Abstract
Controlled actin assembly is crucial to a wide variety of cellular processes, including polarity establishment during early development. The recently discovered actin mesh, a structure that traverses the Drosophila oocyte during mid-oogenesis, is essential for proper establishment of the major body axes. Genetic experiments indicate that at least two proteins, Spire (Spir) and Cappuccino (Capu), are required to build this mesh. The spire and cappuccino genetic loci were first identified as maternal effect genes in Drosophila. Mutation in either locus results in the same phenotypes, including absence of the mesh, linking them functionally. Both proteins nucleate actin filaments. Spir and Capu also interact directly with each other in vitro, suggesting a novel synergistic mode of regulating actin. In order to understand how and why proteins with similar biochemical activity would be required in the same biological pathway, genetic experiments were designed to test whether a direct interaction between Spir and Capu is required during oogenesis. Indeed, data in this study indicate that Spir and Capu must interact directly with one another and then separate to function properly. Furthermore, these actin regulators are controlled by a combination of mechanisms, including interaction with one another, functional inhibition and regulation of their protein levels. Finally, this work demonstrates for the first time in a multicellular organism that the ability of a formin to assemble actin filaments is required for a specific structure.
Keywords: Spire, Cappuccino, Drosophila, Actin, Oogenesis
INTRODUCTION
Drosophila oogenesis requires several distinct actin-based structures. Some are present throughout the entire process, whereas others are only evident during specific stages of development. A loosely organized actin structure that traverses the Drosophila oocyte during mid-oogenesis, called the actin mesh, was recently discovered (Dahlgaard et al., 2007). Disappearance of the mesh late in oogenesis coincides with the onset of a dynamic process called fast, microtubule-dependent cytoplasmic streaming (Dahlgaard et al., 2007; Gutzeit and Koppa, 1982). Two proteins, Spire (Spir) and Cappuccino (Capu), are required for proper establishment and maintenance of this mesh. Their mammalian counterparts are likewise necessary for a mesh-like actin structure in mouse oocytes, suggesting conservation of function (Pfender et al., 2011).
The spire and cappuccino genetic loci were first identified as maternal effect genes in Drosophila. Mutation of either locus disrupts both the anterior/posterior and dorsal/ventral axes of the oocyte and causes female sterility (Manseau and Schüpbach, 1989). However, the mechanism of action remains poorly understood. It was previously confirmed that Capu, a member of the formin-family of actin nucleators, nucleates actin polymerization (Quinlan et al., 2005). In addition, Spir can nucleate actin (Quinlan et al., 2005). Spir is the founding member of a third class of nucleators, the WH2 nucleators (in addition to formins and the Arp2/3 complex) (Qualmann and Kessels, 2009). According to FlyBase, there are two ‘full-length’ Spir isoforms, SpirA and SpirB, which differ by only 29 residues in a poorly conserved region of the ∼1000 amino acid gene product (Fig. 1). Among the five additional isoforms, two variants that have been studied are SpirD and SpirC. SpirD encompasses the first 585 residues of the full-length gene. It contains the kinase inactive domain (KIND), which is necessary and sufficient to bind directly to Capu (Ciccarelli et al., 2003; Quinlan et al., 2007; Vizcarra et al., 2011). SpirD also contains four tandem actin-monomer-binding WASP homology 2 (WH2) domains, the source of the actin nucleation activity of Spir (Quinlan et al., 2005). In addition to nucleating, these tandem WH2 domains can sever actin filaments, albeit weakly, and sequester actin monomers (Bosch et al., 2007; Chen et al., 2012). SpirC begins with a distinct ∼100 residue sequence fused to the C-terminal ∼525 residues of full-length Spir. SpirC contains potential signaling and lipid-binding domains (Kerkhoff et al., 2001; Otto et al., 2000). SpirC cannot nucleate actin independently (Dahlgaard et al., 2007; Rosales-Nieves et al., 2006). SpirD and SpirC have been proposed to work in trans (Liu et al., 2009; Rosales-Nieves et al., 2006). In this study, I asked whether they are sufficient to rescue a spir null separately or together, and found that only full-length Spir (SpirB) produced robust fertility.
Fig. 1.
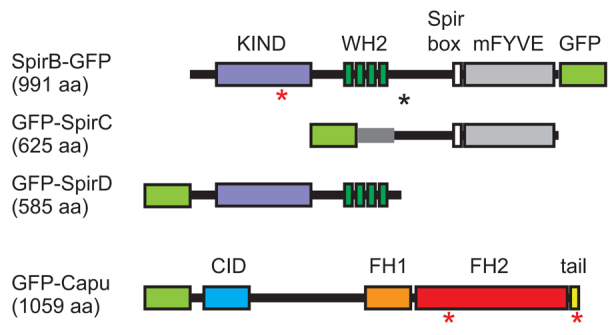
Domain organization of Spir and Capu. SpirB-GFP, GFP-SpirC, GFP-SpirD and GFP-Capu. Numbers in parentheses are the lengths of the gene products, not including the GFP. Major domains indicated are: KIND, kinase non-catalytic C-lobe domain (purple); WH2, Wiscott-Aldrich homology 2 motif (dark green); Spir box (white); mFYVE, modified Fab1/YOTB/Vac1/EEA1 zinc-binding domain (gray); CID, Capu inhibitory domain (blue); FH1, formin homology 1 (orange); FH2, formin homology 2 (red); tail (yellow). The black asterisk indicates the alternative splicing site that results in a 29 residue difference between SpirA and SpirB. The red asterisks indicate the point mutations used in this paper: SpirB(Y232K) in the KIND domain, Capu(I706A) in the FH2 domain and Capu(L1048A) in the tail. A light-green box is drawn where the GFP tag was added to each construct.
The fact that both Spir and Capu can nucleate actin polymerization led me to question why proteins with similar biochemical activity might be required in the same biological pathway. Spir and Capu interact directly with one another in vitro, suggesting a novel synergistic mode of producing new actin filaments (Quinlan et al., 2007; Quinlan and Kerkhoff, 2008; Rosales-Nieves et al., 2006). The available data lead to a clear hypothesis: Spir and Capu collaborate to build the actin mesh, which regulates processes critical to oogenesis, including microtubule organization, RNA localization and the timing of fast streaming initiation. Here, I describe the results of experiments that test the hypothesis that Spir and Capu interact directly to build the actin mesh. Indeed, I found that Spir and Capu must physically interact with one another to function when expressed at physiological levels; furthermore, they also separate to work properly. The data indicate that these actin regulators are controlled by a complex combination of mechanisms. In addition to the requirement of a reversible interaction, my data suggest that both may be functionally inhibited (i.e. turned off) and that their protein levels are temporally regulated. Finally, I demonstrate that the ability of a formin to assemble actin filaments is required for a specific structure in a multicellular animal, namely the actin mesh.
MATERIALS AND METHODS
Drosophila stocks
The following mutants were obtained from the Bloomington Stock Center: spir1 and capu1 (Manseau and Schüpbach, 1989); Df(2L)Exel6046 (Exelixis); UASp-GFP-SpirC and UASp-GFP-SpirD (Rosales-Nieves et al., 2006); and nos-GAL4-vp16 (Van Doren et al., 1998). GFP-CapuΔN was kindly provided by Daniel St Johnston (Dahlgaard et al., 2007). New SpirB-GFP transgenes were generated by inserting the coding region of spir (CG10076-RB) or the coding region with a point mutation created by QuikChange mutagenesis, between the KpnI and SpeI sites of pTIGER (Ferguson et al., 2012) with GFP inserted between the BamHI and XbaI sites. Primers were: forward, 5′-catagcggtaccatgacggagcaccaggccgag-3′; reverse, 5′-catagcactagtcttccagacgggcgtcag-3′. The Y232K mutagenesis primer was 5′-ggaaatcgtcaaaaccgccttggattac-3′. The GFP-Capu trangenes were generated by inserting the coding region of capu (CG3399-RA), or the coding region with a point mutation created by QuikChange mutagenesis, between the BamHI and XbaI sites of pTIGER with GFP inserted between the KpnI and SpeI sites. Primers were: forward, 5′-cgcggatccatggccttgcagctaggc-3′; reverse, 5′-catagctctagatcattagttcttgctacgcctcatc-3′. The L1048A mutagenesis primer was 5′-ccggacggatttcggcgaaggagcgcatgctg-3′ and the I706A mutagenesis primer was 5′-cgaaatgtgggcgcaatctggcggag-3′. Transgenic flies with these plasmids inserted into the attP2 site were made by Genetic Services (Groth et al., 2004). w1118 was used as wild type.
Fertility assays
Approximately 100 test females were crossed to 40 wild-type males and kept on apple plates for 2 nights at 25°C. Flies were pre-cleared on a fresh plate with yeast paste for at least 1.5 hours, the plate was changed and eggs laid over the next 3 hours were collected. Typically, 150-250 eggs were laid in this time period. Eggs were transferred to a fresh plate and stored at 25°C. The number of eggs that hatched after 24 hours was recorded. Each trial was repeated three times with independent crosses.
Microscopy and staining
The localization of GFP fusions, visualization of cytoplasmic flows and the actin mesh were performed on a Leica SPE I inverted confocal microscope. Flies were kept at 25°C and fed yeast paste for ∼24 hours before an experiment. The actin mesh was stained as described by Dahlgaard et al. (Dahlgaard et al., 2007) with modifications. Briefly, ovaries were dissected, teased apart and fixed in 10% paraformaldehyde/PBS (pH 7.4) for a total of less than 10 minutes. Samples were stained with 1 μM AlexaFluor647-phalloidin (so GFP could be examined in the same egg chambers) dissolved in 0.3% Triton X-100/PBS for 25 minutes at room temperature. Samples were then washed extensively and mounted in ProLong Gold. Images were recorded within 24 hours of staining because phalloidin staining quality degraded over time, as has been reported (Dahlgaard et al., 2007). Flies were treated with latrunculin A, as described previously (Dahlgaard et al., 2007).
Immunofluorescent staining of GFP fusions was performed by dissecting ovaries in cold IMADS (Robinson and Cooley, 1997), and fixing in 1-4% paraformaldehyde dissolved in 100 mM lysine and 10 mM sodium phosphate (pH 7.2). Samples were stained with 1:1000 rabbit anti-GFP (Invitrogen) overnight at 4°C and 1:5000 Cy3 goat anti-rabbit (Jackson ImmunoLabs) for 2 hours at room temperature along with AlexaFluor647-phalloidin counterstain.
Fluid flow analysis
Live oocytes were dissected under halocarbon 700 oil and excited with a 488 nm laser to examine GFP-fusion localization. To track fluid flow, images of autofluorescent yolk granules excited at 405 nm were collected every 5 seconds for 5 minutes. Standard deviation ‘z’-projections were created in Fiji for demonstration of relative motion (Schindelin et al., 2012).
Cytoplasmic streaming velocities were determined from confocal images using custom-built particle image velocimetry (PIV) (Raffel et al., 1998) implemented in Python using NumPy/SciPy (Jones et al., 2001). Briefly, PIV involves measurement of displacement of regions of images (called interrogation windows) from one frame to the next. The velocity corresponding to a given interrogation window is the displacement divided by the time gap between the two frames.
PIV was performed on each movie for 30 frames with a time gap of 10 seconds between frames. The prescription for measurement of streaming velocities for each movie is as follows.
A region of interest (ROI) was specified by constructing a polygon around the periphery of the oocyte by hand using the polygon selection tool in Fiji (Schindelin et al., 2012). Only interrogation windows in the ROI were considered, and the same ROI was used for each frame.
The frames of the movie were preprocessed for PIV using intensity capping (Shavit et al., 2007). All pixels in a given frame that had an intensity above I_max were set to I_max. I_max was chosen to be the mean pixel intensity plus two standard deviations of the pixel intensity for the given frame.
PIV was performed on each image pair using 64×64 pixel interrogation windows, corresponding to ∼17.2×17.2 μm. Frequency domain correlation was used. The position of maximal correlation was determined to subpixel accuracy using a three-point Gaussian estimator (Raffel et al., 1998). There was no overlap of the interrogation windows and no desampling of the images.
Outliers were detected using the normalized median test (Westerweel and Scarano, 2005), with a threshold value of 2, neighborhood radius of 1 pixel and noise threshold level of 0.1. Outliers were replaced by the median of the neighborhood values.
Using the displacements from the first iteration to estimate displacements, PIV was again performed on 32×32 pixel (≈8.6×8.6 μm) interrogation windows with no overlap. Subpixel resolution, and outlier detection and correction were performed as for the first pass.
Using the displacements from the second iteration, PIV was again performed on 32×32 pixel interrogation windows with 50% overlap, giving velocity vectors every 16 pixels (≈4.3 μm). Subpixel resolution and outlier detection and correction were performed as for the first two passes.
After the streaming velocities were determined, the speeds of all interrogation windows in a given movie were collated and the 95th percentile speed was determined. Histograms of streaming speeds for typical stage 9 and 11 oocytes are shown in supplementary material Fig. S1A.
Immunoblots
Levels of Spir and Capu in whole ovaries versus stage 11 egg chambers were detected as follows: whole ovaries of 10-15 flies were dissected in ice-cold IMADS buffer. Samples were transferred to a 500 μl tube, buffer was siphoned off and replaced with 20 μl PBS, 0.1% NP-40 and a protease inhibitor cocktail. Ovaries were crushed and centrifuged at 16,000 g at 4°C for 10 minutes. The supernatant was transferred to a fresh tube. Twenty-five to 30 stage 11 egg chambers were dissected from ovaries in cold IMADS. Only 10 μl of buffer were added to these samples after IMADS was removed. SDS sample buffer was added and each sample was boiled for no more than 5 minutes. Samples were loaded on a 7.5% SDS-PAGE gel based on the results of protein concentration assays. Transfer was optimized to ensure small and large proteins were captured. PVDF membrane was used and transferred at 100 V for 1 hour, in Tris/glycine buffer with 10% methanol and 0.01% SDS. Antibodies against Spir-KIND and Capu-FH2 have been described previously (Quinlan et al., 2007). Antibodies specific for shorter forms of Spir were kindly supplied by S. Parkhurt (Liu et al., 2009). Samples were detected by ECL, stripped and reprobed. Three independent replicates were performed, alternating which protein was detected first. Relative amounts of protein were determined by densitometry using GAPDH or tubulin as a loading control. The loading control was always detected last.
RESULTS
Full-length Spir is required for female fertility
I used a rescue strategy to study Spir in Drosophila oogenesis. There are seven putative alternative splice variants of the spir gene described in FlyBase. Two of these, SpirC and SpirD, have been studied in oogenesis (Dahlgaard et al., 2007; Rosales-Nieves et al., 2006). These isoforms have minimal overlapping sequence (Fig. 1) and were proposed to act in trans to coordinate the actin and microtubule cytoskeletons in Drosophila oocytes (Rosales-Nieves et al., 2006). Later, this group presented immunoblot evidence that SpirC and SpirD are the dominant protein products expressed in the ovary, in contrast to the higher full-length Spir message levels they detected by northern blot analysis (Liu et al., 2009). However, full-length Spir was detected in excess over SpirD in similar immunoblot experiments (Quinlan et al., 2007). To resolve this discrepancy, I asked which splice variants were sufficient to produce fertile female flies. To do so, GFP-SpirC and GFP-SpirD expression were driven individually and together using the germline-specific promoter nanos-GAL4-vp16 in a spir mutant background (spir1/Df(2L)Exel6046). This study included three assays: (1) examination of the actin mesh at stage 9, when the mesh should be present, and stage 11, when it normally disappears; (2) measurement of cytoplasmic streaming rates at stage 9, when fluid flows are slow, and stage 11, when the flows are normally coordinated and fast; and (3) assessment of fertility by quantifying egg-hatching success.
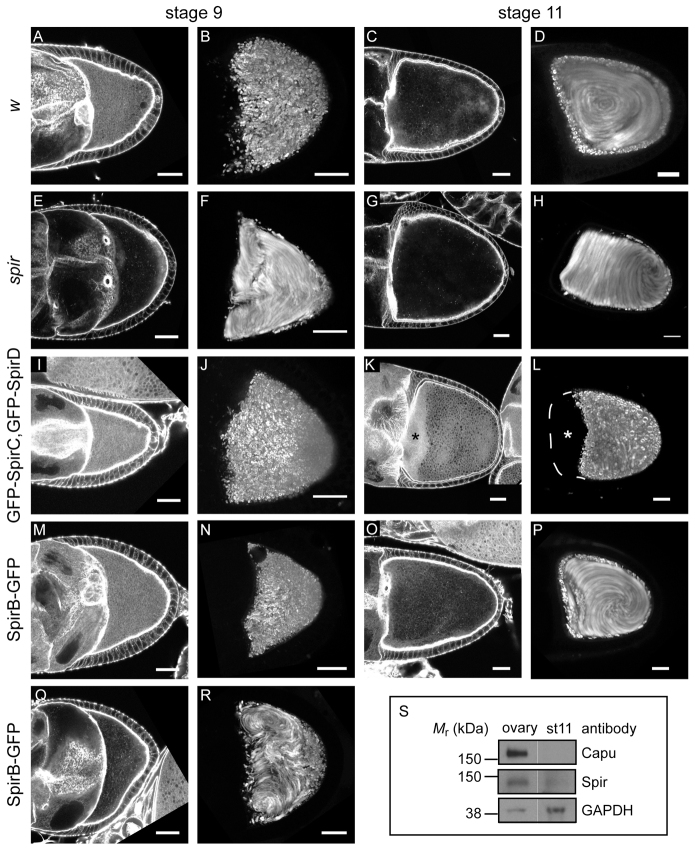
Female flies lacking wild-type Spir are sterile (Manseau and Schüpbach, 1989). Their eggs do not hatch; their oocytes lack actin mesh at any stage; and they exhibit premature onset of fast cytoplasmic streaming (Table 1; Fig. 2A-H) (Dahlgaard et al., 2007; Theurkauf, 1994). Dahlgaard et al. (Dahlgaard et al., 2007) reported that GFP-SpirC expression in a mutant spir background did not rescue the mesh, nor did it prevent premature streaming, as one might expect, as SpirC does not have actin nucleating activity. I observed similar phenotypes (data not shown) and found that GFP-SpirC expression does not produce any viable eggs, consistent with the idea that the actin mesh and/or correct timing of fast streaming are necessary for early development (Table 1). Dahlgaard et al. (Dahlgaard et al., 2007) found that GFP-SpirD expression results in oocytes with an actin mesh and that premature fast streaming is prevented. My results match theirs (data not shown) but when fertility was measured, only 12% of the eggs laid by these flies hatched (Table 1), suggesting that the actin mesh is not sufficient for rescue of fertility. Oocytes expressing both SpirC and SpirD also had an actin mesh and premature streaming was prevented (Fig. 2I,J). Expression of the two constructs did not improve fertility when compared with expression of SpirD alone. In fact, fewer embryos hatched (6% versus 12% hatched, Table 1). Thus, SpirC does not regulate or cooperate with SpirD in trans. Dahlgaard et al. (Dahlgaard et al., 2007) observed a mesh that persists in stage 11 oocytes and later when GFP-SpirD was expressed under control of the nanos driver. This group also saw layering of nurse cell cytoplasm upon dumping, with yolk granules retained at the posterior of the oocyte. I observed the same phenotypes when both GFP-SpirC and GFP-SpirD were present (Fig. 2K,L). Together, these data suggest that simply building an actin mesh is not sufficient; its timely removal is also important to the progression of oogenesis.
Table 1.
Fertility assays
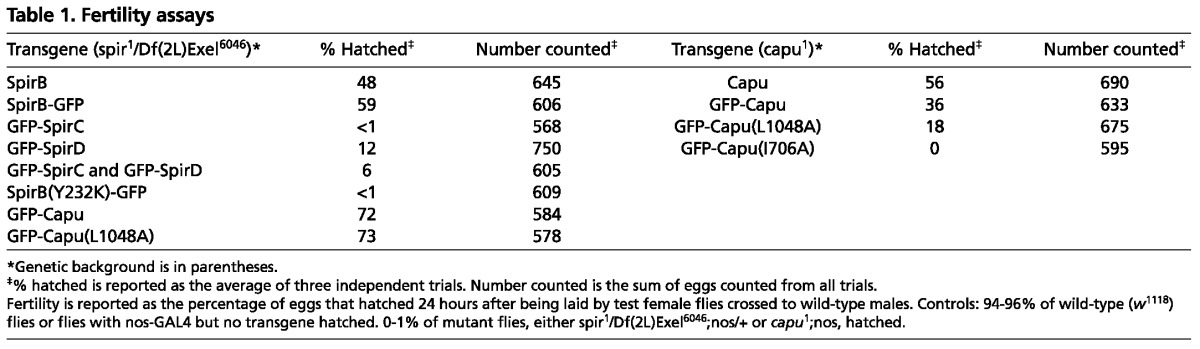
Fig. 2.
Full-length Spir is required to rescue spir mutant. (A,C,E,G,I,K,M,O,Q) Stage 9 (A,E,I,M,Q) and 11 (C,G,K,O) egg chambers stained with AlexaFluor647-phalloidin to examine the actin mesh in transgenic flies. (B,D,F,H,J,L,N,P,R) Standard deviation projections of autofluorescent yolk granule positions over 5 minutes for typical stage 9 (B,F,J,N,R) and stage 11 (D,H,L,P) oocytes from the same fly lines. Egg chambers are from w1118 controls (A-D), spir1/Df(2L)Exel6046;+/nos flies (E-H), spir1/Df(2L)Exel6046;GFP-SpirC,GFP-SpirD/nos flies (I-L) or spir1/Df(2L)Exel6046;SpirB-GFP/nos flies (M-R). Approximately 30% of the SpirB-GFP-expressing egg chambers lacked an actin mesh or exhibited premature streaming, as represented in Q and R. The asterisks in K and L highlight the layering of nurse cell cytoplasm and yolk granule-rich ooplasm that results when the actin mesh is not removed. Oocytes are oriented with posterior towards the right. Scale bars: 30 μm. (S) Immunoblots to determine relative Spir and Capu levels in whole-ovary lysates versus stage 11 (st 11) egg chambers. Bands are from the same blot, moved together for presentation. Spir and Capu levels in stage 11 egg chambers are less than 10% of the levels in whole oocytes, based on averages from three repeats. GAPDH is shown as a loading control. Molecular masses (Mr) indicated are based on full-range rainbow molecular weight markers (GE Healthcare).
When I examined Spir expression in Drosophila ovaries by immunoblot, I detected full-length Spir in excess over SpirD (Quinlan et al., 2007). Based on this observation and on the fact that expression of the shorter isoforms failed to produce fertile female flies, I asked whether a full-length splice variant was sufficient to produce fertility? Flies expressing SpirB-GFP were mostly fertile: 59% of eggs hatched (Table 1). Consistent with this result, most of the eggs expressing SpirB-GFP had an actin mesh during mid-oogenesis, and they had proper temporal control of streaming (Fig. 2M-P). A small fraction exhibited premature streaming, which agrees well with the fertility data (4 of 17 stage 9 oocytes; Fig. 2Q,R; Fig. 3M). Importantly, the mesh had gone by stage 11, as is seen in wild-type flies (Fig. 2C,O), and fast streaming appeared normal (Fig. 2D,P; Fig. 3N). Controls with untagged SpirB suggest that the GFP tag is not the major source of infertility in these experiments. In fact, SpirB-GFP performed slightly better (59% versus 48% hatched, Table 1). The nanos-GAL4-vp16 system produces GAL4 at higher levels in the earliest stages of development (stages 1-4), at lower levels during mid stages and increases again from stage 8 or 9 on (Hudson and Cooley, 2010). SpirB-GFP levels track the known GAL4 expression pattern, suggesting that protein turnover is relatively rapid (supplementary material Fig. S2A). This pattern is different from that observed by immunofluorescence and immunoblot (Quinlan et al., 2007 and below), potentially explaining why the rescue is not closer to 100%.
Fig. 3.
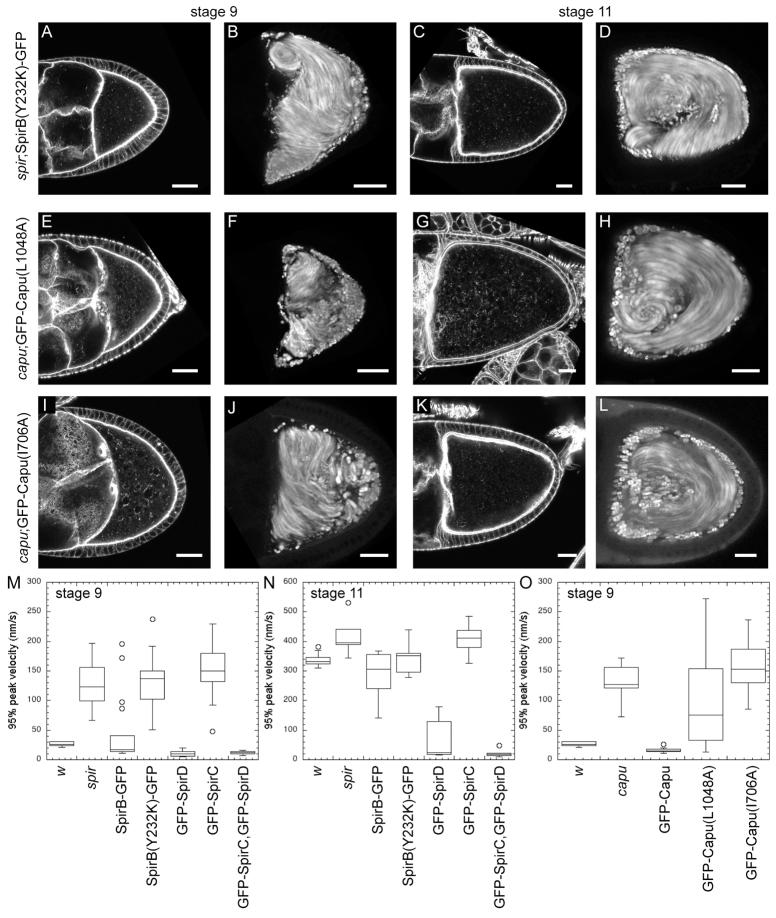
Direct interaction between and separation of Spir and Capu during oogenesis. Stage 9 and 11 egg chambers stained with AlexaFluor647-phalloidin and standard deviation projections of autofluorescent yolk granule motion over 5 minutes. (A-D) Genotype of egg chambers is spir1/Df(2L)Exel6046;SpirB(Y232K)-GFP/nos. SpirB(Y232K) cannot bind Capu. (E-H) Genotype of egg chambers is capu1;GFP-Capu(L1048A)/nos. Capu(L1048A) cannot bind Spir. Egg chambers expressing intermediate levels of protein are shown (see supplementary material Fig. S4 for a range of expression levels). (I-L) Genotype of egg chambers is capu1;GFP-Capu(I706A)/nos. Capu(I706A) cannot stimulate actin assembly. Oocytes are oriented with posterior towards the right. Scale bars: 30 μm. (M-O) Box and whisker plots of streaming velocities. n≥10 for each stage 9 phenotype analyzed; n≥5 for each stage 11 phenotype analyzed. The bottom and top of the boxes are the first and third quartiles, and the band inside the box is the median. Individual points are outliers.
Emmons et al. (Emmons et al., 1995) found that treating flies with an actin depolymerizing factor, cytochalasin D, causes premature streaming. Dahlgaard et al. (Dahlgaard et al., 2007) drew a link between the mesh and regulation of streaming by confirming that another actin depolymerizing factor, latrunculin A, caused the actin mesh to disappear. They also found that the actin mesh was stabilized by expression of a constitutively active version of Capu, GFP-CapuΔN, but not when GFP-SpirD was expressed. If SpirB were the active isoform of Spir, it might better stabilize the mesh than SpirD. Indeed, oocytes from flies expressing SpirB-GFP which were also treated with latrunculin A, retained some mesh (supplementary material Fig. S3). The presence of mesh was variable, probably reflecting expression levels similar to those seen in rescue experiments. Despite this variability, the difference between wild-type and flies expressing SpirB-GFP was marked, supporting my claim that SpirB is the active isoform in oocytes.
In summary, SpirB-GFP activity, as gauged by the presence or absence of the mesh, is regulated in a manner that GFP-SpirD is not and nanos-driven expression of SpirB-GFP compensates well for loss of endogenous Spir. Based on these results, the remainder of the experiments were performed with SpirB-GFP and mutants of this full-length splice variant.
Regulation of Spir and Capu
Because removal of the mesh and onset of fast streaming around stage 11 correlated with increased fertility, I examined the temporal control of Spir and Capu protein levels. Previous immunofluorescence experiments indicated that endogenous Spir was expressed throughout mid-oogenesis (detected in oocytes as early as stage 5) and that protein levels dropped after stage 10b, when the mesh disappears and fast streaming begins (Quinlan et al., 2007). Because no Capu antibody was available that works for immunofluorescence and because this method becomes less reliable in later stages of oocyte development, I used immunoblots to examine protein levels. Spir and Capu levels were compared in whole ovaries, which contain all stages of development, with protein levels in stage 11 egg chambers. Using an antibody that detects the Spir-KIND domain and, therefore, SpirA/B and SpirD, I found that endogenous levels of full-length Spir decrease dramatically in stage 11 egg chambers, consistent with the immunofluorescence results (Fig. 2S). As before, only low levels of SpirD were detected compared with full-length Spir (Quinlan et al., 2007). No stage-dependent difference in levels of this isoform were found (data not shown). I observed a decrease in Capu levels similar to the decrease in full-length Spir levels (Fig. 2S). Both full-length Spir and Capu protein levels in stage 11 egg chambers are, on average, less than 10% of those in the whole ovary. Thus, one mode of regulating Spir and Capu activity is through controlling protein levels.
Direct interaction between Spir and Capu is required
I used mutagenesis in the rescue scheme to determine whether a direct interaction between Spir and Capu is necessary during oogenesis. Previously, a point mutation was described in the Spir-KIND domain (Y232K) that abolishes interaction between Spir and Capu in binding assays and in pyrene actin polymerization assays (Vizcarra et al., 2011). In the same studies, SpirD with this mutation fails to colocalize with full-length Capu in tissue culture (S2) cells (Vizcarra et al., 2011). I, therefore, asked, what happens when I replace wild-type Spir with SpirB(Y232K)-GFP in flies? This construct failed to rescue the mutant phenotype in every aspect assayed. Flies expressing SpirB(Y232K)-GFP were sterile, had no detectable actin mesh during mid-oogenesis and consistently exhibited premature streaming (Fig. 3A,B, Table 1). Furthermore, streaming velocities were indistinguishable from the spir mutant (Fig. 3M). SpirB(Y232K)-GFP did not have a detectable effect on later stages of oogenesis in that, as usual, no mesh was visible and fast streaming was observed at stage 11 (Fig. 3C,D,N). Direct physical interaction between Spir and Capu is, therefore, required for them to function correctly in Drosophila oogenesis.
I also tested a complementary point mutation in Capu that ablates the Spir-Capu interaction. No other interactions with the Spir-KIND domain are known of, but, in principle, the Y232K mutation in Spir could have other effects. I first confirmed that GFP-Capu was sufficient to rescue capu mutants. As reported by Dahlgaard et al. (Dahlgaard et al., 2007), GFP-Capu expression rescues the capu mutant background in that the mesh is present and streaming does not begin prematurely in 100% of egg chambers examined (supplementary material Fig. S2B; Fig. 3O). I also tested its ability to rescue fertility, finding that 36% of eggs hatched (Table 1). These results were compared with unlabeled Capu, which showed improved rescue of fertility (56% hatched, Table 1). Thus, the GFP has a negative effect, despite the ability of GFP-Capu to contribute to actin mesh production and control of streaming onset. These data suggest that Capu may play additional roles during oogenesis that are more sensitive to the presence of the GFP tag. It was important to be able to gauge expression levels in individual egg chambers expressing Capu mutants; therefore, I used GFP-Capu, as the dynamic range of fertility (0% to 36%) was sufficient to distinguish between wild-type Capu and mutants, which is borne out by the following data.
Several individual mutations in the Capu-tail are sufficient to prevent Spir and Capu from interacting in binding assays, pyrene actin polymerization assays or colocalizing in S2 cells (Vizcarra et al., 2011). I chose the L1048A mutation because it does not block autoinhibition of Capu-stimulated actin assembly (Bor et al., 2012) or alter the ability of Capu to bind microtubules or actin filaments (B. Bor, E. Roth-Johnson and M.E.Q., unpublished). GFP-Capu(L1048A) expression resulted in 18% hatching, half of the rescue level produced by wild-type GFP-Capu but more than the <1% hatching produced by SpirB(Y232K)-GFP. Results from GFP-Capu(L1048A)-expressing flies were sensitive to expression levels. The range of expression was similar to that observed with GFP-Capu but the effect was different. Although low levels of wild-type GFP-Capu were sufficient to prevent premature streaming, low levels of GFP-Capu(L1048A) were not (supplementary material Fig. S4A,B). The majority of egg chambers had intermediate expression of GFP-Capu(L1048A) and these also failed to rescue (Fig. 3E-H; supplementary material Fig. S4B). At higher expression levels, oocytes did have a mesh and did not stream prematurely, consistent with the partial rescue of fertility (supplementary material Fig. S4). However, even at the highest expression levels, the mesh was not as dense as that created by intermediate levels of wild-type GFP-Capu, which can interact with Spir (supplementary material Fig. S4C,D). Further evidence that the rescue phenotype reflected expression level comes from measurement of the premature streaming speed. These flies had a broad range of streaming velocities and were the only oocytes that had velocities intermediate to wild type and mutants (Fig. 3O). In sum, rescue by GFP-Capu(L1048A), which cannot interact with Spir, is successful only at the highest expression levels of this mutant protein. I hypothesized that this result was consistent with a previous report that expression of wild-type GFP-Capu could partially compensate for loss of Spir (Dahlgaard et al., 2007). Supporting this idea, the two constructs, wild type and mutant, produced the same amount of viable eggs in a mutant spir background (72 and 73%; Table 1). Although results with GFP-Capu(L1048A) were less straightforward than SpirB(Y232K)-GFP, they support my conclusion that the Spir-Capu interaction is required under normal conditions, i.e. at physiological concentrations of Spir and Capu.
Spir-Capu complex separates
Can Spir and Capu act individually or are they part of an obligate complex during oogenesis? Spir-KIND inhibits both the nucleation and elongation activity of Capu (Quinlan et al., 2007; Vizcarra et al., 2011). Given these data, if Spir and Capu do not separate in the oocyte, the actin assembly activity of Capu would not be necessary. A mutation in the FH2 domain of Capu, I706A, based on mutagenesis experiments with Bni1 (Xu et al., 2004), virtually abolishes both the nucleation and elongation activities of Capu (Quinlan et al., 2007) (C. L. Vizcarra and M.E.Q., unpublished). I, therefore, introduced this mutation [GFP-Capu(I706A)] into flies lacking wild-type Capu. These flies were sterile (0% hatched, Table 1). I also examined the actin mesh and streaming of oocytes expressing GFP-Capu(I706A) (Fig. 3I-L). As expected, based on the fertility defect, the mesh is not detectable. Likewise, fast streaming begins early. The phenotype is highly penetrant, given that the speed of premature streaming is indistinguishable from the capu mutant flies (Fig. 3O). Thus, a functional FH2 domain is required for the activity of Capu: that is, by polymerizing actin through its FH2 domain, Capu builds the actin mesh. Based on the combination of the results presented here, i.e. actin assembly activity of Capu is required, and our earlier biochemical observation, i.e. Spir inhibits this activity, I proposed that Spir and Capu alternately bind and separate to function properly during oogenesis.
I then asked whether the localization patterns of Spir and Capu are consistent with the conclusion that they alternately interact and function separately in the oocyte? Indeed, my findings agree with this model. Dahlgaard et al. (Dahlgaard et al., 2007) examined GFP-Capu localization in live oocytes and found that it was diffuse with some enrichment at the oocyte/nurse cell interface. I observed a similar localization pattern (Fig. 4A,B) but when expression is lower in a nurse cell adjacent to the oocyte, the GFP-Capu level was also lower at that part of interface (Fig. 4B, arrowhead). To determine whether the cortical enrichment is predominantly in the nurse cells as opposed to one cortical surface of the oocyte, stage 11 egg chambers were examined when a layer of follicle cells separates the nurse cells from the oocyte. At this stage no enrichment of GFP-Capu at the cortex of the oocyte is detected (Fig. 4C). Instead, GFP-Capu is uniformly distributed throughout the oocyte. Endogenous Spir is enriched at the oocyte cortex (Quinlan et al., 2007). The antibody used to make this observation does not distinguish between SpirA, B or D, so I examined the localization of SpirB-GFP using native GFP fluorescence. It was found to be enriched in the oocyte cortex of live samples, consistent with our earlier immunofluorescence results (Fig. 4E). Because SpirB-GFP expression is low, this observation was confirmed in fixed samples using an anti-GFP antibody (Fig. 4F). SpirB-GFP is enriched in the oocyte cortex and nurse cell borders. I also investigated whether localization of Spir and Capu were dependent upon one another. In contrast to findings in tissue culture cells, no apparent change in GFP-Capu localization was detected in the absence of Spir, nor any changes in SpirB-GFP localization in the absence of Capu (Fig. 4D,H) (Quinlan et al., 2007; Vizcarra et al., 2011). In summary, although Spir and Capu localizations do overlap, there are also notable differences: both are enriched at nurse cell edges; Spir is enriched around the entire oocyte cortex, while Capu is more concentrated at the oocyte/nurse cell interface; and Capu is present at higher concentrations than Spir throughout the ooplasm. These observations fit the model of Spir and Capu interacting but also separating.
Fig. 4.
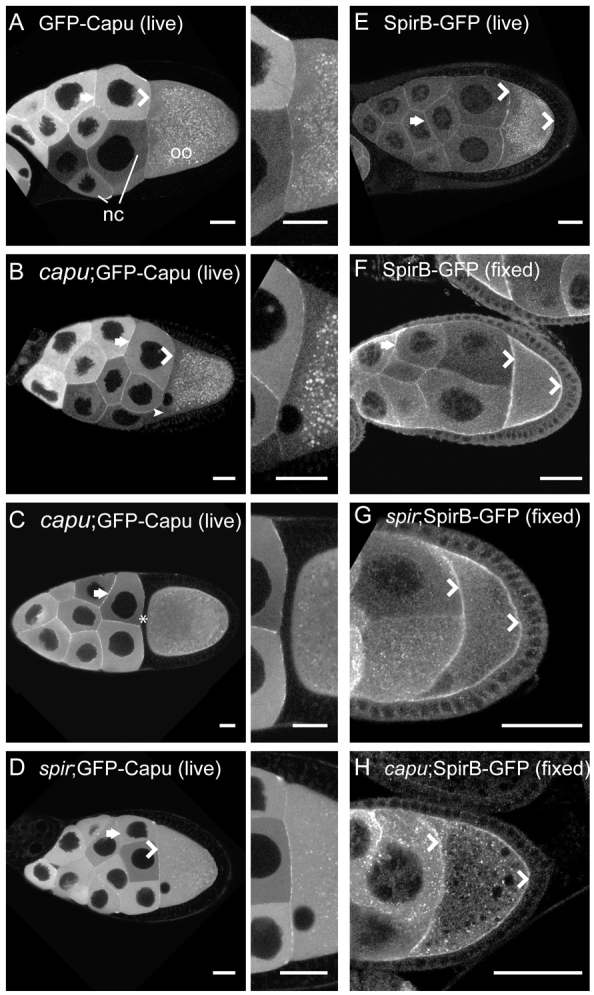
Spir and Capu localization. (A-D) Capu localization in live egg chambers is enriched at the cortex of the nurse cells (arrows), at the interface between the nurse cells and oocyte (chevrons), and is diffuse in both nurse cells and oocytes, as previously described (Dahlgaard et al., 2007). Punctae in the oocytes are autofluorescent yolk granules. No difference in this pattern is apparent whether or not GFP-Capu is expressed in a wild-type background (A) versus capu (B) or spir (D) mutant background. Adjacent images are ∼3× enlargements of the nurse cell/oocyte interface. (B) The small arrowhead indicates a region at the nurse cell/oocyte interface with lower intensity adjacent to a nurse cell with low GFP-Capu expression level. (C) No enrichment in the oocyte cortex closest to the nurse cells is visible in stage 11 egg chambers (asterisk). (E) Live egg chamber expressing SpirB-GFP, which is enriched at the cortex of the oocyte (chevrons), as well as those of the nurse cells (arrow) and appears punctate in the cytoplasm. Many of the punctae are autofluorescent yolk granules, based on their absence in fixed images. (F) Immunofluorescent image of a fixed egg chamber expressing SpirB-GFP, stained for GFP localization shows similar localization to the live egg chamber in E. (G,H) SpirB-GFP localization is unchanged in spir and capu mutant backgrounds. Egg chambers are oriented with posterior towards the right. nc, nurse cells; oo, oocyte. Scale bars: 30 μm.
DISCUSSION
By combining quantitative analysis of fertility with examination of the actin mesh and temporal control of fast streaming, I gained several insights into how Spir and Capu function during oogenesis. In summary, full-length SpirB, among several putative splice variants, is sufficient for oogenesis in a spir mutant background, while the shorter isoforms, SpirC and SpirD, are not, even when both are present. Spir and Capu are required to work together during oogenesis, through a direct interaction and a likely subsequent separation. A functional Capu-FH2 domain is required to build the actin mesh; however, it is not possible to distinguish whether Capu must nucleate, act as an elongation factor or both. Building the actin mesh is a crucial aspect of the function of Spir and Capu, but proper removal of the mesh is also important. Finally, temporal control of Spir and Capu protein levels and, perhaps, activity underlie regulation of the mesh and most likely fluid flows.
The non-zero fertility of flies expressing SpirB or SpirD when compared with SpirC indicates that the ability of the former two isoforms to build the actin mesh is crucial. Both SpirD and Capu expression produced an actin mesh in 100% of the egg chambers observed but fertility levels that were markedly lower than wild type. In addition, the fact that SpirB rescues fertility better than SpirD (or Capu) suggests that building the mesh is not the whole story. The difference observed between flies expressing SpirB versus SpirD may be due to whether or not the mesh disappears and fast streaming begins after stage 10b. Based on the polar localization of markers such as Par-1, the major body axes are thought to be established as early as stage 7 (Doerflinger et al., 2006). Several other classical polarity factors, such as oskar and bicoid, accumulate at the posterior and anterior ends of the oocyte shortly thereafter. However, functionally significant accumulation of such factors continues in later stages in a fast-streaming-dependent manner (Forrest and Gavis, 2003; Sinsimer et al., 2011). Thus, the inability to downregulate SpirD, either its protein levels or its activity, results in a persistent mesh that prevents fast flows and may be the cause of impaired fertility in these flies. Together, these data suggest that regulation of Spir, and thereby the mesh, is mediated by sequence present in the C-terminal half of Spir and that regulation requires an intact protein as opposed to two separate halves. What is the mode of regulation? I observed a dramatic decrease in protein levels of stage 11 egg chambers. Perhaps SpirB is exclusively regulated by degradation in the oocyte. Alternatively, SpirB activity may be inhibited in addition to being controlled at the protein level, much as Capu is auto-inhibited (Bor et al., 2012). At this time it is not possible to distinguish whether the control is pre- or post-translational. However, while degradation of actin nucleators as a regulatory mechanism is unusual, it is not unprecedented: mDia2 is degraded in a cell cycle-dependent manner (DeWard and Alberts, 2009).
Spir and Capu must interact directly in vivo. That this interaction is required to build the actin mesh fits well with the original genetic data implicating them in the same biological pathway (Manseau and Schüpbach, 1989). Based on these findings and on previously published work, I envision a cycle of interaction and separation, as illustrated in Fig. 5. Capu is depicted as being auto-inhibited (Fig. 5A) (Bor et al., 2012). Less is known about full-length Spir. The modified FYVE domain in the C terminus may dimerize Spir and drive it to membranes (Kerkhoff et al., 2001). I demonstrated here that Spir and Capu must interact during oogenesis (Fig. 5B). I propose that Spir-KIND binding to the Capu-tail competes with auto-inhibitory interactions (Fig. 5A,B) (Vizcarra et al., 2011; Bor et al., 2012). Spir binds actin monomers within this complex but Capu cannot nucleate or elongate when bound to Spir. Spir, Capu and actin could form a ‘pre-nucleation’ complex (Fig. 5B,C) (Quinlan et al., 2007; Vizcarra et al., 2011). Because a functional Capu-FH2 domain is essential to build the actin mesh, I propose that Spir is released, perhaps handing off an actin nucleus (Fig. 5C,D). Then, Capu would be free to act as an elongation factor, processively moving with and accelerating growth at the barbed end of the new actin filament (Bor et al., 2012). How elongation is terminated is not shown. One possibility is that Spir binds Capu again, knocking it off the end of the filament, resulting in a complex ready to form another new filament (Fig. 5C) (Bor et al., 2012; Vizcarra et al., 2011). Some or all of these interactions could be regulated by additional unknown factors.
Fig. 5.
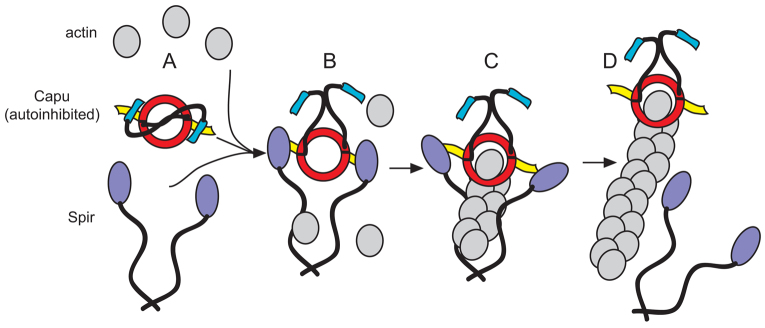
Model of Spir/Capu interaction. (A) Capu on its own is expected to be auto-inhibited (Bor et al., 2012). Spir on its own may be a monomer or dimer. If it is an in vivo nucleator, it should be be regulated, perhaps also by auto-inhibition. (B) This paper demonstrates that Spir and Capu must interact during oogenesis. Spir-KIND binding to the Capu-tail competes with autoinhibitory interactions (Vizcarra et al., 2011; Bor et al., 2012). (C) Spir binds actin monomers within this complex, but Capu cannot nucleate or elongate when bound to Spir, resulting in a ‘pre-nucleation’ complex (Quinlan et al., 2007; Vizcarra et al., 2011). (D) Because a functional Capu-FH2 domain is essential to build the actin mesh, Spir is proposed to be released, perhaps handing off an actin nucleus. Then, Capu is free to act as an elongation factor, processively moving with and accelerating growth at the barbed end of the new actin filament (Bor et al., 2012). How elongation is terminated is not shown. One possibility is that Spir could bind Capu again, knocking it off the end of the filament, resulting in a complex that is ready to form another new filament (Vizcarra et al., 2011; Bor et al., 2012). Some or all of these interactions are probably regulated by additional unknown factors.
This work clearly demonstrates that Spir and Capu interact in vivo but the issue of why they must come together remains open. The nucleation of Spir is enhanced when bound to Capu, but its peak nucleation activity is weaker than that of Capu alone (Quinlan et al., 2007). Perhaps Spir enhances the activity of Capu in the complex cellular environment, in a manner that is not readily apparent with purified proteins. Consistent with this idea, the mesh built independently of Spir, even at the highest levels of Capu(L1048A) expression, was not as dense as that typically observed in wild-type flies. Recent advances in our understanding of other models of interactions between actin assembly factors also fit this idea. Nucleation by the yeast formin Bni1 is enhanced by its co-factor Bud6 in the presence and absence of profilin (Graziano et al., 2011). Although Bud6 does not nucleate independently, it does bind actin monomers. This activity and binding to the tail end of Bni1 are required to stimulate this formin (Tu et al., 2012). Similarly, the formin mDia1 binds directly to APC, another potent actin nucleator (Okada et al., 2010). Their interaction enables the pair to nucleate in the presence of profilin and capping protein, conditions that block spontaneous polymerization and strongly dampen nucleation by most formins (Breitsprecher et al., 2012; Okada et al., 2010). Furthermore, APC and mDia1 separate after nucleation, as is proposed for Spir and Capu (this study). In the case of mDia1/APC, mDia1 moves processively with the fast growing barbed end of the filament and APC remains at the site of nucleation, presumably associating with the pointed end. A similar model was proposed for Spir and Capu (Quinlan and Kerkhoff, 2008). Capu, like mDia1 and other formins, is a processive elongation factor (Bor et al., 2012); whether or not Spir remains associated with the nucleation site or pointed end of filaments is an unresolved issue (Quinlan et al., 2005; Bosch et al., 2007; Stroupe et al., 2009; Ito et al., 2011). Thus, although there are distinct differences between Bud6, APC and Spir, there are compelling similarities and they may all play similar roles as formin nucleation-promoting factors.
Is the actin mesh the sole structure built by Spir and Capu? Premature streaming in the absence of the mesh is presumed to be causative in the polarity defect that leads to infertility. However, data comparing GFP-Capu and untagged Capu (this study) indicate that some other structure may be involved. That is, GFP-Capu expressing flies all had a mesh but the fertility levels were low, and lower than those with untagged Capu. Consistent with this idea, recent findings suggest that Spir and Capu may also build a structure at the posterior of the Drosophila oocyte that is important for establishing the anterior/posterior axis (Chang et al., 2011; Tanaka et al., 2011). These data implicate Mon2, a Golgi-endosomal marker, and Oskar as Spir/Capu interacting proteins. Many questions arise from these findings, such as: do Mon2 and Oskar act solely at the oocyte posterior or are they important for regulating other structures built by Spir and Capu, including mesh formation? However, neither is linked to premature streaming. Thus, there is more to learn about the roles of Spir and Capu and how they are regulated during oogenesis.
Supplementary Material
Acknowledgments
I thank members of the Quinlan lab and O. Akin for helpful discussions and close reading of the manuscript. I especially thank Justin Bois for invaluable technical help with PIV analysis; and Batbileg Bor, Elizabeth Roth-Johnson and William Silkworth for support when I needed it most.
Footnotes
Funding
This work was supported by a National Institutes of Health (NIH)-National Institute of General Medical Sciences (NIGMS) grant [R01 GM096133], by a Burroughs-Wellcome Fund Career Award in the Biomedical Sciences, by a grant from the Center for the Study of Women (UCLA) and by a March of Dimes Foundation Grant [#5-FY10-81] to M.E.Q. Deposited in PMC for release after 12 months.
Competing interests statement
The author declares no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.097337/-/DC1
References
- Bor B., Vizcarra C. L., Phillips M. L., Quinlan M. E. (2012). Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol. Biol. Cell 23, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Le K. H., Bugyi B., Correia J. J., Renault L., Carlier M. F. (2007). Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell 28, 555–568 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D., Jaiswal R., Bombardier J. P., Gould C. J., Gelles J., Goode B. L. (2012). Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. W., Nashchekin D., Wheatley L., Irion U., Dahlgaard K., Montague T. G., Hall J., St Johnston D. (2011). Anterior-posterior axis specification in Drosophila oocytes: identification of novel bicoid and oskar mRNA localization factors. Genetics 188, 883–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. K., Sawaya M. R., Phillips M. L., Reisler E., Quinlan M. E. (2012). Multiple forms of Spire-actin complexes and their functional consequences. J. Biol. Chem. 287, 10684–10692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F. D., Bork P., Kerkhoff E. (2003). The KIND module: a putative signalling domain evolved from the C lobe of the protein kinase fold. Trends Biochem. Sci. 28, 349–352 [DOI] [PubMed] [Google Scholar]
- Dahlgaard K., Raposo A. A., Niccoli T., St Johnston D. (2007). Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWard A. D., Alberts A. S. (2009). Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J. Biol. Chem. 284, 20061–20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H., Benton R., Torres I. L., Zwart M. F., St Johnston D. (2006). Drosophila anterior-posterior polarity requires actin-dependent PAR-1 recruitment to the oocyte posterior. Curr. Biol. 16, 1090–1095 [DOI] [PubMed] [Google Scholar]
- Emmons S., Phan H., Calley J., Chen W., James B., Manseau L. (1995). Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 9, 2482–2494 [DOI] [PubMed] [Google Scholar]
- Ferguson S. B., Blundon M. A., Klovstad M. S., Schüpbach T. (2012). Modulation of gurken translation by insulin and TOR signaling in Drosophila. J. Cell Sci. 125, 1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest K. M., Gavis E. R. (2003). Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13, 1159–1168 [DOI] [PubMed] [Google Scholar]
- Graziano B. R., DuPage A. G., Michelot A., Breitsprecher D., Moseley J. B., Sagot I., Blanchoin L., Goode B. L. (2011). Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol. Biol. Cell 22, 4016–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit H., Koppa R. (1982). Time-lapse film analysis of cytoplasmic streaming during late oogenesis of Drosophila. J. Embryol. Exp. Morphol. 67, 101–111 [Google Scholar]
- Hudson A. M., Cooley L. (2010). Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. J. Cell Biol. 188, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Narita A., Hirayama T., Taki M., Iyoshi S., Yamamoto Y., Maéda Y., Oda T. (2011). Human spire interacts with the barbed end of the actin filament. J. Mol. Biol. 408, 18–25 [DOI] [PubMed] [Google Scholar]
- Jones E., Oliphant T., Peterson P., et al. (2001). SciPy: Open source scientific tools for Python. http://www.scipy.org/
- Kerkhoff E., Simpson J. C., Leberfinger C. B., Otto I. M., Doerks T., Bork P., Rapp U. R., Raabe T., Pepperkok R. (2001). The Spir actin organizers are involved in vesicle transport processes. Curr. Biol. 11, 1963–1968 [DOI] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E., Parkhurst S. M. (2009). Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 136, 2849–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau L. J., Schüpbach T. (1989). cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 3, 1437–1452 [DOI] [PubMed] [Google Scholar]
- Okada K., Bartolini F., Deaconescu A. M., Moseley J. B., Dogic Z., Grigorieff N., Gundersen G. G., Goode B. L. (2010). Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J. Cell Biol. 189, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto I. M., Raabe T., Rennefahrt U. E., Bork P., Rapp U. R., Kerkhoff E. (2000). The p150-Spir protein provides a link between c-Jun N-terminal kinase function and actin reorganization. Curr. Biol. 10, 345–348 [DOI] [PubMed] [Google Scholar]
- Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. (2011). Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21, 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., Kessels M. M. (2009). New players in actin polymerization—WH2-domain-containing actin nucleators. Trends Cell Biol. 19, 276–285 [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Kerkhoff E. (2008). Actin nucleation: bacteria get in-Spired. Nat. Cell Biol. 10, 13–15 [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D. (2005). Drosophila Spire is an actin nucleation factor. Nature 433, 382–388 [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007). Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffel M., Willert C. E., Kompenhans J. (1998). Particle Image Velocimetry: A Practical Guide. Berlin: Springer-Verlag; [Google Scholar]
- Robinson D. N., Cooley L. (1997). Examination of the function of two kelch proteins generated by stop codon suppression. Development 124, 1405–1417 [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves A. E., Johndrow J. E., Keller L. C., Magie C. R., Pinto-Santini D. M., Parkhurst S. M. (2006). Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavit U., Lowe R. J., Steinbuck J. V. (2007). Intensity Capping: a simple method to improve cross-correlation PIV results. Exp. Fluids 42, 225–240 [Google Scholar]
- Sinsimer K. S., Jain R. A., Chatterjee S., Gavis E. R. (2011). A late phase of germ plasm accumulation during Drosophila oogenesis requires lost and rumpelstiltskin. Development 138, 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe M. E., Xu C., Goode B. L., Grigorieff N. (2009). Actin filament labels for localizing protein components in large complexes viewed by electron microscopy. RNA 15, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kato Y., Matsuda K., Hanyu-Nakamura K., Nakamura A. (2011). Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development 138, 2523–2532 [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E. (1994). Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science 265, 2093–2096 [DOI] [PubMed] [Google Scholar]
- Tu D., Graziano B. R., Park E., Zheng W., Li Y., Goode B. L., Eck M. J. (2012). Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc. Natl. Acad. Sci. USA 109, E3424–E3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L., Lehmann R. (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243–246 [DOI] [PubMed] [Google Scholar]
- Vizcarra C. L., Kreutz B., Rodal A. A., Toms A. V., Lu J., Zheng W., Quinlan M. E., Eck M. J. (2011). Structure and function of the interacting domains of Spire and Fmn-family formins. Proc. Natl. Acad. Sci. USA 108, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerweel J., Scarano F. (2005). Universal outlier detection for PIV data. Exp. Fluids 39, 1096–1100 [Google Scholar]
- Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.