Background: DNA gyrase catalyzes ATP-dependent negative DNA supercoiling.
Results: Deletion of the acidic C-terminal tail causes stronger DNA bending by the B. subtilis gyrase C-terminal domains and accelerated DNA-stimulated ATPase and supercoiling activities of gyrase.
Conclusion: The C-tail down-regulates negative supercoiling by B. subtilis gyrase.
Significance: The C-tail is a versatile element that differentially regulates the activity of different gyrases.
Keywords: DNA Gyrase, DNA Topology, Enzyme Mechanisms, Protein Conformation, Single Molecule Biophysics, ATP-dependent Conformational Changes, C-terminal Domain, Negative Supercoiling, Regulation
Abstract
Gyrase is a type II DNA topoisomerase that introduces negative supercoils into DNA in an ATP-dependent reaction. It consists of a topoisomerase core, formed by the N-terminal domains of the two GyrA subunits and by the two GyrB subunits, that catalyzes double-stranded DNA cleavage and passage of a second double-stranded DNA through the gap in the first. The C-terminal domains (CTDs) of the GyrA subunits form a β-pinwheel and bind DNA around their positively charged perimeter. As a result, DNA is bound as a positive supercoil that is converted into a negative supercoil by strand passage. The CTDs contain a conserved 7-amino acid motif that connects blades 1 and 6 of the β-pinwheel and is a hallmark feature of gyrases. Deletion of this so-called GyrA-box abrogates DNA bending by the CTDs and DNA-induced narrowing of the N-gate, affects T-segment presentation, reduces the coupling of DNA binding to ATP hydrolysis, and leads to supercoiling deficiency. Recently, a severe loss of supercoiling activity of Escherichia coli gyrase upon deletion of the non-conserved acidic C-terminal tail (C-tail) of the CTDs has been reported. We show here that, in contrast to E. coli gyrase, the C-tail is a very moderate negative regulator of Bacillus subtilis gyrase activity. The C-tail reduces the degree of DNA bending by the CTDs but has no effect on DNA-induced conformational changes of gyrase that precede strand passage and reduces DNA-stimulated ATPase and DNA supercoiling activities only 2-fold. Our results are in agreement with species-specific, differential regulatory effects of the C-tail in gyrases from different organisms.
Introduction
The integrity of chromosomal DNA is vitally important for all organisms. Cellular processes such as replication, recombination, and transcription are affected by the supercoiling state of the DNA. Topoisomerases are enzymes that catalyze the interconversion of different DNA topoisomers by cleaving one (type I) or both (type II) strands of the DNA and by transporting a second single- or double-stranded segment, provided in cis or in trans, through the gap in the first (for a review, see Ref. 1). DNA gyrase is a unique topoisomerase mainly found in bacteria but also in archaea and plants that can negatively supercoil DNA in an ATP-dependent reaction (2). Gyrase also catalyzes nucleotide-independent DNA relaxation and decatenation in vitro, albeit with much lower efficiency (3, 4). The active enzyme is formed by the association of a GyrA dimer with two GyrB subunits (5, 6). Catalysis of DNA topology changes by gyrase is believed to occur by a strand passage mechanism (see Fig. 1; Ref. 7, for a review, see Ref. 8) according to the two-gate model put forward for type II topoisomerases (7, 9, 10). The catalytic cycle is initiated by binding and cleavage of the so-called gate-DNA segment (G-segment)2 to the DNA-gate at the interface of the GyrA and GyrB subunits (11) (see Fig. 2, A and B). The DNA-gate harbors two DNA cleavage sites encompassing the catalytic tyrosine residues (12) and binding sites for Mg2+ ions (13). Binding of ATP to the N-terminal domains of GyrB induces dimerization and closure of the so-called N-gate (see Fig. 1) (14–16), thereby capturing a second DNA duplex, the transfer-DNA segment (T-segment; see Fig. 1) (14), in a cavity delimited by the GyrB subunits. The T-segment is believed to be transported through the gap in the cleaved G-segment by sequential opening and closing of the DNA- and the C-gates in GyrA (see Figs. 1 and 2B) (17) in a tightly controlled process linked to ATP hydrolysis (18). ADP and phosphate release results in resetting of the enzyme for subsequent supercoiling cycles (15, 19, 20).
FIGURE 1.
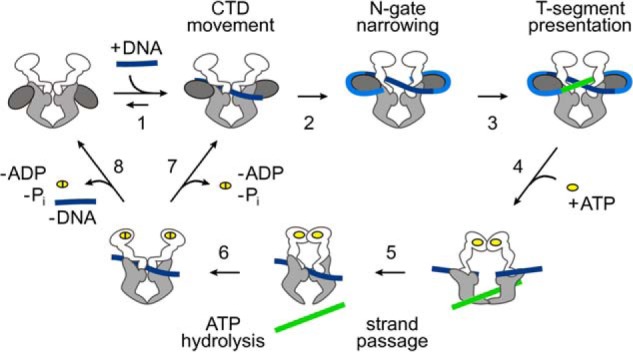
Conformational changes in the catalytic cycle of gyrase. Schematic depiction of the strand passage mechanism for ATP-dependent negative DNA supercoiling by gyrase (for a review, see Ref. 8). Gyrase is shown as a schematic (see Fig. 2 for details) with GyrB in white, the GyrA N-terminal domain in light gray, and the GyrA CTD in dark gray. In the first step, the G-segment (dark blue) binds at the DNA-gate (1), leading to an upward movement of the CTDs. DNA wrapping (blue) by the CTDs leads to N-gate narrowing (2). The CTDs ensure the correct positioning of the T-segment (green) above the G-segment in the upper cavity (3). ATP binding (4) then leads to N-gate closure and T-segment capture. DNA-gate opening allows for strand passage (5), and the T-segment can then leave the bottom cavity upon C-gate opening. ATP hydrolysis (6) leads to reopening of the N-gate and allows for subsequent catalytic cycles without (7) or with (8) dissociation of the DNA from the DNA-gate.
FIGURE 2.
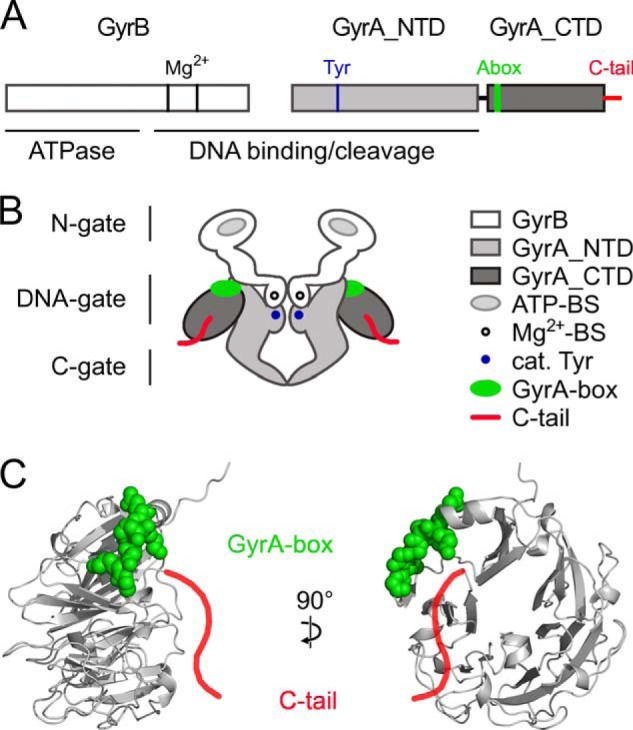
DNA gyrase and the GyrA C-tail. A, primary structure of B. subtilis gyrase. GyrB and N- and C-terminal domains of GyrA are shown in white and light and dark gray, respectively. The C-tail is shown in red. The GyrA-box (Abox), the catalytic (cat.) tyrosine, and the magnesium ion-binding sites are depicted in green, blue, and black, respectively. The ATPase domain on GyrB and the DNA-binding region on both GyrB and GyrA are marked. B, schematic depiction of gyrase. The color code is the same as in A. The ATP-binding sites (ATP-BS) on GyrB are shown in light gray. The dimerization interfaces, the N-gate (GyrB), the DNA-gate (GyrB and GyrA), and the C-gate (GyrA), are labeled on the left side. C, side (left) and front views (right) of a homology model of the GyrA_CTD of B. subtilis using the GyrA_CTD of X. campestris as a template (Ref. 31; Protein Data Bank code 3L6V). The GyrA_CTD forms a six-bladed β-pinwheel structure. The C-tail (red) is drawn schematically as an extension to the C terminus of the model, and the GyrA-box is shown in space-filling representation (green). Mg2+-BS, Mg2+-binding sites.
A highly conserved C-terminal domain (CTD), connected to the N-terminal domain (NTD) of GyrA by a flexible linker (see Fig. 2), is an important determinant for the DNA supercoiling activity of gyrase (21, 22), and deletion of the CTDs converts gyrase into a regular type II topoisomerase that catalyzes ATP-dependent relaxation (23). The CTD consists of six segments of short, antiparallel β-sheets (blades) forming a spiral or a cylindrical β-pinwheel fold with a diameter of ∼50 Å (Fig. 2C) (24, 25). The isolated CTD binds DNA via an elongated patch of positively charged amino acids around its perimeter, resulting in DNA bending and introduction of positive writhe (22, 24). In gyrase, the CTDs associate with DNA adjacent to the G-segment that extends from the DNA-gate on the enzymatic core (26, 27). Contact formation between these DNA regions and the CTDs is accompanied by a repositioning of the CTDs, aligning them with the DNA-gate (26). The geometry imposed on the DNA by the CTDs directs the DNA exiting from the CTDs toward the N-gate in an orientation that allows the flanking region to serve as a T-segment for strand passage (15, 26, 28). Establishing this strand crossing is merely an effect of DNA binding and manifests in stabilization of a positive supercoil. Strand passage, coupled to the nucleotide cycle of gyrase, then leads to conversion into a negative supercoil (29, 30).
A conserved, 7-amino acid motif within the β-pinwheel of the CTD, the GyrA-box, is located in a loop connecting the first blade of the pinwheel to blade 6 (Fig. 2, A–C) (31). The GyrA-box is a hallmark feature of gyrase (32) and is essential for DNA supercoiling (33). Its deletion reduces DNA bending and affects the communication between DNA-gate, CTDs, and N-gate, leading to a decreased DNA-stimulated ATPase activity, a failure to narrow the N-gate in response to DNA binding, and supercoiling deficiency (28).
Although the β-pinwheel, including the GyrA-box, is shared by all CTDs, in many GyrA homologs, the CTD contains an acidic C-terminal tail (C-tail; Fig. 2, A–C) with significant divergence in length and amino acid composition. It generally contains a high number of acidic residues and lacks positively charged residues (34). GyrA from Escherichia coli contains a longer tail (34 amino acids) and many acidic residues (18 amino acids), whereas the ortholog from Bacillus subtilis has a shorter tail with fewer negatively charged residues (13 and 9 amino acids, respectively). GyrA from Borrelia burgdorferi completely lacks a C-terminal extension. The C-tail has not been resolved in crystal structures of CTDs from different organisms and has even impeded crystal formation, pointing toward significant flexibility (25, 31, 35). The C-tail of the E. coli gyrase CTD prevents DNA binding to the isolated CTD. In the context of gyrase, however, the C-tail confers a high DNA supercoiling efficiency, and it has been suggested that it interacts with other parts of the enzyme. The C-tail appears to be an important regulatory element that allows for fine-tuning of gyrase activity (34).
Here we present a detailed analysis of the role of the C-tail of B. subtilis gyrase in individual steps of the supercoiling cycle. We investigated the flexibility of the C-tail, its contribution to DNA binding and bending, its effect on DNA- and nucleotide-induced conformational changes of gyrase preceding strand passage, and its effect on DNA-induced ATP-hydrolysis and ATP-dependent DNA supercoiling. The C-tail reduces bending of DNA bound to the CTDs but otherwise has little effect on other events before strand passage. We do not find any indication for an interaction of the C-tail with the remainder of gyrase. Notably, the C-tail leads to a reduction in DNA-stimulated ATP hydrolysis and in ATP-dependent DNA supercoiling, consistent with its function as a repressive element for gyrase activity. Thus, the C-tail might differentially modulate the activity of gyrases from different organisms depending on its length and charge.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
To generate B. subtilis GyrA lacking the C-terminal 13 amino acids 809NEEDENEEEQEEV821, the codon for Asn-809 was exchanged to a TAA stop codon by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA) using gyrA (coding for GyrA (36)), gyrA_CTD (coding for the isolated CTD of GyrA, GyrA_CTD (28)), or gyrBA (coding for a fusion protein of GyrB and GyrA, GyrBA (15)) as a template. Cysteines were introduced into the GyrA_CTD at positions 757 and at the C terminus (extension by Cys-822) by site-directed mutagenesis using gyrA_CTD as a template.
Primers were 5′-GCTTTAGTTG AGAAATAAGA AGAAGATGAG AATG-3′ (N809stop_for), 5′-CATTCTCATC TTCTTCTTAT TTCTCAACTA AAGC-3′ (N809stop_rev), 5′-GCAGTGAAAG CTACTTGTGG TGAAGAGGAT CTAATG-3′ (K757C_for), 5′-CATTAGATCC TCTTCACCAC AAGTAGCTTT CACTGC-3′ (K757C_rev), 5′-GAACAAGAAG AAGTGTGTTA ACTTGCGGCC GCATAATG-3′ (stop822C_for), and 5′-CATTATGCGG CCGCAAGTTA ACACACTTCT TCTTGTTC-3′ (stop822C_rev). The underlined parts indicate the altered codons.
Peptides, Proteins, and DNA Substrates
A fluorescently labeled peptide with the sequence of the 13-amino acid C-terminal tail of B. subtilis GyrA was synthesized by Genosphere Biotechnologies (Paris, France) (purity higher than 95% according to HPLC-MS). The peptide carries an ϵ-aminofluorescein-modified lysine at the C terminus for fluorescence measurements. The lyophilized peptide was dissolved in 200 mm Tris/HCl, pH 8.0 (25 °C) and stored at −20 °C.
GyrB, GyrA, heterodimeric GyrA, GyrBA, and GyrA_CTD were purified from recombinant E. coli BL21(DE3), BL21(DE3) RP, or Rosetta(DE3) as described before (15, 26, 28, 36). Briefly, proteins were purified from crude extracts by Ni2+-nitrilotriacetic acid affinity chromatography, ion exchange chromatography and size exclusion chromatography, flash frozen, and kept at −20 °C. Proteins were stored in 50 mm Tris/HCl, pH 7.5 (4 °C), 300 mm NaCl, 10 mm MgCl2, and 2 mm β-mercaptoethanol. Protein concentrations were determined photometrically from the absorption at 280 nm using the following extinction coefficients: ϵ280 = 42,750 m−1 cm−1 (GyrA), ϵ280 = 17,420 m−1 cm−1 (GyrA_CTD), ϵ280 = 52,720 m−1 cm−1 (GyrB), and ϵ280 = 95,470 m−1 cm−1 (GyrBA).
For fluorescent labeling, 30 μm GyrA_CTD_K757C or GyrA_CTD_822C were incubated with Alexa Fluor 488 (Alexa488)-maleimide in 50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 10 mm MgCl2, and 1 mm tris(2-carboxyethyl)phosphine at 25 °C for 1 h. Non-reacted dye was removed by size exclusion chromatography using illustra Microspin G-25 columns (GE Healthcare). Heterodimeric GyrA comprising a wild-type subunit and a subunit carrying the T140C/K594C mutations and homodimeric GyrBA_S7C were fluorescently labeled with Alexa488 (donor) and Alexa Fluor 546 (Alexa546) (acceptor) maleimide dyes as described before (15, 26). Labeling efficiencies were determined as described (26).
Negatively supercoiled pUC18 plasmid was purified from E. coli XL1-Blue cells using the Qiagen Giga prep kit (Qiagen, Hilden, Germany). Relaxed plasmid was obtained by incubation of negatively supercoiled pUC18 with gyrase as described (15). Unmodified and fluorescently labeled 40- (28) and 60-nucleotide (37) DNAs were purchased from Purimex (Grebenstein, Germany). Double-stranded DNAs were prepared by annealing complementary strands in a thermocycler. The 40-bp DNA serves as a wrapping substrate for the CTDs (24, 28), and the 60-bp DNA serves as a gate-DNA that binds to the gyrase DNA-gate (37).
Fluorescence Anisotropy Measurements
Dissociation constants of gyrase-DNA complexes were determined in fluorescence anisotropy titrations. 100 nm 40-bp DNA carrying Alexa488 as a fluorescent probe (λex = 494 nm, λem = 518 nm) in 50 mm Tris/HCl, pH 7.5, 40 mm NaCl, 10 mm MgCl2, and 10% (v/v) glycerol was titrated with GyrA_CTD as described previously (28). 200 or 50 nm 60-bp DNA carrying Alexa546 (λex = 555 nm, λem = 570 nm) in 50 mm Tris/HCl, pH 7.5, 100 mm KCl, and 10 mm MgCl2 was titrated with GyrA or GyrBA, respectively, and data were analyzed with a 1:1 binding model according to Equation 1 as described previously (37).
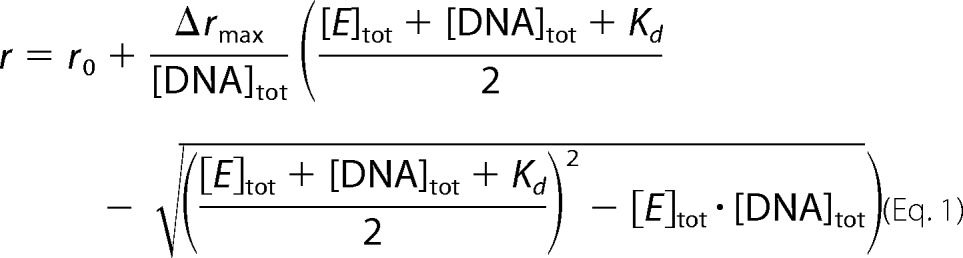 |
r is the measured anisotropy, r0 denotes the anisotropy measured for free DNA, and Δrmax is the anisotropy increase upon saturation. [E]tot and [DNA]tot are total enzyme and DNA concentrations, respectively, and Kd is the dissociation constant.
Anisotropy measurements with Alexa488-labeled GyrA_CTD (50 nm dye, λex = 494 nm, λem = 518 nm) were performed in 50 mm Tris/HCl, pH 7.5, 10 mm MgCl2, and varying concentrations of NaCl at 25 °C. Samples were equilibrated in this buffer for 120 s prior to fluorescence measurements.
Anisotropy measurements with 200 nm fluorescein-labeled peptide were performed in the presence of GyrA and GyrBA with and without C-tail in 50 mm Tris/HCl, pH 7.5, 100 mm NaCl, and 10 mm MgCl2 (λex = 494 nm, λem = 518 nm). Samples were equilibrated at 25 °C for 120 min prior to fluorescence measurements.
DNA Bending
Bending and binding of DNA by GyrA_CTD as a function of NaCl concentration were measured by FRET and fluorescence anisotropy using 200 nm 40-bp DNA that carried Alexa488 and Alexa546 dyes on opposite ends (28) in the absence and presence of 50 μm CTD in 50 mm Tris/HCl, pH 7.5, 10 mm MgCl2, and varying concentrations of NaCl. Samples were incubated at 25 °C for 120 s prior to measurements. Alexa488 fluorescence was excited at 460 nm to minimize direct acceptor excitation, and emission spectra were recorded between 480 and 700 nm. Subsequently, fluorescence anisotropy was measured by selective excitation of Alexa546 at 555 nm, and fluorescence was detected at 570 nm. To determine FRET efficiencies, fluorescence spectra were normalized by the sum of fluorescence intensities at 518 and 570 nm, and the apparent FRET efficiency, EFRET,app, was calculated from donor and acceptor fluorescence intensities (ID,518 nm and IA,570 nm, respectively) (Equation 2).
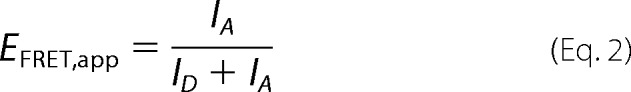 |
Single Molecule FRET Experiments
Single molecule FRET (smFRET) experiments were performed on a home-built confocal microscope with an excitation wavelength of 475 nm as described previously (37). 50 pm (donor concentration) labeled GyrA was incubated with 8 μm GyrB, 20 nm negatively supercoiled or relaxed pUC18 plasmid, and 2 mm adenosine 5′-(β,γ-imido)triphosphate (ADPNP; Sigma-Aldrich or Jena Bioscience GmbH, Jena, Germany) if present. Reaction mixtures were equilibrated for 5 min, and experiments were performed at 37 °C.
Data analysis was restricted to fluorescence events exceeding 100 photons. Donor and acceptor intensities were corrected for background, fluorescence cross-detection, different detection efficiencies, and quantum yields as well as direct acceptor excitation as described previously (38). Correction parameters for heterodimeric GyrA/GyrA_T140C/K594C and homodimeric GyrBA_S7C have been determined previously (15, 26). Data analysis is described in detail in Ref. 39.
Supercoiling and Relaxation Reactions
ATP-dependent introduction of negative supercoils into DNA by gyrase and gyrase lacking the C-tail was performed with 15 nm relaxed pUC18 plasmid in 50 mm Tris/HCl, pH 7.5, 100 mm KCl, and 10 mm MgCl2 with 800 nm GyrB, and GyrA and ATP concentrations as indicated. Reaction components were mixed on ice, then incubated at 37 °C, and reactions were stopped by addition of 10 mm EDTA, pH 8.0, 1% (w/v) SDS, and 10% (v/v) glycerol. Reaction products were separated on a 1.2% (w/v) agarose gel in 0.5× TBE buffer (110 mm Tris, 90 mm boric acid, and 2.5 mm EDTA, pH 8.0) in the absence or presence of 10 μg/ml chloroquine. Electrophoresis was performed at 5.5 V/cm for 120 min, and DNA was stained with ethidium bromide and visualized by UV transillumination.
Steady-state ATP Hydrolysis
ATP hydrolysis by GyrBA was measured in a coupled enzymatic assay where ATP hydrolysis is coupled to the oxidation of NADH to NAD+, monitored as a decrease of absorption at λ = 340 nm as described before (15, 37). Reactions were performed with 200 nm GyrBA in the absence and 50 nm GyrBA in the presence of 100 nm negatively supercoiled pUC18 plasmid at 37 °C in 50 mm Tris/HCl, pH 7.5, 100 mm KCl, 10 mm MgCl2, and varying concentrations of ATP. Initial reaction velocities were calculated from the decrease in A340, and plots of these rates as a function of ATP concentration were analyzed according to the Michaelis-Menten equation (Equation 3) to obtain kcat and Km values.
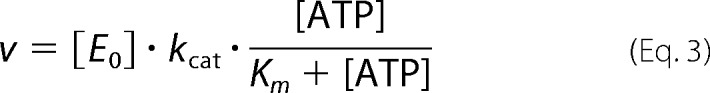 |
[ATP] and [E0] are nucleotide and total enzyme concentrations. v, Km, and kcat are the observed reaction velocity, the Michaelis-Menten constant, and the turnover number for ATP.
RESULTS
The GyrA C-terminal Acidic Tail in B. subtilis Gyrase Does Not Associate with the CTD or Other Parts of Gyrase
A recent study has identified the C-terminal tail of E. coli gyrase as a regulatory element and has shown that its deletion severely impacts the DNA supercoiling activity (34). The E. coli GyrA C-tail, formed by the C-terminal 34 amino acids that have not been resolved in x-ray structures of the GyrA_CTD (25, 34), is enriched in negatively charged residues. According to sequence alignments and a homology model of the B. subtilis GyrA_CTD (created using the Xanthomonas campestris GyrA_CTD as a template (31); Protein Data Bank code 3L6V), the C-tail of the B. subtilis GyrA_CTD can be assigned as the C-terminal 13 amino acids, containing 9 negatively charged residues. In E. coli gyrase, partial deletion of the C-tail reduces the ability to introduce writhe into DNA accompanied with a loss of ATP-dependent supercoiling activity (34). Conversely, the isolated GyrA_CTD lacking the full C-tail or its proximal part (called “insert” by the authors) was shown to bind and wrap DNA, which was not the case for the full-length CTD (34). These findings suggest an interaction of the C-tail with other parts of gyrase, thereby positively influencing one or more steps of the supercoiling reaction. Given its high negative charge, it can be envisaged that the C-tail might mimic DNA and compete for DNA-binding sites on gyrase either on the C-terminal domain of GyrA, GyrA_NTD, or GyrB.
To investigate potential interactions of the C-tail with the GyrA_CTD, we used an isolated CTD and labeled a cysteine residue added at the C terminus (GyrA_CTD_822C) with Alexa488 for fluorescence anisotropy measurements (Fig. 3A). The anisotropy of the Alexa488 attached to the C-tail is low with 0.096 ± 0.004 at a NaCl concentration of 30 mm (Fig. 3B), pointing toward significant flexibility of the dye and the C-tail and arguing against strong interactions of the C-tail with the CTD. With increasing ionic strength, the anisotropy gradually decreases and levels off at 0.063 ± 0.005 at NaCl concentrations higher than 500 mm. This change in anisotropy is small but reproducible and significant. The anisotropy decrease may hint at even less interaction of the C-tail with the rest of the CTD at increased ionic strength (Fig. 3B) where electrostatic interactions are weakened. To test whether the loss of anisotropy might be caused by a change in local flexibility of the dye, the same experiment was performed with the CTD lacking the C-tail that was fluorescently labeled on the β-pinwheel (GyrA_CTD_Δtail_K757C) (Fig. 3A). The anisotropy of the Alexa488 moiety attached to the CTD body is 0.144 ± 0.004 at 30 mm NaCl and decreases to 0.123 ± 0.004 over the concentration range from 500 to 1200 mm NaCl (Fig. 3B). The values are considerably higher than those observed for Alexa488 attached to the C-tail in agreement with a rigid β-pinwheel and a higher flexibility of the C-tail. The dependence of the anisotropy values on NaCl concentrations is similar in both cases, suggesting only minor interactions between the C-tail and the CTD.
FIGURE 3.
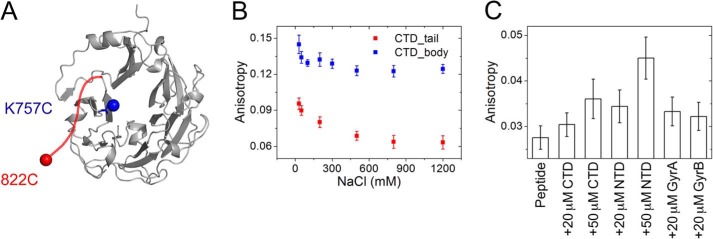
Interaction of the GyrA C-tail with gyrase subunits. A, homology model of the GyrA_CTD of B. subtilis using the GyrA_CTD of X. campestris as a template (Ref. 31; Protein Data Bank code 3L6V). Introduced cysteine residues are shown in blue (K757C) and red (Cys-822). B, steady-state anisotropies of Alexa488 attached to the C terminus of the CTD (red; Cys-822) and to the CTD body in CTD_Δtail (blue; K757C) as a function of NaCl concentration. Error bars denote anisotropy fluctuations. C, steady-state anisotropies of Alexa488_C-tail in the presence of various concentrations of GyrA_CTD_Δtail (denoted CTD), GyrA_NTD (NTD), GyrA_Δtail (GyrA), and GyrB. Error bars indicate the S.D. of the measured anisotropy.
We also tried to monitor a possible interaction between the CTD body and the C-tail by following the anisotropy of a fluorescein-labeled peptide of the C-tail sequence with the CTD lacking the C-tail. At 100 mm NaCl, the anisotropy of the fluorescein attached to the peptide is very low with a value of 0.028 ± 0.003. Addition of 20 and 50 μm GyrA_CTD lacking the C-tail induces hardly any increase in anisotropy with values of 0.030 ± 0.003 (20 μm) and 0.036 ± 0.004 (50 μm) (Fig. 3C). These values are still very low and not in agreement with complex formation of the peptide with the CTD. Thus, an interaction between the C-tail and the CTD body must be very weak if present at all. We next tested whether the C-tail may interact with GyrA_NTD. Addition of 20 and 50 μm GyrA_NTD to the fluorescein-labeled peptide resulted in anisotropy values of approximately 0.034 ± 0.004 and 0.045 ± 0.005, pointing toward a weak, if any, interaction (Fig. 3C). Similarly, the addition of 20 μm GyrA lacking the C-tail or 20 μm GyrB to the peptide caused no significant increase of the anisotropy (0.033 ± 0.003 with GyrA and 0.032 ± 0.003 with GyrB) (Fig. 3C). Thus, these experiments provide no evidence for binding of the C-tail to DNA-binding (or other) sites on gyrase, although they do not exclude a weak association with GyrA and GyrB.
Effect of the C-tail on DNA Binding and Bending by the C-terminal Domain
The isolated CTD of E. coli GyrA encompassing the C-tail does not bind DNA. Partial or complete deletion of the C-tail leads to an increase in DNA affinity and the introduction of writhe into DNA (34). To investigate DNA binding and bending of the GyrA_CTD from B. subtilis, we produced the full-length CTD (amino acids 500–821) and a CTD lacking the C-tail (amino acids 500–808; CTD_Δtail) and determined dissociation constants of CTD-DNA complexes in the absence and presence of the C-tail using a 40-bp DNA end-labeled with Alexa488. Kd values were 4.0 ± 0.5 (CTD_Δtail) and 8.3 ± 1.7 μm (C-tail present) (Fig. 4A), demonstrating that the CTD binds DNA approximately 2-fold more strongly when the C-tail is deleted. Thus, the C-tail slightly interferes with DNA binding to the CTD.
FIGURE 4.
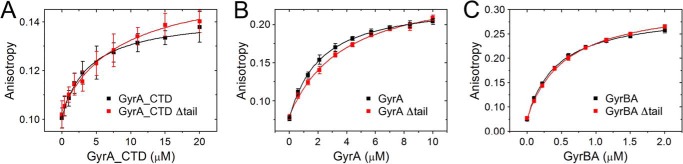
DNA binding by GyrA_CTD, GyrA, and gyrase. A, anisotropy of a 40-bp wrapping DNA titrated with GyrA_CTD (black) and GyrA_CTD_Δtail (red). B and C, anisotropy of a 60-bp gate-DNA titrated with GyrA and GyrBA, respectively. Data for proteins with and without the C-tail are shown in black and red, respectively. Error bars denote anisotropy fluctuations.
DNA bending at low ionic strength has previously been shown by the CTDs from B. burgdorferi (24) and for the B. subtilis CTD (28). To monitor the effect of the C-tail on DNA bending, we used a 40-bp DNA labeled with Alexa488 at one end and Alexa546 at the other end (Fig. 5A) that has previously been established to measure wrapping and DNA bending by an increase in FRET (24, 28). As the bend, but not binding, is lost at increasing salt concentrations (28), we measured DNA bending and binding in parallel as a function of NaCl concentration. First, fluorescence spectra of the double-labeled DNA were recorded to determine FRET efficiencies as a measure for bending. Exemplary spectra recorded at 40 mm NaCl in the absence or presence of CTD are shown in Fig. 5B, and the salt dependence of the FRET efficiency is shown in Fig. 5C. Subsequently, the fluorescence anisotropy of the Alexa546 fluorophore was determined as a measure for binding (Fig. 5D). A CTD concentration of 50 μm was used to ensure saturating conditions.
FIGURE 5.
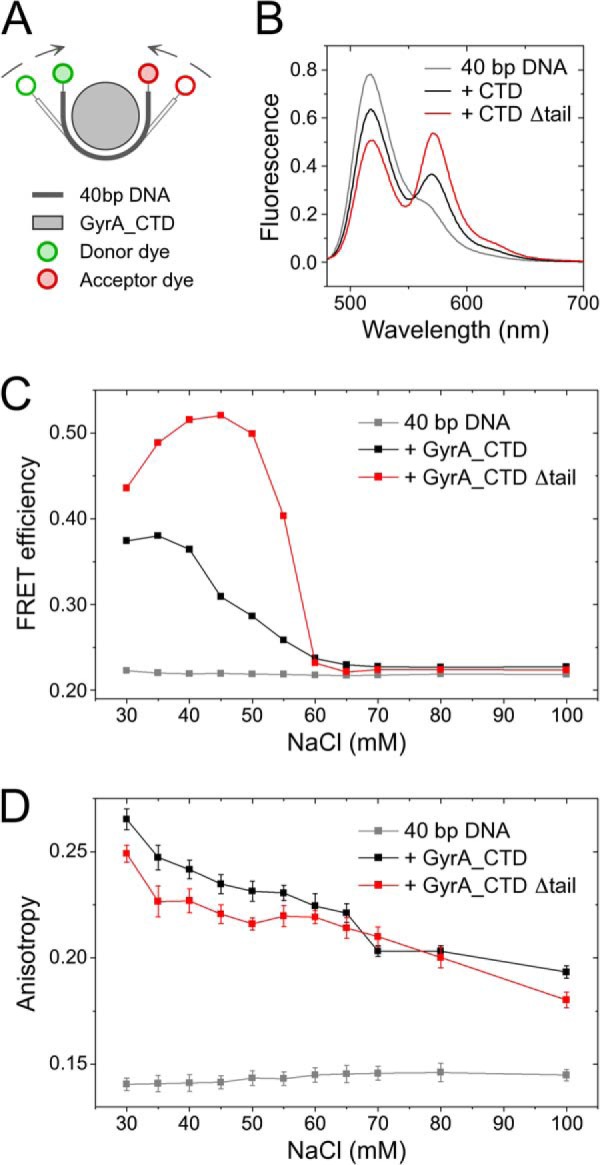
DNA binding and bending by GyrA_CTD. A, schematic of a DNA fluorescently labeled with Alexa488 (green; donor) and Alexa546 (red; acceptor) dyes on opposite ends. Binding can be followed as an increase in Alexa546 fluorescence anisotropy (D), and bending of the 40-bp DNA results in a reduced end-to-end distance in the DNA detectable by an increase in FRET efficiency (B and C). B, fluorescence spectra for the double-labeled DNA in the absence (gray) and presence of 50 μm GyrA_CTD (black) or GyrA_CTD_Δtail (red) at 40 mm NaCl. C, FRET efficiency as a function of the NaCl concentration derived from the fluorescence spectra of the double-labeled 40-bp DNA in the absence (gray) and presence of 50 μm GyrA_CTD (black) or GyrA_CTD_Δtail (red). Lines connecting data points are guides to the eye. D, anisotropy of the Alexa546 fluorescence for the double-labeled 40-bp DNA as a function of NaCl concentration in the absence (gray) and presence of 50 μm GyrA_CTD (black) or GyrA_CTD_Δtail (red). Error bars indicate the S.D. of the measured anisotropies. Lines connecting data points are guides to the eye.
At a NaCl concentration of 30 mm, the fluorescence anisotropy of 0.141 ± 0.003 for the free DNA increased to 0.265 ± 0.005 and 0.249 ± 0.004 when the CTD with or without the C-tail, respectively, was added, consistent with DNA binding. In both cases, the anisotropies decreased with increasing NaCl concentration (Fig. 5D). For DNA bound to the CTD containing the C-tail, a decrease from approximately 0.265 ± 0.005 to 0.193 ± 0.003 was observed in a NaCl concentration range from 30 to 100 mm. In experiments with the CTD lacking the C-tail, anisotropy values were generally slightly lower and decreased from 0.249 ± 0.004 to 0.180 ± 0.004 in the same NaCl concentration range. In control experiments with only DNA, the anisotropy did not change significantly as a function of the NaCl concentration with values between 0.140 and 0.146 (Fig. 5D). The anisotropy measurements show that DNA is bound to the CTDs with and without the C-tail at salt concentrations below 100 mm. Thus, the C-tail does not seem to significantly affect DNA binding to the CTD.
We have previously measured bending of the 40-bp DNA by the CTD at different NaCl concentrations (28). To be able to detect small differences between DNA bending by the CTD with and without the C-tail, we followed bending of the 40-bp DNA by both CTDs in parallel. Fluorescence spectra of the Alexa488- and Alexa546-labeled DNA show high donor and low acceptor signals both at low and high salt concentrations, corresponding to apparent FRET efficiencies of 0.22, that are consistent with linear double-stranded DNA. In the presence of the CTD lacking the C-tail, the acceptor fluorescence increases compared with free DNA at NaCl concentrations between 30 and 60 mm (Fig. 5C). The FRET efficiency reaches a maximum value of approximately 0.52 at 45 mm NaCl in agreement with a smaller interdye distance and thus with bending of the DNA. In the concentration range of 50–65 mm NaCl, the FRET efficiency decreases sharply and maintains a constant value of approximately 0.22 (Fig. 5C) at higher NaCl concentrations that is identical to the value for extended DNA in the absence of the CTD. Thus, the CTD lacking the C-tail still binds but does not significantly bend DNA at NaCl concentrations above 65 mm.
For the CTD carrying the C-tail, FRET efficiencies of 0.37–0.38 at low NaCl concentrations gradually decreased between 45 and 65 mm NaCl and leveled off at values of around 0.22, corresponding to the free, extended DNA (Fig. 5C). Thus, the loss of bending at higher NaCl concentrations is similar in both proteins and independent of the C-tail, whereas different FRET efficiencies at lower NaCl concentrations indicate differences in bending under these conditions. The lower FRET efficiencies of DNA bound to the CTD containing the C-tail are consistent with a lower degree of bending when the C-tail is present and point to interference of the C-tail with bending.
DNA Binding to GyrA and Gyrase
The C-tail does not interfere with DNA binding to the C-terminal domain of GyrA. We now investigated binding of gate-DNA to GyrA and gyrase using a 60-bp DNA labeled with Alexa546 that we have previously validated as a model gate-DNA for gyrase (37). This DNA binds to full-length GyrA and to GyrA lacking the C-tail. The Kd value for the GyrA-DNA complex is 2.0 ± 0.1 μm, and for the complex with GyrA lacking the C-tail, it is slightly higher (3.6 ± 0.3 μm; Fig. 4B). Gate-DNA binding to gyrase was measured using a cysteine-free GyrBA fusion protein in which the B and A subunits are connected by the short amino acid linker GAP and Cys residues are exchanged (GyrB, C58S/C414S; GyrA, C350L). It has been shown previously that homodimeric (GyrBA)2 retains the enzymatic activity of wild-type gyrase (15). The dissociation constants of the gate-DNA complexes were similar in the presence and absence of the C-tail with Kd values of 436 ± 15 (GyrBA_Δtail) and 555 ± 19 nm (GyrBA), pointing toward a slight increase in DNA affinity when the C-tail is absent (Fig. 4C).
Altogether, there is a small increase in DNA affinity for the isolated CTD, GyrA, and GyrBA lacking the C-tail. Thus, the C-tail in B. subtilis gyrase has only a subtle inhibitory effect on DNA binding to gyrase despite its high negative charge.
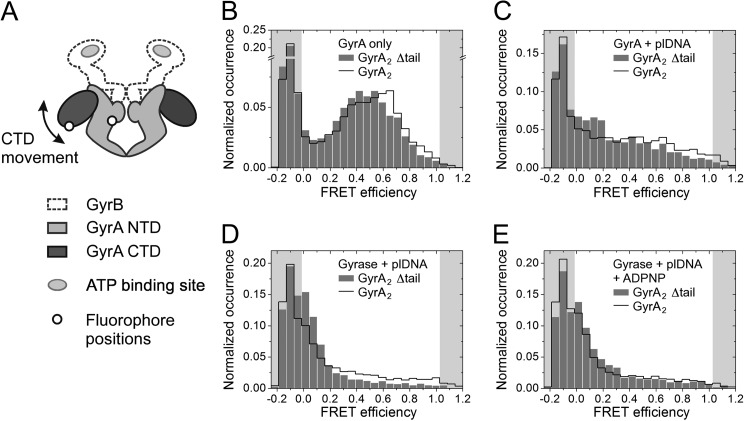
Gyrase Lacking the C-tail Shows DNA-induced CTD Movement
We have previously presented single molecule FRET experiments monitoring DNA-induced movement of the C-terminal domains relative to the N-terminal domain of GyrA (26). A GyrA heterodimer lacking native cysteines (C350L) but containing cysteines for fluorescent labeling in the NTD (T140C) and the CTD (K594C) in one protomer was labeled with Alexa488 and Alexa546 fluorophores (Fig. 6A). In the absence of ligands, this donor-/acceptor-labeled GyrA shows a low FRET efficiency (EFRET of 0.4–0.6), consistent with a CTD position close to the NTDs (Fig. 6B). Addition of a DNA sufficiently long to bind to the DNA-gate on the NTD and to contact the CTD induces a broad FRET efficiency distribution, suggesting increased flexibility of the CTDs (Fig. 6C). Gyrase shows a decrease in FRET efficiency (EFRET close to 0) that reflects the DNA-induced upward and sideways movement of the CTDs upon binding of this DNA (Fig. 6D). The FRET efficiency remains unchanged when ADPNP, a non-hydrolyzable ATP analog, is added to the DNA complex (Fig. 6E).
FIGURE 6.
CTD movement in response to binding of GyrB, DNA, and nucleotide. A, schematic depicting gyrase with one wild-type GyrA and one mutant GyrA protomer with one Cys residue on the N-terminal domain and another on the C-terminal domain that are attachment sites for fluorescent labels. B–F, smFRET histograms for heterodimeric GyrA labeled with donor and acceptor dyes. Data for the wild-type GyrA and GyrA_Δtail are depicted with a black line and gray bars, respectively. smFRET histograms show GyrA in the absence of ligands (B), GyrA in the presence of 50 nm relaxed plasmid (C), and gyrase in the presence of 50 nm relaxed plasmid (D) and with 2 mm ADPNP added (E). plDNA, plasmid DNA.
We now tested whether the C-tail affects the DNA-induced movement of the CTDs. smFRET histograms for donor-/acceptor-labeled heterodimeric GyrA lacking the C-tails in both protomers resemble those of the full-length GyrA dimer with intermediate FRET efficiencies (0.4–0.5) in agreement with a position of the CTDs close to the NTDs (Fig. 6B). The slightly lower FRET efficiency in the absence of the C-tail suggests a minor increase in interdye distance. Binding of DNA to GyrA lacking the C-tail leads to a broad FRET histogram (Fig. 6C). In the gyrase-DNA complex, the FRET efficiency is reduced, indicating that the DNA-induced CTD movement takes place when the C-tail is absent (Fig. 6D). Again, the FRET efficiency is slightly smaller compared with the gyrase-DNA complex containing the C-tail, pointing to minor conformational differences. As with gyrase containing the C-tail, addition of ADPNP does not affect the FRET efficiency (Fig. 6E).
Thus, the DNA-induced movement of the CTDs in gyrase does not depend on the C-tail. The minor influence of the C-tail on the position of the CTD might be caused by a weak interaction of the tail with the enzymatic core (see above).
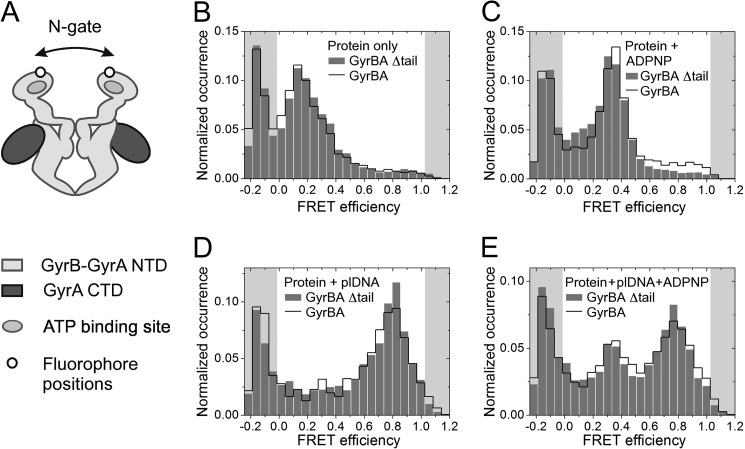
Effect of the C-tail on DNA-induced N-gate Narrowing and Nucleotide-induced N-gate Closure
Previous smFRET experiments have shown that binding of DNA to the DNA-gate and wrapping around the CTDs causes a narrowing of the N-gate (15). Deletion of the GyrA-box, a short conserved motif in the CTDs that is a hallmark feature of gyrases (32, 33), or of the entire CTD abolishes the DNA-induced conformational change of the N-gate (15, 28). The failure to narrow the N-gate in response to DNA binding when the GyrA-box is absent has been linked to different DNA bending, leading to differences in the relative position of G- and T-segments (8, 28) (Fig. 1D). We have shown above that the C-tail also affects bending of DNA by the CTDs, and therefore, we now investigated whether a deletion of the C-tail influences conformational changes at the N-gate.
smFRET experiments were performed with GyrBA to prevent subunit dissociation at picomolar concentrations (15). FRET histograms for donor-/acceptor-labeled GyrBA_S7C (Fig. 7A) show a FRET maximum at ∼0.15, indicative of an open N-gate, both in the presence and absence of the C-tail (Fig. 7B). Upon addition of ADPNP to GyrBA with or without the C-tail, the FRET efficiency increases to 0.35, and the distribution narrows in agreement with a well defined, decreased interdye distance caused by dimerization of the ATP-binding subunits and closure of the N-gate (Fig. 7C). Thus, the C-tail does not affect the nucleotide-induced closure of the N-gate that leads to T-segment capture.
FIGURE 7.
Conformational change in the gyrase N-gate induced by DNA and nucleotide binding. A, schematic of homodimeric GyrBA with cysteines introduced at the N terminus of the GyrB subunit for fluorescent labeling. B–E, smFRET histograms for donor-/acceptor-labeled GyrBA (black line) and GyrBA_(A)Δtail (gray bars). B, GyrBA in the absence of ligands; C, GyrBA in the presence of 2 mm ADPNP; D, GyrBA in the presence of 50 nm relaxed plasmid; E, GyrBA in the presence of relaxed plasmid and ADPNP. plDNA, plasmid DNA.
Upon addition of relaxed plasmid DNA to GyrBA, a shift to a high FRET efficiency of 0.8 was observed in the presence and absence of the C-tail. Thus, the DNA-induced narrowing of the N-gate still takes place in the absence of the C-tail (Fig. 7D). Adding ADPNP to the GyrBA-DNA complex results in a bimodal FRET efficiency distribution with well defined maxima at 0.3 and 0.8, corresponding to gyrase populations with narrowed (EFRET = 0.8) and closed N-gates (EFRET = 0.3). The ratio of these species has been linked to T-segment presentation (15). The FRET efficiencies of these two species as well as their relative populations are virtually identical in the presence and absence of the C-tail (Fig. 7E).
Overall, gyrase lacking the C-tail thus shows the same ligand-induced conformational changes prior to strand passage as the full-length protein. Thus, small changes in DNA bending by the CTDs in the absence of the C-tail are not reflected in altered DNA wrapping and T-segment presentation.
DNA-stimulated ATP Hydrolysis Is Increased in the Absence of the C-tail
The basal ATP hydrolysis activity of gyrase is strongly stimulated in the presence of DNA, resulting in lower Km,ATP and increased kcat values (15). DNA-stimulated ATPase activity is significantly reduced in gyrase lacking the GyrA-box or the entire GyrA_CTD as a result of differences in (or lack of) DNA wrapping (15, 28). To test whether the slight differences in DNA wrapping in the absence of the C-tail are transmitted to the N-gate of gyrase, we compared the steady-state ATPase activities of GyrBA and GyrBA lacking the C-tail in coupled enzymatic assays (Fig. 8). The ATP turnover numbers are virtually identical in the absence of DNA with kcat values of 0.638 ± 0.016 s−1 in the presence and 0.633 ± 0.019 s−1 in the absence of the C-tail, respectively. The Michaelis-Menten constants are also identical with Km,ATP values of 1.6 ± 0.1 (with C-tail) and 1.7 ± 0.1 mm (without C-tail). Thus, the C-tail does not have an effect on the intrinsic ATPase activity of gyrase (Fig. 8).
FIGURE 8.
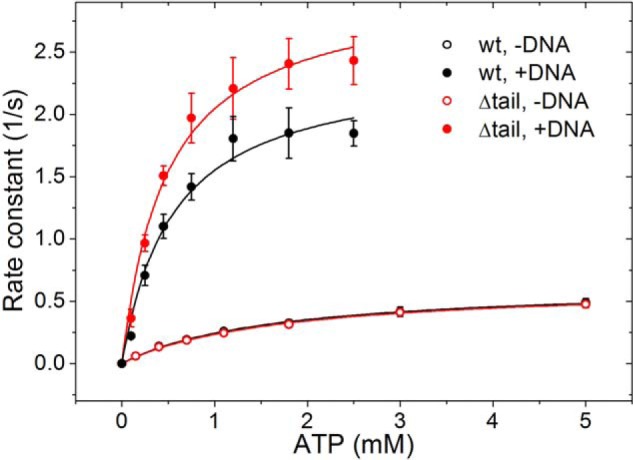
Steady-state ATPase activity of GyrBA. Rate constants of ATP hydrolysis as a function of the ATP concentration for full-length GyrBA (black) and GyrBA lacking the GyrA C-tail (red) in the absence (open circles) and presence of negatively supercoiled plasmid (filled circles). Error bars are S.D. from triplicate experiments.
In the presence of DNA, the ATP hydrolysis rate of GyrBA is 2.40 ± 0.16 s−1, whereas the enzyme lacking the C-tail exhibits a slightly higher kcat of 3.03 ± 0.14 s−1. Thus, GyrBA shows a DNA stimulation of the ATP hydrolysis by 3.8- (with C-tail) and 4.8-fold (without C-tail). In other words, coupling between DNA binding and nucleotide turnover is more efficient in the absence of the C-tail than in the full-length enzyme. The Km values for ATP in the presence of DNA are 0.55 ± 0.11 and 0.48 ± 0.07 mm, respectively (Fig. 7), and thus identical within the error of the experiment.
Deletion of the GyrA C-terminal Tail Affects the Degree of Supercoiling by Gyrase
To investigate the effect of the GyrA C-tail on DNA supercoiling by gyrase and on individual steps of the catalytic cycle, we compared the ATP-dependent supercoiling activities of gyrase containing wild-type GyrA or GyrA_Δtail at GyrA concentrations from 0 to 400 nm (Fig. 9A). Gyrase lacking the C-tail exhibits robust ATP-dependent DNA supercoiling activity, demonstrating that the C-tail is dispensable for B. subtilis gyrase activity in vitro. However, although wild-type gyrase completely supercoils 15 nm plasmid at concentrations of 75–120 nm, gyrase lacking the C-tail shows the same activity already at 45–75 nm (Fig. 9A). When supercoiled species are resolved by electrophoresis in the presence of 10 μg/ml chloroquine (Fig. 9B), it becomes evident that the supercoiling density at the end of the reaction is independent of the presence of the C-tail. When the supercoiling reaction was followed over time (0–30 min) at concentrations close to single turnover conditions (25 nm GyrA and 15 nm plasmid), supercoiling catalyzed by wild-type gyrase was completed within 14–20 min, whereas gyrase lacking the C-tail completed the reaction within 9–14 min (Fig. 9C). Altogether, we conclude that the increased supercoiling activity of gyrase lacking the C-tail is due to an approximately 2-fold higher turnover number.
FIGURE 9.
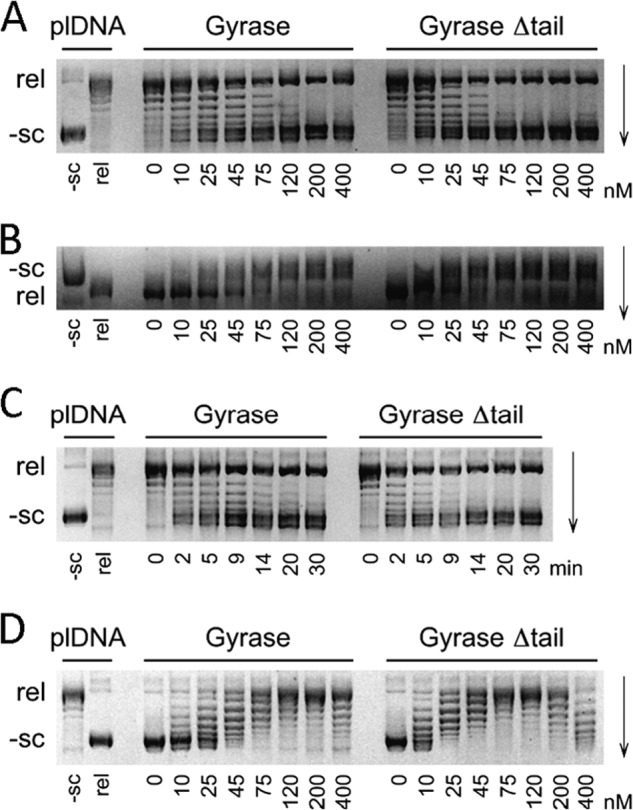
ATP-independent relaxation and ATP-dependent DNA supercoiling by gyrase. Negatively supercoiled (−sc) and partially relaxed (rel) plasmids (plDNA) are labeled on the left. A, supercoiling of 15 nm relaxed plasmid by gyrase containing GyrA, GyrA_Δtail, and 800 nm GyrB in the presence of 2 mm ATP. B, same reaction as in A but with electrophoretic separation in the presence of 10 μg/ml chloroquine. C, kinetics of supercoiling of 15 nm relaxed plasmid by 25 nm GyrA and 800 nm GyrB in the presence of 2 mm ATP. Incubation times in min are indicated below the lanes. D, relaxation of 15 nm negatively supercoiled plasmid by gyrase containing GyrA, GyrA_Δtail, and 800 nm GyrB in the absence of ATP. In A, B, and D, the concentrations of GyrA in nm are indicated below the lanes.
In addition to its ATP-dependent DNA supercoiling activity, gyrase also catalyzes nucleotide-independent DNA relaxation. When we compared relaxation activities of wild-type gyrase and gyrase lacking the C-tail, twice the concentration of wild-type gyrase compared with enzyme lacking the C-tail was needed for completing the relaxation reaction (Fig. 9D). This finding suggests a similar (inhibitory) contribution of the GyrA C-terminal tail to DNA supercoiling and relaxation.
In summary, the C-tail of gyrase reduces DNA bending and moderates ATP hydrolysis and DNA supercoiling. The results support a general role of the C-tail as a regulatory element and a subtle modulator of interdomain communication of gyrase.
DISCUSSION
Gyrase has been characterized extensively over more than three decades both from a structural and a functional point of view. The C terminus of the GyrA subunit, an appendix to the β-pinwheel fold of the C-terminal domain that varies in sequence and length, has received little attention until recently when its modulatory effect on DNA wrapping and supercoiling E. coli gyrase activity was discovered (34). As the C-tail is not conserved among gyrases, it has been proposed to modulate gyrase function in a species-dependent manner (34).
Here we have characterized the effect of the C-tail of B. subtilis GyrA on individual steps of the gyrase supercoiling cycle. We show that the C-tail does not strongly affect DNA affinity of GyrA_CTD, GyrA, or gyrase. Similar to the C-tail of E. coli gyrase, the B. subtilis C-tail modulates the degree of DNA bending by the isolated GyrA_CTD. However, in the context of gyrase, these local alterations of DNA bending do not affect the DNA-induced movement of the GyrA CTDs relative to the enzymatic core or alter the conformational behavior of the N-gate in response to DNA wrapping and T-segment presentation. Notably, deletion of the C-tail results in higher DNA-induced ATP hydrolysis and ATP-dependent DNA supercoiling activities, consistent with the C-tail acting as a repressing element that moderately down-regulates B. subtilis gyrase activity.
The C-tail Modulates DNA Bending around the C-terminal Domains in Gyrase
We have previously demonstrated that the isolated CTD of B. subtilis GyrA can bend DNA (28). Bending and wrapping of DNA by the CTDs ensure that DNA protruding from the DNA-gate is redirected to the gyrase core where it serves as a T-segment for strand passage, and therefore, the CTD is indispensable for DNA supercoiling by gyrase (23, 26). The GyrA-box, a positively charged 7-amino acid consensus sequence located on a loop connecting the first blade of the β-pinwheel fold to blade 6, is a hallmark feature of gyrases (32). The GyrA-box is crucial for DNA bending and wrapping, and deletion causes a loss of DNA supercoiling activity by gyrase (28, 33).
The GyrA C-tail has been proposed as a second element on the E. coli CTD that influences DNA binding and wrapping (34) and supercoiling. In contrast to the E. coli CTD, the CTD of B. subtilis gyrase already binds DNA when the C-tail is present with a Kd in the micromolar range, and DNA binding is virtually identical when the C-tail is deleted. Bending of the DNA bound by the B. subtilis gyrase CTDs is more pronounced when the C-tail is deleted in line with the C-tail as an inhibitory element for DNA bending. E. coli gyrase contains a longer tail (34 amino acids) with more acidic residues (18 amino acids) than the B. subtilis gyrase. Although the effect of the C-tail on bending has not directly been studied with E. coli gyrase, it has been shown that the CTD does not induce writhe when the C-tail is present but is capable of doing so when either the first part (called insert by the authors) or the complete tail is deleted (34). The GyrA_CTD from Mycobacterium tuberculosis has a shorter tail (18 amino acids) with fewer acidic residues (5 amino acids), rendering it comparable with the C-tail of B. subtilis gyrase. This CTD also wraps DNA in the presence of the C-tail (35). Finally, the CTD of B. burgdorferi naturally lacks a C-tail and induces strong DNA bending (24). In all CTDs studied so far, strong bending of DNA and wrapping is an intrinsic property of the CTD conferred by the DNA-binding site along the rim of the β-pinwheel (24, 27). The presence of an acidic C-tail of more than 13–18 amino acids appears to generally result in decreased DNA bending and wrapping possibly because the C-tail influences the geometry of DNA bound to the CTD via nonspecific repulsive electrostatic interactions. It is interesting to note that most CTDs studied to date bind DNA and induce bending even in the presence of a C-tail with the E. coli CTD being the only exception so far.
The C-tail Does Not Influence DNA-induced Conformational Changes in Gyrase
We have previously dissected DNA-induced conformational change of gyrase prior to strand passage (for reviews, see Refs. 7 and 8). A DNA-induced upward movement of the CTDs relative to the GyrA_NTD occurs when short DNA binds to the DNA-gate and contacts the CTDs (26). DNA-induced N-gate narrowing requires longer DNA that is wrapped around the CTDs in the correct geometry for strand passage (15, 28). As the C-tail affects the degree of bending of the DNA bound to the CTDs, it was tempting to speculate that it may impact on these DNA-dependent conformational changes. However, the CTDs still swing out upon DNA binding when the C-tail is deleted, showing that the C-tail does not interfere with the formation of primary contacts of the DNA that emanates from the DNA-gate with the CTDs. Deletion of the GyrA-box, although altering DNA bending, similarly does not affect displacement of the CTDs (28). However, different bending and wrapping by gyrase lacking the GyrA-box interfere with N-gate narrowing and T-segment capture (28). Interestingly, deletion of the C-tail does not affect the DNA-induced narrowing of the N-gate or the response of the gyrase-DNA complex with a narrowed N-gate to ADPNP binding. Altered DNA bending and wrapping therefore do not necessarily completely abrogate communication from the DNA-gate to the N-gate or interfere with T-segment capture.
DNA Supercoiling by Different Gyrase Homologs Is Differentially Regulated by the GyrA C-tail
Although the C-tail does not show significant influence on any individual step in DNA supercoiling up to strand passage, the enzyme lacking the C-tail shows a 2-fold higher velocity of ATP-dependent DNA supercoiling and of DNA relaxation in the absence of ATP. Thus, the C-tail may be involved in the regulation of DNA supercoiling at the step of strand passage or a subsequent step of the catalytic cycle. Because the C-tail does not seem to bind strongly to the remainder of B. subtilis gyrase, the regulatory effect might be due to electrostatic repulsion. We have not found any indication of interactions of the C-tail with the CTDs or other gyrase domains, although our results do not exclude that C-tails of other gyrases may interact with distant parts of the enzyme.
ATPase activities in the absence of DNA are virtually identical, but the kcat value for DNA-stimulated ATP hydrolysis is increased upon C-tail deletion, again identifying the C-tail as a (very moderate) repressive element. The increase in DNA bending by the CTDs lacking the tail may allow for more efficient T-segment presentation, resulting in increased DNA-stimulated ATP turnover and ATP-dependent DNA supercoiling. Thus, the C-tail down-regulates the overall enzymatic activity by slowing down ATP turnover and strand transfer.
For E. coli gyrase, removal of approximately the first half of the tail (insert) led to a 50-fold loss in both DNA supercoiling and relaxation activities (34) and lower supercoiling densities at the end point of the reaction. DNA-stimulated ATP hydrolysis was just slightly accelerated upon tail deletion. These results pointed to a role of the C-tail in E. coli gyrase in positive coupling of ATP turnover to strand passage and DNA supercoiling (34). From a comparison of the limited data on C-tails, their lengths, and effects on gyrase activity, it can be speculated that shorter C-tails might generally be involved in fine-tuning of gyrase activity, whereas longer C-tails with potentially longer reach and more negative charges seem to be necessary for more severe influence.
The hitherto available data support the hypothesis that the C-tail modulates gyrase activity in a species-dependent manner, adapting the enzymatic activity to the special needs of the respective organism. A regulatory effect might be brought about by interactions of the C-tail with DNA (repulsion), with other parts of gyrase, or even with other cellular factors yet to be identified. Given the wide range of C-tail sequences and lengths, more biochemical and biophysical evidence from other gyrase homologs is needed to understand the spectrum of effects of the C-tails on gyrase activity.
Acknowledgments
We thank Airat Gubaev for designing and creating GyrBA mutants lacking the C-tail and contributions to smFRET experiments on N-gate narrowing and Jessica Guddorf for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft.
- G-segment
- gate-DNA segment
- T-segment
- transfer-DNA segment
- CTD
- C-terminal domain
- C-tail
- acidic C-terminal tail
- Δtail
- CTD lacking the C-tail
- GyrBA
- fusion protein of GyrB and GyrA
- NTD
- N-terminal domain
- smFRET
- single molecule FRET
- ADPNP
- adenosine 5′-(β,γ-imido)triphosphate
- Alexa488
- Alexa Fluor 488
- Alexa546
- Alexa Fluor 546.
REFERENCES
- 1. Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 2. Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 73, 3872–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Higgins N. P., Peebles C. L., Sugino A., Cozzarelli N. R. (1978) Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc. Natl. Acad. Sci. U.S.A. 75, 1773–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kreuzer K. N., Cozzarelli N. R. (1980) Formation and resolution of DNA catenanes by DNA gyrase. Cell 20, 245–254 [DOI] [PubMed] [Google Scholar]
- 5. Klevan L., Wang J. C. (1980) Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an α2β2 protein core. Biochemistry 19, 5229–5234 [DOI] [PubMed] [Google Scholar]
- 6. Sugino A., Higgins N. P., Cozzarelli N. R. (1980) DNA gyrase subunit stoichiometry and the covalent attachment of subunit A to DNA during DNA cleavage. Nucleic Acids Res. 8, 3865–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roca J., Wang J. C. (1994) DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77, 609–616 [DOI] [PubMed] [Google Scholar]
- 8. Gubaev A., Klostermeier D. (2014) The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage. DNA Repair 16, 23–34 [DOI] [PubMed] [Google Scholar]
- 9. Roca J., Berger J. M., Harrison S. C., Wang J. C. (1996) DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl. Acad. Sci. U.S.A. 93, 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roca J. (2004) The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 279, 25783–25788 [DOI] [PubMed] [Google Scholar]
- 11. Morais Cabral J. H., Jackson A. P., Smith C. V., Shikotra N., Maxwell A., Liddington R. C. (1997) Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388, 903–906 [DOI] [PubMed] [Google Scholar]
- 12. Horowitz D. S., Wang J. C. (1987) Mapping the active site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 262, 5339–5344 [PubMed] [Google Scholar]
- 13. Noble C. G., Maxwell A. (2002) The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 318, 361–371 [DOI] [PubMed] [Google Scholar]
- 14. Ali J. A., Orphanides G., Maxwell A. (1995) Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry 34, 9801–9808 [DOI] [PubMed] [Google Scholar]
- 15. Gubaev A., Klostermeier D. (2011) DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proc. Natl. Acad. Sci. U.S.A. 108, 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wigley D. B., Davies G. J., Dodson E. J., Maxwell A., Dodson G. (1991) Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629 [DOI] [PubMed] [Google Scholar]
- 17. Williams N. L., Maxwell A. (1999) Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry 38, 13502–13511 [DOI] [PubMed] [Google Scholar]
- 18. Tingey A. P., Maxwell A. (1996) Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 24, 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baird C. L., Harkins T. T., Morris S. K., Lindsley J. E. (1999) Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. U.S.A. 96, 13685–13690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harkins T. T., Lewis T. J., Lindsley J. E. (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry 37, 7299–7312 [DOI] [PubMed] [Google Scholar]
- 21. Reece R. J., Maxwell A. (1989) Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264, 19648–19653 [PubMed] [Google Scholar]
- 22. Reece R. J., Maxwell A. (1991) The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 19, 1399–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kampranis S. C., Maxwell A. (1996) Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 93, 14416–14421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corbett K. D., Shultzaberger R. K., Berger J. M. (2004) The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 101, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruthenburg A. J., Graybosch D. M., Huetsch J. C., Verdine G. L. (2005) A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J. Biol. Chem. 280, 26177–26184 [DOI] [PubMed] [Google Scholar]
- 26. Lanz M. A., Klostermeier D. (2011) Guiding strand passage: DNA-induced movement of the gyrase C-terminal domains defines an early step in the supercoiling cycle. Nucleic Acids Res. 39, 9681–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papillon J., Ménétret J. F., Batisse C., Hélye R., Schultz P., Potier N., Lamour V. (2013) Structural insight into negative DNA supercoiling by DNA gyrase, a bacterial type 2A DNA topoisomerase. Nucleic Acids Res. 41, 7815–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lanz M. A., Klostermeier D. (2012) The GyrA-box determines the geometry of DNA bound to gyrase and couples DNA binding to the nucleotide cycle. Nucleic Acids Res. 40, 10893–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirkegaard K., Wang J. C. (1981) Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell 23, 721–729 [DOI] [PubMed] [Google Scholar]
- 30. Liu L. F., Wang J. C. (1978) DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell 15, 979–984 [DOI] [PubMed] [Google Scholar]
- 31. Hsieh T. J., Yen T. J., Lin T. S., Chang H. T., Huang S. Y., Hsu C. H., Farh L., Chan N. L. (2010) Twisting of the DNA-binding surface by a β-strand-bearing proline modulates DNA gyrase activity. Nucleic Acids Res. 38, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward D., Newton A. (1997) Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 26, 897–910 [DOI] [PubMed] [Google Scholar]
- 33. Kramlinger V. M., Hiasa H. (2006) The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J. Biol. Chem. 281, 3738–3742 [DOI] [PubMed] [Google Scholar]
- 34. Tretter E. M., Berger J. M. (2012) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: I. a nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J. Biol. Chem. 287, 18636–18644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tretter E. M., Berger J. M. (2012) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II. the shape of the GyrA subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency. J. Biol. Chem. 287, 18645–18654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Göttler T., Klostermeier D. (2007) Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA. J. Mol. Biol. 367, 1392–1404 [DOI] [PubMed] [Google Scholar]
- 37. Gubaev A., Hilbert M., Klostermeier D. (2009) The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc. Natl. Acad. Sci. U.S.A. 106, 13278–13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Theissen B., Karow A. R., Köhler J., Gubaev A., Klostermeier D. (2008) Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 105, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andreou A. Z., Klostermeier D. (2012) Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 511, 75–109 [DOI] [PubMed] [Google Scholar]