Abstract
The development and rapid spread of chloroquine resistance (CQR) in Plasmodium falciparum have triggered the identification of several genetic target(s) in the P. falciparum genome. In particular, mutations in the Pfcrt gene, specifically, K76T and mutations in three other amino acids in the region adjoining K76 (residues 72, 74, 75 and 76), are considered to be highly related to CQR. These various mutations form several different haplotypes and Pfcrt gene polymorphisms and the global distribution of the different CQR- Pfcrt haplotypes in endemic and non-endemic regions of P. falciparum malaria have been the subject of extensive study. Despite the fact that the Pfcrt gene is considered to be the primary CQR gene in P. falciparum , several studies have suggested that this may not be the case. Furthermore, there is a poor correlation between the evolutionary implications of the Pfcrt haplotypes and the inferred migration of CQR P. falciparum based on CQR epidemiological surveillance data. The present paper aims to clarify the existing knowledge on the genetic basis of the different CQR- Pfcrt haplotypes that are prevalent in worldwide populations based on the published literature and to analyse the data to generate hypotheses on the genetics and evolution of CQR malaria.
Keywords: malaria, chloroquine, Pfcrt gene, haplotypes, evolution
Chloroquine (CQ): a drug of choice for malaria treatment - Malaria is an infectious disease that has been present in the tropics for much of history. It varies widely in epidemiology and clinical manifestation and is responsible for an estimated 216 million clinical episodes and approximately 655,000 deaths per year, of which approximately 90% occur in Africa (WHO 2011). The variability in the spectrum of malarial diseases is the result of several factors, including the distribution of the two primary species of malaria parasites ( Plasmodium falciparum and Plasmodium vivax ), their levels of susceptibility to antimalarial drugs, the distribution and efficiency of mosquito vectors, climate and other environmental conditions and the behaviour and level of acquired immunity of the exposed human populations (Bloland 2001). Due to the lack of an effective vaccine, malaria is currently incurable and thus its case management depends solely on anti-malarials (WHO 1973, 1984). In the Western world, the first anti-malarial used to treat human malaria was quinine, which is extracted from the bark of the cinchona tree and was described as early as 1632 (Baird et al. 1996). In Chinese medicine, the use of Artemisia annua (qinghao) plants for the treatment of intermittent fever/malaria was described as early as 283-343 AD. Around 340 AD, in Hong Ge’s Handbook of Prescriptions for Emergency Treatment , a cold extraction method of qinghao was described for the treatment of intermittent fevers (Klayman 1985). Although primaquine and quinacrine were produced after World War I (1914-1918) and remained effective for malaria treatment for a period of time, the intense demand for other anti-malarials led to the discovery of CQ by Bayer in Germany (Thompson & Werbel 1972).
CQ, a 4-aminoquinoline derivative of quinine, was first synthesised in 1934 ( Thompson & Werbel 1972 ) and has since become the most widely used antimalarial drug. Historically, CQ was used to combat malaria in 1946 after the Second World War. Since then, it has been considered to be the drug of choice for the treatment of non-severe, un-complicated malaria and for chemoprophylaxis. Apart from certain toxic side effects, such as retinal and psychiatric symptoms, cardiac disorders, respiratory depression, neurological problems and severe gastro-intestinal irritation ( Telgt et al. 2005 ), certain unique properties, including high efficacy, wide distribution, ready availability, quick metabolism, inexpensiveness and high therapeutic index ( Payne 1987 ), made CQ the drug of choice for treating malaria ( Coatney 1963 ). Over the years, CQ has proven to be one of the most successful and important drugs ever deployed against malaria, especially in the highly endemic areas of Africa, where the malaria parasite P. falciparum infects nearly every child ( Wellems & Plowe 2001 ). This efficiency of CQ also prompted the World Health Organization (WHO) to spearhead projects and establish large mass drug administration programs using CQ ( Litsios 1996 , WHO 2002 ). The introduction of CQ near the end of Second World War brought dramatic new power to malaria control programs ( Wellems et al. 2009 ) and these efforts further reduced the incidence of malaria in most of the endemic regions in the world. However, the malaria eradication campaign started by the WHO in the 1950s excluded Africa, the continent with the highest burden of malaria and focused on the rest of the world. As such, by the late 1950s and early 1960s, malaria was eradicated in most of the Western world and was reduced to its historically lowest level in Asia and the Americas, but remained at approximately the same level in Africa ( Talisuna et al. 2004 ).
Mechanism of action of CQ - CQ acts on the endolysosomal system of malaria parasites, causing morphologic changes and haemoglobin accumulation in endocytic vesicles ( Fitch 2004 , Ecker et al. 2012 ). Being alkaline in nature, CQ accumulates in high concentrations within the digestive vacuole (DV) of the parasite and raises its pH. Because the DV is acidic in pH, the deprotonation of CQ renders the DV alkaline ( Orjih et al. 1994 , Ecker et al. 2012 ). CQ then induces the rapid clumping of the malarial pigment and eventually inhibits the parasitic enzyme haeme polymerase, which normally converts the toxic ferric haeme (ferriprotoporphyrin IX) into the non-toxic haemozoin (5-haematin). This inhibition results in the accumulation of toxic ferric haeme, leading to lysis and, ultimately, parasite death ( Roepe 2009 , Ecker et al. 2012 ). Studies have suggested that the mechanism of action of CQ relies heavily on the accumulation of high concentrations of the drug ( Fitch 2004 , Ecker et al. 2012 ).
CQ resistance (CQR) is a major hurdle to malaria control - The tremendous success of CQ and its heavy use for almost 12 years ( Wongsrichanalai et al. 2002 ) led to the development of resistance in P. falciparum during the late 1950s ( Maberti 1960 , Moore & Lanier 1961 , Young & Moore 1961 , Reyes 1981 , Peters 1987 ). The contribution of the extensive use and misuse of CQ to the selection of resistant parasites became particularly evident during the Global Malaria Eradication Campaign, which was launched by the WHO in 1955. CQR was implicated in the spread of malaria to new areas and the re-emergence of malaria in areas where the disease was previously eradicated due to population movement ( Bloland 2001 , Tatem & Smith 2010 ). CQR was reported for the first time at the Thailand-Cambodia border in 1957 and the Venezuela-Colombia border in 1959 and eventually spread to other countries throughout the world ( Wernsdorfer & Payne 1991 , Mehlotra et al. 2001 , Ridley 2002 ). Moreover, several recent molecular epidemiological studies have identified at least six independent origins of CQR from different regions of the world ( Mehlotra et al. 2008 , Wellems et al. 2009 ). Despite the suggested multiple independent origins of CQR, CQR parasites share some common phenotypes, such as increased 50% inhibitory concentration (IC 50 ), chemosensitisation, reduced CQ accumulation, low pH in the DV and similar genetic mutations ( Jiang et al. 2006 ). Drug pressure in the field is also considered to be an essential prerequisite for the development of resistance ( Wellems 2002 , Plowe 2009 ). However, the rate at which drug resistance spreads and how the resistant mutants survive in nature are still a matter of investigation ( Talisuna et al. 2004 , Anderson & Roper 2005 , Hyde 2005 ). Several models, including the degree of drug use, drug elimination half-life, host heterogeneity ( Hastings et al. 2002 ), parasite biomass ( Hastings & D’Alessandro 2000 ), parasite fitness ( Walliker et al. 2005 ), malaria transmission intensity ( Hastings & Watkins 2005 ), host immunity and intrahost dynamics ( Hastings 1997 ), were developed to better understand this drug resistance ( Talisuna et al. 2003 , 2004 , 2007 ). Because CQR parasites have been experimentally shown to have greater fitness potential in CQ environments than CQS parasites ( Walliker et al. 2005 ), the resistant parasites were able to spread and establish themselves throughout the P. falciparum malaria-endemic zones. Following the emergence and spread of CQR, the drug policies of many countries were revised and several new drugs were introduced in the field either as single agents or in combination therapies. Gradually, P. falciparum developed resistance to nearly all anti-malarials in use ( Table I ), although the geographical distribution of resistance to any single-agent antimalarial drug varies greatly ( Bloland 2001 , Mita & Tanabe 2012 ). Like CQ, the extensive deployment of other antimalarial drugs also placed tremendous selection pressure on P. falciparum to evolve mechanisms of resistance ( Anderson 2009 ). Additionally, cross-resistance and the genetic plasticity of the parasite contributed to CQR ( White 2004 ). Despite the prevalence of CQR in P. falciparum , CQ still remains the drug of choice for the treatment of non-severe P. falciparum and non- P. falciparum infections in many malaria-endemic countries and its several unique properties make it advantageous over all other anti-malarial drugs. Despite the introduction of several therapies to treat complicated and non-complicated malaria, CQ still has a prominent place in malaria treatment. Thus, the development of CQR poses a great hurdle to malaria control measures and has contributed to rollbacks in malaria programmes ( Talisuna et al. 2004 ).
TABLE I. Year wise occurrence of chloroquine (CQ) resistance worldwide.
| Country | Year of CQ resistance reported |
|---|---|
| Asia | |
| Thailand | 1959 |
| Cambodia | 1962 |
| Vietnam | 1962 |
| Malaysia | 1962 |
| Myanmar | 1969 |
| Bangladesh | 1970 |
| Nepal | 1972 |
| India | 1973-1984 |
| Indonesia | 1973-1980 |
| Philippines | Early 1970s |
| Papua New Guinea | 1976 |
| Solomon Islands | 1980 |
| Vanuatu | 1980 |
| Iran | 1983 |
| Sri Lanka | 1984 |
| Africa | |
| Kenya | 1978 |
| Tanzania | 1978 |
| Comoros Islands | Early 1980’s |
| Madagascar | Early 1980’s |
| Uganda | Early 1980’s |
| Zambia | Early 1980’s |
| Malawi | Early 1980’s |
| Angola | Mid 1980’s |
| Namibia | Mid 1980’s |
| Nigeria | Mid 1980’s |
| Benin | Mid 1980’s |
| Togo | Mid 1980’s |
| Ghana | Mid 1980’s |
| Senegal | Mid 1980’s |
| Gambia | Mid 1980’s |
| South America | |
| Venezuela | 1959 |
| Columbia | 1959 |
| Brazil | 1961 |
| Guyana | 1969 |
| Suriname | 1972 |
| Ecuador | 1976 |
| Peru | 1980 |
| Bolivia | 1980 |
Genetics of CQR in P. falciparum - Identification of the Pfcrt gene - Soon after CQR P. falciparum isolates were found to be widespread in malaria-endemic zones, the mutagenic basis of CQR was made evident by several clinical and epidemiological studies ( Wellems et al. 1991 , Fidock et al. 2000 ). Phenotypic studies involving genetic crosses between CQR and CQ-sensitive (CQS) strains further supported the hypothesis of the genetic basis of CQR ( Wellems et al. 1991 ) and genetic loci on chromosome 13 ( Pfnhe 1 gene) ( Fig. 1 ) and chromosome 5 ( Pfmdr 1 gene) ( Fig. 1 ) were proposed to be associated with higher IC 50 values in the progeny of genetic crosses ( Wellems et al. 1991 , Ferdig et al. 2004 ). However, a direct association between Pfnhe 1 gene mutations and CQR could not be established. Instead, Pfnhe1 mutations were found to correlate well with quinine in several studies ( Cooper et al. 2002 , Hayton & Su 2004 , 2008 ). Mutations in Pfmdr1 , which encodes a homolog of the human multidrug resistance p -glycoprotein ( PfPgh 1), were also found to be associated with CQR ( Djimde et al. 2001 , Mu et al. 2003 , Duraisingh & Cowman 2005 , Sidhu et al. 2005 , Valderramos & Fidock 2006 ), but the contribution of Pfmdr1 in modulating CQR remains debatable ( Hayton & Su 2004 , 2008 ). It was later established that CQR is inherited as a single locus in a genetic cross between the CQR Dd2 (Indochina) and CQS HB3 (Honduras) clones and this locus was identified to be the single determinant of CQ sensitivity ( Wellems et al. 1991 , Su et al. 1997 , Fidock et al . 2000 ). The CQR phenotype was further mapped to a 48-Kb chromosomal locus harbouring the highly interrupted gene Pfcrt ( P. falciparum CQR transporter) ( Figs 1 , 2 ). This gene is present on chromosome 7, spans 3.1 kb and has 13 exons ranging in size from 45-269 bp. It produces a 1,275-bp cDNA that encodes the 424-amino acid 48.6-kDa Pf CRT protein, which has 10 transmembrane domains (TMDs) ( Wellems et al. 1991 , Su et al. 1997 , Fidock et al . 2000 , Bray et al. 2005 ). Further evidence establishing Pfcrt as a CQR determinant came from studies of culture-adapted field isolates, which showed that CQR P. falciparum isolates had extensive linkage disequilibrium (LD) surrounding a 36-Kb segment of Pfcrt ( Wootton et al. 2002 ).
Fig. 1. schematic representation of the three genes, Pfnhe 1, Pfmdr 1 and Pfcrt , respectively, associated with chloroquine resistance in Plasmodium falciparum . The red boxes depict exons.
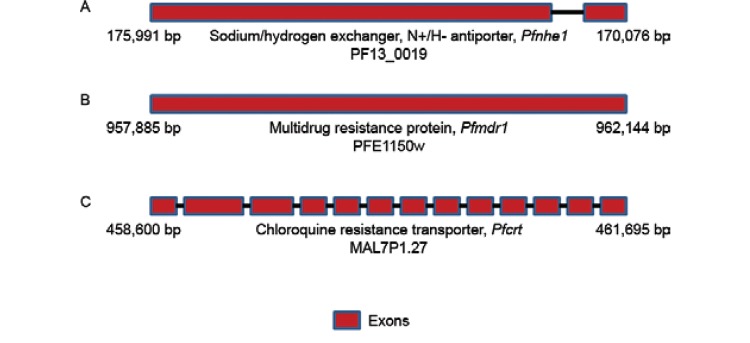
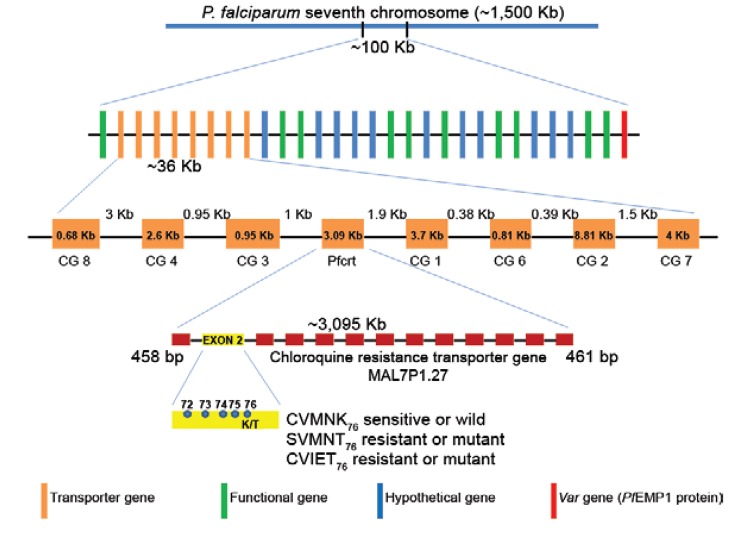
Fig. 2. : location of the ~100 Kb segment present in the seventh chromosome of Plasmodium falciparum harbouring the transporter genes, Pfcrt and var gene. Further ~36 Kb segment is highlighted encompassing the eight transporter genes including the Pfcrt gene and a more schematic view of the Pfcrt gene with its 13 exons and the K76T mutation is highlighted. The five amino acids present from 72-76 position in exon 2 characterise the resistant (CVIET and SVMNT) and sensitive (CVMNK) chloroquine resistance Pfcrt haplotypes.
Putative functional role of PfCRT in P. falciparum - The endogenous function of Pf CRT remains unknown, but its transmembrane structure and cellular location suggest that it is involved in the transport of critical metabolites, such as drugs and maintains the pH balance in the DV of P. falciparum ( Dzekunov et al. 2000 , Bennett et al. 2004 , Ecker et al. 2012 ). Other potential roles for Pf CRT include the expulsion of amino acids resulting from haemoglobin digestion from the DV and indirect involvement in maintaining H + balance in the DV ( Jiang et al. 2008 ). The roles of other transporters, such as Pf VP2, the Ca +2 /H + antiporter VCX1, PFE0785c and ATPase/synthase (PF11_0412 and PFC0840w), which might also play crucial roles in CQR, have also been well documented ( Jiang et al. 2008 ). Moreover, phylogenetic analyses predict Pf CRT to be a member of the drug/metabolite transporter superfamily of electrochemical potential driven transporters, thus supporting its hypothesised roles in P. falciparum ( Martin & Kirk 2004 ).
PfCRT, glutathione (GSH) and the human immune response - Human immune responses play an important role in shaping the ability of the host to resolve drug-resistant infections harbouring mutant Pfcrt ( Djimde et al. 2003 ), as failures in treatment are generally associated with specific polymorphisms in the parasite genome or gene copy number ( Picot et al. 2009 ). A low level of CQR P. falciparum and acquired protective immunity can explain why CQ treatment is able to successfully cure some infections harbouring mutant Pfcrt parasites in semi-immune individuals ( Wellems & Plowe 2001 , Djimde et al. 2003 ) and why, at other times, the immune response allows a relatively ineffective drug to clear an infection without any therapy ( Schofield & Mueller 2006 , Greenhouse et al. 2009 ). Altered intracellular levels of GSH have been shown to cause a corresponding shift in CQ susceptibility in P. falciparum ( Ginsburg et al. 1998 ). Additional indirect evidence has suggested a potential link between CQR and GSH ( Ginsburg & Golenser 2003 ), which originated from the observation that the Pfmrp gene, which is localised to the parasite surface, is disrupted in CQR ( Raj et al. 2009 ). Moreover, a recent report found that Pf CRT homologs in Arabidopsis thaliana mediate GSH transport and stress tolerance when assayed in Xenopus oocysts ( Maughan et al. 2010 ).
The K76T mutation in PfCRT: a key factor? - Sequence comparisons of Pf CRT in CQR and CQS P. falciparum have identified several mutations, among which the mutation of residue 76 (wild type K to the mutant T) could be directly associated with CQR. This finding was confirmed by allelic exchange studies ( Fidock et al. 2000 , Sidhu et al . 2002 , Lakshmanan et al. 2005 ). Other additional single nucleotide polymorphisms (SNPs) present in exons 2, 3, 4, 6, 9, 10 and 11 of the Pfcrt gene have also been proposed to have some association with CQR. Similarly, at the protein level, approximately 32 mutations in the 10 α-helical TMD of Pf CRT have been reported to be associated with CQR. Studies have suggested that these mutations might epistatically interact with the K76T mutation and might also evolve to maintain homeostasis ( Fidock et al. 2000 , Wootton et al. 2002 ). However, these mutations have been casually associated with resistance in vitro and in vivo and even with altered drug accumulation ( Sanchez et al. 2003 , 2004 , 2005 ). Regardless of the exact knowledge of these mutations, in general, it was found that parasites that are resistant to CQ and that bear mutations in Pf CRT accumulate less CQ due to either active energy-dependent CQ efflux ( Krogstad et al. 1987 , Sanchez et al. 2003 ) or the passive efflux of diprotic CQ ( Sanchez et al. 2010 ).
Compensatory mutations within Pfcrt and Pfmdr1 - Recent studies have suggested that Pf CRT mutations may affect parasite survival by switching back to their CQS form in the absence of drug pressure, perhaps owing to the low fitness properties of the resistant Pf CRT in competing against the sensitive Pf CRT, as observed in Malawi, Kenya and Hainan ( Kublin et al. 2003 , Mita et al. 2003 , 2004 , Wang et al. 2005 , Laufer et al. 2006 , Mwai et al. 2009 ). On the contrary, some parasite lines ( e.g ., FCB and Dd2) grow well in vitro, even in the absence of drug pressure, suggesting the presence of potential compensatory mutations within Pf CRT. These compensatory changes within Pf CRT may not fully restore the biological functions of the protein and further changes in other parts of the genome may be required ( Jiang et al. 2008 ). This compensatory role is believed to be played by the Pfmdr1 gene. This hypothesis is supported by the fact that strong LD was found between variants of both the Pfcrt and Pfmdr1 genes ( Duraisingh et al. 2000 , Adagut & Warhurst 2001 , Duraisingh & Refour 2005 , Mu et al. 2005 , Sutar et al. 2011 ). Furthermore, Pfmdr1 was observed to be non-randomly associated with the mutant Pfcrt gene and directly related to CQR to improve parasite fitness ( Ekland & Fidock 2007 ). Both genes also combine in a region-specific manner to create higher levels of drug resistance ( Sa et al. 2009 ). However, the precise role of the Pfmdr1 gene in the efflux mechanism of CQ in P. falciparum is still unclear ( Krogstad 1990 , Krogstad et al. 1992 ). It has been proposed that copy number variations influence Pfmdr1 expression in response to CQ and mefloquine selection or mutations in Pfcrt , suggesting a direct association between these two genes ( Cowman et al. 1994 , Price et al. 2004 , Anderson et al. 2005 , Duraisingh & Cowman 2005 , Hayton & Su 2008 ). Moreover, a low copy number Pfmdr1 in P. falciparum also increases its susceptibility to other drugs ( Sidhu et al. 2006 ). Thus, it seems that Pfcrt has a causal effect on CQR, while Pfmdr1 acts as a secondary modulator ( Babiker et al. 2001 , Ngo et al. 2003 , Holmgren et al. 2006 , Jiang et al. 2006 ). Surprisingly, apart from Pfmdr1 , no other gene has been found to be associated with CQR, although quantitative CQ responses differ in CQR and CQS strains, even when the Pfcrt and Pfmdr1 genes remain unchanged. This finding indicates that the level of the CQ response may be influenced by additional genes ( Foote et al. 1990 , Reed et al. 2000 , Mu et al. 2003 ). Moreover, studies on culture-adapted isolates that harbour the mutant Pfcrt gene reported low CQ IC 50 values that failed to meet the standard criteria for CQR, providing indubitable evidence that mutant Pfcrt is insufficient to confer CQR to all genetic backgrounds, even though the strains showed high CQ tolerance and recrudescence under CQ pressure ( Valderramos et al. 2010 ).
Pfcrt mutations and haplotypes: global distribution of different haplotypes - Amino acid polymorphisms have been found in exon 2 of the Pfcrt gene at residues 72, 74, 75 and 76 in P. falciparum isolates, suggesting that they may be involved in the genetic characterisation of CQR and CQS ( Fig. 2 ). Accordingly, whereas the C 72 V 73 M 74 N 75 K 76 haplotype is considered to be CQS, parasites with polymorphisms at any of these amino acid positions are considered to be CQR ( Awasthi et al. 2011 , 2012 ) ( Fig. 2 ). For CQR P. falciparum , two principal haplotypes, with the amino acid sequences C 72 V 73 I 74 E 75 T 76 and S 72 V 73 M 74 N 75 T 76 ( Awasthi et al. 2011 , 2012 ) ( Fig. 2 ), are widely distributed. Based on nucleotide sequence data, the SVMNT haplotype is further categorised as either S agt VMNT or S tct VMNT, a di-nucleotide polymorphism at codon 72, in which the sequence is changed from AGT to TCT. However, this nucleotide change does not lead to an amino acid substitution, as both AGT and TCT code for serine ( Mehlotra et al. 2008 ). Due to the widespread yet structured present-day distribution across P. falciparum -endemic zones across the globe, these two haplotypes are hypothetically considered to be CQR mother haplotypes and the 19 minor haplotypes are believed to have been derived from them ( Awasthi et al. 2011 , 2012 ) ( Table II ). While it has been established that CVIET and SVMNT are widely prevalent, whether all of the other minor haplotypes were derived from these two or evolved independently is still an open question. It appears that these multiple resistant haplotypes may have evolved independently, but that only some of them have been able to selectively sweep through populations. In addition to the accepted five foci of origin for CQR P. falciparum , specifically, CVIET (Southeast Asia and Africa), S agt VMNT (Asia, South America and Tanzania), S tct VMNT (South America and Angola), CVMET (Colombia) and CVMNT (South America and the Philippines), a sixth focus has been described in India and Iran ( Mehlotra et al. 2008 , Zakeri et al. 2008 , Wellems et al. 2009 ). An in-depth description of the distribution of the different haplotypes in the three malaria-endemic continents (Asia, Africa and South America) is discussed below.
TABLE II. Various derived (minor) chloroquine resistance (CQR) Pfcrt haplotypes with their reported countries and relevant references.
| Derived/minor haplotypes | Reported countries of CQR | References |
|---|---|---|
| CVIDT | Cambodia, India, Angola, Madagascar, Indochinese Peninsula, Vietnam, China, Philippines | Lim et al. (2003), Huaman et al. (2004), Cooper et al. (2005), Randrianarivelojosia et al. (2006), Keen et al. (2007), Rason et al. (2007), Yang et al. (2007), Niang et al. (2008), Gama et al. (2010), Takahashi et al. (2012) |
| CVIKT | Indonesia, Papua New Guinea | Mehlotra et al. (2001, 2008), Nagesha et al. (2003), Huaman et al. (2004), Cooper et al. (2005) |
| SVIET | Indonesian Papua New Guinea, Congo, Central Africa Republic | Nagesha et al. (2003), Plummer et al. (2004), Menard et al. (2006), Niang et al. (2008) |
| SVIEK | Central Africa Republic | Menard et al. (2006) |
| CVIEK | Central Africa Republic, Sudan | Menard et al. (2006), Summers et al. (2012) |
| CVMNT | PNG, Cambodia, India, Brazil, Peru, Ecuador, Columbia, Philippines, Angola, Iran, India | Cortese et al. (2002), Lim et al. (2003), Nagesha et al. (2003), Vieira et al. (2004), Mittra et al. (2006), Echeverry et al. (2007), Keen et al. (2007), Pati et al. (2007), Restrepo et al. (2008), Zakeri et al. (2008), Gama et al. (2010), Mixon-Hayden et al. (2010), Takahashi et al. (2012) |
| SVMIT | Guyana | Plummer et al. (2004), Cooper et al. (2005), Menard et al. (2006), Takahashi et al. (2012) |
| SVMET | Columbia, Central Africa Republic | Plummer et al. (2004), Menard et al. (2006) |
| SVMDT | Philippines | Hatabu et al. (2009), Takahashi et al. (2012) |
| RVMNT | Guyana | Plummer et al. (2004), Cooper et al. (2005) |
| CVMET | Columbia | Echeverry et al. (2007), Yang et al. (2007) |
| CVMNN | Indonesia | Huaman et al. (2004), Cooper et al. (2005) |
| CVTNT | Cambodia | Lim et al. (2003), Durand et al. (2004) |
| CVINT | Central Africa Republic, Angola | Menard et al. (2006), Gama et al. (2010) |
| CVMHT | Philippines | Yang et al. (2007) |
| CVMDT | Angola, Philippines | Hatabu et al. (2009), Gama et al. (2010), Takahashi et al. (2012) |
| CVIEA | Thailand (cloneJ9) | Chaijaroenkul et al. (2011), Summers et al. (2012) |
| CVIEI | Laboratory strain (106/1-I) | Cooper et al. (2002), Summers et al. (2012) |
| CVIEN | Laboratory strain (106/1-N) | Cooper et al. (2002), Summers et al. (2012) |
Distribution of the CQR-Pfcrt haplotypes in Asia - The distribution of the CQR- Pfcrt haplotypes presents a unique pattern in Asia, particularly in Southeast Asia [Cambodia, Thailand, Bangladesh, Laos, Indochina, Indonesia, Philippines, Papua New Guinea (PNG), East Timor Islands, Solomon Islands and Vanuatu] and South Asia (India, Pakistan, Sri Lanka and Iran). The CVIET mother haplotype is proposed to have originated at the Thailand-Cambodia border in Southeast Asia ( Mehlotra et al. 2001 , Wootton et al. 2002 ). In Thailand and Bangladesh, the CVIET haplotype is the major reported haplotype ( Hatabu et al. 2005 , Takahashi et al. 2012 ) and, very recently, a new haplotype, CVIEA, was also observed in Thailand ( Chaijaroenkul et al. 2011 ). In Cambodia, apart from CVIET, three more derived haplotypes (CVIDT, CVTNT and CVMNT) have also been reported ( Lim et al. 2003 ). In Laos, CVIET and SVMNT haplotypes have been reported, with the latter having a relatively higher frequency ( Dittrich et al. 2005 ). Interestingly, the Philippine Islands were found to be dominated by SVMNT and its derived haplotypes (CVMNT and CVMHT) ( Chen et al. 2003 , Yang et al. 2007 ), with a recent report indicating the distribution of the CVIET-derived haplotype CVIDT ( Huaman et al. 2004 ). The Pfcrt haplotypic view of PNG is quite unusual; despite its geographic proximity to the Southeast Asian focus of the resistance-carrying CVIET haplotype, the CQR parasites in this country harbour haplotypes that are similar to the SVMNT haplotype, which originated from South America ( Chan et al. 2012 ). Multi- locus microsatellite studies have also illustrated a greater evolutionary affinity between P. falciparum isolates from PNG and Southeast Asia, as opposed to South America, which further emphasises the unexpected nature of the Pfcrt polymorphism findings ( Mehlotra et al. 2008 ). However, apart from the Pfcrt substitutions, S agt VMNT and S tct VMNT have been associated with a different genetic background in PNG and South America, respectively and as such, it has been argued that PNG most likely represents another independent focus of CQR ( Chan et al. 2012 ). In PNG, apart from the S agt VMNT haplotype, which occurs at appreciable frequency, two CVIET-derived haplotypes (SVIET and CVIKT) have also been reported with minor frequency in Indonesian Papua (West New Guinea) ( Nagesha et al. 2003 , DaRe et al. 2007 , Takahashi et al. 2012 ). In the East Timor Islands, Solomon Islands and Vanuatu, the SVMNT haplotype is prevalent ( Tanabe et al. 2004 , Sakihama et al. 2006 , Almeida et al. 2009 , Mita et al. 2009 , Takahashi et al. 2012 ). In Indonesia, apart from the highly frequent SVMNT haplotype, a new haplotype, CVMNN, was found to be frequent in Lombok and Irian Jaya ( Huaman et al. 2004 ). Most interestingly, India has a mixture of many CQR- Pfcrt haplotypes, which are primarily dominated by SVMNT, but also show appreciable frequencies of CVIET, CVMNT and CVIDT ( Vinayak et al. 2003 , 2006 , Vathsala et al. 2004 , Mittra et al. 2006 , Keen et al. 2007 Pati et al. 2007 , Bharti et al. 2009 , Mixon-Hayden et al. 2010 , Awasthi et al. 2011 , Sutar et al. 2011 , Lumb et al. 2012 ). In Pakistan, Iran and Sri Lanka, the S agt VMNT haplotype is reported at appreciable frequencies and is believed to have been imported from India ( Zakeri et al. 2008 , Zhang et al. 2011 , Rawasia et al. 2012 ). A very recent study from Yemen and Saudi Arabia confirmed the major presence of the CVIET haplotype ( Al-Hamidhi et al. 2013 ).
Distribution of the CQR-Pfcrt haplotypes in Africa - The Pfcrt CQR haplotypic view in Africa is completely biased towards the CVIET haplotype, owing to the wide usage of CQ and amodiaquine (AQ) drugs in many African countries ( Djimde et al. 2010 ). To date, in most sub-Saharan African countries, including Comoros, Senegal, Gabon, Djibouti, Cameroon, Gambia, Niger, Ivory Coast, Ghana, Nigeria, Kenya, Mali, the Dominican Republic of Congo, Guinea Bissau, Mozambique, Benin, Zambia, Rwanda, Burundi, Tanzania, the Republic of South Africa, Sudan, Congo, Madagascar, Malawi and Uganda, the CVIET haplotype is the only CQR- Pfcrt haplotype that has been reported in high frequency ( Cooper et al. 2005 , Ariey et al. 2006 , Randrianarivelojosia et al. 2006 , Severini et al. 2006 , Juliano et al. 2007 , Nsobya et al. 2007 , Mehlotra et al. 2008 , Niang et al. 2008 , Bob et al. 2010 , Gadalla et al. 2010 , Takahashi et al. 2012 ). However, in Tanzania, the SVMNT haplotype is present at an appreciable frequency ( Alifrangis et al. 2006 ). In Congo and Madagascar, two CVIET-derived haplotypes are also present, SVIET in Congo and CVIDT in Madagascar ( Severini et al. 2006 , Rason et al. 2007 ). Interestingly, the Central African Republic shows the CVIET haplotype at an appreciable frequency, along with six derived haplotypes (SVIET, SVIEK, CVIEK, CVMNT, SVMET and CVINT) in low frequencies ( Menard et al. 2006 ). Interestingly, a recent study reported the presence of the S tct VMNT haplotype in a very high frequency in Angola, with a low frequency of CVIET and three derived haplotypes (CVMNT, CVINT and CVMDT) with relatively lower frequencies ( Gama et al. 2010 ). Furthermore, high CQR- Pfcrt haplotype diversity and the emergence of the S agt VMNT haplotype in Cameroon have recently been reported ( Mbenda & Das 2013 ). Thus, in general, while all of the African countries were found to be dominated by the CVIET Pfcrt -CQR haplotype, Angola, Tanzania, Cameroon and the Central Africa Republic were exceptions. It seems probable that the SVMNT haplotype found in Angola, Tanzania, Cameroon and the Central African Republic might have originated in South America and the Western Pacific. Because Angola and Cameroon are located on the Southwest coast of Africa, these countries might have received the CQR Pfcrt migrants of P. falciparum from South America due to frequent travellers between Brazil and Africa ( Gama et al. 2010 , Ecker et al. 2012 ) and due to an increase in the use of AQ in Africa either alone or in combination with artesunate ( Summers et al. 2012 ). Regardless, the presence of seven different haplotypes (CVIET, SVIET, SVIEK, CVIEK, CVMNT, SVMET and CVINT) in the Central African Republic might be explained with a better knowledge of the drug combinations administered in this region to date ( Menard et al. 2006 ), as the increasing use of AQ in Africa poses the threat of a selective sweep of highly AQ and CQ-resistant parasites with Pfcrt and Pfmdr1 mutations that are as advantaged and persistent as in South America ( Sa et al. 2009 ).
Distribution of the CQR-Pfcrt haplotypes in South America - South America is thought to be one of the six foci of origin of CQR P. falciparum , as the CQR- Pfcrt haplotype S tct VMNT was first reported at the Colombia-Venezuela border ( Mehlotra et al. 2001 , 2008 ) and is still highly prevalent across the continent. The high prevalence of the S tct VMNT haplotype in South American countries is attributed to many factors, such as (i) the absence of CQ pressure, (ii) the wide usage of AQ, (iii) region-specific differences in drug usage, (iv) a reduced rate of polyclonal infections and (v) the absence of competitive wild type parasites ( Sa et al. 2009 , Sa & Twu 2010 , Ecker et al. 2012 ). The highly prevalent S tct VMNT haplotype is reported to be the sole haplotype in Bolivia. In Brazil, Venezuela and Peru, the S tct VMNT haplotype is present in high frequency, along with the CVIET haplotype, at an appreciable frequency ( Sa & Twu 2010 ). In contrast, Ecuador and Guyana are completely dominated by the SVMNT-derived haplotype CVMNT, with some incidences of S tct VMNT ( Griffing et al. 2010 ). In Colombia, the S tct VMNT haplotype was reported initially, but has been replaced by the CVMNT haplotype ( Restrepo et al. 2008 ). Apart from these frequent CQR Pfcrt haplotypes, three other low-frequency haplotypes, SVMIT and RVMIT in Guyana (Plummer et al. 2004) and CVMET across the Amazon Basin, have also been reported ( Cortese et al. 2002 , Vieira et al. 2004 , Echeverry et al. 2007 , Pineda et al. 2008 ). In general, the haplotypic view in South America suggests that the S tct VMNT haplotype and its derivatives are predominant, with the CVIET haplotype also being present in Brazil and Venezuela ( Londono et al. 2009 ). The CVIET haplotype has only been rarely reported in South America and was most likely imported from Africa, as most of the parasites in Brazil have the typical SVMNT allele. In Haiti, most of the parasites have the CVMNK allele and CVIET is rare. In this context, it is important to recognise that in Central American countries, including Haiti, CQ remains as the primary drug for the treatment of P. falciparum malaria ( Londono et al. 2009 ). In response to the rise in anti-malarial drug resistance in the Amazon and in South American countries, a surveillance network named Amazon Network for the Surveillance of Antimalarial Drug Resistance was created, with the primary responsibilities of formulating drug policies, monitoring drug resistance and promoting the suitable use of drugs within the continent ( Gama et al. 2011 ).
Pfcrt haplotypes and the origin and spread of CQR: any correlation? - The putative origin and spread of CQR P. falciparum was mainly inferred by epidemiological surveillance data ( Wernsdorfer & Payne 1991 , Wernsdorfer 1994 , Anderson 2009 ). Thus, the current distribution patterns of CQR P. falciparum are primarily based on this inference and are dependent on the time of the report of CQR P. falciparum in any endemic country. Accordingly, three different models based on CQR prevalence data in three separate malaria-endemic zones, Southeast Asia, Africa and South America ( Awasthi et al. 2012 ), have been suggested. According to the first model, CQR P. falciparum possibly originated independently in Southeast Asia (Thailand-Cambodia border) and South America (Venezuela-Colombia border) during 1957 and 1959, respectively. By 1980, CQR P. falciparum populated a maximum number of Asian countries ( Table I ). Similarly, in South America, Peru and Bolivia reported incidences of CQR P. falciparum in 1980 ( Table I ). In Africa, CQR P. falciparum was reported relatively late. The first report came from Kenya in 1978 and by the early 1990s, CQR P. falciparum isolates were found in almost all African countries. Thus, by the end of the 1980s and in the early 1990s, almost all of the malaria-endemic countries worldwide had some form of CQR P. falciparum .
Since the discovery of the distinct genetic lineages of Southeast Asian (CVIET), South American (S tct VMNT) and Southeast Asian and Asian (S agt VMNT) Pfcrt , the epidemiological observations of rare origin and contiguous spread have been interpreted as evidence of a rare and complex underlying genetic mechanism of CQR ( Plowe 2009 , Awasthi et al. 2012 ). Some early studies on the molecular epidemiology of CQR suggested that resistant malaria arose both focally and locally in direct response to CQ drug pressure ( Wernsdorfer & Payne 1991 , Wernsdorfer 1994 ). Moreover, it has been suggested that CQR Pfcrt haplotypes resulting from amino acid changes at positions 72-76 are strongly associated with the geographic region-restricted evolution of P. falciparum resistance to CQ and that these haplotypes are good estimators for predicting evolution and geographical spread of resistance, as other polymorphisms outside these positions have no clear geographical association with CQR ( Mita et al. 2009 , Mita & Tanabe 2012 ).
The differential distribution of the most frequently found CQR Pfcrt haplotypes offers opportunities to track the movement of these haplotypes, creating a haplotypic view across continents and to indirectly infer the spread of CQR P. falciparum . Recently conducted studies in both worldwide and Indian populations have clearly revealed that such patterns can be inferred from several CQR Pfcrt haplotypes, thus offering the opportunity to correlate these patterns with the epidemiological surveillance data on CQR P. falciparum parasites ( Awasthi et al. 2011 , 2012 ). Accordingly, the CVIET haplotype populated all of Southeast Asia by the early 1970s and reached India by 1973. This haplotype moved out of Asia and into Africa and this fact is well correlated with the epidemiological data ( Awasthi et al. 2011 , 2012 ). Alternatively, the CVIET haplotype might have moved from the Southeast Asian countries across the Pacific to South America, which is reflected by the presence of the CVIET haplotype in Venezuela and Brazil, although it is only present in a small percentage ( Contreras et al. 2002 , Cortese et al. 2002 , Griffing et al. 2010 ). Alternatively, the presence of the CVIET haplotype in Brazil and Venezuela ( Vieira et al. 2004 ) may be due to its movement from geographically close African countries ( Awasthi et al. 2011 , 2012 ). Very similarly, the S tct VMNT haplotype originated in South America, while the S agt VMNT haplotype originated in PNG ( Mehlotra et al. 2008 ). These haplotypes first spread locally within the respective continents before migrating to other malaria-endemic regions. As a result, the S tct VMNT haplotype moved eastward to reach West African countries, evidenced by the fact that the S tct VMNT haplotype is found at an appreciable frequency in Angola and in low frequency in Tanzania ( Alifrangis et al. 2006 , Gama et al. 2010 ). Within Asia, the S agt VMNT haplotype, which originated in PNG, established itself quite successfully in many places, over-dominating the original haplotype, CVIET, especially in India, Pakistan, Sri Lanka, PNG, the Philippines and Iran. It seems that Iran received this haplotype (and CVIET) from India and Pakistan ( Awasthi et al. 2011 , 2012 , Rawasia et al. 2012 ). Additionally, the CVIET haplotype could have spread to Yemen and Saudi Arabia from either Iran or Africa ( Al-Hamidhi et al. 2013 ). However, the global spread of CQR P. falciparum , as inferred from the epidemiological surveillance data, does not completely correlate with the inferred movements of the CQR Pfcrt haplotypes ( Awasthi et al. 2012 ). In turn, the routes inferred by the CQR Pfcrt haplotype data correlate well with the intercontinental usage of anti-malarials and the migration and successful establishment of CQR P. falciparum in different parts of the world ( Awasthi et al. 2012 ). For example, in places such as South America, AQ and CQ, which were not in use for the past several years, have resulted in the complete fixation of the S tct VMNT haplotype ( Sa et al. 2009 ). On the contrary, dramatic changes have been observed after discontinued drug pressure in certain African countries and Southeast Asia. In the absence of drug pressure, the SVMNT haplotype provides equal fitness to P. falciparum (as in the presence of drug pressure) in comparison to the CVIET haplotype ( Sa et al. 2009 ). Furthermore, CVIET haplotype-bearing P. falciparum are known to revert back to the CQS (CVMNK) type ( Kublin et al. 2003 , Mita et al. 2003 , 2004 ), whereas SVMNT-bearing P. falciparum do not ( Fidock et al. 2000 ). For example, a region of Malawi that is known for highly prevalent CQR was re-populated with drug-sensitive parasites within 10 years after CQ use was stopped ( Kublin et al. 2003 ). A similar recovery of CQS P. falciparum populations was recently reported in Kenya and has also been observed in China ( Wang et al. 2005 , Mwai et al. 2009 ). These changes in the absence of drug pressure have also been explained by fitness costs that are carried by CQ-resistant mutants ( Laufer et al. 2006 ). However, such a selective disadvantage has been less apparent in South America, where CQS parasites have not replaced their CQ-resistant counterparts. A satisfactory explanation for this difference between the Southeast Asian/African and South American forms of CQR has not been proposed ( Sa et al. 2009 ). This contention also supports the hypothesis that approximately 70% of the total Pfcrt -CQR haplotypes in Southeast Asia and South America are S agt VMNT and S tct VMNT, respectively ( Awasthi et al. 2011 , 2012 ).
Evolutionary puzzle of the Pfcrt gene in India - India is a country where malaria is highly endemic and where CQR P. falciparum is widely prevalent ( Sharma 2007 , Singh et al. 2009 ). CQR P. falciparum was first detected as early as 1973 in the Asom state of India ( Sehgal et al. 1973 ). Genetic studies of CQR Pfcrt revealed the presence of four major haplotypes (CVIET, SVMNT, CVMNT and CVIDT) in India, with the SVMNT haplotype populating the majority of the Indian states compared to the CVIET haplotype ( Awasthi et al. 2011 ). In India, the predominant distribution of the SVMNT haplotype, compared to the minimal presence of the CVIET haplotype, is quite puzzling. Because India is geographically closer to Southeast Asia (Thailand, Cambodia, Bangladesh, Laos) than to PNG and Oceania, it is expected that India should share its haplotypic status with Southeast Asia (high frequency of the CVIET haplotype). However, in reality, India shares its CQR- Pfcrt haplotypic status with PNG, Indonesia and Oceania, which harbour the S agt VMNT haplotype. Furthermore, based on the distribution of the four haplotypes (S agt VMNT, CVIET, CVMNT and CVIDT), the two following routes of possible migration of Pfcrt haplotypes (SVMNT and CVIET) into India have been hypothesised: (i) while the S agt VMNT haplotype originated in PNG, it travelled through PNG ↔ Indonesia ↔ the Philippines ↔ Malaysia ↔ Andaman and the Nicobar Islands and then entered mainland India through the east coastal state of Odisha and the S tct VMNT haplotype originated in South America, travelled through South America ↔ PNG ↔ Indonesia ↔ the Philippines ↔ Malaysia ↔ Andaman and the Nicobar Islands and reached India via Odisha. Similarly, (ii) the CVIET haplotype from the Thailand-Cambodia border travelled from Thailand and Cambodia through Myanmar to populate Mizoram and other Northeastern Indian states and reached as far as Karnataka (Southern India) ( Awasthi et al. 2011 ). A recent microsatellite variation study of the Pfcrt gene and its adjacent sequences in the Indian population suggested that the CQR- Pfcrt haplotypes might have originated in Southeast Asia and spread into Eastern India and other parts of this country through the Northeastern regions ( Mallick et al. 2013 ). Although these routes were inferred from in-depth population genetic analyses of the currently available data on the CQR Pfcrt haplotypes, the complexity of the prevalence and distribution of the S agt VMNT haplotype has confounded the overall scenario of the distribution of the CQR Pfcrt haplotypes in India ( Vathsala et al. 2004 ), as India and Iran have also been labelled as the sixth focus of origin of CQR P. falciparum parasites ( Mehlotra et al. 2008 , Zakeri et al. 2008 , Wellems et al. 2009 ).
Another interesting and puzzling issue is the evolutionary course of the Pfcrt gene in India. It is widely known from global genetic diversity studies of CQR isolates that because the Pfcrt gene is responsible for an important function in P. falciparum and is targeted by natural selection, it is described under the “selective sweep” model ( Clark 2002 , Wootton et al. 2002 ). This model perfectly fits the explanation of the origin and subsequent proliferation of CQR malaria parasites across the globe ( Wootton et al. 2002 , Mu et al. 2010a , Volkman et al. 2012 ). However, genetic diversity data on the Pfcrt gene from CQR P. falciparum in India do not conform to this evolutionary model ( Mittra et al. 2006 , Vinayak et al. 2006 , Das & Dash 2007 ). Although a very recent study provided evidence on the role of natural selection in the evolution of the Pfcrt gene in India ( Mixon-Hayden et al. 2010 ), the inferences of this study are unclear for the two following reasons: (i) the aims of the study were to correlate cerebral malaria with drug resistance gene polymorphisms and thus, the study contains sample bias and (ii) the study analysed only a single population from central India ( Mixon-Hayden et al. 2010 ). Considering that India is a vast country with a variable climate and malaria epidemiology ( Singh et al. 2009 ), the mystery of Pfcrt gene evolution needs to be resolved by deep sampling and finer evolutionary analyses.
Is Pfcrt the sole candidate for CQR? - To visualise the relevance of the genetic basis of any drug resistance from a public health perspective, an absolute correlation between genotype and phenotype is essential. In this respect, Pfcrt has not met all of the requirements for determining this gene as the sole agent of CQR. In fact, several studies have indicated that it is unclear if the Pfcrt gene is directly and solely associated with CQR P. falciparum . For example, (i) not all of the CQR P. falciparum isolates were found to bear the K76T mutation in the Pfcrt gene and vice versa ( Vinayak et al. 2003 ), (ii) the Pfcrt homologue in P. vivax ( Pvcrt-o ) is not associated with CQR in P. vivax ( Martin & Kirk 2004 ), (iii) the K76T mutation is not sufficient for the transport of CQ via Pf CRT, which is consistent with the view that one or more other Pf CRT mutations act in concert with K76T to confer CQR ( Summers et al. 2012 ), (iv) a strong LD was observed in the ~40-Kb region surrounding the Pfcrt gene in chromosome 7 ( Mu et al. 2010a ), supporting the fact that the observed genetic patterns in the Pfcrt gene could merely reflect the role of evolutionary force in hitherto uncharacterised gene(s) that have a direct association with CQR P. falciparum ( Gupta et al. 2010 , Mu et al. 2010a ) and (v) a strong association was observed between Pfcrt and the adjoining var gene in the VarS4 region of the P. falciparum genome ( Fowler et al. 2006 ). This final observation ( Fowler et al. 2006 ) corroborates the findings of Mu et al. (2010a) , clearly reflecting the importance of the ~100-Kb region of chromosome 7 in the P. falciparum genome ( Fig. 2 ) rather than the Pfcrt gene alone. Furthermore, unlike the global pattern depicting the role of natural selection in the evolution of the Pfcrt gene ( Wootton et al. 2002 ), the Pfcrt gene in Indian P. falciparum does not seem to follow the same pattern, which could be due to a shift in the target of selection ( Das & Dash 2007 ). In addition, the poor correlation between the CQR epidemiological surveillance data and the CQR Pfcrt haplotypes ( Awasthi et al. 2012 ) weakens the contention that Pfcrt is the sole controller of CQR.
Conclusion and future prospects - The current genetic understanding of CQR P. falciparum not only has provided several meaningful insights and enhanced the knowledge pertaining directly to malaria research, but also has advanced the academic understanding of how a single gene and a single amino acid mutation can significantly affect gross phenotypic characteristics. Because human infectious diseases are difficult to control, mainly due to the development of drug-resistant pathogens and successful environmental adaptation, the detailed genetic understanding of CQR P. falciparum may prove to be a model that can be applied to other infectious disease systems. In this regard, enormous amounts of genetic data on the Pfcrt gene in global P. falciparum have been generated and several genetic, epidemiological and evolutionary hypotheses have been proposed and tested. Furthermore, association studies between the drug response (IC 50 values) and SNPs in different candidate genes have identified several associations between the Pfcrt gene and CQR.
However, despite this wealth of knowledge, it is unclear if one can reliably consider Pfcrt to be the sole gene that is responsible for CQR in P. falciparum . Several studies in global P. falciparum have suggested that Pfcrt is the primary determinant of CQR. At the same time, enough empirical evidence has disputed this hypothesis ( Su et al. 1997 , Basco & Ringwald 1999 , 2001 , Durand et al. 1999 ), supporting the presence of secondary determinants of CQR ( Ecker et al. 2012 ). While the role of the Pfcrt gene cannot entirely be negated, with increasing bodies of evidence from several genome wide association studies and quantitative trait loci analyses of genetic crosses, it is reasonable to hypothesise a role for other gene(s) in conferring CQR in P. falciparum ( Wootton et al. 2002 , Kidgell et al. 2006 , Volkman et al. 2007 , Mu et al. 2010a , Ecker et al. 2012 ). For example, a recently conducted genome scan of global P. falciparum isolates reported important genomic information on the genetic basis of antimalarial resistance ( Mu et al. 2010b ). In particular, a 100-Kb region located in chromosome 7 of the P. falciparum genome ( Fig. 2 ) was found to have very low recombination activity (Gupta et al. 2010, Mu et al. 2010a ). This region contains eight transporter genes (CG1, CG2, CG3, CG4, Pfcrt , CG6, CG7, CG8) ( Fig. 2 ). Furthermore, evidence for the tight linkage between genes located in this chromosomal region has also been documented ( Fowler et al. 2006 ). Taken together, these data support the hypothesis that other gene(s) located within this linked genetic block on chromosome 7 in P. falciparum might also play a role in conferring CQR, either alone or in close functional associations with the Pfcrt gene. Specifically, one of these eight transporter genes ( Fig. 2 ), CG2 , when placed downstream of the Pfcrt , has been shown to be phenotypically associated with CQR (Su et al. 1997, Basco & Ringwald 1999 , 2001 , Durand et al. 1999 ). However, the association between CQR and the CG2 genotype is not sufficient to completely justify an exclusive role for the CG2 gene in CQR ( Gupta et al. 2010 ).
Based on the currently available data, it seems that the ~100-Kb region in chromosome 7 in the P. falciparum genome holds the key for the determination of CQR ( Gupta et al. 2010 , Mu et al. 2010a ). Considering the dubious role of Pfcrt and the possible involvement of other nearby transporter genes, further evolutionary genetic studies ( Stephan 2010 ) in this 100-Kb region could provide novel insights into the genetic basis of the P. falciparum drug resistance mechanisms ( Gupta et al. 2010 ) and identify previously unknown genes that may be involved in determining CQR in P. falciparum . Further functional validation of such novel genes could possibly clarify the genetic determinants of CQR P. falciparum . This clarification will not only provide new directions for malaria research and further our understanding of the molecular epidemiology of P. falciparum malaria, but also contribute to the development of new genetic control measures for malaria. This increased understanding could also improve the management of other human infectious diseases that are dominated by drug-resistant pathogens.
ACKNOWLEDGEMENTS
To Prof Wolfgang Stephan’s Lab, Ludwig’s Maximilians University, Munich, Germany, where GA and AD were academic visitors, for the initiation of writing this paper, to the Journal of Cell Sciences, UK, and Boehringer Ingelheim Fonds, Germany, for providing travel fellowships to GA, to the ICMR, for Senior Research Fellowship, to the Department of Biotechnology, Govt of India, for providing Overseas Associateship to AD, to Prof W Stephan, for providing excellent facilities and support in his lab, to Ms Hueggette Gaelle Ngassa Mbenda, for help in Pfcrt haplotype data collection from Africa, to the anonymous reviewers, for their helpful and critical comments.
REFERENCES
- Adagut IS, Warhurst DC. Plasmodium falciparum : linkage disequilibrium between loci in chromosomes 7 and 5 and chloroquine selective pressure in Northern Nigeria. Parasitology. 2001;123:219–224. doi: 10.1017/s0031182001008344. [DOI] [PubMed] [Google Scholar]
- Al-Hamidhi S, Mahdy MA, Al-Hashami Z, Al-Farsi H, Al-Mekhlafi AM, Idris MA, Beja-Pereira A, Babiker HA. Genetic diversity of Plasmodium falciparum and distribution of drug resistance haplotypes in Yemen. 244Malar J. 2013;12 doi: 10.1186/1475-2875-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifrangis M, Dalgaard MB, Lusingu JP, Vestergaard LS, Staalsoe T, Jensen AT, Enevold A, Rønn AM, Khalil IF, Warhurst DC, Lemnge MM, Theander TG, Bygbjerg IC. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–1741. doi: 10.1086/504269. [DOI] [PubMed] [Google Scholar]
- Almeida AD, Arez AP, Cravo PVL, do Rosário VE. Analysis of genetic mutations associated with anti-malarial drug resistance in Plasmodium falciparum from the Democratic Republic of East Timor. 59Malar J. 2009;8 doi: 10.1186/1475-2875-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ. Mapping the spread of malaria drug resistance. PLoS Med. 2009;6: doi: 10.1371/journal.pmed.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Nair S, Qin H, Singlam S, Brockman A, Paiphun L, Nosten F. Are transporter genes other than the chloroquine resistance locus ( pfcrt ) and multidrug resistance gene ( pfmdr ) associated with antimalarial drug resistance? Antimicrob Agents Chemother. 2005;49:2180–2188. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Roper C. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 2005;94:269–280. doi: 10.1016/j.actatropica.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Ariey F, Fandeur T, Durand R, Randrianarivelojosia M, Jambou R, Legrand E, Ekala MT, Bouchier C, Cojean S, Duchemin JB, Robert V, Le Bras J, Mercereau-Puijalon O. Invasion of Africa by a single pfcrt allele of South East Asian type. 34Malar J. 2006;5 doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi G, Prasad GB, Das A. Population genetic analyses of Pfcrt haplotypes reveal the evolutionary history of chloroquine-resistant malaria in India. Int J Parasitol. 2011;41:705–709. doi: 10.1016/j.ijpara.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Awasthi G, Satya GBK, Das A. Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem Inst Oswaldo Cruz. 2012;107:129–134. doi: 10.1590/s0074-02762012000100018. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr 1. J Infect Dis. 2001;183:1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- Baird JK, Nalim MFS, Basri H, Masbar S, Leksana B, Tjitra E, Dewi RM, Khairani M, Wignall FS. Survey of resistance to chloroquine by Plasmodium vivax in Indonesia. Trans R Soc Trop Med Hyg. 1996;90:409–411. doi: 10.1016/s0035-9203(96)90526-x. [DOI] [PubMed] [Google Scholar]
- Basco LK, Ringwald P. Chloroquine resistance in Plasmodium falciparum and polymorphism of the cg2 gene. J Infect Dis. 1999;180:1979–1986. doi: 10.1086/315150. [DOI] [PubMed] [Google Scholar]
- Basco LK, Ringwald P. Point mutations in the Plasmodium falciparum cg2 gene, polymorphism of the kappa repeat region and their relationship with chloroquine resistance. Trans R Soc Trop Med Hyg. 2001;95:309–314. doi: 10.1016/s0035-9203(01)90247-0. [DOI] [PubMed] [Google Scholar]
- Bennett TN, Kosar AD, Ursos LM, Dzekunov S, Sidhu ABS, Fidock DA, Roepe PD. Drug resistance-associated PfCRT mutations confer decreased Plasmodium falciparum digestive vacuolar pH. Mol Biochem Parasitol. 2004;133:99–114. doi: 10.1016/j.molbiopara.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Bharti PK, Alam MT, Boxer R, Shukla MM, Gautam SP, Sharma YD, Singh N. Therapeutic efficacy of chloroquine and sequence variation in Pfcrt gene among patients with falciparum malaria in central India. 3340Trop Med Int H ealth. 2009;15 doi: 10.1111/j.1365-3156.2009.02425.x. [DOI] [PubMed] [Google Scholar]
- Bloland PB. Drug resistance in malaria. WHO/CDS/CRS/DRS; Geneva: 2001. 32 [Google Scholar]
- Bob NS, Diop BM, Renaud F, Marrama L, Durand P, Tall A, Ka B, Ekala MT, Bouchier C, Mercereau-Puijalon O, Jambou R. Parasite polymorphism and severe malaria in dakar (Senegal): a west African urban area. 5PLoS ONE. 2010;23 doi: 10.1371/journal.pone.0009817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray PG, Martin RE, Tilley L, Ward SA, Kirk K, Fidock DA. Defining the role of Pf CRT in Plasmodium falciparum chloroquine resistance. Mol Microbiol. 2005;56:323–333. doi: 10.1111/j.1365-2958.2005.04556.x. [DOI] [PubMed] [Google Scholar]
- Chaijaroenkul W, Ward SA, Mungthin M, Johnson D, Owen A, Bray PG, Na-Bangchang K. Sequence and gene expression of chloroquine resistance transporter ( pfcrt ) in the association of in vitro drugs resistance of Plasmodium falciparum. 42Malar J. 2011;10 doi: 10.1186/1475-2875-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CW, Spathis R, Reiff DM, McGrath SE, Garruto RM, Lum JK. Diversity of Plasmodium falciparum chloroquine resistance transporter ( pfcrt ) exon 2 haplotypes in the Pacific from 1959 to 1979. PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Kyle DE, Pasay C. Pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob Agents Chemother. 2003;47:3500–3505. doi: 10.1128/AAC.47.11.3500-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG. Malaria variorum. Nature. 2002;418:283–285. doi: 10.1038/418283a. [DOI] [PubMed] [Google Scholar]
- Coatney GR. Pitfalls in a discovery: the chronicle of chloroquine. Am J Trop Med Hyg. 1963;12:121–128. doi: 10.4269/ajtmh.1963.12.121. [DOI] [PubMed] [Google Scholar]
- Contreras CE, Cortese JF, Caraballo A, Plowe CV. Genetics of drug-resistant Plasmodium falciparum malaria in the Venezuelan state of Bolivar. Am J Trop Med Hyg. 2002;67:400–405. doi: 10.4269/ajtmh.2002.67.400. [DOI] [PubMed] [Google Scholar]
- Cooper RA, Ferdig MT, Su XZ, Ursos LM, Mu J, Nomura T, Fujioka H, Fidock DA, Roepe PD, Wellems TE. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- Cooper RA, Hartwig CL, Ferdig MT. Pfcrt is more than the Plasmodium falciparum chloroquine resistance gene: a functional and evolutionary perspective. Acta Trop. 2005;94:170–180. doi: 10.1016/j.actatropica.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Cortese JF, Caraballo A, Contreras CE, Plowe CV. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 2002;186:999–1006. doi: 10.1086/342946. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr 1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci USA. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRe JT, Mehlotra RK, Michon P, Mueller I, Reeder J, Sharma YD, Stoneking M, Zimmerman PA. Microsatellite polymorphism within pfcrt provides evidence of continuing evolution of chloroquine-resistant alleles in Papua New Guinea. 34Malar J. 2007;6 doi: 10.1186/1475-2875-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Dash AP. Evolutionary paradigm of chloroquine-resistant malaria in India. Trends Parasitol. 2007;23:132–135. doi: 10.1016/j.pt.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Dittrich S, Alifrangis M, Stohrer JM, Thongpaseuth V, Vanisaveth V, Phetsouvanh R, Phompida S, Khalil IF, Jelinek T. Falciparum malaria in the north of Laos: the occurrence and implications of the Plasmodium falciparum chloroquine resistance transporter ( pfcrt ) gene haplotype SVMNT. Trop Med Int Health. 2005;10:1267–1270. doi: 10.1111/j.1365-3156.2005.01514.x. [DOI] [PubMed] [Google Scholar]
- Djimde AA, Barger B, Kone A, Beavogui AH, Tekete M, Fofana B, Dara A, Maiga H, Dembele D, Toure S, Dama S, Ouologuem D, Sangare CP, Dolo A, Sogoba N, Nimaga K, Kone Y, Doumbo OK. A molecular map of chloroquine resistance in Mali. FEMS Immunol Med Microbiol. 2010;58:113–118. doi: 10.1111/j.1574-695X.2009.00641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde AA, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. A molecular marker for chloroquine resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Djimde AA, Doumbo OK, Traore O, Guindo AB, Kayentao K, Diourte Y, Niare-Doumbo S, Coulibaly D, Kone AK, Cissoko Y, Tekete M, Fofana B, Dicko A, Diallo DA, Wellems TE, Kwiatkowski D, Plowe CV. Clearance of drug-resistant parasites as a model for protective immunity in Plasmodium falciparum malaria. Am J Trop Med Hyg. 2003;69:558–563. [PubMed] [Google Scholar]
- Duraisingh MT, Cowman AF. Contribution of the pfmdr 1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Refour P. Multiple drug resistance genes in malaria - from epistasis to epidemiology. Mol Microbiol. 2005;54:874–877. doi: 10.1111/j.1365-2958.2005.04748.x. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, von Seidlein LV, Jepson A, Jones P, Sambou I, Pinder M, Warhurst DC. Linkage disequilibrium between two chromosomally distinct loci associated with increased resistance to chloroquine in Plasmodium falciparum. Parasitology. 2000;121:1–7. doi: 10.1017/s0031182099006022. [DOI] [PubMed] [Google Scholar]
- Durand R, Gabbett E, Di Piazza JP, Delabre JF, Le Bras J. Analysis of κ and ω repeats of the cg2 gene and chloroquine susceptibility in isolates of Plasmodium falciparum from sub-Saharan Africa. Mol Biochem Parasitol. 1999;101:185–197. doi: 10.1016/s0166-6851(99)00073-0. [DOI] [PubMed] [Google Scholar]
- Durand R, Jafari S, Vauzelle J, Delabre JF, Jesic Z, Le Bras J. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol Biochem Parasitol. 2001;114:95–102. doi: 10.1016/s0166-6851(01)00247-x. [DOI] [PubMed] [Google Scholar]
- Durand V, Berry A, Sem R, Glaziou P, Beaudou J, Fandeur T. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter ( Pfcrt ) and their relationship to chloroquine resistance in vitro. Mol Biochem Parasitol. 2004;136:273–285. doi: 10.1016/j.molbiopara.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Dzekunov SM, Ursos LM, Roepe PD. Digestive vacuolar pH of intact intraerythrocytic P. falciparum either sensitive or resistant to chloroquine. Mol Biochem Parasitol. 2000;110:107–124. doi: 10.1016/s0166-6851(00)00261-9. [DOI] [PubMed] [Google Scholar]
- Echeverry DF, Holmgren G, Murillo C. Polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am J Trop Med Hyg. 2007;77:1034–1038. [PubMed] [Google Scholar]
- Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland EH, Fidock DA. Advances in understanding the genetic basis of antimalarial drug resistance. Curr Opin Microbiol. 2007;10:363–370. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naudé B, Deitsch KW, X-z Su, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falcipa rum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch CD. Ferriprotoporphyrin IX, phospholipids and the antimalarial actions of quinoline drugs. Life Sci. 2004;74:1957–1972. doi: 10.1016/j.lfs.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- Fowler EV, Chavchich M, Chen N, Peters JM, Kyle DE, Gatton ML, Cheng Q. Physical linkage to drug resistance genes results in conservation of var genes among west pacific Plasmodium falciparum isolates. J Infect Dis. 2006;194:939–948. doi: 10.1086/506619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla NB, Elzaki SE, Mukhtar S, Warhurst DC, El-Sayed B, Sutherland CJ. Dynamics of pfcrt alleles CVMNK and CVIET in chloroquine-treated Sudanese patients infected with Plasmodium falciparum. Malar J. 2010;974 doi: 10.1186/1475-2875-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama BE, Lacerda MVG, Daniel-Ribeiro CT, Ferreira-da-Cruz MF. Chemoresistance of Plasmodium falciparum and Plasmodium vivax parasites in Brazil: consequences on disease morbidity and control. Mem Inst Oswaldo Cruz. 2011;106(Suppl. I):159–166. doi: 10.1590/s0074-02762011000900020. [DOI] [PubMed] [Google Scholar]
- Gama BE, Pereira-Carvalho GAL, Kosi FJIL, Oliveira NKA de, Fortes F, Rosenthal PJ, Daniel-Ribeiro CT, Ferreira-da-Cruz MF. Plasmodium falciparum isolates from Angola show the StctVMNT haplotype in the pfcrt gene. 174Malar J. 2010;9 doi: 10.1186/1475-2875-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg H, Famin O, Zhang J, Krugliak M. Inhibition of glutathione dependent degradation of heme by chloroquine and amodiaquine as a possible basis for their antimalarial mode of action. Biochem Pharmacol. 1998;56:1305–1313. doi: 10.1016/s0006-2952(98)00184-1. [DOI] [PubMed] [Google Scholar]
- Ginsburg H, Golenser J. Glutathione is involved in the antimalarial action of chloroquine and its modulation affects drug sensitivity of human and murine species of Plasmodium. Redox Rep. 2003;8:276–279. doi: 10.1179/135100003225002907. [DOI] [PubMed] [Google Scholar]
- Greenhouse B, Slater M, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Clark TD, Staedke SG, Kamya MR, Hubbard A, Rosenthal PJ, Dorsey G. Decreasing efficacy of antimalarial combination therapy in Uganda is explained by decreasing host immunity rather than increasing drug resistance. J Infect Dis. 2009;199:758–765. doi: 10.1086/596741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing S, Syphard L, Sridaran S, McCollum AM, Mixson-Hayden T, Vinayak S, Villegas L, Barnwell JW, Escalante AA, Udhayakumar V. Pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother. 2010;54:1572–1579. doi: 10.1128/AAC.01243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta B, Awasthi G, Das A. Malaria parasite genome scan: insights into antimalarial resistance. Parasitol Res. 2010;107:495–499. doi: 10.1007/s00436-010-1917-8. [DOI] [PubMed] [Google Scholar]
- Hastings IM. A model for the origins and spread of drug resistant malaria. Parasitology. 1997;115:133–141. doi: 10.1017/s0031182097001261. [DOI] [PubMed] [Google Scholar]
- Hastings IM, D’Alessandro U. Modeling a predictable disaster: the rise and spread of drug-resistant malaria. Parasitol Today. 2000;16:340–347. doi: 10.1016/s0169-4758(00)01707-5. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM. Intensity of malaria transmission and the evolution of drug resistance. Acta Trop. 2005;94:218–229. doi: 10.1016/j.actatropica.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM, White NJ. The evolution of drug-resistant malaria: the role of drug elimination half-life. Philos Trans R Soc Lond B Biol Sci. 2002;357:505–519. doi: 10.1098/rstb.2001.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatabu T, Iwagami M, Kawazu S, Taguchi N, Escueta AD, Villacorte EA, Rivera PT, Kano S. Association of molecular markers in Plasmodium falciparum crt and mdr1 with in vitro chloroquine resistance: a Philippine study. Parasitol Int. 2009;58:166–170. doi: 10.1016/j.parint.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Hatabu T, Kawazu S, Kojima S, Sato K, Singhasivanon P, Looareesuwan S, Kano S. In vitro susceptibility and genetic variations for chloroquine and mefloquine in Plasmodium falciparum isolates from Thai-Myanmar border. Southeast Asian J Trop Med Public Health. 2005;36(Suppl. 4):S73–S79. [PubMed] [Google Scholar]
- Hayton K, X-z Su. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr Drug Targets Infect Disord. 2004;4:1–10. doi: 10.2174/1568005043480925. [DOI] [PubMed] [Google Scholar]
- Hayton K, X-z Su. Drug resistance and genetic mapping in Plasmodium falciparum. Curr Genet. 2008;54:223–239. doi: 10.1007/s00294-008-0214-x. [DOI] [PubMed] [Google Scholar]
- Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Björkman A. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr 1 86Y. Infect Genet Evol. 2006;6:309–314. doi: 10.1016/j.meegid.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Huaman MC, Yoshinaga K, Suryanatha A, Suarsana N, Kanbara H. Polymorphisms in the chloroquine resistance transporter gene in Plasmodium falciparum isolates from Lombok, Indonesia. Am J Trop Med Hyg. 2004;71:40–42. [PubMed] [Google Scholar]
- Hyde JE. Drug-resistant malaria. Trends Parasitol. 2005;21:494–498. doi: 10.1016/j.pt.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Joy DA, Furuya T, X-z Su. Current understanding of the molecular basis of chloroquine-resistance in Plasmodium falciparum. J Postgrad Med. 2006;52:271–276. [PubMed] [Google Scholar]
- Jiang H, Patel JJ, Yi M, Mu J, Ding J, Stephens R, Cooper RA, Ferdig MT, X-z Su. Genome-wide compensatory changes accompany drug-selected mutations in the Plasmodium falciparum crt gene. PLoS ONE. 2008;3: doi: 10.1371/journal.pone.0002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano JJ, Kwiek JJ, Cappell K. Minority-variant Pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13:873–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, Johnson JR, Le Roch K, Sarr O, Ndir O, Mboup S, Batalov S, Wirth DF, Winzeler EA. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathol. 2006;2: doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen J, Farcas GA, Zhong K, Yohanna S, Dunne MW, Kain KC. Real-time PCR assay for rapid detection and analysis of PfCRT haplotypes of chloroquine-resistant Plasmodium falciparum isolates from India. J Clin Microbiol. 2007;45:2889–2893. doi: 10.1128/JCM.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klayman DL. Qinghaosu (artemisinin): an antimalarial drug from China. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- Krogstad DJ. Chloroquine resistance not linked to mdr -like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Krogstad DJ, Gluzman IY, Herwaldt BL, Schlesinger PH, Wellems TE. Energy dependence of chloroquine accumulation and chloroquine efflux in Plasmodium falciparum. Biochem Pharmacol. 1992;43:57–62. doi: 10.1016/0006-2952(92)90661-2. [DOI] [PubMed] [Google Scholar]
- Krogstad DJ, Gluzman IY, Kyle DE, Oduola AM, Martin SK, Milhous WK, Schlesinger PH. Efflux of chloroquine from Plasmodium falciparum : mechanism of chloroquine resistance. Science. 1987;238:1283–1285. doi: 10.1126/science.3317830. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimdé AA, Kouriba B, Taylor TE, Plowe CV. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Bray PG, Verdier-Pinard D, Johnson DJ, Horrocks P, Muhle RA, Alakpa GE, Hughes RH, Ward SA, Krogstad DJ, Sidhu ABS, Fidock DA. A critical role for PfCRT K76T in Plasmodium falciparum verapamil reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- Lim P, Chy S, Ariey F, Incardona S, Chim P, Sem R, Denis MB, Hewitt S, Hoyer S, Socheat D, Merecreau-Puijalon O, Fandeur T. Pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob Agents Chemothe 2003:87–94.: doi: 10.1128/AAC.47.1.87-94.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios S. The tomorrow of malaria. Pacific Press; Wellington: 1996. 181 [Google Scholar]
- Londono BL, Eisele TP, Keating J, Bennett A, Chattopadhyay C, Heyliger G, Mack B, Rawson I, Vely JF, Désinor O, Krogstad DJ. Chloroquine-resistant haplotype Plasmodium falciparum parasites, Haiti. Emerg Infect Dis. 2009;15:735–740. doi: 10.3201/eid1505.081063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb V, Madan R, Das MK, Rawat V, Dev V, Khan W, Khan H, Sharma YD. Differential genetic hitchhiking around mutant pfcrt alleles in the Indian Plasmodium falciparum population. J Antimicrob Chemother. 2012;7:600–608. doi: 10.1093/jac/dkr532. [DOI] [PubMed] [Google Scholar]
- Maberti S. Development of resistance to pyrimethamine. Presentation of 15 cases studied in Trujillo, Venezuela. Arch Venez Med Trop Parasitol Med. 1960;3:239–259. [PubMed] [Google Scholar]
- Mallick PK, Sutton PL, Singh R, Singh OP, Dash AP, Singh AK, Carlton JM, Bhasin VK. Microsatellite analysis of chloroquine resistance associated alleles and neutral loci reveal genetic structure of Indian Plasmodium falciparum. Infect Genet Evol. 2013;19:164–175. doi: 10.1016/j.meegid.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RE, Kirk K. The malaria parasite’s chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol Biol Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- Maughan SC, Pasternak M, Cairns N, Kiddle G, Brach T, Jarvis R, Haas F, Nieuwland J, Lim B, Müller C, Salcedo-Sora E, Kruse C, Orsel M, Hell R, Miller AJ, Bray P, Foyer CH, Murray JA, Meyer AJ, Cobbett CS. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, Pf CRT, are required for glutathione homeostasis and stress responses. Proc Natl Acad Sci USA. 2010;107:2331–2336. doi: 10.1073/pnas.0913689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbenda HGN, Das A. Occurrence of multiple chloroquine-resistant Pfcrt haplotypes and emergence of the S(agt)VMNT type in Cameroonian Plasmodium falciparum. J Antimicrob Chemother. 2013 doi: 10.1093/jac/dkt388. [DOI] [PubMed] [Google Scholar]
- Mehlotra RK, Fujioka H, Roepe PD. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with Pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci USA. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Mattera G, Bockarie MJ, Maguire JD, Baird JK, Sharma YD, Alifrangis M, Dorsey G, Rosenthal PJ, Fryauff DJ, Kazura JW, Stoneking M, Zimmerman PA. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52:2212–2222. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D, Djalle D, Yapou F, Manirakiza A, Talarmin A. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Bangui, Central African Republic. Am J Trop Med Hyg. 2006;74:205–210. [PubMed] [Google Scholar]
- Mita T, Kaneko A, Lum JK, Bwijo B, Takechi M, Zungu IL, Tsukahara T, Tanabe K, Kobayakawa T, Bjorkman A. Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg. 2003;68:413–415. [PubMed] [Google Scholar]
- Mita T, Kaneko A, Lum JK, Zungu IL, Tsukahara T, Eto H, Kobayakawa T, Bjorkman A, Tanabe K. Expansion of wild type allele rather than back mutation in pfcrt explains the recent recovery of chloroquine sensitivity of Plasmodium falciparum in Malawi. Mol Biochem Parasitol. 2004;135:159–163. doi: 10.1016/j.molbiopara.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Mita T, Tanabe K. Evolution of Plasmodium falciparum drug resistance: implications for the development and containment of artemisinin resistance. Jpn J Infect Dis. 2012;65:465–475. doi: 10.7883/yoken.65.465. [DOI] [PubMed] [Google Scholar]
- Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Mittra P, Vinayak S, Chandawat H, Das MK, Singh N, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J Infect Dis. 2006;193:1304–1312. doi: 10.1086/502979. [DOI] [PubMed] [Google Scholar]
- Mixon-Hayden T, Jain V, McCollum AM, Poe A, Nagpal AC, Dash AP, Stiles JK, Udhayakumar V, Singh N. Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob Agents Chemother. 2010;54:997–1006. doi: 10.1128/AAC.00846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DV, Lanier JE. Observations on the two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. 1961;10:5–9. doi: 10.4269/ajtmh.1961.10.5. [DOI] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3: doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, Subramanian G, Aravind L, Cooper RA, Wootton JC, Xiong M, X-z Su. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- Mu J, Myers RA, Jiang H, Liu S, Ricklefs S, Waisberg M, Chotivanich K, Wilairatana P, Krudsood S, White NJ, Udomsangpetch R, Cui L, Ho M, Ou F, Li H, Song J, Li G, Wang X, Seila S, Sokunthea S, Socheat D, Sturdevant DE, Porcella SF, Fairhurst RM, Wellems TE, Awadalla P, X-z Su. Plasmodium falciparum genome wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet. 2010a;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Seydel KB, Bates A, X-z Su. Recent progress in functional genomic research in Plasmodium falciparum. Curr Genomics. 2010b;11:279–286. doi: 10.2174/138920210791233081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, Sasi P, Marsh K, Borrmann S, Mackinnon M, Nzila A. Chloroquine resistance before and after its withdrawal in Kenya. 106Malar J. 2009;8 doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagesha HS, Casey GC, Rieckmann KH, Fryauff DJ, Laksana BS, Reeder JC, Maguire JD, Baird JK. New haplotypes of the Plasmodium falciparum chloroquine resistance transporter ( pfcrt ) gene among chloroquine-resistant parasite isolates. Am J Trop Med Hyg. 2003;68:398–402. [PubMed] [Google Scholar]
- Ngo T, Duraisingh M, Reed M, Hipgrave D, Biggs B, Cowman AF. Analysis of pfcrt , pfmdr 1, dhfr and dhps mutations and drug sensitivities in Plasmodium falciparum isolates from patients in Vietnam before and after treatment with artemisinin. Am J Trop Med Hyg. 2003;68:350–356. [PubMed] [Google Scholar]
- Niang M, Marrama L, Ekala MT. Accumulation of CVIET Pfcrt allele of Plasmodium falciparum in placenta of pregnant women living in an urban area of Dakar, Senegal. J Antimicrob Chemother. 2008;62:921–928. doi: 10.1093/jac/dkn299. [DOI] [PubMed] [Google Scholar]
- Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob Agents Chemother. 2007;51:3023–3025. doi: 10.1128/AAC.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjih AU, Ryerse JS, Fitch CD. Hemoglobin catabolism and the killing of intraerythrocytic Plasmodium falciparum by chloroquine. Experientia. 1994;50:34–39. doi: 10.1007/BF01992046. [DOI] [PubMed] [Google Scholar]
- Pati SS, Mishra S, Mohanty S, Mohapatra DN, Sahu PK, Priyadarshi N, Kumar S, Sharma SK, Tyagi PK, Chitnis CE, Das BS. Pfcrt haplotypes and in-vivo chloroquine response in Sundergarh district, Orissa, India. Trans R Soc Trop Med Hyg. 2007;101:650–654. doi: 10.1016/j.trstmh.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- Peters W. Chemotherapy and drug resistance in malaria. 2nd ed. Academic Press; London: 1987. 542 [Google Scholar]
- Picot S, Olliaro P, Monbrison F de, Bienvenu AL, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. 89Malar J. 2009;8 doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda ER, Arango E, do Rosário VEMA, Cravo P. Studies on antimalarial drug susceptibility in Colombia, in relation to Pfmdr1 and Pfcrt. Parasitology. 2008;135:547–553. doi: 10.1017/S0031182008004307. [DOI] [PubMed] [Google Scholar]
- Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 2009;1035:11–14. doi: 10.1016/j.trstmh.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer WB, Pereira LMP, Carrington CVF. Pfcrt and pfmdr1 alleles associated with chloroquine resistance in Plasmodium falciparum from Guyana, South America. Mem Inst Oswaldo Cruz. 2004;99:389–392. doi: 10.1590/s0074-02762004000400008. [DOI] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj DK, Mu J, Jiang H, Kabat J, Singh S, Sullivan M, Fay MP, McCutchan TF, X-z Su. Disruption of a Plasmodium falciparum multidrug resistance-associated protein ( PfMRP ) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem. 2009;284:7687–7696. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randrianarivelojosia M, Fidock DA, Belmonte O, Valderramos SG, Mercereau-Puijalon O, Ariey F. First evidence of pfcrt mutant Plasmodium falciparum in Madagascar. Trans R Soc Trop Med Hyg. 2006;100:826–830. doi: 10.1016/j.trstmh.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rason MA, Andrianantenaina HB, Ariey F, Raveloson A, Domarle O, Randrianarivelojosia M. Prevalent Pfmdr1 N86Y mutant Plasmodium falciparum in Madagascar despite absence of Pfcrt mutant strains. Am J Trop Med Hyg. 2007;76:1079–1083. [PubMed] [Google Scholar]
- Rawasia WF, Sridaran S, Patel JC, Abdallah J, Ghanchi NK, Barnwell JW, Escalante AA, Udhayakumar V, Beg MA. Genetic backgrounds of the Plasmodium falciparum chloroquine resistant transporter ( pfcrt ) alleles in Pakistan. Infect Genet Evol. 2012;12:278–281. doi: 10.1016/j.meegid.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Restrepo E, Fonseca JC, Maestre A. Plasmodium falciparum : high frequency of pfcrt point mutations and emergence of new mutant haplotypes in Colombia. Biomedica. 2008;28:523–530. [PubMed] [Google Scholar]
- Reyes S. Malarial infections with Plasmodium falciparum resistant to chloroquine treatment. The situation in Brazil (1960-1981) Rev Bras Malariol Doencas Trop. 1981;33:109–130. [PubMed] [Google Scholar]
- Ridley RG. Medical need, scientific opportunity and the drive for anti-malarial drugs. Nature. 2002;415:686–693. doi: 10.1038/415686a. [DOI] [PubMed] [Google Scholar]
- Roepe PD. Molecular and physiologic basis of quinoline drug resistance in Plasmodium falciparum malaria. Future Microbiol. 2009;4:441–455. doi: 10.2217/fmb.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa JM, Twu O. Protecting the malaria drug arsenal: halting the rise and spread of amodiaquine resistance by monitoring the Pf CRT SVMNT type. 374Malar J. 2010;9 doi: 10.1186/1475-2875-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakihama N, Ohmae H, Bakote’e B, Kawabata M, Hirayama K, Tanabe K. Limited allelic diversity of Plasmodium falciparum merozoite surface protein 1 gene from populations in the Solomon Islands. Am J Trop Med Hyg. 2006;74:31–40. [PubMed] [Google Scholar]
- Sanchez CP, Dave A, Stein WD. Transporters as mediators of drug resistance in Plasmodium falciparum. Int J Parasitol. 2010;40:1109–1118. doi: 10.1016/j.ijpara.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, McLean JE, Rohrbach P, Fidock DA, Stein WD, Lanzer M. Evidence for a pfcrt -associated chloroquine efflux system in the human malarial parasite Plasmodium falciparum. Biochemistry. 2005;44:9862–9870. doi: 10.1021/bi050061f. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, McLean JE, Stein W, Lanzer M. Evidence for a substrate specific and inhibitable drug efflux system in chloroquine resistant Plasmodium falciparum strains. Biochemistry. 2004;43:16365–16373. doi: 10.1021/bi048241x. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, Stein W, Lanzer M. Trans stimulation provides evidence for a drug efflux carrier as the mechanism of chloroquine resistance in Plasmodium falciparum. Biochemistry. 2003;42:9383–9394. doi: 10.1021/bi034269h. [DOI] [PubMed] [Google Scholar]
- Schofield L, Mueller I. Clinical immunity to malaria. Curr Mol Med. 2006;6:205–221. doi: 10.2174/156652406776055221. [DOI] [PubMed] [Google Scholar]
- Sehgal PN, Sharma MID, Gogai S. Resistance to chloroquine in falciparum malaria in Assam state, India. J Comm Dis. 1973;5:175–180. [Google Scholar]
- Severini C, Menegon M, Sannella AR, Paglia MG, Narciso P, Matteelli A, Gulletta M, Caramello P, Canta F, Xayavong MV, Moura IN, Pieniazek NJ, Taramelli D, Majori G. Prevalence of pfcrt point mutations and level of chloroquine resistance in Plasmodium falciparum isolates from Africa. Infect Genet Evol. 2006;6:262–268. doi: 10.1016/j.meegid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Sharma VP. Battling the malaria iceberg with chloroquine in India. 105Malar J. 2007;6 doi: 10.1186/1475-2875-6-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Valderramos SG, Fidock DA. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Mishra N, Awasthi G, Dash AP, Das A. Why is it important to study malaria epidemiology in India? Trends Parasitol. 2009;24:228–235. doi: 10.1016/j.pt.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Stephan W. Detecting strong positive selection in the genome. Mol Ecol Resour. 2010;10:863–872. doi: 10.1111/j.1755-0998.2010.02869.x. [DOI] [PubMed] [Google Scholar]
- X-z Su, Kirkman LA, Fujioka H, Wellems TE. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- Summers RL, Nash MN, Martin RE. Know your enemy: understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell Mol Life Sci. 2012;69:1967–1995. doi: 10.1007/s00018-011-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutar SKD, Gupta B, Ranjit M, Kar SK, Das A. Sequence analysis of coding DNA fragments of pfcrt and pfmdr-1 genes in Plasmodium falciparum isolates from Odisha, India. Mem Inst Oswaldo Cruz. 2011;106:78–84. doi: 10.1590/s0074-02762011000100013. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Tanabe K, Tsukahara T, Dzodzomenyo M, Dysoley L, Khamlome B, Sattabongkot J, Nakamura M, Sakurai M, Kobayashi J, Kaneko A, Endo H, Hombhanje F, Tsuboi T, Mita T. Large scale survey of novel genotypes of Plasmodium falciparum chloroquine resistance gene Pfcrt. 92Malar J. 2012;11 doi: 10.1186/1475-2875-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talisuna AO, Bloland P, D’Alessandro U. History, dynamics and public health importance of malaria parasite resistance. Clin Microbiol Rev. 2004;17:235–254. doi: 10.1128/CMR.17.1.235-254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talisuna AO, Langi P, Mutabingwa TK, Van Marck E, Speybroeck N, Egwang TG, Watkins WW, Hastings IM, D’Alessandro U. Intensity of transmission and spread of gene mutations linked to chloroquine and sulphadoxine-pyrimethamine resistance in falciparum malaria. Int J Parasitol. 2003;33:1051–1058. doi: 10.1016/s0020-7519(03)00156-5. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Okello PE, Erhart A, Coosemans M, D’Alessandro U. Intensity of malaria transmission and the spread of Plasmodium falciparum -resistant malaria: a review of epidemiologic field evidence. Am J Trop Med Hyg. 2007;77:170–180. [PubMed] [Google Scholar]
- Tanabe K, Sakihama N, Kaneko A. Stable SNPs in malaria antigen genes in isolated populations. 493Science. 2004;303 doi: 10.1126/science.1092077. [DOI] [PubMed] [Google Scholar]
- Tatem AJ, Smith DL. International population movements and regional Plasmodium falciparum malaria elimination strategies. Proc Natl Acad Sci USA. 2010;107:12222–12227. doi: 10.1073/pnas.1002971107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgt DS, van der Ven AJ, Schimmer B, Droogleever-Fortuyn HA, Sauerwein RW. Serious psychiatric symptoms after chloroquine treatment following experimental malaria infection. Ann Pharmacother. 2005;39:551–554. doi: 10.1345/aph.1E409. [DOI] [PubMed] [Google Scholar]
- Thompson PE, Werbel LM. Antimalarial agents: chemistry and pharmacology. Academic Press Inc; New York: 1972. 395 [Google Scholar]
- Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderramos SG, Valderramos JC, Musset L, Purcell LA, Mercereau-Puijalon O, Legrand E, Fidock DA. Identification of a mutant Pfcrt -mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 2010;6: doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vathsala PG, Pramanik A, Dhanasekaran S, Devi CU, Pillai CR, Subbarao SK, Ghosh SK, Tiwari SN, Sathyanarayan TS, Deshpande PR, Mishra GC, Ranjit MR, Dash AP, Rangarajan PN, Padmanaban G. Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter ( Pfcrt ) gene haplotype SVMNT in P. falciparum malaria in India. Am J Trop Med Hyg. 2004;70:256–259. [PubMed] [Google Scholar]
- Vieira PP, Ferreira MU, Alecrim MDAC, Alecrim WD, Silva LH da, Sihuincha MM, Joy DA, Mu J, X-z Su, Zalis MG. Pfcrt polymorphism and the spread of chloroquine resistance in Plasmodium falciparum populations across the Amazon basin. J Infect Dis. 2004;190:417–424. doi: 10.1086/422006. [DOI] [PubMed] [Google Scholar]
- Vinayak S, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. Prevalence of the K76T mutation in the pfcrt gene of Plasmodium falciparum among chloroquine responders in India. Acta Trop. 2003;87:287–293. doi: 10.1016/s0001-706x(03)00021-4. [DOI] [PubMed] [Google Scholar]
- Vinayak S, Mittra P, Sharma YD. Wide variation in microsatellite sequences within each Pfcrt mutant haplotype. Mol Biochem Parasitol. 2006;147:101–108. doi: 10.1016/j.molbiopara.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Neafsey DE, Schaffner SF, Park DJ, Wirth DF. Harnessing genomics and genome biology to understand malaria biology. Nat Rev Genet. 2012;13:315–328. doi: 10.1038/nrg3187. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, Mboup S, Duraisingh MT, Lukens A, Derr A, Stange-Thomann N, Waggoner S, Onofrio R, Ziaugra L, Mauceli E, Gnerre S, Jaffe DB, Zainoun J, Wiegand RC, Birren BW, Hartl DL, Galagan JE, Lander ES, Wirth DF. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Walliker D, Hunt P, Babiker H. Fitness of drug-resistant malaria parasites. Acta Trop. 2005;94:251–259. doi: 10.1016/j.actatropica.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Wang X, Mu J, Li G, Chen P, Guo X, Fu L, Chen L, Su X, Wellems TE. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People’s Republic of China. Am J Trop Med Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- Wellems T, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci USA. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems TE. Plasmodium chloroquine resistance and the search for a replacement antimalarial drug. Science. 2002;298:124–126. doi: 10.1126/science.1078167. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Hayton K, Fairhurst RM. The impact of malaria parasitism: from corpuscles to communities. J Clin Invest. 2009;119:2496–2505. doi: 10.1172/JCI38307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems TE, Plowe CV. Chloroquine resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- Wernsdorfer WH. Epidemiology of drug resistance in malaria. Acta Trop. 1994;56:143–156. doi: 10.1016/0001-706x(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Wernsdorfer WH, Payne D. The dynamics of drug resistance in Plasmodium falciparum. Pharmacol Ther. 1991;50:95–121. doi: 10.1016/0163-7258(91)90074-v. [DOI] [PubMed] [Google Scholar]
- White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Chemotherapy of malaria and resistance to antimalarials: report of a WHO scientific group. WHO Tech Rep Ser 529; 1973. pp. 1–121. [PubMed] [Google Scholar]
- World Health Organization Advances in malaria chemotherapy. WHO Tech Rep Ser. 1984;711:1–218. [PubMed] [Google Scholar]
- World Health Organization Southern Africa Malaria Control Programme. 2002. [Google Scholar]
- World Health Organization World malaria report 2011 , WHO Global Malaria Programme. WHO; Geneva: 2011. 278 [Google Scholar]
- Wongsrichanalai C, Pickard AL, Wernsdorfer WH. Epidemiology of drug resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, X-z Su. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang Z, Sun X. Molecular analysis of chloroquine resistance in Plasmodium falciparum in Yunnan Province, China. Trop Med Int Health. 2007;12:1051–1060. doi: 10.1111/j.1365-3156.2007.01882.x. [DOI] [PubMed] [Google Scholar]
- Young MD, Moore DV. Chloroquine resistance in Plasmodium falciparum. Am J Trop Med Hyg. 1961;10:317–320. doi: 10.4269/ajtmh.1961.10.317. [DOI] [PubMed] [Google Scholar]
- Zakeri S, Afsharpad M, Kazemzadeh T, Mehdizadeh K, Shabani A, Djadid ND. Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2008;78:633–640. [PubMed] [Google Scholar]
- Zhang JJ, Senaratne TN, Daniels R, Valim C, Alifrangis M, Amerasinghe P, Konradsen F, Rajakaruna R, Wirth DF, Karunaweera ND. Distribution pattern of Plasmodium falciparum chloroquine transporter ( pfcrt ) gene haplotypes in Sri Lanka 1996-2006. 811Am J Trop Med Hyg. 2011;85 doi: 10.4269/ajtmh.2011.11-0167. (Fre). [DOI] [PMC free article] [PubMed] [Google Scholar]