Abstract
G4 motifs are greatly enriched near promoters, suggesting that quadruplex structures may be targets of transcriptional regulation. Here we show, by ChIP-Seq analysis of human cells, that 40% of the binding sites of the transcription-associated helicases, XPB and XPD, overlap with G4 motifs. The highly significant overlap of XPB and XPD binding sites with G4 motifs cannot be explained by GC-richness or parameters of the genomewide analysis, but instead suggests that these proteins are recruited to quadruplex structures that form in genomic DNA (G4 DNA). Biochemical analysis demonstrates that XPD is a robust G4 DNA helicase, and XPB binds to G4 DNA. XPB and XPD are enriched near the transcription start site (TSS) at 20% of genes, especially highly transcribed genes. XPB and XPD enrichment at G4 motifs characterizes specific signaling pathways and regulatory pathways associated with specific cancers. These results identify new candidate pathways for therapies targeted to quadruplexes.
The G4 sequence motif, G3N1−xG3N1−xG3N1−xG3, confers the ability to form G-quadruplex or G4 DNA. In the G-quadruplex structure, four guanines hydrogen bond to form a planar ring, a G-quartet, and stacking of the hydrophobic G-quartets stabilizes the quadruplex structure1,2. Specific G4 motifs and G-quadruplex structures have been shown participate in critical cellular processes including telomere maintenance, recombination and replication3. However, the presence of a G4 sequence motif only identifies potential for formation of a quadruplex structure, and relatively little is known about which of the many G4 motifs in the human genome form quadruplex structures in a living cell, or how this relates to genomic function. Genomewide analysis provides one potentially powerful approach to address that question.
Regions flanking the transcription start site (TSS) and at the 5' end of first introns are particularly enriched in G4 motifs, as are some genes, most notably oncogenes4–7. G-quadruplex structures form spontaneously upon transcription of regions bearing G4 motifs in vitro or in vivo8. This suggested that some of the G4 motifs near the TSS form quadruplex structures that contribute to transcriptional regulation, a possibility that has stimulated efforts to identify those regulatory factors and to develop therapeutic small molecule ligands that can bind to quadruplexes and stimulate or inhibit their regulatory activities9,10. Some of these efforts have focused on specific G4 motifs at promoters of genes of immediate clinical interest, such as the oncogenes c-MYC or c-KIT11,12.
Quadruplex structures may present challenges to both transcription and replication. Unresolved quadruplexes may block progression of RNA polymerase II and thereby inhibit gene expression13,14. They may also promote genomic instability. ChIP-Seq has shown that γ-H2AX, an endogenous marker for double-strand breaks, accumulates at genes bearing G4 motifs in human cells treated with the G4 ligand pyridostatin, and that accumulation is transcription-dependent, consistent with transcription-induced formation of quadruplexes15.
The enrichment of G4 motifs near the TSS and evidence for transcription-induced formation of quadruplex structures suggests that factors that recognize and resolve G-quadruplexes might be associated with the general transcription apparatus. TFIIH is a general transcription factor, with functions in transcription initiation and nucleotide excision repair (NER)16–19. TFIIH contains 11 subunits, two of which are the highly conserved superfamily 2 helicases, XPB and XPD20,21. XPB and XPD are absolutely essential in human cells and in most eukaryotes. Their importance and the wide range of pathways they may regulate is evident from the diverse spectrum of symptoms evident in the three human genetic diseases identified with specific mutant alleles of XPB and XPD22,23. Xeroderma pigmentosum (XP) is characterized by defective NER, UV-sensitivity and predisposition to cancer, especially skin cancer and melanoma. Trichothiodystrophy (TTD) is characterized by a broad spectrum of symptoms, including hair abnormalities, ichtyosis, physical and mental retardation, and progeria. Cockayne syndrome (CS) is characterized by developmental defects, progeria and neurological degeneration.
The functions of XPB and XPD in human cells cannot be directly tested, as their essential functions preclude the use of standard mutation or depletion approaches. Nonetheless, XPD defines a small family of helicases with three paralogs in human cells, FANCJ, CHL1 and RTEL121, and analysis of these highly related proteins provides a very useful insight into possible function, as FANCJ and CHL1 are bona fide G4 DNA helicases, and RTEL1 is very likely to unwind G4 DNA. FANCJ associates with the replication apparatus and unwinds quadruplexes that might otherwise inhibit replication24–27. RTEL1 appears to perform an analogous function at the telomeric repeats28, and CHL1 unwinds G4 DNA and promotes chromosome segregation29. The G4 helicase activities of the XPD paralogs raised the possibility that XPD might bind to quadruplex structures or function as a G4 helicase.
We have tested the possibility that XPD and/or XPB might recognize or resolve G-quadruplex structures by mapping genomewide associations of these factors by ChIP-Seq in human cells; and by complementary biochemical analysis to define the activities of these highly conserved proteins in vitro. Here we show that 40% of XPB and XPD binding sites overlap with G4 motifs (p=0); that XPD binds quadruplexes and has robust G4 helicase activity; and that XPB specifically binds to but does not unwind G4 DNA. XPB and XPD are associated with 1.8% of the G4 motifs in the human genome, identifying those G4 motifs as potential sites of formation of G-quadruplex structures in living cells. XPB and XPD enrichment at G4 motifs characterizes specific signaling pathways and regulatory pathways associated with specific cancers, suggesting that binding to quadruplex structures may recruit XPB and/or XPD to these pathways. These results support the view that quadruplexes may form in human promoter regions and function as targets of transcriptional regulation by XPB and XPD.
RESULTS
XPB and XPD are greatly enriched at G4 motifs
We mapped genomewide associations of XPB and XPD in human cells by ChIP-Seq, using highly specific commercial monoclonal antibodies (Supplementary Results, Supplementary Fig. 1) to target the endogenous, wild-type proteins in HT1080 cells, a fibrosarcoma line with stable diploid karyotype. After mapping paired-end fragments, peaks for XPB and XPD were calculated using Model Based Analysis for ChIP-Seq (MACS)30. Using a p-value cutoff of 1e-15, we identified 21,554 XPB peaks and 14,570 XPD peaks. The average lengths of XPB peaks were 890 bp (median = 833 bp), and of XPD peaks were 998 bp (median = 937 bp). Many peaks directly overlapped: 22.4% of XPB peaks and 39.2% of XPD peaks.
We searched for overlaps of XPB or XPD peaks with the G4 consensus motif, G≥3N1–12G≥3N1–12G≥3N1–12G≥3, which includes all G4 motifs with loops from 1–12 nt in length. This loop length was chosen because analyses of G4 motifs near replication origins in human cells31 suggested that biologically relevant motifs may be characterized by loop lengths longer than the 1–7 nt used in many previous analyses, and 1–12 nt loops are well within the range for stable G-quadruplex formation in vitro32. The human genome contains 722,264 G4 motifs as defined by this criterion, which cover 0.9% of genomic sequence. Remarkably, 40% of XPB and XPD peaks overlapped G4 motifs (p=0; Table 1). Genomewide mapping showed that a highly significant fraction of XPB and XPD peaks mapped within 1 kb of TSS (16%; p=0), as predicted for components of TFIIH (Supplementary Fig. 2). Graphs of the distribution of XPB and XPD peaks within 1 kb of the TSS identified strong peaks near the initiation site, as predicted by the functions of these factors as components of TFIIH (Supplementary Fig. 3a). Calculation of the distance between the points of highest enrichment (summits) of XPB and XPD peaks showed that enrichment was frequently coincident (Supplementary Fig. 3b). Binding sites that did not map to the TSS were distributed among RefSeq genes (43.4% of XPB and 43.6% of XPD peaks) and within regions not classified as RefSeq genes (39.6% of XPB and 39.2% of XPD peaks), which include non-transcribed regions with regulatory functions as well as many transcribed non-coding RNAs. Overlap of XPB and XPD peaks with G4 motifs was comparable among annotated genes and non-RefSeq regions (Supplementary Table 1).
Table 1.
XPB and XPD peaks are significantly enriched for G4 motifs.
| Region | Coverage of Region | Percent of Peaks with G4 | Percent of G4 Motifs in Peaks | Binomial p-value | ||
|---|---|---|---|---|---|---|
| G4 Motifs | Peak Sets | |||||
| Whole Genome | 0.90% | XPB | 0.67% | 40.49% | 1.88% | 0 |
| XPD | 0.51% | 42.95% | 1.40% | 0 | ||
| TSS ± 1 kb | 4.06% | XPB | 4.47% | 63.81% | 0.55% | 0 |
| XPD | 3.33% | 65.42% | 0.39% | 0 | ||
| >1 kb from TSS | 0.82% | XPB | 0.58% | 35.71% | 1.33% | 0 |
| XPD | 0.44% | 38.28% | 1.01% | 0 | ||
A reported p-value of 0 is indicative of a result that is very unlikely to occur by chance, and reflects a calculated result below the minimum value that can be represented by the analysis software (p≤2–1024). We performed additional controls to establish that this overlap did not reflect details of the analysis. (1) To ensure that the enrichment of XPB and XPD at G4 motifs did not reflect affinity of these factors for regions of high GC content, rather than G4 motifs, we counted the instances of G4 motif overlaps with randomly selected regions of equal GC content to XPB and XPD peaks. We found that XPB and XPD peaks were far more likely to contain G4 motifs than regions with matched GC content (>18-fold more near the TSS (±1 kb), and >13-fold more elsewhere; Table 2 and Supplementary Table 2). Thus, enrichment at G4 motifs is independent of GC content of XPB and XPD peaks. (2) To ensure that results were independent of the size of the XPB or XPD peaks identified by ChIP-Seq, the search for overlaps was repeated using more stringent criteria, by confining the search to regions within 50 bp of the site of maximal binding (peak summits) rather than including entire peaks (p<1e-23; Supplementary Table 1). With this constraint, XPB and XPD enrichment at G4 motifs was highly significant both near the TSS (±1 kb; p<1e-46), and elsewhere (p<1e-54; Table 2). (3) To ensure that statistical significance did not reflect the 1–12 nt loop size used in the algorithm that identified G4 motifs, we performed the same G4 enrichment tests but scoring only motifs with 1–7 nt loops, which reduces the total number of sites identified as G4 motifs approximately 2-fold. Results retained very high statistical significance (p<1e-92; Table 2 and Supplementary Table 1).
Table 2.
Tests of G4 enrichment parameters show that significance of enrichment of G4 motifs in XPB and XPD peaks is robust with regards to GC content, peak length, and G4 motif specifications.
| Region | ChIP | Random Matched GC% | Reduced Peak Size | 1–7 nt Loops | |||
|---|---|---|---|---|---|---|---|
| % Random with G4 | ChIP / Random | % Peaks with G4 | Binomial p-value | % Peaks with G4 | Binomial p-value | ||
| TSS±1 kb | XPB | 3.37% | 18.94 | 10.88% | 7.17e-59 | 38.45% | 0 |
| XPD | 3.34% | 19.58 | 11.33% | 7.64e-47 | 39.89% | 0 | |
| >1 kb from TSS | XPB | 2.62% | 13.61 | 6.47% | 5.19e-80 | 18.07% | 5.96e-310 |
| XPD | 2.39% | 16.00 | 6.67% | 7.79e-55 | 19.98% | 2.00e-243 | |
We conclude that the overlap of XPB and XPD motifs with G4 motifs, at the TSS and elsewhere, is extremely unlikely to reflect a chance occurrence, and cannot be explained by affinity for G-rich regions rather than G4 motifs, or parameters of the genomewide analysis. Instead, it is very likely to reflect an important aspect of genomic biology. The most straightforward functional explanation would be that either or both of these factors binds to G-quadruplex structures, as tested below.
XPD unwinds G4 DNA
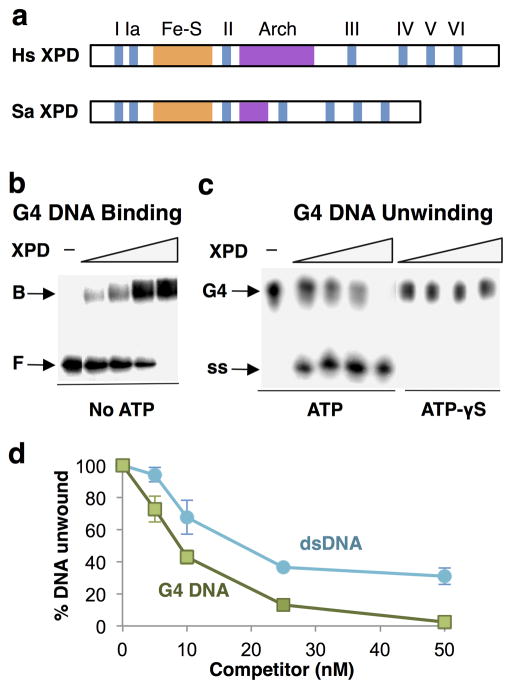
XPD is a 5’-3’ helicase, and its helicase, Fe-S and Arch domains are conserved from archeae to humans (Fig. 1a). XPD from the thermophilic crenarchote, Sulfolobus acidocaldarius (SaXPD) shares helicase, Fe-S and Arch domains with human XPD (HsXPD) and is highly related in sequence (26% identical, 46% similar; BLASTP search), and it has been used extensively for biochemical and structural analyses20,21,33–35. To ask if XPD binds or unwind G4 DNA, we took advantage of this and analyzed the recombinant thermophilic enzyme expressed in E. coli. This enabled us to study protein purified from extracts that had been heat-treated to denature endogenous proteins, eliminating potential artifacts caused by contamination with other G-quadruplex binding proteins. Purified recombinant SaXPD bound G4 DNA (Fig. 1b) and exhibited robust G4 DNA unwinding activity in the presence of ATP but not ATP-γS (Fig. 1c). G4 DNA competed G4 DNA unwinding activity more effectively than a DNA duplex bearing a 5’ overhang previously shown to be an excellent XPD unwinding substrate34 (Fig. 1d). XPD is therefore a robust ATP-dependent G4 DNA helicase.
Figure 1. XPD is a robust G4 DNA helicase.
(a) Domain structure of human (Hs) and S. acidocaldarius (Sa) XPD, showing conserved helicase motifs I–VI and Fe-S and Arch domains. (b) Gel mobility shift assay of binding by XPD (0, 25, 50, 100 and 200 nM) to 32P-G4 DNA. Arrows indicate bound (B) and free (F) DNA. (c) Assay of unwinding of 32P-G4 DNA by XPD (0, 25, 50, 100 and 200 nM) in the presence of 1 mM ATP (left) or 1 mM ATP-γS (right). Arrows indicate G4 DNA substrate and single-stranded (ss) products of unwinding. (d) Competition of 32P-G4 DNA unwinding by indicated amount of unlabeled G4 DNA or dsDNA.
XPB binds G4 DNA
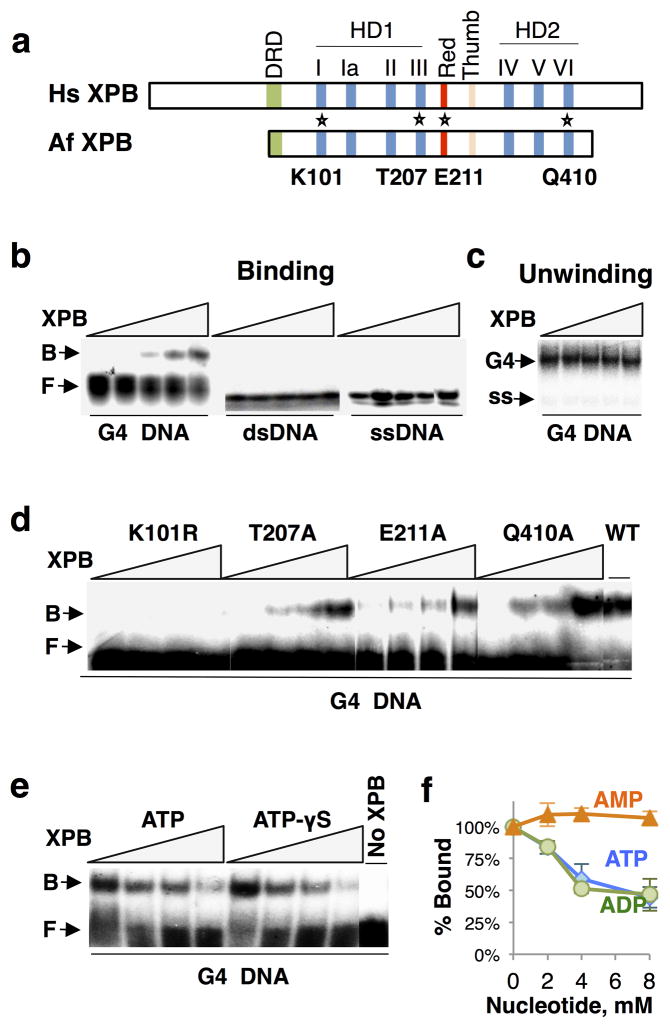
XPB is a 3'-5' helicase. XPB from the thermophile Archaeoglobus fulgidus (AfXPB) shares conserved helicase, DRD, Red and Thumb domains with the human enzyme (HsXPB; Fig. 2a), and is highly related in sequence (27% identical, 44% similar; BLASTP search). As with XPD, we took advantage of conservation and assayed the activity of recombinant thermophilic enzyme, expressed in E. coli and purified from extracts that had been heat-treated to eliminate possible artifacts caused by contaminants with G-quadruplex binding activity. We found that AfXPB bound to G4 DNA in vitro, but not to dsDNA or ssDNA (Fig. 2b). AfXPB did not unwind G4 DNA (Fig. 2c).
Figure 2. XPB binds but does not unwind G4 DNA, and binding is ATP-sensitive.
(a) Domain structure of human (Hs) and A. fulgidus (Af) XPB, showing conserved helicase domains (HD1 and HD2), helicase motifs I–VI, damage recognition domain (DRD), Red and Thumb domains. Positions of point mutations analyzed are indicated with asterisks. (b) Gel mobility shift assay of binding by XPB (0, 4, 8, 16 and 32 nM) to 32P-end-labeled G4 DNA, dsDNA and ssDNA DNA. Arrows indicate bound (B) and free (F) DNA. (c) Assay of unwinding of 32P-G4 DNA by XPB (0, 4, 8, 16 and 32 nM). (d) Gel mobility shift assay of binding by indicated AfXPB point mutants (0, 25, 50 and 100 nM) or wild-type XPB (WT, 25 nM) to 32P-labeled G4 DNA. (e) Gel mobility shift assay of binding by XPB (50 nM) to 32P-labeled G4 DNA in the presence of ATP or ATP-γS (0, 2, 4 and 8 mM). (f) Quantification of inhibition of XPB binding to G4 DNA by ATP, ADP or AMP.
A point mutation abolishes XPB G4 DNA binding activity
The specificity of AfXPB binding to G4 DNA relative to dsDNA or ssDNA was striking, and led us to ask whether G4 DNA binding might be altered by XPB mutation. We tested four mutants associated with human disease, activity of which has been characterized in human cells and also in the yeast, Saccharomyces pombe. The XPB ATPase activity is essential for both NER and promoter opening, and AfXPB K101R is mutant at the highly conserved lysine in the Walker A ATPase (motif I) essential to ATP hydrolysis, corresponding to human XPB (HsXPB) K346R and S. pombe (SpXPB) K354R mutations17,36–40. AfXPB E211A is mutant at a conserved glutamate in the RED domain, where mutation in human XPB (HsXPB E473A) affects both transcription and NER40; and AfXPB T207A and AfXPB Q410A mutations affect helicase activity but not NER, corresponding to HsXPB T469A and HsXPB Q639A and SpXPB T478A and SpXPB Q647A38,39. The AfXPB K101R mutation abolished G4 DNA binding, but mutations at other positions had modest if any effect (Fig. 2d). This is the first point mutation in any protein shown to abolish G4 DNA binding activity. The dramatic effect of this mutation is unlikely to be due to deleterious consequences to overall structure, because similar mutations at the conserved lysine in the Walker A ATPase motif of a number of proteins have been shown to affect ATPase activity without causing larger-scale structural changes34,39,40.
The dramatic effect of the XPB K101R mutation in the ATPase active site suggested that ATP might regulate G4 DNA binding. Binding of wild type AfXPB to G4 DNA was indeed reduced in the presence of 2 mM ATP, and essentially abolished by 8 mM ATP (Fig. 2e). ATP-γS similarly inhibited G4 DNA binding, so inhibition did not depend upon ATP hydrolysis (Fig. 2e). Binding was also inhibited by ADP, but not by AMP, with a Ki of approximately 4 mM (Fig. 2f). These results suggest that XPB binding to quadruplex structures may be modulated or regulated by ATP.
XPB and XPD bind near the TSS of highly expressed genes
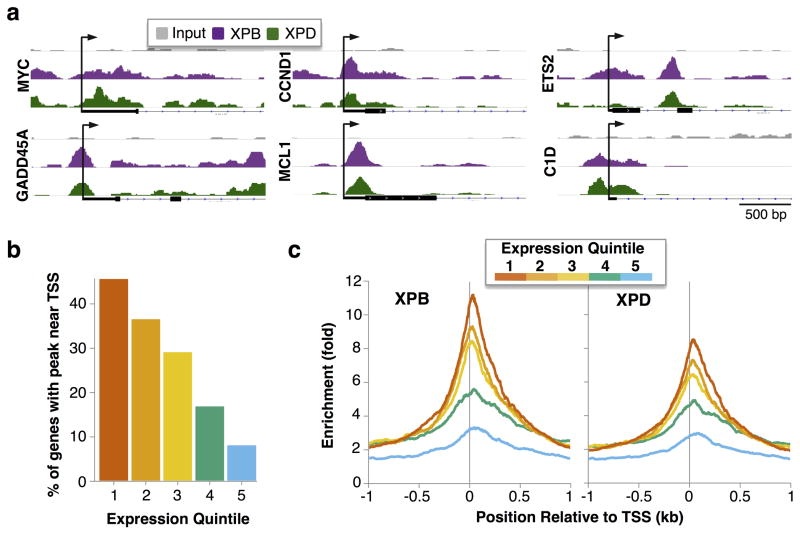
Maps of associations of XPB and XPD at six representative individual genes showed that these factors are enriched near the TSS (Fig. 3a). We asked if XPB and XPD association correlated with gene expression, by first assigning expressed genes to five equal quintiles based on expression level as determined by microarray analysis (Supplementary Data Set 1), then determining the fraction of genes in each quintile at which XPB or XPD were enriched near the TSS (±1 kb; Supplementary Data Set 2). Over 40% of the genes in the top expression quintile were enriched for XPB or XPD, and fewer in the lower quintiles, with enrichment apparent at <10% of genes in the lowest expression quintile (Fig. 3b). A plot of the average enrichment of XPB and XPD at each position relative to the TSS, for genes in each expression quintile, showed that XPB and XPD were most highly enriched near TSSs of the most highly expressed genes (Fig. 3c). XPB and XPD enrichment near the TSS correlated closely with gene expression levels, and nearly half the genes in the top quintile of expression were stably associated with XPB and/or XPD. Retention of these factors following initiation of one round of transcription may enable immediate reinitiation that maintains promoter activity. We note that over half (60%) of the most highly expressed genes were not highly enriched for either XPB or XPD. Thus, although XPB and XPD binding correlated with gene expression at many genes, high level gene expression does not cause or require continuous stable association of these factors.
Figure 3. Enrichment of XPB and XPD at TSS is correlated with transcription.
(a) Selected fragment overlap maps near individual TSS. (b) Percent of genes in each expression quintile with enrichment of XPB or XPD near the TSS (±1 kb). TSSs were binned into quintiles based on gene expression (1 to 5, high to low), as determined by exon array analysis. (c) Fold enrichment of XPB and XPD fragment overlaps relative to Input near the TSS (±1 kb) for genes in each expression quintile.
G4 binding may recruit XPB and XPD to specific pathways
We used the Genomic Regions Enrichment of Annotations Tool (GREAT)41 to query XPB and XPD peaks that contained G4 motifs within 50 bp of the peak summit to determine whether XPB and XPD peaks are overrepresented in regulatory domains from genes in diverse ontologies, including regulatory domains up to 1 Mb from the TSS. This analysis identified highly significant associations of XPB peaks containing G4 motifs with genes that respond to stimulation by EGF or fibronectin; with genes upregulated in response to UV; with genes involved in ATR, RAC1, and RhoA signaling pathways, which are frequently dysregulated in cancer (Table 3); with targets of C-MYC, HIF-1 and P53 (Table 3; Supplementary Data Set 3); and with other genes associated with specific cancers (Supplementary Data Set 3). Similarly significant results were obtained if GREAT analysis was restricted to regulatory domains within 200 kb of the TSS (Supplementary Table 3). These results suggest that XPB and/or XPD may be recruited to specific genes or to genes in specific pathways via interactions with G-quadruplex structures, in which case these pathways may be candidates for therapies directed to quadruplex targets.
Table 3.
G4 motifs enriched for XPB and XPD correlate with cancer-associated ontologies. Selected GREAT ontology results for XPB and XPD peaks that contain G4 motifs.
| Database | Ontology | Peaks containing G4 motifs | |
|---|---|---|---|
| XPB | XPD | ||
| MSigDB | EGF stimulation | **** | ** |
| Fibronectin stimulation | **** | *** | |
| UVB up-regulation | **** | **** | |
| Pathway Commons | ATR signaling | ** | |
| RAC1 signaling | ** | ||
| RhoA signaling | ** | ||
| C-MYC | * | ** | |
| HIF-1α | * | ||
| p53 | ** | ||
Binomial p-value:
<1e-4;
<1e-8;
<1e-12;
<1e-16.
DISCUSSION
The results reported here provide strong evidence that G-quadruplex structures are targets of XPB and XPD in human cells. Genomewide analysis showed that approximately 40% of XPB and XPD peaks overlapped G4 motifs (p=0), and that XPB and XPD were especially enriched at a subset of highly expressed genes. Biochemical analysis showed that both XPB and XPD bind to G4 DNA, and that XPD is a robust G4 helicase. A p-value approaching 0 is a compelling indicator of significance of a finding. Overlap of sites of XPB and XPD association cannot be explained by G-richness of binding sites, or by parameters of the genomewide analysis.
These results suggest that XPB and XPD bind to quadruplex structures formed at some promoters to regulate transcription. The G4 helicase activity of XPD was predicted by analogy with its paralogs, FANCJ, RTEL1 and CHL1. One way in which XPD may function is by resolving quadruplex structures that might otherwise impair transcription or replication, analogous to the function of its paralog, FANCJ, in replication26,27. The G4 DNA binding activity of XPB was not as clearly anticipated. The XPB/G4 DNA interaction appears to involve a region of the protein closely associated with ATPase activity, as binding was abolished by a lysine to arginine mutation in the Walker A box ATPase active site, or by incubation with ATP or ADP, raising the possibility that ATP or ADP might regulate binding in vivo. The colocalization of XPB and XPD as part of the larger TFIIH complex in eukaryotes may provide a platform for coordinated binding and unwinding of G4 structures. Though XPD is dispensable for the transcription initiation functions of TFIIH, it is required for DNA repair, and may be required for unwinding of G4 structures that are recognized by XPB.
The binding targets of XPB and XPD comprise 1.8% of all G4 motifs in the human genome (Table 1). Helicases are moving targets, so a map of genomewide associations of a wild-type G4 helicase may underestimate the fraction of sites bound. Genomewide associations of another G4 helicase, S. cerevisiae Pif1, were previously determined using a mutant derivative that lacked helicase activity in order to avoid that potential complication42. XPD and Pif1 helicases share a conserved ATPase domain, and the K264A mutation used to inactivate S. cerevisiae Pif1 for those experiments was at the same invariant active site lysine as the AfXPB K101R mutation that totally abolished XPB G4 DNA binding (Fig. 2d). This suggests that Pif1 and XPB depend on distinct structural domains to bind to G4 DNA. No structure has yet been determined of a G4 helicase/G4 DNA complex, and this is an important challenge for the future.
GREAT analysis identified unanticipated enrichment of XPB and XPD at G4 motifs near specific pathways associated with disease, particularly cancer. These results have novel implications for understanding how mutations in XPB and XPD may contribute to the genetic disease, xeroderma pigmentosum (XP). The highly significant association (p<1e-12) of XPB and XPD at genes upregulated following UV irradiation, when most transcription is shut down43, suggests a role in regulation of the complex transcriptional response to UV, which could contribute to the development of skin cancer characteristic of XP patients. Similarly, associations of XPB and XPD at genes in the RAC1 pathway, which is associated with melanoma44–46, raises the possibility that transcriptional dysregulation may contribute to the development of this cancer, which occurs at 100-fold elevated frequency in XP patients. More generally, GREAT analysis (Supplementary Data Set 3) identified additional disease-associated pathways at which XPB and XPD are likely regulators. Some of these may be candidates for future development of drugs targeted to quadruplexes.
Why are XPB and XPD enriched at some G4 motifs and not others? Transition of a G4 motif in duplex DNA to a four-stranded G-quadruplex structure requires local DNA denaturation, which can be caused by transcription, replication, or local changes in superhelicity. All G4 motifs are unlikely to simultaneously adopt the quadruplex configurations necessary for XPB and XPD binding. In addition, quadruplex structures are very diverse, with conformation determined by lengths of runs of G's and of loops of non-G’s between G-runs, parallel or anti-parallel orientation of the strands, and orientation of the bases in the G-quartets47. XPB and XPD may bind to only some quadruplex structural conformations. Such structural selectivity would enable XPB and XPD to be targeted to specific sites, genes and pathways.
The map of genomewide associations of the G4 DNA binding proteins, XPB and XPD, represents an important step toward deciphering the significance of G4 motifs and the biological roles of quadruplexes. The quadruplex structures that participate in key processes can be thought of as comprising the “G4 genome”, analogous to the transcriptome, proteome or metabolome3. In this view, DNA functions not only as a simple linear alphabet but also a more complex architecture. The ChIP-Seq analysis reported here establishes a functional map of the G4 genome, by locating G4 motifs that are bound by two of the factors shown to interact with G-quadruplex structures in vitro. It should be possible to extend our understanding of the G4 genome by analogous analysis of other endogenous proteins that bind to G4 DNA, including RecQ family helicases such as human BLM and WRN. Additional landmarks in the G4 genome may be mapped using other tools, including small molecule ligands that cause endogenous proteins to stably associate with genomic sites15 or quadruplex-binding antibodies48,49.
ONLINE METHODS
ChIP-Seq
HT1080 cells were cultured in Minimum Essential Medium Eagle, Alpha Modification (MEM_) with 5% fetal bovine serum, penicillin and streptomycin. ChIP and library preparation were performed essentially as described previously50. ChIP was carried out with anti-XPB antibodies (NB100–61060, Novus Biologicals) at 1:200, and anti-XPD antibodies (sc-20696, Santa Cruz Biotechnology) at 1:50; and Western blotting with anti-XPB antibodies (sc-293, Santa Cruz Biotechnology) and anti-XPD antibodies (H00002068-M01, Novus Biologicals) at 1:800. The anti-XPB and anti-XPD antibodies both effectively and specifically recognized their respective targets (Supplementary Fig. 1). For library preparation, a second round of size selection was performed following 10 cycles of preamplification to exclude molecules <200 bp. Libraries were sequenced to 101 nt in paired-end mode using an Illumina Hi-Seq 2000 (J. Shendure, University of Washington).
ChIP-Seq alignment, peak calling, and identification of peak summits
Raw ChIP-Seq reads were trimmed to 60 nt, and filtered to remove pairs in which at least one mate had more than 3 positions with a PHRED score below 20. The remaining high-quality reads were mapped to the UCSC build hg19 (GRCh37) human genome using Bowtie v0.12.7 with the settings -n 0, -m 1, and --best in paired-end mode51. Paired-end alignments mapped to the genome comprised 64,279,152 from Input, 17,199,527 from XPB ChIP, and 21,976,361 from XPD ChIP. Peak locations were called using Model-based Analysis for ChIP-Seq (MACS) v1.4 with default settings and a p-value cutoff of 1e-1530. A randomly selected subset of the Input alignments equal in number to the XPB and XPD alignment sets were used as negative controls for peak calling. MACS identified 21,555 XPB and 14,570 XPD peaks genomewide.
Mapped paired-end reads were converted to BEDGraph format by calculating the number of overlapping fragments at each position in the genome using a Perl script. For rapid analysis of genomic targets, the BEDGraph files were converted to binary format using the UCSC bedGraphToBigWig tool52. “Summits” were identified using a Perl script that utilized the BioPerl Bio::DB::BigWig module53 to find the position with the highest fragment overlap within each peak called by MACS.
Comparison of XPB and XPD to G4 motifs and RefSeq genes
The hg19 human genome in FASTA format and the RefSeq gene table were downloaded from the UCSC genome browser database54. Regions ±1 kb from the TSS were selected using Perl scripts. The Cis-regulatory Element Annotation Tool (CEAS) was used to calculate enrichment near many parts of RefSeq genes55. G4 motifs were identified using QuadParser56 with settings for 4 or more runs of 3 or more Gs or Cs with 1–12 nt or 1–7 nt loops between runs (GC 3 4 1 12 and GC 3 4 1 7, respectively), and results in DAS format were converted to BED format using Perl scripts. The longer search allowed loops between G-runs of 1–12 nt in length to capture both short intrastrand G4 structures that readily form in vitro32 and longer structures that may form in vivo31,57. Perl scripts were then used to count the number of times G4 motifs were found in XPB or XPD peaks, the number of times peak summits overlapped regions near TSS, and the number of overlaps between XPB and XPD peaks. Binomial tests were performed using a locally installed version of the GREAT Calculate Binomial P-Value tool41. Images of individual TSS were captured using Integrated Genomics Viewer v2.3.558.
Deconvolution of GC% enrichment and G4 motif enrichment
A pool of 250,000 randomly selected regions was generated by concatenating 12 copies of the 21,554 XPB peaks and randomly reassigning chromosome and start locations while keeping length constant. The resulting regions were trimmed to 250,000 entries, and the sequences of each region were retrieved from the hg19 genome using the UCSC twoBitToFa tool52. A Perl script was used to calculate the GC% for each random region and each XPB and XPD peak. Perl scripts were also used to flag regions and peaks that overlapped regions within 1 kb of TSS and G4 motifs. To determine if XPB and XPD GC% differed from randomly selected regions, all 250,000 random regions were used to calculate expected GC% for four categories: Regions within 1 kb of the TSS with or without G4 and Regions >1 kb from the TSS with or without G4. T-tests were performed using R (http://www.r-project.org/) to determine if the mean GC% among XPB or XPD peaks differed from the expected distribution found among randomly selected regions. This analysis showed that XPB and XPD peaks have greater average GC content than random regions (t-test p-values < 1e-23; Supplementary Table 2), with the exception of peaks near TSS that also overlap G4 motifs, which have significantly lower GC content than random regions (p-values < 1e-25; Supplementary Table 2). We then randomly sampled a subset of the 250,000 random regions that was equal in number to XPB or XPD peaks and had matching GC% (rounded to the nearest whole percentage point). We counted the number of random regions in each TSS/G4 category as described above, and calculated the fold enrichment of XPB and XPD peaks relative to the random regions as the percent of peaks in each overlap category that contained G4 motifs divided by the percent of randomly selected regions in each category that contained G4 motifs (Supplementary Table 2).
Expression arrays
Four biological replicates of total RNA from actively dividing HT1080 cells were harvested and purified using TRIzol (Invitrogen). At least 6 μg from each replicate were shipped to Ocean Ridge Biosciences for further processing and analysis using the Affymetrix GeneChip Human Exon 1.0 ST array platform. Results from core transcript clusters were normalized using an RMA algorithm, and filtered based on detection of RMA-normalized Exon-level probe sets. Of 22,011 Core Transcript Clusters, 14,767 were detected as expressed (>50% of exons detected), and were used for subsequent analysis. For quintile expression analysis, the normalized, log2 expression values for each gene were averaged across all 4 replicates, and ranked from highest to lowest expression score; and genes divided into 5 equal groups of 2,953 genes based on those scores (Supplementary Data Set 1). Locations of gene TSS ±1 kb were selected using RefSeq table entries with matching gene names.
Ontology enrichment analysis
Regions ±50 bp of peak summits that overlapped G4 motifs were identified by comparing G4 locations to summit region locations (Supplementary Table 2). XPB and XPD peaks identified with this overlap were submitted to the GREAT v2.0.2 server with default settings for the hg19 genome41. The top 10 results in the GO Biological Process, Disease Ontology, Pathway Commons, MSigDB Perturbation, and MSigDB Predicted Promoter Motifs categories are presented in Supplementary Dataset 3.
Expression vectors, protein expression and purification
Cloned A. fulgidus XPB (from John Tainer, Scripps) was expressed in E. coli BL21 (DE3) cells in the pET-28b expression vector at 28°C and purified as described37. Lysates were pre-cleared by DEAe-cellulose chromatography and protein batch purified by Ni-affinity (Sigma) and Q-Sepharose Fast Flow (GE Healthcare). Cloned S. acidocaldarius XPD (from Malcolm White, St. Andrews University, Scotland) was expressed in E. coli in the pET-28c vector and purified as described59. Fractions were concentrated using Amicon centrifugal concentrators, MW cutoff of 30,000 kD. Purified XPD was aliquoted for single use, aliquots flash frozen in liquid nitrogen then topped off with argon gas for storage. Similarity to human proteins was determined by a Standard Protein BLAST search (blastp at http://blast.ncbi.nlm.nih.gov/) using the human XPB or XPD sequence to query A. fulgidus and S. acidocaldarius protein databases for the best possible match.
All point mutations were made using Quikchange XL site-directed mutagenesis kit (Stratagene), with the following oligonucleotides and their complements:
XPB K101R: 5’-TCCAACAGGCTCAGGAAGAACTCACGTTGCAATG
XPB T207A: 5’-CTGGGACTGACGGCAGCTTTCGAAAGGGAGG
XPB Q410A: 5’-GCGAGGGAGTACATCGCGCGACTCGGGAGAAT
XPB E211A: 5′-CAACTTTCGAAAGGGCGGATGGAAGGCATGA
DNA substrates and enzyme assays
G4 DNA was formed from the TP oligonucleotide, 5’-TGGACCAGACCTAGCAGCTATGGGGGAGCTGGGGAAGGTGGGAATGTGA, a prototypical binding substrate derived from the murine S2a heavy chain switch region sequence2,60, by incubation of 1 mg/ml oligonucleotide in TE, 0.9 M NaCl and 0.1 M KCl overnight at 60°C followed by native gel purification, then stored in TE containing 40 mM KCl. ssDNA was formed from the oligonucleotide 5’-AGAATGGAGCAGGTGCGCTGCGAGCAGTG. dsDNA was formed by annealing the ssDNA substrate to its complement. Substrates for binding and unwinding assays were 5’-32P end-labeled; competitor DNAs were unlabeled.
Binding was analyzed in 20 μl reactions containing 25 mM Tris-HCl (pH 7.9), 50 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 100 μg/ml BSA, 0.5 nM 5’-32P-labeled DNA, and indicated quantities of enzyme for 15 min at 45°C (XPD) or 20 min at 50°C (XPB), followed by addition of 5% v/v glycerol and resolution by electrophoresis on 5% polyacrylamide gels in 1XTBE containing 10 mM KCl. XPB DNA binding was not dependent upon Mg++, and this cation was omitted from reactions analyzing the effects of ATP and other adenine nucleotides on binding to avoid chelating nucleotide. Helicase assays were carried out in 20 μl reactions containing 20 mM MES, pH 6.5, 1 mM DTT, 1 mM Mg-ATP, 100 μg/ml BSA; monovalent cation (40 mM NaCl, 5 mM KCl) was included upon addition of enzyme (0–200 nM) and DNA substrate (5 nM). After incubation for 15 min at 45°C (XPD) or 20 min at 50°C (XPB), reactions were terminated by addition of 5 μl Stop (10 mM Tris-Cl, pH 8.0, 5 mM EDTA and 2 mg/ml Proteinase K); incubated for 10 min at 37°C; and products resolved by 6% native polyacrylamide gel electrophoresis in 1X TBE containing 40 mM KCl. Labeled DNAs were visualized by Storm phosphorimager and quantified by ImageQuant software (GE healthcare). Uncropped versions of all gels are available as Supplementary Fig. 4.
Data and scripts
Sequencing data and processed files (including MACS peak-finder output) for XPB and XPD ChIP-seq are available at Gene Expression Omnibus (GEO) study GSE44849. All scripts required to replicate analysis of G4, ChIP-seq, and expression data analyses are available at http://code.google.com/p/graylt-2013/.
Supplementary Material
Acknowledgments
Research reported in this manuscript was supported by National Institutes of Health National Cancer Institute P01 CA077852. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank our Program Project colleagues for active discussions; Malcolm White (St. Andrews University, Scotland) for the XPD expression construct; John Tainer (Scripps Research Institute, La Jolla, CA) for the XPB expression construct; Arnold Bailey for help with sequencing library preparation and Choli (Charlie) Lee and Jay Shendure for sequencing of ChIP-seq libraries on an Illumina Hi-Seq 2000 (University of Washington, Seattle, WA).
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
Contributions
L.T.G. performed ChIP-Seq and expression array experiments and analyzed those results. A.C.V. and J.E. performed biochemical studies of XPB and XPD. L.T.G., A.C.V., J.E., and N.M. conceived the study and designed experiments. L.T.G. and N.M. interpreted the data and wrote the manuscript.
References
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eddy J. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Z, Zhao Y, Li N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009;37:6784–6798. doi: 10.1093/nar/gkp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy J, Maizels N. Selection for the G4 DNA motif at the 5' end of human genes. Mol Carcinog. 2009;48:319–325. doi: 10.1002/mc.20496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.la Faverie de AR, et al. Nucleic acids targeted to drugs: SELEX against a quadruplex ligand. Biochimie. 2011;93:1357–1367. doi: 10.1016/j.biochi.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 11.González V, Hurley LH. The C-terminus of nucleolin promotes the formation of the c- MYC G-quadruplex and inhibits c- MYC promoter activity. Biochemistry. 2010;49:9706–9714. doi: 10.1021/bi100509s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei D, Parkinson GN, Reszka AP, Neidle S. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012;40:4691–4700. doi: 10.1093/nar/gks023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez REL, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nature Chemical Biology. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksenych V, Coin F. The long unwinding road: XPB and XPD helicases in damaged DNA opening. Cell Cycle. 2010;9:90–96. doi: 10.4161/cc.9.1.10267. [DOI] [PubMed] [Google Scholar]
- 17.Egly JM, Coin F. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 2011;10:714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 18.Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 20.Fan L, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White MF. Structure, function and evolution of the XPD family of iron-sulfur-containing 5‘-->3’ DNA helicases. Biochem Soc Trans. 2009;37:547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto S, Egly JM. Trichothiodystrophy view from the molecular basis of DNA repair/transcription factor TFIIH. Hum Mol Genet. 2009;18:R224–30. doi: 10.1093/hmg/ddp390. [DOI] [PubMed] [Google Scholar]
- 23.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 24.London TBC, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Shin-ya K, Brosh RM. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkies P, et al. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39:1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM. Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem. 2012;287:1007–1021. doi: 10.1074/jbc.M111.276022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besnard E, et al. Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 32.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Research. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubaele S, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudolf J, Rouillon C, Schwarz-Linek U, White MF. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucleic Acids Res. 2010;38:931–941. doi: 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 37.Fan L, et al. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat Struct Mol Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 39.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Oksenych V, de Jesus BB, Zhovmer A, Egly JM, Coin F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 2009;28:2971–2980. doi: 10.1038/emboj.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paeschke K, Capra JA, Zakian VA. DNA Replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LHF. DNA damage response and transcription. DNA Repair (Amst) 2011;10:743–750. doi: 10.1016/j.dnarep.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 44.Bauer NN, Chen YW, Samant RS, Shevde LA, Fodstad O. Rac1 activity regulates proliferation of aggressive metastatic melanoma. Exp Cell Res. 2007;313:3832–3839. doi: 10.1016/j.yexcr.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr Opin Struct Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–6. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam EYN, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun. 2013;4:1796–8. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray LT, Fong KK, Pavelitz T, Weiner AM. Tethering of the conserved piggyBac transposase fusion protein CSB-PGBD3 to chromosomal AP-1 proteins regulates expression of nearby genes in humans. PLoS Genet. 2012;8:e1002972. doi: 10.1371/journal.pgen.1002972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stajich JE, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–9. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 56.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Decorsière A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3'-end processing and function during DNA damage. Genes Dev. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJR, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 60.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.