mouse (lower panel) show the severe spinal deformity compared to its Hip1r−/−; Hip1+/+ littermate. (D) Radiographs of 6-week-old male littermates. The Hip1r−/−; Hip1null/null mouse (lower panel) show the severe spinal deformity compared to its Hip1r−/−; Hip1+/null littermate.
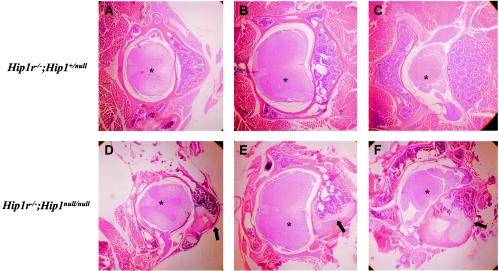
The development of kypholordosis in the absence of both HIP1 and HIP1r was dramatically accelerated compared to the 4-month onset in HIP1-deficient mice; Hip1r−/−; Hip1null/null mice exhibited noticeable spinal deformities as early as 2 weeks of age, and all exhibited the deformity by weaning at 3 weeks of age. Hematoxylin and eosin staining of decalcified paraffin-embedded spine sections made from Hip1r−/−; Hip1null/null and Hip1r−/−; Hip1+/null littermates at 13 weeks of age demonstrated skeletal disorganization in the Hip1r−/−; Hip1null/null mouse in both the thoracic (Fig. 8D) and lumbar (Fig. 8E and F) regions. In contrast, the spine histology of the Hip1r−/−;Hip1+/null littermate was normal (Fig. 8A to C). The vertebral bodies of the Hip1r−/−; Hip1null/null mouse displayed asymmetry with encroachment of cartilage into regions normally occupied by bone marrow (Fig. 8D to F, arrows). Similar to the HIP1-deficient mice, no apparent histological abnormalities in the spinal cord were observed in the double-knockout spine (Fig. 8) (17, 19a).
FIG. 8.
Histology of spinal deformity in Hip1r−/−; Hip1null/null mice. (A to C) Hematoxylin and eosin staining of Hip1r−/−; Hip1+/null decalcified spinal cross-sections at 13 weeks of age. (D to F) Hematoxylin and eosin staining of Hip1r−/−; Hip1null/null decalcified spinal cross-sections at 13 weeks of age showing skeletal disorganization (arrows). Thoracic (A and D) and lumbar (B, C, E, and F) sections are shown. Note the abnormal asymmetric vertebral body with cartilage encroaching the bone marrow (arrows) and normal spinal cord (✽).
DISCUSSION
In the present study, we examined the consequences of Hip1r deletion in the mouse. Northern and Western blot analysis indicated that there was no expression of the HIP1r mRNA or protein in Hip1r−/− mice. By 1 year of age, mice lacking HIP1r showed no gross abnormalities. Histological analysis of multiple tissues, including the skin, which has high HIP1r levels, did not show differences in the development or organization of major organs. Furthermore, analysis of cells derived from Hip1r−/− mice showed normal responses in the regulation of growth factor receptor levels upon ligand stimulation and receptor-mediated endocytosis. Mice with deletion of both HIP1 and HIP1r exhibited accelerated onset of spinal abnormalities and dwarfism compared to HIP1-deficient mice.
The lack of an apparent phenotype in Hip1r−/− mice compared to a complex phenotype in the Hip1 knockout mice highlights the idea that, although HIP1r and HIP1 share many functional domains, they may have distinct cellular functions. Mutation of Hip1 has shown a role for the HIP1 protein in the normal development of spermatogenic progenitors (21), as well as some primitive hematopoietic progenitors and possibly the eye (19a). Furthermore, absence of HIP1 can result in a spinal phenotype by 4 months of age associated with gait ataxia and tremor (17, 19a). An obvious interpretation of the differences seen between the Hip1 and Hip1r knockout mice is that HIP1 or other functionally related proteins are able to compensate for the loss of HIP1r, resulting in normal development and endocytic functions. In contrast, in the Hip1 knockouts, HIP1r and/or other proteins may not compensate for the loss of HIP1 in all tissues that normally express HIP1, resulting in the complex abnormalities observed in these knockout mice. In support of this compensation hypothesis, deletion of both Hip1 and Hip1r results in a more severe phenotype than Hip1 deletion alone, with dwarfism and kypholordosis occurring very early in adult development. This indicated that, although HIP1r cannot fully compensate for absence of HIP1 as seen by the complex phenotype of the Hip1−/− mouse, partial functional compensation by HIP1r is indeed present.
We have found that overexpression of HIP1 or HIP1r in a transient system results in a prolonged half-life of growth factor receptors such as the EGFR and PDGF β R after ligand stimulation (10). Conversely, transfection of a dominant-interfering mutant lacking the ANTH domain resulted in destabilization of growth factor receptors. Using similar ligand stimulation assays of MEFs and keratinocytes derived from Hip1r−/− mice, we found that absence of HIP1r does not significantly affect the half-life of the EGFR or PDGF β R after ligand stimulation. Since HIP1 and HIP1r are able to heterodimerize, it is possible that the dominant-interfering mutants disrupt the functions of both HIP1 and HIP1r or other proteins with which they associate. Other interpretations would be that HIP1r, although participating in endocytic function, is not absolutely necessary for endocytosis or that compensatory cellular pathways take over in the chronic Hip1r knockout milieu. Consistent with the lack of effect of HIP1r deficiency on growth factor receptor half-life, we show here that internalization of neither EGF nor transferrin was disrupted in Hip1r−/− MEFs. This indicates that HIP1r is not required for the internalization phase of either regulated or constitutive endocytosis as measured by EGF or transferrin uptake, respectively. It has recently been shown that Sla2p is required for the productive conversion of endocytic membrane patches to invaginations and vesicles (12). Actin and endocytic proteins are still able to associate in yeast with deletion of Sla2p, suggesting that Sla2p may regulate, rather than enable, these interactions (12). Whether HIP1 and/or HIP1r play similar roles in mammalian cells remains to be determined. Continuing these studies utilizing Hip1r−/−; Hip1null/null MEFs, keratinocytes, and tissues will help to elucidate the functional importance of the HIP1 proteins in clathrin-mediated vesicle trafficking. Further work analyzing the mechanism of phenotypic acceleration in the Hip1r/Hip1 double-knockout mice, conditional knockout mice, and primary cell lines derived from the various mutant murine derived tissues will help to establish the physiologic role of the HIP1 family of proteins in endocytic processes, as well as other functions of which we are currently unaware.
Supplementary Material
Acknowledgments
We thank Incyte Genomics for assistance with mapping and construction of the targeting vector and the University of Michigan transgenic animal core for assistance with blastocyst injections and ES cell growth. We also thank members of the Ross lab for continual scientific input and comments on the manuscript.
This study was supported by NIH grants RO1 CA82363-01A1 and RO1 CA098730-02 (T.S.R). T.S.H. is supported by the University of Michigan Cancer Biology training grant, and T.S.R. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (award DRS-22).
Footnotes
Supplemented material for this article may be found at http://mcb.asm.org
REFERENCES
- 1.Anonymous. 2004. Mouse genome database. Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine. [Online.] http://www.informatics.jax.org.
- 2.Chopra, V. S., M. Metzler, D. M. Rasper, A. E. Engqvist-Goldstein, R. Singaraja, L. Gan, K. M. Fichter, K. McCutcheon, D. Drubin, D. W. Nicholson, and M. R. Hayden. 2000. HIP12 is a non-proapoptotic member of a gene family including HIP1, an interacting protein with Huntingtin. Mamm. Genome 11: 1006-1015. [DOI] [PubMed] [Google Scholar]
- 3.Dlugosz, A. A., A. B. Glick, T. Tennenbaum, W. C. Weinberg, and S. H. Yuspa. 1995. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 254: 3-20. [DOI] [PubMed] [Google Scholar]
- 4.Engqvist-Goldstein, A. E., M. M. Kessels, V. S. Chopra, M. R. Hayden, and D. G. Drubin. 1999. An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J. Cell Biol. 147: 1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engqvist-Goldstein, A. E., R. A. Warren, M. M. Kessels, J. H. Keen, J. Heuser, and D. G. Drubin. 2001. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J. Cell Biol. 154: 1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ford, M. G., I. G. Mills, B. J. Peter, Y. Vallis, G. J. Praefcke, P. R. Evans, and H. T. McMahon. 2002. Curvature of clathrin-coated pits driven by epsin. Nature 419: 361-366. [DOI] [PubMed] [Google Scholar]
- 7.Ford, M. G., B. M. Pearse, M. K. Higgins, Y. Vallis, D. J. Owen, A. Gibson, C. R. Hopkins, P. R. Evans, and H. T. McMahon. 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291: 1051-1055. [DOI] [PubMed] [Google Scholar]
- 8.Hirst, J., A. Motley, K. Harasaki, S. Y. Peak Chew, and M. S. Robinson. 2003. EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol. Biol. Cell 14: 625-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtzman, D. A., S. Yang, and D. G. Drubin. 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae . J. Cell Biol. 122: 635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyun, T. S., D. S. Rao, D. Saint-Dic, L. Evan Michael, P. D. Kumar, S. V. Bradley, I. F. Mizukami, K. I. Oravecz-Wilson, and T. S. Ross. 2004. HIP1 and HIP1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via ENTH domains. J. Biol. Chem. 279: 14294-19306. [DOI] [PubMed]
- 11.Itoh, T., S. Koshiba, T. Kigawa, A. Kikuchi, S. Yokoyama, and T. Takenawa. 2001. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291: 1047-1051. [DOI] [PubMed] [Google Scholar]
- 12.Kaksonen, M., Y. Sun, and D. G. Drubin. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115: 475-487. [DOI] [PubMed] [Google Scholar]
- 13.Kalthoff, C., S. Groos, R. Kohl, S. Mahrhold, and E. J. Ungewickell. 2002. Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol. Biol. Cell 13: 4060-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman, M. H. 1992. The atlas of mouse development, p. 387. Academic Press, Ltd., London, United Kingdom.
- 15.Legendre-Guillemin, V., M. Metzler, M. Charbonneau, L. Gan, V. Chopra, J. Philie, M. R. Hayden, and P. S. McPherson. 2002. HIP1 and HIP12 display differential binding to F-actin, AP2, and clathrin: identification of a novel interaction with clathrin-light chain. J. Biol. Chem. 277: 19897-19904. [DOI] [PubMed] [Google Scholar]
- 16.Metzler, M., V. Legendre-Guillemin, L. Gan, V. Chopra, A. Kwok, P. S. McPherson, and M. R. Hayden. 2001. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J. Biol. Chem. 276: 39271-39276. [DOI] [PubMed] [Google Scholar]
- 17.Metzler, M., B. Li, L. Gan, J. Georgiou, C. A. Gutekunst, Y. Wang, E. Torre, R. S. Devon, R. Oh, V. Legendre-Guillemin, M. Rich, C. Alvarez, M. Gertsenstein, P. S. McPherson, A. Nagy, Y. T. Wang, J. C. Roder, L. A. Raymond, and M. R. Hayden. 2003. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 22: 3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills, I. G., G. J. Praefcke, Y. Vallis, B. J. Peter, L. E. Olesen, J. L. Gallop, P. J. Butler, P. R. Evans, and H. T. McMahon. 2003. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 160: 213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra, S. K., N. R. Agostinelli, T. J. Brett, I. Mizukami, T. S. Ross, and L. M. Traub. 2001. Clathrin- and AP-2-binding sites in HIP1 uncover a general assembly role for endocytic accessory proteins. J. Biol. Chem. 276: 46230-46236. [DOI] [PubMed] [Google Scholar]
- 19a.Oravecz-Wilson, K. I., M. J. Kiel, L. Li, D. S. Rao, D. Saint Dic, P. D. Kumar, M. M. Provot, K. D. Hankenson, V. N. Reddy, A. Lieberman, S. S. Morrison, and T. S. Ross. 2004. Huntingtin interacting protein 1 mutations lead to abnormal hematopoiesis, spinal defects and cataracts Hum. Mol. Genet. 13: 851-867. [DOI] [PubMed] [Google Scholar]
- 20.Rao, D. S., S. V. Bradley, P. D. Kumar, T. S. Hyun, D. Saint-Dic, K. Oravecz-Wilson, C. G. Kleer, and T. S. Ross. 2003. Altered receptor trafficking in Huntingtin interacting protein 1-transformed cells. Cancer Cell 3: 471-482. [DOI] [PubMed] [Google Scholar]
- 21.Rao, D. S., J. C. Chang, P. D. Kumar, I. Mizukami, G. M. Smithson, S. V. Bradley, A. F. Parlow, and T. S. Ross. 2001. Huntingtin interacting protein 1 Is a clathrin coat binding protein required for differentiation of late spermatogenic progenitors. Mol. Cell. Biol. 21: 7796-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao, D. S., T. S. Hyun, P. D. Kumar, I. F. Mizukami, M. A. Rubin, P. C. Lucas, M. G. Sanda, and T. S. Ross. 2002. Huntingtin-interacting protein 1 is overexpressed in prostate and colon cancer and is critical for cellular survival. J. Clin. Investig. 110: 351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rees, D. J., S. E. Ades, S. J. Singer, and R. O. Hynes. 1990. Sequence and domain structure of talin. Nature 347: 685-689. [DOI] [PubMed] [Google Scholar]
- 24.Seki, N., M. Muramatsu, S. Sugano, Y. Suzuki, A. Nakagawara, M. Ohhira, A. Hayashi, T. Hori, and T. Saito. 1998. Cloning, expression analysis, and chromosomal localization of HIP1R, an isolog of huntingtin interacting protein (HIP1). J. Hum. Genet. 43: 268-271. [DOI] [PubMed] [Google Scholar]
- 25.Sibilia, M., A. Fleischmann, A. Behrens, L. Stingl, J. Carroll, F. M. Watt, J. Schlessinger, and E. F. Wagner. 2000. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102: 211-220. [DOI] [PubMed] [Google Scholar]
- 26.Stahelin, R. V., F. Long, B. J. Peter, D. Murray, P. De Camilli, H. T. McMahon, and W. Cho. 2003. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 278: 28993-28999. [DOI] [PubMed] [Google Scholar]
- 27.Todaro, G., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17: 299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waelter, S., E. Scherzinger, R. Hasenbank, E. Nordhoff, R. Lurz, H. Goehler, C. Gauss, K. Sathasivam, G. P. Bates, H. Lehrach, and E. E. Wanker. 2001. The Huntingtin interacting protein HIP1 is a clathrin and α -adaptin-binding protein involved in receptor-mediated endocytosis. Hum. Mol. Genet. 10: 1807-1817. [DOI] [PubMed] [Google Scholar]
- 29.Wasiak, S., V. Legendre-Guillemin, R. Puertollano, F. Blondeau, M. Girard, E. de Heuvel, D. Boismenu, A. W. Bell, J. S. Bonifacino, and P. S. McPherson. 2002. Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 158: 855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wesp, A., L. Hicke, J. Palecek, R. Lombardi, T. Aust, A. L. Munn, and H. Riezman. 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae . Mol. Biol. Cell 8: 2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann, E. M., L. Li, Y. T. Hou, M. Cannon, G. M. Christman, and K. N. Bitar. 1997. IGF-I induces collagen and IGFBP-5 mRNA in rat intestinal smooth muscle. Am. J. Physiol. 273: G875-G882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.