Abstract
A new type of signaling network element, called cancer signaling bridges (CSB), has been shown to have the potential for systematic and fast-tracked drug repositioning. On the basis of CSBs, we developed a computational model to derive specific downstream signaling pathways that reveal previously unknown target–disease connections and new mechanisms for specific cancer subtypes. The model enables us to reposition drugs based on available patient gene expression data. We applied this model to repurpose known or shelved drugs for brain, lung, and bone metastases of breast cancer with the hypothesis that cancer subtypes have their own specific signaling mechanisms. To test the hypothesis, we addressed specific CSBs for each metastasis that satisfy (i) CSB proteins are activated by the maximal number of enriched signaling pathways specific to a given metastasis, and (ii) CSB proteins are involved in the most differential expressed coding genes specific to each breast cancer metastasis. The identified signaling networks for the three types of breast cancer metastases contain 31, 15, and 18 proteins and are used to reposition 15, 9, and 2 drug candidates for the brain, lung, and bone metastases. We conducted both in vitro and in vivo preclinical experiments as well as analysis on patient tumor specimens to evaluate the targets and repositioned drugs. Of special note, we found that the Food and Drug Administration-approved drugs, sunitinib and dasatinib, prohibit brain metastases derived from breast cancer, addressing one particularly challenging aspect of this disease.
Introduction
Drug repositioning benefits significantly from the systematic investigation of the mechanism of action of drugs against a new disease indication. Our previous work developed a new type of signaling network elements, called cancer signaling bridges (CSB), to investigate underlying signaling mechanisms systematically (1). CSBs are able to extend the known canonical signaling pathways (2–4) to proteins whose coding genes have a close relationship with cancer genetic disorders (5, 6) or, in brief, cancer proteins. Each CSB is a specific instance of a network motif (7), that is, recurrent and statistically significant sub-graphs or patterns, in the protein–protein interaction (PPI) network. To further ensure that the CSBs are able to link many previously unrelated cancer proteins to a known signaling pathway of interest, the CSBs were defined as those network motif instances whose proteins include at least one protein in a signaling pathway and at least one cancer protein outside the signaling pathway. As an example, a CSB comprises four proteins, BRCA1, GRB2, HSPA8, and NPM1 with four protein–protein interactions, BRCA1<>HSPA8, BRCA1<>NPM1, GRB2<>HSPA8, and GRB2<>NPM1. The coding gene of the NPM1 protein is found mutated in acute promyelocytic leukemia, but its signaling mechanism remains unclear. Using this CSB, we can expand the NPM1 to the EGF pathway through the linkage of GRB2 or E2F transcription factor network through the linkage of BRCA1.
The identified CSBs enable drug repositioning based on transcriptional response data and has been evaluated in drug repositioning studies against breast cancer, prostate cancer, and promyelocytic leukemia cells (1). However, similar to many other available drug repositioning methods, such as those using gene signatures to address the similarities between drugs (8) or the associations between drugs and diseases (9, 10), our previously reported drug repositioning method relies on the availability of transcriptional response data.
Alternative methods of drug repositioning aim to reconstruct disease-specific networks or pathways from the common gene expression profiles without any drug treatment information. The key proteins identified in the networks or pathways may serve as potential drug targets (11–13). A common problem for these methods is that they are restrictive in finding reliable drug target candidates from generally known or canonical signaling pathways, obtained from either publicly available databases, such as Kyoto Encyclopedia of Genes and Genomes (4) and Reactome (14), or commercially available databases, such as TransPath (Bio-Base Inc), MetaBase (GeneGo Inc.), and Ingenuity Pathway Analysis (Ingenuity Systems Inc.). For example, the casual reasoning method (12) only takes into account upstream signaling proteins whereas the pathway pattern-based approach (13) simply employs the information on known pathways directly to address disease relationships. These methods are incapable of studying subtypes of the same cancer or different cancers sharing common pathways as they fail to explore specific mechanisms of action that are unknown to the existing databases. In addition, the efforts on upstream signaling proteins cannot discern the detailed downstream differences on the signaling mechanisms among cancer subtypes or cancers sharing similar signaling pathways. Repositioning drugs for these specific cancers warrants fresh approaches to derive their specific signaling pathways.
The CSBs are a powerful means to derive the specific signaling mechanisms for cancer subtypes. Different from the common upstream signaling pathways, more than half of the CSB proteins are located in the nucleus and serve as downstream signaling molecules. Our study showed that the CSBs are mostly cancer-specific and play important roles as the targets of anticancer drugs (9). These properties indicate the merits of CSBs in identifying specific cancer signaling mechanisms for individual cancer subtypes.
Following our strategy of integrating computational biology and experimental biology to address cancer problems, we extended the concept of CSBs and developed a computational model to reposition drugs for cancer subtypes on the basis of the common gene expression profiles without any drug treatment information. The hypothesis behind our model is that individual cancer subtypes have their own specific signaling mechanisms. We applied the integrative cancer biology-based drug repositioning method to brain, lung, and bone metastases of breast cancer. The results confirmed the hypothesis in that the signaling networks for the three metastases include different sets of proteins and drug targets. Accordingly, we repositioned drug candidates for the three metastases of breast cancer to individual sets of drug targets identified. Both in vitro and in vivo experiments validate the targets and identify two known drugs for new indication in brain metastasis of breast cancer.
Materials and Methods
RNA isolation and gene-expression profiling
Core biopsies from primary tumors of 52 patients of breast cancer were obtained for Affymetrix Human Genome U133 GeneChip cDNA array analysis. This study was approved by the Institutional Review Board at The Methodist Hospital. The data analysis approach was described previously in our publication (15).
Human breast tumor microarray datasets
Microarray data from four cohorts of breast tumors that include patients with brain, lung, and bone relapse were used in the analysis. The EMC-192 (16) cohort includes 16 cases of brain relapse, 21 cases of lung relapse, and 53 cases of bone relapse. The EMC-286 (17) cohort includes 10 cases of brain relapse, 25 cases of lung relapse, and 69 cases of bone relapse. The MSK-82 (18) cohort includes five cases of brain relapse, 14 cases of lung relapse, and 14 cases of bone relapse. The TMH-52 cohort is a treatment-refractory triple negative breast tumor brain relapsed cohort that includes 11 cases of brain relapse. The TMH-52 cohort microarray data is available at NCI GEO with ID: GSE 46928.
The computational modeling of specific downstream signaling pathways
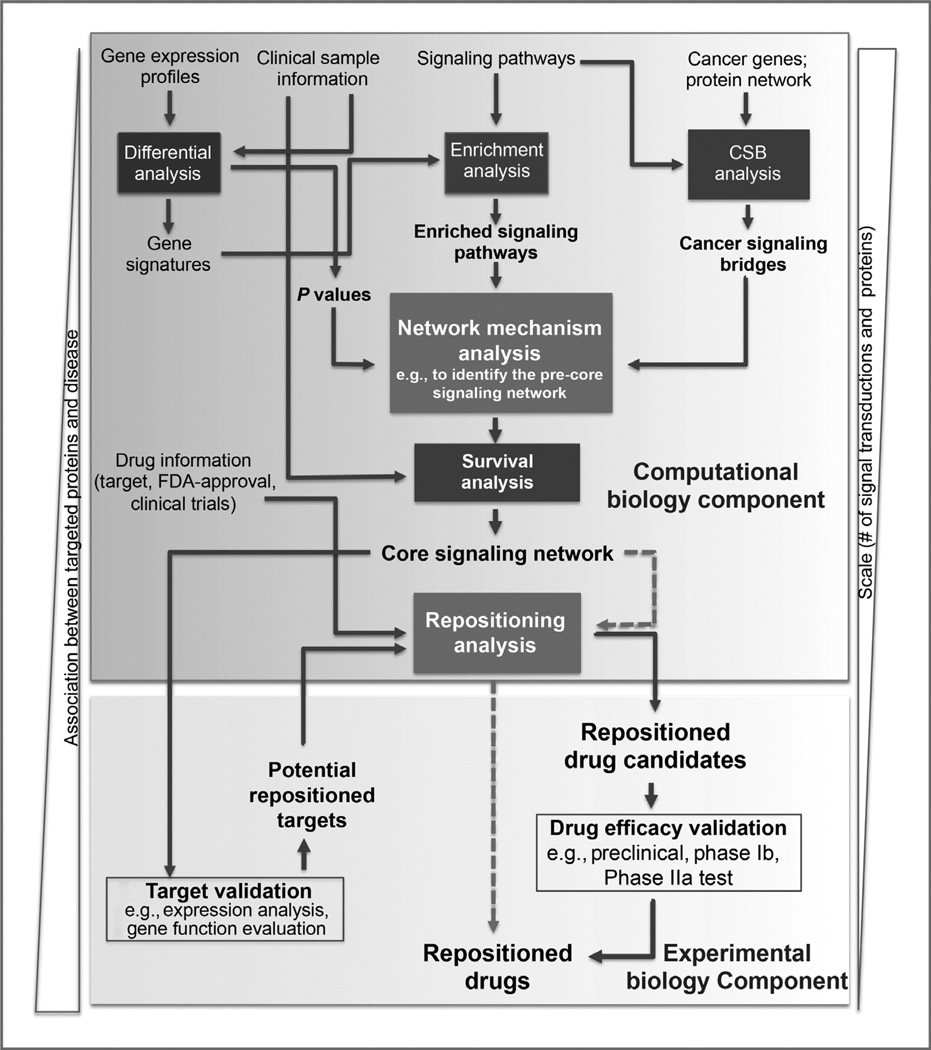
The drug repositioning pipeline comprises six computational analysis modules and two experimental modules: differential analysis, enrichment analysis, CSB analysis, network mechanism analysis (the computational modeling), survival analysis, and repositioning analysis; and target and drug efficacy validation (Fig. 1). All analysis modules enable the extensive integration of gene expression profiles and clinical survival information of patients, known signaling pathways, and drug information of targets, clinical trials, and Food and Drug Administration (FDA)-approval statuses.
Figure 1.
An overview of the technical pipeline for cancer drug repositioning. The pipeline comprises six modules for the computation biology analyses (Differential analysis, Enrichment analysis, CSB analysis, Survival analysis, Signaling network analysis, and Repositioning analysis) and two modules for experimental biology analyses (Target validation and Drug efficacy validation). The Signaling network analysis, which is the core of the computational component, is to refine the general signaling networks to the core signaling network that is specific to the cancer of interest. The Differential analysis and Enrichment analysis modules provide the differential genes and enriched signaling pathways for the mathematical model (see Materials and Methods), whereas the CSB analysis supplies the essential cancer signaling network for the mathematical model. The Survival analysis enables further narrowing of the signaling networks down to the core signaling network based on the metastasis-free survival times of patients. The Repositioning analysis identifies the repositioned drug candidates from the available drug information, integrating with the two experimental biology modules, Target validation and Drug efficacy validation.
Differential analysis and enrichment analysis
Differential and enrichment analyses aim to address the differentially expressed genes (gene signatures) from gene expression profiles of patients and discover enriched signaling pathways associated with the identified genes signatures. Two commercial software packages were used to implement the analyses: Partek (http://www.partek.com/) and Ingenuity Pathway Analysis (http://www.ingenuity.com/).
CSB analysis
CSBs are specific instances of network motifs in the PPI network, acting as the important signaling elements between known signaling pathways and cancer proteins associated with cancer genetic disorders. For a detailed definition, please see Supplementary Methods section 1 and reference (1). A relatively high-quality CSB set was filtered out for this drug repositioning study in which the proteins physically interact and the PPIs are verified by at least two experiments (Supplementary Methods).
Signaling network analysis
By integrating the outputs from the differential and the enrichment analyses, this analysis component addressed the signaling networks from CSBs. The network mechanism analysis can be represented by a mathematical model, that is, multiobjective optimization model, as shown in equations (1–5). The output of this analysis, that is, presignaling network, involves the maximal number (defined by the first objective parameter, (a) of signaling pathways and includes the most differentially expressed genes in the gene signatures (defined by the second objective parameter, (b). To satisfy the survival analysis requirement on the protein paths instead of single genes, we used a heuristic strategy to solve the proposed mathematical model. The strategy first decomposes the CSBs into identical length protein paths by using an enumeration method, and then, for each path, it computes the terms goal1 and goal2. Thus, the parameters, (a) and (b) are helpful to prioritize the paths by considering two criteria: (i) the number of enriched signaling pathways and (ii) the statistical significance of differentially expressed genes. In this study, we used a path length l of 5; the parameterization process of (a) and (b) is shown in Section 2 of Supplementary Methods.
| (1) |
| (2) |
| (3) (4) (5) |
where x is a variable vector denoting all of the proteins of CSBs; each xi(i = 1, 2, ⋯, N) equals either 1 or 0, indicating whether the protein i is kept in the output of the model (core signaling network); and Aij is an adjacent matrix representing the interactions between the CSB proteins. Equations 3 and 4 are used to ensure the connectivity of the core signaling network. Equation (1) is used to ensure the first criterion: including the maximal number of enriched signaling pathways in the output; Si is a signaling pathway set for protein i that includes the enriched signaling pathways that take protein i as their signaling components; I(xi) is an indicator with the same value as xi; and the term goal1 denotes the average number of enriched signaling pathways of an output network. Similarly, equation 2 satisfies another criterion: involving the most differentially expressed genes in the output; P valuei is the output of the statistical analysis of differential analysis; the term goal2 stands as the average statistical significance of the coding genes of an output network; and a and b are two parameters to control the scale of the output network, playing important roles in the following analyses.
Survival analysis
The survival analysis component aims to integrate the outputs of the signaling network analysis component with the available clinical information of the patients, for example, metastasis-free survival time, to further refine the identified presignaling networks. To filter out the high confidence proteins paths, we conducted hierarchical clustering on the paths and Kaplan–Meier survival analysis on the survival times of the patients, iteratively.
To do hierarchical clustering on the expression profiles of protein paths rather than those of individual genes, we applied our previously developed network biomarker method (19) to transform the expression profiles from the gene probe-level to the path-level:
| (6) |
where path = [probe1, probe2, ⋯, probel]; probei is any probe of the coding genes of the ith protein in the protein path; Iprobei is a vector of the gene expression values of probei across all of the patients considered; P valuei is the P value of the statistical analysis of the differential analysis; and Ipath is the merged gene expression profile defined by its component probes.
After the hierarchical clustering on the expression profiles of the paths, a heuristic cut-tree algorithm was used to focus on the relevant paths. The cut-tree algorithm can cut the clustering tree into relatively small branches. The heuristic algorithm is used to remove each branch and conduct Kaplan– Meier survival analysis based on the remaining branches, including a portion of the paths. The statistical significance in the Kaplan–Meier survival analysis can help to determine which branch should be removed from the paths. Hierarchical clustering, cut-tree algorithm, and Kaplan–Meier survival analysis were implemented in R (http://www.r-project.org/), using the hclust and survival packages.
A remaining problem is evaluating the outputs of the cut-tree algorithm. After running the cut-tree algorithm on the paths, the output is composed of a set of subtrees after removing the branches iteratively. Each subtree can be characterized by a three-dimensional cube with statistical P value of Kaplan–Meier survival analysis, path number, and classification on the patients (Supplementary Fig. S7 and Supplementary Methods sections 3 and 4). The subtree with relatively low P value, small number of paths, and better classification becomes the final output of this algorithm, and the corresponding paths are entered into the finalized signaling network.
Repositioning analysis
The signaling network derived from the survival analysis is highly associated with the metastasis- free survival time of patients and helps to illustrate specific signaling interactions associated with specific cancers. In addition, these signaling networks may delineate the underlying unknown signaling mechanisms such that the drug repositioning analysis can be conducted to search for known drugs targeting the signaling networks.
Because drug repositioning typically focuses on existing drugs or shelved assets that have passed at least phase I clinical trials, we developed a Web-accessible database, DrugMap Central, to support efficient query and navigation during the drug repositioning studies. DrugMap Central integrates information of chemical structures and properties with targets of known drugs or compounds, as well as accesses online clinical trial updates and FDA-approval information (20).
RT2-PCR array
Customized RT2-PCR Array from the SuperArray Bioscience Corporation was used to examine the expression of the 31 genes in the brain metastasis of breast cancer (BCBM) signaling network. Housekeeping genes ACTB, GAPDH, and RPLP0 were included on the array to normalize the RNA amounts. The data analysis approach used was described previously in our publication (21). We used 20 patient specimens because (i) the tissues were available for the analysis, and (ii) all 20 patients have 0-month metastasis-free survival time, that is, when they presented to the doctors, they were diagnosed with both primary tumor and metastatic tumor.
Immunohistochemistry
Immunohistochemistry (IHC) was conducted on 5 µm formalin-fixed, paraffin-embedded tissue sections from BCBM patients (n = 29; The Methodist Hospital Tissue Bank). This study was approved while waivers of consent were granted by the Institutional Review Board at The Methodist Hospital. The staining for p-RET [Phospho-Ret (Tyr905) antibody, 1:100, Santa Cruz Biotechnology, Inc. cat # sc-57431] and p-FYN [Phospho-Fyn (Tyr416) 1:100, Santa Cruz Biotechnology, cat # sc-16848] on the total 29 samples and scoring the IHC stains were conducted by TMHRI Pathology Core. An H score was calculated by multiplying the fraction of positively stained tumor (percentage) by staining intensity (0, 1+, 2+, or 3+; ref. 22). Membranous immunoreactivity was scored (0 and 1+ indicates negative; 2+, indeterminate; and 3+, positive for overexpression), and the percentage of tumor cells staining positive was visually estimated.
Drugs and cell lines
Sunitinib and dasatinib were purchased from LC Laboratory. Sunitinib was dissolved in 0.1 mol/L citrate buffer (pH 4.7) at a stock concentration of 3 mg/mL. Dasatinib was dissolved at 10 mmol/L in 100% dimethyl sulfoxide (DMSO). The human breast cancer brain metastatic sublines MB231-Br-HER2+, MB231Br-vector (MB231-Br-HER2-), and CN34-Br were previously described (16, 23, 24). Primary human brain microvessel endothelial cells (HBMEC) and astrocytes (NHA) were purchased from Cell Systems Corporation and cultured in the suggested media. All cell lines used in this study were regularly authenticated by morphologic observation and tested for absence of Mycoplasma contamination (MycoAlert, Lonza Rockland).
RNA interference-mediated knockdown cell lines
Knockdown of FYN and RET were achieved by short hairpin RNA (shRNA) Lentiviral Particles (sc-35425-V for FYN, sc-36404-V for RET, Santa Cruz Biotechnology), which is a pool of concentrated viral particles containing three target-specific constructs that encode 19 to 25 nt (plus hairpin) shRNA designed to knock down FYN and RET gene expression (Supplementary Materials). Lentiviral Particles containing a shRNA construct encoding a scrambled sequence were used as control shRNA (sc-108080, Santa Cruz Biotechnology). The efficiency of the knockdown was confirmed by Western immunoblot analysis; antibodies for Ret (sc-167) and FYN (sc-434) were purchased from Santa Cruz Biotechnology. Cell lines with a transduction rate of more than 70% were used for further studies.
Western blotting and antibodies
Cell or tissue lysates and immunoblotting analysis were done as described previously (24). The antibodies are described in Supplementary Materials. Densitometric analysis was conducted using ImageJ software.
In vitro blood–brain barrier assay
The analysis on cancer cell lines penetrating in vitro blood– brain barrier (BBB) was conducted as described by Paula and colleagues (16), with 105 HBMECs placed in the top chamber of the inserts and human primary astrocytes on the counter side of the insert membrane.
In vivo animal experiments
Animal procedures were conducted in accordance with the guidelines of Institutional Animal care and Use Committee and the regulations of the Animal Research and Comparative Medicine Committee of The Methodist Hospital Research Institute. Female BALB/c nude mice (6–7 weeks old; Charles River Laboratories) were anesthetized with isoflurane/O2, and injected in the left cardiac ventricle with cancer cell lines (1.75 × 105 cells in 0.1 mL serum-free medium). The animals were then randomly divided into five treatment groups: vehicle, low-dose sunitinib (40 mg/kg), high-dose sunitinib (80 mg/kg), low-dose dasatinib (25 mg/ kg), and high-dose dasatinib (50 mg/kg). Treatment started three days after cell injection. Both sunitinib and dasatinib were administered orally once daily for 28 days. The other batches of mice were injected with the FYN or RET shRNA knockdown cell lines, and appearance of brain metastasis was monitored every 3 days by IVIS200 bioluminescent imaging. The body condition was monitored once every day, and the mice were euthanized by CO2 asphyxiation if there were signs of neurologic impairment or if the body condition score was 2 or less.
The animals were euthanized 6 hours after last treatment. The whole brain was subjected to enhanced green fluorescent protein (EGFP) fluorescence imaging to detect the presence of the injected cells (Maestro 420 In Vivo Spectral Imaging System, Cambridge Research and Instrumentation). The data acquisition and processing software was used to unmix images of fluorescence from multiple sources (Nuance Technology). After fluorescence imaging, some of the mouse brains were used for histologic and pathologic studies, and others were used for examining the EGFP expression in brain lysates.
Mouse brain sections (10 µm thick) were cut serially. One section from every 100 µm was stained with hematoxylin and eosin (H&E) and immunohistochemistry for p-RET, p-FYN, Ki67 (1:200, Abcam cat # ab66155), and CD31 (1:200, Abcam cat #ab9498). The whole-section montage images were acquired by an Olympus BX61 upright microscope (TMHRI Advanced Cellular and Tissue Microscope Core Facility), and an image analysis algorithm was developed to automatically quantify the number of large lesions in the montage H&E gray-scale images, as described previously (24).
Statistical methods
The Kaplan–Meier method and log-rank test were used to compare differences among survival curves for the shRNA knockdown study. For in vivo drug treatment study, the normality of each group data was examined with the Jarque–Bera test first, and then Levene's F test was used to test multigroup homogeneity of variance (HOV). If the groups enrolled followed the HOV, a Student t test was used for analyzing experimental data. Otherwise, a Wilcoxon rank sum test was executed. For the in vitro study results, ANOVA was deployed for the data analysis. For in vitro BBB cell transmigration assay analysis, a one-way ANOVA was conducted, by cell line (231-Br-HER2-, 231-Br-HER2+), with compound doses or different compounds specified as the factor. For ANOVA of the IHC data, a binomial distribution for the outcome variable and a logit link function were used. Higher order effects (e.g., three-way interactions) were dropped from the model if P was greater than 0.05. Survival analysis was implemented by an R package "Survival." Statistical significance was defined as P less than 0.05. All statistical tests were two-sided. All analyses were conducted with SAS statistical software (version 9.1; SAS Institute, Cary, NC).
Results
The computational modeling of specific downstream signaling pathways
In our computational model, we assume that different metastases have their own specific downstream signaling pathways. In this article, "upstream" signaling pathways refer to those signal transductions starting from ligands and receptors near the cell membrane, whereas "downstream" signaling pathways refer to the signals ending inside nucleus. Further study on the CSBs indicates that more than half of CSB proteins locate in the nucleus (Supplementary Fig. S1A). The new computational model presented in this article addresses the specific downstream signaling pathways for each metastasis type from the CSBs by considering two criteria: (i) these downstream signaling pathways are activated by the maximal number of upstream signaling pathways; and (ii) they involve most differentially expressed genes specific to the metastasis of interest (see Materials and Methods).
To enable computational modeling, we need the following inputs to the model: the downstream signaling network identified from CSBs; the activated upstream signaling pathways; and the differential genes for each of the metastasis studied. Our model contains a number of computational analyses, including differential analysis for differential genes, enrichment analysis for activated upstream signaling pathways, and CSB analysis for candidate downstream signaling pathways (Fig. 1).
Differential analysis, enrichment analysis, and CSB analysis
Differential and enrichment analyses were used to identify the differential genes and the activated upstream signaling pathways for each metastasis type using the patients' gene expression profiles (16–18) and known signaling pathways (2–4). The most differentially expressed genes (P < 0.001, Student t test) were identified between patients who had tissue specific-metastasis and other patients in the same cohort. The differential genes identified were used for pathway enrichment analysis (Fisher exact two-tailed test). For brain, lung, and bone metastasis of breast cancer, we addressed eight, eight, and 21 activated upstream signaling pathways, as shown in Supplementary Tables S1.
The CSB analysis was used to define the candidate downstream signaling pathways. The CSBs are essentially network motifs that connect upstream signaling pathway proteins to cancer-specific proteins whose coding genes are closely related to genetic cancer disorders (ref. 1; Supplementary Methods). We filtered out a set of high-quality CSBs, including 673 proteins and 1,759 physical PPIs, in which each interaction was verified by at least two independent experimental methods (Supplementary Methods).
Computational modeling
The presented computational model takes the results of the differential analysis, enrichment analysis, and CSB analysis as inputs to derive the specific downstream signaling pathways for each metastasis type by using a multiple-objective optimization model (see Materials and Methods). We make use of two objectives in the multiple-objective optimization model to implement the two computational modeling criteria (see Materials and Methods). The heuristic strategy decomposed the network of CSBs into identical-length protein paths and then applied the two aforementioned criteria to obtain specific downstream signaling pathways (Supplementary Methods). Subsequently, we identified 98-, 144-, and 134-protein downstream signaling networks for brain, lung, and bone metastases of breast cancer (Supplementary Figs. S1B–S1D).
Correlation of the downstream signaling pathways with patients' survival times and repositioning drugs based on the derived high-confidence downstream signaling mechanisms
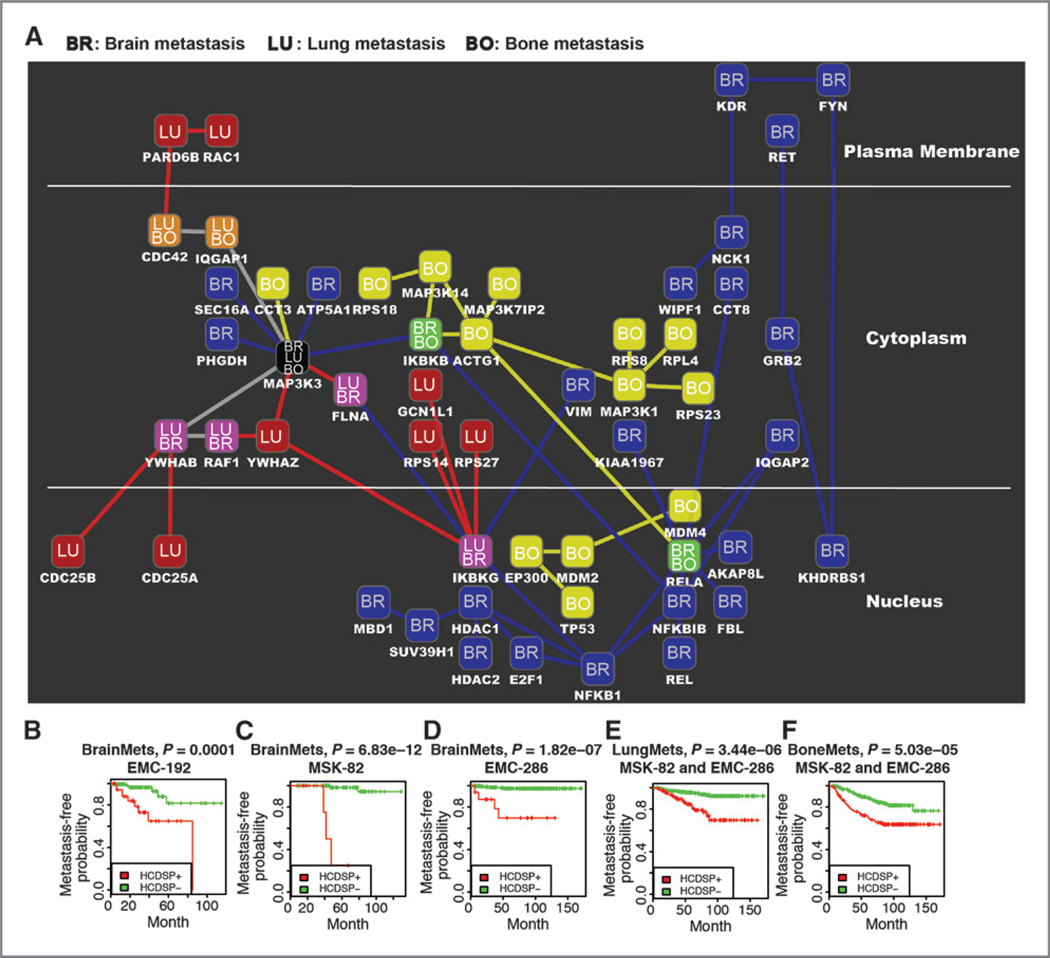
To improve the confidence of the identified downstream signaling pathways, we correlated them with patient survival times. This survival analysis aims to use clinical information to refine the derived downstream signaling pathways. The strategy employed in this module iteratively conducted both hierarchical clustering on the decomposed protein paths of the downstream signaling pathways and Kaplan–Meier survival analysis on the metastasis-free survival times of patients. To satisfy the hierarchical clustering, the network biomarker method (19) was applied to converge the gene probe-level expression to the path-level (see the equation 6 in Materials and Methods). Following the hierarchical clustering on the paths, a heuristic cut-tree algorithm was designed to narrow down the paths to achieve the best results for Kaplan–Meier survival analysis (see Supplementary Methods). Those paths remaining in the cut-tree algorithm with the best statistical significance after the Kaplan–Meier survival analysis are high-confidence downstream signaling pathways (HCDSP). The HCDSPs for brain, lung, and bone metastases contained 31, 15, and 18 proteins, verifying our hypothesis that different metastases have their own specific downstream signaling pathways (Fig. 2A). Distinguished as BR (brain), LU (lung), and BO (bone), the downstream signaling mechanisms between these three metastasis types are different from each other.
Figure 2.
Signaling networks for brain, lung, and bone metastases of breast cancer. A, the signaling networks for the three metastatic sites are denoted by the labels in the nodes as well as the colors in the nodes and the interactions. Brain, blue BR; lung, red LU; and bone, yellow BO. The shared nodes are represented by the labels BR/LU, BR/BO, LU/BO, and BR/LU/BO. The artwork was generated by using Cytoscape (47). B, the Kaplan–Meier curves for metastasis-free survival on the basis of brain, bone, or lung HCDSP path-level values in the indicated cohorts of breast tumors. High-confidence downstream signaling pathway (HCDSP) contains the protein paths remaining in the cut-tree algorithm with the best statistical significance in the Kaplan–Meier survival analysis. The strategy employed in the survival analysis iteratively conducted both hierarchical clustering on the decomposed protein paths of the downstream signaling pathways and Kaplan–Meier survival analysis on the metastasis-free survival times of patients. The cut-tree algorithm helps to classify the patients into HCDSP+ and HCDSP− by cutting the tree in the hierarchical clustering into branches. The HCDSP+ means the patient samples are in the branch with indicator 1, whereas HCDSP− indicates the patient samples are in the remaining branches with indicator −1, where 1 and −1 are defined by the classification based on branches in the cut-tree algorithm. For more details, please see the cut-tree algorithm in Supplementary Methods.
We used different cohorts to apply the cut-tree algorithm in the Kaplan–Meier survival analysis. For lung and bone metastases, we considered the combined cohort, EMC-368 (MSK-82 and EMC-286), whereas for brain metastasis, we selected the individual cohorts, MSK-82, EMC-192, and EMC-286. We designed a special study for brain metastasis because its sample numbers in each of the MSK-82, EMC-192, and EMC-286 cohorts were smaller than those for lung and bone metastases. For the cut-tree algorithm, a small number of samples are much easier to be clustered together. Therefore, by only considering the combined cohort, EMC-368 (MSK-82 and EMC-286), the confidence for brain metastasis would be lower than that for lung or bone metastasis. Therefore, we applied the cut-tree algorithm on these three cohorts individually.
The HCDSPs were paired with the targets of known drugs, including existing drugs, discontinued or "shelved" assets that, nevertheless, passed phase I clinical trials, drug candidates currently under development for other conditions, and experimental drugs (see Materials and Methods). The repositioned drug candidates for brain, lung, and bone are shown in Table 1, and Supplementary Tables S4 and S5, respectively.
Table 1.
List of drugs of breast cancer brain metastasis that were repositioned and their BBB permeability
| Drug target | Drug name | Approval status |
Approved indications | "Rule of Five" for CNS drugsa |
BBB permeability |
|---|---|---|---|---|---|
| HDAC1, HDAC2 | Vorinostat | Approved | Cutaneous T-cell lymphoma | ✓ | 2009, Clin Can Res (26) |
| KDR, RET | Sunitinib | Approved | Gastrointestinal stromal tumor (GIST); advanced renal cell carcinoma (RCC); advanced pancreatic neuroendocrine tumors (pNET) | ✓ | 2009, Lancet Oncol (45) |
| KDR | Cabozantinib(XL184) | Phase II/III | Breast cancer (phase II); metastatic prostate cancer (phase II); carcinoid tumor (phase II); pancreatic neuroendocrine tumor (phase II); thyroid cancer (phase III); glioblastoma multiforme (phase II); carcinoma, non-small cell lung (phase II); astrocytic tumors (phase II) | ✓ | 2011, J Clin Oncol (28) |
| RET | Imatinib | Approved | Chronic myeloid leukemia (CML) | ✓ | |
| RAF1, KDR | Sorafenib | Approved | Hepatocellular carcinoma; renal cell carcinoma | ✓ | |
| Pazopanib | Approved | Renal cell carcinoma | ✓ | 2010, Clin Can Res (27) | |
| FYN | Dasatinib | Approved | Chronic myeloid leukemia (CML); Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) | ✓ | 2008, Blood (44) |
| NFKB1 | Dexamethasone | Approved | Allergic states; dermatologic diseases; endocrine disorders; gastrointestinal diseases; hematologic disorders; neoplastic diseases; nervous system; ophthalmic diseases; renal diseases; respiratory diseases; rheumatic disorders | ✓ | |
| Thalidomide | Approved | Multiple myeloma; erythema nodosum leprosum | ✓ | ||
| Pranlukast | Phase III | Chronic sinusitis | ✓ | ||
| GRB2 | Pegademase | Approved | Adenosine deaminase (ADA) deficiency | ||
| IKBKB | Auranofin | Approved | Rheumatoid arthritis | ||
| Arsenite | Not approved | ||||
| VIM | Acetate ion | Not approved | |||
| PHGDH | NADH | Not approved |
The "Rule of Five" is so named because all the essential physical properties of compounds are parameters of five. According to this rule, a good absorption and permeability are likely if: molecular weight is ≤500; oil/water distribution coefficient (LogP) is ≤5; hydrogen bond donors ≤5 (expressed as the sum of OHs and NHs); hydrogen bond acceptor ≤10 (expressed as the sum of Ns and Os); and number of rotatable bonds ≤10.
Lung-, bone-, and brain-metastasis signaling networks of HCDSPs and repositioned drugs
The HCDSP for brain metastasis includes 31 proteins, of which 26 (84%) are unique for brain metastasis, three (10%) are shared with lung metastasis, four (13%) are shared with bone metastasis, and one (3%) is shared with both lung and bone metastases. The gene expression data used for the brain metastasis study included three public breast tumor cohorts, EMC-192, MSK-82, and EMC-286, as well as our own TMH-52 breast tumor brain metastasis cohort. We implemented the signaling analysis on the TMH-52 cohort first and then applied the survival analysis on the EMC-192, MSK-82, and EMC-286 cohorts (Fig. 2B–D).
The HCDSP for lung metastasis contains 15 proteins, of which 8 (53%) are specific to lung and 4 (27%), 2 (13%), and 1 (7%), respectively, appear in the signaling networks of brain, bone, and both. Both the signaling analysis and survival analysis were conducted on the MSK-82 + EMC-286 combined cohort. Similarly, the signaling network for bone metastasis involved 18 proteins, in which 13 (72%) are unique, and 2 (11%), 2 (11%), and 1 (5%) are found in the signaling networks of brain, lung, and both of them. The Kaplan–Meier survival analysis results for these two signaling networks are shown in Fig. 2E and F.
The HCDSP in Fig. 2A provides 12, four, and two drug targets for brain, lung, and bone metastases of breast cancer. The 12 brain metastasis targets allowed repositioning of 15 drug candidates, including 10 that were FDA approved, 2 in phase II clinical trials, and 3 without any clinical information (Table 1). The four lung metastasis targets repositioned 9 candidate drugs, containing 3 approved and 6 without any clinical information (Supplementary Table S1). The two bone metastasis targets repurposed 2 candidate drugs, 1 approved, and 1 without any clinical information (Supplementary Table S1).
Signaling mechanism of the repositioned drug targets of brain metastasis of breast cancer
Brain metastasis is designated as an unmet medical need by the United States FDA. We, thus, focused on BCBM for examining expression of signaling molecules in clinical samples, and conducting functional validation in in vitro and in vivo models.
Among the 15 repositioned drug candidates for brain metastasis listed in Table 1, the chemical structures of 10 drugs satisfy the "rule of five" (Table 1, Supplementary Fig. S2; ref. 25), indicating their potentials to permeate the central nervous system (CNS). Three of those 10 drugs, vorinostat (26), pazopanib (27), and XL184 (28), have shown efficacies on inhibiting brain metastases in recent independent preclinical or clinical studies. Dexamethasone is, currently, often used in the clinic to improve brain metastasis symptoms, and thalidomide is used in combination with temozolomide or radiation therapy for treatment of brain metastasis (29, 30). Two of the 10 drugs, sunitinib (approved for treating advanced renal cell carcinoma and gastrointestinal stromal tumors) and dasatinib (approved for treating chronic myelogenous leukemia), with no reports connecting them to brain metastasis of breast cancer, were selected via their targets in the signaling network (RET and KDR for sunitinib, and FYN for dasatinib) and validated in vitro and in vivo.
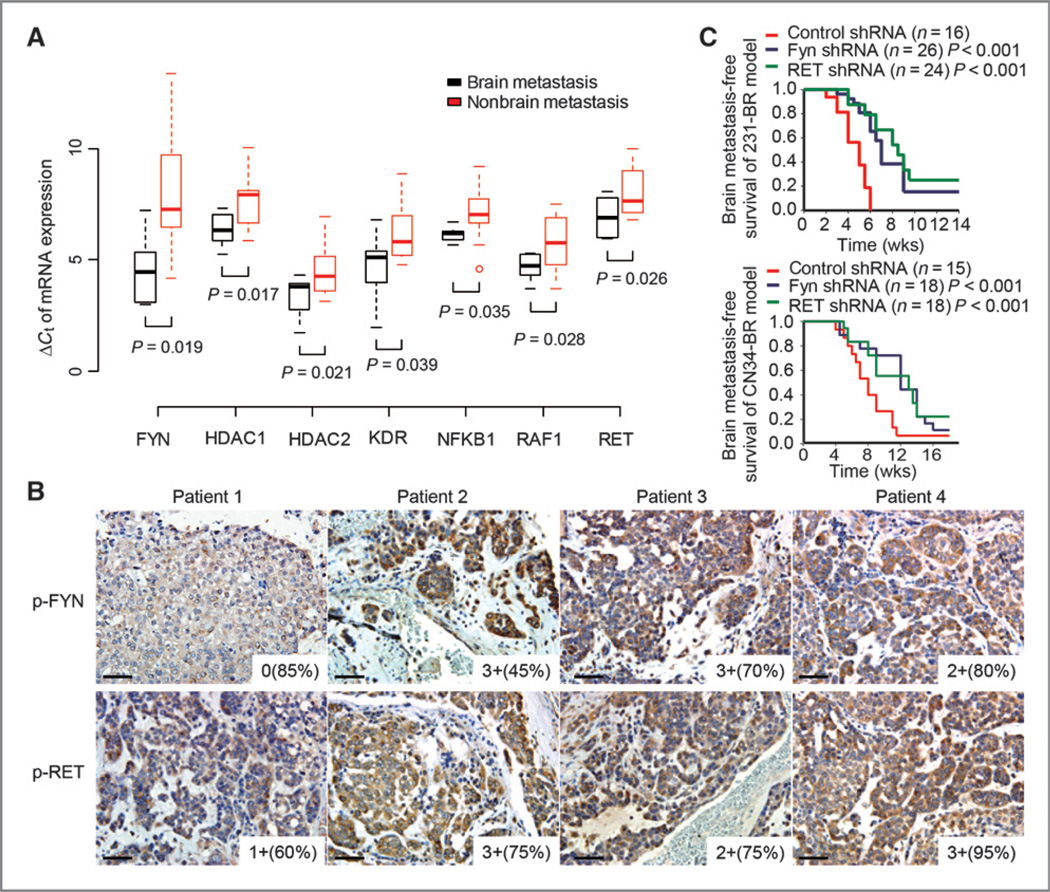
Real-time quantitative reverse transcription PCR and IHC experiments were used to examine the gene and protein-level expressions of the known drug targets in brain metastasis signaling pathways. We confirmed that the codinggenes of seven drug targets, FYN, HDAC1, HDAC2, KDR, NFKB1, RAF1, and RET, are significantly overexpressed in the primary breast tumors of 11 brain-relapsed patients by real-time quantitative RT-PCR (P < 0.05, Student t test), as shown in Fig. 3A. We then investigated the levels of protein activity for both FYN and RET in 29 brain metastatic tumors of breast cancer, of which 4 are matched samples with both primary and brain tumors. Functional FYN (phospho-FYN Tyr 416) and RET (phosph-RET Tyr 905) were detected as positive (H score 10–300, see Materials and Methods) in 25 (86%) and 23 (79%) of the 29 brain metastatic tumors, respectively (Fig. 3B and Supplementary Table S3).
Figure 3.
Validation of the identified brain metastasis targets. A, measurements of seven drug-target genes, FYN, HDAC1, HDAC2, KDR, NFKB1, RAF1, and RET, in primary tumors of brain relapsed patients by the 31-gene low-density real-time quantitative RT-PCR array. Primary breast tumor tissues from 20 patients with breast cancer were used for this quantitative RT-PCR array study; 11 of these patients had brain relapse. To compare expression profiles between specimens, a geometric averaging of three reference genes (ACTB, GAPDH, and RPLP0) was used for normalization as previously described (48). The average expression of the mean of the three reference genes is CT = 22.98. The Spearman rank correlation between the normalized Affymetrix data and quantitative RT-PCR array data were significantly positive for 24 of 31 (77.42%) of the genes. B, representative immunohistochemical stains of phosphorylated-FYN (p-FYN) and phosphorylated-RET (p-RET) on brain metastases tissue sections of patients of breast cancer. Images were taken under ×20 objective. Scale bar, 50 µm. Staining intensity and cellularity are indicated at the right bottom corner of each image. C, Kaplan–Meier curves for brain metastasis-free survival of mice injected with the indicated cell lines expressing shRNA vector control or shRNA targeting FYN or RET. P values were determined by log rank test.
RET protein is a proto-oncogene tyrosine-protein kinase receptor, and RET mutations are rarely identified in breast cancer (31). It is involved in cellular mechanisms including cell proliferation, neuronal navigation, cell migration, and cell differentiation upon binding with glial cell-derived neurotrophic factor family ligands. FYN is unique among Src family kinases as it is up regulated in multiple cancers (32, 33). Selective targeting of FYN has been proven to be especially effective given the role of FYN in tumor invasion and metastasis (34). The RET gene is located at chromosome 10q11, and FYN is at 6q21. Copy number alterations at these regions occur in breast cancer (35), suggesting that increased copy number contributes to the higher expression of RET and FYN in breast tumors and brain metastasis.
To test the functionality of the signaling proteins on brain metastasis development, RNA interference-mediated knockdown of FYN or RET expression was examined in the brain metastatic breast cancer cell lines MDA-MB231-BR (231-BR; refs. 16, 24) and CN34-BR (Supplementary Fig. S3A; ref. 6). Both significantly decreased the in vivo brain metastatic activities of the cell lines: MDA-MB231-BR (P < 0.001, log-rank test) and CN34-BR (P < 0.05, log-rank test; Fig. 3C).
Effects of repositioned drugs on brain metastasis of breast cancer
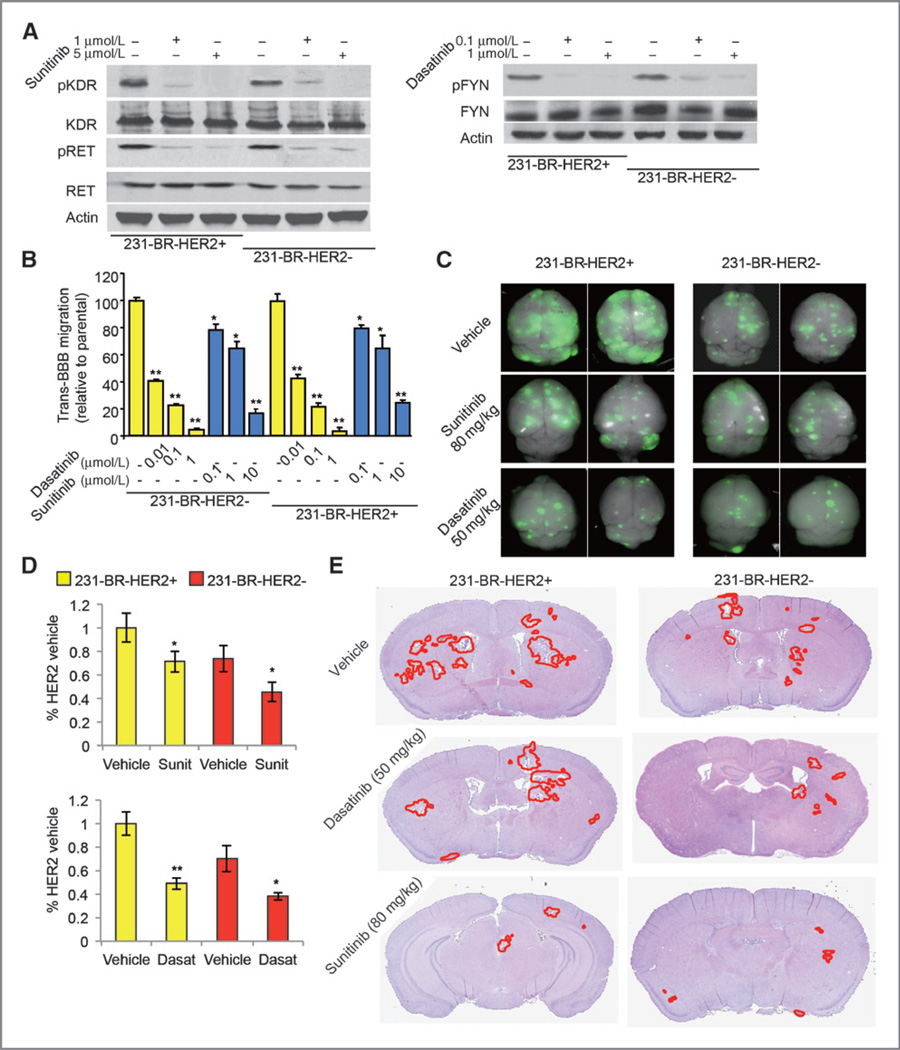
Both HER2+ and HER2− breast cancer brain metastasis models were used for the in vitro and in vivo experiments (see Materials and Methods). We established these two models based on the results from the survival analysis that the brain metastasis signaling network is significantly associated with the brain relapse within both HER2+ and HER2− early-stage tumors (P < 0.01, Supplementary Fig. S4). In vitro drug treatment studies revealed that treatments with either sunitinib or dasatinib inhibit the transmigration of 231-BR-HER2+ and 231-BR-HER2− cells through the in vitro BBB (Fig. 4A and B, Supplementary Fig. S3B).
Figure 4.
Effects of sunitinib and dasatinib treatment on brain metastases. A, target inhibition by sunitinib and dasatinib on the indicated cell lines and conditions. B, in vitro BBB transmigration activity of the indicated cell lines and conditions. The number of transmigrated cells relative to the parental cell lines is plotted with n = 12. *, P < 0.05; **, P < 0.01 versus vehicle. P values were determined by one-way ANOVA. C, sunitinib and dasatinib inhibit brain metastatic colonization of 231-BR cells examined by ex vivo whole-brain imaging. The 231-BR-HER2+ or 231-BR-HER2− cells, with a retrovirus transduction expressing EGFP, were injected into the left ventricle of BALB/c nude mice. Three days after injection, sunitinib, dasatinib, or vehicle was administered by once-daily oral gavage for 28 days. Brains dissected at necropsy were imaged using a Maestro 420 Spectral Imaging System to detect the presence of EGFP-expressing metastases derived from the injected 231-BR cells. Representative dorsal whole-brain images from two mice in each treatment group are shown. D, sunitinib and dasatinib inhibit brain metastases in two 231-BR models examined by EGFP Western blot on whole-brain lysates of the animals. *, P < 0.05 versus vehicle; **, P < 0.01 versus vehicle. E, representative H&E staining images of the whole-brain sections to show the inhibition on brain metastatic loci by sunitinib and dasatinib treatment.
In the in vivo study, both sunitinib (40 and 80 mg/kg) and dasatinib (25 and 50 mg/kg) showed antibrain metastasis efficacies when measuring the EGFP-positive tumor cells in the whole mouse brain and the metastatic lesions in the histologic sections (Table 2) upon effectively inhibiting the expressions of their respective targets: p-RET (target of sunitinib) and p-FYN (target of dasatinib; Supplementary Fig. S5A). There were fewer EGFP-positive loci in the drug-treated groups compared with the vehicle control in both models (Fig. 4C). This was confirmed by Western blot analysis using anti-EGFP antibody (P < 0.05, Student t test, Fig. 4D). The number of large metastatic lesions (>50 µm2) and micrometastases (≤50 mm2) in H&E-stained histologic sections were quantified by the image analysis algorithm described previously (see Fig. 4E and Table 2; ref. 24). For the mouse model of 231-BR-HER2-, 80 mg/kg sunitinib treatment decreased the number of large metastases by 33% (P = 0.03, Student t test), whereas 50 mg/kg dasatinib treatment decreased the number of large metastases by nearly 47% (P = 0.02, Student t test) and the number of micrometastases by 54% (P = 0.009, Student t test). Similarly, for the 231-BR-HER2+ mouse model, those treated with 80 mg/kg sunitinib decreased the number of large metastasis by approximately 30% (P = 0.02, Student t test), whereas 50 mg/kg dasatinib decreased the number of metastasis by 55% in large metastases (P = 0.02, Student t test) and 61% in micrometastases (P = 0.01, Student t test). These results indicate that dasatinib effectively inhibits formation of both the large and micro metastases formation, and sunitinib predominantly inhibits the outgrowth of large metastases in brains of both animal models.
Table 2.
Sunitinib and dasatinib inhibit brain metastases in preclinical models
| 231-Br-HER2+ model | 231-Br-HER2− model | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | N | Large metastases mean (95% CI) |
P | Micro metastases mean (95% CI) |
P | N | Large metastases mean (95% CI) |
P | Micro metastases mean (95% CI) |
P |
| Vehicle | 18 | 20 (18–23) | ref | 293.6 (255.1–332.1) | ref | 19 | 15 (12–17) | ref | 218.3 (185.3–251.3) | ref |
| Sunitinib 80 mg/kg | 17 | 14 (12–15) | 0.02 | 199.8 (175.7–223.9) | 0.05 | 16 | 10 (9–11) | 0.03 | 150.2 (128.2–177.2) | 0.06 |
| Sunitinib 40 mg/kg | 16 | 16 (14–18) | 0.05 | 228.1 (199.8–256.4) | 0.07 | 17 | 12 (10–14) | 0.08 | 156.1 (131.1–181.1) | 0.05 |
| Dasatinib 50 mg/kg | 14 | 9 (7–10) | 0.02 | 113.2 (89.4–137) | 0.01 | 16 | 8 (6–8) | 0.02 | 99.5 (89.2–109.8) | 0.009 |
| Dasatinib 25 mg/kg | 15 | 13 (11–14) | 0.04 | 175.7 (147.9–203.5) | 0.04 | 18 | 9 (7–10) | 0.03 | 144.6 (132.1–157.1) | 0.012 |
NOTE: Mice were injected with 1.75 × 105 231-Br-HER2+ or 231-Br-HER2− cells through the left ventricle and treated with sunitinib, dasatinib, or vehicle starting three days later. The treatment was carried out once daily for 28 days. Numbers of metastases were determined in 10-step sections from each mouse brain described in Materials and Methods. CI, confidence interval. For each group of data, the normality was examined with the Jarque–Bera test first, and then Levene F test was used to test multigroup HOV. If the groups enrolled followed the HOV, the Student t test was used for analyzing experimental data; if not, the Wilcoxon rank test was executed. All P values are two-sided.
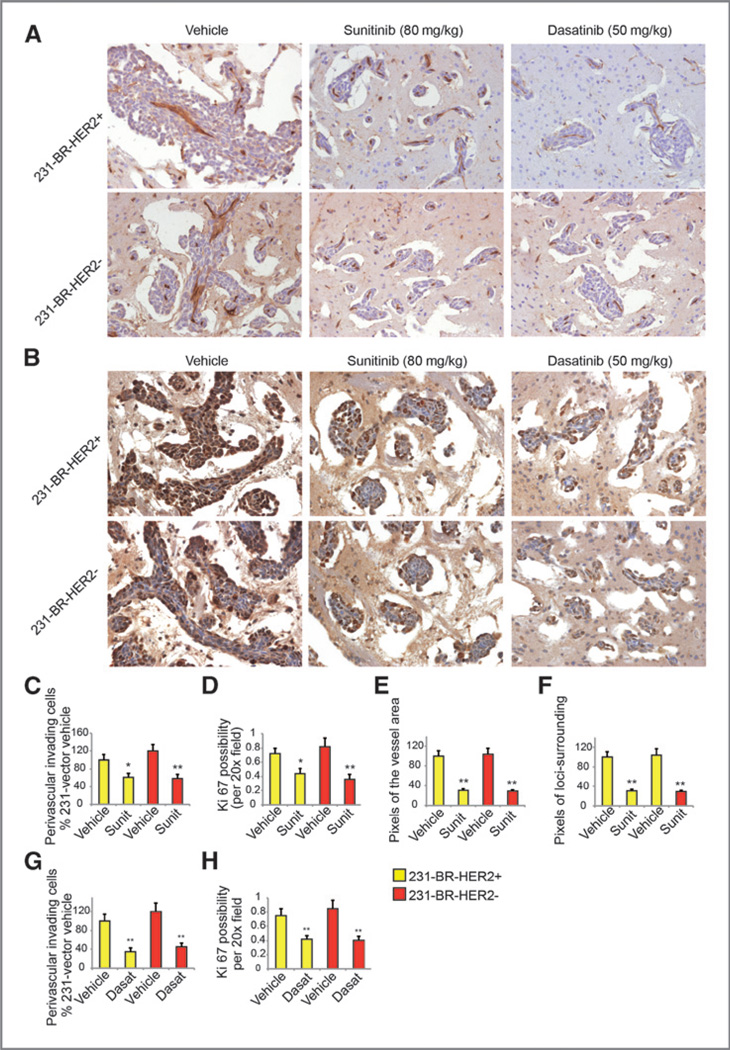
Sunitinib or dasatinib treatment decreased the percentage of proliferative (Ki67-positive) cells in the tumor sections, as well as the perivascular invasive cells on both xenograft models (P < 0.05, Student t test, Fig. 5A–D, G, and H) through Akt, MEK1/2, and p70S6K signaling (Supplementary Fig. S3C). Sunitinib treatment led to fewer enlarged and tortuous vessels within the metastasis lesions compared with the vessels in the vehicle-treated metastatic lesions (P < 0.05, Student t test, Fig. 5E and Supplementary Fig. S5D and S5E). Although the density of microvessels in large metastatic lesions did not change, sunitinib treatment significantly decreased the lesion-surrounding edema areas (P < 0.05, Student t test, Fig. 5F, Supplementary Fig. S5B and C). These results indicate that the mechanism of action of sunitinib in decreasing large brain metastases is through induction of vessel normalization, and the mechanism for both sunitinib and dasatinib in decreasing metastases may be through antitumor cell extravasation and proliferation.
Figure 5.
Mechanism of sunitinib and dasatinib on brain metastases. A and B, representative IHC of CD31 (A) and Ki67 (B) staining on the brain sections. Images were taken under ×20 objective. The brown signals are from the sections with positive staining. C, D, G, and H, quantification of the perivascular invading cells and possibility of Ki67 expression. E and F, quantification of the pixels of the vessel area and pixels of the loci-surrounding edema area. *, P < 0.05 versus vehicle; **, P < 0.01 versus vehicle.
Discussion
We developed a CSB-based computational model to uncover or derive the downstream signaling mechanism of a cancer of interest. In contrast to the prevalent gene–signature-based approaches for drug repositioning or network analysis, the proposed computational model introduces the notion of signaling networks to indicate the heterogeneity and complexity of downstream signaling pathways of cancer, and in our application, breast cancer metastases. The identified high-confidence downstream signaling mechanisms are instrumental for identifying repositioned drug candidates for the three metastatic types of breast cancer illustrated in this article. In addition, a detailed study was implemented on the unmet need of brain metastasis, showing the efficacies of the repositioned drugs in prohibiting metastatic colonization in in vivo experiments.
The HCDSPs contain few genes in the previously published gene signatures of brain, lung, and bone metastasis of breast cancer (16, 18, 36). We found only one gene (PHGDH) for brain, one gene (PARD6B) for lung, and no genes for bone metastases in the comparisons (P < 10e-10, Fisher exact two-tailed test). We reason that our analysis takes advantage of the enriched signaling pathways defined by differentially expressed genes, but is not limited to the known gene signatures comprised of differential genes. The HCDSPs focus on downstream signaling proteins in the CSBs instead of those upstream. Moreover, the survival analysis in our study filters out most clinically relevant genes rather than differential ones. Clearly, the benefits of using such a complicated computational approach over a more brute force experimental one are cost-effectiveness and elimination of investigator bias.
The identification of individualized signaling networks opens a new vista to address the heterogeneity and complexity of three metastases of breast cancer, as illustrated by the shared and unique proteins of HCDSPs in Fig. 1. MAP3K3 was identified as the shared signaling factor for metastases in general and is involved in the mitogen-activated protein/extracellular signal–regulated kinase (MEK) signaling pathway, which, in turn, is important to breast cancer metastasis (37). In addition, we identified that RET and FYN were upregulated in brain metastasis. Both have been reported to regulate brain function via adhesion mediated signaling or neuronal navigation and migration (38, 39). They are overexpressed in the brain metastatic tumors and the drugs down-regulating their functions were able to inhibit the development of brain metastasis.
A phase II clinical trial of sunitinib is now underway to evaluate progression-free survival and response rate in the CNS metastases of breast cancer at our hospitals (NCT00570908, www.clinicaltrials.gov). The HSDSP of brain metastasis signaling network are significantly associated with the status of brain relapse in estrogen receptor negative (ER−), progesterone receptor negative (PR−), and HER2− breast tumors receiving adjuvant therapy (P < 0.01; Supplementary Fig. S4). Thus, the identified known-drug targets in the HSDSP are useful to stratify patients into target-positive subtypes for clinical trials. Sunitinib failed clinical trials on breast cancer as there has been no biomarker selection for patients. New trials on RET-overexpressing patients with breast cancer are warranted. Sunitinib inhibits the outgrowth of brain metastatic cells by repressing the MEK/ERK and mTOR–S6K signals downstream of RET (Supplementary Fig. S3C). Combinatory inhibition of these two signaling pathways has been shown to efficiently prohibit brain metastases in animal models (24, 40). In addition, the antibrain-metastasis role of sunitinib benefits from its anti-angiogenesis role. Normalizing tumor vessels would also have the potential to improve the quality of life of patients with breast cancer as the interstitial pressure and edema can be reduced (41).
Dasatinib targeting of the Src family FYN inhibits the formation of brain metastatic colonization up to 50% compared with the untreated mice. Dasatinib was developed as a dual BCR–ABL and Src family tyrosine kinase inhibitor. There is a significant increase in ABL activity in the MDA-MB231 cell line, and it is highly sensitive to dasatinib (42). FYN mRNA can be upregulated by BCR–ABL1 fusion gene oxidative stress in chronic myelogenous leukemia cells (33). Thus, clarifying the ABL7 mechanism on different brain metastasis models will further help prioritize patient selection for dasatinib treatment.
Clinically, the usual dose of sunitinib for patients is 50 mg/d and for dasatinib it is 100 mg/d. Converting these doses to mice (43), the clinically relevant dose on mice for sunitinib is approximately 10 and 20 mg/kg for dasatinib. However, sunitinib and dasatinib have limited CNS penetration (44, 45). In our pharmacokinetic study on the xenograft model, the brain accumulation was measured at 25% to 28% for sunitinib and 7% to 11% for dasatinib (Supplementary Fig. S6). Recent data on metastatic renal cell carcinoma showed that increased exposure to sunitinib appears well tolerated and is associated with improved clinical outcome, resulting in maintenance of antitumor activity (46). According to these results, to achieve effective concentration in CNS for treatment of brain metastases, appropriate higher doses of these drugs on patients with brain metastasis are suggested with consideration to the balance between efficacy and toxicity. Alternatively, newer formulations of these drugs may be considered to increase the CNS penetration.
The new downstream disease-specific signaling pathway may facilitate the repositioning of targeted drugs. The pipeline and computational tools will be publically released for the scientific community in the future.
Supplementary Material
Acknowledgments
The authors thank Drs. Patricia Steeg and Joan Massague for providing the MB231-Br and CN34-Br cell lines, Dr. Neal Copeland for providing valuable comments on the article, and Drs. Rebecca Danforth and James Mancuso for proofreading. Allmicroscopic imaging studies were conducted at The Methodist Hospital Research Institute's Advanced Cellular and Tissue Microscope Core Facility directed by S.T.C. Wong.
Grant Support
This work is funded by NIH grants U54 CA149196, NIH R01 CA139976, and NIH R01 CA121225, a John S. Dunn Research Foundation grant (S.T.C. Wong), and an NIH U54 CA149196 pilot project grant (H. Zhao). The authors acknowledge the computational time funding support from the Texas Advanced Computing Center (TACC; Project ID: TG-MCB110130) at The University of Texas in Austin and BlueBioU (IBM POWER 7 Bioscience Computing Core) at Rice University to access their powerful super-computing resources.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
H. Zhao, G. Jin, and S.T.C. Wong have ownership interests, including a patent pending for molecular diagnostics and methods for predicting brain metastasis of breast cancer. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: H. Zhao, G. Jin, K. Cui, Y. Fan, A. Rodriguez, J. Chang, S.T.C. Wong
Development of methodology: H. Zhao, G. Jin, D. Ren, J. Chang, S.T.C. Wong
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): H. Zhao, D. Ren, A. Rodriguez, S.T.C. Wong
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): H. Zhao, G. Jin, K. Cui, D. Ren, P. Chen, S. Wong, F. Li, Y. Fan, J. Chang, S.T.C. Wong
Writing, review, and/or revision of the manuscript: H. Zhao, G. Jin, S. Wong, F. Li, Y. Fan, J. Chang, S.T.C. Wong
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K. Cui, D. Ren, T. Liu, S.T.C. Wong
Study supervision: J. Chang, S.T.C. Wong
References
- 1.Jin G, Fu C, Zhao H, Cui K, Chang J, Wong ST. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 2012;72:33–44. doi: 10.1158/0008-5472.CAN-11-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou S, Chang J, LaFever L, Tang W, Johnson EL, Hu J, et al. Identification of dAven, a Drosophila melanogaster ortholog of the cell cycle regulator Aven. Cell Cycle. 2011;10:989–998. doi: 10.4161/cc.10.6.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug–target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 7.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 8.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2011;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shats I, Gatza ML, Chang JT, Mori S, Wang J, Rich J, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2011;71:1772–1780. doi: 10.1158/0008-5472.CAN-10-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson Reuters. White paper. Knowledge-based drug repositioning to drive R&D productivity. 2012 Sep; http://thomsonreuters. com/business-unit/science/subsector/pdf/knowledge-based-drug-repositioning- to-drive-rd-productivity.pdf. [Google Scholar]
- 12.Chindelevitch L, Ziemek D, Enayetallah A, Randhawa R, Sidders B, Brockel C, et al. Causal reasoning on biological networks: interpreting transcriptional changes. Bioinformatics. 2012;28:1114–1121. doi: 10.1093/bioinformatics/bts090. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Agarwal P. A pathway-based view of human diseases and disease relationships. PLoS ONE. 2009;4:e4346. doi: 10.1371/journal.pone.0004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 16.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 18.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin G, Zhou X, Wang H, Zhao H, Cui K, Zhang XS, et al. The knowledge-integrated network biomarkers discovery for major adverse cardiac events. J Proteome Res. 2008;7:4013–4021. doi: 10.1021/pr8002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu C, Jin G, Gao J, Zhu R, Ballesteros-villagrana E, Wong STC. DrugMap Central (DMC), an on-line query and visualization tool to facilitate drug repositioning studies. Bioinformatics. 2013;29:1834–1836. doi: 10.1093/bioinformatics/btt279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez AA, Makris A, Wu MF, Rimawi M, Froehlich A, Dave B, et al. DNA repair signature is associated with anthracycline response in triple negative breast cancer patients. Breast Cancer Res Treat. 2011;123:189–196. doi: 10.1007/s10549-010-0983-z. [DOI] [PubMed] [Google Scholar]
- 22.Andersen JN, Sathyanarayanan S, Di Bacco A, Chi A, Zhang T, Chen AH, et al. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Sci Transl Med. 2010;2:43ra55. doi: 10.1126/scitranslmed.3001065. [DOI] [PubMed] [Google Scholar]
- 23.Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res. 2001;16:1486–1495. doi: 10.1359/jbmr.2001.16.8.1486. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Cui K, Nie F, Wang L, Brandl MB, Jin G, et al. The effect of mTOR inhibition alone or combined with MEK inhibitors on brain metastasis: an in vivo analysis in triple-negative breast cancer models. Breast Cancer Res Treat. 2011;131:425–436. doi: 10.1007/s10549-011-1420-7. [DOI] [PubMed] [Google Scholar]
- 25.Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRx. 2005;2:541–553. doi: 10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmieri D, Lockman PR, Thomas FC, Hua E, Herring J, Hargrave E, et al. Vorinostat inhibits brain metastatic colonization in a model of triple-negative breast cancer and induces DNA double-strand breaks. Clin Cancer Res. 2009;15:6148–6157. doi: 10.1158/1078-0432.CCR-09-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gril B, Palmieri D, Qian Y, Smart D, Ileva L, Liewehr DJ, et al. Pazopanib reveals a role for tumor cell B-Raf in the prevention of HER2+ breast cancer brain metastasis. Clin Cancer Res. 2011;17:142–153. doi: 10.1158/1078-0432.CCR-10-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon MS, Vogelzang NJ, Schoffski P, Daud A, Spira AI, O'Keefe BA, et al. Activity of cabozantinib (XL184) in soft tissue and bone: results of a phase II randomized discontinuation trial (RDT) in patients (pts) with advanced solid tumors. J Clin Oncol. 2011;29:3010. [Google Scholar]
- 29.Hwu WJ, Lis E, Menell JH, Panageas KS, Lamb LA, Merrell J, et al. Temozolomide plus thalidomide in patients with brain metastases from melanoma: a phase II study. Cancer. 2005;103:2590–2597. doi: 10.1002/cncr.21081. [DOI] [PubMed] [Google Scholar]
- 30.Hwu WJ, Krown SE, Menell JH, Panageas KS, Merrell J, Lamb LA, et al. Phase II study of temozolomide plus thalidomide for the treatment of metastatic melanoma. J Clin Oncol. 2003;21:3351–3356. doi: 10.1200/JCO.2003.02.061. [DOI] [PubMed] [Google Scholar]
- 31.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 32.Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: a novel molecular target in cancer. Cancer. 2010;116:1629–1637. doi: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ban K, Gao Y, Amin HM, Howard A, Miller C, Lin Q, et al. BCR-ABL1 mediates up-regulation of Fyn in chronic myelogenous leukemia. Blood. 2008;111:2904–2908. doi: 10.1182/blood-2007-05-091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav V, Denning MF. Fyn is induced by Ras/PI3K/Akt signaling and is required for enhanced invasion/migration. Mol Carcinog. 2011;50:346–352. doi: 10.1002/mc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 37.Reddy KB, Nabha SM, Atanaskova N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003;22:395–403. doi: 10.1023/a:1023781114568. [DOI] [PubMed] [Google Scholar]
- 38.Fukai I, Hussey RE, Sunder-Plassmann R, Reinherz EL. A critical role for p59(fyn) in CD2-based signal transduction. Eur J Immunol. 2000;30:3507–3515. doi: 10.1002/1521-4141(2000012)30:12<3507::AID-IMMU3507>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 39.Venteo S, Bourane S, Mechaly I, Sar C, Abdel Samad O, Puech S, et al. Regulation of the Na,K-ATPase gamma-subunit FXYD2 by Runx1 and Ret signaling in normal and injured non-peptidergic nociceptive sensory neurons. PLoS ONE. 2012;7:e29852. doi: 10.1371/journal.pone.0029852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nie F, Yang J, Wen S, An YL, Ding J, Ju SH, et al. Involvement of epidermal growth factor receptor overexpression in the promotion of breast cancer brain metastasis. Cancer. 2012;118:5198–5209. doi: 10.1002/cncr.27553. [DOI] [PubMed] [Google Scholar]
- 41.Carmeliet P, Jain RK. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev. 2011;10:417–427. doi: 10.1038/nrd3455. [DOI] [PubMed] [Google Scholar]
- 42.Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, et al. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/"triple-negative" breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 43.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 44.Porkka K, Koskenvesa P, Lundan T, Rimpilainen J, Mustjoki S, Smykla R, et al. Dasatinib crosses the blood-brain barrier and is an efficient therapy for central nervous system Philadelphia chromosome-positive leukemia. Blood. 2008;112:1005–1012. doi: 10.1182/blood-2008-02-140665. [DOI] [PubMed] [Google Scholar]
- 45.Gore ME, Szczylik C, Porta C, Bracarda S, Bjarnason GA, Oudard S, et al. Safety and efficacy of sunitinib for metastatic renalcell carcinoma: an expanded-access trial. Lancet Oncol. 2009;10:757–763. doi: 10.1016/S1470-2045(09)70162-7. [DOI] [PubMed] [Google Scholar]
- 46.Buti S, Donini M, Lazzarelli S, Passalacqua R. A new modified schedule of sunitinib for metastatic renal cell carcinoma: a retrospective analysis. Acta Biomed. 2012;83:88–94. [PubMed] [Google Scholar]
- 47.Barsky A, Gardy JL, Hancock RE, Munzner T. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007;23:1040–1042. doi: 10.1093/bioinformatics/btm057. [DOI] [PubMed] [Google Scholar]
- 48.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.