Abstract
The uropathogenic Escherichia coli strain 536 carries at least five genetic elements on its chromosome that meet all criteria characteristic of pathogenicity islands (PAIs). One main feature of these distinct DNA regions is their instability. We applied the so-called island-probing approach and individually labeled all five PAIs of E. coli 536 with the counterselectable marker sacB to evaluate the frequency of PAI-negative colonies under the influence of different environmental conditions. Furthermore, we investigated the boundaries of these PAIs. According to our experiments, PAI II536 and PAI III536 were the most unstable islands followed by PAI I536 and PAI V536, whereas PAI IV536 was stable. In addition, we found that deletion of PAI II536 and PAI III536 was induced by several environmental stimuli. Whereas excision of PAI I536, PAI II536, and PAI V536 was based on site-specific recombination between short direct repeat sequences at their boundaries, PAI III536 was deleted either by site-specific recombination or by homologous recombination between two IS100-specific sequences. In all cases, deletion is thought to lead to the formation of nonreplicative circular intermediates. Such extrachromosomal derivatives of PAI II536 and PAI III536 were detected by a specific PCR assay. Our data indicate that the genome content of uropathogenic E. coli can be modulated by deletion of PAIs.
Pathogenicity islands (PAIs) represent distinct large chromosomal regions that contribute to the evolution of bacterial pathogens (17). Characteristically, (i) they can be found in pathogenic strains but not or only rarely in nonpathogenic variants, (ii) they are inserted at the 3′ end of tRNA genes and carry (often many) virulence genes, (iii) their G+C content differs from that of the rest of the bacterial chromosome, (iv) they are associated with (sometimes cryptic) fragments of mobile genetic elements such as integrase genes or transposase genes, and in most cases, (v) they are flanked by insertion elements or direct repeats (DRs). Furthermore, some PAIs have the tendency to be deleted from the chromosome.
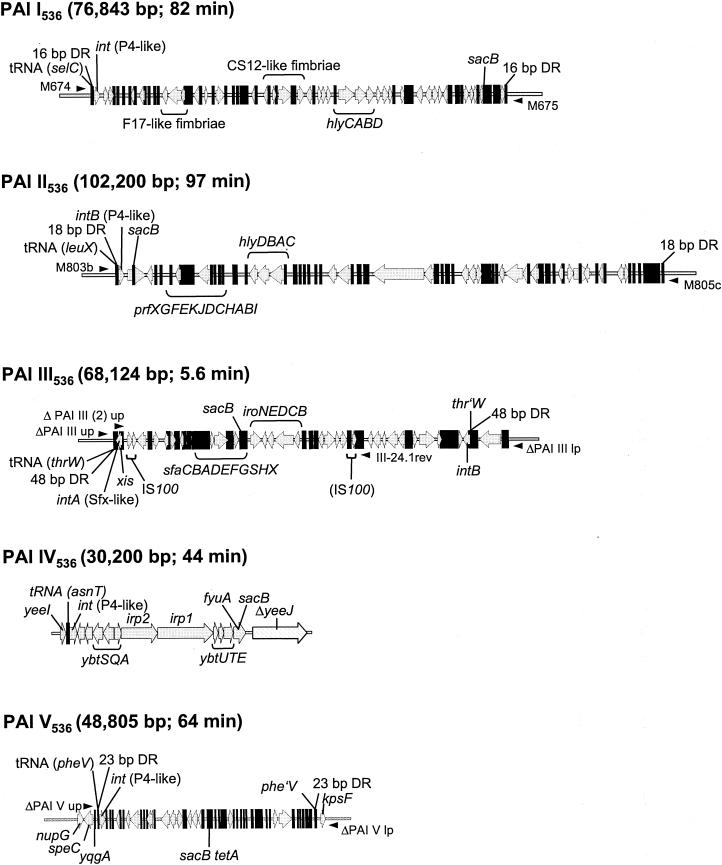
The uropathogenic Escherichia coli (UPEC) strain 536 (O6:K15:H31), which was originally isolated from a patient suffering from a urinary tract infection, is one of the best-characterized model organisms for the study of PAIs. The ongoing sequence project of the genome of this strain has revealed that it carries at least five of these genetic elements (PAI I536 to PAI V536), which are inserted at different sites of the chromosome and exhibit the main features of PAIs (14, 41; G. Schneider, U. Dobrindt, H. Brüggemann, G. Nagy, B. Janke, G. Blum-Oehler, G. Gottschalk, L. Emody, and J. Hacker, unpublished data). These PAIs carry many of the so far known virulence determinants of E. coli 536 (Fig. 1). They encode two α-hemolysin gene clusters (PAI I536 and PAI II536), P-related fimbriae (PAI II536), S-fimbriae (PAI III536), and the salmochelin and yersiniabactin siderophore systems (PAI III536 and PAI IV536, respectively). Interestingly, the genetic structure of PAI IV536 is identical to the core element of the so-called high-pathogenicity island (HPI) of Yersinia species. PAI I536 to PAI V536 are associated with the tRNA genes selC, leuX, thrW, asnT, and pheV, respectively. In general, the above-mentioned tRNA genes can be considered hotspots for the integration of foreign DNA into the prokaryotic chromosome, since they have been described as insertion sites for bacteriophages, conjugative transposons, and several PAIs in other bacterial species (17, 22, 23, 37).
FIG. 1.
Organization of PAI I536 to PAI V536. Localizations of important virulence genes, PAI-associated tRNA genes, integrase genes, DR regions flanking the PAIs, and the insertion site of the counterselectable marker sacB are indicated. With the exception of PAI IV536, the latter was introduced into noncoding regions of the corresponding PAI. Small black arrows symbolize oligonucleotides used for exclusion PCRs. Sizes of the PAIs and their relative locations in the chromosome (in minutes corresponding to the E. coli K-12 chromosome) are also given. Abbreviations: int, integrase genes; hly, hemolysin determinants; prf, P-related fimbrial adhesin-encoding genes; xis, excisionase gene; sfa, S-fimbriae-encoding genes; iro, salmochelin gene cluster; ybt, irp, and fyuA, genes encoding yersiniabactin biosynthesis and uptake; yeeJ, nupG, speC, and yqgA, chromosomal genes.
The PAIs of E. coli 536 are associated with integrase genes that exhibit the highest homology to the int genes of coliphage P4 and the Shigella flexneri phage SfX (Fig. 1). Our data indicate that all open reading frames encoding these integrases seem to be functional (B. Hochhut, G. Balling, and J. Hacker, unpublished data). Furthermore, with the exception of PAI IV536, the islands are flanked by DRs of different sizes (Fig. 1) (14; Schneider et al., unpublished). These flanking repeat regions correspond to the left and right end junctions (attL and attR) that result from the integration of phage DNA into the prokaryotic chromosome. Therefore, it is very likely that comparable to the insertion-excision mechanism of bacteriophages, insertion and deletion of PAIs is mediated by the respective PAI-encoded integrase and functions via site-specific recombination between the flanking DRs. The close association of virulence relevant genes and large unstable DNA regions such as PAIs is interesting for two reasons. First, the excision of PAIs and their potential propagation could contribute to genome plasticity and bacterial evolution. Second, deletion of PAIs could play an important role during the transition from an acute to a chronic phase of infection (6, 17).
Instability of PAI I536 and PAI II536 has been discovered previously on the basis of the formation of nonhemolytic colonies due to the loss of both islands via site-specific recombination between their flanking DRs (6). Furthermore, instability of PAIs is a phenomenon that has been described for other organisms as well, e.g., the cag PAI of Helicobacter pylori, the HPI of Yersinia pestis and Yersinia pseudotuberculosis, the locus of enterocyte effacement (LEE) of rabbit-specific enteropathogenic E. coli (REPEC) strain 84/110-1, and the Shigella resistance locus of S. flexneri (7, 9, 43, 44). PAI-negative cells appear in the REPEC strain 84/110-1 and in S. flexneri 2a with frequencies of around 10−6 to 10−5, which are comparable to previously obtained data for PAI I536 and PAI II536 (43, 44). Other PAIs, like the Vibrio cholerae PAI or the Staphylococcus aureus PAI (S. aureus PAI 1), have not only the tendency to be deleted from the chromosome but both are also transmissible to recipient organisms by transducing phages (25, 32, 39).
In this study, the instability of a complete set of PAIs from one strain was analyzed in detail for the first time. The deletion rate of PAI I536 to PAI V536 was determined by using the island-probing approach, which is based on insertion of the counterselectable marker sacB from Bacillus subtilis into the islands (34, 38). Furthermore, the influence of different environmental conditions such as low or elevated temperature, osmotic stress, or depletion of nutrients on the stability of PAIs was investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) Miller broth, M9 minimal medium (40), or artificial urine (AU). AU was prepared according to the instructions of Jackson et al. (24) but with a reduced CaCl2 concentration of 1 mM. For solid medium, agar was added to a final concentration of 1.5% (wt/vol). If necessary, the medium was supplemented with 7% sucrose, 5% sheep blood, 100 μg of ampicillin ml−1, 10 μg of tetracycline ml−1, or 20 μg of chloramphenicol ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| 536 | Wild-type UPEC strain (O6:K15:H31) | 4 |
| 536-21 | 536 ΔPAI I536 ΔPAI II536 | 18 |
| 536 PAI I536::sacB | sacB inserted into PAI I536 | This study |
| 536 PAI II536::sacB | sacB inserted into PAI II536 | 28 |
| 536 PAI III536::sacB | sacB inserted into PAI III536 | This study |
| 536 PAI IV536::sacB | sacB inserted into PAI IV536 | This study |
| 536 PAI V536::sacB tetA | sacB tetA inserted into PAI V536 | This study |
| 536 attBλ::sacB bla | sacB inserted into attBλ | This study |
| 536 ΔrecA PAI III536::sacB | ΔrecA | This study |
| 536 ΔrecA PAI V536::sacB tetA | ΔrecA | This study |
| DH5α | F− ΔlacU169 (φ80 lacZΔM15) recA1 hsdR17 | |
| SM10λpir | F−recA::RP4-2-Tc::Mu λpir Knr | 29 |
| Plasmids | ||
| pBluescript II KS(−) | bla, cloning vector | Stratagene |
| pGEM-T Easy | bla, T/A cloning vector | Promega |
| pGP704 | bla oriR6KmobRP4 | 29 |
| pCVD442 | bla oriR6KmobRP4sacB | 30 |
| pASK75 | bla tetR, expression vector | 42 |
| pBR322 | bla tetA, cloning vector | New England Biolabs |
| pBMM3 | pGP704 derivative for insertion of sacB into PAI I536 | This study |
| pBMM6 | pGP704 derivative for insertion of sacB into PAI III536 | This study |
| pGP704 fyuA::sacB | pGP704 derivative for insertion of sacB into PAI IV536 | Gift from E. Carniel, Institut Pasteur |
| pKS-sacB tetA | pBluescript II KS(−) sacB tetA | This study |
| pLDR8 | neo, int expression vector, Ts | 11 |
| pLDR9 | bla neo, cloning vector to integrate DNA into attBλ | 11 |
| pKD46 | bla repA101(Ts) araC araBρ γ β exo | 10 |
| pKD3 | cat with flanking FRT sites, template plasmid | 10 |
| pCP20 | bla, carries yeast FLP recombinase gene, Ts | 10 |
| pPAI II-CI | bla, positive control for detection of PAI II536-specific CIs | This study |
| pPAI III-CI (1) | bla, positive control for detection of PAI III536-specific CIs of deletion type I | This study |
| pPAI III-CI (2) | bla, positive control for detection of PAI III536-specific CIs of deletion type II | This study |
Oligonucleotides.
A list of oligonucleotides used in this study is available at http://www.uni-wuerzburg.de/infektionsbiologie. Oligonucleotides specific for PAI III536, PAI IV536, and the capsule region of E. coli 536 have been published previously (12, 13, 26). Oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany) or Sigma-ARK (Steinheim, Germany).
Labeling of E. coli 536-specific PAIs and attBλ with the counterselectable marker sacB.
For labeling of PAI I536, a noncoding 2.3-kb EcoRI fragment of this island was isolated from a cosmid clone (14) and subcloned into the SmaI site of pBluescript II KS(−). The construct was linearized with SphI, blunted, and ligated with a 2.6-kb PstI fragment of pCVD442 carrying the sacB gene and its cis-acting regulatory locus sacR, which had also been blunted. The complete 4.9-kb insert was finally cut out with XhoI and SacI. The ends were filled in, and the fragment was cloned into the EcoRV restriction site of the suicide vector pGP704, resulting in the vector pBMM3. For labeling of PAI III536, a noncoding region of this island was amplified with primer pair PAI III 6.4-1376/PAI III 6.4-3543. The 2.2-kb PCR product was cloned into EcoRV/SmaI-digested pBluescript II KS(−), and the resulting construct was linearized with PstI by cutting only in the center of the PAI III536-specific insert. The 2.6-kb PstI fragment of pCVD442 carrying the sacB gene was cloned into this restriction site. The entire 4.8-kb insert was cut out with BamHI and XhoI, the ends were blunted, and the fragment was ligated into the EcoRV restriction site of pGP704, resulting in the vector pBMM6. For labeling of PAI IV536, we used the vector pGP704 fyuA::sacB. The plasmids pBMM3, pBMM6, and pGP704 fyuA::sacB were transferred separately into E. coli 536 by conjugation, and double-crossover mutants corresponding to 536 PAI I536::sacB, 536 PAI III536::sacB, and 536 PAI IV536::sacB, respectively, were selected by screening for colonies with an ampicillin- and sucrose-sensitive phenotype. For labeling of PAI V536, tetA from pBR322 was amplified with primers Tet3 and Tet2, and the tetA promoter/operator region from plasmid pASK75 (42) was amplified with oligonucleotides tetp/o and Tet4. Both fragments were assembled in a recombinant PCR (21). The resulting 1.4-kb PCR fragment was cloned into the SmaI restriction site of pBluescript II KS(−). The construct was linearized with XbaI, the ends were blunted and ligated with the sacB gene that had been amplified with the primer pair sacB-161/XhoI and sacB-1975 from pCVD442. In the resulting vector pKS-sacB tetA, both genes are inserted consecutively in the same orientation. A subsequent PCR with pKS-sacB tetA as a template and the primer pair PAI Vmut 400 and PAI Vmut 499 generated a 3.3-kb PCR product that was used for electroporation of E. coli 536/pKD46 by following the protocol of Datsenko and Wanner (10). E. coli 536 PAI V536::sacB tetA was selected by screening for resistance to tetracycline and sensitivity to sucrose. For the insertion of sacB into the λ attachment site attBλ, the 2.6-kb PstI fragment of pCVD442 encoding the sacB gene was cloned into the PstI restriction site of pLDR9 (11). A NotI fragment containing the sacB gene, the attP site, and the bla gene was recircularized and subsequently introduced by electroporation into E. coli 536/pLDR8 by following the protocol of Diederich et al. (11). E. coli strain 536 attBλ::sacB bla was isolated by screening for resistance to ampicillin and sensitivity to sucrose.
Construction of ΔrecA mutants.
ΔrecA mutants were constructed with a one-step chromosomal gene inactivation technique (10). The cat gene of pKD3 was amplified with the primer pair recA_cat1-recA_cat2, and the resulting PCR product was electroporated into E. coli 536/pKD46. Mutants with a replacement deletion of recA were selected on agar plates containing chloramphenicol. The cat cassette was subsequently removed as described previously (10).
Preparation and manipulation of DNA.
Plasmid DNA and chromosomal DNA were isolated according to standard protocols (40). Recombinant DNA manipulations were carried out with enzymes supplied by Amersham or New England Biolabs according to the manufacturer's instructions and standard procedures (1). DNA was introduced into E. coli K-12 derivatives by transformation with CaCl2 competent cells (40). E. coli 536 was transformed by electroporation. Cells were grown to mid-log phase, repeatedly washed with ice-cold water, and resuspended in 10% (vol/vol) glycerol to a cell density of ∼3 × 1010 cells ml−1. Electroporation was performed with a Bio-Rad gene pulser at 2.5 kV, 25 μF, and 600 Ω in 2-mm-gap electroporation cuvettes.
PCR analysis and DNA sequencing.
PCR mixes contained 1 μl of cell lysate or 500 ng of chromosomal DNA as a template in a total volume of 50 μl. All PCRs were carried out with Taq polymerase from Qiagen (Hilden, Germany) according to the manufacturer's manual in an Eppendorf or Biometra thermocycler. For sequencing reactions, PCR products were cloned into pBluescript II KS(−) or pGEM-T Easy. DNA sequencing was done with the BigDye system (PE Biosystems, Inc.) and ABI-377 automated DNA sequencers (Applied Biosystems, Weiterstadt, Germany). Sequences were analyzed with the BLASTN and BLASTX programs (National Center for Biotechnology Information).
Determination of deletion rates of sacB-labeled PAIs.
Overnight cultures (15 h, 37°C) of sacB-labeled derivatives of E. coli 536 were diluted 1:100 in 30 ml of LB Miller, AU, or M9 minimal medium. Bacteria were incubated at 37, 42, or 20°C in an orbital shaker at 220 rpm, and samples were taken during the late lag, mid-log, early stationary, and late stationary phases. To determine the rate of spontaneously occurring sucrose resistant (Sucr) colonies, 536 attBλ::sacB bla was treated accordingly. Serial dilutions were plated on LB agar and LB agar supplemented with 7% sucrose to determine the CFU and the number of Sucr cells. Agar plates were incubated at 20°C for at least 48 h. The deletion rate of PAIs was calculated as the quotient of Sucr cells and CFU. All data are mean values of results from at least six independent experiments.
Postexperimental screening of Sucr colonies.
To test whether sucrose resistance resulted from excision of the respective PAI, samples of Sucr colonies were analyzed for the presence or absence of PAI-specific nucleotide sequences. Cells derived from 536 PAI I536::sacB and 536 PAI II536::sacB were tested phenotypically for their hemolytic activity on LB agar plates containing 3 to 5% sheep blood. For PAI III536 and PAI IV536, cells were screened by PAI-specific PCRs. For PAI V536, cells were further analyzed on LB plates containing tetracycline. For the control (536 attBλ::sacB bla), cells were tested on LB plates containing ampicillin.
PCR assays for detection of deletion of PAI I536 to PAI V536 (exclusion PCR).
Representative numbers of Sucr colonies were tested in a PCR assay specific for the chromosomal att site after deletion of the respective PAI. This assay is based on the usage of primer pairs binding to chromosomal regions flanking the island (Fig. 1). As the distance between primer binding sites is too large when the PAI is integrated in the chromosome, a PCR product was only amplified after excision of the island. Deletion of PAI I536 was detected with the primer pair M674-M675 (6), deletion of PAI II536 was detected with M803b-M805c (28), deletion of PAI III536 was detected with ΔPAI III up-ΔPAI III lp (type I deletion) and ΔPAI III (2) up-III-24.1rev (type II deletion), and deletion of PAI V536 was verified with ΔPAI V up-ΔPAI V lp.
Detection of CIs by PCR.
For the detection of circular intermediates (CIs), total DNA preparations were adjusted to a concentration of 200 ng μl−1 and sheared by vortexing and freezing. The PCRs contained 2 μl of 10× reaction buffer, 0.2 μl of 20 mM deoxynucleoside triphosphates, 0.2 μl of each primer (100 pmol μl−1), 0.2 μl of Taq polymerase (Qiagen), and 200 to 500 ng of DNA in a total volume of 20 μl. PAI II536-specific CIs were detected with the primer pair leu2 and concat1. Intermediates that resulted from excision of PAI III536 were detected with the primer pairs PAI III-ci (1) upnest-PAI III-ci (1) lpnest (type I deletion) and PAI III-ci (2) up-PAI III-ci (2) lp (type II deletion). As negative controls, the ΔPAI I536 ΔPAI II536 derivative 536-21 and E. coli K-12 DH5α were included in the assay. As positive controls, plasmids were constructed that carried, as inserts, the left and right ends of the respective PAI arranged in the orientation corresponding to the CIs. Defined copy numbers of these controls were employed in the assay. To verify that reaction mixes contained equal amounts of DNA, an internal region of the 16S rDNA was also amplified with the primer pair rrsA up-rrsA lp. All PCRs were carried out in an Eppendorf thermocycler (50 cycles of 30 s at 95°C, 30 s at 60°C, and 1 to 2 min at 68°C). Ten-microliter aliquots of the PCR samples were analyzed by submarine gel electrophoresis on a 1% (wt/vol) agarose gel.
RESULTS
PAI I536 to PAI V536 are deleted with different frequencies.
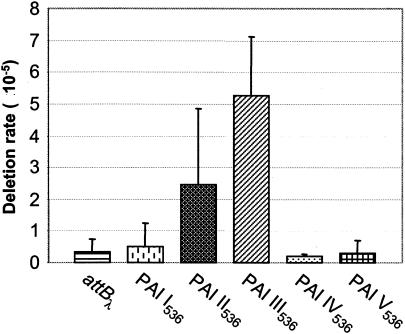
When the deletion rate of sacB-labeled PAIs of E.coli 536 was quantified after growth overnight in LB medium at 37°C, it was found that they exhibit different stabilities (Fig. 2). Analysis of strains 536 PAI II536::sacB and 536 PAI III536::sacB revealed that these islands are relatively unstable because Sucr colonies were generated with average frequencies of 2 × 10−5 to 3 × 10−5 and 5 × 10−5, respectively. Postexperimental screening of these colonies demonstrated that the majority of Sucr colonies resulted from deletion of the corresponding PAI and that the frequency of Sucr colonies was nearly equivalent to the number of PAI-negative cells in the culture. In contrast, Sucr colonies of 536 PAI I536::sacB, 536 PAI IV536::sacB, and 536 PAI V536::sacB tetA appeared with frequencies that were about 10-fold lower than the deletion rates of PAI II536 and PAI III536 and similar or only slightly increased compared to the rate of spontaneous mutations in sacB integrated into the λ att site (Fig. 2). This rate was determined with frequencies of 3 × 10−6 to 4 × 10−6 and was similar to previously published observations (5). Additional analyses (see Materials and Methods) revealed that about 50 to 60% of Sucr colonies derived from 536 PAI I536::sacB and 30 to 40% of Sucr colonies derived from sacB-labeled PAI V536, respectively, resulted from deletion events of the entire islands or parts of them, whereas in case of PAI IV536, sucrose resistance was based on spontaneous mutations in sacB rather than on deletion of this PAI.
FIG. 2.
Deletion rates of PAI I536 to PAI V536. E. coli 536 derivatives with sacB-labeled PAI I536 to PAI V536 and the control (536 attBλ::sacB bla) were grown overnight in LB medium at 37°C. Deletion rates were calculated as the quotient of Sucr cells divided by CFU. The data are mean values of the results from at least six independent experiments.
In summary, PAI I536-, PAI II536-, PAI III536-, and PAI V536-negative cells arose with frequencies of ∼2 × 10−6, 2 × 10−5, 5 × 10−5, and ∼1 × 10−6, respectively, while PAI IV536 was stable.
PAI I536, PAI II536, PAI III536, and PAI V536, but not PAI IV536, are deleted by site-specific recombination.
Analysis of nonhemolytic clones of E. coli 536 has previously revealed that PAI I536 and PAI II536 can be deleted by site-specific recombination between their flanking DR sequences (16 and 18 bp, respectively). Similar to lambdoid phage excision, one copy of the flanking DRs was deleted with the PAI during excision while the other remained in the chromosome (6). To analyze whether Sucr and PAI-negative colonies of sacB-labeled E. coli 536 derivatives also resulted from precise excision of the respective islands, exclusion PCR assays were designed. They were based on the amplification of the chromosomal junction region after deletion of a PAI. PCRs with primer pairs binding specifically to the flanking regions of PAI I536 and PAI II536, respectively, confirmed that these two islands were deleted site specifically from the chromosome of Sucr and PAI-negative cells.
In contrast to PAI I536 or PAI II536, PAI IV536 is not flanked by DRs. As described previously, E. coli 536 only carries a 17-bp sequence at the 3′ end of asnT that is identical to the 17-bp DRs flanking the HPI in Y. pseudotuberculosis, whereas the repeat at the 3′ end of the island is missing (14, 41). Therefore, it was not too surprising that no precise deletion mutants of PAI IV536 were found. Instead, postexperimental screening of Sucr colonies revealed that deletion of this island occurred very rarely and affected either internal parts of PAI IV536 or the entire island with flanking regions of the chromosome. These data confirmed that PAI IV536 could be considered a stably integrated island.
For PAI III536 and PAI V536, it has been postulated that the associated tRNA gene at the 5′ end and a truncated tRNA gene at the 3′ end terminate these islands (Fig. 1). PAI V536 is flanked by the intact tRNA gene pheV (76 bp) and a partial tRNA gene phe′V (22 bp). phe′V corresponds to the 3′ end of pheV but carries a deletion of 1 bp compared to the sequence of pheV (Schneider et al., unpublished). To test whether deletion of PAI V536 was mediated by recombination between these two repeats, sucrose-resistant and tetracycline-sensitive (Sucr Tets) colonies derived from E. coli strain 536 PAI V536::sacB tetA were analyzed in a PAI V536-specific exclusion PCR. A product of the expected size was amplified from circa 45% of the examined colonies. Cloning and sequencing of this product confirmed that site-specific recombination between pheV and phe′V had caused deletion of the entire PAI V536. The regions preceding and following pheV in PAI V536-negative cells corresponded to the sequences localized immediately upstream and downstream of the PAI in the wild-type strain, indicating that no codeletion of neighboring chromosomal DNA regions had occurred during excision of PAI V536. After deletion, the copy of pheV remaining in the chromosome carries the 1-bp deletion introduced by phe′V during site-specific recombination between the flanking DRs. Since a second gene for a phenylalanine tRNA, pheU, is encoded by the E. coli chromosome, it remains unclear whether the mutated pheV is functional.
Sucr Tets colonies that had been negative in the PAI V536-specific exclusion PCR contained imprecise deletions of the PAI. This was verified by further PAI V536-specific PCRs. In contrast to the above-described site-specific recombination between pheV and phe′V, the detected deletions seemed unspecific, as they encompassed regions of different sizes surrounding the insertion site of the sacB tetA cassette and sometimes even larger areas, including DNA from the core chromosome. However, the frequency of their occurrence was about 10-fold higher than the rate of spontaneous deletions of stable chromosomal regions determined by postexperimental screening of Sucr colonies derived from E. coli strains 536 PAI IV536::sacB and 536 attBλ::sacB bla (data not shown). Sequence comparison of the regions located upstream and downstream of the sacB tetA insertion site in PAI V536 revealed no regions of significant identity that could be responsible for site-specific or homologous recombination leading to the observed deletion types. Hence, they might be caused by the selective pressure for Sucr colonies, but the underlying mechanism for their occurrence has to be investigated.
Similar to PAI V536, PAI III536 is flanked by an intact tRNA gene (thrW, 76 bp) at the 5′ end and a truncated tRNA gene (thr′W) at the 3′ end (14). The DR at the 3′ end encompasses the last 46 bp of thrW and the first 2 bp following downstream in the PAI. With 48 bp, the DRs of PAI III536 are much larger than those flanking PAI I536, PAI II536, or PAI V536 (Fig. 1), which might at least in part account for the higher deletion rate of this island. To investigate whether deletion of PAI III536 is the result of site-specific recombination between thrW and thr′W, we used Sucr colonies derived from E. coli strain 536 PAI III536::sacB as templates in an exclusion PCR with the primer pair ΔPAI III up-ΔPAI III lp (Fig. 1). To our surprise, a product of the expected size was only amplified from 41% of the Sucr cells (Table 2; Fig. 3). Determination of the nucleotide sequence of the PCR product demonstrated that loss of the entire PAI III536 with a size of ∼68 kb was driven by site-specific recombination between thrW at the 5′ end and its truncated copy at the 3′ end. Additional PAI III536-specific PCRs that covered the complete PAI with short intervals verified this result. Thus, this deletion process, which will be referred to as a type I deletion, is restricted to the island itself. It is similar to the deletion mechanism of PAI I536, PAI II536, and PAI V536 and does not affect neighboring sequences of the core chromosome.
TABLE 2.
Distribution of PAI III536 deletion types
| Strain | Conditions | % Type Ia | % Type IIb |
|---|---|---|---|
| 536 PAI III536::sacB | LB, 37°C | 41 | 59 |
| 536 ΔrecA PAI III536::sacB | LB, 37°C | 100 | 0 |
| 536 PAI III536::sacB | M9, 37°C | 73 | 27 |
| 536 PAI III536::sacB | LB, 42°C | 67 | 33 |
| 536 PAI III536::sacB | LB + 2% NaCl, 37°C | 65 | 35 |
| 536 PAI III536::sacB | LB, 20°C | 71 | 29 |
| 536 PAI III536::sacB | AU, 37°C | 100 | 0 |
Percentage of Sucr colonies that were positive in PCR assays with primer pair ΔPAI III up/ΔPAI III lp.
Percentage of Sucr colonies that were positive in PCR assays with primer pair ΔPAI III (2) up/III-24.1rev.
FIG. 3.
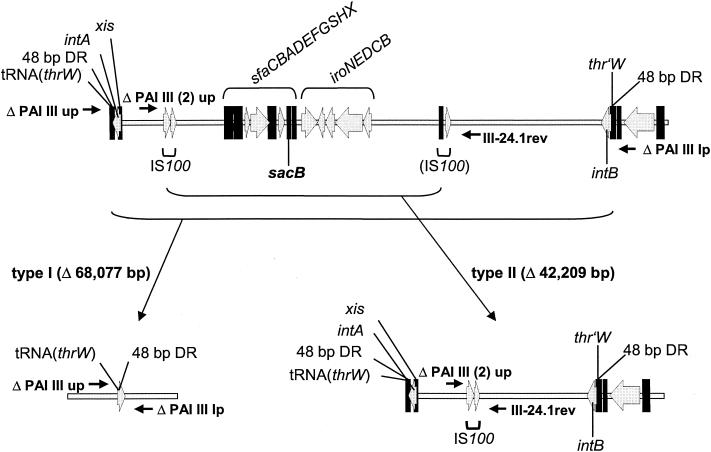
Deletion types of PAI III536. The gene organization in the two types of deletion mutants is shown. Localizations of important virulence genes, PAI-associated integrase genes, direct repeat regions flanking the PAI, IS100 copies, and the insertion site of the counterselectable marker sacB are indicated. Small arrows symbolize primer pairs used for exclusion PCRs. For definitions of abbreviations, see the legend to Fig. 1.
An internal part of PAI III536 can be deleted by homologous recombination.
A screening of the remaining 59% of Sucr colonies derived from E. coli 536 PAI III536::sacB with PCRs covering the island revealed that in contrast to PAI V536 no imprecise deletion had occurred. Instead, all samples tested seemed to have lost the same portion of the island, indicating a second deletion type that covered a shorter part in the center of the PAI including the sacB insertion site (type II deletion). Starting from both ends of PAI III536, nearly 4 kb of the 5′ end and 23 kb of the 3′ end were still present. Sequence comparison of the supposed boundaries revealed the existence of an IS100 element (positions 2124 to 4077 of PAI III536; GenBank accession no. X16664) and a cryptic IS100 element (positions 44870 to 46287) (Fig. 3). The intact copy is nearly identical to the IS100 element of Y. pestis, but a mismatch in the 5′ inverted repeat may have rendered it stable. However, a 1,418-bp region of identical nucleotides in both PAI III536-specific insertion elements could have been the basis for recombination and loss of the internal part of the PAI. A second exclusion PCR assay with primers that were flanking the two IS100 copies was designed to confirm this assumption (Fig. 3). PCR products of the expected size could be amplified from all Sucr colonies that had been negative in the first exclusion PCR assay specific for type I deletion of PAI III536. Cloning and sequencing of the PCR product finally demonstrated that a full-length copy of IS100 remained in the chromosome while the cryptic IS100 element was deleted together with nearly 42 kb of PAI III536.
Whereas the first deletion type of PAI III536 was the result of recombination between 48-bp DRs, type II deletion resulted from recombination between larger regions of sequence identity. This suggested that the second deletion type of PAI III536 depended on RecA, a protein involved in homologous recombination and DNA repair in E. coli. Therefore, a ΔrecA mutant of E. coli strain 536 PAI III536::sacB was constructed, and Sucr colonies were isolated. They arose with slightly lower rates than that of the wild-type strain, suggesting that one of the two deletion types required RecA. To determine which mechanism was affected, Sucr colonies were analyzed in the exclusion PCR assays specific for both deletion types. In contrast to the wild-type strain, where the deletion types appeared with similar frequencies, only type I deletions were detected in the ΔrecA mutant (Table 2). This indicated that deletion type II of PAI III536 was based on homologous recombination between the two IS100 copies, whereas deletions of type I did not require RecA.
RecA is not required for deletion of PAI V536.
Whereas RecA was required for internal deletions of PAI III536, it was not necessary for the site-specific excision of either PAI I536, PAI II536, or the entire PAI III536 (see above) (6, 28). To examine the influence of RecA on the deletion of PAI V536, we constructed a ΔrecA derivative of 536 PAI V536::sacB tetA. The deletion rate of PAI V536 in this strain was comparable to that of the wild-type, but an increase of spontaneous Sucr colonies was observed, probably due to the accumulation of mutations in a strain that is deficient in DNA repair. PCR analysis of Sucr Tets colonies revealed that both precise and imprecise excision of PAI V536 still occurred. However, compared to the wild-type strain, the distribution was altered in the ΔrecA mutant and a slight increase of site-specific deletions was found (data not shown). Therefore, similar to PAI I536 and PAI II536, the site-specific excision of PAI V536 was RecA independent, whereas the imprecise deletion of this island seemed to be moderately influenced by RecA.
Detection of CIs of PAI II536 and PAI III536.
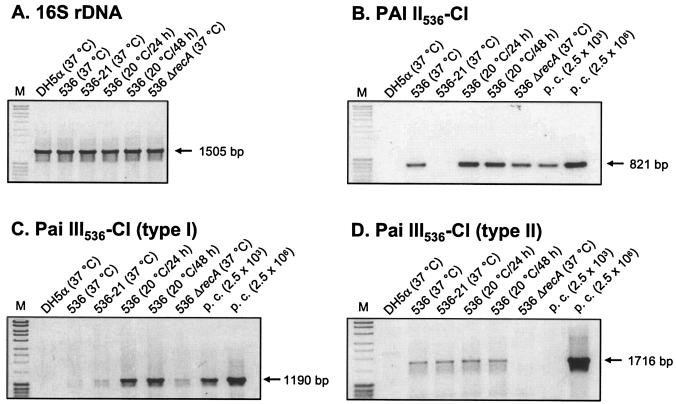
Comparable to the excision mechanism of bacteriophages, PAIs are thought to exist at least transiently as CIs after excision from the chromosome. Since the islands of E. coli 536 contain apparently no origin of replication, it has been hypothesized that CIs get lost upon cell division when they fail to reintegrate into the chromosome. In this study, we tried to detect circular intermediates of E. coli 536-specific PAIs by a sensitive PCR assay. The primer pairs used for these experiments were oriented towards the right and left PAI-chromosome junctions. A PCR product was only amplified after excision and formation of a CI when the primer binding sites were oriented towards each other. The expected products were amplified with primer pairs specific for CIs resulting from site-specific excision of PAI II536 and PAI III536, respectively (Fig. 4B and C). Cloning and sequencing of the PCR fragments confirmed that the CIs contained one copy of the PAI-flanking DRs and that this sequence separated the former ends of the island, now oriented towards each other. Formation of the CIs was independent of RecA, as the amount of PCR product derived from a ΔrecA mutant was comparable to that of the wild type (Fig. 4B and C). CIs corresponding to type II deletion of PAI III536 driven by homologous recombination could also be detected in this assay (Fig. 4D) and were verified by nucleotide sequencing. As expected, no specific product was amplified from a ΔrecA mutant, confirming our previous results.
FIG. 4.
Detection of CIs. Ethidium bromide-stained 1% agarose gels of PCR products amplified from total DNA preparations were isolated from cultures grown for 15 h at 37°C unless indicated differently. (A) 16S ribosomal DNA (rDNA)-specific control PCR. (B) PCR to detect CIs of PAI II536. (C) PCR to detect CIs resulting from excision of PAI III536 by site-specific recombination (type I). (D) PCR to detect CIs resulting from excision of PAI III536 by homologous recombination (type II). Sizes of the PCR products are indicated. M, 1-kb ladder; p.c., defined copy numbers of plasmid controls.
In contrast to PAI II536 and PAI III536, CIs of PAI I536 or PAI V536 could not be detected even if the amount of template DNA or the number of PCR cycles were increased. Therefore, CIs could only be detected for the two islands with a relatively high deletion rate, and the assay was not sensitive enough to detect the probably low numbers of PAI I536- and PAI V536-specific CIs.
Deletion of PAI II536 and PAI III536 is inducible by environmental conditions.
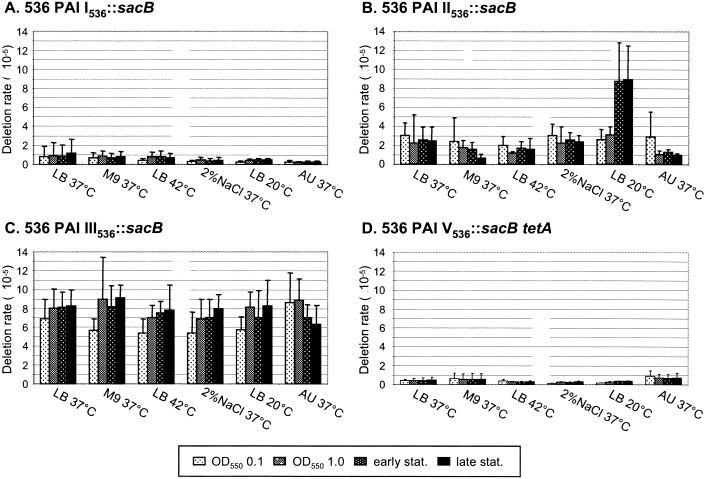
We were interested in whether instability of PAIs in UPEC is influenced by environmental stimuli, such as low or elevated temperature, osmotic stress, nutrient limiting conditions, or growth in AU, which mimics conditions faced by the bacteria in the human host. Therefore, the relative amount of Sucr colonies was calculated at characteristic time points when cells were grown under different conditions. While an increased amount of Sucr cells indicated novel deletion events of a particular PAI, stable values during the entire growth curve pointed out that the fraction of PAI-negative cells in the culture was unchanged and that deletion of the respective island was not stimulated under the indicated condition.
Whereas none of the tested stimuli affected deletion rates of PAI I536 and PAI V536 (Fig. 5A and D), E. coli strain 536 PAI II536::sacB responded to growth in LB medium at 20°C with a threefold-higher frequency of Sucr colonies compared to the deletion rate at 37°C (Fig. 5B) (28). More than 95% of the Sucr colonies showed reduced hemolytic activity, indicating complete loss of PAI II536. A more-detailed analysis of the deletion rate of PAI II536 between the mid-log and early stationary phases revealed that excision of this island was induced during the transition from the logarithmic to the stationary phase, i.e., when maximum cell densities (optical density at 550 nm > 2.0) were reached. Furthermore, higher amounts of PAI II536-specific CIs were detected in cells grown at low temperatures for 24 and 48 h than in cells grown at 37°C, thereby reflecting an induced excision rate of PAI II536 at 20°C (Fig. 4B). However, during stationary-phase growth, no further deletion events seemed to occur. Both stimuli, low temperature and high cell density, were responsible for the increased number of PAI II536-negative cells, since a similar effect was not observed when bacteria were grown in LB medium at 37 and 42°C (Fig. 5B) or when the culture reached the stationary phase at an optical density at 550 nm of <2.0 (data not shown). Similarly, high salt concentrations, nutrient limitation, or growth in AU did not induce deletion of PAI II536. However, when 536 PAI II536::sacB was grown in M9 minimal medium, the deletion rate of PAI II536 was decreased in the late stationary phase (Fig. 5B). This could point out that PAI II536-negative cells are deficient in growth in M9 minimal medium and are outcompeted by the wild type, but the difference between values was not statistically significant. So far, growth differences between PAI-negative and PAI-positive cells have not been observed. Finally, sequence analysis of PAI II536 revealed no genes whose deletion would account for growth deficiency.
FIG. 5.
Influence of environmental factors on deletion of PAI I536, PAI II536, PAI III536, and PAI V536. All strains were cultivated under the indicated conditions. Samples were taken during late lag, mid-log, early stationary, and late stationary phase. The deletion rate was calculated as the quotient of Sucr cells divided by CFU. All data are the results from at least six independent experiments. Evaluation of data by paired t test revealed that the increased deletion rate of PAI II536 in LB at 20°C is statistically significant (P < 0.5).
Similar to PAI I536 and PAI V536, the deletion rate of PAI III536 seemed to be unaffected by the tested environmental conditions. Only slightly increased numbers of Sucr colonies could be observed during growth in M9 minimal medium, in LB medium at low or high temperatures, or under salt stress (Fig. 5D). However, a closer analysis of the distribution of PAI III536-specific deletion types revealed a difference (Table 2). Whereas the basic ratio of type I to type II was unaltered when cells were grown in LB medium at 37°C, deletion type I was increased to more than 60% of the Sucr cells after incubation at 42°C or in presence of 2% NaCl. In M9 minimal medium or after growth at 20°C, this effect was even more pronounced and the ratio of deletion types changed to circa 70 to 30%. Comparable to PAI II536, the increase of type I deletion in cells grown at low temperatures could be confirmed with the PCR assay designed to detect CIs of PAI III536 that originate from site-specific excision (Fig. 4C). Finally, when cells were grown in AU, only Sucr colonies that had lost PAI III536 by type I deletion events were isolated (Table 2). In summary, PAI III536 was not only the island with the highest deletion rate under all tested conditions but it was also susceptible to all environmental stimuli tested in vitro. The observed shift in the distribution of PAI III536-specific deletion types was the result of either induced type I deletion or decreased type II deletion. Since PAI III536 is deleted by site-specific and homologous recombination, an influence of the environmental conditions on recA and the PAI III536-associated integrase genes has to be evaluated in future experiments.
DISCUSSION
The capacity of bacteria to modulate their genome structure is an important feature for adapting to changing environmental conditions. Genome flexibility also has an impact on the evolution of new bacterial pathogens. The acquisition of new traits by horizontal gene transfer is one of the driving forces in the emergence of new bacterial variants, but point mutations, gene rearrangements, or loss of genetic information also play crucial roles (15, 27, 31). In this context, it has been interesting to find that instability seems to be a characteristic of PAIs in particular pathogens, including UPEC (6, 16, 33). Furthermore, other PAIs have been identified that share the tendency to be deleted from the bacterial chromosome (2, 7, 9, 34, 43, 44). In this study, we applied the island-probing approach to analyze for the first time the (in)stability of all so far identified PAIs of one pathogenic strain in detail.
The deletion rates of 10−6 to 10−5 that were obtained for PAI I536 to PAI V536 were similar to the rates previously reported for other PAIs (34, 43, 44). With the exception of PAI III536, where a second deletion mechanism based on homologous recombination between a complete and a truncated IS100 copy was detected, excision and subsequent deletion of PAIs in E. coli 536 is mediated by site-specific recombination between short flanking DR sequences. It can be speculated whether the length of the flanking DRs is one reason for the observed differences in the stability of E. coli 536-specific PAIs. However, a direct connection between the size of flanking repeats and excision frequency can be ruled out by comparing PAI I536 and PAI II536, which are both flanked by DRs of similar length (16 and 18 bp, respectively) but are deleted with rates differing nearly 10-fold. In case of PAI V536, the mismatch in the DR located at the 3′ end of the island (phe′V) may decrease the efficiency of site-specific recombination, resulting in a relatively low deletion rate. Interestingly, the identified mismatch is also present in the 3′ DR of the LEE in REPEC strain 84/110-1 (43). Similar to PAI V536, the relatively low deletion rate of this island may be ascribed to the imperfect DR. As it is unlikely that the same mismatch developed independently and spontaneously by point mutation in the 3′ DR of both islands, the REPEC LEE and PAI V536, it can be speculated that it has been already present in a precursor of the two islands. Finally, PAI IV536 is stably integrated into the chromosome of strain 536, apparently due to deletion of one of the flanking DR sequences, as it has also been reported for Y. enterocolitica Ye8081 (2, 41). In contrast, HPIs of Y. pestis and Y. pseudotuberculosis are relatively unstable. In Y. pestis, the HPI core element is deleted together with the flanking pigmentation segment by homologous recombination between two IS100 elements comparable to type II deletion of PAI III536, and in Y. pseudotuberculosis, deletion of the HPI core region is due to site-specific recombination between 17-bp DRs (7, 8).
Besides the length and integrity of the flanking DRs, the levels and activities of PAI-associated integrases probably contribute to the frequency of excision of PAIs in E. coli 536. It has been shown that P4-like integrases of the REPEC LEE or the HPI of Y. pestis mediate site-specific integration of small artificial DNA fragments corresponding to an attP site into all asn tRNA loci or into pheU, respectively (35, 43). Preliminary data indicate that integrases of PAIs in E. coli 536 also trigger deletion of their islands. Therefore, it will be interesting to investigate their activity in more detail in future experiments.
Our data support the hypothesis that stabilization of PAI I536 and PAI V536 is an ongoing process, since both islands have decreased deletion rates compared to PAI II536 and PAI III536. It has been hypothesized that PAIs have been acquired by horizontal gene transfer followed by integration into the bacterial genome (15, 17). If their genetic features turn out to be advantageous for the host organism, they are subjected to selective pressure, thereby favoring mutations that render the islands stable. Examples are the HPI of Y. enterocolitica or some islands of Salmonella enterica that seem to have lost all former traits of mobility (2, 17, 36).
It has been speculated that the loss of virulence determinants may play a crucial role during the transition from an acute state of disease to chronic infection (6, 17). Therefore, in addition to the mechanisms underlying the deletion of PAIs in E. coli 536, we were particularly interested in examining the influence of environmental conditions on their stability. We confirmed previous results that deletion of PAI II536 is significantly induced at 20°C (28) and found that both low temperature and high cell density contribute to this effect. In contrast, other stimuli tested, such as growth at high temperature, salt stress, depletion of nutrients, or growth in AU did not change the deletion rate of PAI II536. One habitat, where bacteria are exposed to low temperature as well as a high density of cells, is the complex community of naturally occurring biofilms. Therefore it can be speculated that such growth conditions facilitate excision and mobilization of PAIs, e.g., by transducing phages that may be induced within biofilms. Interestingly, the mobile islands V. cholerae PAI and S. aureus PAI 1 are both transmissible to recipient organisms via phage transduction, and it would be interesting to analyze whether their transfer rates are also modulated by environmental parameters. In the case of PAI III536, the deletion rate was not significantly altered by environmental stimuli, but comparable to PAI II536, growth at low temperatures affected the excision of this island and resulted in a shift of PAI III536-specific deletion types to site-specific recombination (deletion type I). The same effect could be observed when cells were grown in M9 minimal medium, at high temperature, or under salt stress, and it was most pronounced in AU, indicating that modulation of PAI stability could be important in the environment and in the host during infections.
It is believed that loss of PAIs is preceded by excision and circularization of the respective islands (6). Detection of CIs of PAI II536 and PAI III536 with a specific PCR assay confirmed this assumption. Even though a precise quantification of CIs was not possible with this assay, the results corroborated the findings that environmental stimuli have an impact on the deletion rate of at least two PAIs of E. coli 536. Excision of PAIs generates less-virulent variants, and it remains to be studied in more detail which role deletion of PAIs plays in natural habitats of UPEC strains. In contrast to PAI II536 and PAI III536, no modulation of the deletion rate and no CIs were found in the case of PAI I536 and PAI V536, again suggesting that these islands are relatively stably inserted into the chromosome.
The basis for enhanced deletion of PAI II536 and PAI III536 is yet unknown. It can be speculated that island-associated integrase genes are differentially regulated in response to environmental stimuli, thereby altering the frequency of excision. As the elevated deletion rate of both islands is closely connected to high cell density, quorum sensing may play a role for this effect. It has recently been shown that the time when autoinducer 2 of the quorum-sensing circuit in E. coli is produced overlaps with the time point when deletion of PAI II536 and PAI III536 is induced (3, 19). Additionally, the alternative sigma factor of the stationary growth phase (σs) is another candidate that may play a role in the increased deletion of PAIs in E. coli 536 (20).
Future experiments will focus on the role of integrases and on the impact of environmental stimuli during deletion of PAIs. Finally, it remains to be studied in more detail which role deletion of PAIs plays in natural habitats of UPEC strains.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB479), the Fonds der Chemischen Industrie, and the Bayerische Forschungsstiftung.
We thank E. Carniel for kindly providing plasmid pGP704 fyuA::sacB, G. Schneider for the nucleotide sequence of PAI V536, and A. Siegl for technical assistance.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith. 1991. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bach, S., C. Buchrieser, M. Prentice, A. Guiyoule, T. Msadek, and E. Carniel. 1999. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect. Immun. 67:5091-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, B. L. 2002. Small talk. Cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 4.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 6.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchrieser, C., R. Brosch, S. Bach, A. Guiyoule, and E. Carniel. 1998. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 30:965-978. [DOI] [PubMed] [Google Scholar]
- 8.Buchrieser, C., M. Prentice, and E. Carniel. 1998. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol. 180:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration into the λ attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 12.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Fünfstück, and J. Hacker. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U., and J. Reidl. 2000. Pathogenicity islands and phage conversion: evolutionary aspects of bacterial pathogenesis. Int. J. Med. Microbiol. 290:519-527. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, J., L. Bender, M. Ott, J. Wingender, B. Lund, R. Marre, and W. Goebel. 1990. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8:213-225. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 18.Hacker, J., S. Knapp, and W. Goebel. 1983. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154:1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardie, K. R., C. Cooksley, A. D. Green, and K. Winzer. 2003. Autoinducer 2 activity in Escherichia coli culture supernatants can be actively reduced despite maintenance of an active synthase, LuxS. Microbiology 149:715-728. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, N.Y.
- 22.Hochhut, B., K. Jahreis, J. W. Lengeler, and K. Schmid. 1997. CTnscr94, a conjugative transposon found in enterobacteria. J. Bacteriol. 179:2097-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou, Y.-M. 1999. Transfer RNAs and pathogenicity islands. Trends Biochem. Sci. 24:295-298. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaolis, D. K. R., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 26.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 35:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Middendorf, B., G. Blum-Oehler, U. Dobrindt, I. Mühldorfer, S. Salge, and J. Hacker. 2001. The pathogenicity islands (PAIs) of the uropathogenic Escherichia coli strain 536: island probing of PAI II536. J. Infect. Dis. 183:S17-S20. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 31.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 32.O'Shea, Y. A., and E. F. Boyd. 2002. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol. Lett. 214:153-157. [DOI] [PubMed] [Google Scholar]
- 33.Ott, M. 1993. Dynamics of the bacterial genome: deletions and integrations as mechanisms of bacterial virulence determination. Zentbl. Bakteriol. 278:457-468. [DOI] [PubMed] [Google Scholar]
- 34.Rajakumar, K., C. Sasakawa, and B. Adler. 1997. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun. 65:4606-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rakin, A., C. Noelting, P. Schropp, and J. Heesemann. 2001. Integrative module of the high-pathogenicity island of Yersinia. Mol. Microbiol. 39:407-415. [DOI] [PubMed] [Google Scholar]
- 36.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter, W. D., P. Palm, and S. Yeats. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyrat, J. M., V. Pelicic, B. Gicquel, and R. Rappuoli. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 66:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruzin, A., J. Lindsay, and R. P. Novick. 2001. Molecular genetics of SaPI1-a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 41:365-377. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schubert, S., A. Rakin, D. Fischer, J. Sorsa, and J. Heesemann. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409-414. [DOI] [PubMed] [Google Scholar]
- 42.Skerra, A. 1994. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151:131-135. [DOI] [PubMed] [Google Scholar]
- 43.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 44.Turner, S. A., S. N. Luck, H. Sakellaris, K. Rajakumar, and B. Adler. 2001. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183:5535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]