Abstract
Stem cell behavior is regulated by extrinsic signals from specialized microenvironments, or niches, and intrinsic factors required for execution of context-appropriate responses to niche signals. Here we show that function of the transcriptional regulator longitudinals lacking (lola) is required cell autonomously for germline stem cell and somatic cyst stem cell maintenance in the Drosophila testis. In addition, lola is also required for proper execution of key developmental transitions during male germ cell differentiation, including the switch from transit amplifying progenitor to spermatocyte growth and differentiation, as well as meiotic cell cycle progression and spermiogenesis. Different lola isoforms, each having unique C-termini and zinc finger domains, may control different aspects of proliferation and differentiation in the male germline and somatic cyst stem cell lineages.
Keywords: Drosophila, Spermatogenesis, Germline stem cell, Spermatogonia, Spermatocyte, Transcription
Introduction
Adult stem cells self-renew and produce differentiated progeny that replenish and repair tissues throughout life. Stem cell behavior is influenced by extracellular signals produced in specialized microenvironments, or niches. Although niche signals are beginning to be identified in a variety of adult stem cell systems, the intrinsic factors, including transcription and chromatin regulators, that enable stem cells and their descendants to interpret and respond appropriately to niche signals remain largely unknown. Drosophila spermatogenesis has emerged as a model system for genetic analysis of the mechanisms that program self-renewal and maintenance of adult stem cells, as well as proliferation and differentiation of their progeny (Fig. 1A). Two adult stem cell populations, germline stem cells (GSCs) and somatic cyst stem cells (CySCs), reside at the apical tip of the testis, attached to a plug of somatic support cells called the hub. Spermatogenesis initiates with the oriented division of a GSC, normally producing one daughter cell that maintains contact with the hub and self-renews stem cell identity and a second daughter cell, the gonialblast (Gb), which exits the niche and initiates spermatogonial differentiation. CySCs self-renew and give rise to post-mitotic somatic cyst cells (Gonczy and DiNardo, 1996), two of which encapsulate each nascent Gb to form a cyst.
Fig. 1.
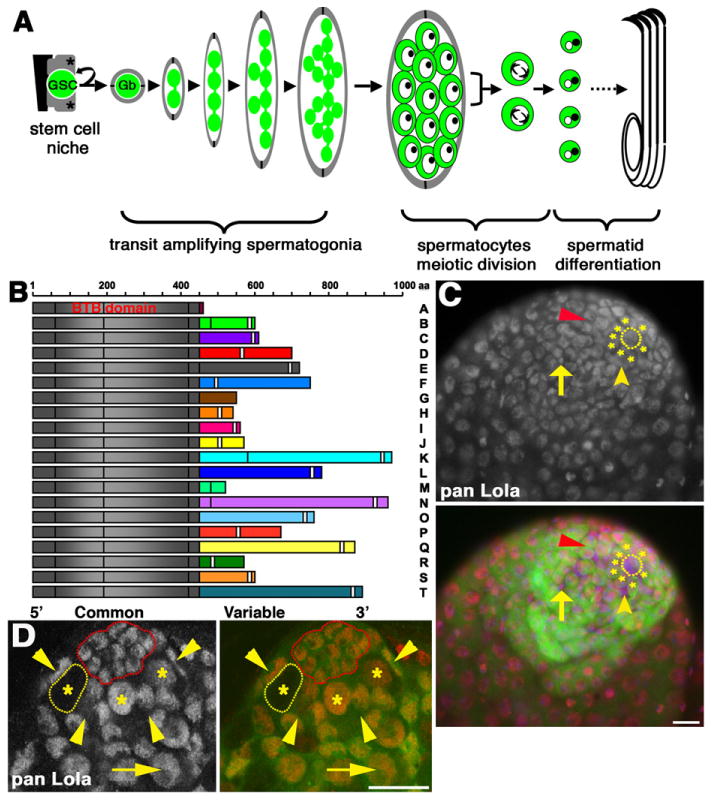
lola encodes a family of BTB-Zf transcriptional regulators expressed in germline and somatic cells in the testis: (A) Diagram of Drosophila spermatogenesis. Green: GSC lineage. Gray: CySC lineage. Asterisks: CySCs. Black: hub. Gb: gonialblast, encysted by two cyst cells (gray). Primary spermatocytes: large germ cells with prominent nucleoli (dark dots in nuclei). For simplicity, the division products of one spermatocyte are shown. (B) Lola protein isoforms A–T (N termini at left). Gray: Common N-terminus with BTB domain. Colored: Alternately spliced C-terminal exons. White bars: zinc finger motifs. C2HC-C2H2 zinc fingers motifs: B, C, D, F, H, J, K, L, O, P, Q, R, T. C2HC-C2HC zinc finger motifs: I and N. C2H2 motif: E, S. Isoforms A, G and M do not have predicted zinc finger domains. (C) Wildtype testis stained with pan Lola antibody. Red: Lola. Green: nos-Gal4, UAS-tubulin-GFP. Blue: DAPI. Dotted outline: hub. Asterisks: GSCs. Red arrowhead: spermatogonia. Yellow arrowhead: CySC. Arrow: Cyst cell. Scale bar: 20 μm. (D) lolaORE76 mosaic testis at 2 days post clone induction stained with pan Lola antibody. Red: Lola. Green: Ubi-nls-GFP (clonal marker). Yellow outline: homozygous mutant lolaORE76 GSC. Red outline: hub. Asterisks: GSCs. Arrowheads: CySC nuclei. Arrow: spermatogonia. Scale bar: 15 μm.
Several key cell state transitions occur during differentiation in the male germline stem cell lineage. The encapsulated Gb undergoes spermatogonial mitotic amplification divisions with incomplete cytokinesis to form a cyst of 16 interconnected germ cells, which subsequently adopts spermatocyte fate. Spermatocytes enter meiotic S phase, grow in size, silence many transcripts expressed in transit amplifying cells, and express transcripts required for the meiotic divisions and spermatid differentiation (reviewed in Fuller, 1993). Upon completion of the meiotic divisions, the resultant 64 haploid spermatids undergo a dramatic series of morphological changes to form mature sperm. Spermiogenesis is largely driven by transcripts expressed in spermatocytes under the control of the testis TAFs (tTAFs), paralogs of the TFIID transcription complex, and the tMAC complex, a spermatocyte specific version of the chromatin regulatory complex, Mip/DREAM (Hiller et al., 2001, 2004; White-Cooper et al., 2000).
An outstanding challenge is to identify the intrinsic regulatory factors that control self-renewal and differentiation in stem cell lineages. Here we report that lola is required cell autonomously for stem cell maintenance, a timely transition from the spermatogonial division program to the spermatocyte cell growth and gene expression program, and progression through the meiotic divisions in the Drosophila testis. The lola locus encodes a family of at least 20 different nuclear proteins, designated Lola-A through T (Goeke et al., 2003), which share a common N-terminal domain containing a Broad complex, Tramtrack, Bric-a-Brac (BTB) protein dimerization motif that is alternately spliced to unique C-terminal exon(s), many of which contain zinc finger domains (Goeke et al., 2003; Horiuchi et al., 2003) (Ohsako et al., 2003; Tweedie et al., 2009) (Fig. 1B). Thirteen lola isoforms have a C-terminal domain with a tandem C2HC-C2H2 zinc finger motif, two have a tandem C2HC-C2HC motif, and two have a single C2H2 zinc finger motif (Goeke et al., 2003). Homo- and hetero dimeric pairings amongst Lola isoforms are likely, since the Lola N-terminus formed homodimers in yeast two hybrid assays (Giot et al., 2003), and Lola-F was shown to complex with at least two different Lola isoforms in embryo extracts (Zhang et al., 2003). In addition, Lola isoforms may pair with other BTB-ZF proteins (Stogios et al., 2005). Analysis of isoform-specific, loss-of-function alleles and RNAi knock-downs suggest stage and cell type specific requirements for lola-B, K, L, N and P isoforms during spermatogenesis.
Materials and methods
Fly stocks and husbandry
Fly stocks were maintained on standard cornmeal-molasses medium. y,w; FRT42D lolaORE76/Cyo is an EMS allele containing a stop codon in the BTB domain that is predicted to be protein null for all lola isoforms (Crowner et al., 2002; Madden et al., 1999; Spletter et al., 2007). FRT42D P{lacW}lola5D2/Cyo is a severe hypomorphic allele containing a P-element insertion upstream of the translational start site that is predicted to affect transcription of all lola isoforms (Giniger et al., 1994). y,w; FRT42D lolaORC4/Cyo is a K-specific, loss-of-function mutant allele that introduces a premature stop codon at codon 771, upstream of both K-specific zinc finger motifs (Goeke et al., 2003). y,w; FRT42D lolaORE119/Cyo, an L-specific loss-of-function allele, contains a missense mutation in the linker sequence between the two L-specific zinc fingers that converts a conserved proline residue at codon 712 into leucine (Goeke et al., 2003). The P712L amino acid substitution is predicted to disrupt interactions with DNA and/or protein binding partners (Goeke et al., 2003), making lolaORE119 a likely null mutation specific for isoform L UAS-Dcr-2 (stock 60007), UAS-dsRNA strains complementary to the lola common domain (stocks 12574 and 12573), and lola-B (stock 41415) were obtained from the VDRC. Genbank accession numbers for different lola protein coding variants are found in Table 1 (Goeke et al., 2003). sa1 and sa2 are described in Hiller et al. (2004) and Lin et al. (1996). aly2 and aly5 are described in Lin et al. (1996) and White-Cooper et al. 2000). Alleles used for TGF-ß pathway clonal analysis were obtained from the Bloomington Stock Center: y w; tkva12FRT40A/SM6a; w; mad1–2FRT40A/Cyo and w; mad8–2FRT40A/Cyo. All other strains were obtained from the Bloomington Stock Center.
Table 1.
Nineteen Lola protein coding variants identified in Goeke et al. (2003) are listed with their corresponding Genbank Accession numbers and Flybase isoform designations.
| Lola Isoform designation (Goeke et al., 2003) | Genbank Accession number | Flybase isoform designation |
|---|---|---|
| A | AB107272 | lola-RM |
| B | AB107273 | lola-RQ |
| C | AB107274 | lola-RL |
| D | AB107275 | lola-RA |
| E | AB107276 | lola-RZ; lola-RS |
| F | AB107277 | lola-RE; lola-RD |
| H | AB107279 | lola-RK |
| I | AB107280 | lola-RF |
| J | AB107281 | lola-RT; lola-RU |
| K | AB107282 | lola-RI |
| L | AB107283 | lola-RB; lola-RC |
| M | AB107284 | lola-RH |
| N | AB107285 | lola-RP |
| O | AB107286 | lola-RJ |
| P | AB107287 | lola-RO |
| Q | AB107288 | lola-RW; lola-RN |
| R | AB107289 | lola-RY |
| S | AB107290 | lola-RX |
| T | AB107291 | lola-RR; lola-RG |
Immunofluorescence
Whole mount immunostaining was performed on testes fixed in 4% formaldehyde in 1 × PBS for 20 min at room temperature. Permeabilization, washes and blocking were performed as described in Hime et al. (1996). Antibodies were diluted into in 1 × PBS with 0.3% BSA and 3% normal goat serum (Jackson Immunoresearch Inc., West Grove, PA, USA); primary incubations were conducted overnight at 4 °C, and secondary incubations were conducted for 90 min at 4 °C. Testes were mounted in Vectashield with DAPI (Vector Labs Burlingame, CA USA). Primary antibodies used in this study include: rabbit polyclonal pan anti-Lola (E. Giniger, NIH, Bethesda, MD, USA, 1:40); rabbit anti-GFP (Invitrogen A-11122, 1:3000); guinea pig anti-Tj (D. Godt, University of Toronto, Toronto, Canada, 1:500); mouse anti-Bam (Developmental Studies Hybridoma Bank (DSHB), 1:10); rabbit polyclonal anti-STAT92E (D. Montell, Johns Hopkins University, Baltimore, MD, USA, 1:1000); rabbit polyclonal anti-Zfh1 (R. Lehmann, Skirball Institute, NY, USA, 1:5000). Immunofluoresence on squashed testes using methanol/acetone fixation was performed as described in Chen et al. (2005) using guinea pig anti-Sa (M. Fuller, Stanford University, Stanford, CA, USA, 1:100). Alexafluor secondary antibodies (Molecular Probes/Invitrogen) were used at 1:1000. Measurement of cell cycle timing by sequential administration of the S-phase analogs EdU and BrdU was performed according to Insco et al. (2009). Images in Fig. 1C were taken using a Zeiss Axioskop; all other images were acquired using a Leica TCS SP2 AOBS confocal system. Images were processed using Adobe Photoshop software.
In situ hybridization
DIG-labeled riboprobes and in situ hybridization on wildtype testes were performed as described in White-Cooper et al. (1998). Constructs for the lola-K and lola-L riboprobes were a gift from S. Goeke and E. Giniger, and are described in Goeke et al. (2003).
Mosaic analysis
Clonal analysis experiments were performed using the FLP/FRT mitotic recombination system (Xu and Rubin, 1993) according to the protocol described in Kiger et al. (2001) on males with the following genotypes: control clones wildtype for lola function: y,w,hs-FLP22; FRT42D, Ubi-GFP/FRT42D. lola null clones: y,w,hs-FLP22; FRT42D, Ubi-GFP/FRT42D lolaORE76. lola hypomorph clones: y,w,hs-FLP22; FRT42D, Ubi-GFP/FRT42D lola5D2. lola-K loss-of-function clones: y,w,hs-FLP22; FRT42D, Ubi-GFP/FRT42D lolaORC4. lola-L loss-of-function clones: y,w,hs-FLP22; FRT42D, Ubi-GFP/FRT42D lolaORE119. Mosaic testes costained with antibodies against GFP and Tj were scored using confocal microscopy at the indicated time points following clone induction. At least 75 testes were scored per genotype, per time point. Definitions for GSC clones: GFP negative, Tj negative cells directly abutting the hub. Differentiating germ cell clones: GFP negative, Tj negative germ cell cysts displaced more than one cell diameter away from the hub. CySC clones: GFP negative, Tj positive cells up to one cell diameter away from the hub. Cyst cell clones: GFP negative, Tj positive cells >2 cell diameters from the hub.
Inducible RNAi in early germ cells
Virgin female w, UAS-Dcr-2; nos-Gal4VP16/TM3Sb flies were crossed to males of the following genotypes: (1) w; UAS-pan lola RNAi (VDRC 12574); UAS-pan lola RNAi (VDRC 12573), (2) w; UAS-lola-L RNAi/Cyo; UAS-lola-L RNAi/TM6b, (3) w; UAS-lola-B RNAi, UAS-lola-B RNAi/Cyo, (4) w; UAS-lola-N RNAi/Cyo; UAS-lola-N RNAi/TM6b, and (5) w; UAS-lola-P RNAi/Cyo; UAS-lola-P RNAi/TM6b. Genotypes scored: nos-Gal4 driver alone: w, UAS-Dcr-2; +/Cyo; nos-Gal4VP16/TM6b. Hairpin alone controls: w, UAS-Dcr-2; UAS-lola RNAi/+; UAS-lola RNAi/+. lola knock-downs: w, UAS-Dcr-2; UAS-lola RNAi/+; UAS-lola RNAi/NG4VP16. Timed lays were performed for 6 h at 25 °C, and progeny were shifted to 30 °C at 24 h post-lay. Testes from two to three day old adults were analyzed in live squashes by phase contrast microscopy and were immunostained with anti-Lola and anti-Tj as described above. Counts of the number of germ cell nuclei in intact cysts of mature spermatocytes were performed as described in Insco et al. (2009).
TUNEL staining
TUNEL staining was performed according to the manufacturer's instructions using the In Situ Cell Death Detection Kit, TMR Red (Roche, Catalog #12 156 792 910).
UAS-lola RNAi transgenic lines
UAS-dsRNA constructs complementary to the isoforms-specific exons of lola-B, L, N, and P were subcloned into the pWIZ vector according to the scheme described in Lee and Carthew (2003), and transgenic fly lines were created using standard methods for P-element mediated germline transformation (Rubin and Spradling, 1982).
Primer sequences:
lola B RNAi (S): 5′–TTACTCTAGACCAGCAGCGGTGGAAATG–3′
lola B RNAi (A): 5′–AATGTCTAGACCTTGTGCACATAGTAGTAG–3′
lola L RNAi (S): 5′–TTACTCTAGATTACGCGTATAGCGGGACTC–3′
lola L RNAi (A): 5′–TTACTCTAGAGTAGACGCAGAATGGGCACT–3′
lola N RNAi (S): 5′–TTACTCTAGAGCAAACGGTCACTCAAACTG–3′
lola N RNAi (A): 5′–AATGTCTAGACAGGTGACGCTTCAGATTG–3′
lola P RNAi (S): 5′–TTACTCTAGACATCTATCCGATCCTTGGC–3′
lola P RNAi (A): 5′–AATGTCTAGACGTAGTCCTCGTCATCCTC–3′
Results and discussion
lola encodes a family of nuclear proteins expressed in germline and somatic cells throughout the Drosophila testis
Staining of wildtype testes with antibodies raised against the common N-terminal domain of Lola revealed that Lola protein(s) were present in early germline and somatic cell nuclei, including GSCs, CySCs and their differentiating progeny (Fig. 1C). Specificity of the antibody for Lola was confirmed by staining mosaic testes containing lola null germline clones. Lola protein was not detected in germ cell cysts homozygous mutant for lola, while neighboring heterozygous or wildtype cysts contained nuclear Lola protein (Fig. 1D).
lola is required cell autonomously for GSC maintenance
Clones homozygous mutant for either of two independently derived, strong loss-of-function lola alleles were induced by FLP/FRT mediated mitotic recombination (Section 2) and the fate of marked clones was monitored over time (Fig. 2A). GSCs homozygous mutant for lola were induced at frequencies comparable to marked wildtype control GSCs, but they were not maintained (Fig. 2B). In contrast, the percentage of testes harboring marked control GSCs remained essentially constant over 2 weeks (Fig. 2B). Both lola mutant alleles behaved identically in this assay, suggesting that the observed GSC maintenance defect was due to loss of lola rather than a secondary lesion on the lola mutant chromosome arms.
Fig. 2.
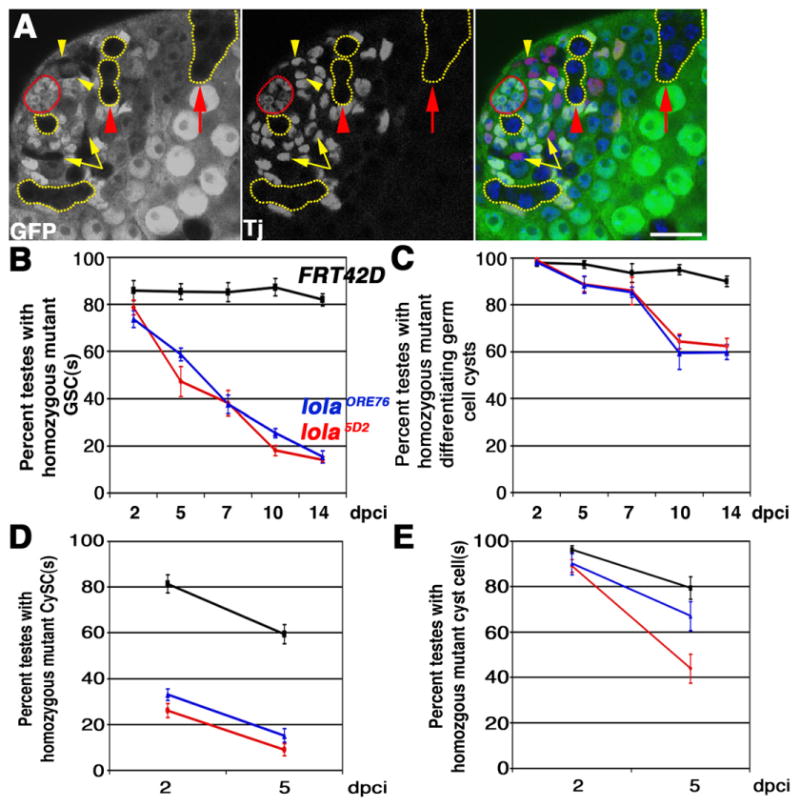
lola is required cell autonomously for GSC and CySC maintenance: (A) lolaORE76 mosaic testis containing clones marked by absence of GFP at 2 days post clone induction (dpci). Red: Tj, somatic nuclei. Green: Ubi-nls-GFP. Blue: DAPI. Red outline: hub. Yellow outlines: Examples of GFP negative germline clones. Note lola mutant GSC abutting the hub. Yellow arrowheads: lola mutant CySCs next to the hub. Yellow arrows: lola mutant cyst cells. Red arrowhead: lola mutant spermatogonial cyst. Red arrow: lola mutant spermatocyte cyst. Scale bar: 20 μm. (B) and (C) Percentage of mosaic testes containing homozygous mutant (B) GSC(s) or (C) spermatogonia and/or spermatocyte cyst(s) from 2 to 14 dpci. (D) and (E) Percentage of mosaic testes containing homozygous mutant (D) CySCs or (E) cyst cell(s) from 2 to 5 dpci. (B)–(E): Black: FRT42D control clones. Blue: lolaORE76 clones. Red: lola5D2 clones. Error bars: standard error of the mean. n≥75 testes per genotype, per time point.
GSCs mutant for lola initiated differentiation, since marked germ cell cysts homozygous mutant for lola were observed at both the spermatogonial and spermatocyte stages (Fig. 2A). The percentage of testes containing differentiating germline clones mutant for lola declined over time, subsequent to and presumably as a consequence of the reduction in lola mutant GSCs (Fig. 2C). Consistent with the hypothesis that loss of lola was not cell lethal, none of the lola5D2 homozygous mutant GSCs (n=10), lolaORE76 homozygous mutant GSCs (n=17), or marked control FRT42D GSCs (n=25) scored were positive for TUNEL staining at 3 days post clone induction (dpci).
Co-expression of UAS-Dcr-2 and UAS-dsRNA hairpins complementary to the lola common domain under control of nos-Gal4 also elicited GSC loss (Table 2). To evaluate efficacy of the knockdown, pan-lola knock-down and hairpin alone testes were stained with antibodies raised against the common N-terminal domain of Lola and antibodies against the somatic cell marker Traffic Jam (Tj). Pan-Lola immunostaining was undetectable in GSCs and spermatogonia in pan-lola knock-down testes (31/31 testes scored), while Pan-Lola staining in the hub, CySCs and cysts cells remained unchanged, as judged by colocalization with Tj (31/31 testes scored; Supplementary Fig. 1A). However, Pan-Lola staining in spermatocytes was detectable in 10/31 of the pan-lola knockdown testes, a result likely explained by poor expression or driving ability of the nos-Gal4VP16 driver in primary spermatocytes. In contrast, Pan Lola staining was detected throughout the germline and in somatic cell nuclei in all of the hairpin alone testes scored (n=19 testes; Supplementary Fig. 1B). These data suggest that GSCs made homozygous mutant for lola by mitotic recombination are not likely to be lost due to cell competition with adjacent heterozygous or homozygous wildtype GSCs.
Table 2.
Inducible RNAi knock-down of lola in early germ cells elicits GSC loss. Live testis squashes were scored by phase contrast microscopy. n=number of testes scored.
| Percentage of testes scored | |||
|---|---|---|---|
|
| |||
| Elongated, coiled morphology | Round morphology | Devoid of early germ cells | |
| nos-Gal4 driver alone (n=18) | 100 | 0 | 0 |
| pan lola hairpin alone (n=69) | 94.2 | 5.8 | 0 |
| pan lola knock-down (n=80) | 83.4 | 16.6 | 26.3 |
| lola-B hairpin alone (n=53) | 100 | 0 | 0 |
| lola-B knock-down (n=76) | 0 | 100 | 100 |
| lola-L hairpin alone (n=74) | 98.6 | 1.4 | 0 |
| lola-L knock-down (n=89) | 100 | 0 | 53.9 |
Clonal analysis revealed that lola may influence CySC maintenance cell autonomously, although even marked control CySCs appeared to have a shorter half life than control GSCs in our experiments. Mosaic testes costained with anti-GFP and the somatic nuclear marker Traffic Jam (Tj) were scored at 2 and 5 dpci for the presence of homozygous mutant CySCs and cyst cells (see Section 2). Far fewer lola mutant CySCs were observed than control CySCs at 2 dpci, and the number of lola mutant CySCs decreased further with time (Fig. 2D). Since perdurance of the nuclear Ubi-GFP clonal marker precluded scoring at time points earlier than 2 dpci, CySC clone induction frequencies were inferred from the percentage of testes harboring marked lola mutant or wildtype control cyst cells at 2 dpci. Cyst cells arise directly from CySC divisions, and normally do not undergo transit amplification divisions. The percentage of testes with homozygous mutant cyst cells at 2 dpci was similar for both lola alleles tested and the control FRT42D chromosome (Fig. 2E), suggesting that lola mutant CySCs were induced and were rapidly lost to differentiation.
lola likely acts in parallel to the JAK-STAT pathway in the testis stem cell niche. In wildtype testes, STAT protein is enriched in GSCs and their immediate progeny, and is not detected in differentiating spermatogonia (Boyle et al., 2007; Issigonis et al., 2009; Leatherman and Dinardo, 2008). Accumulation of STAT protein in GSCs and CySCs is thought to be a read-out of JAK-STAT pathway activation in these cells. STAT was detected in 100% of the lolaORE76 null mutant GSCs scored at 3 dpci (n=30), suggesting that lola is not required for stat92E transcription or maintenance of STAT protein production in GSCs (Fig. 3A). Furthermore, equivalent levels of Zfh-1 protein, a STAT target highly expressed in CySCs (Leatherman and Dinardo, 2008), were observed in lolaORE76 mutant CySCs and neighboring CySCs wildtype for lola function at 2 dpci (n=32 lolaORE76 CySCs scored) (Fig. 3B), suggesting that lola is not required for STAT signal transduction in CySCs. Likewise, lola does not appear to be a direct downstream target of activated STAT in GSCs or CySCs. Chromatin immunoprecipitation experiments were performed on upd over-expressing testes lysates using antibodies against Drosophila STAT, followed by qPCR using primers that spanned predicted STAT consensus binding sites in the lola locus. In three independent experiments, activated STAT was not enriched at predicted target sites in the lola locus relative to non-target sites in a housekeeping gene (data not shown).
Fig. 3.
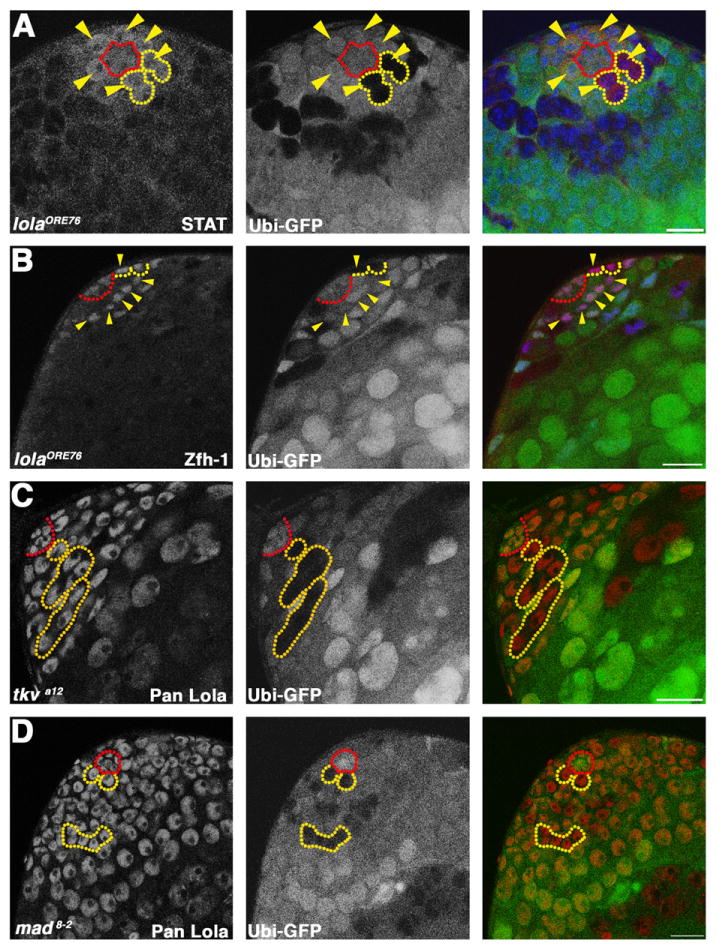
lola likely acts in parallel to the JAK-STAT and TGF-β pathways: (A) lolaORE76 mosaic testis stained with antibodies against STAT. Red: STAT. Green: Ubi-nls-GFP. Blue: DAPI. Red outline: hub. Yellow outlines: homozygous mutant GSC clones. Yellow arrowheads: GSCs positive for STAT. (B) lolaORE76 mosaic testis stained with antibodies against Zfh-1. Red: Zfh-1. Green: Ubi-nls-GFP. Blue: Tj. Red outline: hub. Yellow outlines: homozygous mutant CySC clones. Yellow arrowheads: CySCs expressing Zfh-1. (A) and (B) Scale bars: 15 μm. (C) and (D) tkva12 (C) and mad8–2, (D) mosaic testes at 3 dpci stained with pan Lola antibody. Red: Lola. Green: Ubi-nls-GFP. Red outline: hub. Yellow outlines: tkva12 (C) and mad8–2 (D) homozygous mutant GSCs and spermatogonial cysts. (C) and (D) Scale bars: 20 μm.
Expression of one or more lola isoforms is likely to be TGF-β independent in GSCs. Previous studies showed that activation of the TGF-β pathway is required cell-autonomously in germ cells for GSC maintenance (Kawase et al., 2004; Schulz et al., 2004; Shivdasani and Ingham, 2003). Immunostaining with pan Lola antibody was performed on mosaic testes containing germline clones homozygous mutant for strong loss-of-function alleles of either the Type 1 receptor tkv or the Smad homolog mad at 3 dpci. Lola protein was detected in the nuclei of GSCs homozygous mutant for either tkv (n=13 tkva12 homozygous mutant GSCs scored) or mad (n=22 mad1–2 homozygous mutant GSCs scored; n = 13 mad8–2 homozygous mutant GSCs scored) (Fig. 3C and D).
lola and the onset of spermatocyte differentiation
lola was required cell autonomously in spermatogonia for proper timing of the switch to spermatocyte fate. In Drosophila melanogaster, the Gb undergoes exactly 4 rounds of transit amplification division, producing 16 interconnected germ cells that together exit mitosis and initiate the spermatocyte program of cell growth and meiosis (Fig. 1A). While spermatogonial cysts wildtype for lola function always executed 4 rounds of mitosis prior to adopting primary spermatocyte fate, lola mutant spermatogonial cysts frequently underwent 5 or more mitotic amplification divisions, forming cysts with 32 or more germ cells (Fig. 4A). The over-proliferation phenotype was more severe in lolaORE76 null cysts than in cysts homozygous mutant for lola5D2, a strong hypomorphic allele (Giniger et al., 1994). Germ cell cysts homozygous for lola5D2 usually transitioned to spermatocyte fate after 5 rounds of division, and the cells often adopted the large size and morphological characteristics of spermatocytes (Fig. 4A and C). In contrast, most lolaORE76 null germ cell cysts underwent more than 5 rounds of mitosis, resulting in marked clones filled with small cells that most closely resembled spermatogonia or very early spermatocytes (Fig. 4A and D).
Fig. 4.
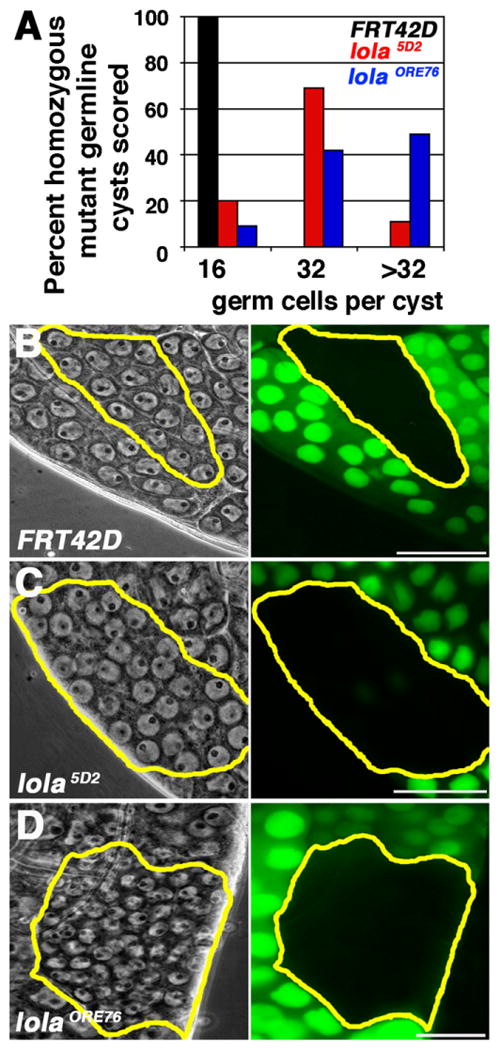
lola is required cell autonomously in the germline to restrict transit amplification divisions: (A) Percentage of homozygous mutant germ cell cysts in the spermatocyte region of the testis with 16, 32, or greater than 32 germ cells per cyst. Black: FRT42D wildtype control clones. Red: lola5D2 clones. Blue: lolaORE76 clones. n=100 cysts in the spermatocyte region per genotype at 4–6 dpci. (B)–(D): FRT42D control (B), lola5D2 (C), and lolaORE76 (D) homozygous germline clones (yellow outlines). Gray: phase contrast. Green: Ubi-nls-GFP clonal marker. (B) FRT42D spermatocyte cyst containing 16 germ cell nuclei (15 visible in focal plane). (C) lola5D2 spermatocyte cyst containing 32 germ cell nuclei (31 visible in focal plane), with size and morphology characteristic of spermatocytes. (D) lolaORE76 spermatocyte cyst containing more than 32 small germ cell nuclei. Scale bars: 20 μm.
The over-proliferation of lola mutant spermatogonia may be due to more rapid cell cycle progression of germ cells in lola mutant cysts than controls. Double pulse labeling experiments indicated that lola may slow cell cycle progression in transit amplifying spermatogonia. Mosaic testes containing lolaORE76 null clones were pulse labeled with the S-phase analog bromodeoxyuridine (BrdU) for 15 min, chased in insect cell culture medium for 7 h, and subsequently labeled with EdU for 5 min prior to fixation and analog detection as described in Insco et al. (2009). Previous studies suggest that the spermatogonial cell cycle lasts longer than 7 h (Insco et al., 2009). EdU-positive germ cell cysts, which were in S-phase at the time of fixation, were scored for BrdU co-labeling. Cysts double positive for BrdU and EdU likely underwent 2 rounds of S-phase during the pulse labeling experiment. None of the EdU-positive cysts wildtype for lola function was positive for BrdU, suggesting that the scored cysts underwent 1 round of S-phase during the experiment (0/30 BrdU+EdU+ control cysts (n=30 EdU+cysts)). In contrast, 20.8% of the lolaORE76 mutant cysts were double positive for BrdU and EdU, suggesting that these cysts likely underwent two rounds of S-phase labeling during the experiment (5/24 BrdU+ EdU+lolaORE76 spermatogonial cysts (n=24 EdU+cysts)). This result suggests that lolaORE76 spermatogonial cysts cycled faster than control cysts wildtype for lola function (p=0.013, Fisher's Exact Test).
Previous studies suggest that both cell cycle length and the accumulation of the differentiation regulator Bam affect the number of transit amplifying divisions that spermatogonia undergo before committing to terminal differentiation (Insco et al., 2009). In wildtype testes, Bam protein begins to accumulate in 4-cell spermatogonial cysts (Insco et al., 2009). The counting defect in lolaORE76 null cysts was not due to a failure to produce Bam, since Bam protein was detected beginning at the 4-cell stage in lolaORE76 spermatogonia (4/4 four cell lolaORE76 mutant cysts were Bam positive, n=18 testes).
lola is required prior to initiation of the aly/tMAC and tTAF spermatocyte-specific gene expression program
Male germ cell cysts null mutant for lola died soon after transitioning to the spermatocyte differentiation program, after down-regulation of Bam protein but prior to the extensive increase in spermatocyte nuclear size and the production of the spermatocyte-specific marker Sa. In wildtype testes Bam protein production reaches a threshold in 8-cell cysts, triggering a cell fate change that results in rapid down-regulation of Bam levels in 16-cell cysts (Insco et al., 2009). Bam protein was not detected in 3/16 16-cell lolaORE76 mutant cysts and 10/13 > 16-cell lolaORE76 mutant cysts (n=18 testes) (Fig. 5A), suggesting that Bam protein levels were downregulated appropriately in lola mutant clones as they initiated the transition to spermatocyte fate.
Fig. 5.
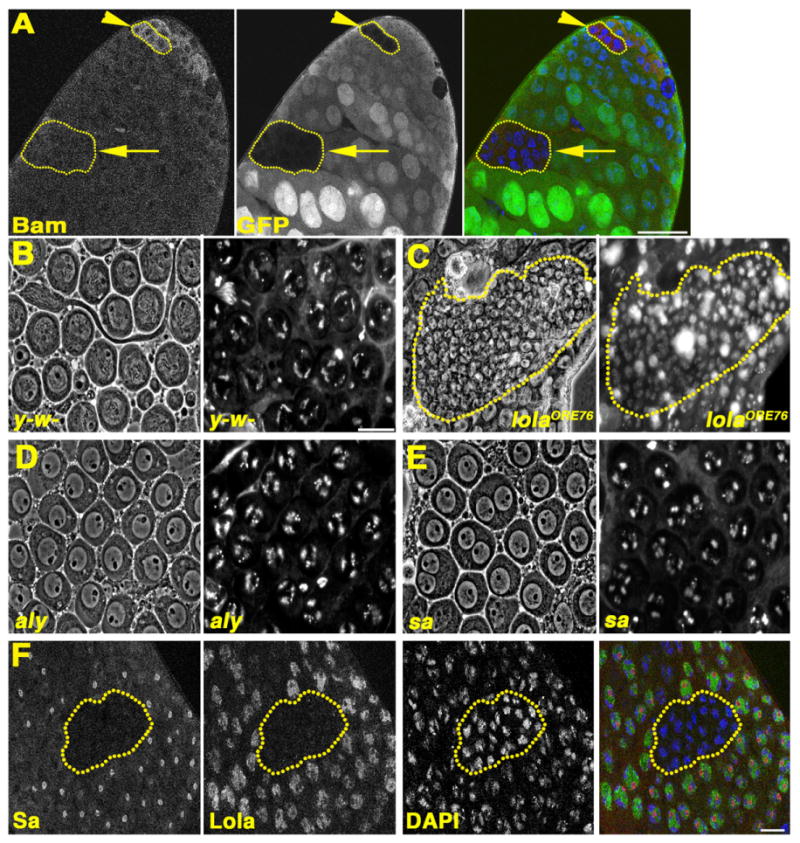
lola mutant spermatogonia execute the switch to spermatocyte fate: (A) lolaORE76 mosaic testis stained with anti-Bam. Red: Bam. Green: Ubi-nls-GFP. Blue: DAPI. Yellow outlines: homozygous mutant germline clones. Arrowhead: lolaORE76 spermatogonia with Bam expressed. Arrow: lolaORE76 cyst in the spermatocyte region lacking Bam. Scale bar: 40 μm. (B)–(E): Live squash preparations stained with Hoechst, showing spermatocytes from (B) y-w-, (C) lolaORE76 mosaic, (D) aly, (E) sa testes. Phase contrast (left) and corresponding fluorescent images (right). Yellow outline in (C) homozygous mutant lolaORE76 cyst. (F) lolaORE76 mosaic testis stained with anti-Sa. Red: Sa. Green: pan Lola. Blue: DAPI. Outline: lolaORE76 mutant germ cell cyst. B-F: Scale bars: 20 μm.
In the overwhelming majority of lolaORE76 null cysts observed, the germ cells neither grew substantially in size nor adopted morphological characteristics of mature spermatocytes. Wildtype mature spermatocytes have large phase light nuclei in which the partially condensed autosomal bivalents form crescent shaped structures near the nuclear periphery (Fig. 5B). lolaORE76 null cysts commonly contained many small germ cells with diffuse chromatin spread throughout nuclei as in spermatogonia or very early spermatocyte cysts (Fig. 5C), even in germ cells where Bam protein was not detected (Fig. 5A).
The testis TAFs (tTAFs) and tMAC complex, cell-type specific homologs of transcriptional control machinery expressed in young spermatocytes, control expression of many genes involved in meiotic cell cycle progression and spermatid differentiation (Hiller et al., 2001, 2004; White-Cooper et al., 2000). In males null mutant for either the tTAF, sa, or the tMAC component, aly, mature spermatocytes with characteristically large nuclei and crescent shaped, partially condensed autosomal bivalents form, but they arrest just prior to the G2/M transition of meiosis I when the partially condensed chromosomes have begun to round up and move inward from the nuclear periphery (Lin et al., 1996) (Fig. 5D and E). In contrast, male germ cell cysts null mutant for lolaORE76 remained small, and did not undergo an extensive increase in spermatocyte nuclear size. Sa protein was not detected in lolaORE76 mutant spermatocyte cysts (n=17 lolaORE76 homozygous mutant germ cell cysts scored), while spermatocyte cysts wildtype for lola function displayed prominent Sa immunostaining in nucleoli and on chromatin (Fig. 5F). In summary, germ cell cysts null mutant for lolaORE76 likely die shortly after the spermatogonia to spermatocyte transition, after the down-regulation of Bam protein but prior to the increase in cell size and formation of testis-specific complexes that coordinate the meiotic division program and spermiogenesis.
Additional evidence that lola function is required in germ cells for normal meiotic cell cycle progression and spermiogenesis came from analysis of germ cells homozygous mutant for the hypomorphic lola5D2 allele, which displayed less severe defects than lolaORE76 null mutant germ cells. At 4 to 6 dpci, comparable percentages of testes harbored marked control germ cell cysts and cysts homozygous mutant for lolaORE76 or lola5D2 in the spermatocyte region (Fig. 6A). However, the percentage of testes containing marked spermatid cysts homozygous mutant for lolaORE76 or lola5D2 was markedly lower than that observed for marked wildtype control cysts (Fig. 6A). Mosaic testes containing lolaORE76 null cysts often had one or more large clusters of refractile cell corpses amongst the spermatocyte cysts, suggesting that lolaORE76 cysts die without undergoing spermatocyte maturation, meiosis or terminal differentiation (Fig. 6B). In contrast, germ cells homozygous mutant for the lola5D2 hypomorphic allele usually grew as large as wildtype spermatocytes, but the cysts typically degenerated without undergoing meiotic divisions (Fig. 6C). Wildtype post-meiotic cysts at the “onion” round spermatid stage consistently have 64 equally sized haploid nuclei, each paired with a phase dark mitochondrial derivative which elongates as the spermatids mature (Fig. 6D). Although rare, when lola5D2 homozygous mutant spermatid cysts were observed, spermiogenesis was always defective (Fig. 6E). Onion-stage spermatids homozygous mutant for the lola5D2 hypomorphic allele displayed either 2 or 4 nuclei of equivalent size per mitochondrial derivative, indicating failure of one or both meiotic divisions, and showed no overt signs of spermatid elongation (Fig. 6E).
Fig. 6.
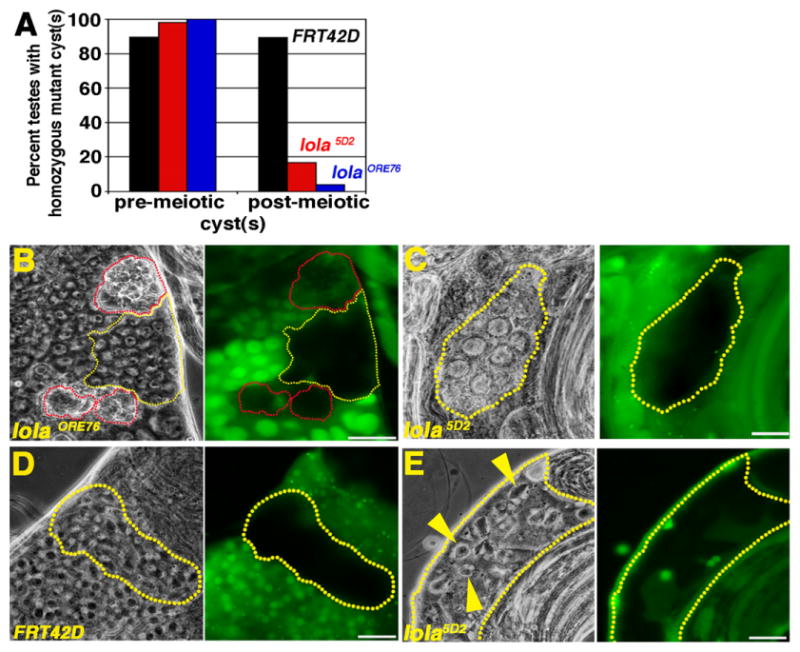
lola is required for progression through the meiotic divisions and spermiogenesis: (A) Percentage of testes with pre-meiotic or post-meiotic control or homozygous mutant germ cell cysts at 4–6 dpci. Black: FRT42D wildtype control. Red: lola5D2 hypomorph. Blue: lolaORE76 null. (B)–(E) Live squash preparations of a lolaORE76 (B), lola5D2 (C) and (E) and FRT42D (D) mosaic testes. Grayscale: phase contrast. Green: Ubi-nls-GFP. Yellow outlines (B)–(E) cysts homozygous mutant for lola (B, C, E) or FRT42D (D). (B) Red outlines: corpses of presumptive lolaORE76 mutant germline cysts. (C) Cyst of degenerating lola5D2 homozygous mutant spermatocytes. (D) FRT42D control spermatid cyst. Each haploid nucleus (white) associated with a mitochondrial derivative (phase dark). (E) lola5D2 homozygous mutant spermatid cyst. Yellow arrowheads: Two or more nuclei (white) were associated with an individual, enlarged mitochondrial derivative (phase dark), indicating failure of one or both meiotic divisions. Scale bars: 20 μm.
Cell-intrinsic requirements for lola in the male GSC lineage may be due to activity of several different lola isoforms
RT-PCR experiments using primers that spanned the splice junction between the lola common domain and each isoform-specific exon revealed that 19 of the splice variants tested (lola A-F, H-T) were expressed in both wildtype testes and upd over-expressing testes, which are highly enriched for cells that most closely resemble GSCs and CySCs (data not shown). Clonal analysis using isoform-specific, loss-of-function alleles for lola-K and lola-L indicated that these isoforms are required for different aspects of lola function in spermatogonia and spermatocytes. Likewise, the effects of inducible, isoform-specific RNAi knock-downs in early germ cells suggested that different requirements exist for lola-B, –N, and –P.
Both lola-K and lola-L were required cell autonomously for GSC maintenance. GSC clones homozygous mutant for either lola-K or lola-L were induced at frequencies comparable to control GSC clones, but the mutant GSCs were not maintained over time (Fig. 7A). lola-L mutant GSCs behaved similarly to lolaORE76 null GSCs, whereas lola-K mutant GSCs exhibited a less severe GSC loss phenotype (Fig. 7A). GSCs mutant for either lola-K or lola-L were lost to differentiation. lola-K mutant cysts were observed at the spermatogonial and spermtatocyte stages, while lola-L mutant cysts were observed at the spermatogonial stages and far from the apical tip of the testis in the spermatocyte region. None of the lola-K or lola-L mutant GSCs scored was positive for TUNEL staining (n=17 lolaORC4 mutant GSCS; n=12 lolaORE119 mutant GSCs). The percentage of testes containing differentiating germline clones mutant for lola-K or lola-L declined over time, as expected from the reduction in homozygous mutant GSCs (Fig. 7B).
Fig. 7.
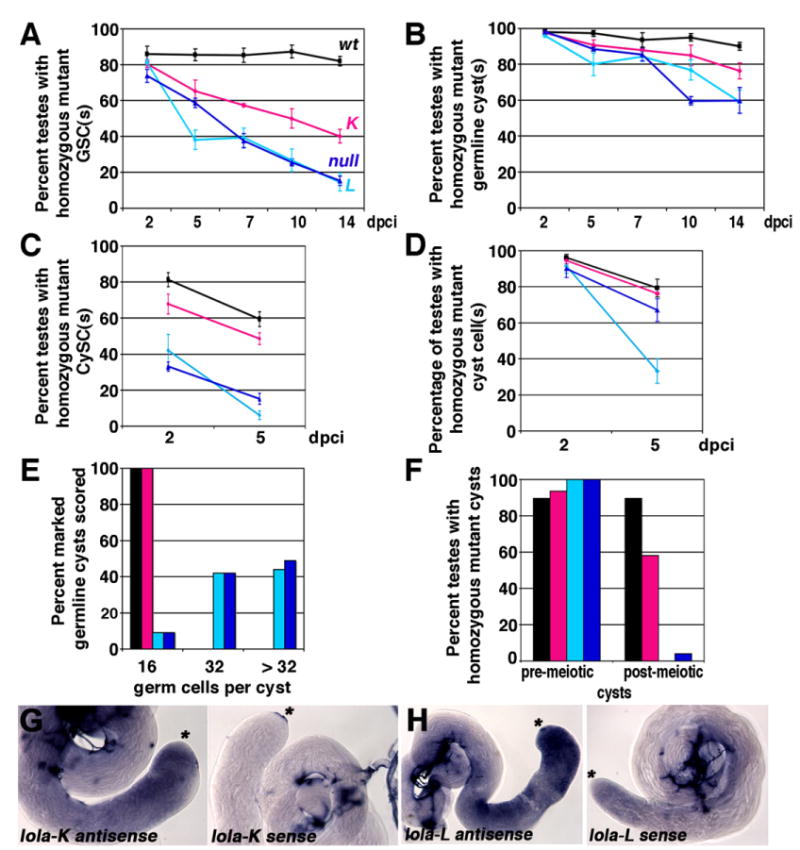
Requirements for lola-K and lola-L differ in the testis: (A) and (B) Percentage of mosaic testes containing homozygous mutant or marked control (A) GSCs or (B) spermatogonial and/or spermatocyte cyst(s) from 2 to 14 dpci. (C) and (D) Percentage of mosaic testes containing homozygous mutant or marked control (C) CySC(s) or (D) cyst cell(s) from 2 to 5 dpci. (E) Percentage of homozygous mutant or marked control or germline cysts in the spermatocyte region with 16, 32, or greater than 32 germ cells per cyst. n=100 cysts scored per genotype at 4–6 dpci. (F) Percentage of mosaic testes containing homozygous mutant or marked control cyst(s) in the spermatocyte region or post-meiotic spermatid cyst(s) at 4–6 dpci. (A)–(F): Black: FRT42D wildtype control clones. Magenta: lolaORC4 (K-specific) clones. Turquoise: lolaORE119 (L-specific) clones. Blue: lolaORE76 (null) clones. (G) and (H) In situ hybridization on wildtype testes with riboprobes complementary to the (G) lola-K specific exons and (H) lola-L specific exon. Left: Anti-sense probe. Right: Sense probe. Asterisks: Apical tip. Stained structures in sense testes are trachea.
Although lola-L mutant CySCs were lost to differentiation as rapidly as lolaORE76 null mutants, the lola-K specific mutation did not cause appreciable loss of CySCs in somatic cell clones (Fig. 7C). Clone induction frequencies in the CySC lineage were comparable for lola-L, lola-K and controls, as assayed by the percentage of mosaic testes harboring marked cyst cell clones at two dpci (Fig. 7D). However, far fewer lola-L mutant CySCs were detected at two dpci than control CySCs, and the trend worsened with time (Fig. 7C). In contrast, lola-K mutant CySCs were maintained over time (Fig. 7C).
The requirements for lola-K and lola-L in differentiating germ cells also appeared to be distinct. lola-L mutant spermatogonial cysts frequently underwent more than four rounds of mitosis, producing cysts of 32 or more small germ cells resembling late spermatogonia or early spermatocytes, similar in appearance to lolaORE76 null cysts (Fig. 7E). In contrast, lola-K mutant cysts always executed four rounds of transit amplifying division, producing cysts that contained 16 mature spermatocytes (Fig. 7E). lola-L was also required cell autonomously in spermatocytes for the meiotic divisions and spermiogenesis (Fig. 7F), whereas lola-K was seemingly dispensable for meiosis and spermatid differentiation (Fig. 7F). Taken together, the lola-L mutant phenotype mimicked the lolaORE76 null phenotype in all respects, suggesting that lola-L activity was required throughout the male GSC lineage. In contrast, lola-K activity was solely required for GSC maintenance. RNA in situ hybridization experiments with probes complementary to the K and L specific exons showed that both isoforms were expressed throughout the germinal proliferation center and primary spermatocytes (Fig. 7G and H). The comparatively weak phenotype seen in lola-K mutant clones may be due to redundancy with lola-T, since the C2HC-C2H2 zinc finger domains in Lola-K and Lola-T are 80% identical and the residues predicted to contact DNA are the same in both isoforms (Goeke et al., 2003). Alternatively, lola-K may be less essential than lola-L in differentiating male germ cells.
Isoform-specific RNAi knock-downs in early germ cells supported the conclusion that different isoforms may be required for specific aspects of lola function during spermatogenesis. Co-expression of UAS-Dcr-2 and UAS-dsRNA hairpins complementary to the lola-L specific coding sequence under control of nos-Gal4 also elicited GSC loss, suggesting that the GSC maintenance defect observed for lola-L mutant clones was indeed due to loss of lola-L function (Table 2). Germline specific knock-down of lola-L altered the number of transit amplifying divisions, producing mature spermatocyte cysts that contained more than 16 germ cells (Table 2), and caused degeneration of mature spermatocyte cysts prior to the G2/M transition of meiosis I (data not shown). As expected, the lola-L specific knock-down phenotypes were similar but less severe than those observed in homozygous mutant lola-L germline clones. lola-B knock-down in early germ cells produced small, round testes devoid of germ cells, suggesting that lola-B is required for GSC maintenance or germ cell survival (Table 2). In contrast, control testes lacking either the nos-Gal4 driver or the UAS-lola-B RNAi hairpins contained germ cells at all stages of development, and were normal in appearance (Table 2). RNAi knock-downs of isoforms N and P produced mature spermatocyte cysts with greater than 16 cells (Table 3), suggesting that each variant was required in spermatogonia for the correct number of transit amplification divisions. Based on analysis of cell number in isolated, intact cysts containing mature spermatocytes, 74% of spermatogonial cysts in lola-N knock-down testes and 10.7% of spermatogonial cysts in lola-P knock-down testes underwent five rounds of mitotic amplification division prior to transitioning to spermatocyte fate. In contrast, control testes lacking either the nos-Gal4 driver or the UAS-dsRNA hairpins always transitioned to the spermatocyte stage after exactly four rounds of mitosis (Table 3).
Table 3.
lola is required in the germline for the correct number of transit amplifying divisions. Live preparations of intact, mature spermatocyte cysts were scored by phase contrast microscopy (see Section 2). n=number of spermatocyte cysts scored.
| Percentage of mature primary spermatocyte cysts containing | |||||
|---|---|---|---|---|---|
|
| |||||
| 8 cells per cyst | 16 cells per cyst | 32 cells per cyst | >32 cells per cyst | Other (cell count not a multiple of 2n) | |
| nos-Gal4 alone (n=45) | 0 | 97.8 | 0 | 0 | 2.2 |
| pan lola hairpin alone (n=57) | 0 | 98.1 | 0 | 0 | 1.9 |
| pan lola knock-down (n=53) | 1.9 | 13.2 | 58.5 | 1.9 | 24.5 |
| lola-L hairpin alone (n=57) | 0 | 98.3 | 0 | 0 | 1.7 |
| lola-L knock-down (n=57) | 0 | 56.1 | 29.9 | 3.5 | 10.5 |
| lola-N hairpin alone (n=56) | 0 | 100 | 0 | 0 | 0 |
| lola-N knock-down (n=58) | 0 | 17.3 | 74.1 | 0 | 8.6 |
| lola-P hairpin alone (n=70) | 0 | 97.1 | 0 | 0 | 2.9 |
| lola-P knock-down (n=56) | 0 | 73.2 | 10.7 | 1.8 | 14.3 |
Conclusions
lola, a gene encoding a family of BTB-ZF transcriptional regulators, is a cell-intrinsic regulator of stem cell state in the testis: GSCs and CySCs homozygous mutant for lola detach from the hub and initiate differentiation. GSCs homozygous mutant for lola are not likely to be lost due to a defect in cell competition with adjacent heterozygous and/or homozygous wildtype GSCs, since pan-lola knock-down in early germ cells also induced GSC loss. In addition, lola is required for several key transitions as cells in the male GSC lineage differentiate. Upon exiting the niche, lola mutant spermatogonia frequently undergo one or more ectopic rounds of mitotic transit amplification division prior to making the cell fate transition from the spermatogonia to spermatocyte state. lola activity may regulate the proliferation rate of transit amplifying spermatogonia, since lola mutant spermatogonial cysts cycled faster than control cysts wildtype for lola function. The shorter cell cycle time in lola mutant germ cell cysts may allow extra round(s) of mitotic division before Bam protein reaches a critical threshold that triggers the switch to spermatocyte fate (Insco et al., 2009). lola null germ cells likely initiate the switch to spermatocyte fate, as cysts of small germ cells that no longer express Bam protein accumulated in the basal region of mosaic testes. However, most lola mutant cysts did not undergo the large increase in cell size typical of spermatocytes, failed to express the tTAF Sa, and ultimately arrested and died prior to undergoing the meiotic divisions or spermiogenesis. Some lola isoforms, like lola-L, appear to be required throughout spermatogenesis, while others, like lola-K, may have stage and/or cell-type specific functions.
lola does not appear to act directly in the JAK-STAT signaling pathway, nor does lola expression appear to be a target of TGF-β signaling in GSCs. lola may play a novel, parallel role, possibly as a transcriptional regulator of GSC and CySC maintenance. Strikingly the BTB-ZF protein PLZF, a transcriptional repressor, is required for maintenance of spermatogonial stem cells in the mouse testis (Buaas et al., 2004; Costoya et al., 2004). Although many characterized BTB-ZF proteins appear to act as dedicated transcriptional repressors (reviewed in Collins et al., 2001; Kelly and Daniel, 2006), it has been proposed that lola may be a bi-functional transcriptional regulator, either activating or repressing target genes in different contexts (Cavarec et al., 1997; Crowner et al., 2002; Spletter et al., 2007).
All five lola isoforms that exhibited a loss-of-function phenotype in the testis have tandem C2HC-C2H2 or C2HC-C2HC zinc finger motifs (Fig. 1B), raising the possibility that these proteins may bind DNA directly. C2HC motifs are predicted to be structurally similar to C2H2 domains, and can function either as DNA, RNA or protein binding domains (Laity et al., 2001). Notably, the Lola-K homolog from Drosophila hydeii was shown to bind to DNA directly (Cavarec et al., 1997). A multiple sequence alignment of the Tramtrack (Ttk) and Lola zinc finger domains revealed that surface residues predicted to contact DNA based on the Tramtrack crystal structure differ for all but two of the Lola isoforms (Goeke et al., 2003), raising the possibility that Lola homo- and/or heterodimers may have different consensus DNA binding motifs, and may regulate distinct sets of target genes. Moreover, at least four alternate transcription start sites have been identified for the lola locus, suggesting a strategy for regulating the relative abundance and/or combinations of lola isoform(s) in a stage or cell-type specific manner (Ohsako et al., 2003). In addition, protein production for certain Lola isoforms may be subject to translational control (Maines et al., 2007; E. Davies, W. Joo and M. Fuller, unpublished observations).
lola may also modify chromatin structure by interacting with histone modifying enzymes and Polycomb group complexes (Ferres-Marco et al., 2006; Zhang et al., 2003). Lola-F bound to the H3S10 kinase JIL-1 in yeast two hybrid assays via sequences in its isoform-specific C-terminal domain (Zhang et al., 2003), and a strong lola loss-of-function mutation dominantly suppressed the embryonic lethality and hypercondensed chromatin phenotype of JIL-1 mutants (Zhang et al., 2003). The antagonistic genetic interaction between JIL-1 and lola suggests that lola may promote heterochromatin formation, and by extension transcriptional silencing, since JIL-1 activity is required for maintenance of euchromatic domains (Zhang et al., 2006). Consistent with these data, strong lola loss-of-function mutations dominantly enhanced the Polycomb (Pc) extra sex comb phenotype in adult flies (Ferres-Marco et al., 2006). Finally, wildtype function of lola was required for chromatin condensation during programmed cell death in late stage oogenesis (Bass et al., 2007).
Intriguingly, different requirements exist for lola in spermato-genesis and oogenesis. In the ovary lola acts as a pro-differentiation factor, and its expression was repressed in GSCs and early cystocytes by the transcriptional repressor Stonewall (Maines et al., 2007). Consistent with these data, clonal analysis revealed that lola was dispensable for female GSC maintenance (J. Maines and D. McKearin, personal communication; E. Davies and M. Fuller, unpublished observations). Bass and colleagues reported that lola function was required for starvation-induced and programmed cell death of the nurse cells during stage 13 of oogenesis, and demonstrated a role for lola-K in this process (Bass et al., 2007). Thus lola function is required during late stage morphogenic events in both the testis and ovary, as loss of lola elicited developmental arrests in both systems. Differences in the severity of the lola-K phenotype in the testis and ovary underscores the notion that lola isoforms exhibit cell-type specific and context-dependent functions during development and tissue homeostasis.
Supplementary Material
Acknowledgments
Many thanks to Maria Spletter, Liqun Luo, Ed Giniger and Kristen Johansen for reagents and helpful discussions on the function of lola in other tissues, and to Hoa Nguyen for help with in situ hybridization experiments and embryo injections to generate UAS-dsRNA transgenic lines for isoform-specific RNAi knock-down experiments. This work was supported by grants from the National Science Foundation, Stanford Graduate Fellowship, and the American Heart Association to E.L.D., the Stanford Graduate Fellowship to J.G.Y.L., and NIH RO1 GM080501 to M.T.F.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2012.11.004.
References
- Bass BP, Cullen K, McCall K. The axon guidance gene lola is required for programmed cell death in the Drosophila ovary. Dev Biol. 2007;304:771–785. doi: 10.1016/j.ydbio.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Cavarec L, Jensen S, Casella JF, Cristescu SA, Heidmann T. Molecular cloning and characterization of a transcription factor for the copia retro-transposon with homology to the BTB-containing lola neurogenic factor. Mol Cell Biol. 1997;17:482–494. doi: 10.1128/mcb.17.1.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT. Science. Vol. 310. New York, NY: 2005. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation; pp. 869–872. [DOI] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Crowner D, Madden K, Goeke S, Giniger E. Development. Vol. 129. Cambridge, England: 2002. Lola regulates midline crossing of CNS axons in Drosophila; pp. 1317–1325. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; New York: 1993. pp. 71–147. [Google Scholar]
- Giniger E, Tietje K, Jan LY, Jan YN. Development. Vol. 120. Cambridge, England: 1994. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila; pp. 1385–1398. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. Science. Vol. 302. New York, N.Y: 2003. A protein interaction map of Drosophila melanogaster; pp. 1727–1736. [DOI] [PubMed] [Google Scholar]
- Goeke S, Greene EA, Grant PK, Gates MA, Crowner D, Aigaki T, Giniger E. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat Neurosci. 2003;6:917–924. doi: 10.1038/nn1105. [DOI] [PubMed] [Google Scholar]
- Gonczy P, DiNardo S. Development. Vol. 122. Cambridge, England: 1996. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis; pp. 2437–2447. [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Development. Vol. 131. Cambridge, England: 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program; pp. 5297–5308. [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109(12):2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Giniger E, Aigaki T. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev. 2003;17:2496–2501. doi: 10.1101/gad.1137303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco ML, Leon A, Tam CH, McKearin DM, Fuller MT. Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0912454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. Science. Vol. 326. New York, N.Y: 2009. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche; pp. 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase E, Wong MD, Ding BC, Xie T. Development. Vol. 131. Cambridge, England: 2004. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis; pp. 1365–1375. [DOI] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Science. Vol. 294. New York, N.Y: 2001. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue; pp. 2542–2545. [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Methods. Vol. 30. San Diego, CA: 2003. Making a better RNAi vector for Drosophila: use of intron spacers; pp. 322–329. [DOI] [PubMed] [Google Scholar]
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Development. Vol. 122. Cambridge, England: 1996. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males; pp. 1331–1341. [DOI] [PubMed] [Google Scholar]
- Madden K, Crowner D, Giniger E. LOLA has the properties of a master regulator of axon-target interaction for SNb motor axons of Drosophila. Dev Biol. 1999;213:301–313. doi: 10.1006/dbio.1999.9399. [DOI] [PubMed] [Google Scholar]
- Maines JZ, Park JK, Williams M, McKearin DM. Development. Vol. 134. Cambridge, England: 2007. Stonewalling Drosophila stem cell differentiation by epigenetic controls; pp. 1471–1479. [DOI] [PubMed] [Google Scholar]
- Ohsako T, Horiuchi T, Matsuo T, Komaya S, Aigaki T. Drosophila lola encodes a family of BTB-transcription regulators with highly variable C-terminal domains containing zinc finger motifs. Gene. 2003;311:59–69. doi: 10.1016/s0378-1119(03)00554-7. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Science. Vol. 218. New York, N.Y: 1982. Genetic transformation of Drosophila with transposable element vectors; pp. 348–353. [DOI] [PubMed] [Google Scholar]
- Schulz C, Kiger AA, Tazuke SI, Yamashita YM, Pantalena-Filho LC, Jones DL, Wood CG, Fuller MT. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics. 2004;167:707–723. doi: 10.1534/genetics.103.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr Biol. 2003;13:2065–2072. doi: 10.1016/j.cub.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Spletter ML, Liu J, Liu J, Su H, Giniger E, Komiyama T, Quake S, Luo L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Dev. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H, Leroy D, MacQueen A, Fuller MT. Development. Vol. 127. Cambridge, England: 2000. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated; pp. 5463–5473. [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Schafer MA, Alphey LS, Fuller MT. Development. Vol. 125. Cambridge, England: 1998. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila; pp. 125–134. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Development. Vol. 117. Cambridge, England: 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues; pp. 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zhang W, Deng H, Bao X, Lerach S, Girton J, Johansen J, Johansen KM. Development. Vol. 133. Cambridge, England: 2006. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila; pp. 229–235. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang Y, Long J, Girton J, Johansen J, Johansen KM. A developmentally regulated splice variant from the complex lola locus encoding multiple different zinc finger domain proteins interacts with the chromosomal kinase JIL-1. J Biol Chem. 2003;278:11696–11704. doi: 10.1074/jbc.M213269200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


