Introduction
The last decade has seen substantial growth in our understanding of mammalian genomes, both at the DNA sequence level and the regulatory mechanisms that modulate their function (The ENCODE Project Consortium, 2012; Lander et al., 2001; Waterston et al., 2002). Advances in genome biology have informed every discipline in biology, including neuroscience. Not only has our understanding of the development and evolution of the nervous system improved in response to the genomic revolution, but it has also reinforced the notion that genetic information governs neural processing and behavior. The recent striking demonstration that genes quantitatively affect distinct behavioral modules in the mouse (Xu et al., 2012), in a fashion similar to what was known for decades in invertebrates (Benzer, 1973), suggests that genetics play a major role in perception, cognition, and behavior of higher organisms. Appreciating the genetic underpinnings of neural processing, without a doubt, will modify our efforts to understand neurological and psychiatric disorders and will provide new approaches for the understanding of the brain.
Upon agreement that genes control behavior, an obvious next question is how the expression of these genes is regulated and coordinated for the generation of a functional nervous system. Seminal experiments performed over half a century revealed how combinations of transcription factors, utilizing the basic principles of synergy and cooperativity that were first described in the lambda phage (Ptashne, 1989), transform spatiotemporal cues to precise orchestration of gene expression and development of the nervous system (Albright et al., 2000). Although these regulatory mechanisms are, by and large, encoded by the genome itself, there are an increasing number of paradigms whereby information encoded in the DNA is overruled by so-called “epigenetic” factors. Although the term “epigenetic” in its original definition assumed heritability, in the case of post-mitotic neurons inheritance of epigenetic information is not applicable; therefore, for the needs of this review, the use of the term epigenetic refers to the Greek etymology of the word, which means “over the genetic information”. Any time we use the term “epigenetic” in this essay, we simply refer to modifications of chromatin and its structure that do not result from changes to the underlying DNA sequence, regardless of heritability. DNA and post-translational histone modifications constitute the best-characterized epigenetic modifications, and their role in neural processes is described in detail in other reviews of this issue. Here, we will focus to a relatively novel axis of epigenetic regulation, which is not directly linked to the epigenetic marks of a genomic locus but instead to its nuclear coordinates. Experiments in various cell types and organisms suggest that the genetic material is a three-dimensional structure defined by topological constraints that may differ between cell types and differentiation states. Thus, the linear order depicted by the genomic coordinates is not necessarily retained in the 3-dimensional nuclear space. Therefore, a gene’s nuclear neighborhood could potentially determine its transcriptional competence or activity, or coordinate the expression of many genes found on separate chromosomes by bringing them in close spatial proximity. Increasingly, evidence suggests nuclear organization does indeed have functional implications and is the subject of regulation. This suggests that the spatial organization of the nucleus, or nuclear architecture, is likely to play an important role in directing cellular differentiation, organismal development, and disease etiology. In this review, we discuss the current understanding of the role that nuclear architecture plays in the developing nervous system. We focus predominantly on the biology of mammalian organisms, but in some cases will include insights observed in other model organisms (e.g. Drosophila melanogaster). We will first provide a general survey of known features of 3-dimensional nuclear organization: higher-order organization of the chromatin fiber, spatial localization of chromosomes and distinct chromatin types, and organization of nuclear processes in nuclear bodies (Figure 1). We aim to provide only a general introduction and refer an interested reader to more extensive reviews on the subject (Bickmore and van Steensel, 2013; Misteli, 2007). We will then highlight examples in which nuclear architecture has been implicated in the development and/or function of neural cell types, emphasizing the potential insights that can be applied to neurogenesis at large.
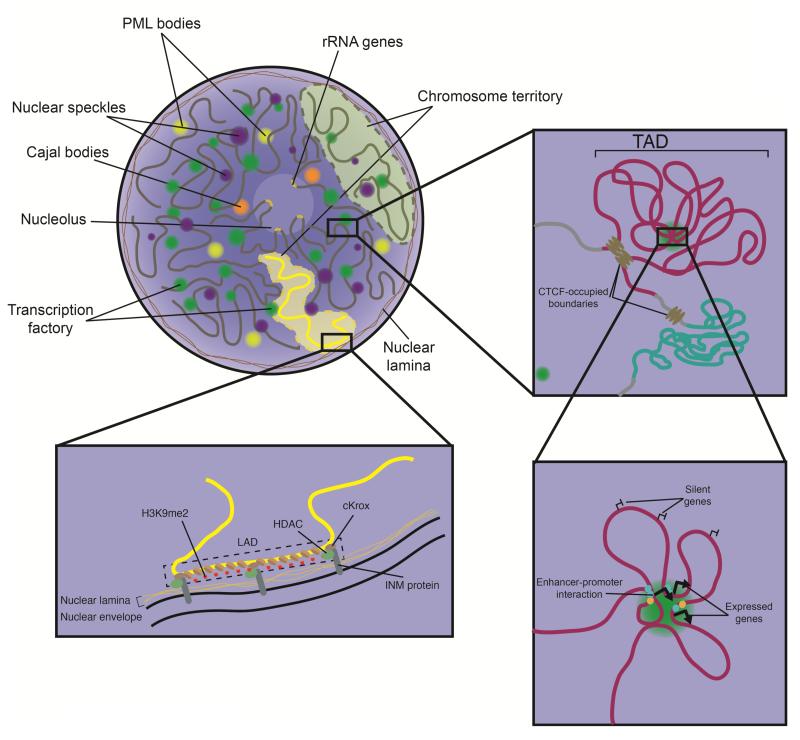
Figure 1.
Organization of chromatin in topological and lamina-associated domains. A. Cartoon model for the higher-order organization of chromatin. A mammalian nucleus shows chromosomes (dark grey lines) confined to distinct territories. rRNA genes are confined within the nucleolus. Cajal bodies are shown juxtaposed to the nucleolus. Nuclear speckles and transcription factories reflect high concentrations of the splicing and transcriptional machinery, respectively. These structures are depleted in nuclear compartments associated with repressed heterochromatin: the nuclear lamina and chromatin associated with the nucleolus. On right, a magnified view of the boxed region shows two topological associated domains (TADs), colored red and green, separated by CTCF-bound boundary regions. Within each TAD are numerous chromosomal interactions; however, few interactions cross boundaries between TADs. On bottom right, further magnification of a transcription factory with a TAD demonstrates close associations between distal regulatory regions and expressed genes. Moreover, numerous expressed genes colocalize in this space and share similar sets of transcription factors (green and orange circles). Silent genes found in between these active genes in linear sequence loop away from the transcription factory and occupy a distinct region. On bottom left, a repressed chromatin domain is associated with the nuclear lamina, known as a lamina-associated domain (LADs). LADs have hallmarks that include high levels of repressive H3K9 methylation. Interactions between inner nuclear membrane proteins, such as emerin and Lap2β, with HDACs and cKrox are also essential for LAD establishment.
Principles of Nuclear Architecture
The nucleus serves as a repository for the DNA content of a cell. Within the cell nucleus, DNA is packaged in chromatin. At its most fundamental level, chromatin is a repeating array of nucleosomes, which may be remodeled, post-translationally modified, or altered through the incorporation of histone variants to modulate this basic structure (for review, see (Bernstein et al., 2007; Campos and Reinberg, 2009; Hargreaves and Crabtree, 2011)). In addition, chromatin is organized into complex but regulated topological hierarchies that determine domains of chromosomal interaction and form the basis of nuclear subcompartments. These aspects of high-order chromatin organization are just coming into view.
Using a number of approaches related to chromosome conformation capture (3C), which uses frequency of ligation between crosslinked and digested genomic DNA to measure chromosomal interactions (Box 1, for review see (de Wit and de Laat, 2012)), multiple groups have investigated higher-order chromatin organization in various model organisms. These studies reveal that chromatin folds into topological associated domains (TADs), regions of widespread intrachromosomal interaction that are isolated from the surrounding chromatin by sharp boundaries. TADs are roughly 0.1-1 megabases (Mb) in size and are themselves organized into much larger interaction domains (Lieberman-Aiden et al., 2009). Furthermore, higher-resolution analysis of genomic loci flanking key developmental genes in embryonic stem cells has identified smaller blocks of interactions within previously identified TADs. These were labeled sub-TADs (Phillips-Cremins et al., 2013). The picture that emerges from these studies is a hierarchical organization of genomic folding where blocks of chromatin self-assemble into interacting globules, which then assemble into still larger domains of chromatin (Lieberman-Aiden et al., 2009).
Box 1.
Chromosome Confirmation Capture (3C)
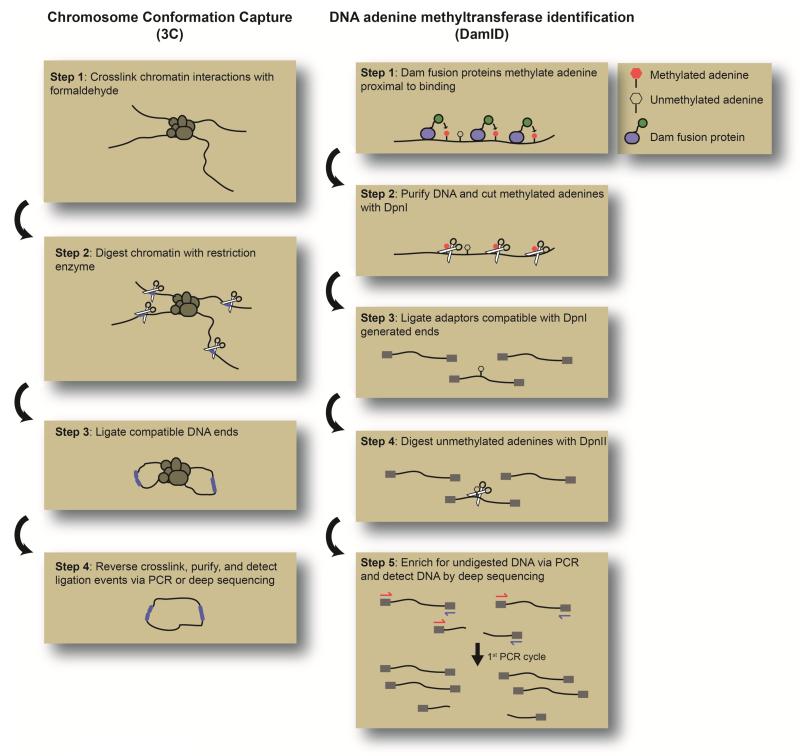
Chromosome confirmation capture is a powerful approach for the detection of interactions between genomic regions. The concept behind 3C is straightforward. Interactions between regions of chromatin are captured by formaldehyde fixation (step 1). The DNA is then digested with a restriction enzyme (step 2) and ligated in conditions that favor intramolecular ligation (step 3). Ligation will generate novel DNA sequences derived from the ligation of compatible restriction ends that are distally located in the linear DNA sequence but in close proximity in the cell nucleus. These ligation products can then be detected by PCR or deep sequencing. In this way, 3C may be used to investigate a single interaction between two genomic regions. This procedure can also be adapted to investigate the genome-wide interactions of a single “bait” locus (called 4C), a fixed number of potential interactions within a large (> 4 Mb) but limited genomic region (5C), or all possible interactions (Hi-C). Furthermore, interactions involving a specific histone mark or transcription factor can be identified by performing immunoprecipitation prior to 3C digestion and ligation. This method, Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET), enriches for those interactions that involve chromatin bearing the histone modification or transcription factor of interest.
DNA adenine methyltransferase identification (DamID)
DamID uses the exogenous expression of the bacterial methyltransferase dam to molecularly label DNA with adenine methylation. The methylated DNA can then be selectively amplified using methylation-sensitive restriction enzyme digestion and PCR amplification or by immunoprecipitating using a methylation-specific antibody. By fusing Dam to a protein of interest, one can identify genomic regions where this protein binds. This makes it possible to study the genomic occupancy of factors for which standard chromatin immunoprecipitation is not available due to lack of quality antibodies. However, because in many cases the Dam fusion protein is continuously expressed and the methylation mark is stable, DamID provides a cumulative read-out of occupancy rather than a snapshot.
TADs are separated from one another by boundary elements that limit the extent of intrachromosomal interactions. These boundaries are distinguished by a set of common features, mainly enrichment for gene promoters, hallmarks of accessible chromatin and transcription, and binding of insulator proteins such as CTCF (Dixon et al., 2012; Hou et al., 2012; Sexton et al., 2012). Some of these features, such as CTCF binding, are often found near transitions between distinct chromatin states (Barski et al., 2007). Indeed, TAD organization often nicely overlaps with domains of histone modifications. This raises the question of whether TAD formation is driven by epigenetic marks or if the boundaries of local chromatin states are a product of TAD organization. In support of the latter hypothesis, establishment of TADs is observed prior to the appearance of histone modification domains during stem cell differentiation (Dixon et al., 2012), which suggests that formation of TADs is independent of the underlying epigenetic modification state. Consistent with this, genetic deletion of histone modifying enzymes disrupts histone modification status but not TAD organization (Nora et al., 2012). These findings suggest that TAD organization may serve as a framework for the establishment and alteration of epigenetic modification.
How does the description of TADs inform our understanding of gene regulation? Interestingly, while TAD boundaries are maintained with differentiation, the frequency of intrachromosomal contacts within TADs is altered (Dixon et al., 2012; Nora et al., 2012). This may reflect dynamic activity of distal regulatory enhancers (Phillips-Cremins et al., 2013). In this way, TADs may represent a coordinated block of chromatin, restricting enhancer-promoter interactions to those found within its boundaries. A gene’s transcriptional activity would then be predominantly regulated by the activity of relevant cis-regulatory modules within its TAD and refined by modulation of the finer chromosomal architecture within this domain (Figure 1). Experimental evidence supports this view; genes within a single TAD were found to have correlated expression changes during differentiation, and genetic deletion of a boundary element led to dysregulation of genes in the neighboring TAD greater than 1 Mb away (Nora et al., 2012). Thus, TADs may be a fundamental organizing unit of DNA and DNA-dependent processes such as transcription.
Chromosome territories and nuclear location
Chromosomes occupy discrete locations within the cell nucleus as opposed to interweaving in the nuclear space. These domains are commonly referred to as “chromosome territories” (CTs) (for review, see (Cremer and Cremer, 2010; Cremer et al., 2006; Lanctôt et al., 2007)). CTs generally demonstrate only limited mobility within the interphase nucleus (Abney et al., 1997; Strickfaden et al., 2010); however, visualization of genetic loci in yeast and flies indicates that within a CT chromatin is highly mobile and displays movement consistent with constrained diffusion (Marshall et al., 1997; Vazquez et al., 2001). Thus, chromatin mobility allows for genomic loci to sample potential interactions within a nuclear neighborhood -- which is presumably critical for the dynamic use of enhancer elements -- while the overall organization of the nucleus is maintained.
The three-dimensional location of genes within a chromosome territory has been suggested to influence or reflect gene expression. This was based on models that suggested that genes within the interior of a CT have limited access to the transcriptional machinery (Cremer and Cremer, 2001). Thus, inactive genes would relocate to the boundary of a CT upon transcriptional activation. However, the relationship between gene position within a CT and transcription status is uncertain. Expressed genes do not necessarily reside at the CT periphery (Küpper et al., 2007; Mahy et al., 2002b), nor is looping out of a CT a requirement for transcriptional activation (Hakim et al., 2011). For example, while the murine Hoxd cluster escapes its CT during activation in the tail bud, no such looping is observed in its expression domain in the limb (Morey et al., 2007). Nevertheless, many loci show a reproducible preference for localization outside of their respective CTs (Chambeyron and Bickmore, 2004; Küpper et al., 2007; Mahy et al., 2002a; Volpi et al., 2000; Williams et al., 2002). 3C-related approaches have consistently noted a propensity for gene rich, active genomic loci to make long-range interchromosomal interactions (Hou et al., 2012; Kalhor et al., 2011; Lieberman-Aiden et al., 2009; Sexton et al., 2012; Simonis et al., 2006), which demonstrates that these regions must have access to other chromosomal regions or territories, perhaps as part of transcription factories (see below). These findings are supported by DNA fluorescence in situ hybridization (FISH) experiments that have observed transcription-dependent chromosomal intermingling (Branco and Pombo, 2006) or localization of gene exons to the periphery of a CT (Boyle et al., 2011). What might regulate localization within a CT is not completely clear, but DNA methylation appears to play a role. For instance, human cells with a mutation in DNMT3B have hypomethylation of a pseudoautosomal region (PAR) on the inactive X chromosome and greater localization of this region to the periphery of the inactive X CT (Matarazzo et al., 2007). Thus, the positioning of genes within a chromosome territory is often linked to transcriptional status, although further investigation is required in order to establish a functional relationship between the two.
In addition to the territorial organization of chromosomes, the nucleus is partitioned into distinct subcompartments or domains, with CTs distributed throughout these domains in a non-random fashion. It is now well established that certain chromosomes tend to populate the nuclear interior while others are found adjacent to the nuclear envelope. Chromosomal location correlates with gene density and chromosome size; small, gene-rich chromosomes tend to localize to a more interior position than large, gene-poor chromosomes (Boyle et al., 2001; Cremer et al., 2001; Croft et al., 1999). The observation that gene-rich chromosomes congregate towards the nuclear interior may reflect a general affinity between transcriptionally active regions, which has been described using DNA FISH (Shopland et al., 2006) and 3C techniques (Hou et al., 2012; Kalhor et al., 2011; Lieberman-Aiden et al., 2009; Sexton et al., 2012; Simonis et al., 2006; Splinter et al., 2011; Yaffe and Tanay, 2011, Li et al., 2012, Chepelev et al., 2012).
The nuclear periphery, in contrast, is populated by domains of repressed heterochromatin that are tethered to the nuclear lamina. Several groups have identified the lamina-associated chromatin (referred to as lamina-associated domains or LADs) using DamID (Box 1, van Steensel and Henikoff, 2000). These studies have revealed LADs as large domains of little transcriptional activity demarcated by sharp boundaries (Guelen et al., 2008; Pickersgill et al., 2006; van Bemmel et al., 2010). LADs lack active chromatin marks and have been found to correlate with some but not all repressive chromatin marks depending on the study, species, and marks investigated (Pickersgill et al., 2006; Guelen et al., 2008; Wen et al., 2009; Peric-Hupkes et al., 2010; Kind et al. 2013). In particular, the repressive histone modification H3K9me2 was found to overlap substantially with LADs in mammalian cells. This histone mark may play a functional role in the recruitment of specific chromatin domains to the lamina. Deletion of the H3K9 methyltransferase G9a disrupted LAD formation in human cells (Kind et al., 2013). In the nematode C. elegans, knockdown of two H3K9 methyltransferases leads to release of transgenic and endogenous DNA sequence from the nuclear lamina (Towbin et al., 2012). In addition, loss of histone deacetylases (Zink et al., 2004; Zullo et al., 2012) or the DNA-binding factor cKrox (Zullo et al., 2012) can lead to disruption of lamina-associated chromatin. Thus, the cooperative activity of both DNA-binding factors and histone modifying enzymes reinforce lamina association (Figure 1).
How do LADs relate to the more general features of chromosomal organization discussed above? Interestingly, LADs share many common features with TADs. For instance, many of the boundaries that delimit LADs from the surrounding chromatin have also been identified as TAD boundaries (Dixon et al., 2012; Nora et al., 2012). This suggests that LADs may simply reflect TADs or collections of TADs that are tethered to the nuclear periphery.
How stable are chromatin-lamina associations and how do these interactions relate to gene expression? Recent studies demonstrate that LADs randomly associate with the lamina after each cell division such that only ~30% of LADs contact the nuclear envelope in a given cell (Kind et al., 2013). In some cases, LADs that do not end up next to the nuclear lamina associate with other repressive compartments, such as the nucleolus (Thomson et al., 2004; Kind et al 2013). When averaged over a population of cells, LADs were mostly conserved between embryonic stem cells, neural precursors, and differentiated astrocytes (Peric-Hupkes et al., 2010); however, some changes in lamina-association were observed, and these correlated with changes in gene expression. For instance, Pcdh9, which is expressed in the brain, is activated during astrocyte differentiation and concomitantly relocated from the nuclear lamina to the interior. Conversely, the cell-cycle gene E2f3 shifts from the interior to the nuclear lamina as neural progenitors differentiated to astrocytes (Peric-Hupkes et al., 2010). This is consistent with microscopic evidence for movement away from the nuclear lamina upon activation of specific genes (Kosak, 2002; Kumaran and Spector, 2004; Williams et al., 2006). Indeed, proximity to the nuclear lamina may be sufficient to suppress gene expression, as forcing genomic loci to the lamina leads to gene silencing in some (Dialynas et al., 2010; Finlan et al., 2008; Reddy et al., 2008; Zullo et al., 2012) though not all (Kumaran and Spector, 2008) cases. Taken together, these findings support a repressive role for the nuclear periphery in the regulation of gene expression.
Nuclear Bodies
Nuclear processes are spatially organized in the nucleus in dense collections of functionally-related factors known as nuclear bodies (for review, see (Dundr and Misteli, 2010; Dundr, 2012))(Figure 1). These include the nucleolus, the most prominent nuclear body that is dedicated to rRNA transcription and ribosome biogenesis (Pederson, 2011), but also Cajal bodies, promyelocytic leukemia protein (PML) bodies, nuclear speckles, and Polycomb bodies. Cajal bodies have been implicated in assembly of spliceosomal snRNPs and are highly enriched in functional RNA species involved in snRNA modification (Nizami et al., 2010). The function of PML bodies remains uncertain but may include the regulation of transcription, DNA replication, and DNA damage (Dundr, 2012; Lallemand-Breitenbach and de The, 2010). Nuclear speckles are accumulations of snRNPs and other splicing factors and may function in recycling the pre-mRNA processing apparatus (Spector and Lamond, 2011). Genes silenced by Polycomb repressive complexes interact with each other at high frequency (Bantignies et al., 2011; Sexton et al., 2012; Tolhuis et al., 2011, Denholtz et al., 2013). In some cell types, these interacting partners are visible by light microscopy as prominent foci of Polycomb proteins (Bantignies et al., 2011). These distinct nuclear subcompartments reflect the complexity in which the constituent parts of the nucleus are ordered to control genome function.
Transcriptional activity occurs in punctuate regions throughout the nucleus in regions commonly referred to as “transcription factories” (for review, see (Edelman and Fraser, 2012)). The nucleolus is a prominent example of this organization in the nucleus. Transcription of the rRNA genes is restricted to the nucleolus, making it, at least in part, a large transcription factory for RNA polymerase (RNAP) I. Transcription of genes by RNAP II also occurs in factories, which have been identified through visualizing nascent RNA (Jackson et al., 1993; Wansink et al., 1993) or components of the transcriptional machinery (Iborra et al., 1996). It is estimated that each factory contains numerous genes, and ChIA-PET studies (see Box 1) have demonstrated extensive interactions between co-expressed gene promoters, (Li et al., 2012, Zhang et al., 2013, Chepelev et al., 2012), which supports the transcription factory hypothesis. This raises the possibility that transcription factories provide an opportunity to spatially coordinate the expression of multiple co-regulated genes. Indeed, interacting genes show some correlation in expression between cell and tissue types (Li et al., 2012, Chepelev et al., 2012). Tumor necrosis factor alpha (TNFα)-responsive genes on distinct chromosomes interact with each other more often than with expressed but non-responsive control genes (Papantonis et al., 2012), and recent studies have provided evidence that these interactions are functionally important for co-regulated gene expression (Fanucchi et al., 2013). Moreover, the active hemoglobin genes Hbb and Hba preferentially associate with other Klf1-regulated genes within the nucleus (Schoenfelder et al., 2010). It is important to note that the existence of spatially segregated transcription factories dedicated to coordination of co-regulated genes is far from established and may be more prevalent for certain transcriptional programs. For instance, a different study found that while human Hbb and Hba are often found in proximity in differentiating erythroblasts, this is due to association with a common nuclear speckle and not sharing of a transcription factory (Brown et al., 2008). Other 3C studies have not identified preferential colocalization of co-regulated genes nor extensive interactions between chromosomes (Hakim et al., 2011; Simonis et al., 2006). The differences between these findings may be explained by distinct factor- or cell type-specific modes of transcriptional control. However, the precise relationship between the well-established organization of transcription into factories and its potential regulatory role in coordinating gene expression programs requires further investigation.
We have explored how chromatin is folded in high-order domains, how chromosomes and distinct chromatin types are partitioned in the interphase nucleus, and how nuclear processes are ordered in distinct nuclear bodies. All of these features contribute to the architecture of the nucleus, which is molded or modified during transitions in cell state. We now consider the modification of nuclear architecture during neurogenesis: how it is altered and how these changes impact the specification, differentiation, and function of neural cell types.
Drosophila Neurogenesis
The fruitfly Drosophila melanogaster is a powerful model to understand neurogenesis. In the fly, neuroblasts give rise to distinct neuronal subtypes based on spatiotemporal cues; a single neuroblast will give rise to a predictable series of neurons and glia during development. This is achieved by restricting the ability of a neuroblast to respond to specification cues and modifying this “competence” for specification over time.
Recent studies have demonstrated that nuclear position of the gene hunchback (hb) modifies the competence of the NB7-1 neuroblast (Kohwi et al., 2013). Prolonged expression of hb in NB7-1 leads to increased numbers of U1/U2 motoneuron subtypes in its neuronal progeny (Isshiki et al., 2001). However, U1/U2 identity, which the authors determined from the expression of endogenous hb (which could be distinguished from ectopic hb) can only be driven in the early-born neurons generated in the first 5 cell divisions of NB7-1. Remarkably, the competence window for U1/U2 identity precisely correlates with relocalization of the endogenous hb gene to the nuclear periphery (Figure 2A). Depletion of nuclear lamins leads to reduced recruitment of the hb gene to the nuclear periphery and extends NB7-1 competence for U1/U2 specification to later in development. Furthermore, prolonged expression of the DNA-binding factor Dan can disrupt recruitment of the hb gene to the nuclear periphery and induce abnormal numbers of U1/U2 motoneurons when hb is also misexpressed, suggesting loss of Dan in NB7-1 neuroblasts restricts their competence by allowing recruitment of the hb locus to a repressive nuclear domain near the nuclear envelope. How localization to the nuclear periphery restricts U1/U2 fate or hb activation was not determined. Nonetheless, these observations provide intriguing evidence for an instructive role for gene positioning in neuronal cell fate.
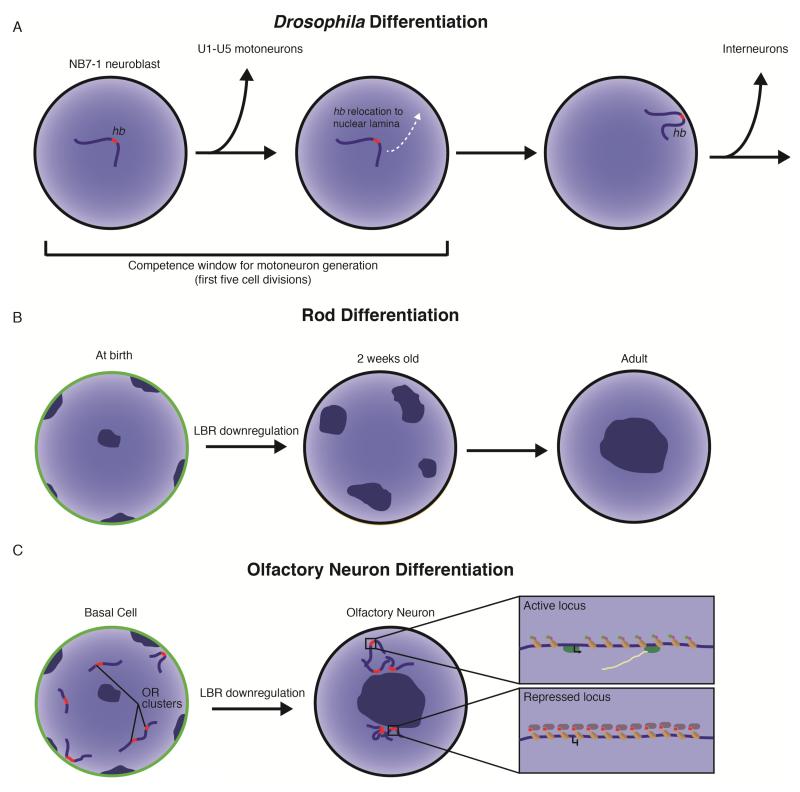
Figure 2.
Nuclear architecture in neurogenesis. A. During Drosophila neurogenesis, relocation of the hunchback (hb) gene marks the end of a competence window where neuroblast NB7-1 can be induced to generate U1/U2 motoneurons by ectopic hb expression. Prior to its first five cell divisions, NB7-1 generates U1-U5 motoneurons sequentially, but ectopic expression of hb can led to additional neurons with U1/U2 identity. After five cell divisions, the hb gene transitions from the nuclear interior to associate with the nuclear lamina, where it is stably silenced and can not promote U1/U2 identity. NB7-1 gives rise to interneurons after hb is recruited to the nuclear lamina. B. At birth, the nuclei of rod photoreceptor cells in nocturnal animals demonstrate conventional organization with chromocenters (dark blue regions) at the nuclear periphery and expression of Lamin B receptor (LBR, green outline). As these cells mature, LBR is downregulated and the chromocenters gradually fuse to leave a single centrally-located chromocenter in the nuclear interior. C. In undifferentiated basal cells, olfactory receptor (OR) genes are dispersed through the nucleus and expression of LBR is robust. In mature olfactory sensory neurons (OSNs), OR genes aggregate near a centrally located chromocenter as LBR expression is lost. Boxes highlight the local chromatin of active and repressed OR alleles. The active OR allele loops out into a nuclear domain of active chromatin and is marked by H3K4me3 (green circles), histone acetylation (purple circles), and transcribing RNA polymerase. The repressed OR genes are enriched for H3K9me3 (red circles) and heterochromatin protein 1β (purple ovals).
Mammalian cells show similar changes in the nuclear positioning of important genes in neurons and other related cell types. For example, glial fibrillary acidic protein (GFAP) is expressed from a single allele in both primary astrocytes and astrocytes differentiated in vitro. The expressed Gfap allele preferentially colocalizes with nuclear speckles and has a more internal nuclear position than the inactive Gfap allele (Takizawa et al., 2008). Mash1 (also known as Ascl1), a basic-helix-loop-helix proneural transcription factor, is induced during neural differentiation of embryonic stem cells. Mash1 transitions from the nuclear periphery to a position in the nuclear interior specifically during neural differentiation, suggesting Mash1 nuclear localization is regulated during this process (Williams et al., 2006). Taken together, these data suggest that the nuclear location of key genes may be an important regulatory mechanism during neural differentiation.
Rod Photoreceptor Cells
The nuclei of rod photoreceptor cells, which make up the outer nuclear layer of the retina, demonstrate an unusual organization of the underlying chromatin. Early microscopic studies of mouse rod nuclei describe a single, compact, centrally-located focus of heterochromatin surrounded by a thin halo of euchromatin (Carter-Dawson and LaVail, 1979). This differs from neighboring cone nuclei and canonical nuclear architecture, where heterochromatin is observed juxtaposed to the nuclear envelope. Recently, Joffe and colleagues (Solovei et al., 2009) revisited the structural organization of rod nuclei and its functional ramifications. They found mature rod nuclei have a tripartite organization: a single chromocenter core in the nuclear interior, LINE-rich heterochromatin surrounding this core, and euchromatin enriched for B1 repeats in a thin shell near the nuclear periphery. This architecture appears gradually over a course of months. At birth, when rod photoreceptor cells are still proliferative, their nuclei demonstrate canonical organization with roughly 12 distinct foci of constitutive heterochromatin abutting the nuclear envelope. These foci then gradually fuse with time to leave 1-2 chromocenters within the nuclear interior by postnatal day 28 (Figure 2B).
Why do mouse rod photoreceptor cells adopt such atypical nuclear organization? Interestingly, a striking correlation between rod nuclear architecture and mammalian behavior was found. Retinas from nocturnal mammals demonstrated rod nuclei with large, centrally located chromocenters, whereas in diurnal mammals, rods displayed the conventional nuclear architecture. Through computer modeling, the authors predicted that the “inside-out” nuclear organization observed in nocturnal mammals modifies the refractive properties of this organelle from predominately light scattering (diurnal mammals) to resembling that of a converging lens. Light must pass through the outer nuclear layer to reach the light-sensitive pigments of rods. Thus, modifying nuclear structure in order to limit distortion of incoming light rays could be adaptive for a life spent foraging or hunting in the dark.
The molecular basis for nuclear reorganization during rod photoreceptor cell maturation appears to be mediated by dynamic regulation of nuclear envelope proteins. Both the lamin B receptor (LBR) and lamin A/C mediate interactions between the nuclear envelope and heterochromatin, and the inside-out nuclear architecture of rod nuclei is linked to absence of both LBR and lamin A/C (Solovei et al., 2013). Mouse rod photoreceptor cells are negative for lamin A/C, but express LBR until roughly 2 weeks after birth. Thus, downregulation of LBR coincides with transformation of rod nuclear organization. Ectopic expression of LBR, but not lamin C, is sufficient to revert rod nuclei to the canonical architecture (Solovei et al., 2013).
Olfactory Sensory Neurons
The olfactory epithelium (OE) is populated by olfactory sensory neurons (OSN), which project axons to the olfactory bulb to relay odorant detection within the nose to processing centers in the brain. Odorants are sensed by a large family of olfactory receptors (ORs)(Buck and Axel, 1991) that are monogenically and monoallelically expressed in each mature OSN in a stochastic fashion throughout the mammalian OE (for review, see (Fuss and Ray, 2009; Mombaerts, 2001; Shykind, 2005). The expressed OR determines the identity of each sensory neuron. OSNs that express the same OR are sensitive to the same odorants and project axons that converge within the same glomeruli in the olfactory bulb. In this way, an odor’s representation in the brain is likely generated by a map of glomerular activation elicited by the combination of ORs stimulated. Thus, singular expression of an OR that is maintained throughout the life of an OSN is essential for the fidelity of smell.
The regulation of OR transcription represents one of the best-characterized examples of epigenetic regulation in the nervous system. OR gene promoters share common transcription factor binding motifs (Clowney et al., 2011), suggesting that their genetic signature is not sufficient to instruct unique expression in single ORs in subpopulations of olfactory neurons. This observation, combined with the monoallelic nature of OR expression (Chess et al., 1994) suggests that the expression of one out of thousands of OR alleles cannot be solely explained by the existence of different transcription factor combinations in different olfactory neurons. Indeed, epigenetic silencing via repressive histone modifications typically found on constitute heterochromatin (H3K9me3 and H4K20me3) appears sufficient for the monogenic and monoallelic expression of OR genes in the olfactory epithelium (Magklara et al., 2011). Moreover, LSD1, a histone demethylase that reverses the epigenetic silencing of OR loci, is necessary for OR gene activation (Lyons et al., 2013). These observations support the notion of epigenetic regulation of OR expression; however, they do not explain how only one out of ~2800 OR alleles is chosen, desilenced, and activated.
The answer to this question may lie in the unusual nuclear organization of olfactory receptor genes (Figure 2C). Despite the existence of nearly 100 chromosomally clustered groups of OR genes in a diploid mouse nuclei, DNA FISH using a complex probe pool that recognizes most OR genes identified only ~5 large OR foci in OSNs; this was not observed in other neuronal cell types nor other developmentally-related cell types in the OE (Clowney et al., 2012). These OR “aggregates” localize adjacent to a few centrally located regions of centromeric and telomeric heterochromatin in an organization similar to that of the rod photoreceptor nuclei of nocturnal mammals (see above). OR clusters located on the same chromosome formed optically indistinguishable foci, suggesting pronounced spatial compaction of OR clusters in cis. Moreover, OR clusters from different chromosomes were frequently found in the same OR aggregate, suggesting heterochromatic OR clusters interact in trans also. These condensed OR foci colocalized with H3K9me3, H4K20me3, and heterochromatin protein 1β (HP1β). Interestingly, the activated OR allele was consistently observed outside of these heterochromatized nuclear regions, instead colocalizing with histone acetylation and RNAP II. Moreover, the active OR allele frequently colocalizes with the enhancer sequence H (Lomvardas et al., 2006), which is essential for expression of only linked OR genes (Fuss et al., 2007; Serizawa et al., 2003). Hence, reorganization of OSN nuclei leads to localization of the active and inactive OR alleles into distinct nuclear compartments with active and repressive chromatin hallmarks, respectively.
As with rod nuclear organization, OSNs regulate their nuclear architecture through modification of the nuclear envelope. LBR is absent in OSNs when compared to globose basal cells, which serve as a progenitor pool for OSN differentiation. Strong evidence for a functional relevance of OR gene aggregation in OSN differentiation comes from experiments where LBR was ectopically expressed in OSNs. This led to disruption of OR aggregation, reduced chromatin compaction, and reduced expression of OR genes. We speculate that reduced OR expression, despite increased accessibility of OR genes, could result from either disruption of essential interchromosomal interactions in LBR-positive OSNs or from a transcriptional squelching effect caused by coexpression of many OR genes in a single OSN. Abnormal axon targeting to olfactory bulb glomeruli, single cell qRT-PCR, and 4C-qPCR experiments from LBR-expressing neurons provide support for both scenarios (Clowney et al., 2012). Thus, an intriguing hypothesis is that the aggregation of silent OR genes near the core of the olfactory nucleus serves two regulatory purposes: It reinforces the epigenetic silencing imposed by heterochromatic histone marks by packaging OR genes in nuclear sub-compartments that preclude the access of transcription factors and the transcriptional machinery; It also assists the robust transcription of a single OR allele by preventing the dilution of OR-specific transcription factors over the ~40MB of the OR subgenome and by bringing the chosen OR allele in spatial proximity to activating regulatory sequences from other OR clusters. In this vein, the notion of cooperative enhancer function, which was described in the case of Hoxd gene expression in the limb (Montavon et al., 2011) and likely occurs in the case of protocadherin gene expression (Monahan et al., 2012), may also be applicable in a system that relies in the singular and robust OR expression.
Neural Activity and Nuclear Architecture
The examples discussed above highlight changes in nuclear architecture that occur during neural differentiation. However, modification in gene localization and nuclear organization has also been observed in response to neuronal activity. In a striking example, cultured hippocampal neurons undergo remodeling of the nucleus in response to bursts of action potentials (Wittman et al., 2009). These nuclei develop infoldings, which the authors propose augment nuclear sensitivity to calcium signaling. Moreover, activity-dependent activation of brain-derived neurotrophic factor (Bdnf) correlates with changes in the nuclear positioning of the Bdnf gene. When rats are given kainate-induced seizures, Bdnf demonstrates reduced localization to the nuclear lamina (Walczak et al., 2013) Interestingly, while Bdnf upregulation peaks after 2 hours and is negligible by 4 weeks after seizure induction, Bdnf remains displaced from the nuclear lamina at this late time point, which could facilitate future activation of Bdnf in response to neuronal activity. In another interesting example, genes that encode subunits of the cytochrome c oxidase enzyme were found to colocalize in the nuclei of neurons using 3C (Dhar et al., 2009). These interactions were augmented by stimulation of neural activity. Finally, activation of some genes (e.g. c-Fos) in response to neural activity results in increased colocalization of these genes with transcription factories (Crepaldi et al., 2013). It is not yet clear how these alterations in genome organization impact long-term gene expression changes and neural activity. However, these findings point to nuclear organization as an additional layer of transcriptional control in activity-induced expressional changes.
Conclusions
The aforementioned studies highlight the intricate 3-dimensional organization of the genome within the nucleus and support the notion that nuclear architecture has functional consequences in a variety of processes ranging from cellular differentiation to the diffraction of light when it passes through the retina. One cannot help but wonder whether other neural processes may use nuclear architecture as a means to regulate gene expression. Given the fact that neurons are post-mitotic and live as long as the organism, it is conceivable that placing genes in transcriptionally active or silent nuclear territories affords stable and long-lasting regulation of their transcriptional state. Thus, exploring whether long-lasting transcriptional changes that coincide with maladaptive neuronal states, such as chronic pain or addiction, are stabilized by nuclear repositioning of relevant genomic loci or widespread nuclear reorganization would be of great interest. Similar processes may also be involved in physiological, long-lasting transcriptional changes, such as those that occur in the hippocampus during memory formation and storage. Beyond these “experiential” changes that may occur in the nuclear architecture of differentiated neurons, it is appealing to hypothesize that the nuclear reorganization that takes place during olfactory neuron differentiation also occurs during the development of other neuronal types. As the striking example of stochastic expression of clustered protocadherin genes demonstrates (Chen et al., 2012; Esumi et al., 2005; Lefebvre et al., 2012), like neurons that receive similar spatiotemporal cues and have common progenitors can have distinct transcription programs. Therefore, employing nuclear architecture as a regulatory mechanism allows neurons with identical transcription factor ensembles to activate and repress different genes based on their relative nuclear location. In other words, nuclear architecture and differential gene positioning could be exploited by the brain for the generation of stochastic gene choices that contribute to the astounding cellular diversity of the nervous system.
Highlights.
-
-
The nucleus is non-randomly organized into compartments and domains that impact genome function.
-
-
Regulation of nuclear architecture accompanies neurogenesis and neuronal activity.
-
-
Nuclear architecture could be a broad epigenetic regulator of neural cell diversity and function.
Box Figure.
Approaches for Studying Nuclear Architecture. 3C and DamID have been widely used to understand chromatin organization and its localization to specific nuclear compartments. Important steps in the workflow of 3C and DamID are shown on the left and right, respectively.
Acknowledgements
We kindly thank members of the Lomvardas lab for input and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright TD, Jessell TM, Kandel ER, Posner MI. Neural science: a century of progress and the mysteries that remain. Cell. 2000;100(Suppl):S1–S55. [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-Dependent Regulatory Contacts between Distant Hox Loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Benzer S. Genetic dissection of behavior. Sci. Am. 1973;229:24–37. doi: 10.1038/scientificamerican1273-24. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The Mammalian Epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011;19:901–909. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of Chromosome Territories in Interphase Suggests Role in Translocations and Transcription-Dependent Associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Green J, Neves, das RP, Wallace HAC, Smith AJH, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J. Comp. Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, Smith C, Thu CA, Tapia JC, Tasic B, et al. Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75:402–409. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear Aggregation of Olfactory Receptor Genes Governs Their Monogenic Expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Research. 2011;21:1249–1259. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;488:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, Hase, von J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Current Opinion in Cell Biology. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Crepaldi L, Policarpi C, Coatti A, Sherlock WT, Jongbloets BC, Down TA, Riccio A. Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories. PLoS Genet. 2013;9:e1003699. doi: 10.1371/journal.pgen.1003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Chromosome Conformation Capture of All 13 Genomic Loci in the Transcriptional Regulation of the Multisubunit Bigenomic Cytochrome c Oxidase in Neurons. Journal of Biological Chemistry. 2009;284:18644–18650. doi: 10.1074/jbc.M109.019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G, Speese S, Budnik V, Geyer PK, Wallrath LL. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–3077. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Biogenesis of Nuclear Bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711–a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. Nuclear bodies: multifunctional companions of the genome. Current Opinion in Cell Biology. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Current Opinion in Genetics & Development. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat Genet. 2005;37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal Contact Permits Transcription between Coregulated Genes. Cell. 2013;155:606–620. doi: 10.1016/j.cell.2013.09.051. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the Nuclear Periphery Can Alter Expression of Genes in Human Cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Ray A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol. Cell. Neurosci. 2009;41:101–112. doi: 10.1016/j.mcn.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hakim O, Sung M-H, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Research. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci. 1996;109(Pt 6):1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. Embo J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nature Biotechnology. 2011;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-Cell Dynamics of Genome-Nuclear Lamina Interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai S-L, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST. Subnuclear Compartmentalization of Immunoglobulin Loci During Lymphocyte Development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper K, Kölbl A, Biener D, Dittrich S, Hase, von J, Thormeyer T, Fiegler H, Carter NP, Speicher MR, Cremer T, et al. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. PML Nuclear Bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661–a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Reviews Genetics. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An Epigenetic Signature for Monoallelic Olfactory Receptor Expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo MR, Boyle S, D’Esposito M, Bickmore WA. Chromosome territory reorganization in a human disease with altered DNA methylation. Proc Natl Acad Sci USA. 2007;104:16546–16551. doi: 10.1073/pnas.0702924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002a;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002b;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the Sequence: Cellular Organization of Genome Function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. How smell develops. Nat Neurosci. 2001;4:1192–1198. doi: 10.1038/nn751. [DOI] [PubMed] [Google Scholar]
- Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proceedings of the National Academy of Sciences. 2012;109:9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, Van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Kohro T, Baboo S, Larkin JD, Deng B, Short P, Tsutsumi S, Taylor S, Kanki Y, Kobayashi M, et al. TNF signals through specialized factories where responsive coding and miRNA genes are transcribed. Embo J. 2012;31:4404–4414. doi: 10.1038/emboj.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How gene activators work. Sci. Am. 1989;260:40–47. doi: 10.1038/scientificamerican0189-40. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature Genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase JV, Stein S, Vincent S, Molloy KR, Kreth G, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shykind BM. Regulation of odorant receptors: one allele at a time. Hum Mol Genet. 2005;14:R33–R39. doi: 10.1093/hmg/ddi105. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, LanctOt C, KOsem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear Architecture of Rod Photoreceptor Cells Adapts to Vision in Mammalian Evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear Speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646–a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJG, Zhu Y, Kaaij LJT, van Ijcken W, Gribnau J, Heard E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 2010;1:284–297. doi: 10.4161/nucl.1.3.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb Domains Are Guided by Chromosome Architecture. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, van Steensel B. The Insulator Protein SU(HW) Fine-Tunes Nuclear Lamina Interactions of the Drosophila Genome. PLoS ONE. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, Dunnen, den JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nature Biotechnology. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr Biol. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Walczak A, Szczepankiewicz AA, Ruszczycki B, Magalska A, Zamlynska K, Dzwonek J, Wilczek E, Zybura-Broda K, Rylski M, Malinowska M, et al. Novel Higher-Order Epigenetic Regulation of the Bdnf Gene upon Seizures. Journal of Neuroscience. 2013;33:2507–2511. doi: 10.1523/JNEUROSCI.1085-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RRE, Azuara V, Perry P, Sauer S, Dvorkina M, Jørgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Williams RRE, Broad S, Sheer D, Ragoussis J. Subchromosomal Positioning of the Epidermal Differentiation Complex (EDC) in Keratinocyte and Lymphoblast Interphase Nuclei ☆. Exp Cell Res. 2002;272:163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- Wittmann M, Queisser G, Eder A, Wiegert JS, Bengtson CP, Hellwig A, Wittum G, Bading H. Synaptic activity induces dramatic changes in the geometry of the cell nucleus: interplay between nuclear structure, histone H3 phosphorylation, and nuclear calcium signaling. Journal of Neuroscience. 2009;29:14687–14700. doi: 10.1523/JNEUROSCI.1160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Coats JK, Yang CF, Wang A, Ahmed OM, Alvarado M, Izumi T, Shah NM. Modular genetic control of sexually dimorphic behaviors. Cell. 2012;148:596–607. doi: 10.1016/j.cell.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. Nature Genetics. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]