Abstract
Inhibition of the signal transducer and activator of transcription 3 (STAT3) signaling pathway has been considered a novel therapeutic strategy to treat human cancers with constitutively active STAT3. In this study, we report the identification of niclosamide, an FDA-approved anthelmintic drug, as a new small-molecule inhibitor of the STAT3 signaling pathway. This compound potently inhibited the activation and transcriptional function of STAT3 and consequently induced cell growth inhibition, apoptosis, and cell cycle arrest of cancer cells with constitutively active STAT3. Our study provides a new promising lead compound with a salicylic amide scaffold for the development of STAT3 pathway inhibitors as novel molecularly targeted anticancer drugs.
Keywords: STAT3 signaling pathway, inhibitor, niclosamide, apoptosis, cell cycle arrest
The signal transducers and activators of transcription (STATs) are a class of transcription factors that regulate fundamental cellular and biological processes, such as cell proliferation, cell survival, immune responses, and angiogenesis, by modulating the expression of specific target genes.1−5 The upstream activators for the STAT pathway include cytokines, growth factors, and other cytoplasmic signaling proteins.1−4 A total of seven different STAT isoforms (STAT1−6) have been identified in mammalian cells. In the last several decades, constitutive STAT activity has been observed and reported to be correlated with oncogenic transformation. STAT3 is frequently overactivated in various human cancer types, including prostate, breast, head and neck cancers, but not in normal epithelial cells.3,6 Persistent activation of STAT3 signaling has been demonstrated to induce cell proliferation and prevent apoptosis in human cancer cells through the misregulation of key proteins, including cell survival proteins [e.g., B-cell lymphoma (Bcl)-xL and myeloid cell leukemia-1 (Mcl-1)], cell cycle regulators (e.g., cyclin D1/D2 and c-Myc), and inducers of angiogenesis such as vascular endothelial growth factor (VEGF) and hypoxia-inducible factor 1 (HIF1). Activated STAT3 is also correlated with resistance to conventional apoptosis-inducing therapies.1−4
The STAT3 protein consists of four functional domains that contribute to its oligomerization, DNA binding, SH2 dimerization, and transactivation, respectively. Upon stimulation by cytokines [such as interleukin (IL) or leukemia inhibitory factor (LIF)] or growth factors [such as epidermal growth factor (EGF)], tyrosine residue 705 (Tyr-705) in the STAT3 SH2 domain is phosphorylated, consequently inducing STAT3 to dimerize, translocate into the nucleus, and induce its binding to specific DNA response elements of target genes.1 Inhibition of STAT3 by antisense, oligonucleotide siRNAs, upstream Janus kinase (JAK), or Src kinase inhibitors or by direct STAT3 inhibitors has been demonstrated to suppress tumor growth and to induce apoptosis in cancer cells. Thus, the STAT3 pathway is considered to be an attractive target for the design of new therapies for human cancers with constitutively active STAT3.6−12
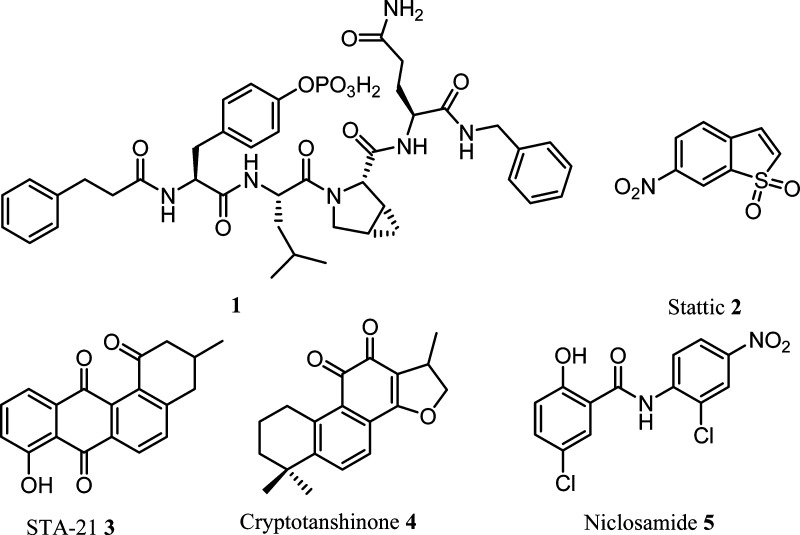
Several classes of small molecules have been identified as selective STAT3 inhibitors using rational design, high throughput screening, or structure-based virtual screening strategies.13−34 Examples included peptidic inhibitor 1,18 synthetic molecules Stattic 2(28) and STA-21 3,26 and the natural product cryptotanshinone 4 (Figure 1).14 However, most of the peptide-based inhibitors suffer from the poor cellular permeability, while nonpeptidic small-molecule STAT3 inhibitors are lack of ideal potency. Most recently, cell-permeable peptidic STAT3 inhibitors were reported from different groups.20,24,25,32,33 Despite these efforts, none of current inhibitors has been developed into a clinical trial.25 It is still highly valuable to identify new STAT3 inhibitors that could be further developed as novel molecularly targeted anticancer drugs. In this paper, we screened a small chemical library containing 1500 clinical drug derivatives and report the identification of niclosamide 5, an FDA approved anthelmintic drug, as a new highly potent small-molecule inhibitor of the STAT3 signaling pathway.
Figure 1.
Examples of reported STAT3 inhibitors and the chemical structure of niclosamide.
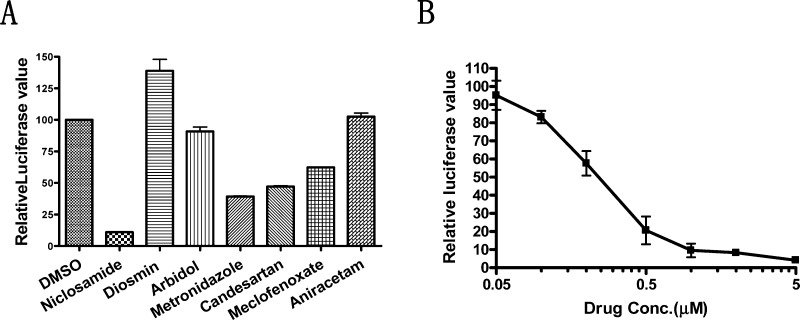
Given the extraordinarily high cost and poor success rate of drug development, repositioning (or repurposing) existing drugs to find new uses for these drugs has become an attractive approach to accelerate the drug development process.35,36 To identify new STAT3 inhibitors with useful pharmacological properties, we screened a small chemical library containing 1500 clinical drug derivatives using a cell-based STAT3-dependent dual luciferase reporter assay.26 HeLa epithelial carcinoma cells were chosen for transfection due to their constitutive overexpression of STAT3,14 and this cell line is frequently used in other transiently transfected luciferase reporter systems. After being transfected flanked with a luciferase reporter driven by a minimal thymidine kinase promoter sequence with seven copies of STAT3 binding sites (pLucTKS3),26 cell lysates showed high luciferase activity. Renilla luciferase was cotransfected as an internal control for normalization. Among all of the compounds tested, niclosamide (5; Figure 1) displayed a remarkable inhibitory effect on STAT3-induced luciferase activity in HeLa cells at 5.0 μM after a 24 h incubation, indicating that niclosamide strongly blocked the binding of STAT3 to pLucTKS3-containing STAT3-binding sites and inhibited the transcriptional function of STAT3. Other compounds, such as the semisynthetic phlebotropic drug diosmin, the antiviral Arbidol, and the anxiolytic aniracetam, did not show obvious inhibitory activity against luciferase activity (Figure 2A). Further evaluation revealed that niclosamide dose dependently inhibited STAT3-dependent luciferase reporter activity with an IC50 of 0.25 ± 0.07 μM (Figure 2B).
Figure 2.
Niclosamide inhibits the STAT3-mediated luciferase reporter activity in HeLa cells. (A) Niclosamide shows the highest inhibitory activity against STAT3 reporter activation among all tested compounds. (B) Niclosamide dose dependently inhibited STAT3 activity. Transit transfections were performed with Lipofectamine 2000. The luciferase reporter activity was evaluated after 24 h of treatment with indicated compounds or different concentrations of niclosamide. Relative luciferase units were the ratio of the absolute activity of firefly luciferase to that of renilla luciferase. Results are representatives of three independent experiments.
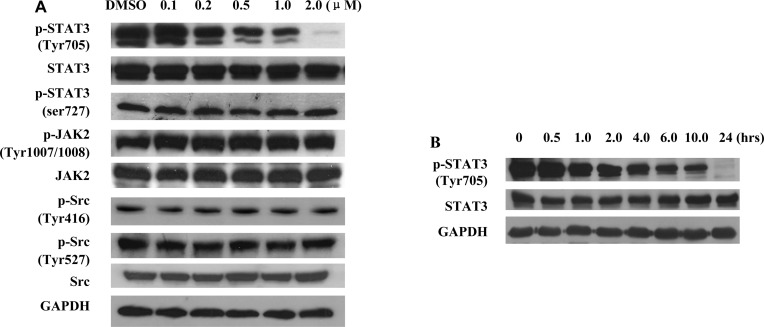
The effect of niclosamide on intracellular STAT3 activation was further investigated to validate the inhibitory activity of niclosamide against the STAT3 pathway. Du145 prostate cancer cells, which also express constitutively active STAT3,14 were treated with niclosamide, and their pTyr-705 STAT3 levels were determined via Western blotting. As shown in Figure 3A, niclosamide dose dependently inhibited the Tyr-705 phosphorylation of STAT3, whereas the total STAT3 protein level remained unchanged after a 24 h incubation. It is also noteworthy that niclosamide did not suppress the phosphorylation of Ser-727, which is another important STAT3 phosphorylation site (Figure 3A). A time−course study revealed that niclosamide obviously inhibited the Tyr-705 phosphorylation of STAT3 within 2.0 h, indicating that this drug might directly block the STAT3 pathway (Figure 3B). The selectivity of niclosamide against STAT3 homologues was also investigated, and our results demonstrated that niclosamide selectively inhibited the phosphorylation of STAT3 and had no obvious inhibition against the activation of other homologues (e.g., STAT1 and STAT5) after a 2 h treatment (Supporting Information). The inhibitory effect of niclosamide on STAT3 was further validated using the HeLa epithelial carcinoma cell model (Supporting Information).
Figure 3.
Niclosamide dose dependently inhibits the phosphorylation of STAT3. (A) Niclosamide exhibits dose-dependent inhibition on STAT3 phosphorylation but has no obvious inhibition on the activation of the upstream proteins JAK2 and Src. Du145 cells were treated with niclosamide with indicated concentrations for 24 h before subjected to Western blot analysis. (B) Niclosamide triggers quick inhibition of STAT3 phosphorylation. Du145 cells were treated with 2.0 μM niclosamide, and Western blot analysis was performed after indicated periods of time.
To determine if the suppressing effect of niclosamide on STAT3 was due to the inhibition of upstream tyrosine kinases, the influence of niclosamide on the JAK1, JAK2, and Src kinases, which are direct activators of STAT3, was also evaluated. Interestingly, niclosamide did not decrease the protein levels of JAK1, p-JAK1, JAK2, p-JAK2, Src, p-Src(416), or p-Src(527) after a 24 or 2 h treatment (Figure 3A and Supporting Information). Further protein kinase profiling analyses demonstrated that the IC50 values for niclosamide to inhibit JAK2 and Src kinases were over 10 μM in vitro. The protein kinase profiling analysis also revealed that niclosamide did not show much inhibition against other protein kinases (e.g., EGFR, VEGFR, and PDGFR etc.), which indicates that niclosamide may inhibit the activation of STAT3 through a kinase-independent pathway (Supporting Information). The SH2 domain of STAT3 protein is essential to its activation and dimeriztion. Therefore, a fluorescence-based binding assay37 was performed to investigate if niclosamide could directly bind to the SH2 domain and therefore block the STAT3 signaling pathway. Our results revealed that niclosamide failed to interrupt the interaction of fluorescence-labeled SH2 peptide with STAT3 protein (data not shown), indicating that it might not directly bind to the SH2 binding site of STAT3.
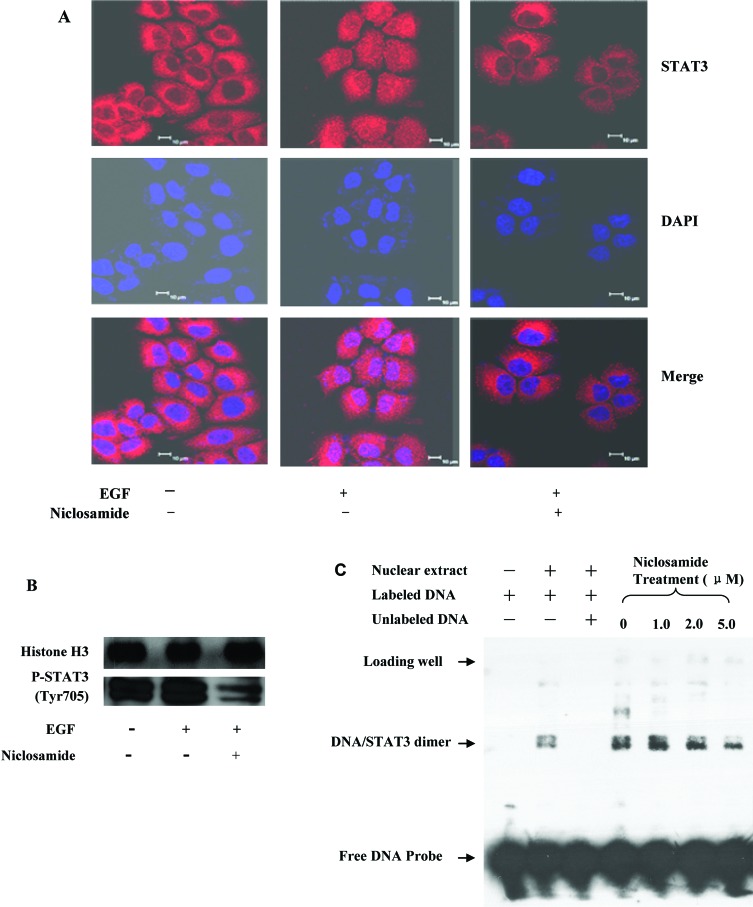
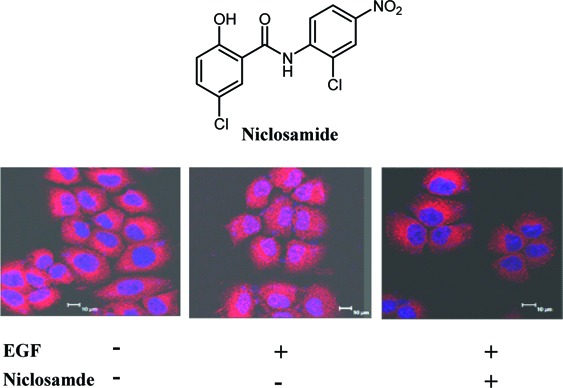
Upon activation, STAT3 forms dimers, translocates into the nucleus, and binds to specific DNA response elements to regulate target gene transcription. Theoretically, a cell permeable small-molecule STAT3 inhibitor would inhibit the nuclear translocation and/or the transcriptional functions of STAT3. An immunofluorescence assay clearly showed that the EGF induced STAT3 nuclear translocation, but this translocation was successfully inhibited after a 2 h treatment with 1.0 μM niclosamide (Figure 4A). The results were further validated by determining the protein level of activated STAT3 via Western blotting with both nuclear extracts and whole cell lysates from niclosamide-treated Du145 cells (Figure 4B and Supporting Information). Furthermore, our electrophoretic mobility shift assay (EMSA) analysis also revealed that although niclosamide did not directly bind to the DNA binding site to inhibit the interaction of STAT3 protein with its consensus DNA elements (Supporting Information), it strongly inhibited activation and nuclear translocation of STAT3 to interfere the DNA binding activity of STAT3 (Figure 4C). Consequently, Western blotting results displayed that the transcriptional function of STAT3 protein was potently inhibited by niclosamide, which led to a significant decrease of the protein levels of downstream target genes, such as cyclin D1, c-Myc, and Bcl-xL (Figure 5).
Figure 4.
(A) Niclosamide inhibits the EGF-induced nuclear translocation of STAT3. Serum-starved Du145 cells were treated with 1.0 μM niclosamide for 2.0 h and then stimulated with 100 ng/mL EGF for 15 min before subjected to immunofluorescence procedure. STAT3 was visualized as red fluorescence, and the nuclei were visualized as purple fluorescence under a confocal laser microscopy. (B) Niclosamide treatment decreases the protein level of p-STAT3 in nucleus. Serum-starved Du145 cells were treated with 1.0 μM niclosamide for 2 h and then stimulated with 100 ng/mL EGF. Nuclear extracts were subjected to Western blot analysis. (C) Niclosamide inhibits DNA binding activity of STAT3 through EMSA assay. Du145 cells were treated with niclosamide for 4 h before nuclear extracts were subjected to EMSA analysis.
Figure 5.
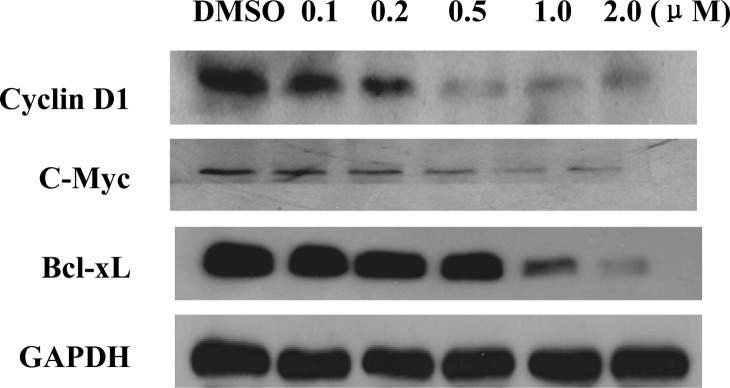
Niclosamide inhibits the transcription of STAT3 downstream genes. Du145 cells were treated with niclosamide with indicated concentrations for 24 h before subjected to Western blot analysis.
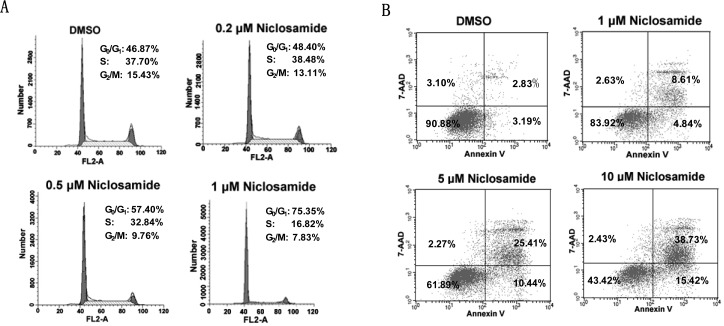
The antiproliferation activity of niclosamide was also evaluated. Our results demonstrated that this drug strongly inhibited the proliferation and colony formation of Du145 cells with IC50 values of 0.7 and 0.1 μM, respectively. Niclosamide also potently inhibited the cellular growth of other cancer cells with constitutively active STAT3 (e.g., HeLa epithelial carcinoma cells, A549 lung adenocarcinoma cells), whereas the compound exhibited relatively low inhibitory potency against cell growth of the other cancer cells with a low level of activated STAT3 (e.g., HT29 colon adenocarcinoma cells, PC3 prostate cancer cells, and A431 epithelial carcinoma cells) (Supporting Information). Flow cytometric analysis revealed that niclosamide dose dependently induced G0/G1 phase arrest and apoptosis of Du145 cancer cells (Figure 6A,B), which may be a consequence of the downregulation of cell survival proteins Bcl-xL, Mcl-1, and cell cycle regulators cyclin D and c-Myc (Figure 5).
Figure 6.
Niclosamide dose dependently induces cell cycle arrest and apoptosis of Du145 cancer cells. (A) Niclosamide causes significant G0/G1 arrest after a 24 h of treatment. (B) Niclosamide elicits apoptosis of Du145 cells in a dose-dependent manner.
In summary, niclosamide, an FDA-approved anthelmintic drug, was identified as a new small-molecule inhibitor of the STAT3 signaling pathway. This drug potently inhibited the activation, nuclear translocation, and transactivation of STAT3 but had no obvious effects on the closely related STAT1 and STAT5 proteins, the upstream JAK1, JAK2, and Src kinases, or other receptor tyrosine kinases. Furthermore, niclosamide inhibited the transcription of STAT3 target genes and induced cell growth inhibition, apoptosis, and cell cycle arrest of cancer cells with constitutively active STAT3. Although niclosamide does not have an ideal pharmarcokinetic profile (i.e., poor oral bioavailability) in humans as an anticestodal drug, it represents a new potent lead compound with salicylic amide scaffold for development of STAT3 pathway inhibitors as new molecularly targeted anticancer drugs. The further structural optimization and extensive mechanism study on niclosamide are undergoing and will be reported in due course.
Acknowledgments
The pLucTKS3 plasmid was generously provided by Prof. Jiayuh Lin from The Ohio State University. The fluorescence-based binding assay was performed in Prof. Shaomeng Wang's group at the University of Michigan (Ann Arbor, MI).
Abbreviations
STAT3, signal transducer and activator of transcription 3; JAK, Janus kinase; Bcl, B-cell lymphoma; Mcl-1, myeloid cell leukemia-1; VEGF, vascular endothelial growth factor; LIF, leukemia inhibitory factor; IL, interleukin; EGF, epidermal growth factor; pLucTKS3, a luciferase promoter driven by a minimal thymidine kinase promoter sequence with seven copies of STAT3 binding sites; EMSA, electrophoretic mobility shift assays.
Supporting Information Available
Typical experimental procedures for the luciferase reporter assay, Western blotting, immunofluorescence assay, cell proliferation inhibition assay, flow cytometric analysis, nuclear protein extraction, EMSA, and the kinase panel profiling results. This material is available free of charge via the Internet at http://pubs.acs.org.
We thank the 100-Talent Program of the Chinese Academy of Sciences (CAS), the Chinese Academy of Sciences Grant (KSCX2-YWR-27), the National Natural Science Foundation (Grant Numbers 21072192 and 90813033), and the National High Technology Research and Development Program (Grant Numbers 2010CB529706 and 2009CB940904) for their financial support.
Supplementary Material
References
- Darnell J. E. Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Bromberg J.; Darnell J. E. Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 2000, 19, 2468–2473. [DOI] [PubMed] [Google Scholar]
- Bowman T.; Garcia R.; Turkson J.; Jove R. STATs in oncogenesis. Oncogene 2000, 19, 2474–2488. [DOI] [PubMed] [Google Scholar]
- Hirano T.; Ishihara K.; Hibi N. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000, 19, 2548–2556. [DOI] [PubMed] [Google Scholar]
- Yu H.; Kortylewski M.; Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [DOI] [PubMed] [Google Scholar]
- Yu H.; Jove R. The STATs of cancer—New molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [DOI] [PubMed] [Google Scholar]
- Darnell J. E. Jr. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2002, 2, 740–749. [DOI] [PubMed] [Google Scholar]
- Turkson J.; Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 2000, 19, 6613–6626. [DOI] [PubMed] [Google Scholar]
- Deng J.; Grande F.; Neamati N. Small molecule inhibitors of stat3 signaling pathway. Curr. Cancer Drug Targets 2007, 7, 91–107. [DOI] [PubMed] [Google Scholar]
- Germain D.; Frank D. A. Targeting the cytoplasmic and nuclear functions of signal transducers and activators of transcription 3 for cancer therapy. Clin. Cancer Res. 2007, 13, 5665–5669. [DOI] [PubMed] [Google Scholar]
- Costantino L.; Barlocco D. STAT 3 as a target for cancer drug discovery. Curr. Med Chem. 2008, 15, 834–843. [DOI] [PubMed] [Google Scholar]
- Fletcher S.; Turkson J.; Gunning P. T. Molecular approaches towards the inhibition of the signal transducer and activator of transcription 3 (Stat3) protein. ChemMedChem 2008, 3, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney K. N.; Hao W.; Xu J.; Gibbons J.; Hucul J.; Roll D.; Brady S. F.; Schroeder F. C.; Clardy J. Phaeosphaeride A, an inhibitor of STAT3-dependent signaling isolated from an endophytic fungus. Org. Lett. 2006, 8, 4067–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. S.; Kim H.-N.; Shin K. D.; Yoon Y. J.; Kim S.-J.; Han D. C.; Kwon B.-M. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 2009, 69, 193–202. [DOI] [PubMed] [Google Scholar]
- Sun J.; Blaskovich M. A.; Jove R.; Livingston S. K.; Coppola D.; Sebti S. M. Cucurbitacin Q: A selective STAT3 activation inhibitor with potent antitumor activity. Oncogene 2005, 24, 3236–3245. [DOI] [PubMed] [Google Scholar]
- For selected examples of peptidic or peptide mimic inhibitors, see ; Chen J.; Nikolosha-Coleska Z.; Yang C.-Y.; Gomez C.; Gao W.; Krajewski K.; Jiang S.; Roller P.; Wang S. Design and synthesis of a new, conformationally constrained, macrocyclic small-molecule inhibitor of STAT3 via ‘click chemistry’. Bioorg. Med. Chem. Lett. 2007, 17, 3939–3942. [DOI] [PubMed] [Google Scholar]
- Gomez C.; Bai L.; Zhang J.; Nikolovska-Coleska Z.; Chen J.; Yi H.; Wang S. Design, synthesis, and evaluation of peptidomimetics containing Freidinger lactams as STAT3 inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 1733–1736. [DOI] [PubMed] [Google Scholar]
- Coleman D. R.; Ren Z.; Mandal P. K.; Cameron A. G.; Dyer G. A.; Muranjan S.; Campbell M.; Chen X.; McMurray J. S. Investigation of the binding determinants of phosphopeptides targeted to the Src homology 2 domain of the signal transducer and activator of transcription 3. Development of a high-affinity peptide inhibitor. J. Med. Chem. 2005, 48, 6661–6670. [DOI] [PubMed] [Google Scholar]
- Siddiquee K. A. Z.; Gunning P. T.; Glenn M.; Katt W. P.; Zhang S.; Schroeck C.; Sebti S. M.; Jove R.; Hamilton A. D.; Turkson J. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem. Biol. 2007, 2, 787–798. [DOI] [PubMed] [Google Scholar]
- Gunning P. T.; Glenn M.; Siddiquee K. A. Z.; Katt W. P.; Masson E.; Sebti S. M.; Turkson J.; Hamilton A. D. Targeting protein-protein interactions: Suppression of Stat3 dimerization with rationally designed small-molecule, nonpeptidic SH2 domain binders. ChemBioChem 2008, 9, 2800–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P. K.; Limbrick D.; Coleman D. R.; Dyer G. A. IV; Ren Z.; Birtwistle J. S.; Xiong C.; Chen X.; Briggs J. M.; McMurray J. S. Conformationally constrained peptidomimetic inhibitors of signal transducer and activator of transcription 3: Evaluation and molecular modeling. J. Med. Chem. 2009, 52, 2429–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P. K.; Ren Z.; Chen X.; Xiong C.; McMurray J. S. Structure-affinity relationships of glutamine mimics incorporated into phosphopeptides targeted to the SH2 domain of signal transducer and activator of transcription 3. J. Med. Chem. 2009, 52, 6126–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva O. A.; Gaponenko V.; Lockett S. J.; Tarasov S. G.; Jiang S.; Michejda C. J.; Perantoni A. O.; Tarasova N. I. Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS Chem. Biol. 2007, 2, 799–809. [DOI] [PubMed] [Google Scholar]
- Mandal P. K.; Liao W. S.-L.; McMurray J. S. Syhthesis of phosphatase-stable cell-permeable peptidomimetic prodrugs that target the SH2 domain of Stat3. Org. Lett. 2009, 11, 3394–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Bai L.; Bernard D.; Nikolovska-Coleska Z.; Gomez C.; Zhang J.; Yi H.; Wang S. Structure-based design of conformationally constrained cell-permeable STAT3 inhibitors. ASC Med. Chem Lett. 2010, 1, 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.; Wang R.; Wang S.; Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 4700–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiquee K.; Zhang S.; Guide W. C.; Blaskovich M. A.; Greedy B.; Lawrence H. R.; Yip M. L. R.; Jove R.; McLaughlin M. M.; Lawrence N. J.; Sebti S. M.; Turkson J. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 7391–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schust J.; Sperl B.; Hollis A.; Mayer T. U.; Berg T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006, 13, 1235–1242. [DOI] [PubMed] [Google Scholar]
- Kim B. H.; Yin C.-H.; Guo Q.; Bach E. A.; Lee H.; Sandoval C.; Jayabose S.; Ulaczyk-Lesanko A.; Hall D. G.; Baeg G.-H. A small-molecule compound identified through a cell-based screening inhibits JAK/STAT pathway signaling in human cancer cells. Mol. Cancer Ther. 2008, 7, 2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Cole D. C.; Chang C.-P. B.; Ayyad R.; Assalin M.; Hao W.; Gibbons J.; Jelinsky S. A.; Saraf K. A.; Park K. Inhibition of the signal transducer and activator of transcription-3 (STAT3) signaling pathway by 4-Oxo-1-phenyl-1,4-dihydroquinoline-3-carboxylic acid esters. J. Med. Chem. 2008, 51, 4115–4121. [DOI] [PubMed] [Google Scholar]
- Xu X.; Kasembeli M. M.; Jiang X.; Tweardy B. J.; Tweardy D. J. Chemical probes that competitively and selectively inhibit Stat3 activation. PLoS ONE 2009, 4, e4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Yue P.; Fletcher S.; Zhao W.; Gunning P. T.; Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interaction and Stat3-dependent tumor process. Biochem. Pharmacol. 2010, 79, 1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S.; Singh J.; Zhang X.; Yue P.; Page B. D.; Sharmeen S.; Zhao M.; Schimmer A. D.; Turkson J.; Gunning P. T. Disruption of transcriptionally active State dimmers with non-phosphorylated, salicylic acid-based small molecules: potent in vitro and tumor cell activities. ChemBiochem 2009, 10, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K.; Masuda Y.; Uehara Y.; Sato H.; Muroya A.; Takahashi O.; Yokotagawa T.; Furuya T.; Okawara T.; Otsuka M.; Ogo N.; Ashizawa T.; Oshita C.; Tai S.; Ishii H.; Akiyama Y.; Asai A.. Identification of a New Series of STAT3 Inhibitors by Virtual Screening. ACS Med. Chem. Lett. 2010, DOI: 10.1021/ml1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong C. R.; Sullivan D. J. New uses for old drugs. Nature 2007, 448, 645–646. [DOI] [PubMed] [Google Scholar]
- Ashburn T. T.; Thor K. B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discovery 2004, 3, 673–683. [DOI] [PubMed] [Google Scholar]
- Schust J.; Berg T. A high-throughput assay for signal transducer and activator of transcription 3. Anal. Biochem. 2004, 330, 114–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.