Abstract
Melanoma is a disease characterized by lesions that activate ERK. Though 70% of cutaneous melanomas harbor activating mutations in the BRAF and NRAS genes, the alterations that drive tumor progression in the remaining 30% are largely undefined. Vemurafenib, a selective inhibitor of RAF kinases, has clinical utility restricted to BRAF mutant tumors. MEK inhibitors, which have shown clinical activity in NRAS-mutant melanoma, may be effective in other ERK pathway-dependent settings. Here, we investigated a panel of melanoma cell lines wild-type for BRAF and NRAS to determine the genetic alteration driving their transformation and their dependence on ERK signaling in order to elucidate a candidate set for MEK inhibitor treatment. A cohort of the BRAF/RAS wild-type cell lines with high levels of RAS-GTP had loss of NF1, a RAS GTPase activating protein. In these cell lines, the MEK inhibitor PD0325901 inhibited ERK phosphorylation, but also relieved feedback inhibition of RAS resulting in induction of pMEK and a rapid rebound in ERK signaling. In contrast, the MEK inhibitor trametinib impaired the adaptive response of cells to ERK inhibition leading to sustained suppression of ERK signaling and significant antitumor effects. Notably, alterations in NF1 frequently co-occurred with RAS and BRAF alterations in melanoma. In the setting of BRAF(V600E), NF1 loss abrogated negative feedback on RAS activation resulting in elevated activation of RAS-GTP and resistance to RAF, but not MEK, inhibitors. We conclude that loss of NF1 is common in cutaneous melanoma and is associated with RAS activation, MEK-dependence and resistance to RAF inhibition.
Keywords: melanoma, NF1, RAS, trametinib, vemurafenib
Introduction
The MAPK (mitogen-activated protein kinase) signaling pathway is a critical regulator of cell growth and cell cycle progression. Receptor tyrosine kinases (RTKs), activated by extracellular mitogens, stimulate the RAS (H-, N- and KRAS) small GTPase proteins. RAS proteins are activated when guanine exchange factors facilitate binding to GTP and inactivated by GTPase activating proteins (GAPs), such as NF1, that facilitate the hydrolysis of GTP to GDP (1). Active RAS facilitates dimerization (2) and activation of RAF (A- B- and CRAF/RAF1) kinases, which in turn activate the MEK1/2 (mitogen-activated protein kinase kinase) and ERK1/2 (extracellular signaling-regulated kinase) kinases. Activated, phosphorylated ERK regulates the transcription of proteins such as cyclin D1 that promote cell cycle progression, transcription factors that promote the transformed phenotype and a network of genes that negatively inhibit pathway output by regulating the activity of RTKs, RAS, and RAF (3).
Alterations resulting in aberrant activation of ERK can be found at almost every level of the MAPK pathway and are common in human tumors. BRAF mutations, particularly at codon 600, are found in many cancers, including 50% of cutaneous melanomas (4). Selective inhibitors of RAF (vemurafenib, dabrafenib) have unprecedented clinical activity in patients whose melanomas harbor BRAF(V600E) mutations, and their use results in a prolongation of progression-free and overall survival (5). NRAS mutations, most commonly at codon 61, have been identified in another 15-20% of melanomas and occur in a mutually exclusive pattern with BRAF(V600E) (4, 6, 7). Treatment options remain limited for these patients and those whose tumors are wild-type for BRAF and NRAS, and the prognosis of such patients is particularly grim with a median overall survival of less than one year (8).
Given the high prevalence of BRAF and NRAS alterations in melanoma, we hypothesized that melanomas including those wild-type for both genes (BRAFWT/RASWT) may exhibit a dependence on ERK pathway activation for maintenance of the transformed phenotype that may be exploited for therapeutic advantage (9, 10). Here, we performed a functional and genomic analysis of BRAFWT/NRASWT melanoma cell lines to determine whether occult MAPK pathway alterations are present in such cells. NF1 alterations that result in RAS activation and MEK dependence were identified in a subset of BRAFWT/NRASWT melanoma cell lines. However, NF1 alterations were not mutually exclusive with RAS and BRAF aberrations. Loss of NF1 in cells co-mutated for BRAF was sufficient to overcome the upstream negative feedback that results in suppression of RAS activation in BRAF(V600E) cells and was sufficient to confer resistance to vemurafenib. Furthermore, NF1 loss and corresponding relief of upstream ERK-dependent negative feedback attenuated the anti-proliferative effects of the selective MEK inhibitor PD0325901 in NF1-null cells. In contrast, trametinib, an allosteric MEK inhibitor recently approved for use in patients with BRAF-mutant melanoma (11), attenuated the phosphorylation of MEK resulting from relief of upstream negative feedback and exhibited greater potency than PD0325901 in NF1-null melanoma cells. In summary, loss of NF1 expression in melanoma results in RAS activation and vemurafenib resistance even in the setting of BRAF mutation. Allosteric MEK inhibitors that impair the adaptive response of cells to ERK inhibition by blocking MEK phosphorylation should be studied in melanoma patients whose tumors harbor NF1 loss.
Materials and Methods
Cell lines and culture conditions
“SK-Mel” cell lines were provided by Taha Merghoub and Alan Houghton (MSKCC). MeWo, Malme3M, A375 and SNF96.2 were purchased from ATCC. M308 was provided by Antoni Ribas (UCLA) and WM3918 by Katherine Nathanson and Meenhard Herlyn (Wistar Institute). Other than A375 and SNF96.2 (grown in Dulbecco's modified Eagle medium), all cell lines were grown in RPMI 1640 as previously described (12).
Genomic studies
Cellular DNA was extracted using the Qiagen DNAeasy Tissue Kit. DNA was analyzed using a mass spectrometry-based fingerprinting assay to validate cell line identity as described previously (13). NRAS (G12A, G12D, Q61K, Q61R, Q61L), BRAF (V600E, V600K, V600R, K601E), and c-KIT (D816V) mutations were detected using a mass spectrometry-based assay (Sequenom) and validated by Sanger sequencing (13). Selected cell lines were screened for mutations and copy number alterations in 279 cancer-associated genes using the IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) assay as has been described (14) and viewed in the Integrated Genomics Viewer (IGV) (15). Genomic data from the TCGA melanoma project was derived from the cBioPortal for Cancer Genomics (http://cbioportal.org).
RNA-seq
RNA was extracted from cells using the RNeasy mini kit (Qiagen, Inc.). Quality assessment, poly-A selection, and sequencing with an Illumina HiSeq 2000 were performed by the Genomics Core Laboratory at MSKCC. All samples had a minimum RNA integrity number (RIN) of 7.0 (16). Sequencing produced 40 to 120 million 75bp reads per sample. FASTQ files were generated using CASAVA 1.8.2 software (Illumina). Low quality bases and adapter sequences were removed with Cutadapt. Trimmed reads were aligned to human genome assembly GRCh37 using Tophat 2.0.8 (17, 18). Gene quantification and differential expression were calculated using Cufflinks 2.1.1 (19). Data visualizations were created with the gplots package for R.
Western blotting
Cells were collected, lysed and blotted as previously described (12). Secondary antibodies were detected using Super Signal (Thermo) and chemiluminesence imaged using a Fuji LAS-4000 imager (GE Lifesciences). Anti-NF1 (SC-67), cyclin D1 (SC-718), KRAS (SC-30), NRAS (SC-519), HRAS (SC-520) and actinin (SC-17829) were from Santa Cruz Biotechnology; Anti-Ras (#1862335) from Thermo Scientific; and anti-pERK (#9101), ERK (#9102), pMEK (#9121), MEK (#9122), and p-CRAF S338 (#9427) from Cell Signaling Technology.
Proliferation assays/FACS analysis
Cell viability was measured by trypan blue incorporation using a Vi-CELL XR 2.03 (Beckman Coulter) as previously described (20) Percent growth was calculated using the equation 100*([Day 5 drug]-[Day 0])/([Day 5 DMSO]-[Day 0]). FACS analysis was performed as previously described (21).
RasGTP assay
GTP-bound Ras was isolated via immunoprecipitation using recombinant Ras binding domain of Raf1 (RAF1-RBD; Active Ras Pull-down and detection kit, Thermo Scientific), according to the manufacturer's instructions. The product was detected using total (pan-RAS) or isoform specific (H-, K-, N-) RAS antibodies.
RNAi studies
siRNA studies used ON-TARGET plus siNF1 SMARTpool and ON-TARGET plus non-targeting siRNA#2. shRNA studies were accomplished using GIPZ shNon-Targeting RNA or TRIPZ Inducible shRNA against NF1. shRNAs (shNF1 #2: CloneID V2THS-260806; shNF1 #4: CloneID V3THS-380114; shRNA#6: CloneID V3THS-380110) were induced with 2μg/mL doxycycline daily for 1 week prior to vemurafenib studies.
Results
NF1 is lost in a subset of RAS-activated BRAFWT/RASWT cell lines
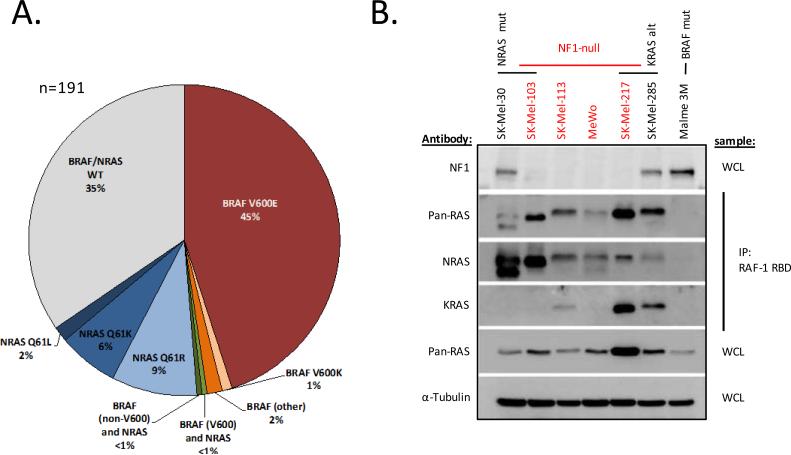
To identify a cohort of BRAFWT/NRASWT melanoma cell lines for genomic and biologic characterization, 191 melanoma cell lines were genotyped for BRAF and NRAS mutations using a mass-spectrometry-based (Sequenom) assay (13, 22). This screen identified 66 cell lines that lacked hotspot mutations in BRAF or NRAS (Fig. 1A, Supplementary Table S1). As this assay was designed to detect only the most common BRAF and NRAS mutations, we further performed Sanger sequencing of BRAF exons 11 and 15 and NRAS exons 2 and 3. This analysis identified BRAF mutations not present in the Sequenom assay in two cell lines (D594G in SK-Mel-264 and N581S in SK-Mel-215; Supplemental Table S2). Direct sequencing of KRAS and HRAS further identified activating mutations in KRAS in two and HRAS in one cell line, respectively (Supplemental Table S2). In summary, 61 cell lines were wild-type for RAS and BRAF.
Figure 1.
NF1-null melanoma cell lines express high levels of activated RAS. A) BRAF and NRAS status of the melanoma cell line panel (n=191). B) Activated RAS protein (RAS-GTP) was quantitated in select melanoma cell lines via immunoprecipitation with the RAS-binding domain of RAF (RAF-1 RBD) followed by immunoblot using pan-RAS and isoform selective NRAS and KRAS antibodies. Expression of NF1, total RAS (pan-RAS) and actinin (as a loading control) were measured by immunoblot from whole cell lysate (WCL). Alt = alteration, mut = mutant, WCL = whole cell lysate.
Given the high prevalence of ERK activation in melanoma (10), we hypothesized that a subset of the BRAFWT/RASWT cohort likely harbored occult alterations within the MAPK pathway that cause RAS to become refractory to negative feedback and thus confer activation of ERK. We thus measured levels of activated, GTP-bound RAS in a subset of the BRAFWT/RASWT cell lines as a surrogate of pathway activation. Similar to human tumors, KRAS- and NRAS-mutant melanoma cancer cell lines exhibit high levels of RAS-GTP whereas BRAF-mutant cell lines have low to undetectable levels of RAS-GTP (Fig. 1B and (3, 23)). RAS was activated to varying levels in the BRAFWT/RASWT melanoma cells, with some expressing levels of activated RAS similar to those present in RAS-mutant cells (Supplementary Fig. S1).
The NF1 gene encodes a protein that functions as the predominant RAS GTPase activating protein (RAS GAP), which suppresses RAS activity and reduces RAS-GTP levels by promoting endogenous RAS GTPase activity. NF1 is inactivated in diverse human cancers (24-27) and would be predicted, if lost, to cause RAS to become refractory to negative feedback. We performed western blot analysis to determine whether loss of NF1 protein expression occurred in, and was correlated with, elevated RAS-GTP levels in BRAFWT/RASWT melanoma cell lines. Complete loss of NF1 expression was noted in five of the BRAFWT/RASWT cell lines, all of which had high levels of RAS-GTP activation (Supplemental Fig. S1). Having previously performed high-resolution DNA copy number profiling (array CGH) on 92 melanoma cell lines (22), we identified a sixth NF1-null cell line that harbored homozygous NF1 gene deletion and concurrent NRAS (Q61R) mutation (SK-Mel-103).
NRAS mutations are significantly more prevalent than other RAS mutations in melanoma even though KRAS mutations are predominant in most other cancers (4). To determine which RAS isoforms were activated in NF1-null melanomas, we assayed activated KRAS, HRAS, and NRAS by performing immunoprecipitation with the RAS binding domain of Raf1 (Raf1-RBD; see methods) followed by RAS isoform-specific immunoblots. All four NF1-null cell lines examined expressed high levels of total active RAS when compared to a BRAF(V600E) control cell line (Fig. 1B and Supplemental Fig. S1). NRAS (Q61K) SK-Mel-30 cells expressed high levels of GTP-bound NRAS, but no detectable levels of activated KRAS, similar to the NRAS (Q61R)/NF1-null SK-Mel-103 line. GTP-bound NRAS was also highly expressed in the other NF1-null cell lines whereas only a subset had concurrent activation of KRAS, including SK-Mel-217, which harbored KRAS gene amplification. Elevated levels of GTP-bound KRAS and NRAS were also detected in the KRAS (G12C) mutant SK-Mel-285 cell line. Levels of activated HRAS were low or undetectable in all the NF1-null melanoma cell lines (Supplemental Fig. S1).
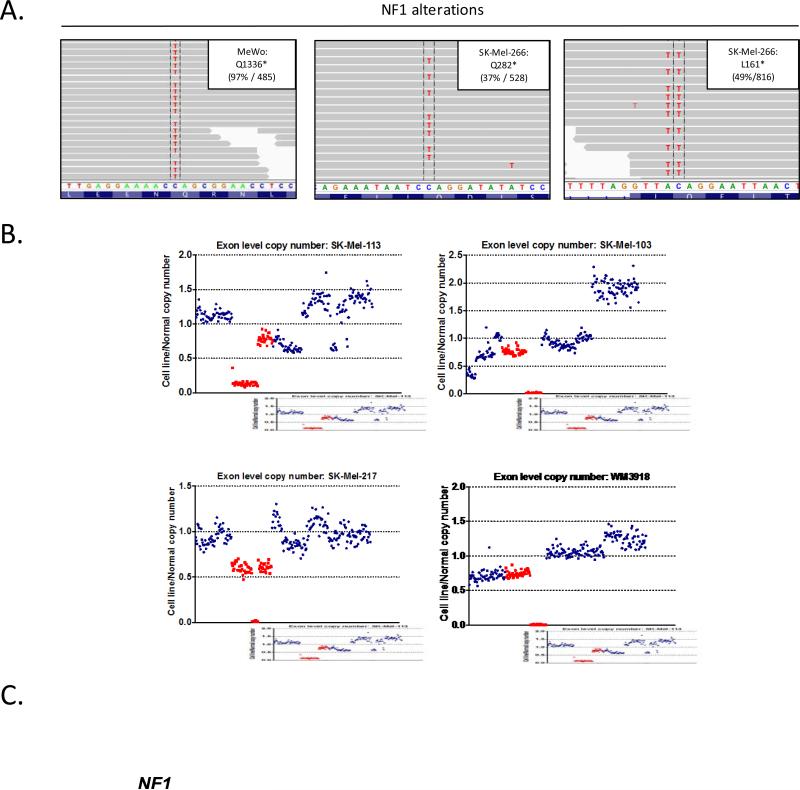
To define the mechanistic basis for the loss of NF1 expression in the melanoma cell lines, we performed next-generation sequencing of 279 genes commonly mutated in human cancer using an exon capture-based approach (IMPACT assay) (14, 28). Two cell lines were found to harbor nonsense mutations in NF1 (Fig. 2A): MeWo, a hemizygous Q1336* mutation, and Sk-Mel-266, L161* and Q282* mutations. The remaining four cell lines had deletions involving the NF1 gene locus: SK-Mel-113, focal homozygous loss of the N-terminal domain; SK-Mel-103 and WM3918, focal homozygous loss of the C-terminal domain, and SK-Mel-217, broad monoallelic loss, as well as a focal, intragenic deletion in the second NF1 allele (Fig. 2B). In sum, genomic alterations sufficient to account for complete loss of NF1 protein expression were identified in all six NF1-null melanoma cell lines.
Figure 2.
The genomic basis of NF1 loss in melanoma cell lines. A) DNA from NF1-null cell lines was analyzed using a capture based, next generation sequencing method (IMPACT). Shown are aligned sequencing reads highlighting select NF1 mutations. Percentages (left) are the ratio of mutant reads over total reads (right). B) Homozygous deletions of NF1 in four melanoma cell lines. Exon-level copy number data is shown for target genes on chromosome 17. C) Summary of mutations and copy-number alterations in NF1-null cell lines by IMPACT and NF1-null melanoma tumors analyzed by TCGA.
Although loss of NF1 was identified in the BRAFWT/RASWT cohort, it was not mutually exclusive with RAS alterations. Notably, concurrent alterations in the NF1 and the RAS genes have also been noted in two recent whole-exome sequencing studies of melanoma tumors including Hodis et al. and the melanoma study performed by the Cancer Genome Atlas (TCGA) working group (Fig. 2C and (29)). CDKN2A and/or TP53 were among the genes most commonly co-altered in the NF1-null melanoma cell lines and tumors, suggesting that these genes may cooperate with NF1 loss in promoting melanomagenesis as has been reported in other cancer types, such as astrocytomas and malignant peripheral nerve sheath tumors (30).
NF1-null cell lines are sensitive to MEK inhibitors that impair the adaptive response of cells to ERK inhibition
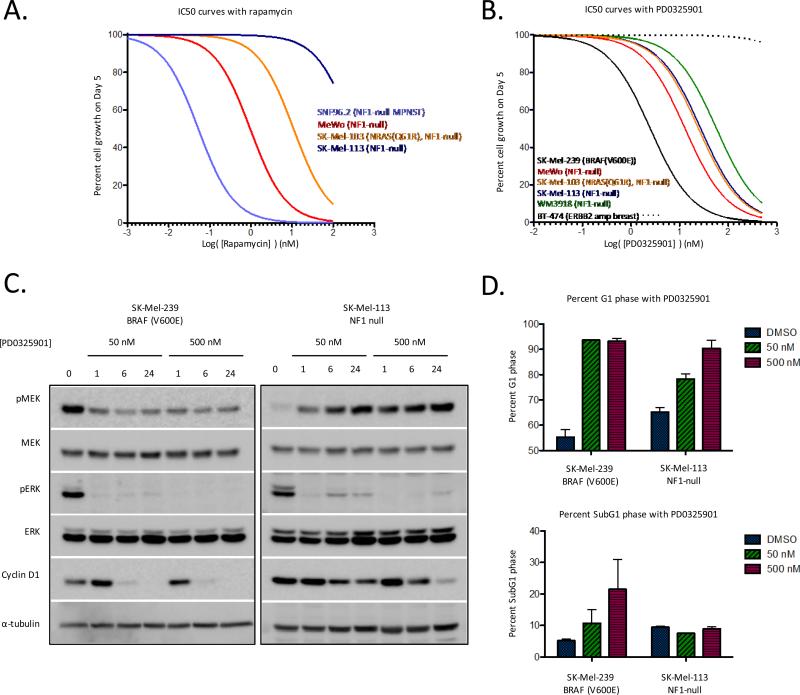
The growth of NF1-null cell lines derived from human malignant peripheral nerve sheath tumors (MPNSTs) and MPNSTs that arise in NF1−/− mice have been shown to be dependent on mTORC1 signaling and exquisitely sensitive to the mTORC1 inhibitor rapamycin (31, 32). To determine whether NF1-null melanomas were also mTORC1-dependent, we treated the NF1-null melanoma cell lines with rapamycin and compared their sensitivity to that of SNF96.2, a representative human NF1-null MPNST cell line (Fig. 3A). The NF1-null melanomas as a group were significantly less sensitive to rapamycin (IC50 ranging from 1 to 286 nM) than SNF96.2 cells (IC50 0.05 nM) suggesting that mTORC1-dependency is not a feature of all NF1-null cells.
Figure 3.
NF1-null melanoma cell lines are MAPK pathway dependent. A) Cells were treated with increasing concentrations of rapamycin for 5 days. Results are percent cell growth as a function of drug concentration (nM). B) Cells were treated with increasing concentrations of the MEK inhibitor PD0325901 for 5 days. Results as in A. C) Cells were treated with 50 or 500 nM PD0325901 for 0, 1, 6 and 24 hours. Phospho- and total levels of MAPK pathway components were determined by immunoblot. D) Cells were treated for 24 hours with 50 or 500nM PD0325901 before undergoing FACS analysis for cell cycle distribution. Error bars are SEM, n=3.
To probe the MEK dependence of NF1-null melanomas, we used PD0325901, a selective allosteric inhibitor of MEK1/2 (Kiapp of 1nM for MEK1 and MEK2) (33). The effect of PD0325901 on the proliferation and survival of four NF1-null melanoma cell lines was compared to that of a MEK inhibitor sensitive BRAF(V600E) melanoma cell line (SK-Mel-239) and to a MEK inhibitor resistant ERBB2 amplified breast cancer cell line (BT-474). The proliferation of all four NF1-null cell lines was inhibited by PD0325901, albeit with IC50s that were 6-20 fold greater than that of the BRAF(V600E) SK-Mel-239 cells (Fig. 3B).
To explore the basis for this differential sensitivity, we assessed the effects of drug exposure on downstream targets of MEK/ERK signaling as a function of concentration and time. Treatment of both BRAF(V600E) and NF1-null cells with 50nM PD0325901 resulted in decreased activation of phosphorylated ERK1/2 (pERK) by 1 hour (Fig. 3C and Supplemental Fig. S2). In BRAF(V600E) cells, suppression of pERK was durable and maintained at 6 and 24 hours and was accompanied by loss of cyclin D1 expression, accumulation of cells in G1 phase, and induction of apoptosis (Fig. 3C,D). In contrast, a partial rebound in pERK activation was apparent by 6 hours in NF1-null cells and was accompanied by a failure of the drug to potently suppress cyclin D1 expression. This rebound in pERK in the NF1-null cells was attenuated with use of a higher concentration of drug (500nM), leading to potent suppression of cyclin D1 expression, maximal accumulation of cells in G1 phase, and inhibition of proliferation (Fig. 3B-D). Together, these data suggest that cyclin D1 expression and cell cycle progression are MEK-dependent in NF1-null melanoma cells, but that rapid rebound in ERK activity may account for the lower sensitivity of NF1-null cells to the MEK inhibitor PD0325901. Induction of cell death was not observed following treatment with the MEK inhibitor in any of the NF1 null cell lines (Fig 3D). Furthermore, co-treatment with PD0325901 and the pan-AKT inhibitor MK2206 did not augment growth inhibition or induce apoptosis as has been shown in colorectal cells with RAS activation (34) (Supplemental Fig. S3).
Resistance to allosteric MEK inhibitors can be induced by upstream pathway hyperactivation (35, 36). We have previously shown that treatment of BRAFWT but not BRAF (V600E) cells with PD0325901 leads to increased activation of phosphorylated MEK (pMEK), which results from relief of upstream ERK-dependent negative feedback (3). Consistent with these prior observations, treatment of NF1-null melanoma cells with PD0325901 resulted in increased activation of phosphorylated MEK (pMEK) (Fig. 3C).
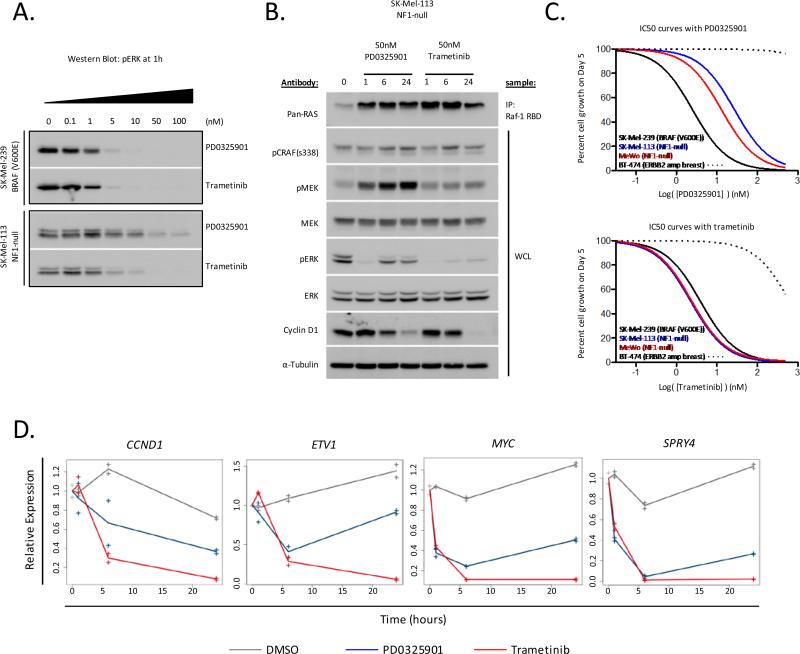
As the induction of pMEK in the NF1-null melanomas paralleled the rebound in pERK activation, we further studied the effects of a second allosteric MEK inhibitor on MEK signaling and cellular proliferation. Trametinib (GSK1120212) has a similar in vitro affinity for MEK1/2 as PD0325901 (IC50s for MEK1 and MEK2 of 0.7 and 0.9nM, respectively), but in contrast to PD0325901, binding of trametinib to MEK blocks its phosphorylation at serine 217 (11). To compare the relative potencies of PD0325901 and trametinib in vivo, we first exposed BRAF(V600E) SK-Mel-239 and NF1-null SK-Mel-113 cells to increasing concentrations of both drugs and assessed the effect of drug treatment on pERK activation at 1 hour. In BRAF(V600E) SK-Mel-239 cells, both drugs were equipotent in their ability to suppress ERK activation at 1 hour (Fig. 4A). In contrast, in NF1-null SK-Mel-113 cells, trametinib was considerably more potent in its ability to suppress pERK activation than either PD0325901 or two additional allosteric MEK inhibitors currently in clinical testing (AZD6244 and MEK162) (37-39) (Fig. 4A and Supplemental Fig. S4). Treatment of NF1-null melanoma cells with either PD0325901 or trametinib resulted in hyperactivation of RAS consistent with relief of upstream negative feedback following inhibition of ERK (Fig. 4B). However, relief of upstream feedback following MEK inhibition was accompanied by a significant increase in the activation of pMEK in PD0325901-treated NF1-null cells, which was attenuated in cells treated with trametinib (Fig. 4B and Supplemental Fig. S4). This attenuation of MEK phosphorylation was unique to trametinib and was not observed with PD0325901, AZD6244, or MEK162 (Supplemental Fig. S4). Furthermore, the resistance of MEK to upstream hyperactivation by RAS in trametinib-treated cells was accompanied by a more durable down-regulation of pERK activation and more potent inhibition of genes whose transcription is dependent upon ERK including CCND1 (cyclin D1), ETV1, MYC and SPRY4 as compared to PD0325901 (Fig. 4B and 4D). Consistent with these biological differences among the MEK inhibitors, the anti-proliferative effects of trametinib were similar in BRAF(V600E) and NF1-null cells, whereas BRAF(V600E) cells exhibited greater sensitivity than NF1-null cells to PD0325901 (Fig. 4C and Supplemental Fig. S4).
Figure 4.
Trametinib durably inhibits ERK activation and proliferation in NF1-null melanoma cell lines. A) Cells were treated with increasing concentrations of PD0325901 or trametinib for 1 hour. pERK was measured by immunoblot. B) Cells were treated with 50nM PD0325901 or 50nM trametinib for 0, 1, 6 and 24 hours. RAS-GTP was quantitated as described in methods. Phospho- and total levels of MAPK pathway components were determined by immunoblot. C) Cells were treated with increasing doses of PD0325901 or trametinib for 5 days. Results are percent cell growth as a function of drug concentration (nM). D) NF1-null SK-Mel-113 cells were treated with DMSO or 50nM PD0325901 or trametinib for 1, 6, or 24 hours. RNA levels were quantitated by RNA-seq. Shown are relative mRNA expression levels as a function of duration of drug exposure for selected ERK output genes.
NF1 loss and resulting RAS activation in BRAF (V600E) melanoma cells confers resistance to allosteric RAF inhibition
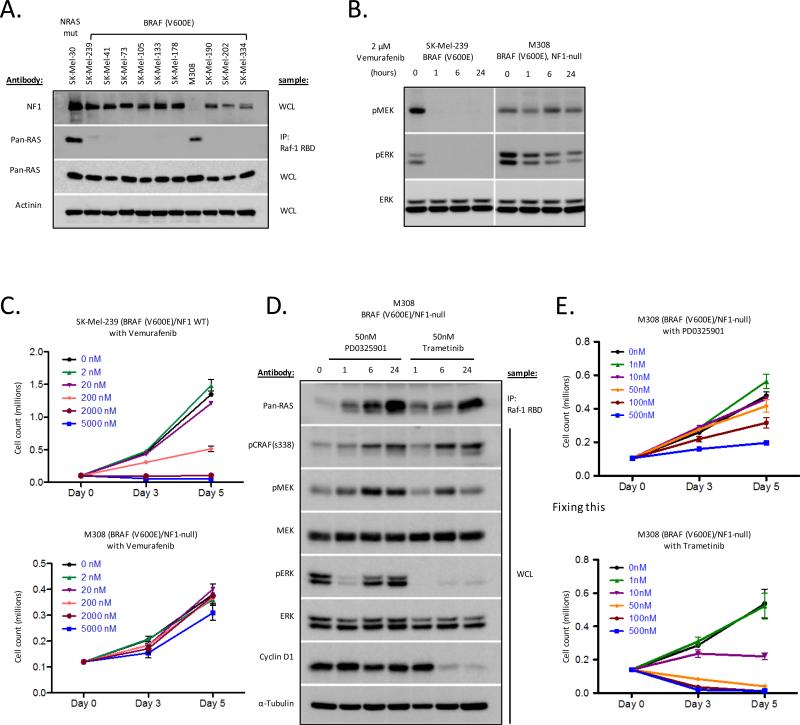
Levels of GTP-bound RAS are low in BRAF(V600E) melanomas as a result of high levels of ERK-dependent negative feedback (23). It was unclear whether loss of NF1 function in this context would be sufficient to overcome the feedback-mediated suppression of RAS activity in BRAF(V600E) cells. We therefore screened a panel of ten BRAF (V600E) melanoma cell lines for loss of NF1 expression and activation of RAS-GTP. Nine of the ten BRAF(V600E) mutant melanoma cell lines expressed low to undetectable levels of RAS-GTP (Fig. 5A). A single BRAF(V600E) cell line (M308) had high levels of RAS-GTP similar to that of an NRAS(Q61K) cell line, and notably, M308 was devoid of NF1 expression by immunoblot (Fig. 5A). Genomic analysis by IMPACT confirmed the presence of a nonsense mutation in the NF1 gene (Q1070*) as the basis for the loss of NF1 protein expression in M308 cells (Supplemental Fig. S5). To determine whether NF1 loss desensitizes BRAF(V600E) cells to RAF inhibition, we assessed the sensitivity of M308 (BRAF (V600E)/NF1-null) cells to vemurafenib. Vemurafenib treatment of SK-Mel-239 (BRAF (V600E)/NF1WT) cells resulted in potent down-regulation of phosphorylated MEK and ERK and inhibition of cell growth in this cell line (Fig. 5B). In contrast, vemurafenib had little effect on levels of phosphorylated MEK and ERK in BRAF (V600E)/NF1-null M308 cells and no effect on cell proliferation (Fig. 5B, C).
Figure 5.
NF1 loss in the context of BRAF(V600E) mutation results in elevated RAS-GTP levels and sensitivity to MEK but not RAF inhibition. A) RAS-GTP was quantitated as described in methods. Mut = mutant, WCL=whole cell lysate. B) Cells were treated with 2 μM vemurafenib for 0, 1, 6, and 24 hours. Phospho- and total levels of MAPK pathway components were determined by immunoblot. C) Cells were treated with increasing concentrations of the RAF inhibitor vemurafenib for 3 or 5 days. Results are cell count as a function of drug concentration over time. Error bars are SEM, n=3. D) Cells were treated with 50nM of PD0325901 or trametinib for 0, 1, 6, and 24 hours. E) Cells were treated with increasing concentrations of PD0325901 or trametinib for 3 or 5 days. Results are cell count as a function of drug concentration over time. Error bars are SEM, n=3.
RAS activation is sufficient to induce vemurafenib resistance in BRAF(V600E) cells (Supplemental Fig. S6). To determine whether NF1 loss activates RAS sufficiently to overcome ERK-dependent negative feedback and induce vemurafenib resistance, we knocked down NF1 expression in BRAF(V600E) mutant A375 cells and assessed levels of RAS activation in the presence and absence of vemurafenib. siRNA and shRNA mediated knockdown of NF1 resulted in induction of RAS-GTP and decreased sensitivity to vemurafenib (Supplemental Fig. S6). These data suggest that loss of NF1 function in BRAF(V600E) cells is sufficient to induce RAS-GTP activation and, as a consequence, vemurafenib resistance (Fig. 5A-C and Supplementary Fig. S6).
To assess the MEK-dependence of the M308 melanoma cells, we determined the effects of PD0325901 and trametinib treatment on ERK activation and cellular proliferation. Analogous to the results seen with these inhibitors in BRAFWT/ NF1-null melanoma cells, exposure of M308 cells to 50nM PD0325901 was insufficient to durably suppress ERK signaling and cell proliferation (Fig. 5D,E). In contrast, treatment of M308 cells with trametinib resulted in durable suppression of pERK activation, potent downregulation of cyclin D1 expression and potent inhibition of cellular proliferation (Fig. 5D,E).
Discussion
Neurofibromatosis Type I is a hereditary disorder caused by germline mutation of the NF1 gene (350 kb genomic DNA; 60 exons; 2818 amino acids). This syndrome is characterized by the formation of benign neurofibromas as well as pigmented café-au-lait spots and Lisch nodules. Individuals with neurofibromatosis are at risk of developing malignant neoplasms at a young age, most commonly optic gliomas, malignant peripheral nerve sheath tumors, and juvenile myelomonocytic leukemia (40). Only recently have somatic alterations in the NF1 gene been implicated in malignancies including gliomas, breast cancers, and leukemias (24-27). The prior underestimation of the prevalence of NF1 alterations in human tumors is likely attributable to the technical challenges previously inherent in sequencing large genes with tumor suppressive function in which mutations throughout the coding region are potentially oncogenic. With the advent of massively parallel next generation sequencing methods, many of these technical challenges have now been overcome.
Here, we report that a significant subset of melanoma cell lines, including those wild-type for BRAF and RAS, exhibit total loss of NF1 protein expression. In all cases, a mutation and/or focal deletion of the NF1 gene, rather than post-transcriptional regulation (41) could be identified as the basis for NF1 loss. In contrast to prior reports (42, 43), all the NF1-null melanoma cell lines expressed levels of active GTP-bound RAS comparable to those found in RAS mutant cells. Notably, NF1 loss was not mutually exclusive with RAS or BRAF mutations. In fact, mutations in NF1 were found to co-associate with additional activating alterations in the RAS/MAP kinase pathway, in particular with exon 11 BRAF mutations (Figure 2C). These BRAF mutations exhibit impaired kinase activity but induce ERK signaling by dimerizing with and activating CRAF (44). As induction of RAS activity through NF1 loss would be predicted to promote the formation of CRAF homo- and heterodimers, NF1 alterations may cooperate with low activity BRAF mutants to induce transformation by further enhancing RAF dimer formation.
Studies using genetically engineered mouse models with melanocyte-targeted deletion of NF1 also suggest that NF1 loss cooperates with BRAF(V600E) mutation to promote melanomagenesis and suggest that this cooperativity results, at least in part, through abrogation of oncogene induced senescence (45). We observed that knockdown of NF1 expression was sufficient to overcome the ERK-dependent feedback suppression of RAS observed in BRAF(V600E) cells. However, loss of both copies of NF1 is likely required to maximally elevate RAS-GTP expression, as a heterozygous splice site mutation in NF1 in A375 cells (45) was not accompanied by NF1 loss or increased RAS-GTP levels (Supplemental Fig. S7). These results suggest that loss of NF1 in BRAF(V600E) melanoma cells may provide a selective advantage, even in the absence of RAF inhibitor exposure, by diminishing the oncogene-induced suppression of RAS mediated by ERK-dependent negative feedback. A secondary consequence of this co-mutation pattern is that such tumors exhibit intrinsic resistance to selective RAF inhibitors. Our studies suggest that partial loss of NF1 function may result in a more pronounced and rapid restoration of RAS signaling following RAF inhibitor therapy. This could result in an attenuation of drug response sufficient to promote the emergence of drug resistant clones.
Efforts to develop clinically useful direct inhibitors of RAS have been unsuccessful to date (46). One alternative pharmacologic strategy for the treatment of tumors with constitutive RAS activation, including those with loss of NF1, is to target the pathways downstream of RAS responsible for maintenance of transformation (Supplemental Fig S8). We observed that in contrast to NF1-null MPNSTs (31, 32), NF1-null melanomas were dependent on ERK pathway activation and not TORC1 for cell cycle progression and cell proliferation. This result indicates that the lineage context within which NF1 is inactivated influences the downstream effector pathways that facilitate RAS-mediated transformation and thus likely dictates the potential utility of targeted pathway inhibitors.
While NF1-null melanomas were dependent upon MEK-ERK activation for cell proliferation, we observed stark differences in the relative potency of allosteric, non-ATP-competitive MEK inhibitors in the NF1-null cohort (11). Specifically, trametinib, which attenuates phosphorylation of MEK by RAF at Serine 217, had greater antitumor effects than PD0325901. Monophosphorylated MEK has only partial activity (11) and the ability of trametinib but not PD0325901 to abrogate the hyperphosphorylation of MEK resulting from relief of upstream negative feedback was associated with more durable inhibition of pERK and cyclin D1 expression and greater anti-proliferative effects. A similar lack of potency was also noted in NF1-null melanoma cells with AZD6244 (Supplemental Fig. S4), a second non-ATP-competitive MEK inhibitor incapable of abrogating RAF phosphorylation of MEK. The inability of AZD6244 to block MEK phosphorylation likely accounts for the partial resistance to MEK inhibition observed following NF1 knockdown in a prior study (47). Our data are also consistent with a recent study suggesting that differences in the cellular potency of MEK inhibitors in KRAS-mutant cells can result from differences in the strength of hydrogen bonding with S212 in MEK, which is critical for blocking feedback induced MEK phosphorylation by wild-type RAF (48). In sum, the data imply that MEK inhibitors that block the phosphorylation of MEK by RAF may have greater clinical activity in tumors with activated RAS, including those with loss of NF1 function. Such inhibitors may, however, have a narrow therapeutic index in patients, as they would be predicted to potently inhibit RAS-dependent ERK signaling in normal tissues.
In summary, NF1 loss is common in cutaneous melanomas. Loss of NF1 is associated with RAS activation, MEK dependence and, in the setting of concurrent BRAF mutation, vemurafenib resistance. Upstream hyperactivation of RAS and RAF resulting from loss of negative feedback following ERK pathway inhibition can result in an attenuation of the anti-tumor activity of allosteric MEK inhibitors. Inhibitors that prevent RAF-mediated phosphorylation of MEK abrogate this adaptive resistance to MEK inhibition and have greater anti-tumor activity in NF1-null cells. With the recent FDA approval of trametinib for the treatment of BRAF-mutant melanomas, these findings have potential therapeutic implications for patients with melanoma and others tumor types with NF1 alterations.
Supplementary Material
Acknowledgements
We thank Alan Houghton for his pioneering work developing the “SK-Mel” cell lines; Jedd Wolchok for insightful comments and collaboration; Katherine Nathanson and Meenhard Herlyn for the “WM” cell lines; the Geoffrey Beene Core, Agnes Viale and the Genomics Core at MSKCC; The Cancer Genome Atlas for allowing pertinent TCGA NF1 results to be incorporated into our analyses; and the patients for contributing tissue for research.
Financial support: This work was funded by the National Institutes of Health; the Melanoma Research Alliance; Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center; Stand Up to Cancer; and the STARR Foundation.
Footnotes
Disclosure of Potential Conflicts of interest: David Solit consults for Pfizer. There are no other potential conflicts of interest.
References
- 1.Downward J. Control of ras activation. Cancer Surv. 1996;27:87–100. [PubMed] [Google Scholar]
- 2.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer research. 2001;61:3595–8. [PubMed] [Google Scholar]
- 3.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N Engl J Med. 2011 doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer research. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 7.Gorden A, Osman I, Gai W, He D, Huang W, Davidson A, et al. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer research. 2003;63:3955–7. [PubMed] [Google Scholar]
- 8.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. The New England journal of medicine. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 9.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nature reviews Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 10.Cohen C, Zavala-Pompa A, Sequeira JH, Shoji M, Sexton DG, Cotsonis G, et al. Mitogen-actived protein kinase activation is an early event in melanoma progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:3728–33. [PubMed] [Google Scholar]
- 11.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 12.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer research. 2010;70:5901–11. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–62. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanrahan AJ, Schultz N, Westfal ML, Sakr RA, Giri DD, Scarperi S, et al. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov. 2012;2:56–67. doi: 10.1158/2159-8290.CD-11-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nusse M, Beisker W, Hoffmann C, Tarnok A. Flow cytometric analysis of G1- and G2/M-phase subpopulations in mammalian cell nuclei using side scatter and DNA content measurements. Cytometry. 1990;11:813–21. doi: 10.1002/cyto.990110707. [DOI] [PubMed] [Google Scholar]
- 22.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31:446–57. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudry-Labis E, Roche-Lestienne C, Nibourel O, Boissel N, Terre C, Perot C, et al. Neurofibromatosis-1 gene deletions and mutations in de novo adult acute myeloid leukemia. Am J Hematol. 2013;88:306–11. doi: 10.1002/ajh.23403. [DOI] [PubMed] [Google Scholar]
- 25.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haferlach C, Grossmann V, Kohlmann A, Schindela S, Kern W, Schnittger S, et al. Deletion of the tumor-suppressor gene NF1 occurs in 5% of myeloid malignancies and is accompanied by a mutation in the remaining allele in half of the cases. Leukemia. 2012;26:834–9. doi: 10.1038/leu.2011.296. [DOI] [PubMed] [Google Scholar]
- 28.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brems H, Beert E, de Ravel T, Legius E. Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. Lancet Oncol. 2009;10:508–15. doi: 10.1016/S1470-2045(09)70033-6. [DOI] [PubMed] [Google Scholar]
- 31.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8573–8. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johannessen CM, Johnson BW, Williams SM, Chan AW, Reczek EE, Lynch RC, et al. TORC1 is essential for NF1-associated malignancies. Current biology : CB. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 33.Sebolt-Leopold JS MR, Omer C, Tecle H, Bridges A, Klohs W, Loi CM, Valik H, Przybranowski S, Meyer M, Leopold WR. The biological profile of PD 0325901: A second generation analog of CI-1040 with improved pharmaceutical potential. Proc Amer Assoc Cancer Res. 2004:45. [Google Scholar]
- 34.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poulikakos PI, Solit DB. Resistance to MEK inhibitors: should we co-target upstream? Sci Signal. 2011;4:pe16. doi: 10.1126/scisignal.2001948. [DOI] [PubMed] [Google Scholar]
- 37.Catalanotti F, Solit DB, Pulitzer MP, Berger MF, Scott SN, Iyriboz T, et al. Phase II trial of MEK inhibitor selumetinib (AZD6244, ARRY-142886) in patients with BRAFV600E/K-mutated melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:2257–64. doi: 10.1158/1078-0432.CCR-12-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel SP, Lazar AJ, Papadopoulos NE, Liu P, Infante JR, Glass MR, et al. Clinical responses to selumetinib (AZD6244; ARRY-142886)-based combination therapy stratified by gene mutations in patients with metastatic melanoma. Cancer. 2013;119:799–805. doi: 10.1002/cncr.27790. [DOI] [PubMed] [Google Scholar]
- 39.Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CM, Queirolo P, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–56. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 40.Legius E, Marchuk DA, Collins FS, Glover TW. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet. 1993;3:122–6. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- 41.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen LB, Fountain JW, Gutmann DH, Tarle SA, Glover TW, Dracopoli NC, et al. Mutations in the neurofibromatosis 1 gene in sporadic malignant melanoma cell lines. Nat Genet. 1993;3:118–21. doi: 10.1038/ng0293-118. [DOI] [PubMed] [Google Scholar]
- 43.Johnson MR, Look AT, DeClue JE, Valentine MB, Lowy DR. Inactivation of the NF1 gene in human melanoma and neuroblastoma cell lines without impaired regulation of GTP.Ras. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5539–43. doi: 10.1073/pnas.90.12.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 45.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338– 49. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Downward J. Targeting RAS signalling pathways in cancer therapy. Nature reviews Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 47.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013 doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.