Abstract
The specificity of signaling through mitogen-activated protein kinase pathways has been attributed to both the control of intensity and duration of signaling and the actions of protein scaffolds. Here we demonstrate that the molecular scaffold KSR1 regulates the intensity and duration of ERK activation to modulate a cell's proliferative and oncogenic potential. Deletion of KSR1 eliminates the prolonged phase of ERK activation induced by platelet-derived growth factor and blocks RasV12-induced transformation. The introduction of KSR1 into KSR1−/− mouse embryo fibroblasts causes a concentration-dependent increase in signaling and transformation, to a maximum at 14 times the wild-type KSR1 expression levels, but inhibits these responses at higher expression levels. An increase in KSR1 expression to levels that are optimal for signaling leads to a threefold increase in proliferative capacity and is coincident with the level of KSR1 expression that maximally associates with all members of the Raf/MEK/ERK cascade. These data reveal that cells contain a reserve proliferative capacity that is accessible by the optimal expression of a noncatalytic signaling component and that altering the expression level of a molecular scaffold can modulate the actions of growth factors and oncogenes.
Mitogen-activated protein (MAP) kinase pathways are implicated in the control of multiple aspects of cell fate, including senescence, proliferation, transformation, differentiation, and apoptosis (7, 30, 34, 47). While the components of these ubiquitous pathways are well established, the means by which they direct cell fate remain unclear. Recently, two ideas for the control of signal outputs from MAP kinase pathways have emerged: the duration and intensity of MAP kinase activation can dictate which targets become phosphorylated (27) and scaffold proteins organize specific MAP kinase cascades to ensure signal specificity (5, 34, 47).
The intensity and duration of ERK activation are critical determinants of ERK's ability to modulate a diverse array of cellular processes (16, 22, 27, 30, 32). In PC12 cells, epidermal growth factor (EGF) induces a transient activation of ERK in the cytoplasm, leading to proliferation. Conversely, nerve growth factor (NGF) induces both a prolonged activation of ERK and the translocation of ERK to the nucleus, leading to differentiation into neurons (22). In mammalian fibroblasts, treatment with EGF induces a transient activation of ERK, whereas treatment with platelet-derived growth factor (PDGF) causes a sustained activation of ERK. This sustained activation leads to phosphorylation of the immediate-early protein c-Fos and to cell cycle progression (16, 27).
Scaffold proteins organize signaling components to determine specificity within MAP kinase cascades (34, 47). Protein scaffolds of kinase cascades can have varied effects on signaling through their pathways. The prototypic MAP kinase scaffold Ste5 is essential for the mating pheromone pathway of budding yeast (11). InaD, a scaffold for Drosophila photoreceptor signaling, is not required, but it greatly enhances the amplitude and kinetics of signaling in this pathway (35).
For mammalian cells, several putative scaffolds for MAP kinase cascades have been identified, including JIP-1 and JIP-2 (10, 46, 50), MP1 (33), and KSR1 (24, 28, 41). While each scaffold has been proposed to enhance signaling through the c-Jun N-terminal kinase, ERK1, or ERK1/2 pathway, respectively, each has been shown to inhibit signaling when it is overexpressed (8, 10, 16, 33, 52). The biological consequences of overexpression of these proteins are consistent with their proposed role as scaffold proteins, since molecular scaffolds of kinase cascades are predicted to affect signal output in a concentration-dependent manner, facilitating signaling to an optimal concentration and inhibiting signaling beyond this optimum (5, 12, 15, 20). KSR1 is a scaffold for the Raf/MEK/ERK kinase cascade (24, 28, 31) and has been proposed to act in such a manner, as low levels of expression lead to increased pathway activity (2, 6, 23, 25, 28, 29, 41, 48) and high levels of expression inhibit signaling through the pathway (4, 6, 8, 17, 37, 52).
Here we examine the role of the molecular scaffold KSR1 in facilitating the intensity and duration of ERK activation to affect cell proliferation and oncogenic transformation. We show that the loss of KSR1 reduces growth factor-induced ERK activation and that the reintroduction of KSR1 into KSR1−/− mouse embryo fibroblasts (MEFs) rescues this deficit in a dose-dependent manner. Furthermore, KSR1 is necessary and sufficient for RasV12-induced transformation, with a dose dependence similar to that seen for signaling. When KSR1 levels are optimized, the cellular response to EGF is converted to a PDGF-like response, including the promotion of prolonged ERK activation and the exit of quiescent cells from G0 into S phase. Optimal levels of KSR1 expression also lead to a threefold increase in the proliferative capacity. These data indicate that altering the expression level of a molecular scaffold can modulate the actions of growth factors and oncogenes.
MATERIALS AND METHODS
Cell culture.
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 0.1 mM minimum essential medium with nonessential amino acids, and 1% penicillin-streptomycin. Cells were incubated at 37°C in 5% CO2.
Construction and production of recombinant retroviruses.
KSR1-FLAG was subcloned from the EcoRI and SalI sites of pCMV5 (17) into the EcoRI and XhoI sites of MSCV-IRES-GFP. An MSCV-IRES-GFP, MSCV-KSR1-IRES-GFP, pBabePuroRasV12, or pBabePuro retroviral vector was cotransfected with an ecotropic packaging vector into 293T cells. At 48 to 72 h posttransfection, viral supernatants were collected and filtered. Viral supernatants were then either stored at −80°C or used immediately to infect cells. Puromycin-resistant cells were selected with 4 μg of puromycin (Sigma)/ml.
Generation of cell lines.
Nonimmortalized MEFs were generated from 13.5-day KSR1−/− and KSR1+/+ embryos as previously described (28). Cells were maintained in culture according to a 3T9 protocol (42) until immortalized populations of cells emerged. For the production of cells with increasing KSR1 expression levels, KSR1−/− or KSR1+/+ MEFs were infected with KSR1-IRES-GFP or the MCSV-IRES-GFP control vector. Fluorescence was detected by flow cytometry and cells were separated according to increasing levels of fluorescence. Cells were excited at 488 nm and separated at 530/20 nm, with the baseline of fluorescence of uninfected cells having a mean intensity of 6 (range, 0 to 15). Postsorted cells were assessed for purity by fluorescence-activated cell sorting (FACS) analysis. Collected pools of cells were grown in culture and assessed for their KSR1 expression level by Western blotting.
Western blot analysis.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Western blots were developed by use of an anti-FLAG M2 (Sigma; 1:3,000), anti-KSR1 (Transduction Laboratories; 1:500), anti-c-Raf (Santa Cruz; 1:1,000), anti-MEK1 (Transduction Laboratories; 1:1,000), anti-MEK2 (Transduction Laboratories; 1:2,500), anti-ERK1 (Santa Cruz; 1:1,000), anti-ERK2 (Santa Cruz; 1:1,000), anti-phospho-ERK1/2 (Cell Signaling; 1:1,000), anti-Ras (Oncogene; 1:1,000), anti-AKT (Cell Signaling; 1:1,000), anti-phospho-AKT (Ser308 [Cell Signaling; 1:1,000] or Thr473 [Cell Signaling; 1:1,000]), anti-Jun (Cell Signaling; 1:1,000), or anti-phospho-Jun (Cell Signaling; 1:1,000) antibody. Anti-mouse and anti-rabbit secondary antibodies conjugated to Alexa Fluor 680 (Molecular Probes; 1:3,000) and IRDye800 (Rockland; 1:3,000), respectively, were used to probe primary antibodies. Protein bands were detected and quantified by Western blotting with the Odyssey system (Li-Cor).
Immunoprecipitation studies.
Cycling cells (70 to 80% confluent) were lysed in either NP-40 lysis buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 mM sodium vanadate, 10 mg of leupeptin/ml, 5 mg of aprotinin/ml) or Empigen lysis buffer (40 mM Tris [pH 7.4], 120 mM NaCl, 0.1% Empigen BB, 10 mM sodium pyrophosphate, 2 mM EGTA, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 mg of leupeptin/ml, 5 mg of aprotinin/ml), and lysates were assayed for protein concentration by the DC protein assay (Bio-Rad). The total protein (1.5 mg) was immunoprecipitated three times with 60 μl of a 1:1 slurry of anti-FLAG-conjugated agarose in phosphate-buffered saline (PBS) (Sigma). Cleared lysates were probed with an anti-KSR1 antibody to ensure that all KSR1 complexes were precipitated.
In-cell Western blotting.
Cells were plated 24 h prior to assay in black-walled 96-well plates so they would be 70 to 80% confluent at the time of the assay. Cells were starved of serum for 4 h in DMEM and then stimulated with either 100 ng of EGF/ml or 25 ng of PDGF/ml in DMEM-1% bovine serum albumin for the indicated times. For termination of the reactions, plates were fixed with 3.7% formaldehyde for 20 min. Cells were then permeabilized by four 5-min washes with 0.1% Triton X-100 and were blocked with Li-Cor Odyssey blocking buffer for 1 h. Anti-pERK1/2 (Cell Signaling no. 9106; 1:100) and anti-ERK1 (Santa Cruz no. sc-93; 1:100) primary antibodies were diluted in a 1:1 mixture of PBS and Li-Cor Odyssey blocking buffer and incubated for 2 h or overnight. Plates were then rinsed three times with PBS-0.1% Tween and incubated with anti-mouse Alexa Fluor 680 (Molecular Probes; 1:100)-conjugated and anti-rabbit IRDye800 (Rockland; 1:100)-conjugated secondary antibodies diluted in a 1:1 mixture of PBS and Li-Cor Odyssey blocking buffer for 1 h. Plates were washed three times with PBS-0.1% Tween, followed by analysis with the Li-Cor Odyssey system.
Proliferation studies.
Cells were seeded at 4 × 104 cells per 35-mm-diameter dish. Triplicate dishes were counted 3 h after seeding to account for plating discrepancies and were then assessed every 24 h for total cell number on a Beckman Coulter counter.
Transformation assays.
MEFs stably expressing RasV12 and KSR1 or control vectors were seeded in 0.32% Nobel agar at 5 × 103 cells per 35-mm-diameter dish to assess anchorage-independent growth or at 105 cells per 10-cm-diameter dish to assess the loss of contact inhibition. Colonies were counted, photomicrographs were taken, and dishes were stained with Wright-Giemsa stain 21 to 28 days after seeding.
BrdU assays.
MEFs were seeded at 20% confluence on either acid-washed coverslips or eight-well chamber slides in DMEM-10% fetal bovine serum (FBS). Four hours later, cells were washed twice with PBS, starved of serum for 72 h, and stimulated with either 100 ng of EGF/ml or 25 ng of PDGF/ml in DMEM-1% bovine serum albumin and 10 μM BrdU (BD Biosciences). A mouse phycoerythrin (PE)-conjugated anti-BrdU antibody (BD Biosciences) was used to detect BrdU incorporation. Cells were counterstained with Hoechst dye to stain nuclei.
RESULTS
KSR1 regulates the time course of ERK activation by growth factors.
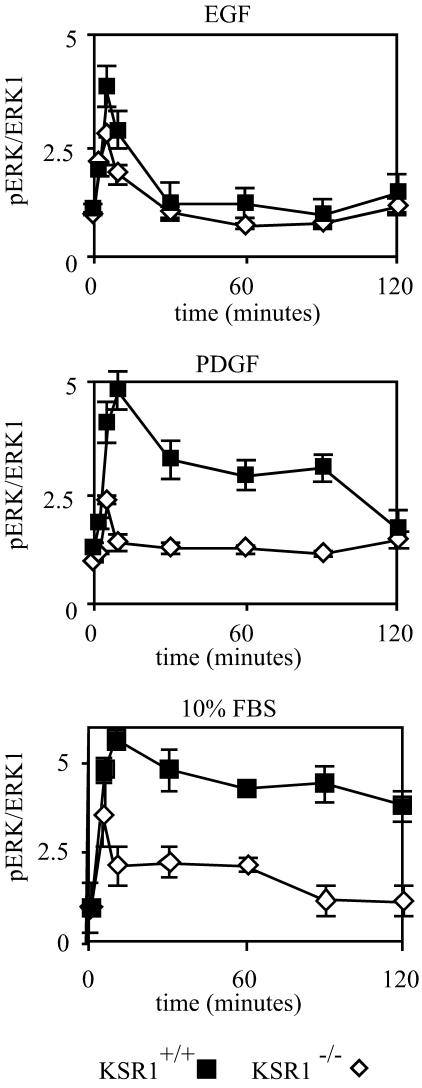
It was previously reported that KSR1 is necessary for the maximal activation of ERKs (28). To further characterize the effect of KSR1 on ERK signaling, we immortalized KSR1−/− and KSR1+/+ MEFs by using a 3T9 protocol to generate stable cell lines. KSR1−/− MEFs show a 50% reduction in EGF-induced ERK activation versus KSR1+/+ MEFs after 5 min of treatment over a range of EGF concentrations (1 to 100 ng/ml), with maximal ERK activation in both cell lines occurring at and above 18 ng/ml (data not shown). The time course of ERK activation by EGF, PDGF, and 10% FBS was assessed. KSR1−/− MEFs showed a decreased maximal activation by EGF, PDGF, and FBS (Fig. 1). Furthermore, the loss of KSR1 resulted in the inhibition of signaling at all time points of stimulation (Fig. 1). Unlike treatment with EGF, which caused only a transient activation of ERK, treatment with PDGF resulted in a sustained activation of ERK (27). KSR1 was necessary for the prolonged ERK activation induced by PDGF and strongly enhanced the prolonged activation of ERK by FBS (Fig. 1).
FIG. 1.
KSR1 controls the intensity and duration of signaling through the ERK kinase cascade. KSR1−/− (diamonds) and KSR1+/+ (squares) MEFs were treated with 100 ng of EGF/ml, 25 ng of PDGF/ml, or 10% FBS for the indicated times, and ERK1/2 phosphorylation levels were determined by in-cell Western blotting for ERK1 and pERK1/2 with a Li-Cor Odyssey system. Data are expressed as ratios of pERK1/2 to ERK1. Standard errors are ±10%.
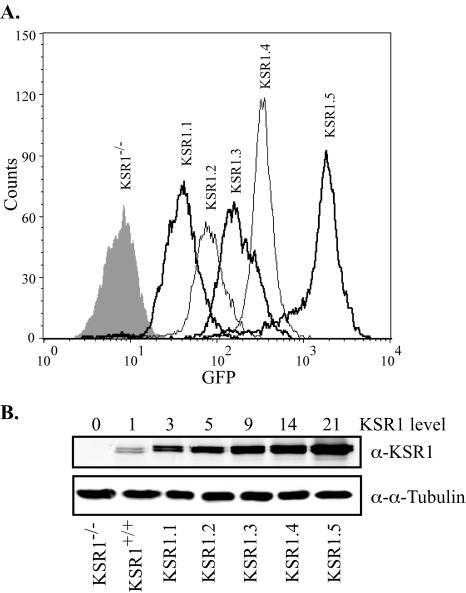
To characterize the biological effects of altered cellular KSR1 concentrations, we generated KSR1−/− MEFs expressing different levels of KSR1 by the introduction of a bicistronic vector encoding KSR1 and green fluorescent protein (GFP) and isolated them by flow cytometry. Cells were gated to generate five populations of cells with increasing GFP expression levels (Fig. 2A). A comparison of KSR1 levels within each population to that of KSR1+/+ MEFs by Western blotting revealed a range of KSR1 expression levels, facilitating an analysis of the KSR1 function in a concentration-dependent manner (Fig. 2B).
FIG. 2.
Generation of KSR1−/− MEFs expressing increasing levels of KSR1. KSR1−/− MEFs were infected with a bicistronic retrovirus encoding KSR1 and GFP or GFP only (control). (A) Infected cells were then subjected to FACS analysis and separated for increasing levels of fluorescence at 530/20 nm. Postsorted cells were assessed for purity by FACS analysis. (B) Collected pools of cells were maintained in culture. Whole-cell lysates containing equivalent amounts of total protein were assessed for KSR1 and α-tubulin expression levels by Western blotting. KSR1 expression levels were quantified with a Li-Cor Odyssey system. Numbers above each lane indicate expression levels compared to that of KSR1+/+ MEFs.
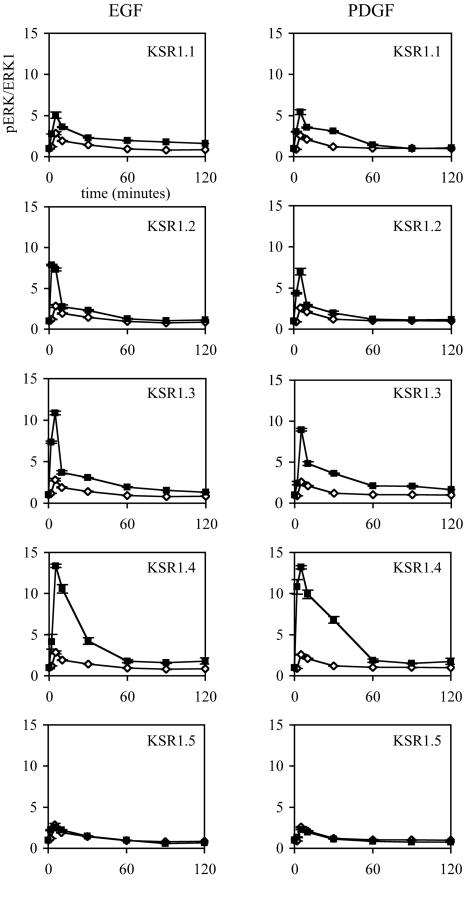
To characterize the role of the KSR1 expression level on signaling through the Raf/MEK/ERK kinase cascade, we assessed the time course of ERK activation in KSR1−/− cells expressing increasing levels of KSR1 (KSR1.1 to KSR1.5) and in vector control cells (GFP). Cells expressing low levels of KSR1 (KSR1.1) showed an increase in both EGF- and PDGF-induced ERK activation compared to control cells (Fig. 3, top panels), confirming that KSR1 can rescue the diminution of signaling seen in KSR1−/− cells. Increasing the level of KSR1 expression increased both the peak activation (Fig. 3) and the duration of activation induced by PDGF and EGF to a maximum in KSR1.4 cells (Fig. 3). Increasing KSR1 expression beyond this level led to a reduction in signaling to levels at or below that of KSR1−/− MEFs (Fig. 3, bottom panels).
FIG. 3.
Expression of KSR1 at 5 to 14 times the endogenous level optimizes ERK activation in KSR1−/− cells expressing KSR1. KSR1−/− MEFs expressing KSR1 at increasing levels (squares) or GFP-only control cells (diamonds) were treated with 100 ng of EGF/ml or 25 ng of PDGF/ml for the indicated times, and ERK1/2 phosphorylation levels were determined as described for Fig. 1. Standard errors are ±10%. The same GFP-only graph is given for comparison at each KSR1 level.
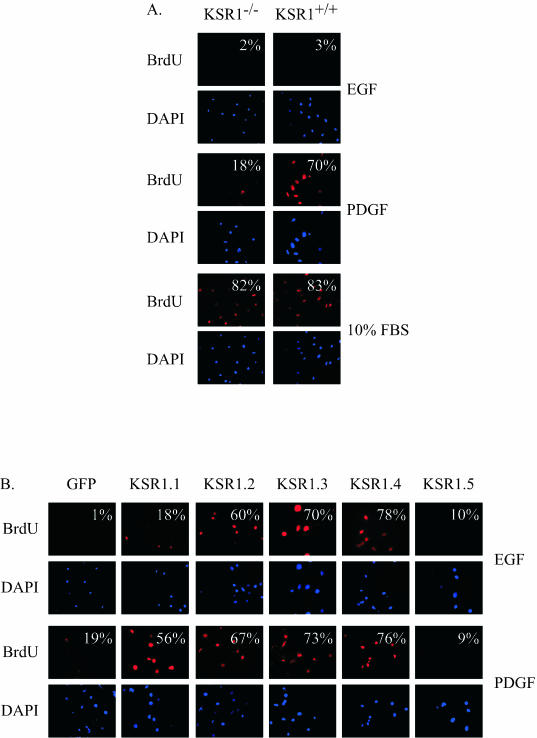
One consequence of prolonged ERK activation is the induction of immediate-early gene products and cell cycle progression (27). We predicted that the loss of KSR1 would impair the ability of PDGF to stimulate cell cycle progression and DNA synthesis. Furthermore, since the reintroduction of KSR1 at optimal concentrations caused a prolonged ERK response to PDGF and EGF, we hypothesized that optimal levels of KSR1 expression may allow EGF to stimulate cell cycle progression. EGF and PDGF were compared to FBS for promoting cell cycle progression and DNA synthesis in KSR1−/− and KSR1+/+ MEFs (Fig. 4A). Ten percent FBS stimulated BrdU incorporation to an equal extent (>80%) in KSR1−/− and KSR1+/+ MEFs. PDGF stimulated BrdU incorporation in 70% of KSR1+/+ MEFs (Fig. 4A). KSR1−/− MEFs had an attenuated response to PDGF treatment, with only 18% of cells incorporating BrdU (Fig. 4A). Unlike PDGF, EGF normally causes a transient ERK activation and is not able to promote cell cycle progression (16, 27) (Fig. 1 and 4A).
FIG. 4.
KSR1 expression controls growth factor-mediated DNA synthesis in quiescent cells. (A) Quiescent KSR1−/− and KSR1+/+ MEFs were treated with 100 ng of EGF/ml, 25 ng of PDGF/ml, or 10% FBS for 20 h and then assessed for BrdU incorporation as described in Materials and Methods. The percentage of BrdU-positive cells versus the total number of stained nuclei is indicated in the upper right corner of each panel. Standard errors are ±20%. (B) Quiescent KSR1−/− MEFs expressing KSR1 at increasing levels or expressing GFP were treated with 100 ng of EGF/ml or 25 ng of PDGF/ml for 20 h, and BrdU incorporation was assessed as for panel A. Standard errors are ±20%.
The reintroduction of KSR1 into KSR1−/− MEFs rescued the deficit in PDGF-stimulated BrdU incorporation in a dose-dependent manner, with maximal BrdU incorporation occurring in KSR1.4 cells (Fig. 4B). Higher levels of expression (KSR1.5) inhibited BrdU incorporation in response to PDGF treatment (Fig. 4B), consistent with the inhibition of ERK activation (Fig. 3). However, optimal levels of KSR1 expression conferred a PDGF-like responsiveness to EGF. The introduction of KSR1 into KSR1−/− MEFs at 5 to 14 times the wild-type expression level (KSR1.2, KSR1.3, and KSR1.4 cells) caused a prolonged ERK activation in response to EGF and allowed EGF to promote DNA synthesis (Fig. 3 and 4B). Thus, KSR1 is necessary for the normal control of growth factor-induced signaling, and the alteration of cellular KSR1 levels can modulate a cell's responsiveness to growth factors.
KSR1 expression regulates the proliferative capacity of MEFs.
As KSR1 expression differentially modulates the ability of growth factor treatment to promote S-phase entry, the growth characteristics of KSR1−/− and KSR1+/+ MEFs were evaluated. Surprisingly, there was no difference in the proliferation of KSR1−/− and KSR1+/+ MEFs in 10% serum (Fig. 5A) even though serum-induced ERK activation was attenuated in KSR1−/− MEFs (Fig. 1). These data indicate that the intensity and duration of serum-stimulated ERK activation in KSR1−/− MEFs are sufficient to sustain normal cell proliferation.
FIG. 5.
KSR1 expression level regulates proliferative capacity of MEFs. MEFs were seeded at 4 × 104 cells per 35-mm-diameter dish in 10% FBS and DMEM. Separate triplicate dishes were assessed for cell number every 24 h on a Beckman Coulter counter. (A) Proliferation in KSR1−/− (diamonds) and KSR1+/+ (squares) MEFs. (B) Proliferation in KSR1−/− MEFs expressing KSR1 at increasing levels or expressing GFP. Closed squares, KSR1.1; open circles, KSR1.2; closed circles, KSR1.3; open triangles, KSR1.4; closed triangles, KSR1.5; open diamonds, GFP. (C) Proliferation in KSR1+/+ MEFs expressing KSR1 at increasing levels. Closed squares, WT0.1; open circles, WT.2; closed circles, WT.3; open triangles, WT.4; closed triangles, WT.5; open diamonds, GFP (WT.0).
The increase in ERK activation caused by optimal KSR1 levels suggested that optimal levels of KSR1 expression would increase the proliferative capacity of cells. There was no increase in the growth rate for KSR1.1 cells compared to GFP controls, confirming that low levels of KSR1 expression do not enhance cell proliferation (Fig. 5B). Increasing the level of KSR1 expression, however, did increase both cell proliferation and the maximal cell density. The expression of KSR1 at five times the wild-type level (KSR1.2) enhanced cell proliferation by 25%. KSR1 expression at 9 (KSR1.3) or 14 (KSR1.4) times the wild-type level enhanced cell proliferation by 200%, but the expression of KSR at higher levels (KSR1.5) reduced the proliferative rate to that seen in KSR1−/− MEFs (Fig. 5B). Cells expressing KSR1 at levels of >21 times the wild-type expression level could not be maintained in culture, suggesting that higher levels of KSR1 prevent cell growth (data not shown). Despite increased proliferative rates, the expression of optimal levels of KSR1 did not cause oncogenic transformation, as measured by anchorage-independent growth or the loss of contact inhibition (data not shown).
It was possible that the enhanced proliferation observed upon expression of exogenous KSR1 was a consequence of aberrant development in KSR1−/− MEFs. To test this possibility, we determined the proliferation rates of five populations of KSR1+/+ MEFs expressing increasing levels of ectopic KSR1 as for KSR1−/− MEFs. As with KSR1−/− MEFs, low levels (three- to fourfold higher than the endogenous level) of ectopic KSR1 did not increase the proliferative rate or maximal cell density of KSR1+/+ MEFs (Fig. 5C). Increasing the ectopic KSR1 expression levels between 5- and 14-fold in KSR1+/+ MEFs did increase the proliferation rate and cell density, with maximal effects observed between 9- and 14-fold higher levels than the endogenous level of KSR1. Interestingly, the proliferation rate of KSR1+/+ MEFs with levels of KSR1 that were 14-fold higher than the endogenous level was not as high as that observed for the null MEFs. Our observation is consistent with the concept that this level of KSR1, when added to endogenous levels, will cause elevated, but not maximal, proliferation. We passaged KSR1.3 and KSR1.4 MEFs for 1 year without resorting either population. Both populations equilibrated at the same level of ectopic KSR1 expression, which we measured to be 12-fold higher than the level of endogenous KSR1 expressed by KSR1+/+ MEFs (data not shown). Thus, the proliferation rate of KSR1+/+ MEFs expressing KSR1 at 14 times the endogenous level would be expected to be lower than that of KSR1−/− MEFs expressing the same level of ectopic KSR1. These data reveal an excess capacity for cell proliferation that can be accessed in the absence of oncogenic transformation.
KSR1 is required for oncogenic transformation by RasV12.
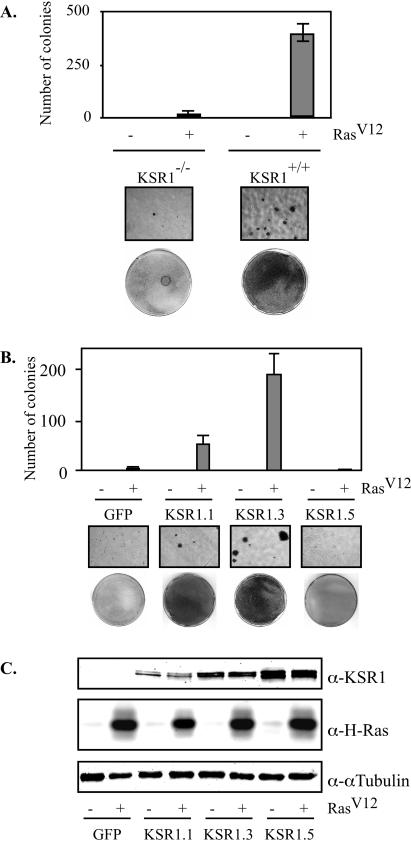
KSR was originally identified in genetic screens of Drosophila melanogaster and Caenorhabditis elegans for genes whose inactivation would revert an activated RasV12 phenotype (19, 38, 40). We tested the oncogenic capacity of RasV12 in mammalian cells lacking KSR1. In comparison to wild-type MEFs, KSR1−/− MEFs showed a 20-fold reduction in RasV12-induced anchorage-independent growth in soft agar (Fig. 6A). Furthermore, colonies detected in KSR1−/− dishes were small (<16 cells) and did not exhibit anchorage-independent growth when transferred to another dish (data not shown). Thus, KSR1 is necessary for RasV12-induced transformation.
FIG. 6.
KSR1 expression regulates oncogenic capacity of RasV12. KSR1−/− and KSR1+/+ MEFs were infected with a recombinant retrovirus encoding RasV12 or a control virus (A). (B) The effect of KSR1 expression level was assessed by infecting KSR1−/− MEFs expressing RasV12 with a bicistronic retrovirus encoding KSR and GFP or GFP only to generate cells with increasing levels of KSR1 expression as described for Fig. 2. Cells were then assessed for transformation by growth on soft agar (top panels), ×10 photomicrographs (middle panels), or focus-forming assays (lower panels) as described in Materials and Methods. (C) Whole-cell lysates used for panel B were assessed for Ras, KSR1, and α-tubulin expression by Western blotting.
To test whether the addition of KSR1 to KSR1−/− MEFs would rescue the loss of transforming capacity, we infected KSR1−/− MEFs expressing RasV12 (Fig. 6A) with a retrovirus encoding the KSR1-IRES-GFP expression vector and sorted them for increasing levels of KSR1 expression as for Fig. 2. A Western blot analysis of KSR1, Ras, and α-tubulin showed that KSR1 expression levels were comparable in RasV12-infected and control cells (Fig. 6C). Cells were then assessed for anchorage-independent growth in soft agar. The expression of KSR1 at low levels (KSR1.1) restored the transforming capacity of KSR1−/− MEFs expressing RasV12 to levels seen in KSR1+/+ MEFs (Fig. 6B). Increasing the KSR1 expression level to nine times the wild-type level (KSR1.3) further enhanced the transforming capacity of RasV12, whereas the expression of KSR1 at higher levels (KSR1.5) completely inhibited transformation, as assessed by the loss of contact inhibition and by growth on soft agar (Fig. 6B). These data indicate that the oncogenic capacity of RasV12 is dependent upon the cellular level of KSR1.
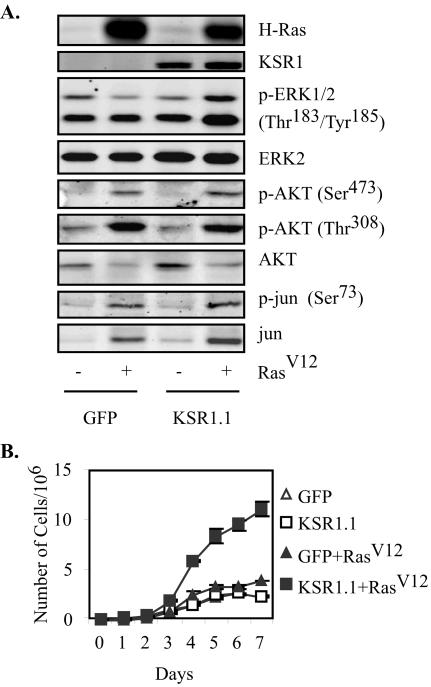
Activated RasV12 causes oncogenic transformation through the induction of multiple effector pathways, including the Raf/MEK/ERK kinase cascade, the phosphatidylinositol 3-kinase (PI 3-kinase)-AKT pathway, and the RalGEF-Ral pathway (14, 18, 43-45, 49). While the RasV12-transformed phenotype uses each of these pathways, experiments with effector loop mutants of Ras have demonstrated that anchorage-independent growth can occur in the absence of Raf activation (14, 18, 43). Since the deletion of KSR1 strongly inhibited RasV12-induced transformation, we determined which Ras effectors were dependent upon KSR1 (Fig. 7A). To assess the activation of the Raf, PI 3-kinase, and RalGEF pathways, we examined the phosphorylation of downstream pathway members in KSR1−/− MEFs expressing RasV12 and KSR1 or in control cells. The expression of KSR1 was necessary for RasV12-induced phosphorylation of ERK, confirming the scaffolding role of KSR1 for this pathway. In contrast, KSR1 expression was not required for phosphorylation of AKT at Ser308 or Thr473 or for phosphorylation of c-Jun at Ser73, indicating that signaling through other RasV12 effector pathways remains intact in KSR1−/− MEFs (1, 9). Since each of these effectors is implicated in the control of cell cycle progression (13), the proliferative capacity of these cells was assessed (Fig. 7B). RasV12 expression in KSR1−/− cells caused a twofold increase in cell proliferation, presumably mediated by the induction of the PI 3-kinase and Ral pathways. The addition of KSR1 at physiologic levels markedly enhanced RasV12-induced proliferation, confirming a role for the Raf/MEK/ERK pathway in this response.
FIG. 7.
KSR1 expression is not required for RasV12-mediated induction of PI 3-kinase or RalGEF-Ral effector pathway. (A) KSR1−/− MEFs expressing KSR1 and RasV12 or control viruses were lysed, and 50 μg of protein was subjected to Western blotting for the indicated proteins or phospho-proteins to assess activation of the Raf/MEK/ERK, PI 3-kinase-AKT, and RalGEF-Ral effector pathways. (B) MEFs expressing GFP only (triangles) or low levels of KSR1 (squares), either without (open) or with (closed) RasV12, were seeded at 4 × 104 cells per 35-mm-diameter dish. Separate triplicate dishes were assessed for cell number every 24 h on a Beckman Coulter counter.
The KSR1 scaffold complex forms at levels of KSR1 that are optimal for signaling and proliferation.
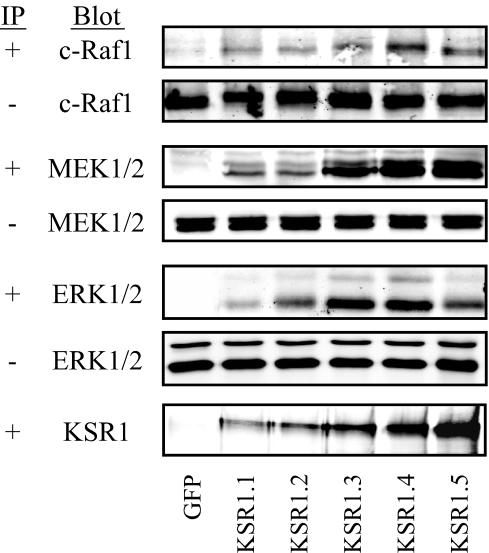
The proposed mechanism for the concentration-dependent effects of molecular scaffolds on signaling pathways is that the scaffold will bring pathway members together until the system is at equilibrium, facilitating signaling. Beyond this equilibrium, however, the scaffold will act to sequester pathway members from each other, inhibiting signaling (3, 5, 12, 20). To examine whether this model explains the effects of altered KSR1 levels on signaling through the Raf/MEK/ERK kinase cascade, we tested the ability of KSR1 expressed at increasing levels to coimmunoprecipitate c-Raf1, MEK1/2, and ERK1/2 in cycling populations of cells (Fig. 8).
FIG. 8.
KSR1-scaffolded complex forms at levels of KSR1 that are optimal for signaling and proliferation. KSR1 immunoprecipitates from 70 to 80% confluent dishes of KSR1−/− MEFs in DMEM plus 10% FBS and expressing GFP only or KSR1 at increasing expression levels (KSR1.1 through KSR1.5) were prepared in anti-FLAG M2-agarose and examined by immunoblotting for c-Raf-1, MEK1/2, ERK1/2, and KSR1. Whole-cell lysates were also probed to verify the uniform expression of each pathway member.
KSR1 was found to interact with Raf, MEK, and ERK in all KSR1-expressing cells. The amount of each signaling component precipitated by KSR1 increased as the level of KSR1 increased, to a maximum at 14 times the wild-type KSR1 expression level (KSR1.4 cells), coincident with the level of KSR1 expression that maximally enhanced ERK activation and cell proliferation in culture (Fig. 8). Increasing KSR1 expression beyond this point (KSR1.5 cells) did not lead to an increase in the amount of coprecipitated Raf, MEK, or ERK (Fig. 8), indicating that a maximal amount of each kinase was precipitated in KSR1.4 cells. Furthermore, the interaction of KSR1 with Raf and ERK was markedly diminished in KSR1.5 cells, suggesting that these interactions are dependent upon the cell's proliferative potential. These data indicate that levels of KSR1 that are optimal for proliferation and signaling are capable of interacting maximally with all components of the kinase cascade. Interestingly, for each scaffold-kinase interaction, KSR1 coprecipitated <5% of the total amount of each kinase within the cell, suggesting that there may be a distinct pool of Raf, MEK, and ERK in the cell for KSR1-dependent functions.
DISCUSSION
The data presented here demonstrate that the molecular scaffold protein KSR1 is required for cell transformation induced by activated RasV12. The extent of RasV12-induced transformation is dependent on the level of KSR1 expression. Incremental increases in the cellular concentration of ectopic KSR1 revealed a reserve capacity for ERK activation, cell proliferation, and transformation that is inaccessible in cells expressing physiological concentrations of KSR1. The cellular concentration of KSR1 that is optimal for cell proliferation interacts maximally with all components of the Raf/MEK/ERK kinase cascade.
Oncogenic potential of RasV12 requires KSR1.
The lack of transformation in KSR1−/− MEFs appears to be due only to a loss of signaling in the Raf/MEK/ERK effector pathway. Signaling through other Ras effector pathways is intact in KSR1−/− cells as measured by RasV12-induced phosphorylation of downstream targets and the enhanced growth rate of KSR1−/− MEFs (Fig. 7). RasV12 expression in KSR1.1 cells caused a synergistic (fivefold) increase in cell proliferation, indicating that KSR1-scaffolded signaling through the Raf/MEK/ERK cascade is necessary for the RasV12 oncogenic phenotype. Interestingly, this enhanced proliferation was also observed without transformation upon the expression of KSR1 to optimal levels in KSR1−/− and KSR1+/+ MEFs in the absence of RasV12 (Fig. 5). This observation implies that, by itself, optimal expression of KSR1 mimics the proliferative input of RasV12, but it also suggests that KSR1 facilitates additional signals specific to RasV12 that are required for anchorage-independent growth and the loss of contact inhibition. These data demonstrate an essential role of KSR1 in oncogenic signaling and suggest a mechanism by which cells may be able to modulate their oncogenic potential by altering KSR1 expression. KSR was originally identified as a positive modifier of RasV12 in Drosophila and C. elegans. Only one previous report (41) was able to show that KSR1 positively affects RasV12-mediated transformation in mammalian cells, whereas others reported inhibitory effects of KSR1 on RasV12-induced transformation (8, 17). The data presented here suggest that the inhibitory effect of ectopic KSR1 observed previously was likely due to levels of KSR1 that are comparable to those observed for KSR1.5 cells. We showed that KSR1 is required for RasV12-induced transformation in cells and that optimal KSR1 expression levels enhance the transformed phenotype. This is consistent with observations that KSR1−/− mice bearing oncogenes have reduced tumorigenesis (21, 28). The deletion of KSR1 doubled the latency of mammary tumorigenesis in middle T transgenic mice, indicating that KSR1-scaffolded signaling contributes to, but is not absolutely required for, tumor development (28). The deletion of KSR eliminated RasV12-mediated skin tumorigenesis (21). Differences between RasV12-induced transformation in KSR1−/− MEFs and tumorigenesis in KSR1−/− mice may be due to differences in the tissue expression of another KSR family member, KSR2. KSR2 was recently cloned in C. elegans, in which RNA interference of KSR2 in a KSR1−/− background led to embryonic lethality (29). KSR2 expression is detectable in KSR1−/− mice, but not in KSR1−/− MEFs (unpublished data).
Endogenous KSR1 is expressed at levels that are suboptimal for signaling and growth in mammalian fibroblasts.
KSR1 affects signaling through the ERK kinase cascade in a concentration-dependent manner. KSR1 enhances signaling as its expression level increases to a maximum at 14 times the wild-type expression level and inhibits signaling beyond this level (Fig. 3). The increase in ERK signaling seen with optimal KSR1 expression levels led us to assess the proliferative capacity of cells expressing increasing levels of KSR1. While the loss of KSR1 had no effect on proliferation, increasing KSR1 expression to levels that were optimal for signaling led to a threefold increase in the rate of cell proliferation (Fig. 5). These data imply that endogenous KSR1 is expressed at levels that are suboptimal for signaling and proliferation. Furthermore, they expose a potential mechanism through which cells may regulate their proliferative capacity by modulating levels of a noncatalytic signaling component. We speculate that cells may be able to draw on this reserve capacity in situations in which high proliferative rates are desirable (e.g., embryogenesis and wound healing). In support of this hypothesis, murine embryonic stem cells have been shown to contain higher levels of KSR1 than do fibroblasts (6). This increased expression may be due to increased transcription and/or translation or decreased degradation of KSR1. Stewart et al. (36) have shown that KSR1 coprecipitates with heat shock proteins and that a disruption of this interaction increases the turnover of KSR1 (36). Further studies of the importance of this interaction as well as the mechanisms controlling KSR1 degradation may be needed to understand the role of KSR1 stability in maintaining cellular function.
PDGF, but not EGF, is sufficient to drive quiescent 3T3 cells into S phase. The ability of PDGF to promote prolonged ERK activation is a major determinant of its ability to promote progression into S phase (16, 27). While recent studies have begun to address the mechanisms that cells use to respond to transient versus sustained ERK activation, limited work has been done to address the upstream signaling components that control the duration of ERK activity. We showed that KSR1 is necessary for PDGF-induced sustained ERK activation and cell cycle entry (Fig. 1 and 4). In KSR1−/− MEFs, PDGF elicits a transient ERK response and allows only a fraction of quiescent MEFs to enter S phase. Conversely, an optimal expression of KSR1 in KSR1−/− MEFs allows EGF alone to elicit a prolonged ERK response and to promote S-phase entry (Fig. 3 and 4). These data suggest that KSR1 is a regulator of a mitogenic signaling cascade that is necessary for cell cycle progression. However, the fact that in culture, KSR1−/− MEFs proliferate in 10% serum at a rate that is indistinguishable from that of wild-type MEFs, despite reduced serum-induced ERK activation, suggests that only moderately prolonged ERK activation (ca. 1 h) is necessary for the induction of cell proliferation. Furthermore, differences in the responsiveness of KSR1−/− MEFs to PDGF versus serum indicate that additional factors in serum can compensate for any limitations in proliferative potential imposed by the deletion of KSR1.
Tightly controlled ERK activity is important for a number of biological processes. PC12 cells can be differentiated into neurons by stimulation with NGF, but not EGF. This correlates with the ability of NGF to stimulate prolonged ERK activation. The ectopic expression of B-KSR1 in PC12 cells allowed EGF to stimulate prolonged ERK activation and neurite outgrowth (25). We hypothesize that the level of B-KSR1 overexpression in that system was similar to the levels of KSR1 expression observed here that promote prolonged ERK activation with EGF in fibroblasts. These observations suggest that any biological effect requiring precise levels of ERK activity may be dependent upon and modulated by KSR1 expression.
Ectopic expression of KSR1 can titrate components of the Raf/MEK/ERK cascade.
Supraphysiologic levels of protein scaffolds have been shown to inhibit signaling through their kinase cascades (6, 10, 17, 33, 51). This loss of signaling is proposed to be due to the titration of pathway members away from each other (3, 5, 12, 15, 20) and has been termed “combinatorial inhibition” (20). The demonstrations that KSR1 interacts with Raf, MEK, and ERK and that maximal interaction is detected in populations of cells expressing KSR1 at levels that are optimal for signaling and proliferation provide experimental evidence for that concept (Fig. 8). As KSR1 expression increased beyond the cellular concentration at which it facilitated signaling, the amounts of precipitated pathway members did not increase, supporting the notion that the components of the Raf/MEK/ERK kinase cascade were being titrated away from one another. In fact, increasing KSR1 expression above optimal levels diminished the amount of Raf and ERK bound to KSR1. One would intuitively predict that high levels of KSR expression should not limit the ability of the scaffold to precipitate its effectors. However, this unexpected observation is consistent with published data demonstrating that the interaction of Raf and ERK with KSR is dependent upon pathway activation (6, 25, 41). These observations lead to the interpretation that as KSR titrates apart the effectors at high scaffold concentrations, the pathway activity declines and the interaction (precipitation) of Raf and ERK with KSR is diminished. The maximal amounts of Raf, MEK, and ERK precipitated by levels of KSR1 that are optimal for proliferation accounted for <5% of the total cellular pool for each protein in the cell. This observation suggests that there are cellular pools of Raf, MEK, and ERK that are inaccessible to KSR1. These pools may be in other subcellular compartments, awaiting signaling events that occur through scaffolds other than KSR1. For example, while KSR1 controls ERK signaling via EGF at the plasma membrane (26), the p14/MP1 scaffold complex is required for ERK signaling on late endosomes (39). Thus, while KSR1 controls some aspects of ERK signaling, there are other ERK scaffolds that may mediate distinct biological functions of ERK.
KSR1 as a protein scaffold.
Heinrich and colleagues (15) suggested three functions for scaffold proteins of kinase cascades. They proposed that scaffold proteins (i) help activate a group of kinases, (ii) could limit the ability of phosphatases to act on bound kinases, and (iii) sequester bound kinases away from the bulk solution to their site of action. We hypothesize that KSR1 fulfills each of these functions.
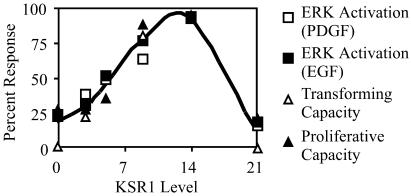
KSR1 is necessary for the maximal activation of ERK by PDGF and EGF, and increasing KSR1 concentrations increase growth factor-induced signaling, proliferation, and transformation to a maximum at 14 times the level of KSR1 expressed in wild-type MEFs and then limit signaling thereafter (Fig. 9). Furthermore, the strong correlation observed between KSR1 expression level and prolonged ERK activation could reflect, in part, the ability of KSR1 to protect ERK from dephosphorylation. Previous studies by our laboratory and others have shown that KSR1 moves to both the plasma membrane and the nucleus, where it can interact with components of the Raf/MEK/ERK cascade (4, 23, 26). Thus, KSR1 may sequester kinases from the bulk solution to specific subcellular compartments.
FIG. 9.
Level of KSR1 expression that maximally associates with members of the Raf/MEK/ERK cascade corresponds precisely with the level of KSR1 expression that is optimal for ERK signaling, proliferation, and transformation. ERK activation by PDGF (open squares) and EGF (closed squares), transforming capacity (open triangles), and proliferative capacity (closed triangles) were plotted as percentages of response versus KSR1 expression levels.
The Raf/MEK/ERK cascade has been implicated in many processes affecting cell fate, including senescence, proliferation, transformation, differentiation, and apoptosis (7, 30, 34, 47). The manipulation of scaffold proteins may explain how this critical kinase cascade can affect such diverse cellular functions. By controlling the magnitude and duration of signaling through ERK, KSR1 aids in the control of both normal proliferation and cell transformation. KSR1 has also been linked to development through the control of Xenopus germinal vesicle breakdown (6, 26, 41) and to the differentiation of PC12 cells into neurons (25). The precise role played by KSR1 in modulating the Raf/MEK/ERK kinase cascade is central to understanding how this pathway affects cell fate.
Acknowledgments
We thank Charles Kuszynski and Linda Wilkie in the UNMC Cell Analysis Facility for their technical expertise in the generation of GFP-expressing cell lines and Eric Gosink and Thomas Smithgall for retroviral vectors. We also thank Gina Razidlo for her critical reading of the manuscript and members of the Lewis lab for their insights and advice.
This research was supported by NIH grants CA90400 and DK52809, the Charlotte Geyer Foundation, the American Diabetes Association (R.E.L.), and grant CA36727-19 to the UNMC/Eppley Cancer Center. R.L.K. was supported by a physician/scientist training fellowship from the American Diabetes Association.
REFERENCES
- 1.Alessi, D. R., M. Andjelkovic, B. Caudwell, P. Cron, N. Morrice, P. Cohen, and B. A. Hemmings. 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15:6541-6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Anselmo, A. N., R. Bumeister, J. M. Thomas, and M. A. White. 2002. Critical contribution of linker proteins to Raf kinase activation. J. Biol. Chem. 277:5940-5943. [DOI] [PubMed] [Google Scholar]
- 3.Bray, D., and S. Lay. 1997. Computer-based analysis of the binding steps in protein complex formation. Proc. Natl. Acad. Sci. USA 94:13493-13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, J. A., D. J. Volle, O. V. Chaika, and R. E. Lewis. 2002. Phosphorylation regulates the nucleocytoplasmic distribution of kinase suppressor of Ras. J. Biol. Chem. 277:5369-5377. [DOI] [PubMed] [Google Scholar]
- 5.Burack, W. R., and A. S. Shaw. 2000. Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 12:211-216. [DOI] [PubMed] [Google Scholar]
- 6.Cacace, A. M., N. R. Michaud, M. Therrien, K. Mathes, T. Copeland, G. M. Rubin, and D. K. Morrison. 1999. Identification of constitutive and Ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol. Cell. Biol. 19:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 8.Denouel-Galy, A., E. M. Douville, P. H. Warne, C. Papin, D. Laugier, G. Calothy, J. Downward, and A. Eychene. 1998. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr. Biol. 8:46-55. [DOI] [PubMed] [Google Scholar]
- 9.de Ruiter, N. D., R. M. Wolthuis, H. van Dam, B. M. Burgering, and J. L. Bos. 2000. Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol. Cell. Biol. 20:8480-8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickens, M., J. S. Rogers, J. Cavanagh, A. Raitano, Z. Xia, J. R. Halpern, M. E. Greenberg, C. L. Sawyers, and R. J. Davis. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277:693-696. [DOI] [PubMed] [Google Scholar]
- 11.Elion, E. A. 1998. Routing MAP kinase cascades. Science 281:1625-1626. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell, J. E., Jr. 2000. What do scaffold proteins really do? Sci. STKE 2000:PE1. [DOI] [PubMed] [Google Scholar]
- 13.Gille, H., and J. Downward. 1999. Multiple Ras effector pathways contribute to G(1) cell cycle progression. J. Biol. Chem. 274:22033-22040. [DOI] [PubMed] [Google Scholar]
- 14.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrich, R., B. G. Neel, and T. A. Rapoport. 2002. Mathematical models of protein kinase signal transduction. Mol. Cell 9:957-970. [DOI] [PubMed] [Google Scholar]
- 16.Jones, S. M., and A. Kazlauskas. 2001. Growth-factor-dependent mitogenesis requires two distinct phases of signalling. Nat. Cell Biol. 3:165-172. [DOI] [PubMed] [Google Scholar]
- 17.Joneson, T., J. A. Fulton, D. J. Volle, O. V. Chaika, D. Bar-Sagi, and R. E. Lewis. 1998. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J. Biol. Chem. 273:7743-7748. [DOI] [PubMed] [Google Scholar]
- 18.Khosravi-Far, R., M. A. White, J. K. Westwick, P. A. Solski, M. Chrzanowska-Wodnicka, L. Van Aelst, M. H. Wigler, and C. J. Der. 1996. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol. Cell. Biol. 16:3923-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornfeld, K., D. B. Hom, and H. R. Horvitz. 1995. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83:903-913. [DOI] [PubMed] [Google Scholar]
- 20.Levchenko, A., J. Bruck, and P. W. Sternberg. 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA 97:5818-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozano, J., R. Xing, Z. Cai, H. L. Jensen, C. Trempus, W. Mark, R. Cannon, and R. Kolesnick. 2003. Deficiency of kinase suppressor of Ras1 prevents oncogenic Ras signaling in mice. Cancer Res. 63:4232-4238. [PubMed] [Google Scholar]
- 22.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179-185. [DOI] [PubMed] [Google Scholar]
- 23.Michaud, N. R., M. Therrien, A. Cacace, L. C. Edsall, S. Spiegel, G. M. Rubin, and D. K. Morrison. 1997. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc. Natl. Acad. Sci. USA 94:12792-12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison, D. K. 2001. KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci. 114:1609-1612. [DOI] [PubMed] [Google Scholar]
- 25.Muller, J., A. M. Cacace, W. E. Lyons, C. B. McGill, and D. K. Morrison. 2000. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol. Cell. Biol. 20:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D. K. Morrison. 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8:983-993. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4:556-564. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, A., W. R. Burack, J. L. Stock, R. Kortum, O. V. Chaika, M. Afkarian, W. J. Muller, K. M. Murphy, D. K. Morrison, R. E. Lewis, J. McNeish, and A. S. Shaw. 2002. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 22:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohmachi, M., C. E. Rocheleau, D. Church, E. Lambie, T. Schedl, and M. V. Sundaram. 2002. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 12:427-433. [DOI] [PubMed] [Google Scholar]
- 30.Pouyssegur, J., V. Volmat, and P. Lenormand. 2002. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem. Pharmacol. 64:755-763. [DOI] [PubMed] [Google Scholar]
- 31.Roy, F., G. Laberge, M. Douziech, D. Ferland-McCollough, and M. Therrien. 2002. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaeffer, H. J., A. D. Catling, S. T. Eblen, L. S. Collier, A. Krauss, and M. J. Weber. 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281:1668-1671. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, K., and C. S. Zuker. 1998. Assembly of the Drosophila phototransduction cascade into a signalling complex shapes elementary responses. Nature 395:805-808. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, S., M. Sundaram, Y. P. Zhang, J. Y. Lee, M. Han, and K. L. Guan. 1999. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol. Cell. Biol. 19:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimoto, T., S. Stewart, M. Han, and K. L. Guan. 1998. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 17:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaram, M., and M. Han. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83:889-901. [DOI] [PubMed] [Google Scholar]
- 39.Teis, D., W. Wunderlich, and L. A. Huber. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3:803-814. [DOI] [PubMed] [Google Scholar]
- 40.Therrien, M., H. C. Chang, N. M. Solomon, F. D. Karim, D. A. Wassarman, and G. M. Rubin. 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83:879-888. [DOI] [PubMed] [Google Scholar]
- 41.Therrien, M., N. R. Michaud, G. M. Rubin, and D. K. Morrison. 1996. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 10:2684-2695. [DOI] [PubMed] [Google Scholar]
- 42.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb, C. P., L. Van Aelst, M. H. Wigler, and G. F. Woude. 1998. Signaling pathways in Ras-mediated tumorigenicity and metastasis. Proc. Natl. Acad. Sci. USA 95:8773-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 45.White, M. A., T. Vale, J. H. Camonis, E. Schaefer, and M. H. Wigler. 1996. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J. Biol. Chem. 271:16439-16442. [DOI] [PubMed] [Google Scholar]
- 46.Whitmarsh, A. J., J. Cavanagh, C. Tournier, J. Yasuda, and R. J. Davis. 1998. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science 281:1671-1674. [DOI] [PubMed] [Google Scholar]
- 47.Whitmarsh, A. J., and R. J. Davis. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23:481-485. [DOI] [PubMed] [Google Scholar]
- 48.Xing, H., K. Kornfeld, and A. J. Muslin. 1997. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr. Biol. 7:294-300. [DOI] [PubMed] [Google Scholar]
- 49.Yang, J. J., J. S. Kang, and R. S. Krauss. 1998. Ras signals to the cell cycle machinery via multiple pathways to induce anchorage-independent growth. Mol. Cell. Biol. 18:2586-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasuda, J., A. J. Whitmarsh, J. Cavanagh, M. Sharma, and R. J. Davis. 1999. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 19:7245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeung, K., T. Seitz, S. Li, P. Janosch, B. McFerran, C. Kaiser, F. Fee, K. D. Katsanakis, D. W. Rose, H. Mischak, J. M. Sedivy, and W. Kolch. 1999. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401:173-177. [DOI] [PubMed] [Google Scholar]
- 52.Yu, W., W. J. Fantl, G. Harrowe, and L. T. Williams. 1998. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr. Biol. 8:56-64. [DOI] [PubMed] [Google Scholar]