Abstract
The organization of the human cerebral cortex has recently been explored using techniques for parcellating the cortex into distinct functionally coupled networks. The divergent and convergent nature of cortico-cortical anatomic connections suggests the need to consider the possibility of regions belonging to multiple networks and hierarchies among networks. Here we applied the Latent Dirichlet Allocation (LDA) model and spatial independent component analysis (ICA) to solve for functionally coupled cerebral networks without assuming that cortical regions belong to a single network. Data analyzed included 1,000 subjects from the Brain Genomics Superstruct Project (GSP) and 12 high quality individual subjects from the Human Connectome Project (HCP). The organization of the cerebral cortex was similar regardless of whether a winner-take-all approach or the more relaxed constraints of LDA (or ICA) were imposed. This suggests that large-scale networks may function as partially isolated modules. Several notable interactions among networks were uncovered by the LDA analysis. Many association regions belong to at least two networks, while somatomotor and early visual cortices are especially isolated. As examples of interaction, the precuneus, lateral temporal cortex, medial prefrontal cortex and posterior parietal cortex participate in multiple paralimbic networks that together comprise subsystems of the default network. In addition, regions at or near the frontal eye field and human lateral intraparietal area homologue participate in multiple hierarchically organized networks. These observations were replicated in both datasets and could be detected (and replicated) in individual subjects from the HCP.
Keywords: Intrinsic Connectivity, MRI, Human Connectome Project, Default Network, Dorsal Attention, Resting-state fMRI
Introduction
Distributed neocortical brain areas form large-scale networks that exhibit complex patterns of divergent and convergent connectivity (e.g., Pandya and Kuypers, 1969; Jones and Powell, 1970; Mesulam 1981; Ungerleider and Desimone, 1986; Goldman-Rakic, 1988; Felleman and Van Essen, 1991). A major challenge in systems neuroscience is to make sense of these connectivity patterns to infer functional organization. In the visual system, connectivity patterns suggest a separation of processing into largely parallel, but interacting, hierarchical pathways (Ungerleider and Desimone, 1986; Felleman and Van Essen, 1991). In contrast, the association cortex comprises networks of widely distributed and densely interconnected areas without rigid hierarchical organization (Goldman-Rakic, 1988; Selemon and Goldman-Rakic, 1988; but see Badre and D'Esposito, 2009).
Resting-state functional connectivity MRI (rs-fcMRI) provides a powerful, albeit indirect, approach to make inferences about human cortical organization (Biswal et al., 1995). Despite its limitations (Buckner et al., 2013), we and others have used functional connectivity to estimate cortical network patterns (e.g., Damoiseaux et al., 2006; Margulies et al., 2007; He et al., 2009; Smith et al., 2009; van den Heuvel et al., 2009; Bellec et al., 2010; Power et al., 2011; Yeo et al., 2011).
The majority of functional connectivity studies have focused on dissociating functionally distinct networks or modules (Greicius et al., 2003; Beckmann et al., 2005; Salvador et al., 2005; Damoiseaux et al., 2006; De Luca et al., 2006; Fox et al., 2006; Dosenbach et al., 2007; Margulies et al., 2007; Seeley et al., 2007; Calhoun et al., 2008; Smith et al., 2009; van den Heuvel et al., 2009; Doucet et al., 2011; Rubinov and Sporns, 2011; Varoquaux et al., 2011; Craddock et al., 2012). Fewer studies have examined the relationships among different functional networks (Sepulcre et al., 2012a; Sporns et al., 2013). For example, Fox et al. (2005) and Fransson (2005) have investigated the antagonistic relationship between the default and task-positive networks. Others (Meunier et al., 2009; Doucet et al., 2011; Lee et al., 2012) have investigated the (spatial) hierarchical relationship across functional networks.
We previously employed a mixture model that relied on a winner-takes-all assumption to map network topography in the human cerebral cortex (Yeo et al., 2011). Each brain region was assigned to a single, best-fit network allowing us to derive connectivity maps that emphasize the interdigitation of parallel, distributed association networks. The key features of this parallel organization are that (1) each association network consists of strongly coupled brain regions spanning frontal, parietal, temporal, and cingulate cortices, and (2) the components of multiple networks are spatially adjacent (Yeo et al., 2011; also see Vincent et al., 2008, Power et al., 2011).
However, it is unlikely that the brain is simply parcellated into a discrete number of nonoverlapping networks (Mesulam 1998). Interactions across networks, as well as the existence of ‘convergence zones’ of regions that participate in multiple networks, are likely important features of brain organization (Pandya and Kuypers, 1969; Jones and Powell, 1970; Mesulam 1998; Beckmann et al., 2005; Bullmore and Sporns, 2009; Spreng et al., 2010; Fornito et al., 2012; Sepulcre et al., 2012b; Power et al., 2013). Relevant to this point, we have observed variability in the goodness of fit of certain regions to their winner-takes-all network (Figures 8 and 10 of Yeo et al., 2011), consistent with the notion that certain brain regions might participate in multiple networks (Beckmann et al., 2005; Sporns et al., 2007; Andrews-Hanna et al. 2010; Leech et al., 2011; Rubinov and Sporns, 2011).
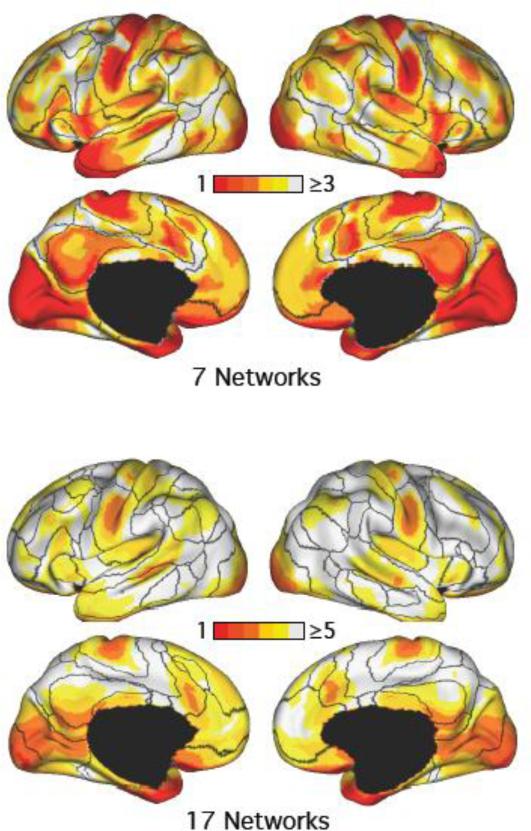
Figure 8.
Pattern of network segregation and overlap replicated across degenerate LDA estimates. The plots show the number of LDA networks each cortical region participates in for the 7-network and 17-network LDA estimates in the GSP dataset, averaged across 100 random initializations. Many association regions participated in multiple networks, while large portions of early sensory and late motor cortices participated in single networks.
Here, we address the possibility of multiple network membership by applying latent Dirichlet allocation (LDA; Blei et al., 2003) and spatial Independent Component Analysis (ICA; Calhoun et al., 2001; Beckman et al., 2004) to examine the topography of overlapping networks. This is an important consideration because network topography may change substantially from our original estimates (Yeo et al., 2011) if constraints are relaxed to permit overlapping networks. Conversely, unbiased estimation of network topography may broadly confirm previous estimates and allow us to investigate the interactions and overlaps among networks.
Materials and Methods
Overview
We applied the LDA model to resting-state data from 1,000 healthy young adults from the Brain Genomics Superstruct Project (GSP), as well as to 12 high quality, high-resolution individual subject datasets from the Human Connectome Project (HCP; Van Essen et al., 2013). The large sample size in GSP and the multiple sessions of individual HCP subjects permitted us to quantify patterns of cortico-cortical coupling that reveal insights into interactions within and across functional networks. Analyses proceeded in four stages. First, we applied the mixture model (Yeo et al., 2011) and LDA model (Blei et al., 2003) to both the GSP and HCP group datasets, in order to examine how cortical network organization changes as regions are permitted to participate in multiple networks (Figure 1). For this analysis, the GSP and HCP datasets were used to provide independent replication samples. Next, we further analyzed several cortical regions participating in multiple sub-networks (Figures 2 to 4). We then exploited the high quality, multi-session HCP data to determine if network organization can be estimated and replicated in individual subjects (Figures 5 and 6). This increased the confidence that the discovered network organization was not merely a consequence of averaging across subjects. Additional control analyses confirmed similar network organization regardless of whether global signal regression was performed during preprocessing (Supplemental Figure 7) and across degenerate (i.e., not highest likelihood) network estimates (Figures 7 and 8).
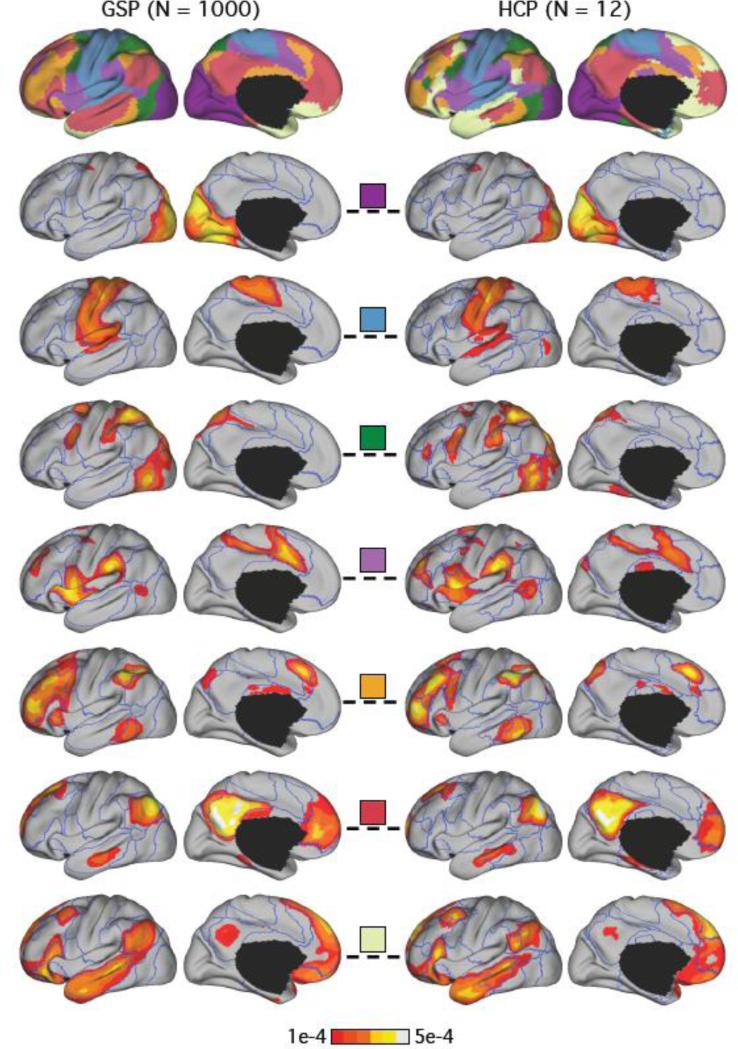
Figure 1.
Large-scale network organization is reliable across datasets (GSP and HCP) and across methods (clustering and LDA). Top row shows the 7-network clustering estimates of the GSP and HCP datasets in the left hemisphere. Overlap (fraction of vertices with same network labels) between GSP and HCP estimates was 78% across the entire cerebral cortex. Five (purple, blue, green, violet and orange) of the seven networks were virtually identical. Bottom rows show the 7-network LDA estimates of the GSP and HCP datasets overlaid on blue boundaries corresponding to the clustering estimates of respective datasets. All seven LDA networks were highly similar across the two group datasets. The mean of the Pearson correlation coefficients of the seven pairs of LDA network estimates was 0.93. The colored squares (center column) indicate correspondences between the LDA and clustering estimates. For example, the LDA estimate in the second row correspond to the purple visual cluster. The percent overlap between the clustering and winner-takes-all LDA estimates in the GSP dataset was 82%, while the overlap in the HCP dataset was 88%.
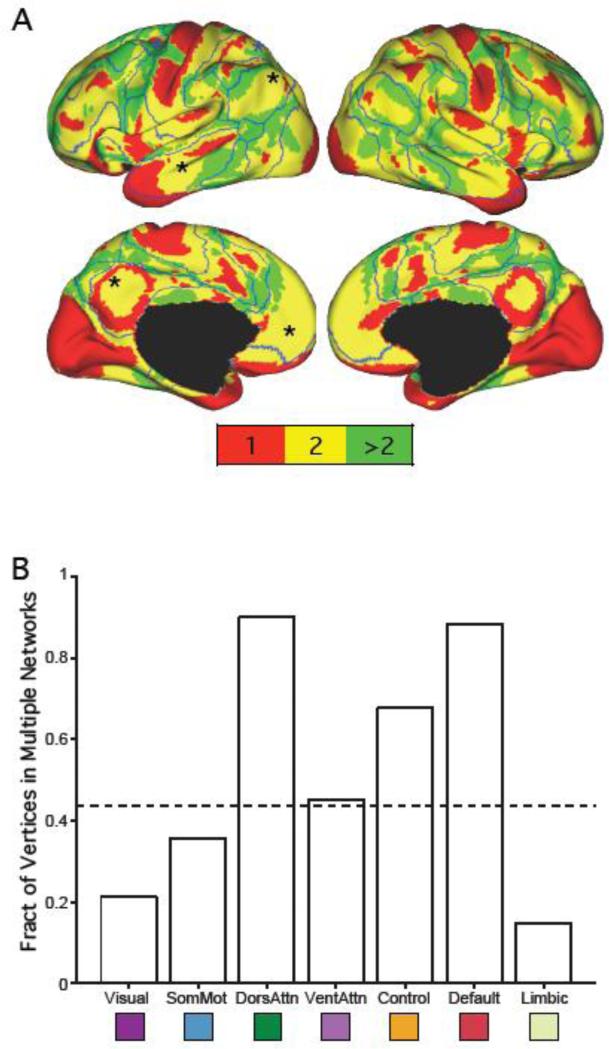
Figure 2.
Regions of association cortex often participate in multiple cortical networks. Early sensory and late motor cortices are involved in fewer networks than many association regions. (A) The colors represent the number of LDA networks each cortical region participates in for the 7-network LDA estimate in the GSP dataset. The number was computed by counting for each vertex the number of networks whose Pr(network | vertex) exceeds 1/7. Boundaries correspond to 7-network clustering solution in GSP dataset. The black asterisks correspond to default network regions that (1) were at least 10mm from both the clustering and winner-takes-all LDA boundaries and (2) participated in multiple networks. The blue asterisks correspond to dorsal attention regions that (1) were at least 10mm from both the clustering and winner-takes-all LDA boundaries and (2) participated in multiple networks. Table 1 reports the MNI coordinates of the six regions. (B) Fraction of vertices within the 7-network clustering estimates participating in multiple networks. Only vertices at least 10mm away from the clustering boundaries are considered. The dotted line indicates that 44% of the vertices (at least 10mm away from clustering boundaries) participated in multiple networks. Visual: Visual network; SomMot: somatomotor network; DorsAttn: dorsal attention network; VentAttn: ventral attention network; Control: frontoparietal control network; Default: default network; Limbic: limbic network.
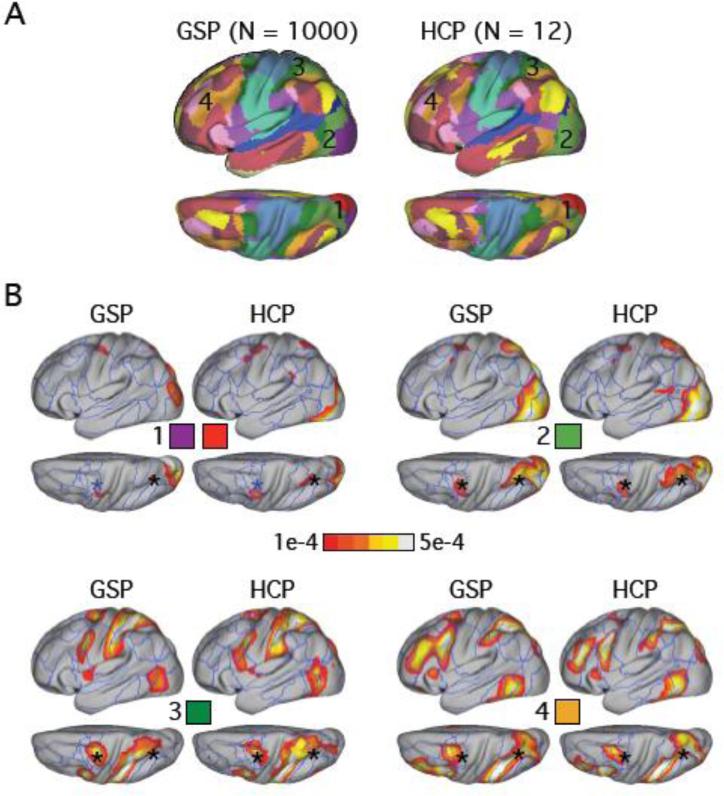
Figure 4.
The lateral intraparietal area (LIP) and frontal eye fields (FEF) participate in multiple subnetworks. (A) 17-network clustering estimates of the GSP and HCP datasets. Only left hemisphere estimates are shown. (B) Four of the 17-network LDA estimates overlapping traditional dorsal attention regions are shown. Panel 1 shows the LDA network that overlaps with the red and purple networks in (A). The LDA networks in the 2nd, 3rd and 4th panels overlap with the light-green, dark-green and orange networks in (A) respectively. The asterisks correspond to LIP and FEF defined in Table 1. An asterisk is colored black if Pr(vertex | network) > 1e-4 at that brain location. .
Figure 5.
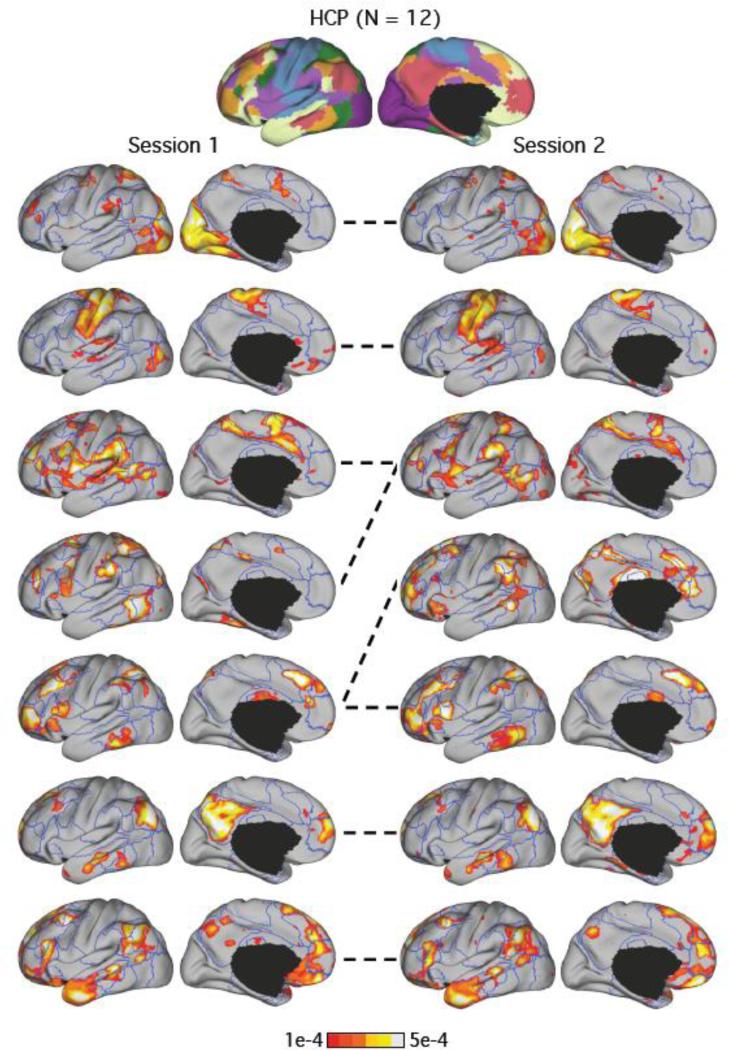
Cortical networks are reliable within individual subjects. LDA of the HCP subject with the best test-retest reliability estimates is displayed. Top row corresponds to the 7-network clustering estimate of the entire HCP dataset. Bottom rows show the 7-network LDA estimates of the subject in two different sessions overlaid on the boundaries of the 7-network clustering estimate. The mean of the Pearson correlation coefficients of the seven pairs of network estimates was 0.75. The dotted lines indicate correspondences between the LDA networks across the two sessions. By visual inspection, the third and fourth networks in session 1 were merged in session 2, while the fifth network in session 1 was split into two in session 2.
Figure 6.
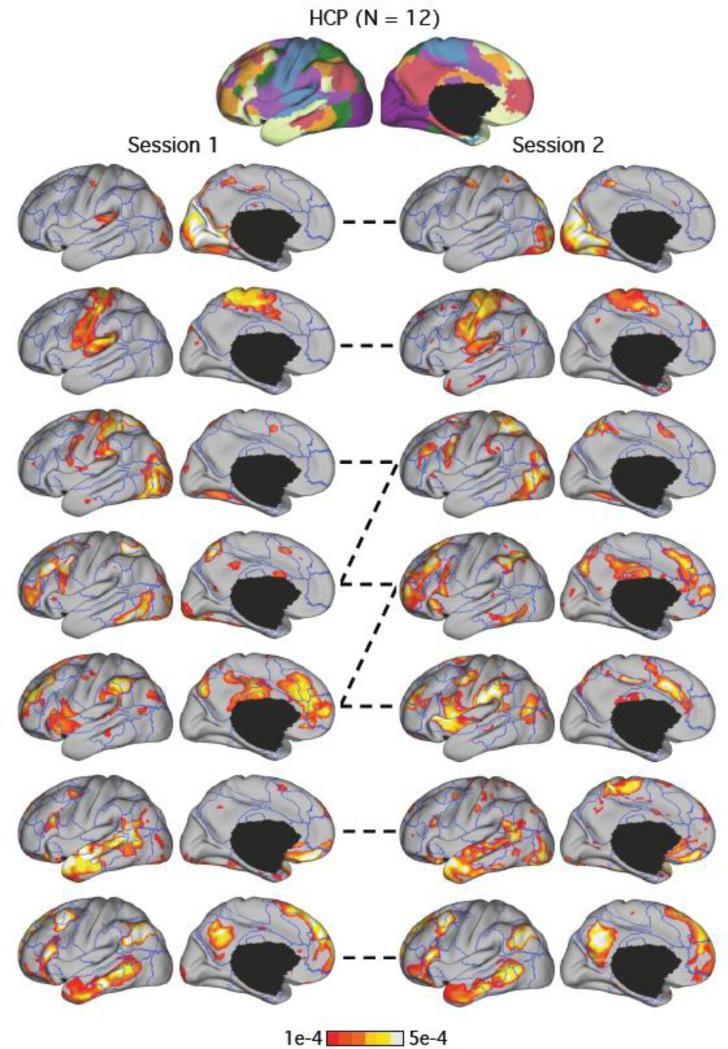
Similar to Figure 6, LDA estimates of the HCP subject with the median test-retest reliability is displayed. Format is identical to Figure 6. The mean of the Pearson correlation coefficients of the seven pairs of network estimates was 0.61. The dotted lines indicate correspondences between the LDA networks across the two sessions. By visual inspection, the third, fourth and fifth networks in sessions 1 and 2 were inter-mixed across the sessions.
Figure 7.
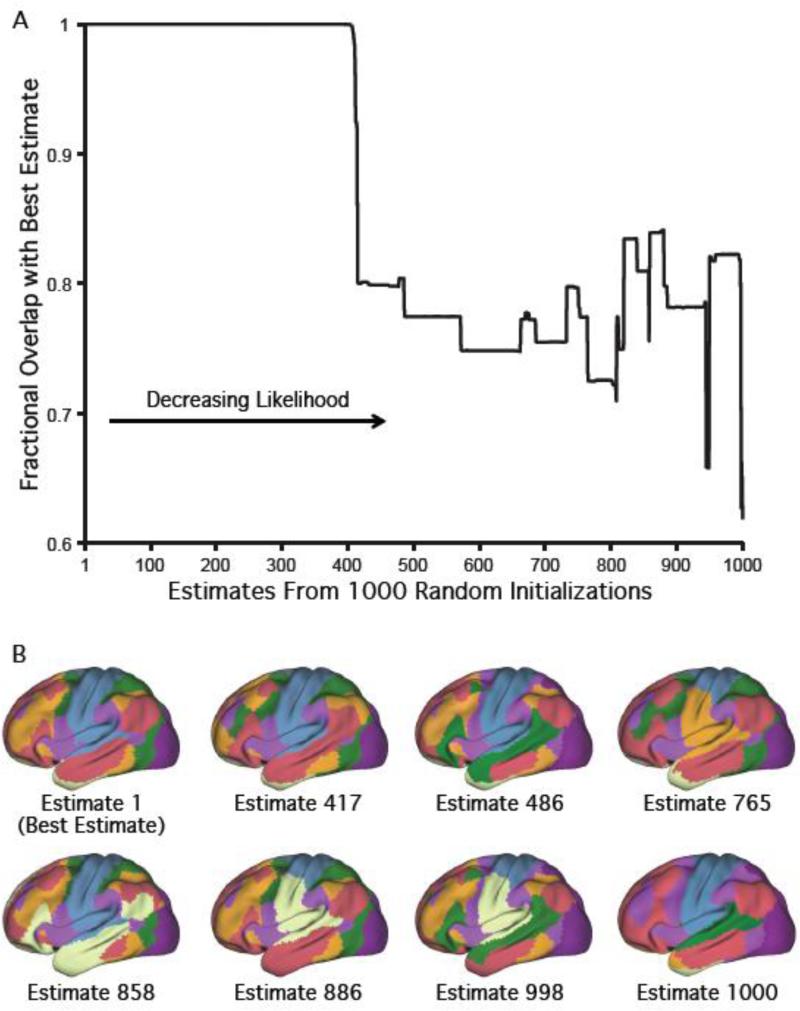
Parallel network structure replicated across degenerate clustering estimates. (A) Overlap between the best 7-network clustering estimate and estimates derived from the 1000 random initializations using the GSP dataset. The clustering estimates were ordered from the highest likelihood (best estimate) to the smallest likelihood. Out of 1000 random initializations, 415 resulted in solutions with at least 90% overlap with the best solution, suggesting the best estimate corresponds to a robust local optimum. (B) “Exhaustive” set of 7-network clustering estimates. The eight clustering estimates were selected to be different from each other and spanned the solution space of the mixture model. Similar to the best estimate, the early sensory and late motor cortices participated in preferentially local networks in all 7 degenerate solutions. Similarly, each association network spanned multiple lobes and the components of multiple networks were spatially adjacent.
Datasets
The GSP subjects were between ages 18-35 (mean age = 21.3; 42.7% male). Participants underwent one or two runs of eyes open rest (EOR). Analyses of the GSP data have been published previously (e.g., Buckner et al., 2011; Yeo et al., 2011; Choi et al., 2012). The HCP subjects were between ages 26-35 (mean age estimate = 30.9; 16.7% male). HCP only provides aggregated data concerning age, hence mean age can only be estimated. HCP participants underwent two runs of passive fixation (FIX) in each of two separate sessions, for a total of four runs (~24 h interval between sessions).
GSP MRI Data Acquisition and Preprocessing
Data were acquired on 3T Tim Trio scanners (Siemens, Erlangen, Germany) using a 12-channel phased-array head coil. Functional data consisted of gradient-echo echo-planar images (EPI) sensitive to blood oxygenation level-dependent (BOLD) contrast. Parameters for the resting data were: repetition time (TR) = 3,000 ms, echo time (TE) = 30 ms, flip angle (FA) = 85°, 3 × 3 × 3 mm voxels, field of view (FOV) = 216, and 47 axial slices collected with interleaved acquisition. Slices were oriented along the anterior commissure-posterior commissure plane. Functional runs lasted 6.2 min (124 time points). Structural data included a multiecho T1-weighted magnetization-prepared gradient-echo (MP-RAGE) image (van der Kouwe et al., 2008).
fMRI processing steps included 1) discarding the first four frames of each run, 2) correcting for slice acquisition-dependent time shifts in each volume with SPM2 (Wellcome Department of Cognitive Neurology, London, UK), and 3) correcting for head motion using rigid body translation and rotation parameters (FSL; Jenkinson et al., 2002; Smith et al., 2004). This was followed by standard functional connectivity preprocessing (Fox et al., 2005; Vincent et al., 2006; Van Dijk et al., 2010). Linear trends over each run were removed and a low-pass temporal filter retained frequencies below 0.08 Hz. Spurious variance was removed using linear regression with terms for head motion, whole brain signal, ventricle signal, white matter signal and their derivatives.
Individual participants’ T1 scans were reconstructed into surface representations using FreeSurfer (http://surfer.nmr.mgh.harvard.edu; Fischl 2012). Functional data were registered to structural images using FreeSurfer's FsFast package (Greve and Fischl, 2009; http://surfer.nmr.mgh.harvard.edu/fswiki/FsFast). The structural preprocessing and structural-functional data alignment steps were described in Yeo et al. (2011). Functional data were projected onto the FreeSurfer surface space (2 mm mesh), smoothed on the surface using a 6 mm full-width half-maximum kernel, and were then downsampled to a 4 mm mesh.
HCP MRI Data Acquisition and Preprocessing
HCP data were part of the HCP initial October 2012 public data release (http://www.humanconnectome.org/data). Data were acquired on a 3T Skyra scanner (Siemens, Erlangen, Germany) using a standard 32-channel head coil. The scanner has a customized SC72 gradient insert and a customized body transmitter coil with 56 cm bore size. The HCP Skyra has the standard set of Siemens shim coils (up to 2nd order). Functional data consisted of gradient-echo EPI sensitive to BOLD contrast. Parameters for the resting data were: TR = 720 ms, TE = 33.1 ms, FA = 52°, 2 × 2 × 2 mm voxels, FOV = 208 × 180 mm, and 72 oblique axial slices alternated between phase encoding in a right to left direction in one run and phase encoding in a left to right direction in the other run (Feinberg et al., 2010; Moeller et al., 2010; Setsompop et al., 2012; Xu et al., 2012). Each functional run lasted 14.55 min (1200 time points). Structural data included a T1-weighted MP-RAGE image. Parameters for the structural scan were as follows: TR = 2400 ms, TI = 1000 ms, TE=2.14 ms, FA = 8°, 0.7 × 0.7 × 0.7 mm voxels and FOV = 224 × 224 mm. More details of the acquisition strategy can be found in Van Essen et al. (2012).
We utilized the fMRI preprocessed data released by the HCP (Glasser et al., 2013). fMRI processing steps included 1) gradient distortion correction (Jovicich et al., 2006), 2) motion correction, 3) distortion correction, 4) registration to the T1 scan (Greve and Fischl, 2009), 5) spline resampling to FSL MNI152 2mm space using FSL FNIRT (Jenkinson et al., 2002; Smith et al., 2004), and 6) intensity normalization to mean of 10000 and bias field correction. This was followed by standard functional connectivity preprocessing as in the GSP dataset. The preprocessed fcMRI data were projected from FSL MNI152 space onto the FreeSurfer surface space (1mm mesh), smoothed using a 6-mm full-width half-maximum kernel and downsampled to a 4mm mesh. The nonlinear mapping between FSL MNI152 volumetric space and FreeSurfer surface space is detailed in Buckner et al. (2011).
Clustering
The method of clustering cerebral cortical data has been previously described (Yeo et al., 2011). Briefly, for each subject, the Pearson's product moment correlation was computed between each surface vertex (N = 18715) and 1175 uniformly distributed cortical regions of interest (ROIs). The “connectivity profile” of each surface vertex was its functional coupling to these ROIs. Each participant's 1175 × 18715 matrix of correlations was binarized to retain the top 10% of correlations before summing across subjects to obtain an overall group estimate P. Therefore, the ith row and jth column of the matrix P was the number of subjects whose correlations between the ith vertex and jth ROI are within the top 10% of correlations (within each individual subject). In other words, each matrix component took on integer values from 0 to 1000 in the GSP dataset. The connectivity profiles were clustered using a mixture of von Mises-Fisher distributions (Lashkari et al., 2010; Yeo et al., 2011). We repeated the clustering algorithm 1000 times with different random initializations and selected the estimate with best model likelihood. For more details, we refer readers to Lashkari et al. (2010) and Yeo et al. (2011). Because our previous analyses (Yeo et al., 2011) identified solutions with 7 and 17 network clusters to be particularly stable, we adopted these for the present study.
Latent Dirichlet Allocation (LDA)
LDA was first introduced in the text mining literature (Blei et al., 2003). The application of LDA to estimate overlapping modules (networks) in graphs has been previously proposed (Zhang et al., 2007). Here, we employed LDA to estimate cortical networks from resting-state fMRI data.
Like spatial independent component analysis (ICA; Beckmann and Smith, 2004), LDA permits a brain region to belong to multiple networks. Both ICA (Beckmann and Smith, 2004) and LDA (Hoffman et al. 2010) seek to factorize a matrix M into a product of two matrices W and H. Because there are an infinite number of ways to factorize a matrix, additional constraints are required.
In the case of spatial ICA applied to fMRI, the spatial weights of the estimated networks (components) are constrained to be independent (Beckman et al., 2005; Calhoun et al., 2001), but can take on negative values. LDA is closely related to non-negative matrix factorization (Hoffman et al. 2010). When applied to resting-state fMRI data in this paper, LDA constrains the spatial weights of the estimated networks to be non-negative (in addition to other constraints). We refer interested readers to Blei et al. (2003) for the probabilistic (and more well-known) interpretation of LDA.
In this work, we used LDA to factorize the 1175 × 18715 matrix P (see the previous “Clustering” section) into the product of two matrices W (1175 × K) by H (K × 18715), where K is the number of networks. The k-th row of the matrix H sums to one over all 18715 vertices of the cerebral cortex. We can interpret the k-th row as the probability that the k-th functional connectivity network includes a particular surface vertex Pr(vertex | k-th network).
To compare the clustering and LDA results, we consider 7 and 17 networks in this paper. The LDA model is estimated with 100 random initializations. The estimate with the best likelihood bound is selected. The LDA code is publicly available1.
Spatial Independent Component Analysis (Spatial ICA)
We applied spatial ICA (FSL melodic 3.10; Beckmann and Smith, 2004) to the surface projected fcMRI-preprocessed data. We estimated 20 ICA components because of precedence (e.g., Smith et al. 2009) and because “20” is close to the number of networks (“17”) we sought in the clustering and LDA models. We also experimented with 17 independent components, as well as applying ICA directly to raw volumetric fMRI data before projecting the spatial components to the surface. The results were similar across these different experiments and so for conciseness, we will focus on the 20 ICA components estimated from surface-projected fcMRI-preprocessed data.
Matching Networks between Datasets and Methods
Since the ordering of the networks (or components) estimated using different methods and datasets is arbitrary, we used the Hungarian matching algorithm (Kuhn 1955) to find the correspondences between networks estimated with different methods and datasets. Essentially, the networks were relabeled so as to maximize the spatial agreement between corresponding networks.
In particular, HCP clustering estimates were reordered to match the GSP clustering estimates by maximizing the number of vertices belonging to corresponding networks across the datasets. Similarly, the HCP LDA estimates were reordered to match the GSP LDA estimates by maximizing the correlation between Pr(vertex | network) of corresponding networks across datasets2.
To match clustering estimates with LDA estimates, we thresholded the LDA estimates to obtain winner-takes-all parcellations. The clustering estimates were then reordered to match the winner-takes-all LDA estimates by maximizing the number of vertices belonging to corresponding (winner-takes-all) networks across methods.
Finally, we found 17 of the 20 ICA components that best correspond to the 17 LDA estimates by maximizing the correlation between Pr(vertex | LDA network) and the un-thresholded, un-normalised Z-score ICA maps of corresponding networks across methods.
Visualization
Clustering, LDA and ICA estimates were transformed from FreeSurfer surface space (Dale et al. 1999; Fischl et al. 1999a; 1999b) to the PALS-B12 surface space for visualization using Caret (Van Essen, 2005). Network colors match Yeo et al. (2011). Lines representing clustering boundaries were smoothed to remove high spatial frequency jaggedness resulting from mapping the data to PALS space.
Comparison of Network Estimates in GSP and HCP Datasets
The 7-network clustering and LDA analyses of the GSP and HCP datasets are shown in Figure 1. Since each LDA network comprises a probability distribution over vertices Pr(vertex | network), we can visualize each distribution as a map over the cortex. Because LDA is a Bayesian model, there is no obvious threshold for displaying the probability maps. Given that there are 18715 vertices and the probability distribution sums to one over all the vertices, any probability larger than 1/18715 is above expectation. Here, we chose a relatively stringent threshold of 1e-4 (roughly two times 1/18715). The LDA estimates in Figure 1 were overlaid on the boundaries of the respective 7-network clustering estimates in the GSP and HCP datasets. The boundaries allowed the comparison between the clustering and LDA estimates.
The 17-network clustering and LDA analyses of the GSP and HCP datasets are shown in Figures 3 and 4. Because of the small number of subjects in the HCP dataset, we will focus our interpretation on the similarities between the HCP and GSP estimates.
Figure 3.
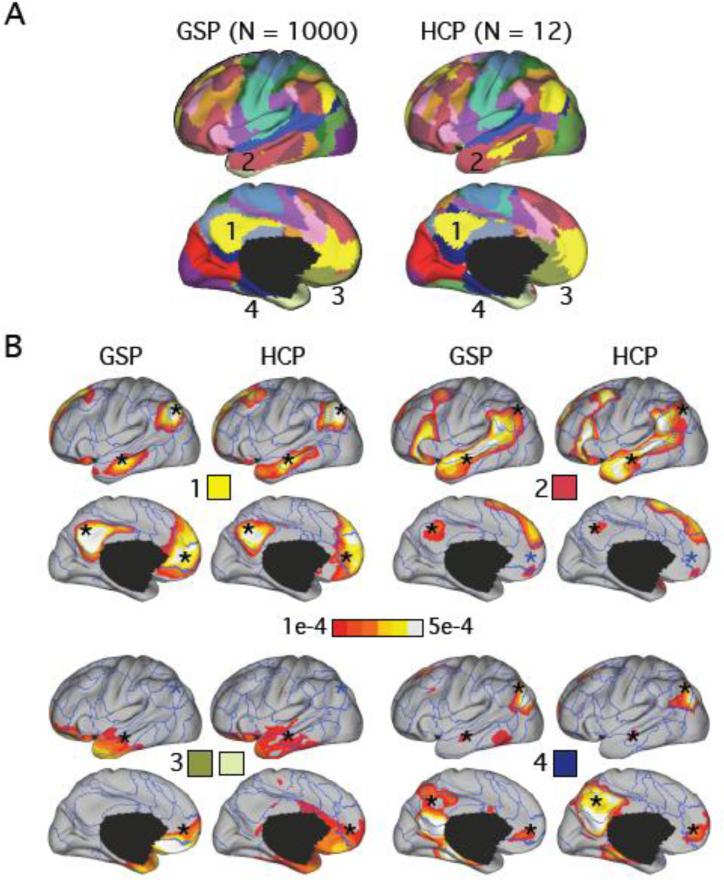
The precuneus (PCUN), lateral temporal cortex (LTC), posterior parietal cortex (PPC) and medial prefrontal cortex (MPFC) participate in multiple subnetworks. (A) 17-network clustering estimates of the GSP and HCP datasets. Only left hemisphere estimates are shown. The percent overlap between the clustering estimates in the two datasets was 69%. (B) Four of the 17-network LDA estimates overlapping traditional default network regions are shown. The LDA networks in the 1st, 2nd and 4th panels overlap with the yellow, red and dark-blue networks in (A) respectively. Panel 3 shows the LDA network that overlaps with the gold and cream networks in (A). The asterisks correspond to the default regions (PCUN, LTC, PPC and MPFC) defined in Table 1. An asterisk is colored black if Pr(vertex | network) > 1e-4 at that brain location.
Supplemental Figures 1 and 2 juxtapose 7 of the 17 corresponding LDA networks and ICA components in the GSP and HCP datasets respectively. To foreshadow the results, ICA and LDA provide very similar network estimates, except for negative spatial weights in the ICA estimates. Both ICA and LDA decompose a signal into a linear combination of component signals (Lee and Seung, 1999). For LDA, only additive combinations are allowed. This is in contrast with ICA, which may allow component signals to cancel each other out via subtractions (Lee and Seung, 1999). Therefore, the non-negativity constraints in LDA are compatible with the intuitive notions of combining parts to form a whole (Lee and Seung, 1999). It may be the case that the negative spatial weights in ICA are functionally meaningful, but assessment is difficult since ground-truth network estimates are not available. Consequently, we chose to focus on the LDA results3.
Robustness to Number of Networks
As discussed in our previous paper (Yeo et al. 2011), the focus on 7-network and 17-network solutions should not be taken to imply that meaningful properties are absent in alternative parcellation schemes. By focusing on both a relatively coarse solution (7 networks) and a fine-resolution solution (17 networks), we hoped to capture broad aspects of the solution space that were consistent within these relative extremes. However, we observed stable results for other network parcellations as well (e.g. 10 &12 networks, see Figure 6 of Yeo et al. 2011). To ensure our present results are robust to the choice of 7 and 17 networks, we estimated 4, 6, 8, 10, 12, 16, 18 and 30 networks by applying clustering and LDA to the GSP dataset (Supplemental Figure 3).
Quantifying Overlap Between Networks
To assess the overlap between networks, we computed Pr(network | vertex) for each vertex by applying Bayes’ rule to Pr(vertex | network) and assuming each network is equally likely, i.e., Pr(network) = 1/K. Since Pr(network | vertex) sums to 1 over the estimated K networks, any probability larger than 1/K is above expectation. For each vertex, we consider the number of networks with Pr(network | vertex) above 1/K as a rough measure of the number of networks the vertex is participating in. Figure 2A shows the map of the number of networks each vertex participates in. For this analysis we focus on the most stable solution – 7-network LDA results for the GSP dataset.
To quantify the distribution of multiple-network participation across the cerebral cortex, we computed the fraction of vertices participating in more than one LDA network for each network of the 7-network clustering estimates (Figure 2B). Vertices within 10mm of clustering boundaries were excluded from this analysis because they might reflect uncertainties in the network estimates. We also computed the fraction of vertices participating in more than one LDA network for each winner-takes-all LDA network (Supplemental Figure 4). Vertices within 10mm of the winner-takes-all LDA boundaries were excluded from this analysis.
To ensure the above analysis was robust to the choice of threshold for Pr(network | vertex), we repeated the analysis using a more liberal threshold of 0.75/K and a more conservative threshold of 1.25/K (Supplemental Figure 5). We also repeated the analysis with 6-network and 8-network LDA estimates to ensure our results were robust to the choice of number of networks (Supplemental Figure 6).
As the results will show, the default and dorsal attention networks have the greatest proportion of regions participating in multiple networks. Therefore, we identified brain regions that (1) were at least 10mm away from both 7-network clustering and winner-takes-all LDA boundaries, (2) participated in multiple networks from the 7-network estimate and either (3a) participated in multiple networks (from the 17-network LDA estimate) that overlapped significantly with the default network or (3b) participated in multiple networks (from the 17-network LDA estimate) that overlapped significantly with the dorsal attention network. We explored the participation of these regions in multiple networks from the 17-network LDA estimates (Figures 3 and 4).
LDA Estimates in Individual Subjects
To ensure the network organization we discovered was not an artifact of intersubject averaging, we applied the 7-network LDA model to individual sessions of each HCP subject. This also allowed us to evaluate the test-retest reliability of the LDA estimates. The LDA estimates of both sessions of subjects with the best (Figure 5) and median (Figure 6) test-retest reliability are juxtaposed.
Global Signal Regression
The regression of global signal during fcMRI preprocessing can introduce negative correlations between brain regions (Fox et al. 2009; Murphy et al. 2009). To ensure the network organization we discovered was robust to variations in fcMRI preprocessing, we randomly selected a subset of 100 subjects from the GSP dataset and processed them without global signal regression. We referred to this data as the non-GSR dataset. We then estimated 7 and 17 networks from this non-GSR dataset using both von Mises-Fisher clustering and LDA (Supplemental Figure 7).
Degenerate High-Likelihood Clustering and LDA Estimates
While we have focused on network estimates with the highest likelihood, there might be degenerate or alternate network estimates with high likelihood values that are topologically distinct from the best estimates (Good et al., 2010). Given that these degenerate estimates have likelihood values close to the best estimate, slight variations of the original models might result in these alternate network estimates becoming the best solutions. Consequently, we employed two different approaches to explore whether the network organization of the degenerate solutions agrees with that of the best estimate (c.f. Rubinov and Sporns, 2011).
First, we assessed whether degenerate estimates exhibited parallel large-scale networks in association cortex by exploring the solution space of the von Mises-Fisher mixture model obtained from 1000 different random initializations. We computed the overlap agreement between each of the 1000 estimates and the best estimate in the GSP dataset (Figure 7A). In addition, we computed an “exhaustive” set of 7-network clustering estimates that are representative of the 1000 random initializations (Figure 7B). We determined this “exhaustive” set as follows. We first initialized the “exhaustive” set to consist only of the best clustering estimate (i.e., estimate with the highest likelihood). We then iterated the following procedure until convergence. At each iteration, among all clustering estimates with less than 80% overlap with the current “exhaustive” set of clustering estimates, we added the clustering estimate with the highest likelihood to the “exhaustive” set. The procedure therefore allowed us to select a subset of clustering solutions that were the most different from each other and spanned the solution space of the mixture model. The 80% overlap threshold was chosen because we found (empirically) that estimates with more than 80% overlap were extremely similar (visually).
Second, to assess whether degenerate estimates exhibited the property that many association regions participated in multiple networks, we computed the number of networks each vertex participates in, averaged across the 100 random initializations of the 7-network and 17-network LDA (Figure 8).
Results
Clustering Estimates of GSP and HCP datasets
The 7-network clustering estimates of the GSP and HCP datasets were similar (top row of Figure 1), with 78% of vertices identically labeled within the entire cerebral cortex. Five of the networks were highly similar across the two datasets, with overlaps between the somatomotor (blue) and visual (purple) networks at more than 90%. Notable differences included portions of the default (red) network in the GSP dataset classified as part of the limbic (cream) network in the HCP dataset. This may relate to differences in signal-to-noise around regions of high susceptibility (which severely influences estimates of the limbic network). The overlap of the 17-network clustering estimates (not shown) between the GSP and HCP datasets was 69%.
LDA Estimates of GSP and HCP datasets
The 7-network LDA estimates of the GSP and HCP datasets were similar (rows 2-8 of Figure 1). The mean of the Pearson correlations of the seven pairs of LDA network estimates was 0.93. The worst pair of networks was correlated at 0.91. The mean pairwise correlation of the 17-network LDA estimates was 0.82. Details of several example networks are discussed below.
ICA Estimates of GSP and HCP datasets
The LDA and ICA estimates were similar in the GSP (Supplemental Figure 1) and HCP (Supplemental Figure 2) datasets. The means of the Pearson correlations between 17 corresponding pairs of networks were 0.72 in the GSP dataset and 0.62 in the HCP dataset. Given the similarities between ICA and LDA (except for the negative spatial weights in the ICA estimates), we will focus on the LDA estimates.
Broad Properties of Clustering and LDA Estimates of Cortical Organization are Similar
A critical question is the degree to which the winner-takes-all and LDA approaches converge on the same basic network organization. The agreement between the 7-network clustering and LDA estimates can be visually appreciated by looking at the correspondences between the LDA estimates and clustering boundaries (rows 2-8 in Figure 1).
The overlap between the 7-network clustering estimates and the winner-takes-all LDA estimates in the GSP dataset was 82%, while the overlap in the HCP dataset was 88%. The overlap between the 17-network clustering estimates and the winner-takes-all LDA estimates in the GSP dataset was 70%, while the overlap in the HCP dataset was 63%.
Robustness to Number of Networks
The focus on 7- and 17-network estimates should not be taken to imply that meaningful properties are absent in alternative parcellation schemes. We found similar agreement between the clustering and LDA estimates for 4, 6, 8, 10, 12, 16, 18 and 30 networks in the GSP dataset; the overlap agreement between the clustering estimates and the winner-takes-all LDA estimates were 98%, 71%, 85%, 77%, 84%, 74%, 64% and 55% respectively. Supplemental Figure 3 shows the clustering estimates for the different number of networks.
The qualitative visualization and quantitative estimates suggest that the broad topographic organization of cortical networks (Yeo et al. 2011) is not an artifact of the requirement that each cortical region must belong to a single network. Nonetheless, differences do emerge from the two approaches, and these are the topic of the remaining results.
Association Cortex Shows More Pervasive Network Participation Than Visual and Somatomotor Cortices
As a first step in visualizing properties of cortical organization revealed by LDA, we plotted the number of networks each cortical region participates in for the 7-network LDA estimates in the GSP dataset (Figure 2A). This number was obtained by counting for each vertex the number of networks whose Pr(network | vertex) exceeded 1/7. What is immediately apparent is that many cortical association regions participate in at least two functional connectivity networks.
To quantify the distribution of multiple-network participation across the cerebral cortex, the fraction of vertices participating in multiple LDA networks for each network of the 7-network clustering estimates is shown in Figure 2B. Vertices within 10mm of clustering boundaries were excluded because they might reflect uncertainties in the network estimations. The dotted line indicates that 44% of the vertices (at least 10mm away from clustering boundaries) participate in multiple networks.
Many cortical association regions participated in at least two functional connectivity networks (Figure 2B). This was in contrast with large portions of early sensory and late motor cortices that participated in a single network. In particular, the dorsal attention network, the ventral attention network, the frontoparietal control network and the default network had above average numbers of cortical vertices participating in multiple networks. In contrast, the visual network, the somatomotor network and the limbic network had below average number of cortical vertices participating in multiple networks.
We obtained consistent results when we computed the fraction of vertices participating in multiple LDA networks for each winner-takes-all LDA network (Supplemental Figure 4) or when we repeated the analyses with different thresholds (Supplemental Figure 5) or different number of networks (Supplemental Figure 6).
Across analyses (Figure 2B and Supplemental Figures 4 to 6), the default and dorsal attention networks had the largest proportions of brain vertices participating in multiple networks. To explore this phenomenon further, we identified brain regions in the left hemisphere that (1) were at least 10mm away from both clustering and winner-takes-all LDA boundaries, (2) participated in multiple networks from the 7-network estimate and (3) participated in multiple networks (from the 17-network LDA estimate) that overlapped significantly with the default network (from the 7-network estimate). Four regions were identified (black asterisks in Figure 2A): precuneus (PCUN), lateral temporal cortex (LTC), posterior parietal cortex (PPC) and medial prefrontal cortex (MPFC). Their MNI coordinates are reported in Table 1. It is worth pointing out that the PCUN region was surrounded by a band of regions participating in only one network, suggesting that its participation in multiple networks was not due to spatial blurring across network boundaries.
Table 1.
Locations of Default and Dorsal Attention Network Regions That Participate in Multiple Networks
| Brain Region | MNI Coordinates | Network | Distance to clustering boundaries | Distance to winner-takes-all LDA boundaries |
|---|---|---|---|---|
| Precuneus (PCUN) | −8, −57, 36 | default | 16mm | 19mm |
| Lateral Temporal Cortex (LTC) | −58, −11, −15 | default | 27mm | 29mm |
| Posterior Parietal Cortex (PPC) | −43, −67, 39 | default | 16mm | 15mm |
| Medial Prefrontal Cortex (MPFC) | −11, 46, −3 | default | 17mm | 10mm |
| Lateral Intraparietal (LIP) area | −28, −61, 60 | dorsal attention | 18mm | 14mm |
| Frontal Eye Fields (FEF) | −26, −6, 48 | dorsal attention | 14mm | 11mm |
We also selected brain regions in the left hemisphere that (1) were at least 10mm away from both clustering and winner-takes-all LDA boundaries, (2) participated in multiple networks from the 7-network estimate and (3) participated in multiple networks (from the 17-network LDA estimate) that overlapped significantly with the dorsal attention network (from the 7-network estimate). Two regions were identified (green asterisks in Figure 2A) and they are putative homologues of macaque areas LIP (lateral intraparietal) and FEF (frontal eye fields) based on a meta-analysis of fMRI literature (Yeo et al., 2011). Coordinates of these two regions are reported in Table 1.
We now explore the participation of the default regions in multiple paralimbic networks and the dorsal attention regions in multiple hierarchically organized networks in the 17-network LDA estimates.
Overlapping Components of Multiple Paralimbic Networks
The 17-network clustering estimates in the GSP and HCP datasets are juxtaposed in Figure 3A. Figure 3B shows four paralimbic networks from the 17-network LDA estimates. The networks overlapped significantly with brain regions typically associated with the default network. The asterisks (Figure 3B) correspond to the default regions defined in Table 1. An asterisk is colored black if Pr(vertex | network) > 1e-4 at that brain location. Therefore both the PCUN and PPC regions preferentially participated in paralimbic networks 1, 2 and 4; the MPFC region preferentially participated in paralimbic networks 1, 3 and 4; the LTC region preferentially participated in all four paralimbic networks.
FEF and LIP are Involved in Multiple Hierarchically Organized Networks
The 17-network clustering estimates in the GSP and HCP datasets are juxtaposed in Figure 4A. Figure 4B shows four networks from the 17-network LDA estimates that overlap significantly with brain regions typically associated with the dorsal attention network. These networks are likely the homologue of the well-studied hierarchical visuomotor pathway in macaques (Maunsell and Van Essen, 1983). The asterisks (Figure 4B) correspond to the dorsal attention regions defined in Table 1. An asterisk is colored black if Pr(vertex | network) > 1e-4. Therefore the LIP-homologue region preferentially participated in all four networks, while the putative FEF region preferentially participated in networks 2, 3 and 4. The visual hierarchy from early visual cortex to superior parietal cortex and frontal eye fields was noted in Yeo et al. (2011) via a series of seed-based functional connectivity analyses. As this example illustrates, LDA detected overlap among functional networks comprising early retinotopic visual areas and other components of this sensory-motor processing stream that were lost when making a winner-take-all assumption.
Cortical Network Structure can be Detected in Individual Subjects
Many research goals require analysis of imaging data at the individual subject level. The HCP dataset provides high quality datasets that are obtained twice in participants over separate days. To ensure the network organization we discovered was not an artifact of averaging across subjects, we analyzed these data to determine whether LDA networks could be detected within individual subjects and whether they would show reliability.
Pairwise correlation of the 7-network LDA estimates between the two sessions of each subject was 0.60 on average. The best subject had a mean pairwise correlation of 0.75. The median subject had a mean pairwise correlation of 0.61. Nine of the twelve subjects had average correlations of at least 0.56, suggesting moderate reliability.
The 7-network LDA estimates of the best and median subjects are shown in Figures 5 and 6. The estimated networks were well replicated across the two sessions even for the median subject. This is consistent with previous assessments of test-retest reliability of resting-state networks (Shehzad et al., 2009; Zuo et al., 2010).
In both subjects (Figures 6 and 7), the visual (row 1), somatomotor (row 2) and paralimbic (rows 6 and 7) networks were well replicated. However, the dorsal attention (green), ventral attention (violet) and frontoparietal control (orange) networks tended to inter-mix across sessions (rows 3 to 5 of Figures 5 and 6). This is not surprising given that the regions from these networks are correlated (Fox et al. 2006; Yeo et al. 2011). For example, dorsal attention regions are known to correlate with ventral attention regions (Fox et al. 2006; Yeo et al. 2011).
Choice of Thresholding LDA Estimates
We should emphasize that the choice of thresholding the LDA networks at 1e-4 was not critical for our analyses. While thresholding Pr(vertex | network) networks was necessary to visualize the results, statistics of agreement among different data-driven approaches and datasets (Figure 1, Supplemental Figures 1 and 2) were computed independent of the threshold. Similarly, statistics for test-retest reliability (Figures 5 and 6) were computed independent of the threshold. Finally, given the relatively stringent threshold of 1e-4 (almost twice the minimal threshold of 1/18715), using a lower threshold would simply confirm our broad point that different cortical regions identified in Table 1 were involved in multiple subnetworks (Figures 3 and 4).
Cortical Network Structure is Invariant to Global Signal Regression
To ensure the network organization we discovered was not an artifact of global signal regression (GSR) during fcMRI preprocessing, we estimated 7 and 17 networks in the non-GSR dataset consisting of 100 GSP subjects processed without global signal regression. The 7-network and 17-network clustering estimates from the non-GSR dataset are shown in Supplemental Figure 7. We found excellent agreement between the network estimates from the non-GSR and full GSP datasets. In particular, the overlap of the 7-network clustering estimates was 95%, while the overlap of the 17-network clustering estimates was 91% across the two datasets. The mean pairwise correlation of the 7-network LDA estimates was 0.99, while the mean pairwise correlation of the 17-network LDA estimates was 0.85 across the two datasets.
Cortical Network Structure is Replicated in Degenerate Network Estimates
Finally, we assessed whether degenerate network estimates reaffirmed the network organization we discovered. Figure 7A plots the overlap between the best 7-network clustering estimate and estimates derived from the 1000 random initializations using the GSP dataset. The clustering estimates were ordered from the highest likelihood (best estimate) to the smallest likelihood. Out of 1000 random initializations, 415 resulted in solutions with overlap of over 90% with the best solution, suggesting the best estimate corresponds to a robust local optimum.
Figure 7B shows the “exhaustive” set of 7-network clustering estimates. The eight clustering estimates were selected to be different from each other and spanned the solution space of the mixture model. Even though the seven “degenerate” clustering estimates do not have the highest likelihoods, they still explained interesting variance in the data. For example, three degenerate solutions fractionate the somatomotor cortex into dorsal-ventral portions, just like in the best 17-network estimate (Figure 3A).
Importantly, while the details of the clustering estimates varied, the broad property that early sensory and late motor cortices appeared to participate in preferentially local networks was also found across all degenerate solutions. Furthermore, (1) each association network spanned multiple lobes and (2) the components of multiple networks were spatially adjacent. Therefore, the broad property that the human association cortex consisted of multiple, parallel networks was found across all degenerate solutions.
Similarly, we found that across 100 random initializations of the 7-network and 17-network LDA (Figure 8), many association regions appeared to participate in multiple networks, while large portions of early sensory and late motor cortices participated in single networks.
Discussion
Complex behaviors are subserved by distributed networks of specialized brain areas (Posner et al., 1988; Distler et al., 1993; Mesulam 1998; Shadlen and Newsome, 2001). In this work, we show that the human association cortex consists of multiple, interdigitated distributed networks in contrast to early sensory and late motor cortices participating in preferentially local networks (Figure 1). Many association regions appear to participate in multiple networks, while large portions of early sensory and late motor cortices participate in single networks (Figure 2; Supplemental Figure 4). We demonstrate examples of overlapping paralimbic networks (Figure 3) and overlapping hierarchically organized networks (Figure 4). This architecture was detected and replicated in individual subjects (Figures 5 and 6). Additional control analyses confirmed similar network organization regardless of global signal regression (Supplemental Figure 7) and across degenerate network estimates (Figures 7 and 8). The overlaps among networks may provide clues to the network interactions that support human cognition.
Association Cortex Comprises Interdigitated, Segregated Large-Scale Circuits
Our findings suggest that the early sensory and late motor cortices participate in preferentially local networks (Figure 1; also see Sepulcre et al. 2010). In contrast, the association cortex comprises interdigitated, partially overlapping large-scale circuits (Figures 1 and 2; Supplemental Figures 1 and 2). The key features of this organization are that (1) each association network consists of strongly coupled brain regions spanning frontal, parietal, temporal, and cingulate cortices, and (2) the components of multiple networks exist adjacent to each other and are partially overlapping.
This organization is replicable across datasets (GSP and HCP) and techniques (clustering, LDA and ICA; Figure 1 and Supplemental Figures 1 and 2). Although details may differ, we can replicate this general organization across different number of estimated networks (Supplemental Figure 3), with and without global signal regression (Supplemental Figure 7) and across degenerate network estimates (Figure 7). Finally, the organization can be detected in individual subjects and replicated across sessions in the HCP dataset (Figures 5 and 6), indicating that this parallel architecture is not an artifact of group averaging.
Given the convergence in cortical network organization across datasets and methods, it is surprising that this parallel organization has not been emphasized in previous ICA analyses. We suspect the main reason is that the winner-takes-all approach allows us to juxtapose the spatial organization of different networks in one single map (top row of Figure 1), as opposed to more sophisticated approaches allowing for multiple memberships (bottom rows of Figure 1). The parallel architecture is easily appreciated in the top row of Figure 1, but is less clear in the bottom rows, even though the estimated networks are extremely similar (88% overlap in the HCP dataset). Furthermore, most previous analyses (e.g., Beckmann et al., 2005; Damoiseaux et al., 2006; De Luca et al., 2006; Smith et al., 2009) visualized the estimated networks (components) using brain slices, making it difficult to appreciate complex topographic relationships.
Estimates of Segregation and Overlap of Functional Connectivity Networks
In general, network overlaps occur throughout association cortex (Figure 2a). These overlaps do not exclusively co-localize with the presence of a border (Figure 2b; Table 1; Supplemental Figure 4). We exclude these boundary regions in our analyses (Figure 2b; Table 1; Supplemental Figure 4) because they may reflect uncertainties in the network estimations (c.f. Power et al., 2013). The default and dorsal attention networks have the largest proportions of brain vertices participating in multiple networks (Figure 2b; Supplemental Figure 4). These results can be replicated with different number of networks (Supplemental Figure 5), different thresholds (Supplemental Figure 6) and across degenerate network estimates (Figure 8).
We will focus on the dorsal attention and default networks in the following sections. However, we should emphasize that regions in other association networks also participate in multiple networks. Our emphasis on the default and dorsal attention networks may simply be due to their large sizes and our exclusion of regions close to network boundaries. This necessarily precludes investigation of the smaller regions of any network, though examples of regions possessing multiple network membership can be found in all networks at the resolution examined.
For example, there are regions in the posterior cingulate sulcus portion of the ventral attention network (area 5Ci; Scheperjans et al. 2008a, 2008b) at least 10mm away from any network boundaries, which also participate in the dorsal attention network (rows 4 and 5 of Figure 1). This is consistent with the correlations (Fox et al., 2006; Yeo et al., 2011) and close functional relationships (Corbetta and Shulman, 2002; Corbetta et al., 2008) between the two networks. Corbetta et al. (2008), as part of a review, noted that a region of right prefrontal cortex overlapped the dorsal and ventral attention systems, providing a candidate region for allowing interactions between the otherwise distinct systems (also see Asplund et al., 2010). The present results suggest the cingulate is also a potential point of interaction with the caveat that, for all such observations, it is difficult to know whether there is true spatial convergence or distinct modules are present below our level of resolution. With this caveat in mind, we now focus our attention on two examples of overlapping networks.
Overlapping parallel paralimbic networks
Numerous lines of evidence from both human imaging and non-human primate anatomical tract tracing suggest the existence of a network of brain regions, known as the default network, involved in internal mentation (for reviews, see Buckner et al., 2008; Binder et al., 2009). These regions include the medial temporal lobe, posterior medial cortex (precuneus and posterior cingulate cortex), medial prefrontal cortex, inferior parietal cortex and lateral temporal cortex.
Recent evidence (Laird et al., 2009; Andrews-Hanna et al, 2010; Leech et al., 2011; Yeo et al., 2011) suggests functional heterogeneity within the default network. In particular, Andrews-Hanna et al. (2010) suggested that the posterior cingulate cortex (PCC) and the (anterior) medial prefrontal cortex (MPFC) form the core of two overlapping default sub-networks. Our current analysis extends Andrews-Hanna's results, suggesting that the precuneus (PCUN), medial prefrontal cortex (MPFC), lateral temporal cortex (LTC) and posterior parietal cortex (PPC) participate in multiple paralimbic networks that overlap traditional default network regions (Figure 3). In particular, our analysis highlights the LTC, which appears to participate in all four paralimbic networks.
Previous results have suggested functional heterogeneity within the posterior medial cortex (Laird et al., 2009; Margulies et al., 2009; Leech et al., 2011; Fornito et al., 2012). Our results also suggest the presence of such heterogeneity. For example, the PCC appears to preferentially participate in paralimbic networks 1 and 4 (Figure 3), while PCUN (asterisk in Figure 3) preferentially participates in paralimbic networks 1, 2 and 4.
While we have focused on its involvement in paralimbic networks (Figure 3), the PCUN region may also participate in a network that prominently includes the specific rostral regions of prefrontal cortex (row 5 in Supplemental Figures 1 and 2), although the involvement is clearer in the LDA estimates than in the ICA estimates. Since the rostral prefrontal cortex is implicated in tasks requiring complex rule application (e.g., Badre and D'Esposito, 2007), this network might be involved in higher-order cognitive control. This may be consistent with heterogeneity of the precuneus previously reported, such as “Cognitive/Associative Central Precuneus” in Marguiles et al. (2009) and “Frontoparietal Network” in Fornito et al. (2012).
Overlapping hierarchical networks in a canonical sensory-motor pathway
While the early sensory and late motor cortices mostly participate in single networks (Figure 2b; Supplemental Figure 4), there are exceptions. For example, overlaps between the sensory and motor networks with the dorsal attention network in association cortex (Figure 4) may be expected from the non-human primate connectivity anatomical studies (Felleman and Van Essen, 1991) and may be involved in the hierarchical flow of information among sensory and motor areas.
The canonical sensory-motor pathway, including retinotopic visual cortex, the MT+ complex, parietal area LIP and the FEF, has been well characterized in the monkey (Maunsell and Van Essen, 1983; Andersen et al. 1990; Colby and Goldberg 1999; Shadlen and Newsome 2001; Gold and Shadlen 2007). The idea is that visual information propagates from early visual areas to MT+, which constrains decision processes arising from interactions with LIP and FEF in the upper stages of the hierarchy. The anatomical connectivity among the areas within this pathway has been extensively explored in nonhuman primates (Maunsell and Van Essen 1983; Felleman and Van Essen, 1991). In the human literature, this pathway has been studied both in relation to spatially directed movements and also in relation to spatial attention, with components of the pathway sometimes referred to as the dorsal attention network (Corbetta and Shulman, 2002).
This visual hierarchy was tested in Yeo et al. (2011) via a series of seed-based functional connectivity (also see Sepulcre et al., 2012b). The seed-based analysis was necessary because the winner-takes-all approach does not capture information about interactions among regions that fall in separate networks (Yeo et al., 2011). In contrast, the LDA estimate of cortical networks detects overlap among functional networks comprising early retinotopic visual areas and other components of this sensory-motor processing stream that were not previously appreciated (Figure 4).
In particular, the estimated human LIP homologue is involved in all four networks in Figure 4, including a network comprising mostly regions in the occipital lobe (network 1), a network comprising mostly regions in occipital and parietal lobes (network 2), a network comprising mostly regions in parietal and frontal lobes (network 3) and a higher association network spanning parietal, temporal and frontal lobes (network 4). Therefore networks 2 and 3 may serve as a bridge between early visual cortex (network 1) and higher association cortex (network 4). The FEF region is preferentially involved in the association networks (Networks 2-4), but less so in the early visual network (network 1), consistent with the fact that FEF occupies a higher position than LIP in the sensory-motor hierarchy (Maunsell and Van Essen 1983; Felleman and Van Essen, 1991).
Parallel Architecture, Network Interactions and Human Behavior
The neurophysiological literature has long recognized the importance of network interactions to cognition (e.g., Buzsáki and Draguhn, 2004; Fries 2005; Akam and Kullman, 2010; Buschman et al., 2012). In recent years there has been increasing interest to use human brain imaging to study how functionally distinct networks interact to support complex behaviors (Spreng et al., 2010; Bassett et al., 2011; de Pasquale et al, 2012; Fornito et al., 2012; Spreng et al., 2013).
Our current work suggests that a particularly robust form of interaction might take place in association cortex (Figures 2, 3 and 4; Supplemental Figure 9). This is consistent with anatomical evidence that heteromodal association regions serve as critical gateways for information processing (Pandya and Kuypers, 1969; Jones and Powell, 1970; Goldman-Rakic 1988; Mesulam 1998). While we have emphasized long-distance interactions by focusing on overlapping regions at some distance from network boundaries (Figures 2b, 3 and 4), the interdigitated topography of networks in association cortex suggests a stereotypic spatial architecture, through which network components can locally interact with adjacent components of other functionally distinct networks (also see Power et al., 2013). In other words, local interactions among networks may occur within recurrent spatial patterns or motifs (Sporns and Kotter, 2004; Bullmore and Sporns, 2009; Rubinov and Sporns, 2010; Power et al., 2011). The expectation is that these local and distant interactions among functionally distinct networks facilitate emergent complex behaviors.
This network organization can be detected and replicated in individual subjects, although the details might vary across sessions and across subjects, especially in the association cortex (Figures 5 and 6). This variability in network organization may underlie individual differences in behavior or even variability in behavior within a subject. This would be consistent with recent results showing that intersubject variability in functional connectivity is the highest in the association cortex, with regions predicting individual differences in cognitive domains predominantly located in regions of high connectivity variability (Mueller et al., 2013).
The distributed association networks converge on cortical regions that are late to develop (Hill et al., 2010) and are expanded in the human brain relative to the macaque brain (Van Essen and Dierker, 2007). Therefore, these distributed association networks have likely been under strong selective pressure to expand in recent hominin evolution. Consequently, even though this parallel architecture may also exist in monkeys (Goldman-Rakic 1988), the details of the interdigitation and overlaps (Figures 1, 3 and 4) may differ from monkeys and contribute to human cognition (see Buckner and Krienen, in press for discussion).
Limitations and Future Work
Tract tracing and physiological studies in monkeys and cats suggest that the cerebral cortex forms spatially organized circuits that include prominent connections with subcortical structures (for review, see Haber 2003; Jones 2007; Strick et al., 2009). In the present work we limit our analysis to the cerebral cortex, and therefore do not consider interactions between the cerebral cortex and subcortical structures. This limitation is partly methodological. Subcortical structures have significantly lower signal-to-noise ratio than the cerebral cortex and so are typically analyzed separately in functional connectivity studies (Di Martino et al. 2008; Zhang et al. 2008; Krienen and Buckner, 2009; Buckner et al., 2011; Choi et al., 2012; Dobromyslin et al., 2012; but see Habas et al., 2009; O'Reilly et al., 2010). Our (unpublished) observations suggest that including subcortical structures with cortical analyses would probably not alter the estimated cortical networks, even though subcortical results might be suboptimal. Consequently, we focus on the cerebral cortex in this paper. We are currently exploring techniques that can more effectively jointly analyze cortical and subcortical structures.
In addition, this work focuses on the spatial relationships among resting-state functional connectivity networks. The parallel architecture and overlaps in hierarchical and paralimbic networks suggest stereotypic spatial motifs in network interactions. However, our (essentially) correlational analyses will miss vital details of the dynamics of network interactions (Chang et al., 2010; de Pasquale et al, 2012; Smith et al., 2012; Hutchison et al., 2013; Allen et al., In Press). Our resting-state analyses will also miss changes in network interactions as a result of task performance (Bassett et al., 2006; Spreng et al., 2010; Bassett et al., 2011; Fornito et al., 2012). These potential issues are outside the scope of this paper and will be addressed in future work.
The network organization we found is robust across datasets (GSP and HCP), preprocessing (with and without global signal regression), number of networks, degenerate/alternate network estimates and techniques (clustering, LDA and ICA). Although details vary, corroboration across the different analyses increases our confidence that the general organization of cortical networks is intrinsic to the data. However, we caution that the agreement among different techniques (clustering, LDA and ICA) may simply result from underlying assumptions common across the techniques. For example, while there are similarities between gradient-based (Cohen et al., 2008) and clustering/modularity-based parcellations (Power et al., 2011; Yeo et al., 2011), the boundaries in certain brain regions (e.g., visual cortex) can be qualitatively different (Wig et al., in press; Buckner and Yeo, under revision). While we suspect the general network organization we discovered would generalize to gradient-based parcellation, we leave this replication (or refutation) to future work.
Conclusions
The human association cortex consists of multiple, interdigitated large-scale networks that, while partially overlapping, possess predominantly parallel organization. This architecture can be detected and replicated in individual subjects. Many, but not all, association regions appear to participate in multiple networks, including those that lie some distance from estimates of network boundaries. The present work suggests that it is possible to consider both the divergent and convergent nature of connectivity in the human cerebral cortex using functional connectivity MRI. Our results suggest that segregation and interdigitation of networks in association cortex may be a true feature of cortical organization and not an artifact of the methods used to estimate their topography.
Supplementary Material
Highlights.
We estimated overlapping cortical networks from resting fMRI data (N=1012).
Many association regions involved in multiple interdigitated, segregated networks.
Many sensory-motor regions involved in single, preferentially local, networks.
Architecture converges across two datasets and three methods.
Results replicable across sessions of individual Human Connectome Project subjects.
Acknowledgements
This work was supported by the National Medical Research Council, Singapore (STaR/0004/2008), Massachusetts General Hospital-University of California, Los Angeles Human Connectome Project (U54MH091665), the Simons Foundation and the Howard Hughes Medical Institute. Data were provided by the Brain Genomics Superstruct Project (Principal Investigators: Randy Buckner, Jordan Smoller, and Joshua Roffman) and by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. This research also utilized resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896 and instruments supported by S10RR021110, S10RR023401 and S10RR019307 from the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital. The authors would like to thank Jesisca Tandi for help with some of the figures in this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Using a different measure (e.g., KL-divergence) achieves the same results.
We note that this negative spatial weight issue is different from the anti-correlations issue discussed in the functional connectivity literature (Fox et al. 2009; Murphy et al. 2009).
References
- Akam T, Kullmann DM. Oscillations and filtering networks support flexible routing of information. Neuron. 2010;67:308–320. doi: 10.1016/j.neuron.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. doi: 10.1093/cercor/bhs352. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RA, Asanuma C, Essick G, Siegel RM. Corticocortical connections of anatomically and physiologically defined subdivisions within the inferior parietal lobule. J Comp Neurol. 1990;296:65–113. doi: 10.1002/cne.902960106. [DOI] [PubMed] [Google Scholar]
- Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. Functional-anatomic fractionation of the brain's default network. Neuron. 2010;65:550–562. doi: 10.1016/j.neuron.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asplund CL, Todd JJ, Snyder AP, Marois R. A central role for the lateral prefrontal cortex in goal-directed and stimulus-driven attention. Nature neuroscience. 2010;13:507–512. doi: 10.1038/nn.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badre D, D'Esposito M. Functional magnetic resonance imaging evidence for a hierarchical organization of the prefrontal cortex. J Cogn Neurosci. 2007;19:2082–2099. doi: 10.1162/jocn.2007.19.12.2082. [DOI] [PubMed] [Google Scholar]
- Badre D, D'Esposito M. Is the rostro-caudal axis of the frontal lobe hierarchical? organization of the frontal lobes. Nature Reviews Neuroscience. 2009;10:659–669. doi: 10.1038/nrn2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Meyer-Linderberg A, Sophie A, Duke T, Bullmore E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc Natl Acad Sci USA. 2006;103:19518–19523. doi: 10.1073/pnas.0606005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci USA. 2011;108:7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellec P, Rosa-Neto P, Lyttelton OC, Benali H, Evans AC. Multi-level bootstrap analysis of stable clusters in resting-state fMRI. Neuroimage. 2010;51:1126–1139. doi: 10.1016/j.neuroimage.2010.02.082. [DOI] [PubMed] [Google Scholar]
- Binder JR, Desai RH, Graves WW, Conant LL. Where is the semantic system? A critical review and meta-analysis of 120 functional neuroimaging studies. Cerebral Cortex. 2009;19:2767–2796. doi: 10.1093/cercor/bhp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Blei DM, Ng AY, Jordan MI, Latent Dirichlet Allocation J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- Buckner RL, Andrews- Hanna JR, Schacter DL. The brain's default network. Ann New York Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM. The evolution of distributed association networks in the human brain. Trends Cogn Sci. doi: 10.1016/j.tics.2013.09.017. in press. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BTT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:2322–2345. doi: 10.1152/jn.00339.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Yeo BTT. Opportunities and limitations of functional connectivity MRI. Nat Neurosci. 2013;16:832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Yeo BTT. Borders, map clusters and supra-areal organization in visual cortex. submitted. [DOI] [PMC free article] [PubMed]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Buschman TJ, Denovellis EL, Diogo C, Bullock D, Miller EK. Synchronous oscillatory neural ensembles for rules in the prefrontal cortex. Neuron. 2012;76:838–846. doi: 10.1016/j.neuron.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Draguhn A. Neuronal oscillations in cortical networks. Science. 2004;304:1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Kiehl KA, Pearlson GD. Modulation of temporally coherent brain networks estimated using ICA at rest and during cognitive tasks. Hum. Brain Mapp. 2008;29:828–838. doi: 10.1002/hbm.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EY, Yeo BT, Buckner RL. The organization of the human striatum estimated by intrinsic functional connectivity. J Neurophysiol. 2012;108:2242–2263. doi: 10.1152/jn.00270.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby CL, Goldberg ME. Space and attention in parietal cortex. Annu Rev Neurosci. 1999;22:319–349. doi: 10.1146/annurev.neuro.22.1.319. [DOI] [PubMed] [Google Scholar]
- Corbetta M, Patel G, Shulman GL. The reorienting system of the human brain: from environment to theory of mind. Neuron. 2008;58(3):306–324. doi: 10.1016/j.neuron.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33(8):1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29:1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- de Pasquale F, Penna SD, Snyder AZ, Marzetti L, Pizzella V, Romani GL, Corbetta M. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron. 2012;74:753–764. doi: 10.1016/j.neuron.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Scheres A, Margulies DS, Kelly AMC, Uddin LQ, Shehzad Z, Biswal B, Walters JR, Castellanos FX, Milham MP. Functional connectivity of human striatum: a resting state FMRI study. Cereb Cortex. 2008;18:2735–2747. doi: 10.1093/cercor/bhn041. [DOI] [PubMed] [Google Scholar]
- Distler C, Boussaoud D, Desimone R, Ungerleider LG. Cortical connections of inferior temporal area TEO in macaque monkeys. J Comp Neurol. 1993;334:125–150. doi: 10.1002/cne.903340111. [DOI] [PubMed] [Google Scholar]
- Dobromyslin VI, Salat DH, Fortier CB, Leritz EC, Beckmann CF, Milberg WP, McGlinchey RE. Distinct functional networks within the cerebellum and their relation to cortical systems assessed with independent component analysis. Neuroimage. 2012;60:2073–2085. doi: 10.1016/j.neuroimage.2012.01.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RAT, Fox MD, Snyder AZ, Vincent JL, Raichle ME, Schlaggar BL, Petersen SE. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci USA. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet G, Naveau M, Petit L, Delcroix N, Zago L, Crivello F, Jobard G, Tzourio-Mazoyer N, Mazoyer B, Mellet E, et al. Brain activity at rest: a multiscale hierarchical functional organization. J Neurophysiol. 2011;105:2753–2763. doi: 10.1152/jn.00895.2010. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed Echo Planar Imaging for sub-second whole brain fMRI and fast diffusion imaging. PLoS One. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999a;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999b;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Harrison BJ, Zalesky A, Simons JS. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc Natl Acad Sci USA. 2012;109(31):12788–12793. doi: 10.1073/pnas.1204185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci USA. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P. Spontaneous low- frequency BOLD signal fluctuations: An fMRI investigation of the resting- state default mode of brain function hypothesis. Human brain mapping. 2005;26:15–29. doi: 10.1002/hbm.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries P. A mechanism for cognitive dynamics: neuronal communication through neuronal coherence. Trends Cogn Sci. 2005;9:474–480. doi: 10.1016/j.tics.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Junqian Xu, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, the WU-Minn HCP Consortium The Minimal Preprocessing Pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold JI, Shadlen MN. The neural basis of decision making. Annu Rev Neurosci. 2007;30:535–574. doi: 10.1146/annurev.neuro.29.051605.113038. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Topography of cognition: parallel distributed networks in primate association cortex. Annu Rev Neurosci. 1988;11:137–156. doi: 10.1146/annurev.ne.11.030188.001033. [DOI] [PubMed] [Google Scholar]
- Good BH, de Montjoye YA, Clauset A. Performance of modularity maximization in practical contexts. Phys. Rev. E. 2010;81:046106. doi: 10.1103/PhysRevE.81.046106. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48:63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas C, Kamdar N, Nguyen D, Prater K, Beckmann CF, Menon V, Greicius MD. Distinct cerebellar contributions to intrinsic connectivity networks. J Neurosci. 2009;29:8586–8594. doi: 10.1523/JNEUROSCI.1868-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber SN. The primate basal ganglia: parallel and integrative networks. J Chem Neuroanat. 2003;26:317–330. doi: 10.1016/j.jchemneu.2003.10.003. [DOI] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y, Evans A. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS One. 2009;4:e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci USA. 2010;107:13135–13140. doi: 10.1073/pnas.1001229107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Blei DM, Bach F. Online learning for latent dirichlet allocation. Adv Neural Inform Process Syst. 2010;23:856–864. [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Penna SD, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, dc Pasquale F, Sporns O, Walter M, Chang C. Dynamic functional connectivity: Promises, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jones EG. The Thalamus. Cambridge Univ. Press; Cambridge, UK: 2007. [Google Scholar]
- Jones EG, Powell TPS. An anatomical study of converging sensory pathways within the cerebral cortex of the monkey. Brain. 1970;93:793–820. doi: 10.1093/brain/93.4.793. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, MacFall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Krienen FM, Buckner RL. Segregated fronto-cerebellar circuits revealed by intrinsic functional connectivity. Cereb Cortex. 2009;19:2485–2497. doi: 10.1093/cercor/bhp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H. The Hungarian Method for the assignment problem. Nav. Res. Logist. Q. 1955;2:83–97. [Google Scholar]
- Laird AR, Eickhoff SB, Li K, Robin DA, Glahn DC, Fox PT. Investigating the functional heterogeneity of the default mode network using coordinate-based meta-analytic modeling. J Neurosci. 2009;29:14496–14505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashkari D, Vul E, Kanwisher N, Golland P. Discovering structure in the space of fMRI selectivity profiles. Neuroimage. 2010;50:1085–1098. doi: 10.1016/j.neuroimage.2009.12.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- Lee MH, Hacker CD, Snyder AZ, Corbetta M, Zhang D, Leuthardt EC, Shimony JS. Clustering of resting state networks. Plos One. 2012;7:e40370. doi: 10.1371/journal.pone.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech R, Kamourieh S, Beckmann CF, Sharp DJ. Fractionating the default mode network: distinct contributions of the ventral and dorsal posterior cingulate cortex to cognitive control. J Neurosci. 2011;31:3217–3224. doi: 10.1523/JNEUROSCI.5626-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Kelly AM, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Mapping the functional connectivity of anterior cingulate cortex. Neuroimage. 2007;37:579–588. doi: 10.1016/j.neuroimage.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos XF, Milham MP, Petrides M. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci USA. 2009;106:20069–20074. doi: 10.1073/pnas.0905314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunsell JHR, Van Essen DC. The connections of the middle temporal visual area (MT) and their relationship to a cortical hierarchy in the macaque monkey. J Neurosci. 1983;3:2563–2586. doi: 10.1523/JNEUROSCI.03-12-02563.1983. 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM. A cortical network for directed attention and unilateral neglect. Ann Neurol. 1981;10:309–325. doi: 10.1002/ana.410100402. [DOI] [PubMed] [Google Scholar]
- Mesulam MM. From sensation to cognition. Brain. 1998;121:1013–1052. doi: 10.1093/brain/121.6.1013. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. Hierarchical modularity in human brain functional networks. Front Neuroinform. 2009;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BTT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. Individual Variability in Functional Connectivity Architecture of the Human Brain. Neuron. 2013;77:586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly JX, Beckmann CF, Tomassini V, Ramnani N, Johansen-Berg H. Distinct and overlapping functional zones in the cerebellum defined by resting state functional connectivity. Cereb Cortex. 2010;20:953–965. doi: 10.1093/cercor/bhp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya DN, Kuypers HG. Cortico-cortical connections in the rhesus monkey. Brain Res. 1969;13:13–36. doi: 10.1016/0006-8993(69)90141-3. [DOI] [PubMed] [Google Scholar]
- Posner MI, Petersen SE, Fox PT, Raichle ME. Localization of cognitive operations in the human brain. Science. 1988;240:1627–1631. doi: 10.1126/science.3289116. [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Lessov-Schlaggar CN, Petersen SE. Evidence for Hubs in Human Functional Brain Networks. Neuron. 2013;79:798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretation. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Weight conserving characterization of complex functional brain networks. Neuroimage. 2011;56:2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman M, Pickard J, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain Cereb. Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Scheperjans F, Eickhoff SB, Hömke L, Mohlberg H, Hermann K, Amunts K, Zilles K. Probabilistic maps, morphometry, and variability of cytoarchitectonic areas in the human superior parietal cortex. Cereb Cortex. 2008a;18:2141–2157. doi: 10.1093/cercor/bhm241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheperjans F, Hermann K, Eickhoff SB, Amunts K, Schleicher A, Zilles K. Observer-independent cytoarchitectonic mapping of the human superior parietal cortex. Cereb Cortex. 2008b;18:846–867. doi: 10.1093/cercor/bhm116. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon LD, Goldman-Rakic PS. Common cortical and subcortical targets of the dorsolateral prefrontal and posterior parietal cortices in the rhesus monkey: evidence for a distributed neural network subserving spatially guided behavior. J Neurosci. 1988;8:4049–4068. doi: 10.1523/JNEUROSCI.08-11-04049.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med. 2012;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Liu H, Talukdar T, Martincorena I, Yeo BTT, Buckner RL. The organization of local and distant functional connectivity in the human brain. PLoS Comput Biol. 2010;6:e1000808. doi: 10.1371/journal.pcbi.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Sabuncu MR, Johnson KA. Network assemblies in the functional brain. Curr Opin Neurol. 2012a;25:384–391. doi: 10.1097/WCO.0b013e328355a8e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Sabuncu MR, Yeo TB, Liu H, Johnson KA. Stepwise connectivity of the modal cortex reveals the multimodal organization of the human brain. J Neurosci. 2012b;32:10649–10661. doi: 10.1523/JNEUROSCI.0759-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadlen MN, Newsome WT. Neural basis of a perceptual decision in the parietal cortex (area LIP) of the rhesus monkey. 2001. [DOI] [PubMed]
- Shehzad Z, Kelly AMC, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP. The resting brain: unconstrained yet reliable. Cereb Cortex. 2009;19:2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney D, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg D, Yacoub EC, Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci USA. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Kotter R. Motifs in brain networks. PLoS Biol. 2004;2:e369. doi: 10.1371/journal.pbio.0020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kötter R. Identification and classification of hubs in brain networks. PLoS One. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Network attributes for segregation and integration in the human brain. Curr Opin Neurobiology. 2013;23:162–171. doi: 10.1016/j.conb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Spreng RN, Stevens WD, Chamberlain JP, Gilmore AW, Schacter DL. Default network activity, coupled with the frontoparietal control network, supports goal-directed cognition. Neuroimage. 2010;53:303–317. doi: 10.1016/j.neuroimage.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng RN, Sepulcre J, Turner GR, Stevens WD, Schacter DL. Intrinsic architecture underlying the relations among the default, dorsal attention, and frontoparietal control networks of the human brain. J Cognitive Neurosci. 2013;25:74–86. doi: 10.1162/jocn_a_00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- Ungerleider LG, Desimone R. Cortical connections of visual area MT in the macaque. J Comp Neurol. 1986;248:190–222. doi: 10.1002/cne.902480204. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RCW, Kahn RS, Hulshoff Pol HE. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp. 2009;30:3127–3141. doi: 10.1002/hbm.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kouwe AJW, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008;40:559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. A population-average, landmark- and surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Penna SD, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SR, Prior F, Schlagger BL, Smith SM, Snyder AZ, Xu J, Yacoub E, the WU-Minn HCP Consortium The Human Connectome Project: A data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, the WU Minn HCP Consortium The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G, Gramfort A, Pedregosa F, Michel V, Thirion B. Multi-subject dictionary learning to segment an atlas of brain spontaneous activity. Information processing in medical imaging. 2011. pp. 562–573. [DOI] [PubMed]
- Vincent JL, Snyder AZ, Fox MD, Shannon BJ, Andrews JR, Raichle ME, Buckner RL. Coherent spontaneous activity identifies a hippocampal parietal memory network. J Neurophysiol. 2006;96:3517–3531. doi: 10.1152/jn.00048.2006. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL. Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J Neurophysiol. 2008;100:3328–3342. doi: 10.1152/jn.90355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Petersen SE. An approach for parcellating human cortical areas using resting-state correlations. Neuroimage. doi: 10.1016/j.neuroimage.2013.07.035. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Moeller S, Strupp J, Auerbach E, Feinberg DA, Ugurbil K, Yacoub E. Highly accelerated whole brain imaging using aligned-blipped-controlled-aliasing multiband EPI. Proc Int Soc Mag Reson Med. 2012;20:2306. [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qiu B, Giles CL, Foley HC, Yen J. An LDA-based community structure discovery approach for large-scale social networks. IEEE Intelligence and Security Informatics. 2007:200–207. [Google Scholar]
- Zhang D, Snyder AZ, Fox MD, Sansbury MW, Shimony JS, Raichle ME. Intrinsic functional relations between human cerebral cortex and thalamus. J Neurophysiol. 2008;100:1740–1748. doi: 10.1152/jn.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. Reliable intrinsic connectivity networks: test-retest evaluation using ICA and dual regression approach. Neuroimage. 2010;49:2163–2177. doi: 10.1016/j.neuroimage.2009.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.