Summary paragraph
Entamoeba histolytica is the causative agent of amoebiasis, a potentially fatal diarrheal disease in the developing world. The parasite was named “histolytica” for its ability to destroy host tissues, which is most likely driven by direct killing of human cells. The mechanism of human cell killing has been unclear, though the accepted model was that the parasites use secreted toxic effectors to kill cells prior to ingestion1. Here we report the surprising discovery that amoebae kill by biting off and ingesting distinct pieces of living human cells, resulting in intracellular calcium elevation and eventual cell death. After cell killing, amoebae detach and cease ingestion. Ingestion of bites is required for cell killing, and also contributes to invasion of intestinal tissue. The internalization of bites of living human cells is reminiscent of trogocytosis (Greek trogo–, nibble) observed between immune cells2–6, but amoebic trogocytosis differs since it results in death. The ingestion of live cell material and the rejection of corpses illuminate a stark contrast to the established model of dead cell clearance in multicellular organisms7. These findings change the paradigm for tissue destruction in amoebiasis and suggest an ancient origin of trogocytosis as a form of intercellular exchange.
Results and Discussion
In the first year of life, approximately one third of infants in an urban slum in Dhaka, Bangladesh are infected with E. histolytica8. Malnourishment and stunting are associated with repeated infections in children9. During infection, amoebae colonize the colon, which can be asymptomatic or result in diarrhea, colitis or extraintestinal disease. Invasive disease is characterized by profound tissue destruction, manifesting as massive intestinal ulceration or fatal extraintestinal abscesses. This is likely driven by the cell killing activity of the amoebae, which are capable of killing epithelial cells, a variety of immune cells and various tissue culture lines. Contact mediated by the amoeba surface Gal/GalNAc lectin is critical, but the molecular mechanism by which cells are killed is unknown. The established paradigm is that the amoebae first kill cells and then ingest dead cell corpses1,10 (Extended Data Fig. 1a).
To elucidate the mechanism for human cell killing, we employed live microscopy to directly examine host-parasite interactions. Surprisingly, we found that immediately after human cell contact, the amoebae internalized distinct “bites” of human cell membrane (Fig. 1a). This was initiated within one minute of human cell contact, and amoebae continued to ingest additional bites thereafter (Video 1). By pre-labeling human cellular compartments, we found that ingested membrane bites contained human cell cytoplasm 90% of the time (Fig. 1a) and mitochondria 31% of the time (Fig. 1b). Pre-labeling of human cells with biotin and streptavidin-gold prior to co-incubation with amoebae further demonstrated that the ingested bites were human-derived (Fig. 1c and Extended Data Fig. 2a). There was pronounced actin polymerization within the amoebae at the site of human cell contact and ingested bites were surrounded by polymerized actin (Extended Data Fig. 2b – 2c). Ingested bites had two membranes, with an inner human cell membrane and an outer amoeba membrane (Extended Data Fig. 2a, 2d).
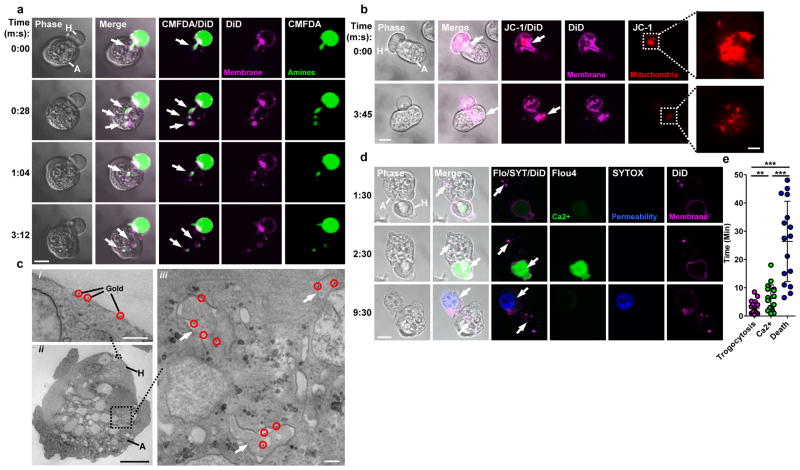
Figure 1. Amoebae internalize human cell bites, preceding human cell death.
a, DiD and CMFDA-labeled human Jurkat cells (H); an amoeba (A) internalizes bites (arrows) over time. Images are representative of three independent experiments. b, DiD and JC-1-labeled human Jurkat cells; mitochondria (arrow) are ingested by the amoeba in a bite. Images are representative of six independent experiments. c, i–ii, Electron microscopy with gold-labeled human Jurkat cells (gold, circles). iii, Bites (arrows) within an amoeba. Images are representative of two independent experiments. d, DiD and Flou4-labeled human Jurkat cells; with SYTOX blue present during imaging. Arrows, amoebic trogocytosis (1:30), [Ca2+]i elevation (2:30) and membrane permeabilization (9:30). Images are representative of 15 independent experiments. e, Timing in 60 cells from 15 independent experiments; shown are data points, means and standard deviations; P-values from t-tests: * < .05, ** < .01, *** < .001. Bars: 10 μm (a, b, d), 2 μm (b, insets), 0.5 μm (c, i, iii) and 5 μm (c, ii).
Another unexpected finding was that human cells were alive while they were being nibbled. Irreversible intracellular calcium elevation occurred in human cells within the first few minutes, typically following detectable biting (Fig. 1d – 1e and Video 2). Despite the fact that numerous bites were physically extracted from human cells by the amoebae, human cells initially retained plasma membrane integrity (Extended Data Fig. 3a). However, human cells were eventually killed as evidenced by loss of plasma membrane integrity11 (Fig. 1d – 1e, Extended Data Fig. 3b and Video 1), degradation of nuclear DNA and loss of mitochondrial potential (Extended Data Fig. 4a – 4b). Interestingly, ingestion of bites ceased once cells were dead (Extended Data Fig. 1b, 3b and Video 1), at which point the amoebae detached from corpses. Ingestion of bites required living cells, since pre-killed human cells were ingested intact (Extended Data Fig. 1c, 4c – 4d). The internalization of bites of living cells is reminiscent of trogocytosis2–6,12 (Greek trogo–, nibble); hence we refer to the E. histolytica process as “amoebic trogocytosis.”
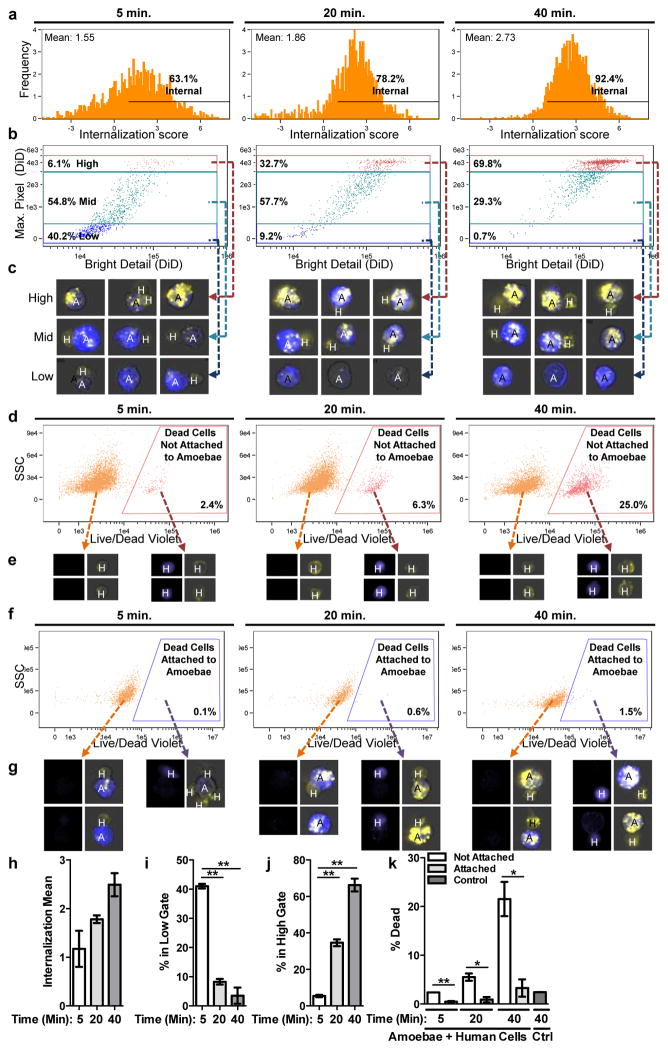
To quantify amoebic trogocytosis and cell death at the population level, we employed imaging flow cytometry (Fig. 2 and Extended Data Fig. 5). We found that internalization of human cellular material increased from 63.1% at 5 minutes to 92.4% at 40 minutes (Fig. 2a, 2h). To determine whether internalized material represented whole cells or bites, we measured the fragmentation of the human cells (Fig. 2b – 2c, 2i – 2j). The population representing high fragmentation (numerous bites) increased over time (Fig. 2b, 2j), from 6.1% at 5 minutes to 69.8% at 40 minutes. In contrast, the population representing low fragmentation (few bites), decreased over time (Fig. 2b, 2i). Thus, population-level quantification demonstrated that amoebic trogocytosis was predominant. The majority of dead human cells were not attached to amoebae (Fig. 2d – 2e, 2k), and very few remained attached to amoebae (Fig. 2f – 2g, 2k), supporting our observation that amoebae cease trogocytosis once cells have been killed (Extended Data Fig. 3b and Video 1).
Figure 2. Amoebic trogocytosis is predominant and specific to live human cells.
Imaging flow cytometry with CMFDA-labeled amoebae (A, blue), DiD-labeled human Jurkat cells (H, yellow) and Live/Dead Violet-labeled dead cells (purple). a, Detection of internalized human material. b, Measurement of fragmentation of internalized human material, gated from low to high. c, Example merged CMFDA, DiD and bright field images. Images are representative of 10,000 images collected for each replicate. d, Detection of dead cells not attached to amoebae. e, Example images; Live/Dead Violet (left panels) and merged CMFDA, DiD and bright field (right panels). Images are representative of 10,000 images collected for each replicate. f, Detection of dead cells attached to amoebae. g, Example images, as in e. Images are representative of 10,000 images collected for each replicate. h – k Means and standard deviations for biological replicates (10,000 events/each). P-values from t-tests: * < .05, ** < .01, *** < .001.
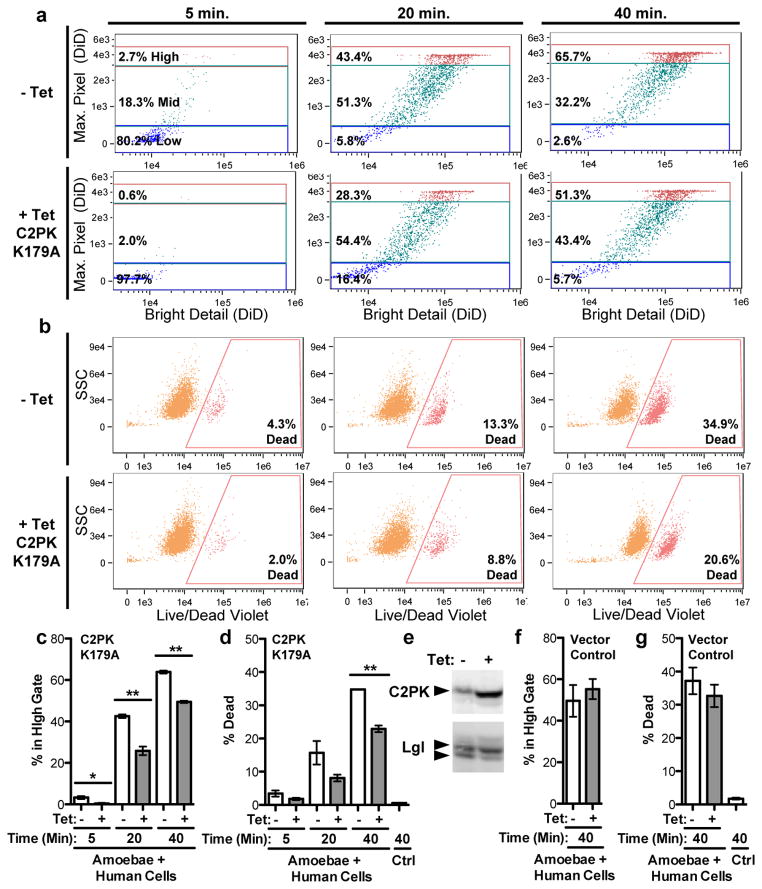
Neither amoebic trogocytosis nor cell killing occurred when cells were artificially brought together by centrifugation and co-incubated at 4 °C instead of 37 °C (Extended Data Fig. 6a – 7f), in agreement with the known inhibition of amoebic cytotoxicity at 4 °C13. Pharmacological inhibition of amoebic phosphatidylinositol 3-kinase (PI3K) signaling using wortmannin also inhibited amoebic trogocytosis and cell death (Extended Data Fig. 6g – 6j). Interference with the dynamics of microfilament formation using cytochalasin D inhibited both amoebic trogocytosis and cell death (Extended Data Fig. 7a – 7f). Tetracycline-inducible over-expression of a kinase-dead point mutant of the amoebic C2 domain-containing protein kinase (EhC2PK)14 reduced both amoebic trogocytosis and cell death (Fig. 3). EhC2PK is an early regulator of phagocytosis in E. histolytica14, thus some initiators of phagocytosis may be common to amoebic trogocytosis. To examine signaling upstream of EhC2PK, we used blocking antibodies to interfere with the amoebic Gal/GalNAc lectin, which has been shown to have a role in initiating cell killing1,15,16 separable from its role in host cell attachment13. An anti-Gal/GalNAc lectin blocking monoclonal antibody directed to epitope 1 that reduces killing while enhancing attachment16, inhibited both amoebic trogocytosis and cell death as compared to a control monoclonal antibody directed to epitope 3 (Extended Data Fig. 7g – 7l). Thus amoebic trogocytosis contributes to human cell killing and requires physiological temperature, actin rearrangements, and Gal/GalNAc lectin, EhC2PK and PI3K signaling.
Figure 3. Amoebic trogocytosis contributes to human cell killing.
a – d, Imaging flow cytometry analysis with amoebae expressing a tetracycline (tet)-inducible copy of EhC2PK K179A. a, Measurement of fragmentation. b, Detection of dead cells. c – d, Means and standard deviations for biological duplicates at each time point (10,000 events/each). e, Western blot demonstrating EhC2PK over-expression in tet-induced cells; Lgl, loading control. Blots are representative of three independent experiments. f – g, Imaging flow cytometry analysis with vector control amoebae; shown are means and standard deviations for biological duplicates (10,000 events/each). P-values from t-tests: * < .05, ** < .01, *** < .001.
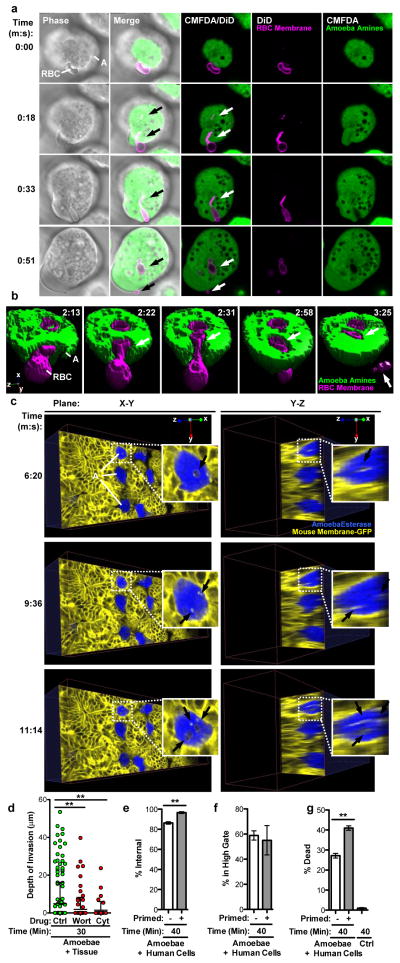
The hallmark of amoebiasis is the detection of amoebae with ingested red blood cells17. Live imaging demonstrated that amoebic trogocytosis occurred during red blood cell ingestion (Fig. 4a – 4b and Videos 3 – 4). We also detected amoebic trogocytosis of human colonic epithelial cells (Extended Data Fig. 8). Intriguingly, amoebic trogocytosis occurred when amoebae were incubated with ex vivo mouse intestinal tissue and monitored in 4-D using two-photon microscopy (Fig. 4c and Videos 5 – 6). Amoebae traversed the intestinal crypts, as has been demonstrated in studies using ex vivo human intestine18. Tissue cell death occurred and amoebic trogocytosis of intestinal enterocytes preceded amoebic invasion into the tissue (Extended Data Fig. 9 – 10 and Videos 7 – 8), further supporting its pathologic relevance. Moreover, inhibition of amoebic trogocytosis by treatment with wortmannin or cytochalasin D significantly blocked tissue invasion (Fig. 4d). Further supporting the relevance of amoebic trogocytosis to host cell destruction, we found that amoebae that had previously undergone amoebic trogocytosis were primed to undergo more ingestion and more cell killing (Fig. 4e – 4g) than amoebae that had not undergone trogocytosis. More of the primed amoebae underwent trogocytosis (Fig. 4e), though they ingested the same number of bites as non-primed amoebae (Fig. 4f), suggesting that priming enhances the initiation of amoebic trogocytosis.
Figure 4. Amoebic trogocytosis occurs with red blood cells, contributes to intestinal invasion, and promotes enhanced cell killing.
a, DiD-labeled red blood cells (RBC) and CMFDA-labeled amoebae (A); ingested bites (arrows) and a fragment remains extracellular (0:51). Bar, 5 μm. Images are representative of three independent experiments. b, Example 3-D reconstruction with DiD-labeled red blood cells and CMFDA-labeled amoebae; biting (arrows) and a fragment remains extracellular (3:25). Images are representative of three independent experiments. c, Planes from 4-D two-photon microscopy with mouse intestinal tissue (yellow); calcein violet-labeled amoebae (blue) are ingesting bites (arrowheads). Images are representative of three independent experiments. d, Amoebae treated with wortmannin or cytochalasin D were incubated with tissue. Invasion depth, means and standard deviations from biological replicates (at least 40 cells/treatment); P-values are indicated. e–g, Amoebae were incubated with or without human Jurkat cells (“primed”), prior to incubation with labeled human Jurkat cells. e, Measurement of internalization. f, Measurement of fragmentation. g, Detection of dead cells. Means and standard deviations for biological duplicates (10,000 events/each). P-values from t-tests: * < .05, ** < .01, *** < .001.
Thus several lines of evidence demonstrate that amoebic trogocytosis contributes to cell killing and is likely to contribute to pathogenesis in vivo. Cell death likely stems from the accumulation of membrane damage. Targeted cells initially retain membrane integrity; hence one bite is not enough to kill. Inhibition experiments demonstrated that a reduction in the number of ingested bites almost completely prevented cell death, suggesting that there is a threshold of tolerated damage. It has previously been reported that amoebae ingest killed cells10, but in these studies the target cells were pre-killed and likely exhibited different characteristics from cells directly killed by the amoebae. Consistent with this idea, we found that pre-killed cells were internalized whole and thus they were recognized differently than endogenously killed cells, which the amoebae ceased ingesting.
The rejection of dead cell corpses is striking and makes a nutritional role for amoebic trogocytosis seem unlikely. Intestinal bacteria may be the primary food source during infection and the energetic costs of full internalization of human cells may outweigh the benefits. The large size of epithelial cells and the tight intercellular connections may make ingestion of entire cells difficult. Amoebic trogocytosis could potentially lower tissue density and create a more spacious environment for amoeboid migration. Ingesting bites could also provide the parasite a means of sensing the environment. Interestingly, amoebae that had undergone amoebic trogocytosis were primed to undergo more trogocytosis, suggesting that there might be “feed-forward” regulation, similar to what has been demonstrated during the ingestion of beads and apoptotic cells19. Thus amoebic trogocytosis and destruction of cells may promote further destruction. Further work will be needed to confirm with certainty that the proposed model for amoebic trogocytosis in cell killing and tissue invasion is applicable to pathogenesis in vivo, and to explore the implications of this process in disease.
Distortion of target cell shape during amoebic ingestion has been previously reported 20,21. We have similarly observed distortion prior to the appearance of bites. We employed a variety of membrane labels, and bites are more readily detected using these tightly localized and bright labels, than with more diffuse probes. Thus it is possible that owing to differences in detection methods, and the dynamic and rapid appearance of bites, they were not previously observed. Since increased red blood cell rigidity reduces cell distortion during ingestion22, it is also conceivable that differences in the deformability of different cell types influence the extent of fragmentation that occurs. Less deformable targets like beads are likely to be ingested whole. Finally, the size of the target cell type determines the amount of material that remains extracellular; very little remains following red blood cell ingestion, while much more remains following Jurkat or epithelial cell ingestion.
Some mechanistic details of trogocytosis between immune cells are beginning to be defined23. A key difference between immune cell trogocytosis and amoebic trogocytosis is that immune trogocytosis does not result in cell death. This may be because immune trogocytosis involves the exchange of fewer bites, and we find that targeted cells can withstand a limited number of bites. Interestingly, there are hints that protozoa like Naegleria fowleri may also be capable of ingesting host cell bites12. Therefore, trogocytosis as a form of intercellular exchange may be more evolutionarily ancient and widespread than is currently appreciated. Finally, amoebic trogocytosis is a potentially promising target for the future development of new therapeutics for amoebiasis, a major neglected disease in the developing world.
Methods
Cell culture
Amoebic trophozoites (HM1:IMSS) and human Jurkat cells (Clone E6-1, ATCC) were cultivated as previously described10. Human Caco-2 cells (ATCC) were cultivated in 5% CO2 at 37 °C in Eagle’s minimum essential medium (Gibco) supplemented with non-essential amino acids, sodium pyruvate, 20% heat-inactivated fetal bovine serum (Gibco), penicillin and streptomycin sulfate. Cells were harvested at 80% confluence by using a rubber policeman. In some experiments, amoebae were stably transfected with a plasmid for tetracycline-inducible expression of EhC2PK K179A14, or as a control, the same plasmid with a bacterial chloramphenicol acetyltransferase (CAT) gene. Following transfection using Attractene reagent (Qiagen), stable transfectants were obtained by selection with hygromycin at 10 μg/ml. To induce protein expression, tetracycline induction was carried out at 20 μg/ml for 36 hours. Jurkat cultures were harvested at mid-log phase and were enriched for viable cells by density gradient centrifugation using Ficoll-Paque PLUS (Amersham) at 800 × g for 10 min at room temperature. Human red blood cells were prepared as previously described24. All co-incubations between amoebae and human cells were conducted in M199 medium (Gibco) not containing phenol red and supplemented with 5.7 mM cysteine, 0.5% bovine serum albumin (Gemini) and 25 mM HEPES (Sigma) pH 6.8, referred to as M199s.
Animals
Animal use protocols were reviewed and approved by the University of Virginia Institutional Animal Care and Use Committee, and were in compliance with PHS and USDA guidelines for laboratory animal welfare. Gt(ROSA)26Sortm4(ACTB-tdTomato,-Egfp)Luo/J mice (The Jackson Laboratory), carrying a floxed gene for a membrane-targeted Tomato fluorescent protein followed by a gene for membrane-targeted enhanced green fluorescent protein (EGFP)25, were crossed to C57BL/6-Tg(Zp3-cre)93Knw/J mice (The Jackson Laboratory) to create mice with membrane-targeted EGFP in all cells26. Tissue for imaging experiments was from female mice that were at least eight weeks in age.
Live confocal imaging
Jurkat cells, Caco-2 cells, human red blood cells or amoebae were washed in M199s and separately labeled with fluorescent dyes prior to confocal imaging. For Jurkat cells, DiD, DiI or DiO (Invitrogen) were used at 5 μM; cells were incubated at 37 °C for 4 minutes followed by 4 °C for 15 minutes and washed twice. CMFDA (Invitrogen) was used to label Jurkat cells at 5 μM at 37 °C for 15 minutes; cells were then washed and incubated in fresh M199s at 37 °C for 10 minutes prior to washing and labeling with DiD as above. JC-1 (Invitrogen) was used to label Jurkat cells at 2.5 μg/μl at 37 °C for 10 minutes prior to washing and labeling with DiD as above. MitoTracker Red CMXRos (Invitrogen) was used to label Jurkats at 40 nM at 37 °C for 15 minutes prior to washing and labeling with DiO as above. Flou4 (Invitrogen) was used to label Jurkat cells at 2.5 μM at room temperature in the dark for 30 minutes prior to washing and labeling with DiD as above. In some experiments, SYTOX blue (Invitrogen) was present in the media during imaging at 200 μM.
To quantify the frequency of internalization of Jurkat cell cytoplasm or mitochondria, we used human cell DiD (or DiO) labeling to identify amoebae that had ingested Jurkat cell bites. We then asked if the DiD (or DiO) positive bites also contained human cell CMFDA labeling to detect cytoplasm in bites, or if they contained human cell JC-1 or MitoTracker Red labeling to detect mitochondria in bites. 90% of amoebae that had internalized DiD-positive bites had ingested CMFDA-labeled human cell cytoplasm (N = 29 amoebae from 3 independent experiments). 31% of amoebae that had internalized DiD- or DiO-positive bites had ingested JC-1- or MitoTracker Red-labeled human cell mitochondria (N = 81 amoebae from 6 independent experiments).
For experiments to monitor mitochondrial potential, JC-1 (Invitrogen) was used to label Jurkat cells at 2.5 μg/μl at 37 °C for 10 minutes prior to washing and labeling with DiD (Invitrogen) at 5 μM; cells were then incubated at 37 °C for 4 minutes followed by 4 °C for 15 minutes and washed twice. For experiments in which pre-killed Jurkat cells were used, cells were labeled with CMFDA (Invitrogen) at 5 μM at 37 °C for 15 minutes; cells were then washed and incubated in fresh M199s at 37 °C for 10 minutes then heat-killed at 55 °C for 15 minutes and washed prior to live imaging. For experiments with Caco-2 cells, CMFDA (Invitrogen) was used at 5 μM at 37 °C for 15 minutes; cells were then washed and incubated in fresh M199s at 37 °C for 10 minutes. Caco-2 cells were then washed and labeled with DiD (Invitrogen) at 5 μM in M199s; cells were incubated at 37 °C for 4 minutes followed by 4 °C for 15 minutes and washed twice. For experiments with human red blood cells, DiD (Invitrogen) was used at 5 μM; cells were incubated at 37 °C for 4 minutes followed by 4 °C for 15 minutes and washed twice. Amoebae and human cells were combined at 1 amoeba to 5 human cells immediately prior to imaging. For experiments in which pre-killed and living human cells were used, cells were combined at 1 amoeba to 5 pre-killed and 5 living human cells. For experiments with red blood cells, cells were combined at 1 amoeba to 10 red blood cells. Cells were imaged in glass bottom 35 mm culture dishes fully filled with media (Mattek) or standard glass slides with coverslips raised by double-sided tape. Confocal images were collected using Zeiss LSM software on a Zeiss LSM 510 inverted microscope equipped with a 100X apochromat oil objective and a heated stage; or using Zeiss Zen software on a Zeiss LSM 700 inverted microscope equipped with a 63X apochromat oil objective and a heated stage.
Immunofluorescence
For imaging polymerized actin with rhodamine-phalloidin, CMFDA (Invitrogen) was used to pre-label amoebae at 200 nM in M199s at 37 °C for 10 minutes; cells were then washed and incubated in fresh M199s at 37 °C for 10 minutes and washed twice. Jurkat cells were washed in M199s and combined with amoebae at 1 amoeba to 5 Jurkat cells and incubated at 37 °C prior to fixation with 4% paraformaldehyde in phosphate buffered saline (PBS) for 30 minutes at room temperature. Samples were kept in solution and permeabilized with 0.5% Triton X-100 (Sigma) and then stained with rhodamine-phalloidin (Cytoskeleton, Inc.) at 100 nM for two hours. For immunofluorescence studies, amoebae and Jurkat cells were washed in M199s and combined at 1:5 and incubated at 37 °C prior to fixation with 4% paraformaldehyde in PBS for 30 minutes at room temperature. Samples were kept in solution and were quenched with 100 mM glycine, permeabilized with 0.2% Triton X-100 and blocked with goat serum (Jackson ImmunoResearch Laboratories, Inc.) and bovine serum albumin (Gemini). Antibody labeling was carried out in solution. The 3F4 anti-Gal/GalNAc lectin monoclonal antibody and an anti-CD3 rabbit polyclonal (Abcam) were used, followed by goat anti-mouse dylight 649 (Jackson ImmunoResearch Laboratories, Inc.) and goat anti-rabbit CY3 (Jackson ImmunoResearch Laboratories, Inc.). For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), Jurkat cells were combined with amoebae in M199S at 1 amoeba to 5 Jurkat cells and incubated at 37 °C for 40 minutes, or Jurkat cells were incubated at 37 °C for 40 minutes in the absence of amoebae. Cells were transferred to ice and labeled with Live/Dead Fixable Red dead cell stain (Invitrogen) at 1 μl per ml for 30 minutes in the dark, and then fixed with 4% paraformaldehyde in PBS for 30 minutes at room temperature. Fixed cells were washed with PBS and then TUNEL labeling was carried out using the APO-BrdU TUNEL Assay Kit (Invitrogen) according to the manufacturer’s directions. Briefly, fixed cells were permeabilized with ice-cold 70% ethanol, washed, and nicked DNA was labeled by using TdT enzyme to incorporate BrdU at 37 °C for 60 minutes, with gentle agitation. Labeled cells were washed and incubated with an antibody conjugated to Alexa Flour 488 directed to BrdU, at room temperature for 30 minutes. All samples were mounted with vectashield H-1000 anti-fading agent and imaged using Zeiss LSM software on a Zeiss LSM 510 inverted microscope equipped with a 100X apochromat oil objective, or using Zeiss Zen software on a Zeiss LSM 700 inverted microscope equipped with a 63X apochromat oil objective.
Electron microscopy
In some experiments, Jurkat cells were pre-labeled with streptavidin-gold prior to incubation with amoebae. Jurkat cells were washed three times in ice-cold PBS pH 8.0 and resuspended at 1×107 cells/ml. Sulfo-NH-SS biotin (Thermo) was reconstituted at 6 mg/ml in water and added to cells at 80 μl/ml. Samples were incubated on ice for 25 minutes with gentle agitation. To quench excess biotin, ice-cold Tris pH 8.0 was then added to 100 mM final and samples were washed three times in ice-cold PBS containing 100 mM Tris pH 8.0. Samples were washed once with ice-cold PBS pH 7.4 and incubated with streptavidin conjugated to 6 nm gold (Electron Microscopy Sciences/Aurion) diluted at 1:3 in ice-cold PBS pH 7.4. Samples were incubated on ice for 30 minutes with gentle agitation. Samples were washed three times with ice-cold PBS pH 7.4 and twice with ice-cold M199s, and then used for co-incubation with amoebae. Amoebae and Jurkat cells were washed in M199s and combined at 1 amoeba to 5 Jurkat cells and incubated at 37 °C prior to fixation with 2% glutaraldehyde/2% paraformaldehyde in 0.1 M sodium phosphate buffer for 60 minutes at room temperature. For experiments without streptavidin-gold labeling, amoebae and Jurkat cells were washed in M199s and combined at 1 amoeba to 5 Jurkat cells and incubated at 37 °C prior to fixation as above. Samples were postfixed with 1% osmium tetroxide and 0.1% potassium ferricyanide (Electron Microscopy Sciences). Dehydration through an ethanol gradient was performed, followed by infiltration and embedment in epon. Sections were cut on a Leica Ultracut Ultramicrotome. Samples without streptavidin-gold labeling were poststained with uranyl acetate and lead citrate, while samples with streptavidin-gold labeling were not poststained. Samples were imaged using a JEOL 1230 transmission electron microscope using ultra high resolution digital imaging.
Imaging flow cytometry analysis
Jurkat cells and amoebae were washed in M199s and separately labeled with fluorescent dyes prior to co-incubation. For experiments using amoebae stably transfected with plasmids for inducible expression of EhC2PK K179A or CAT, cells were grown in the presence or absence of 20 μg/ml tetracycline for 36 hours. CMFDA (Invitrogen) was used to label amoebae at 200 nM in M199s at 37 °C for 10 minutes; cells were then washed and incubated in fresh M199s at 37 °C for 10 minutes and washed twice. Pharmacological interventions or antibody blocking were carried out after CMFDA labeling. Pharmacological interventions were carried out at 37 °C as follows: amoebae were treated with wortmannin (Sigma) at 10 nM, 50 nM, 100 nM or an equal volume of DMSO vehicle; or amoebae were treated with cytochalasin D (Sigma) at 2 μM, 10 μM, 20 μM, or an equal volume of DMSO vehicle. Antibody blocking was carried out at 4 °C, using anti-Gal/GalNAc lectin monoclonal antibodies 3F4 or 7F4 at 10 μg/104 amoebae, as described previously16. For Jurkat cells, sulfonated DiD (Invitrogen) was used at 5 μM in M199s; cells were incubated at 37 °C for 4 minutes followed by 4 °C for 15 minutes and then washed twice. Labeled amoebae and Jurkat cells were combined at 1 amoeba to 5 Jurkat cells in biological replicate or triplicate and incubated at 37 °C for 5, 20 or 40 minutes. Control Jurkat cells were incubated in the absence of amoebae. At each time point, samples were transferred to ice and labeled with Live/Dead Fixable Violet dead cell stain (Invitrogen) at 1 μl per ml for 30 minutes in the dark, and then fixed with 4% paraformaldehyde in PBS for 30 minutes at room temperature. For experiments in which amoebae were pre-incubated with Jurkat cells, amoebae and unlabeled Jurkat cells were combined at a ratio of 1 amoeba to 2 Jurkat cells, or the same amount of amoebae were incubated without Jurkat cells as a control. Samples were incubated 37 °C for 2 hours, at which point, DiD-labeled Jurkat cells were added at a ratio of 1 amoeba to 5 Jurkat cells, and samples were then incubated at 37 °C for 40 minutes, followed by Live/Dead Fixable Violet staining and fixation, as above. All samples were washed twice in PBS and transferred to round-bottom 96-well plates (Corning); plates were sealed with pierceable plate covers (X-Pierce). Data were collected using an AutoSampler on an Amnis ImageStreamX or ImagestreamX Mark II flow cytometer, equipped with a 40X objective. 10,000 – 15,000 events were collected for each sample and data were analyzed using Amnis IDEAS software. In example images presented in Fig. 2, note that images have been cropped to minimize blank space and fluorescence images have been merged with bright field.
Protein preparation and Western blotting
Cells were grown in the presence or absence of 20 μg/ml tetracycline for 36 hours. After washing three times with PBS, protein extracts were prepared by incubation in lysis buffer containing 50 mM Tris-Cl pH 8.3, 150 mM NaCl, 1% NP40 and 1x SigmaFast protease inhibitor cocktail (Sigma). Laemmli sample buffer was added to 1x final concentration and samples were boiled and processed for Western blotting. EhC2PK was detected with a rabbit polyclonal antibody14 and as a loading control, the light chain (Lgl) of the Gal/GalNAc lectin was detected with a mouse monoclonal antibody27. Anti-rabbit and anti-mouse secondary antibodies, conjugated to Cy5 and Cy3 (GE Healthcare) respectively, were used, and fluorescent images were acquired on a GE Typhoon Trio+ Variable Mode Imager.
Ex vivo imaging
Amoebae were labeled with calcein violet (Invitrogen) at 1.6 μM in M199s at 37 °C for 10 minutes and washed twice. Mouse ceca were isolated and gently inverted inside out in order to facilitate washing and access to the lumen. Ceca were washed three times in Hank’s balanced salt solution (Gibco) and then washed once in M199s and transferred to 35 mm petri dishes. Tissue handling was minimal and mucus was not removed. Immediately prior to imaging, 1.2×105 amoebae were added per cecum, and the tissue was gently secured with a glass coverslip supported by silicone grease (Corning). In pilot experiments, SYTOX blue (Invitrogen) was added to 200 μM to verify tissue viability in these conditions with no amoebae present. Cecal samples were imaged using Olympus FLUOVIEW software on an Olympus FLUOVIEW FV1000 two-photon system equipped with with a 20X dipping objective and a heated stage, or a Zeiss LSM 710 two-photon system equipped with a 20X dipping objective and a heated stage. In some experiments, SYTOX orange (Invitrogen) was added to 200 μM to the media during imaging, and samples were imaged using Zeiss Zen software on a Zeiss LSM 700 inverted microscope equipped with a 20X objective and a heated stage. Volocity software (PerkinElmer) was used for 3-D reconstructions and data analysis.
Analysis of tissue invasion
Amoebae were labeled with cell tracker red CMPTX (Invitrogen) at 6 μM in M199s at 37 °C for 15 minutes and washed twice. Pharmacological interventions were carried out after CMPTX labeling at 37 °C for 1 hour as follows: amoebae were treated with wortmannin (Sigma) at 100 nM, cytochalasin D (Sigma) at 20 μM, or an equal volume of DMSO vehicle. Amoebae were washed prior to incubation with tissue. Mouse ceca were isolated and gently inverted inside out in order to facilitate washing and access to the lumen. Ceca were washed three times in Hank’s balanced salt solution (Gibco) and then washed once in M199s and transferred to 35 mm petri dishes. Tissue handling was minimal and mucus was not removed. Ceca were cut into three equal sections, such that tissue from the same cecum was used for each different experimental condition. 5×105 amoebae were added per cecal section, and incubated at 37 °C for 30 minutes in M199s. Samples were fixed with 4% paraformaldehyde in PBS at 4 °C for 24 hours. Samples were then cryoprotected in 30% sucrose overnight, embedded in OCT (Tissue-Tek) and 10 μm sections were obtained by using a cryostat; sections were taken at a defined orientation, such that they represented cross-sections of the tissue. Samples were mounted with vectashield H-1000 anti-fading agent and imaged using Zeiss Zen software on a Zeiss LSM 700 inverted microscope equipped with a 63X apochromat oil objective. In each of two biological replicates per treatment, ten non-consecutive sections were imaged, such that all amoebae that were present in each section were imaged. Prior to quantification of tissue invasion, samples were coded by the assignment of random identifiers; tissue invasion was then scored in the coded samples in a blinded fashion. Tissue invasion was quantified using ImageJ software. Amoebae contacting the epithelium but not invading it were scored as a depth of invasion of 0 μm. For amoebae that had invaded the epithelium, the depth was measured as the minimum distance between the surface of the epithelium and the edge of the amoeba that was closest to the epithelial surface. At least 40 cells from 2 independent experiments were quantified for each treatment. Following quantification of all datasets, the coded samples were unmasked for analysis.
Statistical analysis
Quantitative data are expressed as the mean and standard deviation of the mean. Statistical significance was determined with the unpaired two-tailed Student’s t-test using the program Prism (GraphPad Software, Inc.). Samples analyzed in Fig. 4d had unequal variances (confirmed with the F-test); therefore an unpaired two-tailed Student’s t-test with the Welch correction was performed in this analysis. P-values are indicated on each figure: * < .05, ** < .01, *** < .001.
Supplementary Material
Extended Data Figure 1: Models for ingestion and human cell killing. a, Previous model for amoebic cell killing, in which human cell attachment is followed by host cell killing and then ingestion of the killed cell. b, Model for amoebic trogocytosis in cell killing, in which attachment is followed by ingestion of fragments of human cell material, which leads to host cell death. Following cell killing, amoebae dissociate from the killed cells. c, Model for phagocytosis of pre-killed cells, which are ingested whole.
Extended Data Figure 2: Polymerized actin is detected during amoebic trogocytosis. a, Electron microscopy with human Jurkat cells pre-labeled with biotin and streptavidin-6 nm gold. Human cells (H) were co-incubated with amoebae (A) for 2 minutes. i, low magnification image. ii, membrane-bound bite of gold-labeled (circles) human cell material (arrow) within the amoeba. iii, high magnification image of the amoeba cell membrane, demonstrating the absence of gold labeling. iv, high magnification image demonstrating gold in the human cell membrane but not in the amoeba membrane. Bar, 5 μm (i), 0.5 μm (ii – iv). Images are representative of two independent experiments. b, Electron microscopy imaging of human Jurkat cells co-incubated with amoebae for one minute. i, Demonstration of human cell material contained within polymerized amoeba cytoskeleton (black arrow); note the distorted shape of the human cell as it is pulled into the amoeba (white arrow). ii, A bite of human cell material visible (white arrow) within polymerized amoeba cytoskeleton (black arrow). iii, A bite of human material (white arrow) distal to the targeted human cell is surrounded by polymerized cytoskeleton (black arrow); N, nucleus. Bars, 5 μm. Images are representative of three independent experiments. c, Polymerized actin within the amoebae at the site of human cell attachment. CMFDA-labeled amoebae (green) were co-incubated with human Jurkat cells for 1 minute, and post-stained with rhodamine-phalloidin (red). Polymerized actin within the amoebae is indicated with black arrows. A ring of polymerized actin likely surrounding an ingested bite is indicated with a white arrow. Bars, 5 μm. Images are representative of two independent experiments. d, Immunofluorescence microscopy imaging, with human cells co-incubated with amoebae for five minutes. Shown are images acquired at the indicated z-heights, with the amoeba plasma membrane stained with anti-Gal/GalNAc lectin, the human Jurkat cell plasma membrane stained with anti-CD3 and DAPI stained nuclei. Arrows, human cell bites within amoebae, surrounded by amoebic Gal/GalNAc lectin. Bar, 10 μm. Images are representative of two independent experiments.
Extended Data Figure 3: Ingestion of bites precedes human cell death and ceases after cell death. a – b, Live microscopy with DiD-labeled human Jurkat cells and with SYTOX blue present during imaging. a, Human cells (H) initially retain membrane integrity while amoebae (A) are extensively internalizing bites (arrows), demonstrated by the lack of SYTOX blue uptake. Images are representative of three independent experiments. b, Loss of human cell membrane integrity indicative of cell death at T = 15:20, and disassociation between the amoebae and the dead human cell at T = 16:00. White arrows, amoebae; black arrow, human cell. Bars, 10 μm. Images are representative of three independent experiments.
Extended Data Figure 4: Permeable human cells are not viable and trogocytosis requires human viable cells. a, Detection of 3′OH nicked DNA using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), in conjunction with detection of cell permeability. Amoebae (A) and human Jurkat cells (H) were co-incubated for 40 minutes, or control human cells were incubated in the absence of amoebae. Prior to fixation, cells were labeled with Live/Dead Fixable Red to allow for the detection of membrane permeability. Following fixation, TUNEL was used to allow for the detection of nicked DNA. As indicated by arrows, confocal imaging demonstrates that most permeable human cells (red) also contain nicked DNA (green). Control human cells are not permeable and lack nicked DNA. Images are representative of three independent experiments. b, Detection of mitochondrial potential and membrane permeability using live confocal microscopy. DiD and JC-1-labeled human Jurkat cells were co-incubated with amoebae with SYTOX blue present during imaging. Mitochondrial potential is detected in living, non-permeable human cells (arrows). In contrast, cells that are permeable, as indicated by SYTOX blue staining (arrowheads), lack mitochondrial potential. Images are representative of six independent experiments. c – d, Killed human Jurkat cells were labeled with CMFDA, while live human Jurkat cells were separately labeled with DiD. c, Living and pre-killed human cells were combined at 1:1 and SYTOX blue was present in the media during imaging. SYTOX blue staining confirms that only the pre-killed (green) cells are dead (blue). d, Living and dead human cells were combined with amoebae in the presence of SYTOX blue. DiD-labeled bites (arrows) of living human cells (asterisks) are internalized, while pre-killed cells (arrowheads) are ingested whole, demonstrating that live human cells are required for amoebic trogocytosis. Bars, 10 μm. Images in c-d are representative of three independent experiments.
Extended Data Figure 5: Imaging flow cytometry analysis. Shown is the gating strategy that was used to analyze imaging flow cytometry data, with the percentage of gated events, and number of gated events in parentheses, shown in each case. This example illustrates the gating of the T=40 min. sample shown in Figure 2, with CMFDA-labeled amoebae, DiD-labeled human Jurkat cells, and Live/Dead Violet-labeled dead cells; 10,000 events were collected. 1, In-focus events were gated using a gradient of Brightfield. 2, Events gated in 1 were refined to remove events with more than one cell or group of cells not in contact, i.e. cases where multiple independent events were captured in flow in the same image. 3, Events gated in 2 were gated according to the intensity and aspect ratio of the CMFDA labeled amoebae. These parameters divided the events into “Human Cells Not Attached to Amoebae”, “Amoebae,” and clusters with more than one amoeba. Clusters with more than one amoeba were typically less than 2% of gated events, and were omitted from subsequent analyses due to the difficulty in obtaining independent measurements (i.e. the extent of human cell internalization) on each individual amoeba present in a cluster. 4, Events gated in 3 as “Amoebae” were further examined to determine if human cells were present; events gated as “Amoebae with Human Cells” contained human cells. 5, Human cell positive events gated in 4 were further examined to determine if the human cells were internalized by using the internalization score feature to quantify the overlap between the DiD image and a mask based on the CMFDA image. An internalization score of 1 was used as the cutoff to gate “Internalized Human Cells.” 6, “Internalized Human Cells” from 5 were further examined to determine if the DiD label was intact or fragmented, by measuring the bright detail and maximum pixel intensity of the DiD image. Events were gated as “High,” “Mid,” or “Low” according to the extent of fragmentation. 3a, To assess the viability of human cells in events not containing amoebae, “Human Cells Not Attached to Amoebae” gated in 3 were further examined to measure the intensity of Live/Dead Violet staining and granularity (SSC, side scatter). Dead cells were gated as indicated, to define “Dead Human Cells Not Attached to Amoebae.” 4a, To assess the viability of human cells in events containing amoebae, “Amoebae with Human Cells” gated in 4 were further examined. Dead amoebae were first excluded by measuring the overlap between the Live/Dead Violet image and a mask based on the CMFDA image. “Live Amoebae” were gated as indicated. 4b, Events gated in 4a that contained human cells and live amoebae (“Live Amoebae”) were further examined to identify dead human cells by measuring the intensity of Live/Dead Violet staining and granularity (SSC). Dead human cells were gated as indicated, to define “Dead Human Cells Attached to Amoebae.”
Extended Data Figure 6: Incubation on ice or treatment with Wortmannin inhibits amoebic trogocytosis and human cell killing. a – f, Imaging flow cytometry analysis with amoebae and human Jurkat cells that were incubated at 37 °C (control), or with amoebae and human Jurkat cells that were briefly centrifuged and incubated on ice; cells were co-incubated for 40 minutes. a, Measurement of human cell internalization. b, Measurement of fragmentation of internalized human cellular material, gated from low to high. c, Detection of dead human cells. Shown in d – f are means and standard deviations for biological replicates at each time point (10,000 events/replicate). g – j, Imaging flow cytometry analysis with amoebae treated with Wortmannin. Shown are plots from T = 40 minutes. g, Measurement of fragmentation. h, Detection of dead cells. i – j, means and standard deviations for biological triplicates (15,000 events/replicate). P-values from t-tests: * < .05, ** < .01, *** < .001.
Extended Data Figure 7: Treatment with Cytochalasin D or Anti-Gal/GalNAc lectin blocking antibodies inhibits amoebic trogocytosis and human cell killing. a – f, Imaging flow cytometry analysis with amoebae treated with vehicle (control) or treated with cytochalasin D. Shown are plots from T = 40 minutes. a, Measurement of human cell internalization. b, Measurement of human cell fragmentation. c, Detection of dead human cells. Shown in d – f are means and standard deviations for biological replicates at each time point (10,000 events/replicate). g – l, Imaging flow cytometry analysis with amoebae treated with a control monoclonal antibody16,28 (7F4, control) directed to the amoebic Gal/GalNAc lectin epitope 3 (aa 1082–1138) or treated with a blocking monoclonal antibody16,28 (3F4, blocking) directed to the amoebic Gal/GalNAc lectin epitope 1 (aa 895–998). Mab 3F4 was previously shown to enhance adhesion to human cells, while inhibiting human cell killing16. Shown are plots from T = 40 minutes. g, Measurement of human cell internalization. h, Measurement of human cell fragmentation. i, Detection of dead human cells. Shown in j – l are means and standard deviations for biological replicates at each time point (10,000 events/replicate). P-values from t-tests: * < .05, ** < .01, *** < .001.
Extended Data Figure 8: Amoebic trogocytosis occurs with human Caco-2 colonic epithelial cells. Live confocal microscopy with human Caco-2 cells pre-labeled with DiD and CMFDA. Shown is an amoeba (A) in contact with a human cell (H) at time 0, and bites (arrows) of DiD- and CMFDA-labeled human material over time. Bar, 10 μm. Images are representative of two independent experiments.
Extended Data Figure 9: Amoebic trogocytosis precedes tissue invasion. Live 4-D multiphoton microscopy with amoebae interacting with ex vivo mouse intestinal tissue, demonstrating that amoebic trogocytosis occurs prior to amoebic tissue invasion. Intestinal tissue was from the cecum of a mouse expressing membrane-targeted enhanced EGFP (yellow false color) and amoebae were pre-labeled with calcein violet (blue). a, Time points from one X–Y plane taken from the 3-D reconstruction of the 4-D data. Note that ingestion of bites (arrows) of mouse cells occurs prior to amoebic tissue invasion in the Z-axis. This is visualized as loss of an amoeba (asterisk) from the X–Y plane. b, The same time points as in panel a, shown as 3-D reconstructions. Shown is a subset of the total collected Z-height. The same amoeba (asterisk/arrow) can be seen invading the tissue in the Z-axis. Images are representative of three independent experiments.
Extended Data Figure 10: Cell death occurs during tissue invasion. a, Live confocal microscopy with amoebae interacting ex vivo mouse intestinal tissue, demonstrating that cell death occurs during tissue invasion. Amoebae were pre-labeled with calcein violet (blue) and incubated with mouse intestinal tissue from the cecum of a mouse expressing membrane-targeted enhanced EGFP (green), in the presence of SYTOX Orange. Dead cells are predominantly found adjacent to amoebae, which are undergoing amoebic trogocytosis. Images are representative of three independent experiments. b, Live multiphoton microscopy with amoebae interacting with ex vivo mouse intestinal tissue, demonstrating that the parasites traverse the intestinal crypts, as has been demonstrated in studies using ex vivo human intestine18. Intestinal tissue was from the cecum of a mouse expressing membrane-targeted GFP (green) and amoebae were pre-labeled with calcein violet (blue). Parasites can be seen localized to the center of crypts, shown in cross-section (brackets). Bars, 20 μm. Images are representative of three independent experiments.
Acknowledgments
We thank Jan A. Redick and Stacey J. Guillot for assistance with sample preparation for electron microscopy and David A. Zemo of Olympus for assistance with multiphoton microscopy. We thank the University of Virginia Research Histology Core for assistance with preparation of frozen sections. We thank James E. Casanova, J. David Castle, Joanne Lannigan, Kodi S. Ravichandran and Ronald P. Taylor for helpful discussions. The artwork (Extended Data Fig. 1) was prepared by Anita Impagliazzo. K.S.R. was supported by a Howard Hughes Medical Institute Postdoctoral Fellowship from the Life Sciences Research Foundation, and a Postdoctoral Fellowship from the Hartwell Foundation. N.M.L. was supported by NIH Training Grant AI07046-32. This work was supported by NIH grant 5R01 AI-26649 to W.A.P.
Footnotes
K.S.R. designed, performed and analyzed the experiments. W.A.P. oversaw experimental design and analysis. M.D.S. assisted with Amnis Imagestream experimental design and with collection and analysis of Amnis Imagestream data. N.M.L assisted with isolation and preparation of mouse tissue for ex vivo imaging. S. and A.B. contributed plasmids and antibodies for the study of EhC2PK and contributed to analysis. K.S.R. and W.A.P. wrote the manuscript.
The authors declare that no competing financial interests exist.
References
- 1.Ralston KS, Petri WA., Jr Tissue destruction and invasion by Entamoeba histolytica. Trends Parasitol. 2011;27:254–263. doi: 10.1016/j.pt.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista FD, Iber D, Neuberger MS. B cells acquire antigen from target cells after synapse formation. Nature. 2001;411:489–494. doi: 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- 3.Huang JF, et al. TCR-Mediated internalization of peptide-MHC complexes acquired by T cells. Science. 1999;286:952–954. doi: 10.1126/science.286.5441.952. [DOI] [PubMed] [Google Scholar]
- 4.Hudrisier D, Riond J, Mazarguil H, Gairin JE, Joly E. Cutting edge: CTLs rapidly capture membrane fragments from target cells in a TCR signaling-dependent manner. J Immunol. 2001;166:3645–3649. doi: 10.4049/jimmunol.166.6.3645. [DOI] [PubMed] [Google Scholar]
- 5.Hudson L, Sprent J, Miller JF, Playfair JH. B cell-derived immunoglobulin on activated mouse T lymphocytes. Nature. 1974;251:60–62. doi: 10.1038/251060a0. [DOI] [PubMed] [Google Scholar]
- 6.Joly E, Hudrisier D. What is trogocytosis and what is its purpose? Nat Immunol. 2003;4:815. doi: 10.1038/ni0903-815. [DOI] [PubMed] [Google Scholar]
- 7.Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korpe PS, et al. Breast milk parasite-specific antibodies and protection from amebiasis and cryptosporidiosis in Bangladeshi infants: a prospective cohort study. Clinical infectious diseases. 2013;56:988–992. doi: 10.1093/cid/cis1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mondal D, Petri WA, Jr, Sack RB, Kirkpatrick BD, Haque R. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: evidence from a prospective study. Trans R Soc Trop Med Hyg. 2006;100:1032–1038. doi: 10.1016/j.trstmh.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Huston CD, Boettner DR, Miller-Sims V, Petri WA., Jr Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect Immun. 2003;71:964–972. doi: 10.1128/IAI.71.2.964-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroemer G, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell death and differentiation. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown T. Observations by immunofluorescence microscopy and electron microscopy on the cytopathogenicity of Naegleria fowleri in mouse embryo-cell cultures. J Med Microbiol. 1979;12:363–371. doi: 10.1099/00222615-12-3-363. [DOI] [PubMed] [Google Scholar]
- 13.Ravdin JI, Guerrant RL. Role of adherence in cytopathogenic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J Clin Invest. 1981;68:1305–1313. doi: 10.1172/JCI110377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somlata, Bhattacharya S, Bhattacharya A. A C2 domain protein kinase initiates phagocytosis in the protozoan parasite Entamoeba histolytica. Nat Commun. 2011;2:230. doi: 10.1038/ncomms1199. [DOI] [PubMed] [Google Scholar]
- 15.Ravdin JI, Croft BY, Guerrant RL. Cytopathogenic mechanisms of Entamoeba histolytica. J Exp Med. 1980;152:377–390. doi: 10.1084/jem.152.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saffer LD, Petri WA., Jr Role of the galactose lectin of Entamoeba histolytica in adherence-dependent killing of mammalian cells. Infect Immun. 1991;59:4681–4683. doi: 10.1128/iai.59.12.4681-4683.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Ruiz A, et al. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J Clin Pathol. 1994;47:236–239. doi: 10.1136/jcp.47.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal D, et al. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS Negl Trop Dis. 2009;3:e551. doi: 10.1371/journal.pntd.0000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sateriale A, Vaithilingam A, Donnelly L, Miller P, Huston CD. Feed-forward regulation of phagocytosis by Entamoeba histolytica. Infect Immun. 2012;80:4456–4462. doi: 10.1128/IAI.00671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lejeune A, Gicquaud C. Evidence for two mechanisms of human erythrocyte endocytosis by Entamoeba histolytica-like amoebae (Laredo strain) Biol Cell. 1987;59:239–245. doi: 10.1111/j.1768-322x.1987.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakada-Tsukui K, Okada H, Mitra BN, Nozaki T. Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica. Cell Microbiol. 2009;11:1471–1491. doi: 10.1111/j.1462-5822.2009.01341.x. [DOI] [PubMed] [Google Scholar]
- 22.Lejeune A, Gicquaud C. Target cell deformability determines the type of phagocytic mechanism used by Entamoeba histolytica-like, Laredo strain. Biol Cell. 1992;74:211–216. doi: 10.1016/0248-4900(92)90027-x. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Martin N, et al. T cell receptor internalization from the immunological synapse is mediated by TC21 and RhoG GTPase-dependent phagocytosis. Immunity. 2011;35:208–222. doi: 10.1016/j.immuni.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boettner DR, et al. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 2008;4:e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 26.Williams M, Burdsal C, Periasamy A, Lewandoski M, Sutherland A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev Dyn. 2012;241:270–283. doi: 10.1002/dvdy.23711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy JJ, Weaver AM, Petri WA., Jr Use of monoclonal anti-light subunit antibodies to study the structure and function of the Entamoeba histolytica Gal/GalNAc adherence lectin. Glycoconjugate journal. 1994;11:432–436. doi: 10.1007/BF00731279. [DOI] [PubMed] [Google Scholar]
- 28.Mann BJ. Structure and function of the Entamoeba histolytica Gal/GalNAc lectin. International review of cytology. 2002;216:59–80. doi: 10.1016/s0074-7696(02)16003-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1: Models for ingestion and human cell killing. a, Previous model for amoebic cell killing, in which human cell attachment is followed by host cell killing and then ingestion of the killed cell. b, Model for amoebic trogocytosis in cell killing, in which attachment is followed by ingestion of fragments of human cell material, which leads to host cell death. Following cell killing, amoebae dissociate from the killed cells. c, Model for phagocytosis of pre-killed cells, which are ingested whole.
Extended Data Figure 2: Polymerized actin is detected during amoebic trogocytosis. a, Electron microscopy with human Jurkat cells pre-labeled with biotin and streptavidin-6 nm gold. Human cells (H) were co-incubated with amoebae (A) for 2 minutes. i, low magnification image. ii, membrane-bound bite of gold-labeled (circles) human cell material (arrow) within the amoeba. iii, high magnification image of the amoeba cell membrane, demonstrating the absence of gold labeling. iv, high magnification image demonstrating gold in the human cell membrane but not in the amoeba membrane. Bar, 5 μm (i), 0.5 μm (ii – iv). Images are representative of two independent experiments. b, Electron microscopy imaging of human Jurkat cells co-incubated with amoebae for one minute. i, Demonstration of human cell material contained within polymerized amoeba cytoskeleton (black arrow); note the distorted shape of the human cell as it is pulled into the amoeba (white arrow). ii, A bite of human cell material visible (white arrow) within polymerized amoeba cytoskeleton (black arrow). iii, A bite of human material (white arrow) distal to the targeted human cell is surrounded by polymerized cytoskeleton (black arrow); N, nucleus. Bars, 5 μm. Images are representative of three independent experiments. c, Polymerized actin within the amoebae at the site of human cell attachment. CMFDA-labeled amoebae (green) were co-incubated with human Jurkat cells for 1 minute, and post-stained with rhodamine-phalloidin (red). Polymerized actin within the amoebae is indicated with black arrows. A ring of polymerized actin likely surrounding an ingested bite is indicated with a white arrow. Bars, 5 μm. Images are representative of two independent experiments. d, Immunofluorescence microscopy imaging, with human cells co-incubated with amoebae for five minutes. Shown are images acquired at the indicated z-heights, with the amoeba plasma membrane stained with anti-Gal/GalNAc lectin, the human Jurkat cell plasma membrane stained with anti-CD3 and DAPI stained nuclei. Arrows, human cell bites within amoebae, surrounded by amoebic Gal/GalNAc lectin. Bar, 10 μm. Images are representative of two independent experiments.
Extended Data Figure 3: Ingestion of bites precedes human cell death and ceases after cell death. a – b, Live microscopy with DiD-labeled human Jurkat cells and with SYTOX blue present during imaging. a, Human cells (H) initially retain membrane integrity while amoebae (A) are extensively internalizing bites (arrows), demonstrated by the lack of SYTOX blue uptake. Images are representative of three independent experiments. b, Loss of human cell membrane integrity indicative of cell death at T = 15:20, and disassociation between the amoebae and the dead human cell at T = 16:00. White arrows, amoebae; black arrow, human cell. Bars, 10 μm. Images are representative of three independent experiments.
Extended Data Figure 4: Permeable human cells are not viable and trogocytosis requires human viable cells. a, Detection of 3′OH nicked DNA using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), in conjunction with detection of cell permeability. Amoebae (A) and human Jurkat cells (H) were co-incubated for 40 minutes, or control human cells were incubated in the absence of amoebae. Prior to fixation, cells were labeled with Live/Dead Fixable Red to allow for the detection of membrane permeability. Following fixation, TUNEL was used to allow for the detection of nicked DNA. As indicated by arrows, confocal imaging demonstrates that most permeable human cells (red) also contain nicked DNA (green). Control human cells are not permeable and lack nicked DNA. Images are representative of three independent experiments. b, Detection of mitochondrial potential and membrane permeability using live confocal microscopy. DiD and JC-1-labeled human Jurkat cells were co-incubated with amoebae with SYTOX blue present during imaging. Mitochondrial potential is detected in living, non-permeable human cells (arrows). In contrast, cells that are permeable, as indicated by SYTOX blue staining (arrowheads), lack mitochondrial potential. Images are representative of six independent experiments. c – d, Killed human Jurkat cells were labeled with CMFDA, while live human Jurkat cells were separately labeled with DiD. c, Living and pre-killed human cells were combined at 1:1 and SYTOX blue was present in the media during imaging. SYTOX blue staining confirms that only the pre-killed (green) cells are dead (blue). d, Living and dead human cells were combined with amoebae in the presence of SYTOX blue. DiD-labeled bites (arrows) of living human cells (asterisks) are internalized, while pre-killed cells (arrowheads) are ingested whole, demonstrating that live human cells are required for amoebic trogocytosis. Bars, 10 μm. Images in c-d are representative of three independent experiments.
Extended Data Figure 5: Imaging flow cytometry analysis. Shown is the gating strategy that was used to analyze imaging flow cytometry data, with the percentage of gated events, and number of gated events in parentheses, shown in each case. This example illustrates the gating of the T=40 min. sample shown in Figure 2, with CMFDA-labeled amoebae, DiD-labeled human Jurkat cells, and Live/Dead Violet-labeled dead cells; 10,000 events were collected. 1, In-focus events were gated using a gradient of Brightfield. 2, Events gated in 1 were refined to remove events with more than one cell or group of cells not in contact, i.e. cases where multiple independent events were captured in flow in the same image. 3, Events gated in 2 were gated according to the intensity and aspect ratio of the CMFDA labeled amoebae. These parameters divided the events into “Human Cells Not Attached to Amoebae”, “Amoebae,” and clusters with more than one amoeba. Clusters with more than one amoeba were typically less than 2% of gated events, and were omitted from subsequent analyses due to the difficulty in obtaining independent measurements (i.e. the extent of human cell internalization) on each individual amoeba present in a cluster. 4, Events gated in 3 as “Amoebae” were further examined to determine if human cells were present; events gated as “Amoebae with Human Cells” contained human cells. 5, Human cell positive events gated in 4 were further examined to determine if the human cells were internalized by using the internalization score feature to quantify the overlap between the DiD image and a mask based on the CMFDA image. An internalization score of 1 was used as the cutoff to gate “Internalized Human Cells.” 6, “Internalized Human Cells” from 5 were further examined to determine if the DiD label was intact or fragmented, by measuring the bright detail and maximum pixel intensity of the DiD image. Events were gated as “High,” “Mid,” or “Low” according to the extent of fragmentation. 3a, To assess the viability of human cells in events not containing amoebae, “Human Cells Not Attached to Amoebae” gated in 3 were further examined to measure the intensity of Live/Dead Violet staining and granularity (SSC, side scatter). Dead cells were gated as indicated, to define “Dead Human Cells Not Attached to Amoebae.” 4a, To assess the viability of human cells in events containing amoebae, “Amoebae with Human Cells” gated in 4 were further examined. Dead amoebae were first excluded by measuring the overlap between the Live/Dead Violet image and a mask based on the CMFDA image. “Live Amoebae” were gated as indicated. 4b, Events gated in 4a that contained human cells and live amoebae (“Live Amoebae”) were further examined to identify dead human cells by measuring the intensity of Live/Dead Violet staining and granularity (SSC). Dead human cells were gated as indicated, to define “Dead Human Cells Attached to Amoebae.”
Extended Data Figure 6: Incubation on ice or treatment with Wortmannin inhibits amoebic trogocytosis and human cell killing. a – f, Imaging flow cytometry analysis with amoebae and human Jurkat cells that were incubated at 37 °C (control), or with amoebae and human Jurkat cells that were briefly centrifuged and incubated on ice; cells were co-incubated for 40 minutes. a, Measurement of human cell internalization. b, Measurement of fragmentation of internalized human cellular material, gated from low to high. c, Detection of dead human cells. Shown in d – f are means and standard deviations for biological replicates at each time point (10,000 events/replicate). g – j, Imaging flow cytometry analysis with amoebae treated with Wortmannin. Shown are plots from T = 40 minutes. g, Measurement of fragmentation. h, Detection of dead cells. i – j, means and standard deviations for biological triplicates (15,000 events/replicate). P-values from t-tests: * < .05, ** < .01, *** < .001.
Extended Data Figure 7: Treatment with Cytochalasin D or Anti-Gal/GalNAc lectin blocking antibodies inhibits amoebic trogocytosis and human cell killing. a – f, Imaging flow cytometry analysis with amoebae treated with vehicle (control) or treated with cytochalasin D. Shown are plots from T = 40 minutes. a, Measurement of human cell internalization. b, Measurement of human cell fragmentation. c, Detection of dead human cells. Shown in d – f are means and standard deviations for biological replicates at each time point (10,000 events/replicate). g – l, Imaging flow cytometry analysis with amoebae treated with a control monoclonal antibody16,28 (7F4, control) directed to the amoebic Gal/GalNAc lectin epitope 3 (aa 1082–1138) or treated with a blocking monoclonal antibody16,28 (3F4, blocking) directed to the amoebic Gal/GalNAc lectin epitope 1 (aa 895–998). Mab 3F4 was previously shown to enhance adhesion to human cells, while inhibiting human cell killing16. Shown are plots from T = 40 minutes. g, Measurement of human cell internalization. h, Measurement of human cell fragmentation. i, Detection of dead human cells. Shown in j – l are means and standard deviations for biological replicates at each time point (10,000 events/replicate). P-values from t-tests: * < .05, ** < .01, *** < .001.
Extended Data Figure 8: Amoebic trogocytosis occurs with human Caco-2 colonic epithelial cells. Live confocal microscopy with human Caco-2 cells pre-labeled with DiD and CMFDA. Shown is an amoeba (A) in contact with a human cell (H) at time 0, and bites (arrows) of DiD- and CMFDA-labeled human material over time. Bar, 10 μm. Images are representative of two independent experiments.
Extended Data Figure 9: Amoebic trogocytosis precedes tissue invasion. Live 4-D multiphoton microscopy with amoebae interacting with ex vivo mouse intestinal tissue, demonstrating that amoebic trogocytosis occurs prior to amoebic tissue invasion. Intestinal tissue was from the cecum of a mouse expressing membrane-targeted enhanced EGFP (yellow false color) and amoebae were pre-labeled with calcein violet (blue). a, Time points from one X–Y plane taken from the 3-D reconstruction of the 4-D data. Note that ingestion of bites (arrows) of mouse cells occurs prior to amoebic tissue invasion in the Z-axis. This is visualized as loss of an amoeba (asterisk) from the X–Y plane. b, The same time points as in panel a, shown as 3-D reconstructions. Shown is a subset of the total collected Z-height. The same amoeba (asterisk/arrow) can be seen invading the tissue in the Z-axis. Images are representative of three independent experiments.
Extended Data Figure 10: Cell death occurs during tissue invasion. a, Live confocal microscopy with amoebae interacting ex vivo mouse intestinal tissue, demonstrating that cell death occurs during tissue invasion. Amoebae were pre-labeled with calcein violet (blue) and incubated with mouse intestinal tissue from the cecum of a mouse expressing membrane-targeted enhanced EGFP (green), in the presence of SYTOX Orange. Dead cells are predominantly found adjacent to amoebae, which are undergoing amoebic trogocytosis. Images are representative of three independent experiments. b, Live multiphoton microscopy with amoebae interacting with ex vivo mouse intestinal tissue, demonstrating that the parasites traverse the intestinal crypts, as has been demonstrated in studies using ex vivo human intestine18. Intestinal tissue was from the cecum of a mouse expressing membrane-targeted GFP (green) and amoebae were pre-labeled with calcein violet (blue). Parasites can be seen localized to the center of crypts, shown in cross-section (brackets). Bars, 20 μm. Images are representative of three independent experiments.