Abstract
Deregulation of signaling pathways controlled by protein phosphorylation underlies the pathogenesis of hematological malignancies; however, the extent to which deregulated phosphorylation may be involved in B-cell non-Hodgkin lymphoma (B-NHL) pathogenesis is largely unknown. To identify phosphorylation events important in B-NHLs, we performed mass spectrometry–based, label-free, semiquantitative phosphoproteomic profiling of 11 cell lines derived from three B-NHL categories: Burkitt lymphoma, follicular lymphoma, and mantle-cell lymphoma. In all, 6579 unique phosphopeptides, corresponding to 1701 unique phosphorylated proteins, were identified and quantified. The data are available via ProteomeXchange with identifier PXD000658. Hierarchical clustering highlighted distinct phosphoproteomic signatures associated with each lymphoma subtype. Interestingly, germinal center–derived B-NHL cell lines were characterized by phosphorylation of proteins involved in the B-cell receptor signaling. Of these proteins, phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (PAG1) was identified with the most phosphorylated tyrosine peptides in Burkitt lymphoma and follicular lymphoma. PAG1 knockdown resulted in perturbation of the tyrosine phosphosignature of B-cell receptor signaling components. Significantly, PAG1 knockdown increased cell proliferation and response to antigen stimulation of these germinal center–derived B-NHLs. These data provide a detailed annotation of phosphorylated proteins in human lymphoid cancer. Overall, our study revealed the utility of unbiased phosphoproteome interrogation in characterizing signaling networks that may provide insights into pathogenesis mechanisms in B-cell lymphomas.
Protein phosphorylation is a reversible post-translational modification that modulates stability, enzyme activity, and protein-protein interaction, as well as mediates signal transduction in response to extracellular and intracellular stimuli.1,2 Abnormal protein phosphorylation has been implicated in a wide range of diseases, especially in cancer.3,4 Indeed, recognition and detailed understanding of deregulated phosphorylation-mediated signal transduction pathways have led to the development of targeted therapies with proven utility in the treatment of cancer.5–7
B-cell non-Hodgkin lymphomas (B-NHLs) encompass >30 different types of clonal B-cell proliferations.8 With the exception of a few B-NHLs, in which abnormal kinase activities have been described, such as Janus-activating kinase 2 overexpression in primary mediastinal B-cell lymphoma,9 clathrin-anaplastic lymphoma kinase (ALK) fusion protein in ALK-positive diffuse large B-cell lymphoma,10 or Zap-70 aberrant expression in chronic lymphoid leukemia/small lymphocytic lymphoma,11 the extent to which deregulated phosphorylation may be involved in their pathophysiological characteristics is largely unknown.
Mass spectrometry (MS) instrumentation has become sufficiently sensitive and robust to become an ideal tool for proteomic studies because thousands of proteins can be confidently identified in a single experiment within a relatively short period of time. Tandem mass spectrometry (MS/MS) allows the definitive identification of protein sequences and sites of phosphorylation.12 Such information can be subsequently used to highlight activation status of intracellular signaling pathways. An enrichment of the phosphoproteome is required before conducting MS/MS, mostly because of the low stoichiometry of phosphoproteins. Several methods for enriching proteins/peptides harboring a phosphorylated residue are well described, such as metal oxide affinity chromatography (MOAC).13 Because protein phosphorylation does not occur evenly between serine, threonine, and tyrosine residues (approximate relative ratio of 90, 10, and 0.05, respectively), the exploration of tyrosine phosphorylation requires an additional enrichment through immunopurification with high-affinity anti-phosphotyrosine antibodies (pY-IP).14
In this study, we used a combination of MOAC and pY-IP to perform a shotgun phosphoproteomic analysis of 11 cell lines derived from three B-NHL subtypes: Burkitt lymphoma (BL) and follicular lymphoma (FL), which are derived from germinal center B cells, and mantle-cell lymphoma (MCL), which are derived from naïve B cells. We used a label-free spectral counting method to semiquantify the relative abundance of the phosphorylated peptides/proteins.15 Our phosphoproteomic profiling highlighted the existence of phosphoprotein signatures characteristic of each NHL subtype. To our knowledge, this study provides the first detailed analysis of phosphoproteins with phosphorylation site mapping in human B-cell lymphoma. Furthermore, these results provide a novel perspective on signaling pathways in B-NHLs and emphasize the phosphorylation of the B-cell receptor (BCR) signaling pathway in those arising from the germinal center–derived NHLs.
Materials and Methods
Cell Lines
A total of 11 cell lines were used in this study: three BL cell lines, four transformed FL cell lines, and four MCL cell lines (Supplemental Table S1).
Protein Extraction and Digestion for Phosphoproteomic Analysis
Cells were lysed in buffer containing 9 mol/L urea/20 mmol/L HEPES, pH 8.0/0.1% SDS, and a cocktail of phosphatase inhibitors. Protein (6 mg) was reduced with 4.5 mmol/L dithiothreitol and alkylated with 10 mmol/L iodoacetamide, then digested with trypsin overnight at 37°C using an enzyme/protein ratio of 1:50 (w/w). Samples were desalted on a C18 cartridge (Sep-Pak plus C18 cartridge; Waters, Milford, MA). Each sample was prepared in triplicate.
Phosphopeptide Enrichment
MOAC was performed to enrich phosphorylated peptides and reduce the sample complexity before tyrosine-phosphorylated peptide immunopurification (pY-IP). We used titanium dioxide (TiO2) microparticles (Titansphere Phos-TiO; GL Sciences Inc, Torrance, CA). Briefly, TiO2 microparticles were conditioned with buffer A (80% acetonitrile/0.4% trifluoroacetic acid), then equilibrated with buffer B (75% buffer A/25% lactic acid). Peptides were loaded twice on TiO2 microparticles and washed two times with buffer B and three times with buffer A. Hydrophilic phosphopeptides were eluted with 5% ammonium hydroxide solution, and hydrophobic phosphopeptides were eluted with 5% pyrrolidine solution. The equivalent of 5 mg of protein was further enriched for phosphorylated tyrosine peptides by overnight immunoprecipitation (pY-IP) using a cocktail of anti-phosphotyrosine antibodies [4G10 (Millipore, Billerica, MA); PT-66 (Sigma-Aldrich, St. Louis, MO); p-Tyr-100 (Cell Signaling Technology, Beverly, MA)].
MS Analysis
Ammonium hydroxide and pyrrolidine eluents were dried (SpeedVac, ThermoFisher, Pittsburgh, PA) and reconstituted in 25 μL sample loading buffer (0.1% trifluoroacetic acid/2% acetonitrile). Eluent from phosphotyrosine immunoprecipitation was dried and reconstituted in 35 μL of the loading buffer. A linear trap quadrupole Orbitrap XL (ThermoFisher, Pittsburgh, PA) in line with a high-performance liquid chromatogram Paradigm MS2 (Michrom BioResources, Auburn, CA) was used for acquiring high-resolution MS and MS/MS data. Phospho-enriched peptides were loaded onto a sample trap (Captrap; Bruker-Michrom, Auburn, CA) in line with a nanocapillary column (Picofrit; 75-μm internal diameter × 15-μm tip; New Objective, Woburn, MA) packed in-house with 10 cm of MAGIC AQ C18 reversed-phase material (Michrom BioResources). Two different gradient programs, one each for MOAC and phosphotyrosine immunoprecipitation samples, were used for peptide elution. For MOAC samples, a gradient of 5% to 40% buffer B (95% acetonitrile/1% acetic acid) in a 135- and 5-minute wash with 100% buffer B, followed by 30 minutes of re-equilibration with buffer A (2% acetonitrile/1% acetic acid), was used. For phosphotyrosine immunoprecipitation samples, which were a much less complex mixture of peptides, 5% to 40% gradient with buffer B was achieved in 75 minutes, followed by a 5-minute wash with buffer B and a 30-minute re-equilibration. The flow rate was approximately 0.3 μL/minute. Peptides were directly introduced into the mass spectrometer using a nanospray source. Orbitrap was set to collect one MS scan between 400 and 2000 m/z (resolution of 30,000 at 400 m/z) in an orbitrap, followed by data-dependent collision-induced dissociation spectra on top nine ions in trap quadrupole (normalized collision energy, approximately 35%). Dynamic exclusion was set to two MS/MS acquisitions, followed by exclusion of the same precursor ion for 2 minutes. Maximum ion injection times were set to 300 milliseconds for MS and 100 milliseconds for MS/MS. Automatic gain control was set to 1 × e6 for MS and 5000 for MS/MS. Charge state screening was enabled to discard +1 and unassigned charge states. Technical duplicate data for each of the MOAC elutions (ammonium hydroxide and pyrrolidine) and triplicate data for the phosphotyrosine immunoprecipitation samples were acquired.
Bioinformatics Analysis
RAW files were converted to mzXML using msconvert16 and searched against the Swissprot Human protein database (http://www.uniprot.org) appended with common proteomics contaminants and reverse sequences as decoys. Searches were performed with X!Tandem version 2010.10.01.1 (http://www.thegpm.org/TANDEM) using the k-score plug-in.17,18 For all searches, the following search parameters were used: parent monoisotopic mass error of 50 ppm and fragment ion error of 0.8 Da, allowing for up to two missed tryptic cleavages. Variable modifications were oxidation of methionine, carbamidomethylation of cysteine, and phosphorylation of serine, threonine, and tyrosine. The search results were then post-processed using PeptideProphet (http://peptideprophet.sourceforge.net) and ProteinProphet (http://proteinprophet.sourceforge.net).19–21
Spectral counts were obtained for each cell line using ABACUS.22 Tyrosine-enrichment data were processed through ABACUS separately from the serine and threonine data. ABACUS results were filtered to only retain proteins with a ProteinProphet probability of >0.7. Only phosphorylated peptides with a probability of >0.8 were considered for spectral counting. These parameters resulted in a protein false-discovery rate (FDR) of 0.1 for the serine and threonine data. For the tyrosine enrichment, these ABACUS parameters resulted in a protein FDR of 0.0045. This ABACUS output was used for all subsequent analyses to quantify the relative abundance of phosphorylated peptides/proteins.
Phospho-site localization was performed with an in-house reimplementation of the Ascore algorithm, as described by Beausoleil et al.23 Ascore values represent the probability of detection as the result of chance, with scores >19 corresponding to sites localized with >99% certainty. The classification of general motif classes or sequence categories was identified using a binary decision tree described by Villen et al.24 Briefly, the 13-mer sequences were assigned as follows: proline at +1 (proline-directed phosphopeptide), 5 or more glutamic acid/aspartic acid at +1 to +6 (acidic phosphopeptide), arginine/lysine at −3 (basic phosphopeptide), aspartic acid/glutamic acid at +1/+2 or +3 (acidic phosphopeptide), and ≥2 arginine/lysine at −6 to −1 (basic phosphopeptide). Recurrent phosphorylation site motifs were extracted for three different B-NHL subgroups with the Motif-X algorithm.25 Sequences used for Motif-X were constructed such that the phosphorylated residue was flanked on either side by five amino acid residues. The minimum motif occurrence for Motif-X was set to 20, and only motifs with P < 10−5 were considered.
Hierarchical clustering of the spectral count data were performed separately for each enrichment type (tyrosine versus serine/threonine). Apart from this, the procedure was performed in the same manner on both data sets, as follows. Only proteins with a total spectral count higher than nine in at least one of the cell lines used in this study were retained. After filtering, all protein spectral counts were normalized first to the total spectral counts within each cell line, and then to the total across all cell lines. These normalized values were log2 transformed and underwent hierarchical clustering with a multiscale bootstrap resampling for 10,000 iterations using the pvclust package in R (http://www.r-project.org).26 The resulting dendrogram was then used to represent the clustered log2 normalized data in a heat map.
Immunoprecipitation and Western Blot Analysis
The primary antibodies were anti-phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (PAG1) antibody (Abcam, Cambridge, MA), anti-LYN antibody (Abcam), and anti-phosphotyrosine antibodies (4G10 and PT-66 and p-Tyr-100). For immunoprecipitation, 4 mg of protein lysates was precleared, then incubated overnight at 4°C with 5 μg of antibody previously cross-linked to protein A/G plus agarose.
PAG1 Knockdown
PAG1 was knocked down in the BJAB cell line using the GIPZ lentiviral shRNAmir clone, V2LHS_175609, from Open Biosystems (ThermoFisher). A scrambled GIPZ lentiviral shRNA was used as control. Lentiviral particles were produced by the HEK-293-FT cell line after a FuGene cotransduction of a packaging mix and the lentiviral shRNA. Lentiviral infections of the BJAB cell line were performed in media containing polybrene and selected with 1 μg/mL of puromycin. PAG1 knockdown was assessed by Western blot analysis, performed in triplicate.
In Vitro Proliferation Assay
Cell lines were seeded at a density of 50,000 cells/mL and grown for 48 hours, with or without stimulation by 50 μg/mL anti-IgM antibody (Invitrogen, Grand Island, NY) or 50 μg/mL lipopolysaccharide (Sigma-Aldrich). After 2 hours of incubation with the WST-1 reagent (Roche Diagnostics, Indianapolis, IN), the absorbance was read at 540 nm. Each assay was performed in triplicate.
Colony Formation Assay
BJAB cells, transformed with scramble shRNA or PAG1 shRNA, were incubated for 14 days in a methylcellulose-based media (MethoCult; Stem Cell Technology, Vancouver, BC, Canada). Colonies were stained with iodonitrotetrazolium chloride overnight and then counted under a microscope. Each assay was performed in triplicate.
Results
Identification and Quantification of Phosphorylated Proteins in B-NHLs
To evaluate global phosphoproteomic signatures of B-NHLs, we performed phosphoproteomic profiling of 11 human NHL-derived cell lines of three distinct lymphoma subtypes. To maximize identification of phosphoproteins, we used a two-step approach with an initial MOAC to enrich for peptides carrying phosphorylated serine (p-Ser), threonine (p-Thr), or tyrosine (p-Tyr) residues, followed by a subsequent pY-IP to characterize phosphorylation events occurring at tyrosine residues. Three independent phosphopeptide enrichments were performed for each cell line, and high mass accuracy MS was performed in technical replicates. An experimental design is schematically represented in Supplemental Figure S1. The phosphopeptide enrichment efficiency was computed as the fraction of the peptides containing a phosphorylated residue among the total number of peptides. The mean efficiency of p-Ser/p-Thr peptide enrichment with MOAC was 83.4% (SD, 6.5%), and the mean efficiency of p-Tyr peptide enrichment in pY-IP was 54% (SD, 14.7%). The reproducibility between the biological triplicates ranged from 0.65 to 0.85 (Supplemental Table S2).
In total, 868,578 MS/MS spectra were assigned to 6640 unique phosphorylated peptides. The data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (PRoteomics IDEntifications database) partner repository27 with the data set identifier PXD000658. MS/MS analysis of peptides obtained from the MOAC enrichment led to identification of p-Ser peptides (4752 unique peptides) and/or p-Thr peptides (1292 unique peptides) and p-Tyr peptides (233 unique peptides) with a peptide FDR at 0.032 (Supplemental Figure S2A). MS/MS analysis of peptides from the pY-IP led to identification of 275 unique phosphopeptides composed of 73 p-Ser unique peptides, 49 p-Thr peptides, and 184 p-Tyr peptides with a peptide FDR at 0.045. Combining the data from a sequential enrichment strategy allowed us to considerably improve the identification of p-Tyr proteins from 204 to 388 peptides (Figure 1, A and B); thus, 47.4% of p-Tyr peptides were identified only in pY-IP.
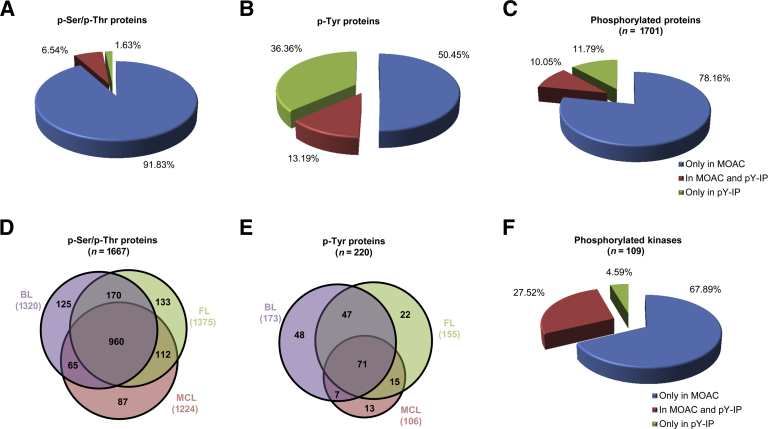
Figure 1.
Overview of identification and quantification results of triplicate phosphoproteome enrichment. A: Distribution of the proteins identified with a p-Ser and/or p-Thr residue between the MOAC enrichments and pY-IP enrichments. Most of the p-Ser/p-Thr proteins were identified in MOAC enrichments. B: Distribution of the proteins identified with a p-Tyr residue between the MOAC enrichments and pY-IP enrichments. The sequential enrichment strategy improved considerably the identification of p-Tyr proteins. C: A total of 1701 phosphorylated proteins were identified only in MOAC enrichment, only in pY-IP, or in both types of enrichment. D: Of the identified phosphoproteins, 109 kinases were found only in MOAC enrichment, only in pY-IP, or in both types of enrichment. E: Venn diagram representing the repartition of proteins identified with p-Ser/p-Thr residues in the three different types of B-NHLs. Of 1667 p-Ser/p-Thr proteins, 960 (57.58%) were common to the three B-NHL types. F: Venn diagram representing the repartition of proteins identified with p-Tyr residues in the three different types of B-NHLs.
A total of 1701 unique phosphoproteins (Figure 1C) were identified, of which 1667 were identified with a p-Ser or a p-Thr residue (Figure 1D) and 220 were identified with a p-Tyr residue (Figure 1E). Of note, 186 proteins were identified with p-Ser/p-Thr and p-Tyr. The complete list of identified proteins is presented in Supplemental Table S3 (MOAC) and Supplemental Table S4 (pY-IP), respectively, and the complete list of identified peptides is presented in Supplemental Table S5 (MOAC) and Supplemental Table S6 (pY-IP), respectively. The total number of unique proteins identified with a p-Ser or p-Thr residue in BL, FL, and MCL cell lines was 1320, 1375, and 1224, respectively. A total of 220 unique proteins were phosphorylated at a tyrosine residue (Figure 1E). The total number of unique proteins identified with a p-Tyr residue in BL, FL, and MCL cell lines was 173, 155, and 106, respectively. There was no significant difference in proportion of p-Ser/p-Thr or p-Tyr phosphoproteins between the three subtypes, BL, FL, and MCL.
Of all identified phosphorylated proteins, 109 were protein kinases (Figure 1F). Seventy-four kinases were phosphorylated only at serine or threonine residues, whereas 30 kinases were phosphorylated at serine, threonine, and tyrosine residues, such as tyrosine-protein kinase Lck, tyrosine-protein kinase Lyn, mitogen-activated protein kinase 3, proto-oncogene tyrosine-protein kinase Src, tyrosine-protein kinase SYK, tyrosine-protein kinase Tec. Five kinases, hepatocyte growth factor receptor, non-receptor tyrosine-protein kinase TYK2, Burton tyrosine kinase, ephrin type A receptor 7, and cyclin-dependent kinase 11A, were phosphorylated only at a tyrosine residue [Supplemental Table S7 (MOAC) and Supplemental Table S8 (pY-IP)]. Of the 90 tyrosine kinases, 14 were found to be phosphorylated: BTK, TYK2, EPHA7, MET, Janus-activating kinase 3, abelson tyrosine-protein kinase 2, protein-tyrosine kinase 2-beta, activated CDC42 kinase 1, SYK, TEC, LYN, SRC, LCK, and abelson tyrosine-protein kinase 1.
Principal Characteristics of Phosphopeptides in B-NHLs
To correctly localize the phosphorylation site within a peptide, we used the Ascore algorithm (Supplemental Tables S5 and S6).23 Of the identified phosphopeptides, 1863 achieved a near certainty (>99%) of localization. Because protein phosphorylation is known to frequently be a step-wise process, where the first event may prime subsequent successive events, we examined the level of phosphorylation of each phosphopeptide, acknowledging the fact that our bottom-up approach is well suited to investigate only phosphorylation events occurring in close linear proximity. In our study, 87.37% of the phosphopeptides identified were found to be singly phosphorylated. Most of the multiply phosphorylated peptides were doubly phosphorylated (Supplemental Figure S2B).
To better understand the kinases involved in the phosphorylation events identified, we next considered the amino acid motifs surrounding the phosphorylated residue.24 Proline-directed sites were most common (39.75%) (Supplemental Figure S2C). When analyzed separately, no significant difference was found between the three NHL subtypes (Supplemental Figure S2C). To further analyze the phosphopeptides, we examined enriched phosphorylation site motifs. Peptide sequences for phosphorylation sites localized with >99% confidence were all aligned, and their length was adjusted to ±6 amino acids from the central position and submitted to the Motif-X algorithm. The results are displayed in Supplemental Figure S3. The five most abundantly enriched p-Ser motifs in each B-NHL subgroup were mainly characterized by the presence of amino acids with negatively charged side groups, such as glutamic acid (E) or aspartic acid (D), in the proximity of a serine residue. The more abundantly enriched p-Thr motifs in each B-NHL subgroup were mainly characterized by the presence of a proline residue. No recurrent p-Tyr motif was found in MCL, whereas BL and FL shared the same EXXpY motif (where X denotes any amino acid).
Distinctive Phosphoproteomic Signatures Discriminate B-NHL Subtypes
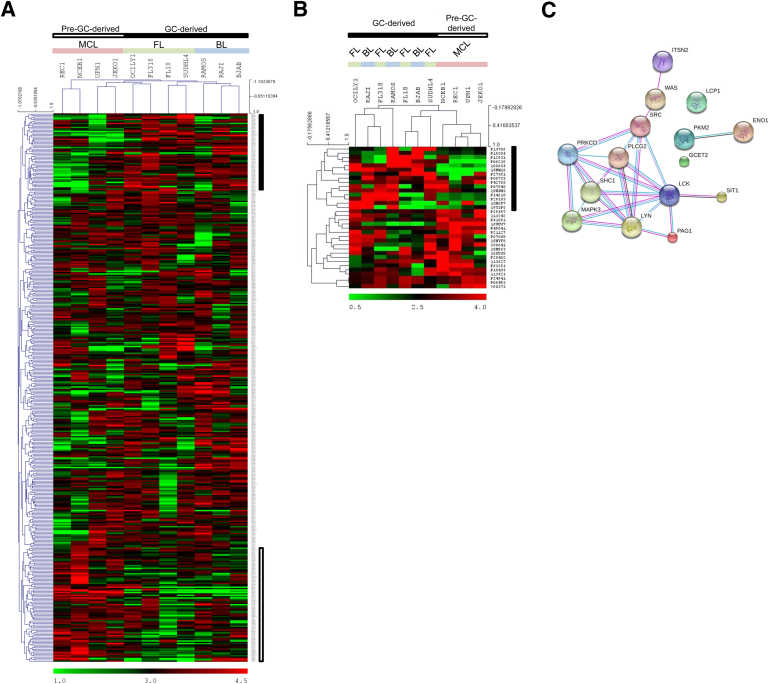
To further define phosphoproteomic signatures for each B-NHL entity, we performed hierarchical clustering using 408 p-Ser/p-Thr phosphoproteins that were selected on the basis of the following criteria: proteins with a total spectral count higher than nine in at least one of the cell lines in the category. As shown in Figure 2A, all germinal center–derived NHLs (BL and FL) are shown in the right half of the heat map, distinct from the pre–GC-derived MCL cell lines. The three BL cell lines (BJAB, Raji, and Ramos) clustered together, whereas the FL cell lines (FL-18, FL-318, OCI-LY-1, and SUD-HL-4) grouped into two clusters. A p-Ser/p-Thr signature of germinal center–derived B-NHLs is highlighted by the vertical black bar on the right side of the heat map, which included transcription factors (eg, TFEB, FOXK1, and NFATC2) that participate in GC B-cell survival,28 and MEF2C, which regulates GC B cells.29 The MCL cell lines NCEB-1 and REC-1 clustered apart from BL and FL cell lines, whereas the two other MCL cell lines (UPN-1 and Jeko-1) clustered with the germinal center–derived B-NHL cell lines. A phospho-signature of pre–germinal center–derived B-NHLs is highlighted by the vertical open bar on the right lower side of the heat map, including DEK, which regulates RNA metabolism, SMAD nuclear-interacting protein 1 (SNIP1), which is involved in cyclin D1 mRNA stability,30 and apoptosis-antagonizing transcription factor (AATF), which is hyperphosphorylated during G1/S phase transition.31
Figure 2.
Two-way hierarchical clustering of phosphorylated proteins. A: Hierarchical clustering of B-NHL cell lines on the basis of p-Ser/p-Thr proteins: the vertical black bar highlights differentially phosphorylated proteins in the GC-derived NHLs, whereas the vertical open bar highlights differentially phosphorylated proteins in the non–GC-derived NHLs. B: Hierarchical clustering of B-NHL cell lines on the basis of p-Tyr proteins: the vertical black bar highlights differentially phosphorylated proteins in the GC-derived NHLs. C: Evidence view of the STRING diagram obtained with proteins highlighted by the vertical black bar in B. Different line colors represent the types of evidence for the association: turquoise, databases; pink, experiments; black, co-experiments; and purple, homology.
Hierarchical clustering analysis of the p-Tyr phosphoproteome using 34 proteins identified by our analysis (Figure 2B) also separated the pre–GC-derived MCL cell lines from the germinal center–derived cell lines. A p-Tyr signature of the GC-derived B-NHLs is highlighted by the vertical black bar (Figure 2B) and was composed of 15 proteins involved in signaling pathways, particularly the BCR signaling. These proteins were analyzed by using the Search Tool for Retrieval of Interacting Genes/Proteins (STRING version 9.05; http://string-db.org)32 at a high confidence score (0.700). The p-Tyr proteins expressed in GC-derived NHLs demonstrate a relatively cohesive group of functionally related proteins (Figure 2C).
BCR Signaling Pathway Is Phosphorylated in GC-Derived B-NHLs
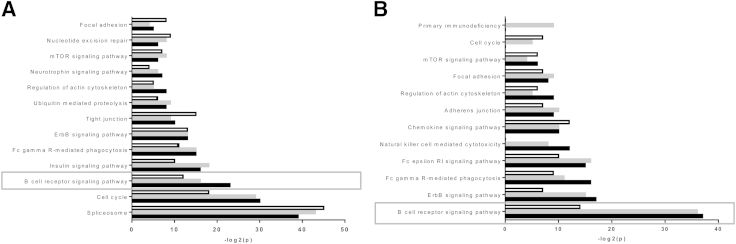
To investigate phosphorylated signaling pathways in the three B-NHL subtypes, we interrogated our data set using the functional annotation tool of DAVID software version 6.7 (http://david.abcc.ncifcrf.gov/summary.jsp).33,34 The logarithm-transformed P values of the top 10 Kyoto Encyclopedia of Genes and Genomes pathways identified in BL, FL, or MCL are plotted in Figure 3. Pathways identified with p-Ser/p-Thr proteins in BL, FL, and MCL indicated that the three B-NHLs shared common phosphorylation of specific pathways, such as the spliceosome, ErbB signaling pathway, ubiquitin-mediated proteolysis, and the mammalian target of rapamycin pathway (Figure 3A). This analysis also underlined the differential phosphorylation of specific pathways, such as the BCR signaling pathway and the insulin signaling pathway in BL and FL cell lines compared with the MCL cell lines. Analysis of p-Tyr proteins similarly highlighted the significant enrichment of proteins involved in the BCR signaling pathway in BL and FL cell lines (Figure 3B).
Figure 3.
Pathways differentially phosphorylated between BL (black bars), FL (gray bars), and MCL (white bars). Kyoto Encyclopedia of Genes and Genomes pathways identified with proteins phosphorylated at a serine or threonine residue (A) and at a tyrosine residue (B). The significance of the identified pathway is represented as −log10(p) for each NHL subgroup.
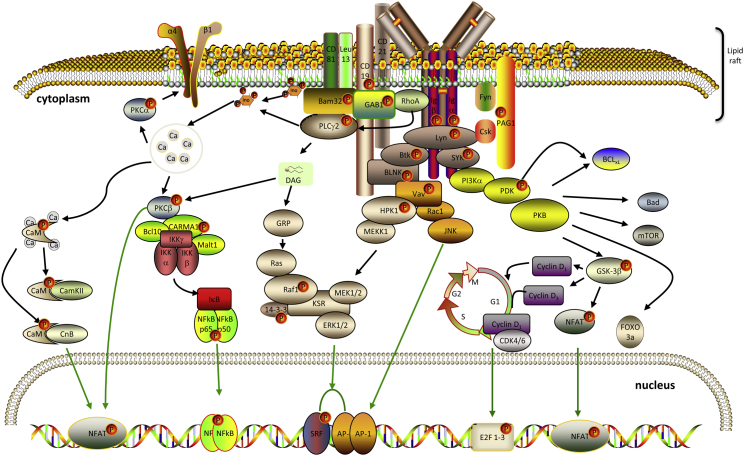
Figure 4 shows a simplified network of proteins involved in the BCR signaling pathway. Several of these proteins (BTK, SYK, CD19, CD22, LCK, LYN, and PAG1) were identified with p-Tyr either only in BL and FL cell lines or with a higher spectral count in BL and FL cell lines compared with MCL cell lines. By using the Motif-X algorithm, we defined EXXpY as the most predominant motif found in the subset of BL and FL phosphorylation sites (Supplemental Figure S4). Of the 26 proteins identified with this EXXpY motif, eight are involved in the BCR signaling pathway (Supplemental Table S9). Interestingly, the PAG1 was identified with the most spectral counts in BL and FL (Supplemental Figure S5). This observation and the implication of PAG1 as a regulator of the BCR signaling pathway35,36 led to subsequent investigation of PAG1 function in GC-derived NHLs.
Figure 4.
Schematic representation of proteins involved in the BCR signaling pathways. A simplified network of proteins involved in the BCR signaling pathway. Several of the proteins (BTK, B-cell linker protein, PAG1, SYK, CD19, CD22, LCK, LYN, PLCγ, and caspase recruitment domain-containing protein 11) have been identified with p-Tyr either only in BL and FL cell lines or with a higher spectral count in BL and FL cell lines compared with MCL cell lines. The p-Tyr proteins are displayed with a small P symbol in a red circle. The design of this figure was aided by materials from ScienceSlides (VisiScience Inc, Chapel Hill, NC).
PAG1 Is Overexpressed and Phosphorylated in GC-Derived NHLs
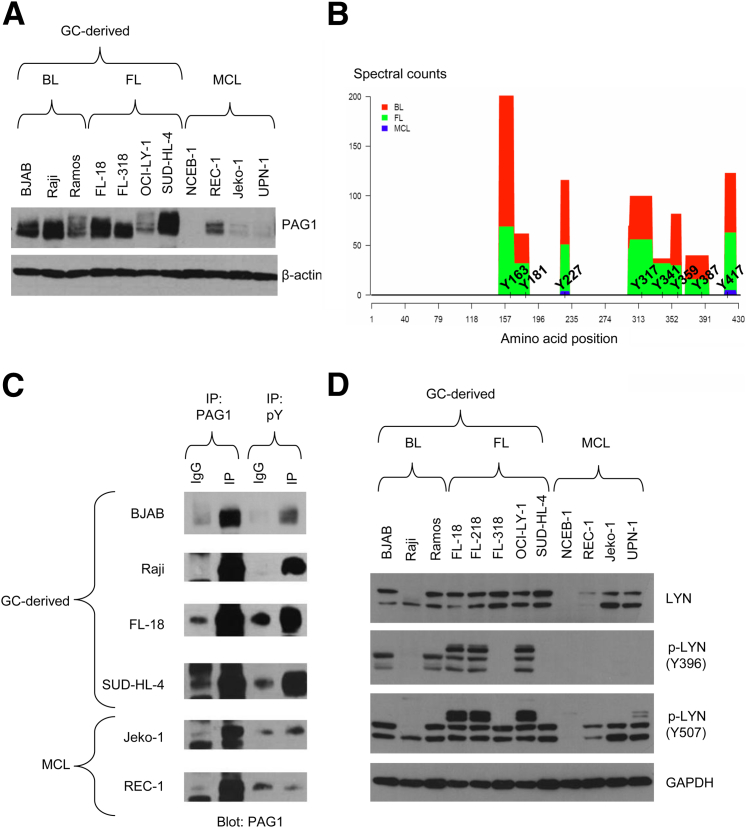
To validate the phosphoproteomic data, we investigated the expression and phosphorylation status of proteins involved in the BCR signaling pathway by Western blot analysis. As shown in Figure 5A, PAG1 is strongly expressed in BL and FL cell lines. In contrast, PAG1 expression is not detectable or weakly expressed in MCL cell lines. By MS, we found that 8 of 10 tyrosine residues, contained in the PAG1 amino acid sequence, were highly phosphorylated in BL and FL, but not in MCL (Figure 5B). To confirm the phosphorylation status of PAG1, we performed immunoprecipitation using either PAG1 or a cocktail of anti-phosphotyrosine antibodies, followed by Western blot analysis for PAG1. These experiments revealed significant enrichment of tyrosine-phosphorylated PAG1 in the four cell lines from GC-derived B-NHLs. Conversely, tyrosine-phosphorylated PAG1 was absent or weakly detectable in MCL cell lines (Figure 5C). These two observations validated the MS results and demonstrated differential expression of tyrosine-phosphorylated PAG1 in the GC-derived lymphoma cell lines compared with the MCL cell lines.
Figure 5.
PAG1 and LYN are overexpressed and phosphorylated in germinal center–derived cell lines. A: PAG1 expression in the 11 cell lines. B: Schematic representation of PAG1 tyrosine residue phosphorylation in different types of B-NHLs. The histogram widths represent the span of phosphopeptides containing a p-Tyr, whereas the histogram heights represent the abundance of the phosphorylation (spectral count). C: Western blot analysis of the immunoprecipitations done with the anti-PAG1 antibody (IP PAG1) or with the cocktail of the three anti-phosphotyrosine antibodies (IP p-Tyr). D: Western blot analysis of LYN expression and its phosphorylation at Y396 (induces the enzymatic activity) and at Y507 (inhibits the enzymatic activity) of cell lines representative of BL, FL, and MCL.
Similarly, we demonstrated that, although LYN is expressed in both GC-derived B-NHLs and MCL, the profile of LYN phosphorylation is significantly different between the GC-derived NHLs and MCL (Figure 5D). Indeed, the balance between the phosphorylation of tyrosine 396, which induces LYN activity, and tyrosine 507, which inhibits LYN activity, is clearly in favor of LYN inactivation in MCL cell lines, but not in GC-derived cell lines.
PAG1 Knockdown Promotes BL Cell Line Proliferation
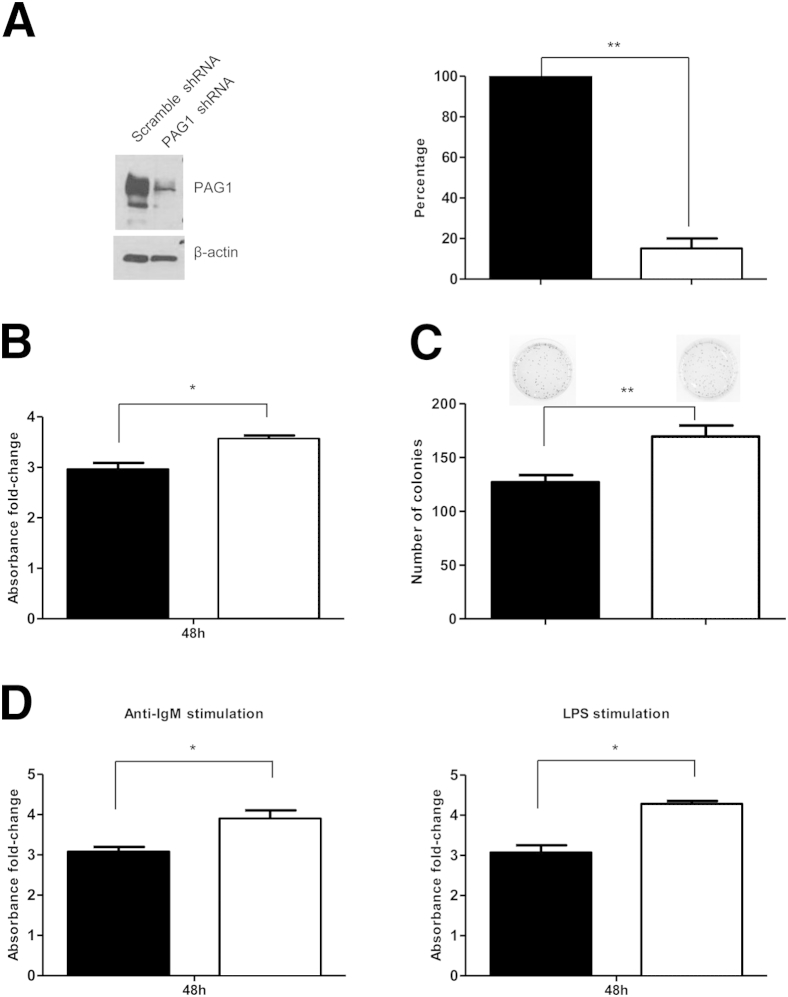
To investigate a possible functional role for PAG1 in GC-derived lymphoma, we performed PAG1 depletion by siRNA-mediated knockdown in a BL-derived cell line, BJAB. Our lentiviral shRNA-mediated PAG1 silencing achieved 85% knockdown of PAG1 protein expression, as demonstrated by Western blot analysis (Figure 6A). PAG1 knockdown in BJAB cells resulted in a significant increase in cell proliferation (Figure 6B). Indeed, after 48 hours, we observed a 1.2-fold increase in proliferation of BJAB cells with PAG1 shRNA (P < 0.05). Moreover, colony formation assays to evaluate the effect of PAG1 knockdown showed significantly more colonies after 14 days in PAG1 knockdown cells (127 colonies for BJAB with scramble shRNA versus 169 colonies for BJAB with PAG1 shRNA; P < 0.01) (Figure 6C).
Figure 6.
PAG1 knockdown results in increased BJAB proliferation and responsiveness to antigen stimulation. BJAB with scramble shRNA, black bars; BJAB with PAG1 shRNA, white bars. A: Western blot analysis control and its histogram of PAG1 knockdown in BJAB cell lines. B: The WST-1 assay demonstrates a significant increase of BJAB proliferation after 48 hours when PAG1 is stably knocked down. C: This result was confirmed by the increase of the number of colonies obtained after 14 days of culture in methylcellulose media. D: Antigenic stimulations of BJAB cells result in a significant increase of proliferation when PAG1 is stably knocked down. ∗P < 0.05, ∗∗P < 0.01. LPS, lipopolysaccharide.
Because PAG1 is involved in the BCR signaling pathway, we evaluated the ability of cells to respond to antigen stimulation when PAG1 was silenced. We demonstrated that PAG1 knockdown significantly increased BJAB response to two different antigen stimulations after 48 hours (Figure 6D). Cell stimulation with an anti-IgM resulted in a 1.3-fold increase of BJAB proliferation when PAG1 was silenced (P < 0.05). Similarly, lipopolysaccharide stimulation resulted in a 1.4-fold increase of BJAB proliferation when PAG1 was silenced (P < 0.05). These results highlighted a possible role for PAG1 in cell response to antigen stimulation.
To investigate the consequences on protein phosphorylation when PAG1 is silenced in BJAB cells, we performed the phosphopeptide enrichments of BJAB with the scramble shRNA and BJAB with PAG1 shRNA in triplicate. Depletion of PAG1 in BJAB cells resulted in the alteration of tyrosine phosphorylation of proteins belonging to either different pathways (redundant) or functionally connected pathways (Supplemental Figure S6A). The most significantly affected pathway was the BCR pathway, with changes in the phosphorylation status of proteins, such as LCK, BTK, SYK, CD19, CD79α/β, and LYN. The STRING diagram displayed the connections between proteins identified with at least a 1.5-fold differential phosphorylation at serine/threonine and/or tyrosine residues (Supplemental Figure S6B). The BCR signaling pathway was one of the three functionally annotated pathways identified, strongly supporting a role for PAG1 in the regulation of BCR signaling.
Discussion
Phosphoproteomic approaches have already demonstrated their ability to discriminate cell lines derived from acute myeloid leukemia, multiple myeloma, and diffuse large B-cell lymphoma.37 In this study, we have used an unbiased approach to analyze the global phosphoproteomic signatures in three different B-cell NHL entities. The unbiased approach permitted identification, semiquantification, and localization of phosphorylated amino acid residues in a sensitive and reproducible manner that facilitates detection of constitutively phosphorylated signaling pathways.
The assembled data set of MOAC and pY-IP enrichments catalogued 6640 phosphorylated peptides corresponding to 1701 phosphorylated proteins. Bioinformatic analyses revealed the existence of specific phosphoproteomic signatures for each lymphoma entity. Specifically, hierarchical clustering analysis revealed distinctive signatures for the MCL and germinal center–derived lymphomas (BL and FL), which correlate with the respective putative cells of origin from which these NHLs are thought to arise. Indeed, MCLs are thought to arise from mature naïve (pregerminal) center B cells, whereas BL and FL putatively arise from germinal center B cells.38,39
The genetic hallmark of MCL is the t(11;14)(q13;q32), resulting in deregulated overexpression of cyclin D1, which controls the cell cycle G1/S phase transition. In phosphoproteomic signatures we identified, SNIP1 was found to be highly phosphorylated in MCL cell lines when compared with FL and BL cell lines. SNIP1 was initially hypothesized to regulate the cyclin D1 promoter, but recent studies revealed that SNP1 affects post-spliced cyclin D1 mRNA, promoting nascent mRNA stability through the SNIP1/SkIP-associated RNA-processing complex, which associates with the 3′-end of the cyclin D1 mRNA.30 A longer half-life of cyclin D1 mRNA contributes to the MCL aggressiveness.40 In addition, the AATF was also found to be phosphorylated in MCL. AATF, inhibitor of the histone deacetylase HDAC1, is hyperphosphorylated during the G1/S phase transition, which leads to the activation of E2F target genes and cell cycle progression.31
Although all MCL cell lines shared the same phosphorylated protein signature and clustered together when p-Tyr proteins were analyzed, two of them (UPN-1 and Jeko-1) were clustered with FL/BL cell lines when p-Ser/p-Thr proteins were analyzed. Jeko-1 harbors a complex karyotype with an amplification at 8q24.21, which includes the MYC gene,41 and UPN-1 harbors a cryptic t(8;14)(q24;q32) and an amplification of the BCL2 gene.42 These additional genetic alterations, which are some of the BL or FL characteristics, might be responsible for their behavior in between MCL and FL/BL, even if we were not able to clearly identify p-Ser/p-Thr proteins, which would be directly correlated to BCL2 or MYC.
Our study revealed phosphosignatures implicating BCR signaling in the GC-derived BL and FL when compared with MCL. Indeed, pathway analysis performed with differentially expressed phosphorylated proteins between these two groups of B-NHLs highlighted the BCR signaling pathway as the highest ranked in germinal center–derived NHL. A role for tonic BCR signaling has already been implicated in another B-NHL known as the activated B-cell–like subtype of diffuse large B-cell lymphoma.43 In our study, we identified 14 proteins with phosphorylated tyrosine residues that participate in the BCR signaling pathway. These included three tyrosine kinases (LYN, SYK, and BTK) that could be therapeutic candidate targets for BL and FL. Indeed, preclinical studies of tyrosine kinase inhibitors against BTK have shown promising results in different cell lines, and a phase 1 clinical study is in progress.44,45 Inhibition of the tyrosine kinase SYK in cell lines has highlighted its role as a potentially promising therapeutic target in diffuse large B-cell lymphoma.46
PAG1 is a lipid raft-anchored adaptor protein whose expression has been observed to be down-regulated in several types of cancers, such as non-small cell lung cancer47 or colorectal cancer.48 In addition, PAG1 overexpression efficiently suppresses c-Src transformation and tumorigenesis,49 supporting a tumor-suppressor role. In B lymphocytes, PAG1 is a negative regulator of the BCR-mediated signaling.50 PAG1 phosphorylation at tyrosine 317 generates a binding site for tyrosine-protein kinase CSK that, in turn, negatively regulates tyrosine-protein kinase Lyn, and tyrosine-protein kinase Fyn, leading to decreased Src kinase–dependent pathway activation.51,52 However, we identified PAG1 with the most phosphotyrosine peptides in both BL and FL cell lines. Immunoprecipitation and Western blot analyses revealed that PAG1 is overexpressed in BL and FL in comparison to MCL, and also highly phosphorylated in these two germinal center–derived NHLs. Previous studies have demonstrated strong immunohistochemical expression of PAG1 in proliferating cells of the germinal center and in lymphomas derived from these cells.53,54 In our study, we demonstrated that PAG1 knockdown increased B-cell proliferation and responsiveness to antigen stimulation, supporting its role as a negative regulator of the BCR signaling pathway. Interestingly, our study revealed that, in addition to Y317 phosphorylation, PAG1 is phosphorylated in vivo at seven additional tyrosine residues, including tyrosine 163 and 181 in BL and FL cell lines. It has been previously demonstrated that PAG1 phosphorylation at these two tyrosines (Y163 and Y181) results in a dual-domain docking module that enhances the affinity of the FYN-PAG1 interaction and renders FYN insensitive to negative regulation by CSK.55 Accordingly, we postulate that differential combinatorial phosphorylation of different tyrosine residues in PAG1 may contribute to the deregulation of the BCR signaling pathway in BL and FL and might be the consequence of the phosphorylation status of different tyrosine residues.
In conclusion, we have generated a comprehensive phosphorylation event data set in three different B-NHL entities, and demonstrated that these entities possess distinctive phosphoproteomic signatures. These signatures highlight the ability of post-translational modification profiling strategies to achieve class discrimination in distinct subtypes of cancer. Our study highlights the phosphorylation of the BCR signaling pathway in B-NHLs derived from the germinal center of secondary lymphatic follicles. Overall, these data reveal the utility of unbiased phosphoproteomic approaches for elucidation of signaling networks involved in the pathogenesis of neoplasia.
Acknowledgments
We thank Hyungwon Choi for helpful discussions and the PRIDE team for their help with data deposition.
Footnotes
Supported by NIH grants R01 DE119249 and R01 CA136905 (K.S.J.E.-J.), R01 CA140806 (M.S.L.), and R01-CA126239 and R01-GM094231 (A.I.N.); the Training Program in Bioinformatics grant 5T32GM070449 (T.W.); and the Fondation pour la Recherche Médicale grant SPE20091117389 (D.R.).
Disclosures: None declared.
Supplemental Data
Experimental work flow with main characteristics of the 11 cell lines used in this study. The phosphoproteome of all cell lines was analyzed in biological triplicate after phosphopeptide enrichments by the combination of metal oxide affinity chromatography (MOAC) and immunoaffinity purification of phosphotyrosine (pY-IP).
Principal characteristics of the identified phosphopeptides. A: Number of phosphopeptides carrying a p-Ser, p-Thr, or p-Tyr identified in eluents from MOAC enrichments or in pY-IP enrichment. B: Distribution of the singly or multiply phosphorylated peptides. C: Classification of phosphorylation sites into the four more general motif classes. The pie chart when all B-NHLs are analyzed together (left panel) and for each subgroup of B-NHLs (right panel).
Phosphorylation-specific motifs using the Motif-X algorithm. For each NHL subtype, the top five enriched motifs are displayed for p-Ser and p-Thr motifs, whereas all p-Tyr motifs are shown.
Recurrent motif of p-Tyr peptides. The Motif-X algorithm was used to generate consensus sequence motifs for p-Tyr sites. The motif EXXpY was identified as the most predominant motif in BL and FL. Table of proteins identified with peptide containing the motif EXXpY. Of 26 proteins, 8 (31%) are involved in the BCR signaling pathway.
Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (PAG1) is highly phosphorylated in BL and FL. Histograms showing proteins binned according to the spectral count of the p-Tyr proteins for BL (A), FL (B), and MCL (C). D: MS/MS spectra of PAG1 p-Tyr peptide (Y227).
PAG1 knockdown results in phosphorylation alterations in the BCR signaling pathway. A: Pathway analysis of the tyrosine phosphorylation alterations after the depletion of PAG1 in BJAB cells. Pathways are ranked according to the P value of their enrichment. The BCR pathway is the more significantly altered pathway after PAG1 knockdown. B: The STRING diagram displays the connections between proteins identified with at least a 1.5-fold differential phosphorylation at serine/threonine and/or tyrosine residues.
References
- 1.Hunter T. Signaling–2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P., Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 4.Harsha H.C., Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4:482–495. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzoli C.G., Baker S., Jr., Temin S., Pao W., Aliff T., Brahmer J., Johnson D.H., Laskin J.L., Masters G., Milton D., Nordquist L., Pfister D.G., Piantadosi S., Schiller J.H., Smith R., Smith T.J., Strawn J.R., Trent D., Giaccone G. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–6266. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Druker B.J., Guilhot F., O’Brien S.G., Gathmann I., Kantarjian H., Gattermann N., Deininger M.W., Silver R.T., Goldman J.M., Stone R.M., Cervantes F., Hochhaus A., Powell B.L., Gabrilove J.L., Rousselot P., Reiffers J., Cornelissen J.J., Hughes T., Agis H., Fischer T., Verhoef G., Shepherd J., Saglio G., Gratwohl A., Nielsen J.L., Radich J.P., Simonsson B., Taylor K., Baccarani M., So C., Letvak L., Larson R.A. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 7.Blanke C.D., Rankin C., Demetri G.D., Ryan C.W., von Mehren M., Benjamin R.S., Raymond A.K., Bramwell V.H., Baker L.H., Maki R.G., Tanaka M., Hecht J.R., Heinrich M.C., Fletcher C.D., Crowley J.J., Borden E.C. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J Clin Oncol. 2008;26:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Vardiman J.W. IARC; Lyon: 2008. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues. [Google Scholar]
- 9.Lenz G., Wright G.W., Emre N.C., Kohlhammer H., Dave S.S., Davis R.E., Carty S., Lam L.T., Shaffer A.L., Xiao W., Powell J., Rosenwald A., Ott G., Muller-Hermelink H.K., Gascoyne R.D., Connors J.M., Campo E., Jaffe E.S., Delabie J., Smeland E.B., Rimsza L.M., Fisher R.I., Weisenburger D.D., Chan W.C., Staudt L.M. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Paepe P., Baens M., van Krieken H., Verhasselt B., Stul M., Simons A., Poppe B., Laureys G., Brons P., Vandenberghe P., Speleman F., Praet M., De Wolf-Peeters C., Marynen P., Wlodarska I. ALK activation by the CLTC-ALK fusion is a recurrent event in large B-cell lymphoma. Blood. 2003;102:2638–2641. doi: 10.1182/blood-2003-04-1050. [DOI] [PubMed] [Google Scholar]
- 11.Rosenwald A., Alizadeh A.A., Widhopf G., Simon R., Davis R.E., Yu X., Yang L., Pickeral O.K., Rassenti L.Z., Powell J., Botstein D., Byrd J.C., Grever M.R., Cheson B.D., Chiorazzi N., Wilson W.H., Kipps T.J., Brown P.O., Staudt L.M. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macek B., Mann M., Olsen J.V. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 13.Larsen M.R., Thingholm T.E., Jensen O.N., Roepstorff P., Jorgensen T.J. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Rush J., Moritz A., Lee K.A., Guo A., Goss V.L., Spek E.J., Zhang H., Zha X.M., Polakiewicz R.D., Comb M.J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 15.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J.M., Yuan J., Bakalarski C.E., Villen J., Kornhauser J.M., Smith B., Li D., Zhou X., Gygi S.P., Gu T.L., Polakiewicz R.D., Rush J., Comb M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenyo D., Beavis R.C. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 18.MacLean B., Eng J.K., Beavis R.C., McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 19.Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 21.Pedrioli P.G. Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol Biol. 2010;604:213–238. doi: 10.1007/978-1-60761-444-9_15. [DOI] [PubMed] [Google Scholar]
- 22.Fermin D., Basrur V., Yocum A.K., Nesvizhskii A.I. Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11:1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beausoleil S.A., Villen J., Gerber S.A., Rush J., Gygi S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 24.Villen J., Beausoleil S.A., Gerber S.A., Gygi S.P. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz D., Chou M.F., Church G.M. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol Cell Proteomics. 2009;8:365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki R., Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 27.Vizcaino J.A., Cote R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J., O’Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J., Kim D.W., Chang W., Choe J., Park C.S., Song K., Lee I. Wnt5a is secreted by follicular dendritic cells to protect germinal center B cells via Wnt/Ca2+/NFAT/NF-kappaB-B cell lymphoma 6 signaling. J Immunol. 2012;188:182–189. doi: 10.4049/jimmunol.1102297. [DOI] [PubMed] [Google Scholar]
- 29.Wilker P.R., Kohyama M., Sandau M.M., Albring J.C., Nakagawa O., Schwarz J.J., Murphy K.M. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol. 2008;9:603–612. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bracken C.P., Wall S.J., Barre B., Panov K.I., Ajuh P.M., Perkins N.D. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruno T., De Angelis R., De Nicola F., Barbato C., Di Padova M., Corbi N., Libri V., Benassi B., Mattei E., Chersi A., Soddu S., Floridi A., Passananti C., Fanciulli M. Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell. 2002;2:387–399. doi: 10.1016/s1535-6108(02)00182-4. [DOI] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L.J., von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi S., Takayama Y., Ogawa A., Tamura K., Okada M. Transmembrane phosphoprotein Cbp positively regulates the activity of the carboxyl-terminal Src kinase, Csk. J Biol Chem. 2000;275:29183–29186. doi: 10.1074/jbc.C000326200. [DOI] [PubMed] [Google Scholar]
- 36.Brdicka T., Pavlistova D., Leo A., Bruyns E., Korinek V., Angelisova P., Scherer J., Shevchenko A., Hilgert I., Cerny J., Drbal K., Kuramitsu Y., Kornacker B., Horejsi V., Schraven B. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J Exp Med. 2000;191:1591–1604. doi: 10.1084/jem.191.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casado P., Alcolea M.P., Iorio F., Rodriguez-Prados J.C., Vanhaesebroeck B., Saez-Rodriguez J., Joel S., Cutillas P.R. Phosphoproteomics data classify hematological cancer cell lines according to tumor type and sensitivity to kinase inhibitors. Genome Biol. 2013;14:R37. doi: 10.1186/gb-2013-14-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeBien T.W., Tedder T.F. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 40.Wiestner A., Tehrani M., Chiorazzi M., Wright G., Gibellini F., Nakayama K., Liu H., Rosenwald A., Muller-Hermelink H.K., Ott G., Chan W.C., Greiner T.C., Weisenburger D.D., Vose J., Armitage J.O., Gascoyne R.D., Connors J.M., Campo E., Montserrat E., Bosch F., Smeland E.B., Kvaloy S., Holte H., Delabie J., Fisher R.I., Grogan T.M., Miller T.P., Wilson W.H., Jaffe E.S., Staudt L.M. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camps J., Salaverria I., Garcia M.J., Prat E., Bea S., Pole J.C., Hernandez L., Del Rey J., Cigudosa J.C., Bernues M., Caldas C., Colomer D., Miro R., Campo E. Genomic imbalances and patterns of karyotypic variability in mantle-cell lymphoma cell lines. Leuk Res. 2006;30:923–934. doi: 10.1016/j.leukres.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 42.M’Kacher R., Farace F., Bennaceur-Griscelli A., Violot D., Clausse B., Dossou J., Valent A., Parmentier C., Ribrag V., Bosq J., Carde P., Turhan A.G., Bernheim A. Blastoid mantle cell lymphoma: evidence for nonrandom cytogenetic abnormalities additional to t(11;14) and generation of a mouse model. Cancer Genet Cytogenet. 2003;143:32–38. doi: 10.1016/s0165-4608(02)00823-3. [DOI] [PubMed] [Google Scholar]
- 43.Davis R.E., Ngo V.N., Lenz G., Tolar P., Young R.M., Romesser P.B. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honigberg L.A., Smith A.M., Sirisawad M., Verner E., Loury D., Chang B., Li S., Pan Z., Thamm D.H., Miller R.A., Buggy J.J. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flemming A. Target discovery: blocking BTK in B-cell disorders. Nat Rev Drug Discov. 2010;9:681. doi: 10.1038/nrd3262. [DOI] [PubMed] [Google Scholar]
- 46.Cheng S., Coffey G., Zhang X.H., Shaknovich R., Song Z., Lu P., Pandey A., Melnick A.M., Sinha U., Wang Y.L. SYK inhibition and response prediction in diffuse large B-cell lymphoma. Blood. 2011;118:6342–6352. doi: 10.1182/blood-2011-02-333773. [DOI] [PubMed] [Google Scholar]
- 47.Kanou T., Oneyama C., Kawahara K., Okimura A., Ohta M., Ikeda N., Shintani Y., Okumura M., Okada M. The transmembrane adaptor Cbp/PAG1 controls the malignant potential of human non-small cell lung cancers that have c-src upregulation. Mol Cancer Res. 2011;9:103–114. doi: 10.1158/1541-7786.MCR-10-0340. [DOI] [PubMed] [Google Scholar]
- 48.Sirvent A., Benistant C., Pannequin J., Veracini L., Simon V., Bourgaux J.F., Hollande F., Cruzalegui F., Roche S. Src family tyrosine kinases: driven colon cancer cell invasion is induced by Csk membrane delocalization. Oncogene. 2010;29:1303–1315. doi: 10.1038/onc.2009.450. [DOI] [PubMed] [Google Scholar]
- 49.Oneyama C., Hikita T., Enya K., Dobenecker M.W., Saito K., Nada S., Tarakhovsky A., Okada M. The lipid raft-anchored adaptor protein Cbp controls the oncogenic potential of c-Src. Mol Cell. 2008;30:426–436. doi: 10.1016/j.molcel.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 50.Janssen E., Zhang W. Adaptor proteins in lymphocyte activation. Curr Opin Immunol. 2003;15:269–276. doi: 10.1016/s0952-7915(03)00044-x. [DOI] [PubMed] [Google Scholar]
- 51.Smida M., Posevitz-Fejfar A., Horejsi V., Schraven B., Lindquist J.A. A novel negative regulatory function of the phosphoprotein associated with glycosphingolipid-enriched microdomains: blocking Ras activation. Blood. 2007;110:596–615. doi: 10.1182/blood-2006-07-038752. [DOI] [PubMed] [Google Scholar]
- 52.Kawabuchi M., Satomi Y., Takao T., Shimonishi Y., Nada S., Nagai K., Tarakhovsky A., Okada M. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature. 2000;404:999–1003. doi: 10.1038/35010121. [DOI] [PubMed] [Google Scholar]
- 53.Svec A., Velenska Z., Horejsi V. Expression pattern of adaptor protein PAG: correlation between secondary lymphatic follicle and histogenetically related malignant lymphomas. Immunol Lett. 2005;100:94–97. doi: 10.1016/j.imlet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Svec A. Expression of transmembrane adaptor protein PAG/Cbp in diffuse large B-cell lymphoma: immunohistochemical study of 73 cases. Pathol Res Pract. 2007;203:193–198. doi: 10.1016/j.prp.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Solheim S.A., Petsalaki E., Stokka A.J., Russell R.B., Tasken K., Berge T. Interactions between the Fyn SH3-domain and adaptor protein Cbp/PAG derived ligands, effects on kinase activity and affinity. FEBS J. 2008;275:4863–4874. doi: 10.1111/j.1742-4658.2008.06626.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental work flow with main characteristics of the 11 cell lines used in this study. The phosphoproteome of all cell lines was analyzed in biological triplicate after phosphopeptide enrichments by the combination of metal oxide affinity chromatography (MOAC) and immunoaffinity purification of phosphotyrosine (pY-IP).
Principal characteristics of the identified phosphopeptides. A: Number of phosphopeptides carrying a p-Ser, p-Thr, or p-Tyr identified in eluents from MOAC enrichments or in pY-IP enrichment. B: Distribution of the singly or multiply phosphorylated peptides. C: Classification of phosphorylation sites into the four more general motif classes. The pie chart when all B-NHLs are analyzed together (left panel) and for each subgroup of B-NHLs (right panel).
Phosphorylation-specific motifs using the Motif-X algorithm. For each NHL subtype, the top five enriched motifs are displayed for p-Ser and p-Thr motifs, whereas all p-Tyr motifs are shown.
Recurrent motif of p-Tyr peptides. The Motif-X algorithm was used to generate consensus sequence motifs for p-Tyr sites. The motif EXXpY was identified as the most predominant motif in BL and FL. Table of proteins identified with peptide containing the motif EXXpY. Of 26 proteins, 8 (31%) are involved in the BCR signaling pathway.
Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (PAG1) is highly phosphorylated in BL and FL. Histograms showing proteins binned according to the spectral count of the p-Tyr proteins for BL (A), FL (B), and MCL (C). D: MS/MS spectra of PAG1 p-Tyr peptide (Y227).
PAG1 knockdown results in phosphorylation alterations in the BCR signaling pathway. A: Pathway analysis of the tyrosine phosphorylation alterations after the depletion of PAG1 in BJAB cells. Pathways are ranked according to the P value of their enrichment. The BCR pathway is the more significantly altered pathway after PAG1 knockdown. B: The STRING diagram displays the connections between proteins identified with at least a 1.5-fold differential phosphorylation at serine/threonine and/or tyrosine residues.