Abstract
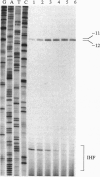
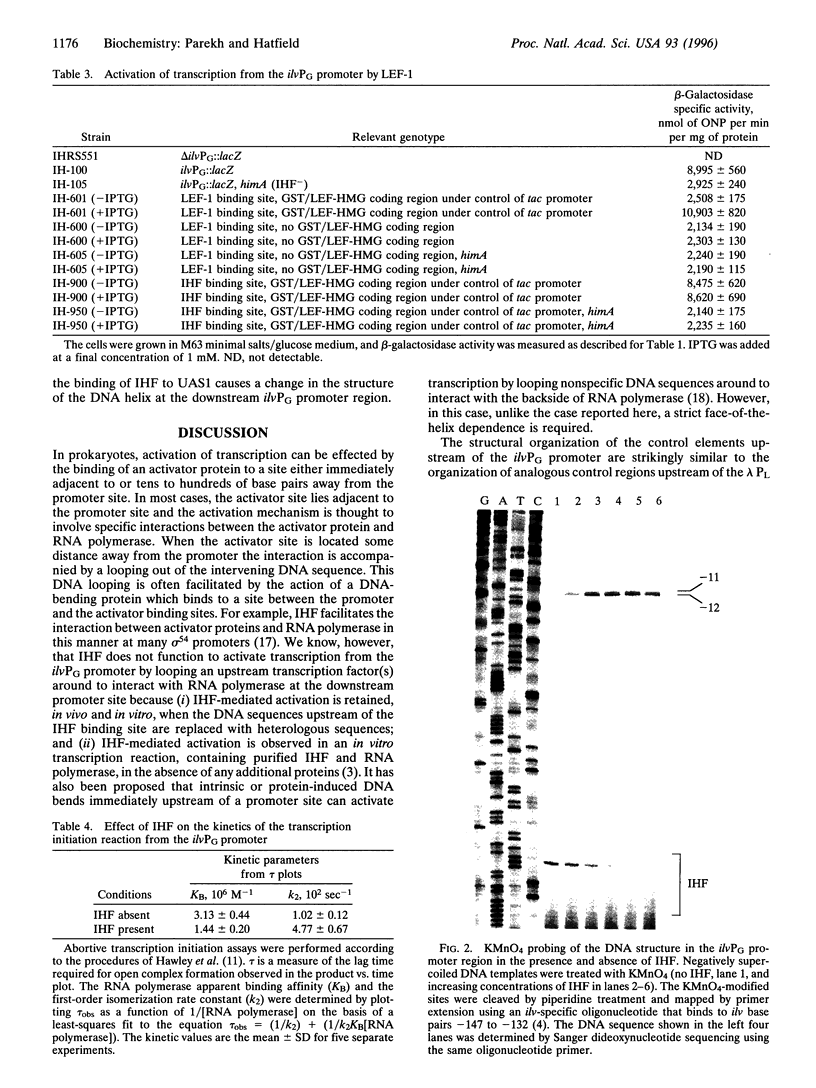
Integration host factor (IHF) is a DNA-bending protein that binds to an upstream activating sequence (UAS1) and, on a negatively supercoiled DNA template, activates transcription from the ilvPG promoter of the ilvG-MEDA operon of Escherichia coli. The transcriptional initiation site of the ilvGMEDA operon is located 92 bp downstream of UAS1. Activation is still observed when the orientation of the upstream IHF binding site is reversed. This manipulation places the IHF binding site on the opposite face of the DNA helix, directs the IHF-induced DNA bend in the opposite direction, and presents the opposite face of the nonsymmetrical, heterodimeric, IHF molecule to the downstream RNA polymerase. Lymphoid enhancer-binding factor, LEF-1, is a DNA-bending, lymphoid-specific, mammalian transcription factor that shares no amino acid sequence similarity with IHF. When the IHF site in UAS1 is replaced with a LEF-1 site, LEF-1 activates transcription from the downstream ilvPG promoter in E. coli as well as it is activated by its natural activator, IHF. These results suggest that specific interactions between IHF and RNA polymerase are not required for activation. The results of DNA structural studies show that IHF forms a protein-DNA complex in the UAS1 region that, in the absence of RNA polymerase, alters the structure of the DNA helix in the -10 hexanucleotide region of the downstream ilvPG promoter. The results of in vitro abortive transcription assays show that IIIF also increases the apparent rate of RNA polymerase isomerization from a closed to an open complex. We suggest, therefore, that IHF activates transcription by forming a higher-order protein-DNA complex in the UAS1 region that structurally alters the DNA helix in a way that facilitates open complex formation at the downstream ilvPG promoter site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Cole J. R., Nomura M. Translational regulation is responsible for growth-rate-dependent and stringent control of the synthesis of ribosomal proteins L11 and L1 in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4129–4133. doi: 10.1073/pnas.83.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Giese K., Amsterdam A., Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991 Dec;5(12B):2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Giladi H., Igarashi K., Ishihama A., Oppenheim A. B. Stimulation of the phage lambda pL promoter by integration host factor requires the carboxy terminus of the alpha-subunit of RNA polymerase. J Mol Biol. 1992 Oct 20;227(4):985–990. doi: 10.1016/0022-2836(92)90514-k. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., McClure W. R. Regulation of open complex formation at the Escherichia coli galactose operon promoters. Simultaneous interaction of RNA polymerase, gal repressor and CAP/cAMP. J Mol Biol. 1992 Mar 5;224(1):15–29. doi: 10.1016/0022-2836(92)90573-3. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Giese K., Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994 Mar;10(3):94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Heck J. D., Hatfield G. W. Valyl-tRNA synthetase gene of Escherichia coli K12. Molecular genetic characterization. J Biol Chem. 1988 Jan 15;263(2):857–867. [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel J. M., Winkelman J. W., Adams C. W., Hatfield G. W. DNA topology-mediated regulation of transcription initiation from the tandem promoters of the ilvGMEDA operon of Escherichia coli. J Mol Biol. 1992 Apr 20;224(4):919–935. doi: 10.1016/0022-2836(92)90460-2. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J., Espinosa M. The RepA repressor can act as a transcriptional activator by inducing DNA bends. EMBO J. 1991 Jun;10(6):1375–1382. doi: 10.1002/j.1460-2075.1991.tb07657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Ross W., Thompson J. F., Newlands J. T., Gourse R. L. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990 Nov;9(11):3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]