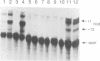
Abstract
We replaced the required TATA element of a yeast gal-his3 promoter by a binding site for GCN4, a protein that normally activates transcription when bound upstream of a TATA element. Surprisingly, GCN4 efficiently activates his3 transcription from wild-type initiation sites, though in a pattern associated with constitutive his3 transcription rather than GCN4 upstream activation through a TATA element. Transcriptional stimulation by GCN4 requires both the DNA-binding domain and the acidic activation function but is not affected by changing the spacing or helical relationship between the GCN4 binding site and the mRNA start sites. GCN4 is not sufficient for this TATA-independent activation; a sequence in the gal fragment distinct from the GAL4 binding sites is also required. Thus, GCN4 functions both when bound upstream of a TATA element and also when bound at the position of a TATA element. In the latter case, we suggest the possibility that GCN4 might be able to stimulate transcription by an alternate mechanism that does not involve a conventional TATA-binding transcription factor.
Full text
PDF

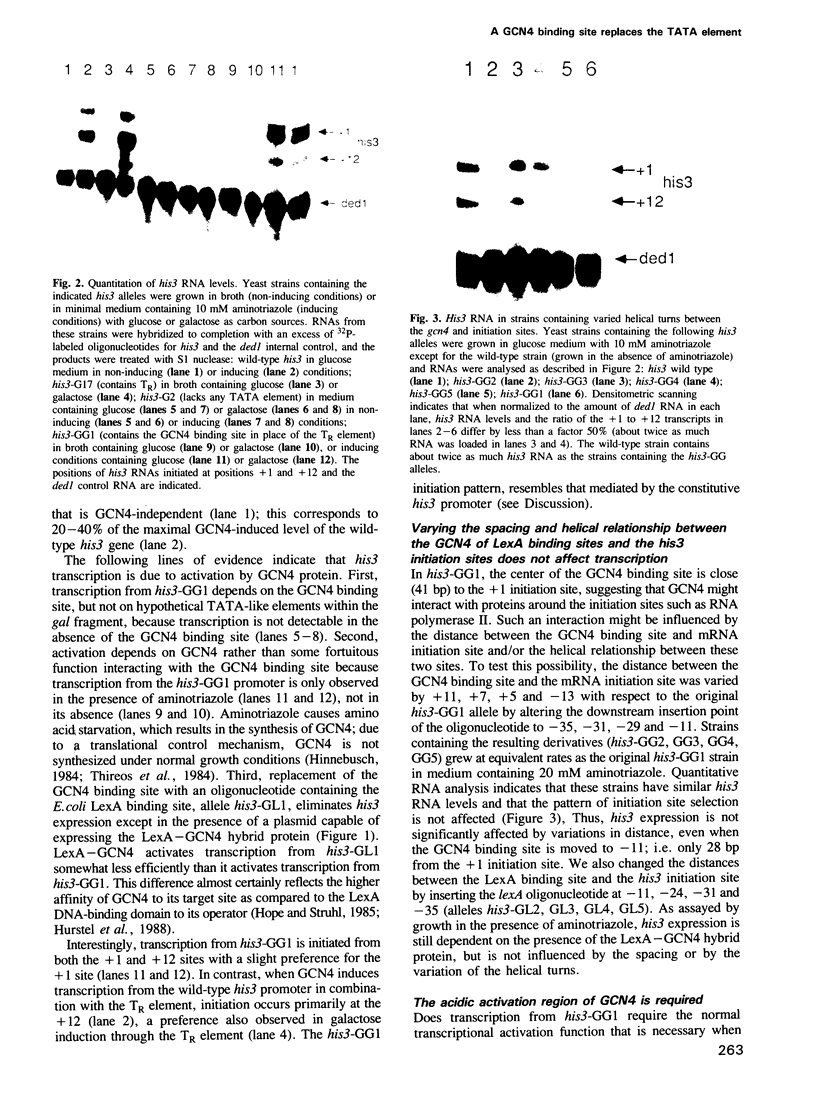





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Halden N. F., Cullen C. R., Corden J. L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988 Jan;8(1):330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Corden J. L., Cadena D. L., Ahearn J. M., Jr, Dahmus M. E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M. E., Kedinger C. Transcription of adenovirus-2 major late promoter inhibited by monoclonal antibody directed against RNA polymerases IIO and IIA. J Biol Chem. 1983 Feb 25;258(4):2303–2307. [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J. T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science. 1985 Nov 1;230(4725):511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homa F. L., Glorioso J. C., Levine M. A specific 15-bp TATA box promoter element is required for expression of a herpes simplex virus type 1 late gene. Genes Dev. 1988 Jan;2(1):40–53. doi: 10.1101/gad.2.1.40. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurstel S., Granger-Schnarr M., Schnarr M. Contacts between the LexA repressor--or its DNA-binding domain--and the backbone of the recA operator DNA. EMBO J. 1988 Jan;7(1):269–275. doi: 10.1002/j.1460-2075.1988.tb02809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988 Jan 29;52(2):161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- McKnight S., Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986 Sep 12;46(6):795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J Mol Biol. 1986 Feb 5;187(3):363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- Melton D. W., Konecki D. S., Brennand J., Caskey C. T. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2147–2151. doi: 10.1073/pnas.81.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Simon M. C., Fisch T. M., Benecke B. J., Nevins J. R., Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988 Mar 11;52(5):723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Chen W., Hill D. E., Hope I. A., Oettinger M. A. Constitutive and coordinately regulated transcription of yeast genes: promoter elements, positive and negative regulatory sites, and DNA binding proteins. Cold Spring Harb Symp Quant Biol. 1985;50:489–503. doi: 10.1101/sqb.1985.050.01.061. [DOI] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Negative control at a distance mediates catabolite repression in yeast. 1985 Oct 31-Nov 6Nature. 317(6040):822–824. doi: 10.1038/317822a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Struhl K. The JUN oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature. 1988 Apr 14;332(6165):649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- West R. W., Jr, Chen S. M., Putz H., Butler G., Banerjee M. GAL1-GAL10 divergent promoter region of Saccharomyces cerevisiae contains negative control elements in addition to functionally separate and possibly overlapping upstream activating sequences. Genes Dev. 1987 Dec;1(10):1118–1131. doi: 10.1101/gad.1.10.1118. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Paluh J. L., van Cleemput M., Moye W. S., Yanofsky C. Nucleotide sequence of Saccharomyces cerevisiae genes TRP2 and TRP3 encoding bifunctional anthranilate synthase: indole-3-glycerol phosphate synthase. J Biol Chem. 1984 Mar 25;259(6):3985–3992. [PubMed] [Google Scholar]