Abstract
The improvement of long-term transplant organ and patient survival remains a critical challenge following kidney transplantation. Proteomics and biochemical profiling (metabolomics) may allow for the detection of early changes in cell signal transduction regulation and biochemistry with high sensitivity and specificity. Hence, these analytical strategies hold the promise to detect and monitor disease processes and drug effects before histopathological and pathophysiological changes occur. In addition they will identify enriched populations and enable individualize drug therapy. However, proteomics and metabolomics have not yet lived up to such high expectations. Renal transplant patients are highly complex, making it difficult to establish cause-effect relationships between surrogate markers and disease processes. Appropriate study design, adequate sample handling, storage and processing, quality and reproducibility of bioanalytical multi-analyte assays, data analysis and interpretation, mechanistic verification and clinical qualification (=establishment of sensitivity and specificity in adequately powered prospective clinical trials) are important factors for the success of molecular marker discovery and development in renal transplantation. However, a newly developed and appropriately qualified molecular marker can only be successful if it is realistic that it can be implemented in a clinical setting. The development of combinatorial markers with supporting software tools is an attractive goal.
Keywords: Biomarkers, metabolomics, proteomics, qualification, renal transplantation, verification
Introduction
Although there has been marked progress in terms of long-term kidney graft survival over the last decades, this improvement has mostly been driven by better short-term graft survival, while long-term attrition is only slowly improving [1]. The reasons for kidney graft loss are multi-factorial and include, but are not limited to, tubular injury due to ischemia-reperfusion injury, acute and subacute persistent rejection (potentially accompanied by destructive multinuclear infiltration and chronic interstitial fibrosis), chronic cellular rejection (with or without antibody component), viral infections, immunosuppressant toxicity, glomerosclerosis, and recurrent glomerulonephritis [2–4]. These may be promoted by donor and recipient risk factors and diseases [2]. Current clinical monitoring of kidney transplant patients is usually based on serum creatinine, blood pressure measurements, therapeutic drug monitoring of immunosuppressants, urinalysis, and in the case of kidney dysfunction, ultrasound and biopsies [2]. Clinical markers such as serum creatinine are less than optimal largely because they only become abnormal in the later stages of kidney injury when therapeutic interventions may be less effective and less likely to result in complete reversal of the injury [5]. It has been shown that up to 30% of grafts with stable creatinine may have “smoldering” rejection [6,7]. All major diseases of renal allografts change patterns over time from early diagnostic lesions to late non-specific patterns [8]. Since the procurement of a kidney biopsy is often guided by a rise of creatinine levels, biopsies are usually taken at a late time point when the disease process has already caused significant damage and is already driven by secondary disease processes such as inflammation and fibrosis. Moreover, markers such as serum creatinine do not reveal information regarding the causation or exact location of said injury. This constitutes a dilemma as treatment should be targeted towards the dominant pathophysiological diagnosis [2].
Current clinical chemistry and biochemistry diagnostics is usually based on a limited set of biomarkers, often only one parameter that is closely correlated with one functional aspect of the organ in question or with a specific disease process. However, there is nor will there ever be a single molecular entity marker such as serum creatinine that captures the function of the kidney in all its complexity. Modern analytical technologies allow for the identification of molecular signatures that confer significantly more information than the measurement of a single parameter, just as a bar code contains more information than a single number. Proteomics and metabolomics complement genomics and are considered phenotypic molecular markers [9].
Proteomics is multi-analyte technology that in an ideal case can assess the complete set of proteins (proteome) in a specific matrix. In analogy, metabolomics is multi-analyte technology that in an ideal case can assess the complete set of small molecules (defined as <1500 Da; metabolome) in a specific matrix (Table 1). For detailed reviews of current metabolomics [9–11] and proteomics technologies please see references [12–15]. In contrast to the genome and proteome that are restricted to an individual organism, the metabolome is an open system and not only measures the downstream products of multiple proteins and genes but also interactions with food, the environment and the gut microbiolome [16]. Metabolomics and proteomics can be viewed as ultra-high throughput clinical chemistry/biochemistry, assessing hundreds and sometimes thousands of metabolites or proteins in a single analytical run [11,16,17].
Table 1. Definitions.
| Biomarker | A characteristic objectively measured and evaluated as an indicator of normal biological processes or pharmacological responses to a therapeutic intervention. Type 0 biomarker: a marker of the natural history of a disease that correlates longitudinally with known clinical indices Type I biomarker: a marker that captures the effects of a therapeutic intervention in context with its mechanism of action Context independent: developed for general clinical and pre-clinical testing Context specific: developed in association with a drug development program and, accordingly, to study and monitor the effects of specific drugs. They may enter the market together with a specific drug as a “companion diagnostic device” [65]. Antecedent: Identifying the risk of developing an illness Screening: screening for subclinical disease Diagnostic: recognizing overt disease Staging: categorizing disease severity Monitoring: assessing disease progression, therapeutic efficacy and adverse effects Prognostic: predicting future disease course/response to therapy |
| Clinical end point | A characteristic or variable that reflects how a patient feels, functions or survives. Intermediate (non-ultimate) end point: a true clinical endpoint, a symptom or measure of function but not the ultimate end-point of the disease Ultimate end point: survival or the rate of other or irreversible morbid events |
| Surrogate end point | A biomarker intended to substitute for a clinical endpoint aiming to predict clinical benefit or harm or lack of benefit or lack of harm on the basis of epidemiological, therapeutic, pathophysiological or other scientific evidence. |
| Metabolome | A quantitative descriptor of all endogenous low-molecular-weight components in a biological sample such as urine or plasma. Each cell type and biological fluid has a characteristic set of metabolites that reflects the organism under a particular set of environmental conditions and that fluctuates according to physiological demands. The metabolome can be divided into the primary metabolome (as controlled by the host genome) and the co-metabolome (dependent on the microbiome). |
| Metabonome | Theoretical combinations, sums and products of the interactions of multiple metabolomes (primary, symbiotic, parasitic, environmental, and co-metabolic) in complex systems. |
| Metabolomics | The comprehensive quantitative analysis of all the metabolites of an organism or a specific biological sample. |
| Metabonomics | The quantitative measurement over time of the metabolic responses of an individual or population to a disease, drug treatment or other challenge. |
| Microbiolome | The consortium of microorganisms, bacteria, protozoa, and fungi that live commensally or symbiotically with a host. |
| Xenometabolome | Characteristic profile of non-endogenous compounds such as drugs, their metabolites and their excipients, dietary components, herbal medicines and environmental exposure. |
| Proteome | The expressed protein and peptide complement of a cell, organ or organism, including all isoforms and post-translational variants. |
| Proteomics | The systematic analysis of proteins for their identity, quantity and function. |
| Genomics | Genomics is the study of the genome (approx. 25,000 genes). It is static and allows for estimate the risk for an individual to develop a disease, may modify the efficacy or tolerability of a drug, or influence its tissue distribution and pharmacokinetics. |
| Transcriptomics | Transcriptomics is also known as functional genomics and is the study of expression patterns of all gene transcripts (approx. 85,000). |
Strategies to maintain transplant function and improve long-term graft survival are important goals of translational research [2]. The development of omics technologies has opened up many new opportunities [18].
As the status of metabolomics and proteomics-based molecular marker development in transplantation has already extensively been reviewed recently [11,19–30], it is our goal to focus here on the basic concepts and challenges rather than on providing another comprehensive and detailed overview of relevant clinical trials.
Opportunities
Cells either directly or indirectly (via extracellular fluid) communicate with body fluids. Cell metabolites, peptides, proteins, and extracellular membrane vesicles (or microparticles) are released from cells or are taken up from body fluids by trans-membrane diffusion or transport, and throughout the death process cells release all of their contents into body fluids. Thus at least to a certain extent, biochemical and protein changes in cells and organs are reflected in body fluids. While tissue samples, biopsies, and certain fluids such as urine (kidney), bile (liver) and cebrospinal fluid (CNS) mainly reflect changes in specific organs and thus are considered “proximal matrices”; plasma samples reflect systemic changes that often cannot be traced back to a certain organ [9]. Such changes of metabolites, peptides and proteins in body fluids, if mechanistically linked to disease processes and drug effects in tissues and organs, have the potential to serve as surrogate markers or biomarkers. A biomarker is defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or pharmacological responses to therapeutic intervention” [31] (for additional relevant definitions please see references [19,31–37] and Table 1). Based on this definition biomarkers have been used all along in clinical diagnostics ranging from the measurement of clinical symptoms such as blood pressure, ECG, over imaging technologies to modern high throughput gene arrays. Since here we discuss technologies that directly or indirectly assess molecular mechanisms, the more focused term “molecular marker” will be used instead of “biomarker”. A molecular marker can consist of the measurement of a single molecular entity but it can also be a set of several molecular entities, a molecular pattern or fingerprint.
Proteomics and metabolomics have the potential to impact and improve renal transplantation on several levels:
Assessment of molecular mechanisms of disease and drug effects in in vitro, animal and clinical studies. This may lead to better understanding of disease mechanisms, drug effects and toxicities as well as to the identification of new therapeutic targets.
Drug development. It is reasonable to expect that the availability of specific and sensitive molecular markers will impact drug development such as that of novel immunosuppressants and immunosuppressive drug regimens as follows [38]: i) faster and more efficient pre-clinical and early clinical development; ii) selection of lead drug candidates with a better therapeutic index; iii) earlier and more sensitive detection of toxic effects; iv) monitoring of pharmacodynamics and toxicodynamics during preclinical and clinical development; v) development of more efficient and predictive animal models; vi) identification of pharmaco- and toxicodynamic mechanisms; vii) access to indications that cannot be assessed by conventional strategies due to time limitations, such as chronic disease processes or disease prevention; viii) guidance of dose finding studies; ix) better long-term safety and efficacy; x) stratification of patient populations during clinical trials; xi) support dose finding studies, xii) identification of “enriched” populations with better chance of therapeutic efficacy and tolerability; xiii) provide new supporting or surrogate study endpoints and xiv) diagnostic tools for clinical management of drugs in clinical practice.
Clinical diagnostics. As discussed above, well-qualified molecular marker protein and/or metabolite patterns yield more detailed and mechanistically relevant information than currently often used single markers, translating into good specificity. The better the specificity of a molecular marker pattern, the more this will reduce non-specific background noise. Reduced background noise, usually, results in better sensitivity. This will allow for detection and identification of changes in molecular signatures associated with disease processes and drug effects in body fluids such as plasma and urine before symptoms and irreversible injury occur. As discussed previously [39,40], the development of a disease or response to drug or toxin can be divided into three major stages: genetic predisposition, biochemical stage and the symptomatic stage. While current diagnostic tools such as serum creatinine and histopathology evaluation of renal biopsies detect disease processes and drug toxicity, usually in the symptomatic stage when injury is often not fully reversible anymore, molecular markers based on protein and/or metabolite patterns have the potential to detect changes already during the biochemical phase when cells start compensating for a challenge and injury and repair are still in equilibrium and pathobiochemical process can often still be fully reversed [39,40]. Thus, it is reasonable to expect that diagnostic strategies based on protein and/or metabolite panels will provide the key to significant improvements, such as enabling individualized and promoting preventive medicine, as well as therapeutic interventions and/or treatment adjustments at a time point when no permanent injury has occurred yet.
Challenges
To better understand the challenges involved in discovering molecular markers and with developing those into clinical diagnostic tools, it is important to briefly revisit proteomics and metabolomics marker development strategies and regulatory requirements.
Non-targeted versus targeted
One of the strengths of omics technologies is that they allow for nonbiased screening of a large number of gene regulation processes, signal transductions, and metabolic pathways. Non-targeted screening technologies have the goal to capture as much unfiltered and nonbiased data as possible. The molecular entities underlying the recorded signals are often unknown. This dramatically minimizes the chance of missing important data or, in other words, reducing the risk of false-negative results. However, as a consequence, the largest problem with non-targeted screening technologies such as proteomics and metabolomics, are false-positive results. Targeted assays are often multi-analyte assays and measure well-defined compounds. Analytical technologies include, but are not limited to, antibody and aptamer arrays, bead immunoassays for proteins and liquid chromatography-mass spectrometry assays (for proteins or metabolites) that allow for the assessment of molecular marker panels ideally in a single run.
Targeted assays are usually semi-quantitative or quantitative and can be validated. Although the quality of the results is much better understood, these assays are limited in terms of their ability to detect unknown effects. Since only selected compounds are measured, targeted analyses are inherently biased.
Regulatory Aspects
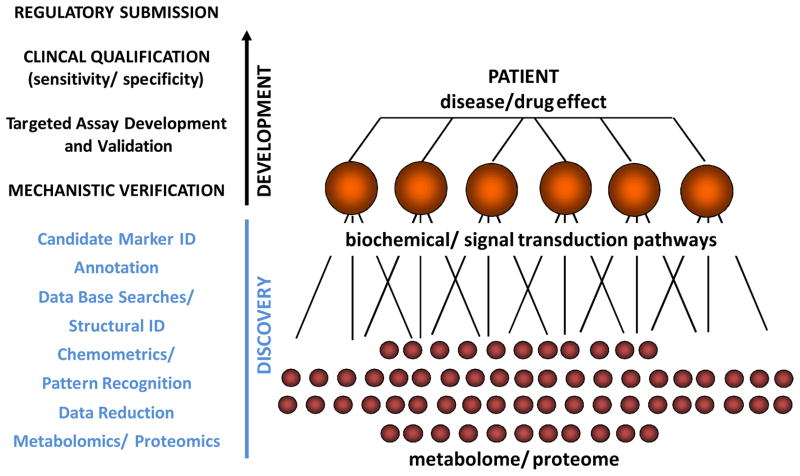
Regulatory agencies have established review structures as well as guidances that outline the biomarker development process [41–47] that can be divided into three stages: Discovery, Verification and Qualification (Figure 1).
Figure 1. Steps During Discovery and Development of Molecular Markers.
Typically discovery involves the non-targeted analysis of the metabolome and/or proteome of the samples. This may result in hundreds and sometimes thousands of data points. The next step is to extract relevant data and to identify candidate surrogate markers of a disease process or drug effect. Making sense of the data is just as important as the bioanalytics and may be even more challenging and time consuming. Chemometrics is defined as the application of mathematical and statistical methods to chemistry. Chemometric analyses are necessary in order to develop statistical pattern recognition models, achieve optimal characterization of the samples and detect biomarkers from diverse, highly dimensional omics datasets [38]. After based on these analyses markers have been selected and their structures have been identified, it is necessary to assess if the markers truly are surrogates for the effect of a disease or drug. Annotation seeks to link the changes in molecular markers to biochemical and cell regulatory pathways. Verification is to establish a mechanistic link between specific markers and a disease process or the pharmacodynamic/toxicodynamic drug effects. Once the candidate markers have been mechanistically verified, their clinical utility in terms of their sensitivity and specificity to detect and/or predict a disease process, clinical outcome or drug response needs to be established (qualification). For this purpose it is often necessary to establish validated bioanalytical assays. Clinical qualification often requires the conduct of prospective multi-center clinical trials. If qualification is successful and a molecular marker shows good specificity and sensitivity, regulatory submission and approval may be required in most cases before the molecular marker can widely be used as a clinical diagnostic tool [64].
Discovery (Figure 1)
Discovery often involves the comparison of samples from a patient population with those of appropriate controls using non-targeted metabolomics and/or proteomics approaches. The typical steps of a non-targeted discovery strategy are described in Figure 1 (please see also references [48–59]). Non-targeted assays result in signal patterns (“fingerprints”) that are analyzed using chemometric methods [49,51,52]. Chemometrics involves statistical pattern recognition models and detection of molecular markers from diverse, highly dimensional omics datasets. More targeted analytical strategies such as protein arrays, bead-based multi-analyte immunoassays (for proteins) or magnetic resonance spectroscopy (MRS, for metabolites) may be used. In contrast to non-targeted assays, the identities of the signals are often already known here. If successful, the result of this process is a molecular marker candidate or a set of candidate molecular markers. As discussed below, it is important to note that this is just the first step towards developing a molecular marker or a molecular marker panel into a diagnostic tool that can be used for drug development or clinical diagnostics.
Verification
Verification mechanistically links the molecular marker to the biochemical process underlying a disease or drug effect. This may range from leveraging pre-existing knowledge to mechanistic in vitro and animal studies such as studies involving knock-out mice. Establishing mechanistic cause-effect relationships between a drug or disease effect and the molecular marker is the core of a robust molecular marker development strategy.
Qualification
Qualification has been defined as “a graded, fit-for purpose evidentiary process linking a biomarker with biology and clinical endpoints” [42] and seeks to establish a link between the molecular marker and clinical outcomes. In addition to determining sensitivity and specificity, a rigorous clinical qualification should also include the assessment of time- and, if applicable, dose-dependency. Receiver operating characteristic (ROC) curves for the definition of sensitivity and specificity [60,61] are basic metrics to assess biomarker performance in comparison with established clinical outcomes parameters [43].
There has been confusion in the literature regarding the term “validation” of a molecular marker. While verification and qualification bridge the results of molecular marker measurements, symptomatic drug effects and disease outcomes, validation focuses on the reliability and performance characteristics of the analytical assay(s) used to measure molecular markers [41,62]. As discussed in detail in reference [63], the validation of molecular marker assays that are often multi-analyte assays of endogenous compounds can be challenging.
When developed into clinical diagnostic tests, for such an in vitro diagnostic device to enter the market, it must comply with a set of rules and regulations and will require regulatory review and approval [64,65].
Challenges
There is no doubt that proteomics and metabolomics are extremely valuable tools in basic mechanistic research. Although an attractive concept that has been lauded by numerous review articles in the literature, the question is why proteomics and metabolomics have not yet resulted in more significant break-throughs in clinical research and have resulted in new and better clinical diagnostic strategies [66]. In this regard it is interesting to note that in a recent review summarizing the current clinical state-of-the art in managing kidney transplant patients, the recommended diagnostic strategy exclusively relied on established clinical markers such as serum creatinine, calculated GFRs, immunosuppressant therapeutic drug monitoring, urinalysis, ultrasound and biopsies, although the short-comings are clearly recognized [2]. This is not only limited to the field of renal transplantation, but has meanwhile been acknowledged as a general problem [67]. A more recent analysis revealed that ten biomarkers of cardiovascular disease that had been studied or were mentioned in more than 6000 publications had no or only limited clinical predictive ability [68]. It was concluded that a paradigm shift in testing and qualifying biomarkers as well as in associated expectations is required. There are many reasons, including all steps of molecular marker development, as to why all this new technology has produced large amounts of data and research, relatively little of which has been translated into useful clinical practice.
Molecular marker discovery
Although non-targeted proteomics and metabolomics technologies are valuable discovery tools and hypothesis-generating technologies, there are methodological limitations. In an ideal world, non-targeted proteomics and metabolomics technologies would be truly unbiased, would capture the complete proteome and metabolome (as this is nowadays possible in the field of genomics), and would allow for at least a semi-quantitative comparison of the proteomes and metabolomes of interest. However, current technologies capture only a part of the metabolome and proteome and therefore produce inherently biased results [69]. Important factors that introduce bias into metabolomics analyses may include, but are not limited to, timing of sample collection, the sample collection procedure, sample processing, stabilization, stability and storage, extraction procedures, dilution of sample, type and number of analytical methods used, preferences of analytical assays for metabolites with certain physico-chemical properties, ion suppression (liquid chromatography-mass spectrometry, LC-MS), derivatization (gas chromatography- mass spectrometry, GC-MS), sensitivity of the assay and range of reliable response [69]. In terms of proteomics the situation is similar [12]. The many computational, chemometric and biostatistical steps required to link changes in metabolite and protein patterns to biochemical and signal transduction pathways have to address multiple challenges such as the very large number of variables (metabolites and/or proteins) that often greatly exceed the number of observations. Most of the hundreds and thousands of data points generated are not relevant to the disease or drug effect, do not convey additional information. This increases the risk of false positive results and may mask valid information. In such settings, classical statistical approaches are inadequate. There is often a severe lack of degrees of freedom generally due to the relatively small sample sizes resulting in lack of statistical power [70–72] and false positives [73,74]. Indeed, most molecular marker discovery studies in kidney transplant patients reported in the literature are limited to between 10–100 subjects. Thus, considering the fact that sometimes hundreds of parameters are screened these studies must be considered underpowered. It is also impossible to adequately statistically power non-targeted clinical discovery studies as it is unknown which molecular makers may be found relevant, and therefore no pertinent information regarding their variability that may serve as the basis for a statistical power analysis is available at the time of study design. The large sparsity of informative variables tends to make metabolomics and proteomics data extremely noisy. Spurious correlations and co-linearities often exist between variables. This may be due to the nature of the data as well as due to dimensionality artifacts [75,76]. In addition, computational challenges arise due to scalability limitations of existing approaches. In this context, the complexity and heterogeneity of metabolites are considerably greater than those of genes and proteins. These problems have been addressed by the development of algorithms for data reduction and filtering, for false discovery rate control, and for high-dimensional statistical modeling strategies [49,52]. However, it is impossible to completely exclude that the selection of the chemometric/bioinformatics strategy may affect the results and conclusions. An overly conservative strategy may reject valid molecular markers (high false negative rate) while a liberal strategy may result in false positive results.
Tightly controlling and analyzing potential confounders such as age, gender, co-medications and diet is critical and can be very challenging in complex patient populations such as renal allograft recipients [77]. For example, it has been shown that age has a significant effect on the urinary proteome [78].
It is critical to take time dependency of the protein and metabolite pattern changes into account. Studies comparing sets of single samples per individual may be difficult to interpret with confidence. Especially during the earlier biochemical stages of a disease process, protein and metabolite pattern changes may vary quickly as the injury progresses. This may include compensatory mechanisms, secondary mechanisms such as oxygen radical formation and damage, changes in cell function and regulation, and the triggering of additional systemic responses, such as immune reactions and inflammation. Different stages during the development of a biochemical injury may be characterized by different sets of markers and this time-dependency and its underlying mechanistic dynamics needs to be understood. Accordingly, the correct timing of sample collection will be critical for the success of a molecular marker development and the later use of a specific molecular marker in clinical trials and as a clinical diagnostic tool. Longitudinal sampling strategies add value to molecular marker-based diagnostic approaches. Fortunately, many of the body fluids analyzed by proteomic and metabolomic studies can be obtained with minimal risk to the patient, and are well-suited to longitudinal studies. Due to the time-dependent changes associated with the development and progression of a disease process, it may be necessary to develop different sets of molecular markers for different stages of a disease process.
Verification
As biomarker discovery studies are usually underpowered, biological plausibility is essential for acceptance and adoption [77]. As discussed above, the verification process mechanistically links a molecular marker to a disease process and molecular mechanism of a drug effect. Obviously, this is problematic if relevant molecular mechanisms are not completely understood yet. Examples are chronic allograft injury and chronic allograft rejection, for which sensitive and predictive molecular markers are considered a big advance [79]. However, both are histological diagnoses that are caused by multiple molecular mechanisms, many of which are either still under discussion or still elusive [79].
Qualification
It is of interest to note that discovery studies are often retrospective studies with samples selected based on already known outcomes. In contrast, qualification studies ideally should be prospective clinical trials, in many cases multi-center trials. Sensitivity and specificity are determined by comparison with current clinical gold standard outcomes. One problem is that organ dysfunction may be caused by numerous distinct underlying biochemical mechanisms that ultimately cause the same symptoms. As aforementioned, several of these distinct and alternate biochemical processes may not even be known yet and may require the development of new classifications within a clinical syndrome. A related problem is that symptomatic injuries caused by different drugs and diseases may ultimately involve the same pathobiochemical and pathological mechanisms such as mitochondrial dysfunction, the formation of oxygen radicals, necrosis, apoptosis, inflammation, and immune reactions. This means that the further a pathological process has progressed, the more difficult it will be to find specific molecular marker changes that can identify the original cause. One of the problems with the gold standard outcome being less specific than the molecular marker is that there is no one-on-one relationship between a molecular marker and the predicted clinical outcome. Several molecular marker patterns that reflect distinct biochemical disease processes that ultimately will lead to the same symptoms will be valid predictors of a single clinical outcome. Such a specific marker pattern will reliably be able to predict a certain clinical outcome, however, sensitivity will be poor since the same outcomes caused by other biochemical processes will be missed. Following current practices and guidances this may lead to the rejection of a valid highly specific molecular marker while ironically a less predictive and specific molecular marker that is a surrogate for later and more common disease processes may be acceptable. Moreover, it should be noted that ROCs that are frequently used to estimate the sensitivity and specificity of molecular markers are a relatively insensitive metric, especially when it comes to their ability to assess the incremental predictive ability of molecular markers when compared to established predictors [80,81].
Another problem is that there is often only poor consensus in terms of definition of the disease endpoint against which a molecular marker needs to be qualified. For example, there are more than 30 different definitions of acute renal failure, now known as acute kidney injury, in the published literature [82,83]. It will be difficult to establish sensitivity and specificity if the gold standard outcome against which molecular markers will be qualified is itself of poor quality. Overall, this raises the question as to whether the current approach of establishing the quality and acceptance of a molecular marker by determination of specificity and sensitivity is a valid approach, or whether these commonly accepted practices and regulatory guidelines will cause valid, sensitive, and specific molecular markers to be abandoned.
Non-targeted diagnostic strategies
The typical molecular marker development strategy as shown in Figure 1 is to reduce the number of molecular markers to a so-called combinatorial marker that is constituted of clearly identified and verified components. However, proteomics and metabolomics technologies can create molecular fingerprints of body fluids such as plasma and urine that are characteristic for diseases and drug effects. It is an attractive idea to build expert systems based on non-targeted/non-biased analytical strategies that will generate a holistic view of a patient’s plasma and urine metabolome/proteome and that in combination with genomics will allow for a systems biology-based approach to medicine. Today, using truly non-targeted screening technologies in decision-making is not yet feasible, mostly because of the complexity of the data generated, the difficulties of validating such assays, the lack of verification and the lack of algorithms to convert this information into robust clinically relevant information.
Current Status
As aforementioned, the status of metabolomics and proteomics-based molecular marker discovery and development has already extensively been reviewed [11,19–30] and is therefore only briefly summarized below.
Metabolite markers
Although an intriguing concept, metabolomics has not yet received as much attention in renal transplantation as has proteomics [11,19–30]. The reason is that bioanalytical metabolomics assays are more challenging as the metabolome covers a wide range of compounds with very different physico-chemical properties, and wide coverage requires the combination of multiple assays [69,84]. Also, since open to the environment and the microbiolome, the metabolome is constantly in flux and thus more complex than the genome and proteome.
Metabolomics studies have assessed the biochemical effects of immunosuppressants and their combinations on the kidney [85–89], the assessment of donor organ quality, storage and ischemia/reperfusion injury as well as relevant therapeutic interventions [90–96], and kidney transplant function in patients [97–101]. Noteworthy is a study in healthy individuals that showed that urine metabolite patterns were already found changed within the first 4 hours after a single 5 mg/kg dose of cyclosporine in its Neoral formulation [89]. This study demonstrates how quickly the kidney responds to a cyclosporine challenge and how sensitively urine metabolite patterns can reflect biochemical changes in the kidney.
Protein markers
As disease processes in the kidney are often focal and not homogenously distributed throughout the organ, needle biopsies are associated with the risk that the regions of interest may be missed. In addition, a biopsy is an invasive procedure. As urine functions as a sink for proteins released by the kidney and can be collected non-invasively and relatively frequently, the concept of a “liquid biopsy” is attractive. As a proximal matrix urine is in direct contact with the transplant kidney. It has been estimated that 30% of the proteins in urine are from plasma and 70% are generated in the kidney [102,103]. Many kidney proteins appearing in urine during injury are either proteins that are usually reabsorbed in the proximal tubuli, are released by cell damage, leak into the urine during inflammation and immune reactions, are tissue matrix break down products, or are repair proteins that are formed and released during the healing process.
In recent years there has been significant progress in the identification of candidate protein markers for the diagnosis of acute and chronic kidney diseases [104–106]. Many of these protein markers are of potential interest for the monitoring of renal transplant patients but will require further study in the appropriate patient populations. Proteomics has been used to study the toxicity of immunosuppressants [87] as well as acute rejection [107] and chronic allograft injury [108] in rat models.
An increasing number of clinical discovery studies assessing the changes of protein pattern changes in peripheral blood, biopsies and in most cases urine caused by chronic allograft injury, acute and chronic rejections after kidney transplantation have been reported [109–128]. These studies are summarized in Table 2. It is of interest to evaluate these studies in light of the aforementioned current principles of molecular marker development. For example, the study published by Quintana et al. [112] relied on single urine samples from 50 subjects: thirty-two patients with chronic allograft dysfunction (14 with interstitial fibrosis and 18 with chronic active antibody mediated rejection) as well as 18 controls (8 stable renal transplant patients and 10 healthy individuals). A matrix-assisted laser desorption/time-of-flight mass spectrometry strategy was used and more than 2000 protein signals were entered into an unsupervised hierarchical cluster analysis. Fourteen protein ions were identified that discriminated between samples from patients with interstitial fibrosis and chronic rejection. However, these protein ions were only identified by their mass/charge ratio and no further attempt was made to identify the underlying proteins. This would have been important as the statistical analysis did not take many of the aforementioned challenges into account, did not correct for co-variates such as age, gender or medications, did not normalize for potential differences in urine concentrations, and did most likely not have sufficient statistical power (>2000 parameters evaluated in 50 subjects). In this case, identification of the proteins that allowed for discrimination of the study groups, would have allowed for further verification of the results based on information in the literature, for hypothesizing cause-effect relationships and for excluding that one or more of the identified candidate protein markers happened to be random noise. In fact, it is interesting to note that in seven [112,115,122–125,128] out of the twenty proteomics studies in renal transplant patients listed in Table 2 candidate marker proteins were not further identified. On the other hand, an especially powerful and promising discovery strategy seems to be the combination of proteomics and genomics (proteogenomics) [110,127,129].
Table 2.
Summary of proteomics studies in renal transplant patients.
| Disease Process | Marker Proteins Identified | Proteomics Platform | References |
|---|---|---|---|
| Chronic allograft injury | β2 microglobulin fragments in urine | SELDI-TOF | [109] |
| Chronic allograft injury | 302 proteins unique to mild and 509 proteins unique to moderate/severe chronic allograft injury in peripheral blood | Proteogenomic approach, MuDPIT, LC-MS | [110] |
| Chronic allograft injury | Peptide ions from uromodulin and kininogen in urine | LC-MS | [111] |
| Chronic allograft injury | 14 signals in urine, protein signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | LC-MS | [112] |
| Chronic allograft injury | 21 proteins in urine, including α-1-antitrypsin, α-1-B-glycoprotein, angiotensinogen, anti-TNF-α antibody light chain, β-2-microglobulin, brevin, heparin sulfate proteoglycan, leucine-rich α-2-glycoprotein-1, and transferrin. | 2D-DIGE, MALDI-TOF, LC-MS | [113] |
| Acute and chronic rejection | ANXA11, integrin α3, integrin β3, TNFα in urine | Microarrays | [114] |
| Acute and chronic rejection | >70 peptide signals in serum, protein signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | MALDI-TOF | [115] |
| Subclinical and clinical acute T-cell mediated rejection | Collagen α(I) and α(III) fragments in urine indicating involvement of matrix metalloproteinase-8 (MMP-8) | CE-MS | [116] |
| Subclinical and acute rejection | 17 polypeptides, one of which was identified as a fragment of collagen α5(IV) in urine | CE-MS | [117] |
| Acute rejection | uromodulin (UMOD), pigment epithelium-derived factor-PEDF (SERPINF1) and CD44 in urine | Shotgun LC-MS, ELISA | [118] |
| Acute rejection | 40 peptide panel, several peptides were identified as degradation products of uromodulin (UMOD) and the collagens COL1A2 and COL3A1, transcriptional changes in corresponding biopsies revealed changes of MMP-7, SERPING1 and TIMP1 | Proteogenomic approach, MALDI-TOF | [119] |
| Acute rejection | Titin, lipopolysaccharide-binding protein, peptidase inhibitor 16, complement factor D, mannose-binding lectin, protein Z-dependent protease, β2 microglobulin, kininogen-1, afamin, serine protease inhibitor, phosphatidyl choline-sterol acyltransferase, sex hormone-binding globulin in plasma | iTRAQ labeling in combination with LC-MS, ELISA | [120] |
| Acute rejection | 66 proteins in peripheral blood including NF-κB, STAT 1 and STAT3 | iTRAQ labeling in combination with LC-MS | [121] |
| Acute rejection | 7 signals in urine, protein signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | SELDI-TOF | [122] |
| Acute rejection | 5 signals in urine, signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | SELDI-TOF | [123] |
| Acute rejection | 45 signals in urine of which 16 were considered molecular marker candidates, signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | SELDI-TOF | [124] |
| Acute rejection | 3 signal clusters in urine, signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | SELDI-TOF | [125] |
| Acute tubular injury | β2 microglobulin fragments in urine | SELDI-TOF | [126] |
| Chronic rejection | >1400 proteins with unique expression profiles in kidney biopsies in biopsies with mild-severe disease compared with normal kidney biopsies | Proteogenomic approach, MuDPIT, LC-MS | [127] |
| BK virus nephropathy | 5 signals in urine, protein signals were characterized based on their mass/charge, but the proteins associated with these signals were not identified | SELDI-TOF | [128] |
Abbreviations: 2D: two-dimensional, CE: capillary electrophoresis; DIGE: differential gel electrophoresis, iTRAQ: isobaric tags for relative and absolute quantification, LC: liquid chromatography, MALDI: matrix assisted laser desorption/ionization, MS: mass spectrometry, MudPIT: multidimensional protein identification technology, SELDI: surface-enhanced laser desorption ionization, TOF: time-of-flight mass spectrometry.
A non-targeted urine proteomics test based on two-dimensional capillary electrophoresis/mass spectrometry is offered for the diagnosis of chronic kidney diseases [130,131], the value of which for managing kidney transplant patients has not fully been established yet. This technology was used in a clinical trial including 39 renal allograft patients in whom urinary peptide spectra were analyzed using capillary electrophoresis- mass spectrometry [116]. Sixteen patients had subclinical acute T-cell-mediated tubulointerstitial rejection and 23 subjects were non-rejection controls. When the results of this training data set was applied to a blinded validation data set including 64 kidney transplant patients, 16 out of 18 subclinical and 10 out of 10 clinical rejections (BANFF grades Ia/Ib), and 28 out of 36 controls without rejection were correctly classified. Altered collagen α(I) and α (III) chain fragments in rejection samples played a key role in the correct classification of patient samples suggesting an involvement of matrix metalloproteinase-8 (MMP-8). [116] A larger clinical trials to assess the ability of urinary peptide patterns as assessed using mass spectrometry to diagnose acute renal allograft rejection, compared to the gold standard allograft biopsy is currently in progress. [132].
In many cases as a result of non-targeted proteomic and genomic discovery studies [133], about two dozen potential markers of acute kidney injury have been identified [12,39,62,81,82,133–139]. The following protein markers have been examined in renal transplant patients: neutrophil gelatinase-associated lipocalin (NGAL) [140–147], kidney injury molecule-1 (KIM-1) [148–152], netrin-1 [153], interleukin-18 [147,154], α and π-glutathione S-transferase (GST) [155], liver fatty-acid-binding protein kidney (L-FABP) [156], N-acetyl-β-d-glucosaminidase (NAG) [144,157,158], β2-microglobulin and cystatin C. Most of these can be measured by ELISA or protein bead-based multi-analyte assays [82], and for some such as NGAL, assays on analytical platforms established in clinical laboratories are either already available [159] or under development. The potential impact of protein kidney dysfunction markers on kidney transplantation is reviewed in references [160,161]. Further systematic clinical development is required to establish the clinical benefit of these markers for testing kidney function in donors and in the management of kidney transplant patients. Thus it is interesting to note that the following protein marker trials after renal transplantation are currently listed in the clinicaltrials.org database: two trials evaluate urinary peptide and protein patterns using a non-targeted mass spectrometry-based approach [132,162] and two trials assess NGAL, IL-18 and/or KIM-1 [163,164]. No trial assessing metabolomics markers was listed.
Conclusions
In summary, to improve the successful discovery of metabolite and protein markers and development into diagnostic tools for the management of renal transplant patients, the following should be taken into consideration:
The goal- combinatorial markers
As discussed before, markers based on a single compound will never be able to monitor complex organs such as the kidney or vascular endothelium as well as complex processes such as allo-immune reactions and inflammation. On the other hand, as of today, non-targeted profiling technologies are challenging in a clinical setting due to complex analyses and software tools required. Combinatorial molecular markers typically consist of 3–10 individual parameters [165]. In general, specific combinatorial biomarker patterns confer significantly more information than a single measurement and can thus be expected to have better specificity and detection power. On the other hand, since a limited number of well characterized markers need to be analyzed, quantitative and validated bioanalytical assays can be developed. Algorithms that truly analyze combinatorial markers as a composite rather than a number of single markers are often missing [77] and need to be developed.
The study design
Most molecular marker and discovery studies listed in Table 2 are based on single samples that were collected for each of the study subjects. However, the strength of combinatorial markers is that they allow for longitudinally following individual renal transplant patients. As mentioned above, disease processes and drug effects may change their metabolite and protein signatures as they pass through different stages. Therefore, it seems reasonable to expect that prospective longitudinal studies that follow patients long-term will be most relevant (for example sample collection pre-transplant and 3, 7, 14 days, 1, 3, 6 and 12 months post-transplant). Such a longitudinal study design also has the added benefit of each patient serving as his/her own control, thus increasing statistical power. A molecular marker discovery and project should always include molecular marker verification and qualification. If the goal is to develop a clinical diagnostic tool, regulatory considerations may have to be taken into account from the beginning of the project.
Study conduct
The quality of study design, adequate sample handling, storage and processing, quality and reproducibility of bioanalytical multi-analyte assays [166–168], data analysis and interpretation are important factors for the success of molecular marker discovery and development in renal transplantation. Especially maintaining and ensuring sample integrity and quality is absolutely critical and seems to be frequently overlooked (the “garbage in – garbage out” principle of bioanalytics). In fact, it is interesting to observe that most of the molecular marker discovery and developments studies listed in Table 2 do not adequately describe measures taken to ensure sample integrity and quality. Improved standards for “omics” procedures, publications, data transparency and standardized tools for sharing data will be critical [168–173].
Translation into a clinical tool for the management of renal transplant patients
It is important to consider that the most sophisticated, sensitive and specific molecular marker will only reluctantly be accepted in clinical practice if it is too complex to implement. Successful translation into and use in clinical practice will hinge on the availability of clinically realistic sample collection and handling procedures, the development and validation of viable bioanalytical strategies, and software tools that allow for data analysis and translation into clinically meaningful information [168]. If urine is used as a matrix, acceptable normalization procedures able to compensate for differences in urine concentration may need to be developed [174]. While for metabolite markers liquid chromatography- mass spectrometry and gas chromatography- mass spectrometry provide universal analytical platforms that allow for the development and validation of clinical multi-analyte assays, the situation for protein multi-analyte assays is more complex. Although bead- and array-based multi-analyte protein assays are available, as antibodies are derived from biological sources, antibody cross-reactivities, manufacturing and batch-to-batch reproducibility can provide challenges. A promising alternative seems to be the development of targeted aptamer-based array chips. Aptamers are highly structured oligonucleotides, which are selected from combinatorial libraries of synthetic nucleic acid by an iterative process referred to as Systematic Evolution of Ligands by Exponential Enrichment (SELEX). [175] They can specifically bind to a wide variety of targets ranging from small organic molecules, proteins to supramolecular structures. In general, they show less cross-reactivity, have a wider linear range than antibodies and can be reproducibly manufactured in compliance with the rules and regulations of good manufacturing practices (GMP).
It is also interesting to note that most molecular marker discovery studies in the field of renal transplantation start with a non-targeted discovery step. However, it may also be attractive to design combinatorial protein and metabolite marker panels leveraging already existing knowledge in the literature. The components of such rationally designed (“logical”) combinatorial markers are usually already well verified and their clinical utility, sensitivity and specificity, and can be tested directly in clinical qualification studies without the necessity for extensive discovery and verification programs with all their risks and challenges.
Today, the management of renal transplant patients still relies on clinical markers such as creatinine the introduction of which as a clinical diagnostic marker is approaching its 100th anniversary [176]. A full molecular marker development program from discovery to regulatory approval is a highly integrated and comprehensive project that requires extensive interdisciplinary expertise, collaborations, and resources [168]. An increased effort and investment seems well worthwhile. As discussed above, first promising steps have been made and clinical combinatorial metabolite and protein-based molecular markers in combination with supporting software can provide much more specific, sensitive and predictive comprehensive patient management solutions. It is reasonable to expect that such management tools will allow for better individualization, will be able detect the development of chronic processes in their early states before they become symptomatic, will provide the basis for more efficient clinical risk evaluation and management strategies, and thus will have a positive effect on long-term outcomes after renal transplantation.
Acknowledgments
Funding: This work was supported by the United States National Institutes of Health, grant R01HD070511
Footnotes
Conflicts of Interest: None
References
- 1.Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. Am J Transplant. 2011;11:450. doi: 10.1111/j.1600-6143.2010.03283.x. [DOI] [PubMed] [Google Scholar]
- 2.Nankivell BJ, Kuypers DR. Diagnosis and prevention of chronic kidney allograft loss. Lancet. 2011;378:1428. doi: 10.1016/S0140-6736(11)60699-5. [DOI] [PubMed] [Google Scholar]
- 3.Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81:643. doi: 10.1097/01.tp.0000190423.82154.01. [DOI] [PubMed] [Google Scholar]
- 4.Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 5.Rosner MH. Urinary biomarkers for the detection of renal injury. Adv Clin Chem. 2009;49:73. doi: 10.1016/s0065-2423(09)49004-8. [DOI] [PubMed] [Google Scholar]
- 6.Nankivell BJ, Chapman JR. The significance of subclinical rejection and the value of protocol biopsies. Am J Transplant. 2006;6:2006. doi: 10.1111/j.1600-6143.2006.01436.x. [DOI] [PubMed] [Google Scholar]
- 7.Chapman JR, O’Connell PJ, Nankivell BJ. Chronic renal allograft dysfunction. J Am Soc Nephrol. 2005;16:3015. doi: 10.1681/ASN.2005050463. [DOI] [PubMed] [Google Scholar]
- 8.Williams WW, Taheri D, Tolkoff-Rubin N, Colvin RB. Clinical role of the renal transplant biopsy. Nat Rev Nephrol. 2012;8:110. doi: 10.1038/nrneph.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wishart DS. Metabolomics: the principles and potential applications to transplantation. Am J Transplant. 2005;5:2814. doi: 10.1111/j.1600-6143.2005.01119.x. [DOI] [PubMed] [Google Scholar]
- 10.Christians U, Albuisson J, Klawitter J, Klawitter J. The role of metabolomics in the study of kidney diseases and in the development of diagnostic tools. In: Edelstein CL, editor. Biomarkers of Kidney Disease. San Diego: Elsevier; 2010. p. 39. [Google Scholar]
- 11.Dieterle F, Riefke B, Schlotterbeck G, Ross A, Senn H, Amberg A. NMR and MS methods for metabonomics. Methods Mol Biol. 2011;691:385. doi: 10.1007/978-1-60761-849-2_24. [DOI] [PubMed] [Google Scholar]
- 12.Christians U, McCrery S, Klawitter J, Klawitter J. The role of proteomics in the study of kidney diseases and in the development of diagnostic tools. In: Edelstein CL, editor. Biomarkers of Kidney Disease. Elsevier; San Diego: 2010. p. 101. [Google Scholar]
- 13.Aebersol R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 14.Domon B, Aebersold R. Mass spectrometry in protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 15.Cravat BF, Simon GM, Yates JR. The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 16.Xu EY, Schaefer WH, Xu Q. Metabolomics in pharmaceutical research and development: metabolites, mechanisms and pathways. Curr Opin Drug Discov Devel. 2009;12:40. [PubMed] [Google Scholar]
- 17.Griffiths WJ, Wang Y. Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem Soc Rev. 2009;38:1882. doi: 10.1039/b618553n. [DOI] [PubMed] [Google Scholar]
- 18.Schnackenberg LK. Global metabolic profiling and its role in systems biology to advance personalized medicine in the 21st century. Expert Rev Mol Diagn. 2007;7:247. doi: 10.1586/14737159.7.3.247. [DOI] [PubMed] [Google Scholar]
- 19.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat Rev Nephrol. 2012;8:22. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 20.Roedder S, Vitalone M, Khatri P, Sarwal M. Biomarkers in solid organ transplantation: establishing personalized transplantation medicine. Genome Med. 2011;3:37. doi: 10.1186/gm253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS. Metabolomics in monitoring kidney transplants. Curr Opin Nephrol Hypertens. 2006;15:637. doi: 10.1097/01.mnh.0000247499.64291.52. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS. Metabolomics: a complementary tool in renal transplantation. Contrib Nephrol. 2008;160:76. doi: 10.1159/000125935. [DOI] [PubMed] [Google Scholar]
- 23.Kienzl-Wagner K, Pratschke J, Brandacher G. Proteomics- a blessing or a curse? Application of proteomics technology to transplant medicine. Transplantation. 2011;92:499. doi: 10.1097/TP.0b013e3182265358. [DOI] [PubMed] [Google Scholar]
- 24.Naesens M, Sarwal M. Molecular diagnostics in transplantation. Nat Rev Nephrol. 2010;6:614. doi: 10.1038/nrneph.2010.113. [DOI] [PubMed] [Google Scholar]
- 25.Sarwal M. De-convoluting the “omics’ for organ transplantation. Curr Opin Organ Transplant. 2009;14:544. doi: 10.1097/MOT.0b013e32833068fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannon RB. Immune monitoring and biomarkers to predict chronic allograft dysfunction. Kidney Int. 2010;119 (supplement):S59. doi: 10.1038/ki.2010.425. [DOI] [PubMed] [Google Scholar]
- 27.Bestard O, Cruzado JM, la Franquesa M, Grinyó JM. Biomarkers in renal transplantation. Curr Opin Organ Transplant. 2010;15:467. doi: 10.1097/MOT.0b013e32833b9ccb. [DOI] [PubMed] [Google Scholar]
- 28.Liang SL, Clarke W. Urine proteomic profiling of biomarkers of acute renal transplant rejection. Methods Mol Biol. 2010;641:185. doi: 10.1007/978-1-60761-711-2_11. [DOI] [PubMed] [Google Scholar]
- 29.Qunitana LF, Bañon-Maneus E, Solé-Gonzalez A, Campistol JM. Urine proteomics biomarkers in renal transplantation: an overview. Transplantation. 2009;88 (3 Supplement):S45. doi: 10.1097/TP.0b013e3181af7cba. [DOI] [PubMed] [Google Scholar]
- 30.Schaub S, Wilkins JA, Nickerson P. Proteomics and renal transplantation: searching for novel biomarkers and therapeutic targets. Contrib Nephrol. 2008;160:65. doi: 10.1159/000125934. [DOI] [PubMed] [Google Scholar]
- 31.Biomarkers Definition Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 32.Puntman VO. How-to-guide on biomarkers: biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad Med J. 2009;85:538. doi: 10.1136/pgmj.2008.073759. [DOI] [PubMed] [Google Scholar]
- 33.Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 34.Goodsaid FM, Frueh FW, Mattes W. Strategic paths for biomarker qualification. Toxicology. 2008;245:219. doi: 10.1016/j.tox.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 35.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol Syst Biol. 2006;2:52. doi: 10.1038/msb4100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng J, Gygi SP. Proteomics: The move to mixtures. J Mass Spectrom. 2001;36:1083. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 38.Christians U, Schmitz V, Klawitter J, Klawitter J. Proteo-metabolomic strategies in the future of drug development. In: Caroli S, Záray G, editors. The Applicability of Analytical Techniques to Clinical Studies. New York: John Wiley and Sons; 2012. [Google Scholar]
- 39.Christians U, Klawitter J, Klawitter J, Brunner N, Schmitz V. Biomarkers of immunosuppressant organ toxicity after transplantation- status, concepts and misconceptions. Expert Opin Drug Metabol Toxicol. 2011;7:175. doi: 10.1517/17425255.2011.544249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JW, Figeys D, Vasilescu J. Biomarker assay translation from discovery to clinical studies in cancer drug development: quantification of emerging protein biomarkers. Adv Cancer Res. 2007;96:269. doi: 10.1016/S0065-230X(06)96010-2. [DOI] [PubMed] [Google Scholar]
- 41.Burckart GJ, Amur S, Goodsaid FM, Lesko LJ, Frueh FW, Huang SM, Cavaille-Coll MW. Qualification of biomarkers for drug development in organ transplantation. Am J Transplant. 2008;8:267. doi: 10.1111/j.1600-6143.2007.02063.x. [DOI] [PubMed] [Google Scholar]
- 42.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther. 2007;81:104. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 43.Goodsaid FM, Frueh FW, Mattes W. Strategic paths for biomarker qualification. Toxicology. 2008;245:219. doi: 10.1016/j.tox.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 44.Lesko LJ, Atkinson AJ., Jr Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Ann Rev Pharmacol Toxicol. 2001;41:347. doi: 10.1146/annurev.pharmtox.41.1.347. [DOI] [PubMed] [Google Scholar]
- 45.Bai JPF, Bell R, Buckman SA, et al. Translational biomarkers: from preclinical to clinical: a report of 2009 AAPS/AAPC biomarker workshop. AAPS J. 2011;13:274. doi: 10.1208/s12248-011-9265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.United States Food and Drug Administration, Center for Drug Evaluation and Research. [accessed 4/5/2012];Manual of Policies and Procedures: Processing and Reviewing Voluntary Genomic Data Submissions (VGDSs). Version 3/16/2005. http://www.fda.gov/downloads/AboutFDA/CentersOffices/CDER/ManualofPoliciesProcedures/ucm073575.pdf.
- 47.European Medicines Agency. Scientific Advice Working Party of CHMP. [accessed 4/5/2012];Qualification of novel methodologies for drug development: guidance to applicants. EMA/CHMP/SAWP/72894/2008 Rev.1, Version 1-9-2012. http://www.emea.europa.eu/docs/en_GB/document_library/Regulatory_and_procedural_guideline/2009/10/WC500004201.pdf.
- 48.Nobeli I, Thornton JM. A bioinformatician’s view of the metabolome. Bioassays. 2006;28:534. doi: 10.1002/bies.20414. [DOI] [PubMed] [Google Scholar]
- 49.Wishart DS. Computational approaches to metabolomics. Methods Mol Biol. 2010;593:283. doi: 10.1007/978-1-60327-194-3_14. [DOI] [PubMed] [Google Scholar]
- 50.Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD. Analytical and statistical approaches to metabolomics research. J Sep Sci. 2009;32:2183. doi: 10.1002/jssc.200900152. [DOI] [PubMed] [Google Scholar]
- 51.Wishart DS. Current progress in computational metabolomics. Brief Bioinform. 2007;8:279. doi: 10.1093/bib/bbm030. [DOI] [PubMed] [Google Scholar]
- 52.Wishart DS. Introduction to cheminformatics. Curr Protoc Bioinformatics. 2007;Chapter 14(Unit 14.1) doi: 10.1002/0471250953.bi1401s18. [DOI] [PubMed] [Google Scholar]
- 53.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 54.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37 (Database issue):D603. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Domon B, Aebersold R. Challenges and opportunities in proteomics data analysis. Mol Cell Proteomics. 2006;5:1921. doi: 10.1074/mcp.R600012-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Ganter B, Zidek N, Hewitt PR, Müller D, Vladimirova A. Pathway analysis tools and toxicogenomics reference databases for risk assessment. Pharmacogenomics. 2008;9:35. doi: 10.2217/14622416.9.1.35. [DOI] [PubMed] [Google Scholar]
- 57.Wheelock CE, Wheelock AM, Kawashima S, et al. Systems biology approaches and pathway tools for investigating cardiovascular disease. Mol Biosyst. 2009;5:588. doi: 10.1039/b902356a. [DOI] [PubMed] [Google Scholar]
- 58.Materi W, Wishart DS. Computational systems biology in drug discovery and development: methods and applications. Drug Discov Today. 2007;12:295. doi: 10.1016/j.drudis.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Leach SM, Tipney H, Feng W, et al. Biomedical discovery acceleration, with applications to craniofacial development. PLoS Comput Biol. 2009;5:e1000215. doi: 10.1371/journal.pcbi.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fawcett T. Introduction to ROC analysis. Pattern Recognition Letters. 2006;27:861. [Google Scholar]
- 61.Parikh CR, Philbrook HT. Statistical considerations in analysis and interpretation of biomarker studies. In: Edelstein CL, editor. Biomarkers of Kidney Disease. San Diego: Elsevier; 2010. p. 39. [Google Scholar]
- 62.Müller PY, Dieterle F. Tissue-specific, non-invasive toxicity biomarkers: translation from preclinical safety assessment to clinical safety monitoring. Expert Opin Drug Metab Toxicol. 2009;5:1023. doi: 10.1517/17425250903114174. [DOI] [PubMed] [Google Scholar]
- 63.Christians U, Klepacki J, Shokati T, Klawitter J, Klawitter J. Mass spectrometry-based multiplexing for the analysis of biomarkers in drug development and clinical diagnostics- how much is too much? Microchem J. 2012 doi: 10.1016/j.microc.2012.02.011. (in print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 65.United States Department of Health and Human Services, Food and Drug Administration. [accessed April 4, 2012];Draft Guidance for Industry and Food and Drug Administration Staff. In vitro Companion Diagnostics Devices. Version July 14, 2011. http://www.fda.gov/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm262292.htm.
- 66.Ioannidis JP. A roadmap for successful applications of clinical proteomics. Proteomics Clin Appl. 2011;5:241. doi: 10.1002/prca.201000096. [DOI] [PubMed] [Google Scholar]
- 67.Editorial What happened to personalized medicine? Nat Biotechnol. 2012;30:1. doi: 10.1038/nbt.2096. [DOI] [PubMed] [Google Scholar]
- 68.Ioannidis JPA, Tzoulaki I. Minimal and null predictive effects of the most popular blood biomarkers of cardiovascular disease. Circ Res. 2012;110:658. doi: 10.1161/RES.0b013e31824da8ad. [DOI] [PubMed] [Google Scholar]
- 69.Christians U, Klawitter J, Hornberger A, Klawitter J. How unbiased is non-targeted metabolomics and is targeted pathway screening the solution? Curr Pharm Biotechnol. 2011;12:1053. doi: 10.2174/138920111795909078. [DOI] [PubMed] [Google Scholar]
- 70.Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J R Statist Soc B. 2004;66:187. [Google Scholar]
- 71.Tong T, Wang Y. Optimal shrinkage estimation of variances with applications to microarray data analysis. J Am Stat Assoc. 2007;102:113. [Google Scholar]
- 72.Wang Y, Ma Y, Carroll RJ. Variance estimation in the analysis of microarray data. J R Statist Soc B. 2009;71:425. doi: 10.1111/j.1467-9868.2008.00690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes analysis of a microarray experiment. J Am Stat Assoc. 2001;96:1151. [Google Scholar]
- 74.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Nat Acad Sci USA. 2001;98:5116. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai T, Lv J. Discussion: The Dantzig Selector: Statistical estimation when p is much larger than n. Annals Stat. 2007;35:2365. [Google Scholar]
- 76.Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. J R Statist Soc B. 2008;70:849. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gieser G, Harigaya H, Colangelo PM, Burckart G. Biomarkers in solid-organ transplantation. Clin Pharmacol Ther. 2011;90:217. doi: 10.1038/clpt.2011.75. [DOI] [PubMed] [Google Scholar]
- 78.Zürbig P, Decramer S, Dakna M, et al. The human urinary proteome reveals high similarity between kidney aging and chronic kidney disease. Proteomics. 2009;9:2108. doi: 10.1002/pmic.200800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takeda A, Horike K, Ohtsuka Y, et al. Current problems of chronic active antibody-mediated rejection. Clin Transplant. 2011;(Suppl 23):2. doi: 10.1111/j.1399-0012.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 80.Cook NR. The use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 81.Siew ED, Ware LB, Ikizler TA. Biological markers of acute kidney injury. J Am Soc Nephrol. 2011;22:810. doi: 10.1681/ASN.2010080796. [DOI] [PubMed] [Google Scholar]
- 82.Vaidya VS, Ferguson MA, Bonventre JV. Biomarkers of acute kidney injury. Annu Rev Pharmacol Toxicol. 2008;48:463. doi: 10.1146/annurev.pharmtox.48.113006.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kellum JA, Levin N, Bouman C, Lameire N. Developing a consensus classification system for acute renal failure. Curr Opin Crit Care. 2002;8:509. doi: 10.1097/00075198-200212000-00005. [DOI] [PubMed] [Google Scholar]
- 84.Psychogios N, Hau DD, Peng J, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmitz V, Klawitter J, Bendrick-Peart J, et al. Metabolic profiles in urine reflect nephrotoxicity of sirolimus and cyclosporine following rat kidney transplantation. Nephron. 2009;111:e80. doi: 10.1159/000209208. [DOI] [PubMed] [Google Scholar]
- 86.Klawitter J, Bendrick-Peart J, Rudolph B, et al. Urine metabolites reflect time-dependent effects of cyclosporine and sirolimus on rat kidney function. Chem Res Toxicol. 2009;22:118. doi: 10.1021/tx800253x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klawitter J, Klawitter J, Kushner E, et al. Association of immunosuppressant-induced protein changes in the rat kidney with changes in urine metabolite patterns: A proteo-metabonomic study. J Proteome Res. 2010;9:865. doi: 10.1021/pr900761m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serkova NJ, Christians U. Biomarkers for toxicodynamic monitoring of immunosuppressants: NMR-based quantitative metabonomics of the blood. Ther Drug Monit. 2005;20:652–656. doi: 10.1097/01.ftd.0000179846.30342.65. [DOI] [PubMed] [Google Scholar]
- 89.Klawitter J, Haschke M, Kahle C, et al. Toxicodynamic effects of ciclosporin are reflected by metabolite profiles in the urine of healthy individuals after a single dose. Br J Clin Pharmacol. 2010;70:241. doi: 10.1111/j.1365-2125.2010.03689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fuller TF, Serkova N, Neimann CU, Freise CE. Influence of donor pretreatment with N-acetylcysteine on ischemia/reperfusion injury in rat kidney grafts. J Urol. 2004;171:1296. doi: 10.1097/01.ju.0000103928.64939.6a. [DOI] [PubMed] [Google Scholar]
- 91.Hauet T, Gibelin H, Godart C, Eugene M, Carretier M. Kidney retrieval conditions influence damage to renal medulla: evaluation by proton nuclear magnetic resonance (NMR) spectroscopy. Clin Chem Lab Med. 2000;38:1085. doi: 10.1515/CCLM.2000.161. [DOI] [PubMed] [Google Scholar]
- 92.Hauet T, Baumert H, Gibelin H, et al. Noninvasive monitoring of citrate, acetate, lactate, and renal medullary osmolyte excretion in urine as biomarkers of exposure to ischemic reperfusion injury. Cryobiology. 2000;41:280. doi: 10.1006/cryo.2000.2291. [DOI] [PubMed] [Google Scholar]
- 93.Hauet T, Baumert H, Gibelin H, Godart C, Carretier M, Eugene M. Citrate, acetate and renal medullary osmolyte excretion in urine as predictor of renal changes after cold ischaemia and transplantation. Clin Chem Lab Med. 2000;38:1093. doi: 10.1515/CCLM.2000.162. [DOI] [PubMed] [Google Scholar]
- 94.Hauet T, Gibelin H, Richer JP, Godart C, Eugene M, Carretier M. Influence of retrieval conditions on renal medulla injury: evaluation by proton NMR spectroscopy in an isolated perfused pig kidney model. J Surg Res. 2000;93:1. doi: 10.1006/jsre.2000.5885. [DOI] [PubMed] [Google Scholar]
- 95.Hauet T, Goujon JM, Tallineau C, Carretier M, Eugene M. Early evaluation of renal reperfusion injury after prolonged cold storage using proton nuclear magnetic resonance spectroscopy. Br J Surg. 1999;86:1401. doi: 10.1046/j.1365-2168.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- 96.Schmitz V, Klawitter J, Bendrick-Peart J, et al. Graft flushing with histidine-tryptophane-ketoglutarate (HTK) followed by extended cold preservation in University of Wisconsin (UW) solution in a rat kidney transplantation model- An improved preservation protocol? Eur J Surg Res. 2006;38:388. [Google Scholar]
- 97.Foxall PJ, Mellotte GJ, Bending MR, Lindon JC, Nicholson JK. NMR spectroscopy as a novel approach to the monitoring of renal transplant function. Kidney Int. 1993;43:234. doi: 10.1038/ki.1993.37. [DOI] [PubMed] [Google Scholar]
- 98.Le Moyec L, Pruna A, Eugène M, et al. Proton nuclear magnetic resonance spectroscopy of urine and plasma in renal transplantation follow-up. Nephron. 1993;65:433. doi: 10.1159/000187525. [DOI] [PubMed] [Google Scholar]
- 99.Rush D, Somorjai R, Deslauriers R, Shaw A, Jeffery J, Nickerson P. Subclinical rejection--a potential surrogate marker for chronic rejection--may be diagnosed by protocol biopsy or urine spectroscopy. Ann Transplant. 2000;5:44. [PubMed] [Google Scholar]
- 100.Wang NJ, Zhou Y, Zhu TY, Wang X, Guo YL. Prediction of acute cellular renal allograft rejection by urinary metabolomics using MALDI-FTMS. J Proteome Res. 2008;7:3597. doi: 10.1021/pr800092f. [DOI] [PubMed] [Google Scholar]
- 101.Wang J, Zhou Y, Xu M, Rong R, Guo Y, Zhu T. Urinary metabolomics in monitoring acute tubular injury of renal allografts: a preliminary report. Transplant Proc. 2011;43:3738. doi: 10.1016/j.transproceed.2011.08.109. [DOI] [PubMed] [Google Scholar]
- 102.Prunotto M, Ghiggeri GM, Brushi M, et al. Renal fibrosis and proteomics: current knowledge and still key questions for proteomic investigation. J Proteomics. 2011;74:1855. doi: 10.1016/j.jprot.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 103.Konvalinka A, Scholey JW, Diamandis EP. Searching for new biomarkers of renal diseases through proteomics. Clin Chem. 2012;58:353. doi: 10.1373/clinchem.2011.165969. [DOI] [PubMed] [Google Scholar]
- 104.Mishak H, Delles C, Klein J, Schanstra JP. Urinary proteomics based on capillary electrophoresis-coupled mass spectrometry in kidney disease: discovery and validation of biomarkers, and clinical application. Adv Chronic Kidney Dis. 2010;17:493. doi: 10.1053/j.ackd.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 105.Spasovski G, Ortiz A, Vanholder R, El Nahas M. Proteomics in chronic kidney diseases: the issues clinical nephrologists need an answer for. Proteomics Clin Appl. 2011;5–6:233. doi: 10.1002/prca.201000150. [DOI] [PubMed] [Google Scholar]
- 106.Mullen W, Delles C, Mischak H, et al. Urinary proteomics in the assessment of chronic kidney disease. Curr Opin Nephrol Hypertens. 2011;20:654. doi: 10.1097/MNH.0b013e32834b7ffa. [DOI] [PubMed] [Google Scholar]
- 107.Dai Y, Lv T, Wang K, Huang Y, Li D, Liu J. Detection of acute renal allograft rejection by analysis of renal tissue proteomics in rat models of renal transplantation. Saudi J Kidney Dis Transplant. 2008;19:952. [PubMed] [Google Scholar]
- 108.Reuter S, Reiermann S, Wörner R, et al. IF/TA-related metabolic changes- proteome analysis of rat renal allografts. Nephrol Dial Transplant. 2010;25:2492. doi: 10.1093/ndt/gfq043. [DOI] [PubMed] [Google Scholar]
- 109.Johnston O, Cassidy H, O’Connell S, et al. Identification of β2-microglobulin as urinary biomarker for chronic allograft nephropathy using proteomics methods. Proteomics Clin Appl. 2011;7–8:422. doi: 10.1002/prca.201000160. [DOI] [PubMed] [Google Scholar]
- 110.Kurian SM, Heilman R, Mondala TS, et al. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4:e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quintana LF, Campistol JM, Alcolea MP, Bañon-Maneus E, Sol-González A, Cutillas PR. Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Mol Cell Proteomics. 2009;8:1658. doi: 10.1074/mcp.M900059-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quintana LF, Solé-Gonzalez A, Kalko SG, et al. Urine proteomics to detect biomarkers for chronic allograft dysfunction. J Am Soc Nephrol. 2009;20:428. doi: 10.1681/ASN.2007101137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bañón-Maneus E, Diekmann F, Carrascal M, et al. Two-dimensional difference gel electrophoresis urinary proteomic profile in the search of nonimmune chronic allograft dysfunction biomarkers. Transplantation. 2010;89:548. doi: 10.1097/TP.0b013e3181c690e3. [DOI] [PubMed] [Google Scholar]
- 114.Srivastava M, Eidelman O, Torosyan Y, Jozwik C, Mannon RB, Pollard HB. Elevated expression levels of ANAX11, integrins β3 and α3, and TNF-α contribute to a candidate proteomic signature in urine for kidney allograft rejection. Proteomics Clin Appl. 2011;5–6:311. doi: 10.1002/prca.201000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sui W, Huang L, Dai Y, Chen J, Yan Q, Huang H. Proteomic profiling of renal allograft rejection in serum using magnetic bead-based sample fractionation and MALDI-TOF MS. Clin Exp Med. 2010;10:259. doi: 10.1007/s10238-010-0094-5. [DOI] [PubMed] [Google Scholar]
- 116.Metzger J, Chatzikyrkou C, Broecker V, et al. Diagnosis of subclinical and clinical acute T-cell-mediated rejection in renal transplant patients by urinary proteome analysis. Proteomics Clin Appl. 2011;5–6:322. doi: 10.1002/prca.201000153. [DOI] [PubMed] [Google Scholar]
- 117.Wittke S, Haubitz M, Walden M, et al. Detection of acute tubulointerstitial rejection by proteomic analysis of urinary samples in renal transplant recipients. Am J Transplant. 2005;5:2479. doi: 10.1111/j.1600-6143.2005.01053.x. [DOI] [PubMed] [Google Scholar]
- 118.Sidgel TK, Kaushal A, Gritsenko M. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin Appl. 2010;4:32. doi: 10.1002/prca.200900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ling XB, Sidgel TK, Lau K, et al. Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J Am Soc Nephrol. 2010;21:646. doi: 10.1681/ASN.2009080876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Freue GV, Sasaki M, Meredith A, et al. Proteomic signatures in plasma during early acute allograft rejection. Mol Cell Proteomics. 2010;9:1954. doi: 10.1074/mcp.M110.000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu D, Zhu D, Xu M, et al. Analysis of transcriptional factors and regulation networks in patients with acute renal allograft rejection. J Proteome Res. 2011;10:175. doi: 10.1021/pr100473w. [DOI] [PubMed] [Google Scholar]
- 122.O’Riordan E, Orlova TN, Mei JJ, et al. Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol. 2004;15:3240. doi: 10.1097/01.ASN.0000145241.83482.68. [DOI] [PubMed] [Google Scholar]
- 123.Clarke W, Silverman BC, Zhang Z, et al. Characterization of renal allograft reception by urinary proteomic analysis. Ann Surg. 2003;237:660. doi: 10.1097/01.SLA.0000064293.57770.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Clarke W. Proteomic research in renal transplantation. Ther Drug Monit. 2006;28:19. doi: 10.1097/01.ftd.0000194500.40021.37. [DOI] [PubMed] [Google Scholar]
- 125.Schaub S, Rush D, Wilkins J, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol. 2004;15:219. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 126.Schaub S, Wilkins JA, Antonovici M, et al. Proteomic-based identification of cleaved urinary beta2-microglobulin as a potential marker for acute tubular injury in renal allografts. Am J Transplant. 2005;5:729. doi: 10.1111/j.1600-6143.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- 127.Nakorchevsky A, Hewel JA, Kurian SM, et al. Molecular mechanism of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J Am Soc Nephrol. 2010;21:362. doi: 10.1681/ASN.2009060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jahnukainen T, Malehorn D, Sun M, et al. Proteomic analysis of urine in kidney transplant patients with BK virus nephropathy. J Am Soc Nephrol. 2006;17:3248. doi: 10.1681/ASN.2006050437. [DOI] [PubMed] [Google Scholar]
- 129.Sarwal MM, Sidgel TK, Salomon DR. Functional proteogenomics- embracing complexity. Semin Immunol. 2011;23:235. doi: 10.1016/j.smim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 130.Jantos-Siwy J, Schiffer E, Brand K, et al. Quantitative urinary proteome analysis for biomarker evaluation in chronic kidney disease. J Proteome Res. 2009;8:268. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- 131. [accessed April 8, 2012]; http://www.diapat.de/DiaPat-Diagnostik/chronic-nephropathies/
- 132.Gwinner W. [accessed September 18, 2012];Diagnosis of acute rejection in renal transplant patients by urine mass spectrometry. http://clinicaltrials.gov/ct2/show/NCT01315067.
- 133.Devarajan P, Williams LM. Proteomics for biomarker discovery in acute kidney injury. Semin Nephrol. 2007;6:637. doi: 10.1016/j.semnephrol.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ozer JS, Dieterle F, Troth S, et al. A panel of urinary biomarkers to monitor reversibility of renal injury and a serum marker with improved potential to assess renal function. Nat Biotechnol. 2010;28:486. doi: 10.1038/nbt.1627. [DOI] [PubMed] [Google Scholar]
- 135.Vaidya VS, Ozer JS, Dieterle F, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol. 2010;28:478. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dieterle F, Perentes E, Cordier A, et al. Urinary clusterin, cystatin C, beta2-microglobulin, and total protein as markers to detect drug-induced kidney injury. Nat Biotechnol. 2010;28:463. doi: 10.1038/nbt.1622. [DOI] [PubMed] [Google Scholar]
- 137.Bonventre JV, Vaidya VS, Schmouder R, Feig P, Dieterle F. Next-generation biomarkers for detecting kidney toxicity. Nat Biotechnol. 2010;28:436. doi: 10.1038/nbt0510-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Devarajan P. Emerging urinary biomarkers in the diagnosis of acute kidney injury. Expert Opin Med Diagn. 2008;2:387. doi: 10.1517/17530059.2.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Edelstein CL, Faubel S. Biomarkers in acute kidney injury. In: Edelstein CL, editor. Biomarkers of Kidney Disease. Elsevier; San Diego: 2010. p. 177. [Google Scholar]
- 140.Magnusson NE, Hornum M, Jørgensen KA, et al. Plasma neutrophil gelatinase associated lipocalin (NGAL) is associated with kidney function in uraemic patients before and after kidney transplantation. BMC Nephrol. 2012;13:8. doi: 10.1186/1471-2369-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bataille A, Abbas S, Semoun O, et al. Plasma neutrophil gelatinase-associated lipocalin in kidney transplantation and early renal function prediction. Transplantation. 2011;92:1024. doi: 10.1097/TP.0b013e318230c079. [DOI] [PubMed] [Google Scholar]
- 142.Ireland R. Transplantation: Urinary NGAL levels in potential deceased kidney donors may be useful in determining donor suitability. Nat Rev Nephrol. 2011;27:364. doi: 10.1038/nrneph.2011.64. [DOI] [PubMed] [Google Scholar]
- 143.Hollmen ME, Kyllönen LE, Inkinen KA, Lalla ML, Merenmies J, Salmela KT. Deceased donor neutrophil gelatinase-associated lipocalin and delayed graft function after kidney transplantation: a prospective study. Crit Care. 2011;15:R121. doi: 10.1186/cc10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hollmen ME, Kyllönen LE, Inkinen KA, Lalla ML, Salmela KT. Urine neutrophil gelatinase-associated lipocalin is a marker of graft recovery after kidney transplantation. Kidney Int. 2011;79:89. doi: 10.1038/ki.2010.351. [DOI] [PubMed] [Google Scholar]
- 145.Hall IE, Yarlagadda SG, Coca SG, et al. IL-18 and urinary NGAL predict dialysis and graft recovery after kidney transplantation. J Am Soc Nephrol. 2010;21:189. doi: 10.1681/ASN.2009030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Malyszko J, Malyszko JS, Mysliwiec M. Serum neutrophil gelatinase-associated lipocalin correlates with kidney function in renal allograft recipients. Clin Transplant. 2009;23:681. doi: 10.1111/j.1399-0012.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- 147.Parikh CR, Jani A, Mishra J, et al. Urine NGAL and IL-18 are predictive biomarkers for delayed graft function following kidney transplantation. Am J Transplant. 2006;6:1639. doi: 10.1111/j.1600-6143.2006.01352.x. [DOI] [PubMed] [Google Scholar]
- 148.Nauta FL, Bakker SJ, van Oeveren W, et al. Albuminuria, proteinuria, and novel urine biomarkers as predictors of long-term allograft outcomes in kidney transplant recipients. Am J Kidney Dis. 2011;57:733. doi: 10.1053/j.ajkd.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 149.Malyszko J, Koc-Zorawska E, Malyszko JS, Mysliwiec M. Kidney injury molecule-1 correlates with kidney function in renal allograft recipients. Transplant Proc. 2010;42:3957. doi: 10.1016/j.transproceed.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 150.Malyszko J, Koc-Zorawska E, Malyszko JS, Mysliwiec M. Kidney injury molecule-1 correlates with kidney function in renal allograft recipients. Transplant Proc. 2010;42:3957. doi: 10.1016/j.transproceed.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 151.Nijboer WN, Schuurs TA, Damman J, et al. Kidney injury molecule-1 is an early noninvasive indicator for donor brain death-induced injury prior to kidney transplantation. Am J Transplant. 2009;9:1752. doi: 10.1111/j.1600-6143.2009.02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van Timmeren MM, Vaidya VS, van Ree RM, et al. High urinary excretion of kidney injury molecule-1 is an independent predictor of graft loss in renal transplant recipients. Transplantation. 2007;84:1625. doi: 10.1097/01.tp.0000295982.78039.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ramesh G, Kwon O, Ahn K. Netrin-1: a novel universal biomarker of human kidney injury. Transplant Proc. 2010;42:1519. doi: 10.1016/j.transproceed.2009.11.040. [DOI] [PubMed] [Google Scholar]
- 154.Hall IE, Doshi MD, Poggio ED, Parikh CR. A comparison of alternative serum biomarkers with creatinine for predicting allograft function after kidney transplantation. Transplantation. 2011;91:48. doi: 10.1097/TP.0b013e3181fc4b3a. [DOI] [PubMed] [Google Scholar]
- 155.Bäckman L, Appelkvist EL, Ringdén O, Dallner G. Glutathione transferase in the urine: a marker for post-transplant tubular lesions. Kidney Int. 1988;33:571. doi: 10.1038/ki.1988.35. [DOI] [PubMed] [Google Scholar]
- 156.Przybylowski P, Koc-Zorawska E, Malyszko JS, Kozlowska S, Mysliwiec M, Malyszko J. Liver fatty-acid-binding protein in heart and kidney allograft recipients in relation to kidney function. Transplant Proc. 2011;43:3064. doi: 10.1016/j.transproceed.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 157.Bone JM, Amara AB, Shenkin A, et al. Calcineurin inhibitors and proximal renal tubular injury in renal transplant patients with proteinuria and chronic allograft nephropathy. Transplantation. 2005;79:119. doi: 10.1097/01.tp.0000146843.23824.93. [DOI] [PubMed] [Google Scholar]
- 158.Marchewka Z, Kuźniar J, Zynek-Litwin M, et al. Kidney graft function in long-term cyclosporine and tacrolimus treatment: comparative study with nephrotoxicity markers. Transplant Proc. 2009;41:1660. doi: 10.1016/j.transproceed.2009.01.116. [DOI] [PubMed] [Google Scholar]
- 159.Grenier FC, Ali S, Syed H, et al. Evaluation of the ARCHITECT urine NGAL assay: assay performance, specimen handling requirements and biological variability. Clin Biochem. 2010;43:615. doi: 10.1016/j.clinbiochem.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 160.Halawa A. The early diagnosis of acute renal graft dysfunction: a challenge we face. The role of novel biomarkers. Ann Transplant. 2011;16:90. [PubMed] [Google Scholar]
- 161.Alkesh J. Biomarkers in kidney transplantation. In: Edelstein CL, editor. Biomarkers of Kidney Disease. Elsevier; San Diego: 2010. p. 233. [Google Scholar]
- 162.Friedewald J. [accessed September 18, 2012];Proteogenomic monitoring and assessment of kidney transplant recipients (“Mini-Kidney”) http://clinicaltrials.gov/ct2/show/NCT01531257.
- 163.Salamzadeh J. [accessed September 18, 2012];Study of the effect of N-acetyl cysteine on the renal graft function biomarkers (IL18, NGAL) http://clinicaltrials.gov/ct2/show/NCT01403506.
- 164.Goilav B. [accessed September 18, 2012];Urinary Kidney Injury Molecule-1 (KIM-1) Excretion as biomarker for injury in kidney transplant recipients. http://clinicaltrials.gov/ct2/show/NCT00805571.
- 165.Koop R. Combinatorial biomarkers: from early toxicology assays to patient population profiling. Drug Discov Today. 2005;10:781. doi: 10.1016/S1359-6446(05)03440-9. [DOI] [PubMed] [Google Scholar]
- 166.Chau CH, Rixe O, McLeod H, Figg WD. Validation of analytic methods for biomarkers used in drug development. Clin Cancer Res. 2008;14:5967. doi: 10.1158/1078-0432.CCR-07-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lee JW, Hall M. Method validation of protein biomarkers in support of drug development or clinical diagnostics/prognosis. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1259. doi: 10.1016/j.jchromb.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 168.Mischak H, Ioannidis JP, Argiles A, et al. Implementation of proteomic biomarkers: making it work. Eur J Clin Invest. 2012;42:1027. doi: 10.1111/j.1365-2362.2012.02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mischak H. How to get proteomics to the clinic? Issues in clinical proteomics, exemplified by CE-MS. Proteomics Clin Appl. 2012 doi: 10.1002/prca.201200027. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 170.Mischak H, Apweiler R, Banks RE, et al. Clinical proteomics: a need to define the field and to begin to set adequate standards. Proteomics Clin Appl. 2007;1:148. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 171.Ioannidis JPA, Khoury MJ. Improving validation practices in “omics” research. Science. 2011;334:1230. doi: 10.1126/science.1211811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Baggerly K. Disclose all data in publications. Nature. 2010;467:401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
- 173.Baggerly KA, Coombes KR. What information should be required to support clinical “omics” publications. Clin Chem. 2011:688. doi: 10.1373/clinchem.2010.158618. [DOI] [PubMed] [Google Scholar]
- 174.Waikar SS, Sabbisetti VS, Bonventre JV. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 2010;78:486. doi: 10.1038/ki.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu Y, Yang X, Wang E. Review: Aptamers in microfluidic chips. Anal Chim Acta. 2010;683:12. doi: 10.1016/j.aca.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 176.Rehberg PB. Über die Bestimmung der Menge des Glomerulusfiltrats mittels Kreatinin als Nierenfunktionsprüfung, nebst einigen Bemerkungen über die Theorien der Harnbereitung. Zentralbl Inn Med. 1929;50:367. [Google Scholar]