Abstract
The past decade has witnessed great advances in our understanding of protein structure-function relationships in terms of the ubiquitous existence of intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs). The structural disorder of IDPs/IDRs enables them to play essential functions that are complementary to those of ordered proteins. In addition, IDPs/IDRs are persistent in evolution. Therefore, they are expected to possess some advantages over ordered proteins. In this review, we summarize and survey nine possible advantages of IDPs/IDRs: economizing genome/protein resources, overcoming steric restrictions in binding, achieving high specificity with low affinity, increasing binding rate, facilitating posttranslational modifications, enabling flexible linkers, preventing aggregation, providing resistance to non-native conditions, and allowing compatibility with more available sequences. Some potential advantages of IDPs/IDRs are not well understood and require both experimental and theoretical approaches to decipher. The connection with protein design is also briefly discussed.
Keywords: intrinsically disordered proteins, protein function, flexibility, protein-protein interaction, molecular recognition, protein design, drug design
Introduction
Being a challenge or amendment to the conventional sequence-structure-function paradigm for proteins, intrinsically disordered proteins (IDPs) have attracted ever-increasing attention during the past decade.1–5 IDPs are intriguing because they do not have ordered structures in the free state under physiological conditions but still possess biological functions. The structural disorder of IDPs and intrinsically disordered regions (IDRs) lies in their distinct amino-acid sequences, that is, they usually have a low hydrophobicity content combined with a high net charge content.1,6,7 Without folded structures, IDPs/IDRs exist in an ensemble of rapidly-changing conformations with a flat free-energy landscape8–10 and exhibit almost unlimited structural heterogeneity.1,11 On the other hand, most IDPs/IDRs undergo a disorder-to-order transition upon binding to their biological partners (i.e., coupled folding and binding),12,13 although some remain disordered even in their bound state.14–17
The structural disorder of IDPs/IDRs enables them to play essential functions that are complementary to those of ordered proteins. Roughly, IDPs/IDRs can be classified into six broad functional classes,18,19 including effectors, scavengers, assemblers, entropic chains, display sites, and chaperones. A detailed bioinformatics analysis of the Swiss-Prot database revealed a positive correlation between IDPs/IDRs and 238 function keywords, the majority of which were related to signaling and regulation of key cellular processes.20 Due to the essential functions of IDPs/IDRs, it is not unexpected that they are abundant in all species.21–24 In particular, the average fraction of disordered residues predicted in eukaryotes is higher than that in prokaryotes, suggesting the importance of IDPs/IDRs in evolution. The abundant existence of IDPs/IDRs and their vital functions, as well as the fact that usual techniques used in characterizing conventional proteins may be not applicable for IDPs/IDRs, make IDPs/IDRs an important subfield of molecular structural biology. In fact, IDPs/IDRs are taking a due place in mainstream studies. For example, predicting disordered regions has become a part of the critical assessment of structure prediction (CASP).25
As IDPs/IDRs perform functions complementary to those of ordered proteins and are persistent in evolution, they might possess some advantages over ordered proteins (and at the same time possess some disadvantages). In this article, we give a brief review of the advantages of IDPs/IDRs provided by their conformational flexibility. Nine possible advantages are summarized and surveyed (Fig. 1): (1) economizing genome and protein resources; (2) overcoming steric restrictions in binding; (3) achieving high specificity with low affinity; (4) increasing binding rate; (5) facilitating posttranslational modifications; (6) enabling flexible linkers; (7) preventing aggregation; (8) providing resistance to non-native conditions; (9) allowing compatibility with more available sequences. Some advantages are self-evident, for example, the potency of acting as flexible linkers is incompatible with ordered structures. Some other advantages, however, are less clear and knowledge on them is still far from complete. It is acknowledged that the current review addresses only a narrow aspect of IDPs' properties. Readers are directed to several excellent reviews1,2,26–30 for a more comprehensive understanding of IDPs.
Figure 1.
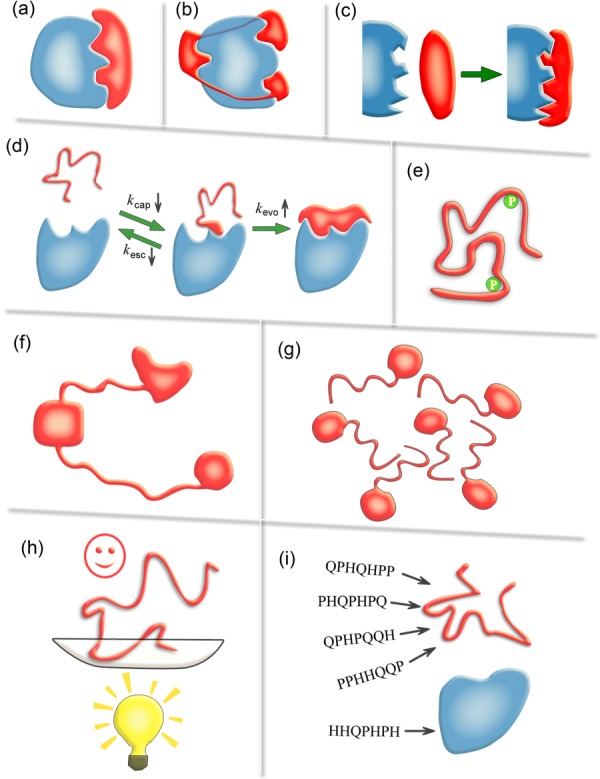
Schematic diagrams illustrating nine possible advantages of IDPs/IDRs. (a) Economizing genome and protein resources: IDPs/IDRs use smaller protein size to afford the same interface area as ordered proteins. (b) Overcoming steric restrictions in binding: IDPs/IDRs can overcome steric restrictions in binding complexes by protruding into partners or wrapping around them in ways difficult for ordered proteins. (c) Achieving high specificity with low affinity: the highly complementary binding interfaces and the unfavorable conformational-entropy changes result in high specificity and low affinity. (d) Increasing binding rate: the kinetic advantage of IDPs in a binding process stems from a faster evolution step from the encounter complex and a slower escaping rate. (e) Facilitating posttranslational modifications: the conformational flexibility of IDPs/IDRs greatly facilitates the exposure of modification sites and their binding to modifying enzymes. (f) Enabling flexible linkers: the lack of ordered structures makes IDPs/IDRs dominant in flexible linkers of proteins which are obviously out of reach of ordered proteins. (g) Preventing aggregation: IDPs/IDRs possess favorable interactions with water and are inherently advantageous in preventing aggregation. (h) Providing resistance to non-native conditions: you cannot unfold what is already unfolded. (i) Allowing compatibility with more available sequences: the sequence space of IDPs/IDRs is expected to be larger than that of ordered proteins.
Advantage 1: Economizing Genome and Protein Resources
The interface area of IDPs in protein-protein complexes is similar in size to that of ordered proteins, but the sequence used to create the same interface area is much shorter for IDPs [Fig. 1(a)].31,32 It originates from the fact that IDPs have extended structures which are stable only in the complexes but not in monomers. As a result, IDPs possess greater interface area per residue than ordered proteins. It has been estimated that the protein size would be two to three times larger if IDPs were required to be as stable as monomers, which would unavoidably exacerbate cellular crowding and increase the sequence size.31 Therefore, IDPs/IDRs are advantageous in saving genome and protein resources. This feature may be more critical for species with small genome size. For example, viruses were shown to have the widest spread of IDPs/IDRs content compared with three domains of life, and Avian carcinoma virus possesses the highest disorder content of 77.3% among 3500 proteomes.21
Intrinsic disorder may also help to reduce the genome size in other mechanisms.27 The number of genes in the human genome is significantly smaller than that of the diverse proteins necessary in higher organisms.27 Alternative splicing is a way to avoid over-large genomes, in which multiple proteins can be produced from a single gene. Regions affected by alternative splicing are frequently biased to be disordered,33 which helps to avoid structural disruption in the spliced proteins. “Moonlighting” is another solution to control the genome size, in which a single protein is capable of carrying out more than one function. IDPs/IDRs provide an important mechanism for moonlighting since they can use the same or overlapping regions to fulfill distinct functions by adopting different conformations upon binding.34 For example, the intrinsically disordered domain of the sulfhydryl oxidase ALR performs dual functions: as a mitochondrial targeting signal in the cytosol and as a crucial recognition site in the disulfide relay system of intermembrane space.17 Another example of multifunctional IDPs is anhydrin which acts as a chaperone and an endonuclease.35
Advantage 2: Overcoming Steric Restrictions in Binding
The high chain flexibility and conformational disorder of IDPs/IDRs enables them to form complementary binding interfaces with their targets more easily. Via coupled folding and binding, IDPs/IDRs can overcome steric restrictions by protruding into the concavities of partners or wrapping around partners in ways difficult for ordered proteins [Fig. 1(b)]. It was found that the partner interfaces of MoRFs (the short segments of IDPs that perform molecular recognition and usually undergo disorder-to-order transitions upon binding) are significantly less flat than those of ordered proteins.36 When measured by the RMSD of interface atoms to the fitting plane, the interface planarity of IDPs partners is 3.76 Å while that of ordered proteins is 2.98 Å.36 The binding modes of IDPs/IDRs are highly diverse and create multifarious unusual complexes: wrappers, chameleons, penetrators, huggers, intertwined strings, long cylindrical containers, connectors, armature, etc.37 For example, the intrinsically disordered calpastatin (endogenous inhibitor of calpain) wraps around and binds to its target on three surfaces to form a flexible wrapper.38
After folding coupled with binding, the interface structure of IDPs/IDRs is still more flexible than that of ordered proteins. The average crystallographic B-factor of interfaces for IDPs (51 Å2) in complex structures was much larger than that of ordered proteins (21 Å2), indicating interface atoms of IDPs remain highly dynamic.39 IDPs/IDRs also have a high content of the poly-l-proline type II (PPII) helix which is markedly more flexible in comparison with α-helix and β-sheet.40,41 In addition to the complete folding upon binding where IDPs/IDRs adopt ordered structures, some IDPs/IDRs experience incomplete folding where a significant part remains disordered in the bound state, forming dynamic or fuzzy complexes.42–44 In the extreme case (random fuzziness), IDPs remain entirely disordered in the bound state and resemble a “binding cloud” where multiple binding sites are dynamically distributed.1,42 An example is the Sic1-Cdc4 complex, where a few binding motifs of Sic1 interact with Cdc4 in a dynamic equilibrium, one at a time, and the parts of Sic1 not interacting with Cdc4 remain disordered.14,45 For the binding with small molecule ligands, IDPs remain disordered and the ligands bind at different sites along the protein chains, so it may be described as ligand clouds around protein clouds.10
Advantage 3: Achieving High Specificity With Low Affinity
Combining high specificity with low affinity is a widely-mentioned advantage of IDPs/IDRs,4,12,46,47 which is a useful pair of properties for a reversible signaling transduction by enabling rapid association/dissociation with the partner without excessive binding strength.
The low affinity of IDPs/IDRs originates from the coupled folding and binding. The ordered (folded) state is unstable for IDPs/IDRs when they are free in solution. In a coupled folding and binding process, the folding increases the free energy while the binding of IDPs/IDRs to their targets decreases the free energy. As a result, the net free-energy change in such a coupled folding-binding process is smaller than that in a pure binding process, leading to a lower affinity. The low affinity allows IDPs to have a high dissociation rate, which can be essential for regulatory and signaling functions. Based on a dataset of protein–protein associations for 35 IDPs and 45 ordered proteins,48 we showed that the distribution of the equilibrium dissociation constants for IDPs is slightly different from that for ordered proteins (Fig. 2), where 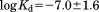 for IDPs and
for IDPs and 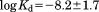 for ordered proteins. In another benchmark dataset, it was found that 62% of the complexes with lower binding affinity contain regions formed via coupled folding and binding.49 The relatively low affinity of IDPs/IDRs also suggested that high-affinity binding proteins can tolerate more structural disorder, consistent with the difference in the content of IDPs/IDRs of prokaryotic and eukaryotic genomes.50 It is noted that although the disorder-to-order transition of IDPs/IDRs decreases the conformational entropy, IDPs/IDRs usually possess more favorable interface interactions than ordered proteins.32,39 Furthermore, the binding free energy is affected by many other factors.51,52 The affinity of either IDPs/IDRs or ordered proteins is highly variable and covers a wide range. So the affinity difference between IDPs/IDRs and ordered proteins applies to average values, not individual cases. Actually, some IDPs exhibit extremely high affinities. For example, the binding affinity of the intrinsically disordered antitoxin CcdA to the gyrase poison CcdB is in the pico-molar range, much higher than that of most ordered proteins.53,54
for ordered proteins. In another benchmark dataset, it was found that 62% of the complexes with lower binding affinity contain regions formed via coupled folding and binding.49 The relatively low affinity of IDPs/IDRs also suggested that high-affinity binding proteins can tolerate more structural disorder, consistent with the difference in the content of IDPs/IDRs of prokaryotic and eukaryotic genomes.50 It is noted that although the disorder-to-order transition of IDPs/IDRs decreases the conformational entropy, IDPs/IDRs usually possess more favorable interface interactions than ordered proteins.32,39 Furthermore, the binding free energy is affected by many other factors.51,52 The affinity of either IDPs/IDRs or ordered proteins is highly variable and covers a wide range. So the affinity difference between IDPs/IDRs and ordered proteins applies to average values, not individual cases. Actually, some IDPs exhibit extremely high affinities. For example, the binding affinity of the intrinsically disordered antitoxin CcdA to the gyrase poison CcdB is in the pico-molar range, much higher than that of most ordered proteins.53,54
Figure 2.
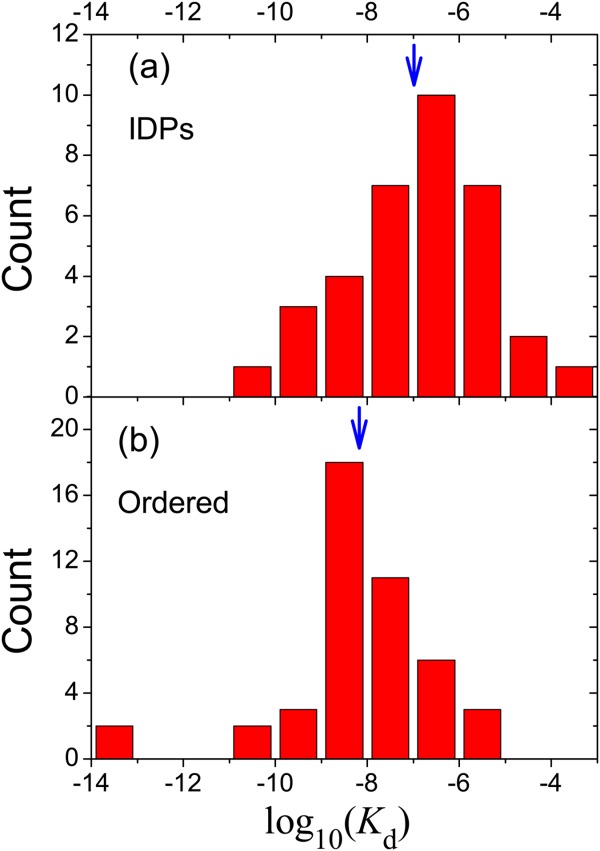
Distribution of binding affinity for (a) IDPs and (b) ordered proteins. The average values of logKd are indicated by arrows. The analysis was based on the dataset compiled in Ref.48.
The high specificity of IDPs/IDRs is generally believed to be related to their interface characteristics.3,4,12,27 The binding specificity is mainly determined by the size and complementarity of the binding interface. Due to the chain flexibility and the coupled folding-and-binding, as mentioned above, the interaction interfaces of IDPs/IDRs to their targets are highly complementary [Fig. 1(c)]. Although IDPs/IDRs usually use short segments in molecular recognitions, the extended structures formed in the complexes enable them to achieve large binding interfaces. Therefore, IDPs/IDRs are expected to exhibit high specificity in molecular recognitions. High specificity is necessary for their critical functions in signal recognition, transduction, and regulation.
However, there is still some debate on the specificity of IDPs/IDRs.47,55 The above reasoning is not consistent with the conventional opinion that more flexible proteins are more promiscuous,47,55 that is, if structural flexibility enables IDPs/IDRs to form complementary interfaces with their cognate targets, the same property could also enable them to fit the surfaces of noncognate targets and thus IDPs/IDRs would lose their interaction specificity. Under interface-interaction perturbations (to mimic the structural difference between cognate and noncognate targets), IDPs/IDRs can either adjust their conformations to maintain the interface complementarity to stabilize the enthalpy or become conformationally more dynamic to stabilize the entropy, and as a result, IDPs/IDRs have a more complete enthalpy–entropy compensation to reduce the influence of the perturbations.55 Indeed, an analysis on the thermodynamic data of interface mutation verified that IDPs had higher enthalpy–entropy compensation than ordered proteins,55 suggesting IDPs to be more malleable.
To clarify the specificity difference between IDPs and ordered proteins, a coarse-grained molecular dynamics (MD) study was conducted to calculate the mutation free energy (ΔΔG) of IDPs and ordered proteins under the same perturbations.55 It was found that ΔΔG for IDPs is smaller than that for ordered proteins by an averaged difference of 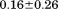 kcal/mol. This difference is smaller than what would be expected from the enthalpy–entropy compensation mentioned above. The reason is the same perturbation caused larger mutation enthalpy (ΔΔH) in IDPs.55 Recently, an analysis of protein-protein complexes from the Protein Data Bank revealed that polar interface interactions make a larger contribution to the specificity in IDPs, and the follow-up computational alanine scanning of both hydrophobic and charged residues confirmed that these mutations caused larger ΔΔH in IDPs.56 Therefore, IDPs are likely to possess high specificity, although maybe not as high as ordered proteins.
kcal/mol. This difference is smaller than what would be expected from the enthalpy–entropy compensation mentioned above. The reason is the same perturbation caused larger mutation enthalpy (ΔΔH) in IDPs.55 Recently, an analysis of protein-protein complexes from the Protein Data Bank revealed that polar interface interactions make a larger contribution to the specificity in IDPs, and the follow-up computational alanine scanning of both hydrophobic and charged residues confirmed that these mutations caused larger ΔΔH in IDPs.56 Therefore, IDPs are likely to possess high specificity, although maybe not as high as ordered proteins.
High specificity may be critical for the biological functions of IDPs/IDRs. On the other hand, since the specificity of IDPs/IDRs may not be as high as that of ordered proteins, IDPs/IDRs may be more promiscuous in molecular recognitions and need to be tightly controlled in cells. Indeed, the abundance of IDPs/IDRs is regulated so that they are available in appropriate amounts and at appropriate times as needed.28,57 The promiscuity of IDPs/IDRs also makes them a major determinant of dosage sensitivity where promiscuous interactions drive pathological changes in response to gene over-expression.58 In the absence of tight regulation of IDPs/IDRs, severe outcomes may result, for example, the occurrence of various diseases.59,60
A closely related characteristic is multiple specificity—the capacity of interacting with many partners.61 The high plasticity and malleability of IDPs/IDRs enable them to bind to multiple partners more readily by changing conformations or interaction regions according to the templates provided by different targets.62,63 The cyclin-dependent kinase (CDK) inhibitor p21, which is a very early example of IDPs, binds to different CDK/cyclin complexes during the cell cycle and the binding diversity is mediated by the adaptability of its LH region.64 The induced structures of the same IDP/IDRs may be different in binding with different partners, for example, the C-terminus of p53 becomes a helix, a sheet, or a coil in different complexes.65 Even when the induced structures of the same IDP/IDRs in different complexes are similar, the utilized anchor residues may be totally different which are important in controlling the specificity.66 Multispecificity is useful for signaling and regulation. IDPs/IDRs are commonly involved in the hubs of protein–protein interaction networks,67–70 where one IDP/IDR may bind to many partners or many IDPs/IDRs bind to one partner. The chaperone function of IDPs is also related to the multispecificity where it is vital to bind multiple aggregation-sensitive client proteins.71 In addition, multispecificity is also a basis of the moonlighting mentioned above.
Advantage 4: Increasing Binding Rate
The binding rate of proteins to their targets is important for diverse processes ranging from enzyme catalysis/inhibition to cellular signaling. Compared with ordered proteins, IDPs/IDRs possess a kinetic advantage, that is, their binding rate is relatively higher.
The first insight into the kinetic advantage of IDPs/IDRs was provided by Shoemaker et al. in terms of the “fly-casting mechanism.”72 By virtue of extended conformations, IDPs/IDRs could bind to their targets weakly at a larger distance (capture radius) from the actual binding site. Then they “reel in” the targets to complete the binding process while simultaneously folding.72 Later, Huang and Liu performed a critical assessment of this mechanism by investigating the inherent influence of chain disorder on the binding kinetics of IDPs/IDRs.48 Via Langevin dynamics simulations with a Gō-like coarse-grained model, it was found that although IDPs/IDRs possess a larger capture radius, the larger capture radius inevitably leads to a slower translational diffusion. As a result, the rate constant for forming the encounter complex actually decreased modestly as the chain flexibility increases, rather than increased as originally proposed. The real source for the kinetic advantage of IDPs/IDRs lies in the second step, where the encounter complex evolves into the final bound state faster and escapes to the unbound state slower than ordered proteins [Fig. 1(d)].48 In addition, the available experimental data on the binding kinetics of IDPs and ordered proteins were compared to show the general difference in real systems.48 On average, IDPs bind two to three times as fast as ordered proteins under the same affinity. It was also noted that the binding rate spans a large range of magnitude so that comparison of a single datum is meaningless.
Protein binding rates are influenced by various factors,73 for example, nonspecific binding and electrostatic interactions. Due to their high chain flexibility, IDPs/IDRs may possess more nonnative interactions in the folding and binding processes than ordered proteins. It was shown that nonnative hydrophobic interactions greatly amplify the kinetic advantages of IDPs/IDRs.74 IDPs/IDRs usually contain a high content of charge residues, and electrostatic interactions were also found to contribute to their kinetic advantage via the electrostatic steering mechanism.75 Evidently, the rate of the intrinsically disordered WASP GBD (the GTPase binding domain of the Wiskott-Aldrich syndrome protein) upon binding its target (Cdc42) is highly dependent on ionic strength in the experimental study.76 In the interactions between IDPs and DNA, it was shown that electrostatic steering and protein flexibility synergistically couple to achieve rapidity in recognition.77 Recently, Ganguly et al. showed that long-range electrostatic interactions not only enhance the encounter rate via the electrostatic steering but also promote the folding-competent topologies in the encounter complexes, allowing rapid formation of short-range native interactions to form the final bound state.78,79
Binding kinetics is important for functions at molecular and system levels.73,80 Association between signaling proteins and their cellular targets should be both fast and transient. The enhanced binding rate of IDPs/IDRs, together with the low affinity, provides an advantageous solution to these needs.47 For the intrinsically disordered translocation domain of Colicin E9, although its binding with TolB is weaker than the competitors, it wins in the competitive recruitment of TolB by its much higher binding rate.81 In nonsense-mediated mRNA decay (NMD), it was suggested that a fly-casting mechanism enabled by long disordered regions in NMD complexes is exploited for effective long-range communication.82
Advantage 5: Facilitating Post-translational Modifications
Posttranslational modifications (PTMs) are an important means to regulate functions of proteins. The conformational flexibility of IDPs/IDRs greatly facilitates exposure of their modification sites and binding to the modifying enzymes [Fig. 1(e)]. The flexibility of IDPs/IDRs also makes it possible for a single enzyme to bind to and modify sites in a wide variety of proteins. In contrast, PTMs on ordered proteins may be restricted by site accessibility. Therefore, IDPs/IDRs facilitate the regulation of cellular processes by PTMs.
Phosphorylation is one of the most important and well-studied PTMs. It dominates the number of experimentally-identified PTMs by an order of magnitude.83 It was found that sequence attributes of regions adjacent to phosphorylation sites are very similar to those of IDPs/IDRs, and such a correlation between the structure disorder and the occurrence of phosphorylation has been employed to improve the prediction of protein phosphorylation sites.84 For Arabidopsis plasma membrane proteins, 30% of the phosphorylation sites are located in long intrinsically disordered regions and 28% of sites are located in shorter disordered regions.85 Phosphorylation of IDPs/IDRs serves as an essential control mechanism in signaling and regulation. A study on the phosphorylation variation during the cell cycle indicated that intrinsically disordered regions tend to contain sites with dynamically varying levels, while ordered regions retain more constant phosphorylation levels.86 Typical examples are eukaryotic cyclin-Cdk inhibitors Sic1 and p27 whose inhibitory activity, stability, and subcellular localization are regulated by phosphorylation on different sites.87,88
Many other PTMs are also associated with IDPs/IDRs. A large-scale analysis of the Swiss-Prot database showed that 17 PTMs keywords (i.e., phosphorylation, amidation, ubiquitination, glycosylation, sulfation, and methylation, etc) are strongly correlated with predicted disorder.89 In comparison with ordered proteins, IDPs/IDRs contain more ubiquitination sites and this observation was used in developing a predictor program for such sites.90,91 The preference of ubiquitination in IDPs/IDRs provides a basis for their efficient regulation via rapid degradation.
Advantage 6: Enabling Flexible Linkers
Acting as entropic chains such as flexible linkers is one of the six broad functional classes of IDPs/IDRs.18,92 The lack of ordered structures of IDPs/IDRs enables them to act as flexible linkers which are obviously out of reach of ordered proteins.
In multidomain proteins, modular domains are often connected by flexible linkers [Fig. 1(f)].12 The primary function of these linkers is to restrict the distance and to enable an orientational freedom of the attached domains. IDRs carry out such functions without undergoing a disorder-to-order transition and can be mimicked reasonably well by the behavior of low-complexity polypeptides. The role of IDRs as flexible linkers was nicely demonstrated in an experimental study on the Escherichia coli tubulin homologue FtsZ.93 When the linker in the E. coli FtsZ was replaced by those from other bacteria, or even from an unrelated IDP (human α-adducin), its function in cell division was found to be unaffected if the length of the new linker was similar to that of the original.93 In the voltage-activated potassium channels Kv, it was found that the intrinsically disordered C-terminal domain behaves as an ideal flexible chain in binding with intracellular scaffold proteins.94
Linking two modular domains or segments via a flexible linker increases the local concentration of one relative to the other as has been widely adopted in biological systems to increase efficiency. This can be used to connect two different functions, for example, the functions of the DNA-binding domain and the activation domain in a DNA transcription factor. In some other cases, this provides a simple way to increase the binding affinity of two linked domains (segments) to a common target.75,95 Furthermore, the binding affinity can be tuned by varying the length of the linker. For example, the variation of linker length in ratiometric fluorescent sensor proteins was shown to tune their Zn(II) binding affinity from picomolar to femtomolar where the effective local concentration was well described by a worm-like chain model.96
Advantage 7: Preventing Aggregation
Uncontrolled aggregation is a constant threat for proteins, and proteins have to acquire sequence and structural adaptations to avoid undesired amyloid-like aggregation.97 The physicochemical properties of the sequences of IDPs/IDRs are generally negatively correlated with those of aggregation-prone sequences.7 For example, most aggregation-prone sequences have high hydrophobicity and low net charge,98,99 while IDPs/IDRs have low hydrophobicity and high net charge.46 So IDP/IDRs sequences are distinct from amyloidogenic peptides,100 and tend to prevent aggregation. The advantage of IDPs/IDRs relative to ordered proteins in preventing amyloid-like aggregation is apparent in examining the packing density of sequences,101 that is, the expected number of neighbor contacts per residue within a given distance (similar to the concept of ligancy in chemistry). A residue with high packing density is more sticking to contact with other residues. It was shown that amyloid proteins possess high expected packing density, and IDPs/IDRs possess low values, while ordered proteins possess moderate values locating between those of amyloids and IDPs/IDRs (Fig. 3).101 It was also shown that ordered proteins contain almost three times as much aggregation nucleating regions as IDPs.102
Figure 3.
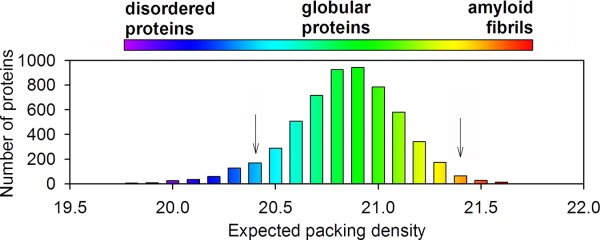
Schematic distribution of the expected packing density for IDPs/IDRs, ordered proteins and amyloid fibrils (adopted from Ref.101).
Attaching disordered regions to ordered proteins or aggregation-prone sequences could protect the latter from aggregation [Fig. 1(g)]. Bioinformatics analysis demonstrated that interaction linear motifs are on average embedded in locally disordered regions, where a typical motif contains about six residues and is surrounded by approximately 20 residues that are intrinsically disordered.103 A simulation with a lattice model showed that small hydrophobic peptides with disordered flanks remain stable under conditions where peptides without flanks tend to aggregate.104 Such a principle was also adopted in artificial protein fusions to effectively prevent aggregation and achieve high soluble protein expression.105 Recently, de novo design was conducted to develop a stable monomeric peptide targeting to the tumor necrosis factor (TNF)-α by increasing the propensity of intrinsic disorder.106
The capability of IDPs/IDRs to prevent aggregation is also essential for their chaperone function. For example, the E. coli protein HdeA experiences a stress-induced order-to-disorder transition to display chaperone activity and binds to other unfolded proteins to prevent their aggregation under the stress conditions.107
Advantage 8: Providing Resistance to Non-Native Conditions
Ordered proteins are usually vulnerable to unfolding. Their structures and functions may be destroyed by various external stresses such as acid, base, inorganic salt, organic solvent, heating and cooling. In contrast, IDPs are more likely to keep stable with respect to various extreme conditions due to their lack of structures. For example, prothymosin α can be boiled for a few days without losing its activity.11 It even displays an atypical “turn out” response where partial structure is induced by heating.108 A positive correlation between disorder content and the resistance to heat shock was shown in a survey on 11 proteins.109 A study on a freezing-induced loss-of-function model of globular-disordered functional protein pairs also confirmed that IDPs are more resistant to cold treatment than ordered proteins.110 Late embryogenesis abundant proteins, which play crucial roles in cellular dehydration tolerance, are mostly IDPs/IDRs.111 The super stability of IDPs/IDRs was exactly summarized by Uversky as “you cannot break what is already broken” [Fig. 1(h)].11
The structural disorder of IDPs/IDRs not only helps them to survive under extreme conditions,112 but is also beneficial for their resistance to environmental perturbations under physiological conditions.39 In a real biological system, proteins perform their functions in complicated environments, which can be affected by various factors. To maintain their activity, proteins should not be overly sensitive to perturbations in cellular conditions.113,114 A coarse-grained simulation revealed that the binding affinity and kinetics of IDPs were less sensitive to the perturbations of temperature and intermolecular interactions than ordered proteins.39 The origin of such robustness was attributed to the capacity of IDPs to adjust their bound conformations to compensate for various perturbations, which was supported by an analysis of protein complex structures. This ability was termed a “buffer effect,”39 which, together with the kinetic advantage discussed above, enables IDPs to conduct their functions in cellular signaling and regulation.
Advantage 9: Allowing Compatibility With More Available Sequences
The protein sequences span only a very small fraction of the whole available sequence space. For ordered proteins, the number of available sequence is greatly reduced by the need to form a unique folded structure while avoiding insoluble aggregates.100,115 For IDPs/IDRs, due to the removal of restrictions posed by forming ordered structure, the available sequence space is expected to be much larger than that of ordered proteins [Fig. 1(i)].1,11 The sequence difference between IDPs/IDRs and ordered proteins can be graphically depicted in the two-dimensional charge-hydropathy (CH) plot where ordered proteins reside in the region with high hydropathy and low net charge (Fig. 4). A comparison of the areas for IDPs and ordered proteins in the CH plot showed that the sequence space of extended IDPs is at least fivefold greater than that of compact soluble proteins (including both “molten globule”-like IDPs and well-folded ordered proteins).11 The difference in available sequence space between IDPs/IDRs and ordered proteins can also be discussed in terms of their sequence designability,4,116 which was defined as the number of sequences coding a protein.117 Without the structural restrictions, IDPs/IDRs may possess more sequence redundancy, resulting in high sequence designability.
Figure 4.
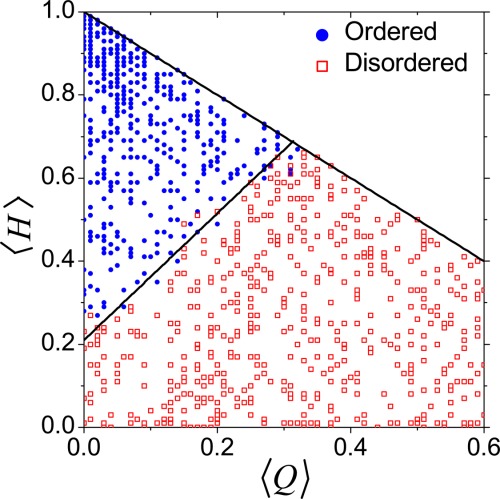
Schematic difference of the areas for IDPs and ordered proteins in the CH plot, where 〈H〉 and 〈Q〉 represent the fraction of hydrophobic and charged residues, respectively (modified from Ref.7).
Proteins were selected by evolution in nature. The greater number of available sequences may contribute some benefit to IDPs/IDRs in the evolution process. During evolution, many mutations are neutral and do not affect the activity of proteins, while rare mutations are advantageous (positive) and will presumably be preserved because they improve or change the protein function in a desired way. An IDP with a given function may on average have more neutral mutations than an ordered protein due to its sequence redundancy, and it may also have more positive mutations to achieve an advantageous phenotype due to the sequence redundancy of the resulting new IDP. The involvement of IDPs/IDRs in organisms may be closely related to their evolution. For flaviviruses, it was shown that the rapid evolutionary dynamics of structural disorder is a potential driving force for their phenotypic divergence.118 In mitochondria, it was shown that the IDPs/IDRs contents are markedly different between those descending from a bacterial ancestor and those being added to the mitochondria more recently, suggesting that the frequency of IDPs/IDRs in mitochondria was due to the evolutionary origin rather than the functional difference of the protein.119
The sequence redundancy of IDPs/IDRs sheds light on the design of IDPs/IDRs. At present, both IDPs design and IDPs-targeted drug design are in their infancy.10,120,121 Structure-based design of ordered proteins has been extensively studied, resulting in some well developed strategies.122 But it is unclear whether these approaches can be applied to design IDPs/IDRs, and rare attempts were conducted in this direction. The lack of a unique folded structure also brings extra hindrance. With an ordered protein, one knows the location and geometry of the active/binding site, which is the basis of conventional rational design. For an IDP, it may be difficult to obtain such knowledge in order to achieve a desired function. On the other hand, IDPs/IDRs possess greater available sequence space than ordered proteins, which would be favorable for the rational IDPs/IDRs design. Recently, Shen et al. have successfully designed a small IDP to inhibiting TNF-α, where the coupled-folding-and-binding complex structure was adopted as a basis in the design.106 It delivered an optimistic hint on the possibility of IDPs/IDRs design.
Some Remarks
The subject of this review is the possible advantages of IDPs, and inevitable bias exists in emphasizing them. In fact, no benefit is free. IDPs indeed possess disadvantages (and even harmfulness). IDPs/IDRs often engage in promiscuous interactions and are associated with various human diseases.58,59 It was found that 79% of cancer-associated proteins contain predicted IDRs of 30 residues or longer.123 In the Swiss-Prot disease category, it was shown that 11 disease-related keywords were strongly correlated with IDPs/IDRs while none correlated with ordered proteins.89 To suppress their harmfulness, the availability of IDPs/IDRs is tightly regulated in cells, which would require extra costs.57 Another apparent disadvantage of IDPs is their susceptibility to proteolysis.27,92,124 To avoid unwanted proteolytic degradation as can occur in many biological environments, some mechanisms may exist to protect IDPs/IDRs in vivo.27,124 Binding with partners can provide effective protection for IDPs/IDRs by hiding the protease-sensitive sites. “Nanny” chaperones were also proposed to protect newly synthesized IDPs/IDRs from degradation by specific interactions.125 In addition, proteases are usually compartmentalized and sequestered in the cell, and their activity is tightly regulated.27,92
Advantages of IDPs/IDRs have been widely discussed in the literature. In the above we have summarized and surveyed nine possible advantages. It is not a complete list, for example, Uversky gave a list with 21 advantages in a recent review.1 It should be noted that many of them lack direct proof. Therefore, they are working concepts and hypotheses rather than established facts. Some advantages seem self-evident, for example, economizing genome/protein resources, overcoming steric restrictions in binding, facilitating posttranslational modifications, and enabling flexible linkers. Some other advantages, however, are complicated and even contradictory, for example, high specificity and compatibility with more available sequences. More work is needed to illuminate these properties of IDPs/IDRs. In this aspect, molecular modeling, especially with coarse-grained models, are powerful tools and extremely useful.126
The discussed advantages are not orthogonal. Many are connected in nature. For example, the ability of overcoming steric restrictions in binding would also facilitate posttranslational modifications.
The possible advantages of IDPs/IDRs over ordered proteins differ in magnitude. For example, the interface area per residue in binding can vary substantially, whereas the average binding rate of IDPs is only two to three times faster than that of ordered proteins when the binding rate itself covers a few orders of magnitude. Therefore, it may be difficult for experimental studies to clearly demonstrate some advantages of disorder. Some conflicting studies on IDPs/IDRs, for example, the role in hubs, the evolution rate, and the role of structural disorder in p53-related diseases, may relate to this.
Conclusion
IDPs/IDRs are distinct from conventional ordered proteins in sequence, structure and function. IDPs/IDRs possess various advantages over ordered proteins, enabling them to perform vital functions in cells and to persist during evolution. Some possible advantages of IDPs/IDRs are not well understood and require both experimental and theoretical approaches to decipher. Such studies would advance our understanding of IDPs/IDRs, and facilitate the development of their application.
Acknowledgments
The authors gratefully acknowledge Dr. Fan Jin and Huaiqing Cao for their insightful discussions. The authors also thank Prof. Dr. Brian Matthews and the anonymous reviewer for their helpful and constructive comments that greatly helped to improve the article.
References
- Uversky VN. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Huang YQ, Liu ZR. Intrinsically disordered proteins: the new sequence-structure-function relations. Acta Phys Chim Sin. 2010;26:2061–2072. [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CR, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Jin F, Liu ZR. Inherent relationships among different biophysical prediction methods for intrinsically disordered proteins. Biophys J. 2013;104:488–495. doi: 10.1016/j.bpj.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Forman-Kay JD. Ensemble modeling of protein disordered states: experimental restraint contributions and validation. Proteins. 2012;80:556–572. doi: 10.1002/prot.23220. [DOI] [PubMed] [Google Scholar]
- Fisher CK, Stultz CM. Constructing ensembles for intrinsically disordered proteins. Curr Opin Struct Biol. 2011;21:426–431. doi: 10.1016/j.sbi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Yu C, Lai LH, Liu ZR. Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins. PLoS Comput Biol. 2013;9:e1003249. doi: 10.1371/journal.pcbi.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013;1834:932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Knott M, Best RB. A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: evidence from molecular simulations. PLoS Comput Biol. 2012;8:e1002605. doi: 10.1371/journal.pcbi.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci USA. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalov AB, Kim WM, Saline M, Stem LJ. The intrinsically disordered cytoplasmic domain of the T cell receptor xi chain binds to the Nef protein of simian immunodeficiency virus without a disorder-to-order transition. Biochemistry. 2008;47:12942–12944. doi: 10.1021/bi801602p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gajda K, Felli IC, Gallo A, Pavelkova A, Kallergi E, Andreadaki M, N Katrakili, C Pozidis, K Tokatlidis. An intrinsically disordered domain has a dual function coupled to compartment-dependent redox control. J Mol Biol. 2013;425:594–608. doi: 10.1016/j.jmb.2012.11.032. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Xie HB, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Pushker R, Mooney C, Davey NE, Jacque JM, Shields DC. Marked variability in the extent of protein disorder within and between viral families. PLoS One. 2013;8:e60724. doi: 10.1371/journal.pone.0060724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztanyi Z, Uversky VN, Obradovic Z, Kurgan L, AK Dunker, J Gough. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domenico T, Walsh I, Tosatto SCE. Analysis and consensus of currently available intrinsic protein disorder annotation sources in the MobiDB database. BMC Bioinformatics. 2013;14:S3. doi: 10.1186/1471-2105-14-S7-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrskyy B, Fidelis K, Moult J, Tramontano A, Kryshtafovych A. Evaluation of disorder predictions in CASP9. Proteins. 2011;79:107–118. doi: 10.1002/prot.23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Mitrea DM, Kriwacki RW. Regulated unfolding of proteins in signaling. FEBS Lett. 2013;587:1081–1088. doi: 10.1016/j.febslet.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- Meszaros B, Tompa P, Simon I, Dosztanyi Z. Molecular principles of the interactions of disordered proteins. J Mol Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, AK Dunker. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chakrabortee S, Meersman F, Schierle GSK, Bertoncini CW, McGee B, Kaminski CF, Tunnacliffe A. Catalytic and chaperone-like functions in an intrinsically disordered protein associated with desiccation tolerance. Proc Natl Acad Sci USA. 2010;107:16084–16089. doi: 10.1073/pnas.1006276107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40:1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- Kiss R, Bozoky Z, Kovacs D, Rona G, Friedrich P, Dvortsak P, Weisemann R, Tompa P, Perczel A. Calcium-induced tripartite binding of intrinsically disordered calpastatin to its cognate enzyme, calpain. FEBS Lett. 2008;582:2149–2154. doi: 10.1016/j.febslet.2008.05.032. [DOI] [PubMed] [Google Scholar]
- Huang YQ, Liu ZR. Smoothing molecular interactions: The “kinetic buffer” effect of intrinsically disordered proteins. Proteins. 2010;78:3251–3259. doi: 10.1002/prot.22820. [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Adzhubei AA, Sternberg MJE, Makarov AA. Polyproline-II helix in proteins: structure and function. J Mol Biol. 2013;425:2100–2132. doi: 10.1016/j.jmb.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M. Fuzziness: linking regulation to protein dynamics. Mol Biosyst. 2012;8:168–177. doi: 10.1039/c1mb05234a. [DOI] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2009;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mittag T, Orlicky S, Choy WY, Tang XJ, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX. Intrinsic disorder: signaling via highly specific but short-lived association. Trends Biochem Sci. 2012;37:43–48. doi: 10.1016/j.tibs.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Liu ZR. Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the “fly-casting” mechanism. J Mol Biol. 2009;393:1143–1159. doi: 10.1016/j.jmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Vreven T, Hwang H, Pierce BG, Weng ZP. Prediction of protein-protein binding free energies. Protein Sci. 2012;21:396–404. doi: 10.1002/pro.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JT, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci USA. 2009;106:19819–19823. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- Wang RX, Lai LH, Wang SM. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput-Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De Greve H, Loris R. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol Cell. 2009;35:154–163. doi: 10.1016/j.molcel.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Drobnak I, De Jonge N, Haesaerts S, Vesnaver G, Loris R, Lah J. Energetic basis of uncoupling folding from binding for an intrinsically disordered protein. J Am Chem Soc. 2013;135:1288–1294. doi: 10.1021/ja305081b. [DOI] [PubMed] [Google Scholar]
- Huang YQ, Liu ZR. Do intrinsically disordered proteins possess high specificity in protein-protein interactions? Chem-Eur J. 2013;19:4462–4467. doi: 10.1002/chem.201203100. [DOI] [PubMed] [Google Scholar]
- Wong ETC, Na D, Gsponer J. On the importance of polar interactions for complexes containing intrinsically disordered proteins. PLoS Comput Biol. 2013;9:e1003192. doi: 10.1371/journal.pcbi.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics. 2009;10:12. doi: 10.1186/1471-2164-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudabadi G, Rajagopalan K, Getzenberg RH, Hannenhalli S, Rangarajan G, Kulkarni P. Intrinsically disordered proteins and conformational noise implications in cancer. Cell Cycle. 2013;12:26–31. doi: 10.4161/cc.23178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr Opin Struct Biol. 2011;21:50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee A, Wallin S. Exploring protein-peptide binding specificity through computational peptide screening. PLoS Comput Biol. 2013;9:e1003277. doi: 10.1371/journal.pcbi.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneva I, Huang YQ, Liu ZR, Wallin S. Binding of two intrinsically disordered peptides to a multi-specific protein: a combined Monte Carlo and molecular dynamics study. PLoS Comput Biol. 2012;8:e1002682. doi: 10.1371/journal.pcbi.1002682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci USA. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Liu ZR. Anchoring intrinsically disordered proteins to multiple targets: Lessons from N-terminus of the p53 protein. Int J Mol Sci. 2011;12:1410–1430. doi: 10.3390/ijms12021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets - the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- Bertolazzi P, Bock ME, Guerra C. On the functional and structural characterization of hubs in protein-protein interaction networks. Biotechnol Adv. 2013;31:274–286. doi: 10.1016/j.biotechadv.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Bardwell JCA, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Liu ZR. Nonnative interactions in coupled folding and binding processes of intrinsically disordered proteins. PLoS One. 2010;5:e15375. doi: 10.1371/journal.pone.0015375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Pang XD, Lu C. Rate constants and mechanisms of intrinsically disordered proteins binding to structured targets. Phys Chem Chem Phys. 2012;14:10466–10476. doi: 10.1039/c2cp41196b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsath L, Dvorsky R, Fiegen D, Carlier MF, Ahmadian MR. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol Cell. 2005;20:313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Levy Y, Onuchic JN, Wolynes PG. Fly-casting in protein-DNA binding: frustration between protein folding and electrostatics facilitates target recognition. J Am Chem Soc. 2007;129:738–739. doi: 10.1021/ja065531n. [DOI] [PubMed] [Google Scholar]
- Ganguly D, Otieno S, Waddell B, Iconaru L, Kriwacki RW, Chen JH. Electrostatically accelerated coupled binding and folding of intrinsically disordered proteins. J Mol Biol. 2012;422:674–684. doi: 10.1016/j.jmb.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D, Zhang WH, Chen JH. Electrostatically accelerated encounter and folding for facile recognition of intrinsically disordered proteins. PLoS Comput Biol. 2013;9:e1003363. doi: 10.1371/journal.pcbi.1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin N, Pei JF, Lai LH. A comprehensive analysis of the influence of drug binding kinetics on drug action at molecular and systems levels. Mol Biosyst. 2013;9:1381–1389. doi: 10.1039/c3mb25471b. [DOI] [PubMed] [Google Scholar]
- Papadakos G, Housden NG, Lilly KJ, Kaminska R, Kleanthous C. Kinetic basis for the competitive recruitment of TolB by the intrinsically disordered translocation domain of Colicin E9. J Mol Biol. 2012;418:269–280. doi: 10.1016/j.jmb.2012.01.039. [DOI] [PubMed] [Google Scholar]
- Kalmar L, Acs V, Silhavy D, Tompa P. Long-range interactions in nonsense-mediated mRNA decay are mediated by intrinsically disordered protein regions. J Mol Biol. 2012;424:125–131. doi: 10.1016/j.jmb.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011;1:90. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nespoulous C, Rofidal V, Sommerer N, Hem S, Rossignol M. Phosphoproteomic analysis reveals major default phosphorylation sites outside long intrinsically disordered regions of Arabidopsis plasma membrane proteins. Proteome Sci. 2012;10:62. doi: 10.1186/1477-5956-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Cox J, Olsen J, Mann M, Frishman D. Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS Comput Biol. 2013;9:e1002842. doi: 10.1371/journal.pcbi.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Lau KY, Sevim V, Tang C. Design principles of the yeast G1/S switch. PLoS Biol. 2013;11:e1001673. doi: 10.1371/journal.pbio.1001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou L, Waddell MB, Kriwacki RW. Mechanism of cell cycle entry mediated by the intrinsically disordered protein p27Kip1. ACS Chem Biol. 2012;7:678–682. doi: 10.1021/cb200487h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HB, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007;6:1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YJK, Lobley AE, Pentony MM, Jones DT. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009;10:50. doi: 10.1186/gb-2009-10-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Gardner K, Moore DA, Erickson HP. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol. 2013;89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidovich E, Orr I, Fass D, Abdu U, Yifrach O. Intrinsic disorder in the C-terminal domain of the Shaker voltage-activated K+ channel modulates its interaction with scaffold proteins. Proc Natl Acad Sci USA. 2007;104:13022–13027. doi: 10.1073/pnas.0704059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX. The affinity-enhancing roles of flexible linkers in two-domain DNA-binding proteins. Biochemistry. 2001;40:15069–15073. doi: 10.1021/bi015795g. [DOI] [PubMed] [Google Scholar]
- van Dongen E, Evers TH, Dekkers LM, Meijer EW, Klomp LWJ, Merkx M. Variation of linker length in ratiometric fluorescent sensor proteins allows rational tuning of Zn(II) affinity in the picomolar to femtomolar range. J Am Chem Soc. 2007;129:3494–3495. doi: 10.1021/ja069105d. [DOI] [PubMed] [Google Scholar]
- Monsellier E, Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo Y, Pons M, Konrat R. Meta-structure correlation in protein space unveils different selection rules for folded and intrinsically disordered proteins. Mol Biosyst. 2012;8:411–416. doi: 10.1039/c1mb05367a. [DOI] [PubMed] [Google Scholar]
- Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Prediction of amyloidogenic and disordered regions in protein chains. PLoS Comput Biol. 2006;2:1639–1648. doi: 10.1371/journal.pcbi.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- Abeln S, Frenkel D. Disordered flanks prevent peptide aggregation. PLoS Comput Biol. 2008;4:e1000241. doi: 10.1371/journal.pcbi.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner AA, Croy CH, Vasanwala FH, Uversky VN, Van YYJ, Dunker AK. Sweeping away protein aggregation with entropic bristles: intrinsically disordered protein fusions enhance soluble expression. Biochemistry. 2012;51:7250–7262. doi: 10.1021/bi300653m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhang CS, Zhang XL, Liu ZR, Lai LH. (in press) Computational design of disordered peptides targeting TNFα.
- Cumberworth A, Lamour G, Babu MM, Gsponer J. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem J. 2013;454:361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P, Myers N, Moscovitz O, Sharon M, Prilusky J, Shaul Y. Thermo-resistant intrinsically disordered proteins are efficient 20S proteasome substrates. Mol Biosyst. 2012;8:368–373. doi: 10.1039/c1mb05283g. [DOI] [PubMed] [Google Scholar]
- Tantos A, Friedrich P, Tompa P. Cold stability of intrinsically disordered proteins. FEBS Lett. 2009;583:465–469. doi: 10.1016/j.febslet.2008.12.054. [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lin SX, Song XW, Liu J, Fu Y, Ge X, Fu XM, Chang ZY, Chen PR. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
- Lee L, Stollar E, Chang JF, Grossmann JG, O'Brien R, Ladbury J, Carpenter B, Roberts S, Luisi B. Expression of the Oct-1 transcription factor and characterization of its interactions with the Bob1 coactivator. Biochemistry. 2001;40:6580–6588. doi: 10.1021/bi010095x. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Lau KF, Dill KA. Theory for protein mutability and biogenesis. Proc Natl Acad Sci USA. 1990;87:638–642. doi: 10.1073/pnas.87.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PD, Pollock DD, Goldstein RA. Evolution of functionality in lattice proteins. J Mol Graph Model. 2001;19:150–156. doi: 10.1016/s1093-3263(00)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Helling R, Tang C, Wingreen N. Emergence of preferred structures in a simple model of protein folding. Science. 1996;273:666–669. doi: 10.1126/science.273.5275.666. [DOI] [PubMed] [Google Scholar]
- Ortiz JF, MacDonald ML, Masterson P, Uversky VN, Siltberg-Liberles J. Rapid evolutionary dynamics of structural disorder as a potential driving force for biological divergence in flaviviruses. Genome Biol Evol. 2013;5:504–513. doi: 10.1093/gbe/evt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Tohsato Y, Sugisawa H, Kohara S, Fukuchi S, Nishikawa I, Nishikawa K. Intrinsically disordered proteins in human mitochondria. Genes Cells. 2012;17:817–825. doi: 10.1111/gtc.12000. [DOI] [PubMed] [Google Scholar]
- Cheng Y, LeGall T, Oldfield CJ, Mueller JP, Van YYJ, Romero P, Cortese MS, Uversky VN, Dunker AK. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Lai LH. Towards structure-based protein drug design. Biochem Soc Trans. 2011;39:1382–1386. doi: 10.1042/BST0391382. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Suskiewicz MJ, Sussman JL, Silman I, Shaul Y. Context-dependent resistance to proteolysis of intrinsically disordered proteins. Protein Sci. 2011;20:1285–1297. doi: 10.1002/pro.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- Chan HS, Zhang ZQ, Wallin S, Liu ZR. Cooperativity, local-nonlocal coupling, and nonnative interactions: Principles of protein folding from coarse-grained models. Annu Rev Phys Chem. 2011;62:301–326. doi: 10.1146/annurev-physchem-032210-103405. [DOI] [PubMed] [Google Scholar]


