Abstract
Chronic antigen-stimulation has been recurrently involved in the earlier stages of monoclonal B-cell lymphocytosis, chronic lymphocytic leukemia and other B-cell chronic lymphoproliferative disorders. The expansion of two or more B-cell clones has frequently been reported in individuals with these conditions; potentially, such coexisting clones have a greater probability of interaction with common immunological determinants. Here, we analyzed the B-cell receptor repertoire and molecular profile, as well as the phenotypic, cytogenetic and hematologic features, of 228 chronic lymphocytic leukemia-like and non-chronic lymphocytic leukemia-like clones comparing multiclonal (n=85 clones from 41 cases) versus monoclonal (n=143 clones) monoclonal B-cell lymphocytosis, chronic lymphocytic leukemia and other B-cell chronic lymphoproliferative disorders. The B-cell receptor of B-cell clones from multiclonal cases showed a slightly higher degree of HCDR3 homology than B-cell clones from mono clonal cases, in association with unique hematologic (e.g. lower B-lymphocyte counts) and cytogenetic (e.g. lower frequency of cytogenetically altered clones) features usually related to earlier stages of the disease. Moreover, a subgroup of coexisting B-cell clones from individual multiclonal cases which were found to be phylogenetically related showed unique molecular and cytogenetic features: they more frequently shared IGHV3 gene usage, shorter HCDR3 sequences with a greater proportion of IGHV mutations and del(13q14.3), than other unrelated B-cell clones. These results would support the antigen-driven nature of such multiclonal B-cell expansions, with potential involvement of multiple antigens/epitopes.
Introduction
B-cell chronic lymphoproliferative disorders (B-CLPD) usually show a monoclonal expansion of a (single) mature-appearing aberrant B-cell clone.1–4 However, patients diagnosed with composite lymphomas and other B-cell chronic lymphocytic leukemias – e.g. chronic lymphocytic leukemia (CLL) – have been reported in the literature for decades, particularly among immunocompromised subjects.5–7 Although early reports considered this phenomenon as a rare event, it might have been underestimated because its detection requires sophisticated multidisciplinary approaches encompassing combined histopathology, cytomorphology, immunophenotypic and cytogenetic techniques and/or molecular analyses of purified cell populations.8 In fact, B-cell neoplasms consisting of two phenotypically distinct populations of clonally unrelated B-lymphocytes coexisting in the same patient (detected either simultaneously or at different time points during follow-up) have been reported in the literature,9,10 with an estimated overall frequency among B-CLPD patients of around 5%.11 Recently, it has also been shown that up to 20% of cases of population-based non-CLL and CLL-like low count monoclonal B-cell lymphocytosis (MBLlo) may also carry two different unrelated B-cell clones.12–14 In addition, data from a small series suggest that the frequency of multiclonality could be particularly high among CLL-like MBL cases in relatives of patients with CLL, (4/6 cases analyzed).12 Altogether, these results support the existence of multiclonality in a significant proportion of both MBL cases and B-CLPD patients.
Multiclonal MBL and B-CLPD cases may consist of expansions of two or more B-cell clones potentially associated with chronic antigen-driven immune responses.15–17 In fact, this is particularly frequent in the earlier stages of MBLlo, which would further support the potential reactive nature of MBL among individuals with normal lymphocyte counts, prior to the stepwise acquisition of genetic alterations and progression to clinical MBL (MBLhi) and CLL.13,14,18,19 If this hypothesis holds true, specific antigenic determinants could potentially be more frequently shared between the coexisting B-cell clones of multiclonal cases than between the expanded B cells in different monoclonal MBL and B-CLPD patients, due to a higher probability of interaction with common immunological determinants. This might even be true when the coexisting clones display clearly distinct immunophenotypic and cytogenetic, as well as clinical, features.20–22
In order to test this hypothesis, in the present study we compared the B-cell receptor (BCR) repertoire and molecular profile, as well as the phenotypic, cytogenetic and hematologic features of CLL-like and non-CLL-like clones (n=228) from multiclonal (n=41 cases) versus monoclonal cases [n=143, including both CLL and CLL-like MBL (n=128), as well as cases of B-CLPD other than CLL and non-CLL-like MBL (n=15)].
Methods
Patients and samples
A total of 184 subjects with one (n=143 monoclonal cases) or two or more (n=41 multiclonal cases) CLL/non-CLL B-CLPD (n=140) and/or CLL-like/non-CLL-like MBL (n=88) B-cell clones, as defined by the World Health Organization 2008 criteria,23 were included. Binet stage24 of CLL subjects was retrospectively collected.
From the 41 multiclonal cases, two (5%) corresponded to healthy individuals with CLL-like MBLlo, eight (19.5%) were CLL-like MBLhi cases, 23 (56%) had CLL and eight (19.5%) had B-CLPD other than CLL; four of these latter cases showed coexistence of either one or two CLL-like MBL B-cell population(s). In 3/41 multiclonal cases, three coexisting B-cell populations were detected. From the 143 monoclonal cases, 13 (9%) corresponded to healthy adults with CLL-like MBLlo, 26 (18%) had CLL-like MBLhi, 89 (62%) had CLL, two (1%) had non-CLL-like MBLlo, two (1%) were non-CLL-like MBLhi cases and 11 (8%) had other B-CLPD. The precise criteria used for the classification of MBLlo and MBLhi are detailed in the Online Supplementary Methods. The age/gender distribution for each diagnostic group is detailed in Online Supplementary Table S1.
Peripheral blood samples were obtained from each subject after written informed consent had been given, and the study was approved by the local ethics committees of the University Hospital of Salamanca and the Blood and Transplantation Center of Coimbra/Portuguese Institute of Blood and Transplantation, in accordance with the Helsinki Declaration of 1975, as revised in 2008.
Immunophenotypic analyses
Immunophenotypic studies to screen for the presence and full characterization of clonal B-cell populations were performed by high-sensitive multiparameter flow cytometry on erythrocytelysed peripheral blood samples, according to previously described procedures18,25–29 which are also detailed in the Online Supplementary Methods. All cases showed a clonal (imbalanced SmIgκ:SmIgλ ratio of >3:1 or <1:3) and/or aberrant CD5+ B-cell population.
Cytogenetic and molecular studies
Cytogenetic analyses were performed by multicolor interphase fluorescence in situ hybridization on slides containing FACS-purified and fixed aberrant B cells, as previously described in detail.18,30 In parallel, patterns of rearrangement of the immunoglobulin heavy chain variable region genes (IGHV) and immunoglobulin κ (IGKV) and λ (IGLV) light chain genes were analyzed for each FACS-purified B-cell clone18,31–32 (see the Online Supplementary Methods section for detailed descriptions). To investigate the level of phylogenetic relationship among IGHV amino acid sequences, a sequence distance tree was built using the neighbor-joining method implemented in the freely available Molecular Evolutionary Genetic Analysis (MEGA) software.33 Two different, coexisting BCR were considered as being phylogenetically related when their IGHV amino acid sequences, going from framework region 1 to HCDR3 (both regions included), showed an identity ≥ 60%. This “identity” threshold was based on previously published concepts about the phylogeny of human IGHV genes based on their amino acid sequences33, and on the minimum identity percentage observed in colocalized sub-branches (presumably with the highest evolutionary relationship33) of the sequence distance tree built in this study (see the Online Supplementary Methods section for more detailed descriptions). HCDR3-alignments were carried out for each multiclonal case whose coexisting B-cell clones showed HCDR3 regions with identical lengths or lengths differing by one amino acid using the bioinformatic tools available at the web services of the European Bioinformatics Institute (EMBL-EBI Cambridge, UK). Through the EMBL-EBI tools, the identical amino acids or those with analogous side-chain polarity per paired intra-case HCDR3-alignment were highlighted (see the Online Supplementary Methods section for more detailed descriptions).
Statistical methods
For all statistical analyses the SPSS software program (SPSS 20.0, IBM SPSS Statistics, IBM, Armonk, NY, USA) was used.
Results
Distribution and immunophenotypic features of B-cell clones
A total of 228 B-cell clones were identified. These corresponded to 143 B-cell clones (89 CLL, 11 non-CLL, 39 CLL-like MBL and 4 non-CLL-like MBL clones) from mono clonal cases and 85 B-cell clones (26 CLL, 14 non-CLL, 40 CLL-like MBL and 5 non-CLL-like MBL clones) from multiclonal cases (Online Supplementary Table S2). The complete immunophenotypic and cytogenetic features of the individual clones of multiclonal cases are summarized in Online Supplementary Table S3.
In 26/41 multiclonal cases, all coexisting B-cell clones showed a CLL-like phenotype, while in 11 of the remaining 15 cases, at least one CLL-like B-cell population coexisting with another non-CLL aberrant B-cell population was identified. In the remaining four cases, two distinct non-CLL-like B-cell clones were found (Online Supplementary Table S3). The distribution of all CLL/non-CLL and CLL-like MBL/non-CLL-like MBL clones analyzed (from all monoclonal and multiclonal cases considered together) in the distinct diagnostic categories was as follows: 27 B-cell clones corresponded to CLL-like MBLlo, 52 to CLL-like MBLhi, 115 to CLL, 5 to non-CLL-like MBLlo, 4 to non-CLL-like MBLhi and 25 to non-CLL B-CLPD (Online Supplementary Table S2). The precise diagnoses of the B-cell clones from patients with B-CLPD other than CLL are specified in the footnote to Online Supplementary Table S2.
Overall size and B-cell receptor features of B-cell clones from multiclonal versus monoclonal cases of monoclonal B-cell lymphocytosis, chronic lymphocytic leukemia, and other B-cell chronic lymphoproliferative diseases
The relative and absolute median number of peripheral blood clonal B cells was significantly lower in multiclonal than in monoclonal cases (13% versus 45% and 2,692 cells/μL versus 9,115 cells/μL, respectively; P=0.001). Of note, the absolute median numbers of CLL-like MBLhi and CLL B-cell clones were also significantly lower in multiclonal than in monoclonal cases (1,254 versus 2,464 cells/μL and 9,113 versus 18,600 cells/μL, respectively; P=0.004 and P=0.02) (Online Supplementary Figure S1). In contrast, the absolute median number of peripheral blood CLL-like MBLlo B-cell clones was significantly higher in multiclonal than in monoclonal cases (79 versus 1 cells/ μL; P=0.002). No significant differences were found in the clone size between non-CLL-like and non-CLL B-cell clones in multiclonal versus monoclonal cases (Online Supplementary Figure S1). In addition, the frequency of CLL-like MBL B-cell clones was significantly higher in multiclonal than in mono clonal cases (47% versus 27%, respectively; P=0.002), whereas the frequency of CLL B-cell clones was higher in monoclonal versus multiclonal subjects (62% versus 31%, respectively; P=0.001). CLL B-cell clones from multiclonal and monoclonal CLL patients showed a similar distribution in Binet stage A versus Binet stages B/C (P>0.05). Of note, non-CLL B-cell clones were present at higher frequencies in multiclonal versus monoclonal cases (17% versus 8%, respectively; P=0.04) (Table 1).
Table 1.
Peripheral blood B-cell counts and BCR features of multiclonal vs. monoclonal B-cell clones from cases of B-cell chronic lymphoproliferative disorders and monoclonal B-cell lymphocytosis.
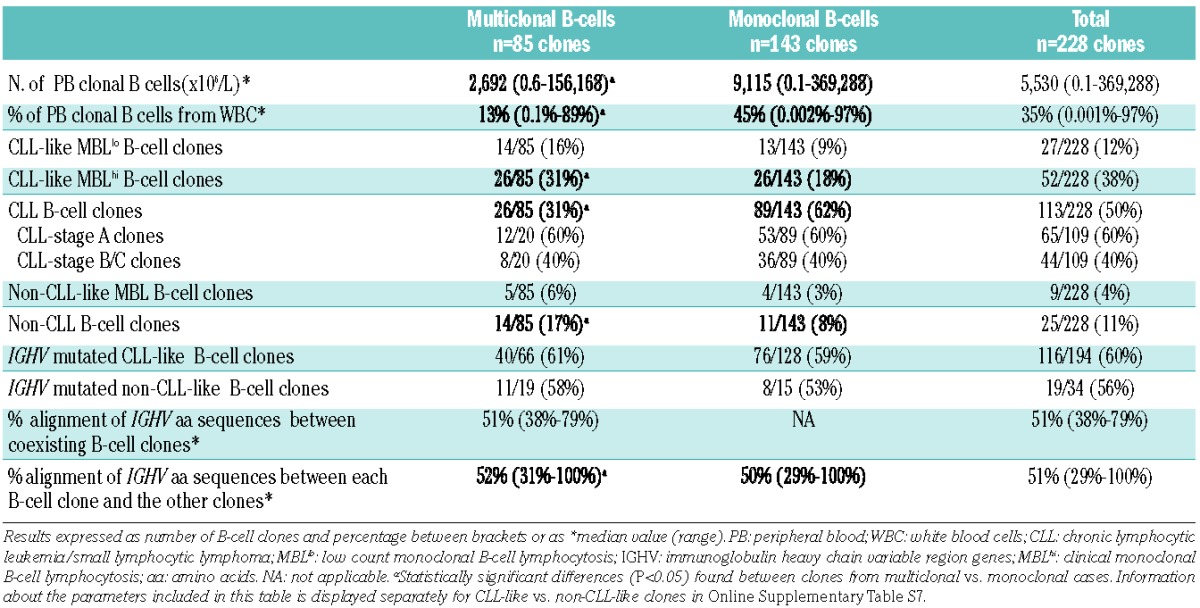
Regarding BCR features, a similar distribution of IGHV-mutated and IGHV-unmutated B-cell clones was found in multiclonal versus monoclonal cases: 51/85 (60%) versus 84/139 (60%) and 34/85 (40%) versus 55/139 (40%), respectively (Table 1). Despite this, the percentage of alignment of IGHV amino acid sequences among B-cell clones from multiclonal cases (n=3,560 two by two comparisons of clonal IGHV amino acid sequence) was slightly higher than that obtained among B-cell clones from monoclonal cases (n=8,891 comparisons): median of 52% versus 50%, respectively (P=0.001; Table 1).
Cytogenetic features of B-cell clones from multiclonal versus monoclonal cases of monoclonal B-cell lymphocytosis and B-cell chronic lymphoproliferative diseases
The frequency of CLL-like MBL and CLL clones from multiclonal cases that showed cytogenetic alterations was significantly lower than that found among CLL-like MBL and CLL clones from monoclonal cases: 27/66 (41%) versus 77/128 (60%), respectively (P=0.02). Likewise, the proportion of CLL-like B-cell clones showing coexistence of two or more cytogenetic alterations was also significantly lower in multiclonal than in monoclonal cases: 8/66 (12%) versus 32/128 (25%; P=0.047). This was specially true among B-cell clones from CLL patients: 2/26 (8%) versus 29/89 (33%), respectively (P=0.03; Table 2).
Table 2.
Cytogenetic features of CLL-like MBLlo, MBLhi and CLL B-cell clones from monoclonal vs. multiclonal cases.
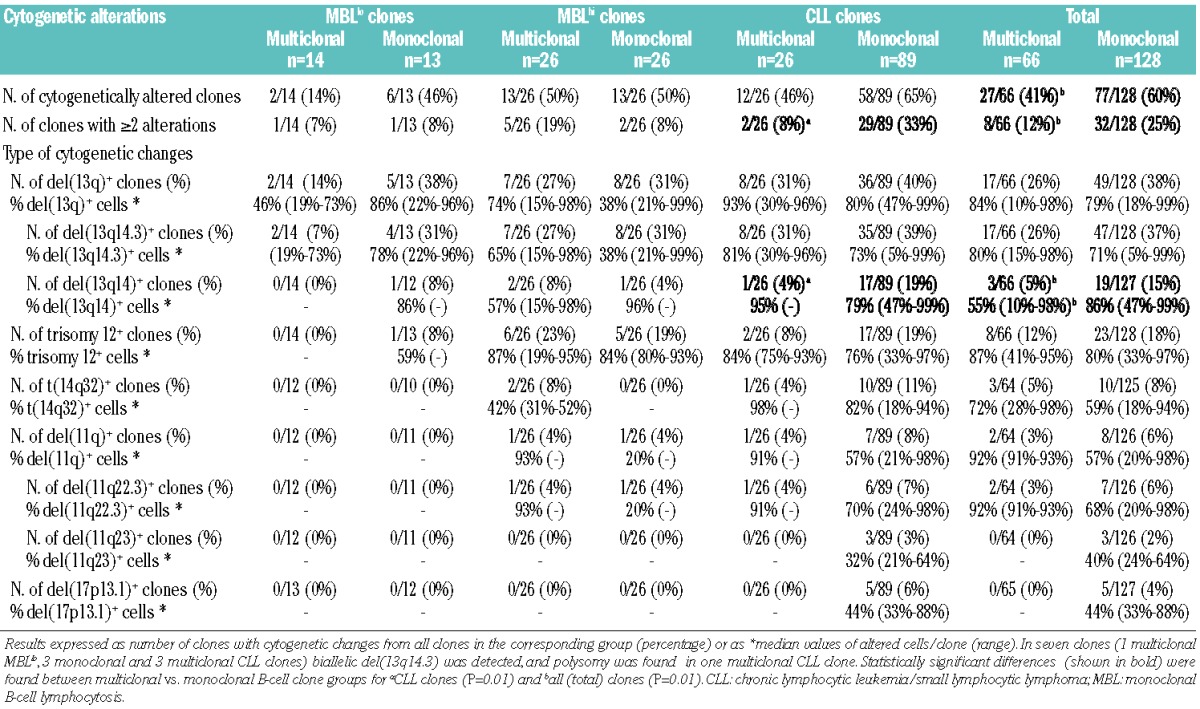
Regarding each specific cytogenetic alteration, only a decreased frequency of CLL-like B-cell clones with del(13q14) involving the RB1 gene and a lower percentage of del(13q14)+ cells was found in multiclonal versus monoclonal cases: the frequency of del(13q14)+ clones was 5% versus 15% with a median of del(13q14)+ cells of 55% versus 86%, respectively (P=0.01; Table 2). Of note, these differences were mostly due to the lower frequency of B-cell clones with del(13q14) (4% versus 19%, P=0.01) found among CLL clones from multiclonal versus monoclonal cases (Table 2).
No statistically significant differences were observed in the cytogenetic patterns of non-CLL B-cell clones from multiclonal versus monoclonal cases, which is probably due to the relatively low number of non-CLL clones included in the study; the precise cytogenetic alterations found in non-CLL/non-CLL-like MBL cases are shown in Online Supplementary Tables S3 and S4. In turn, the overall cytogenetic features of non-CLL like B-cell clones from multiclonal subjects (n=19; 3 non-CLL MBLlo, 2 non-CLL MBLhi, 14 non-CLL B-cell clones) versus monoclonal subjects (n=15; 2 non-CLL MBLlo, 2 non-CLL MBLhi, 11 non-CLL B-cell clones) were similar, as regards both the frequency of cytogenetically altered clones (6/19, 32% and 6/15, 40%) and the percentage of cases with two or more genetic alterations: 2/19 (11%) versus 2/15 (13%) (Online Supplementary Tables S3 and S4).
Molecular characteristics of the B-cell receptors of B-cell clones from multiclonal versus monoclonal cases of monoclonal B-cell lymphocytosis and B-cell chronic lymphoproliferative diseases
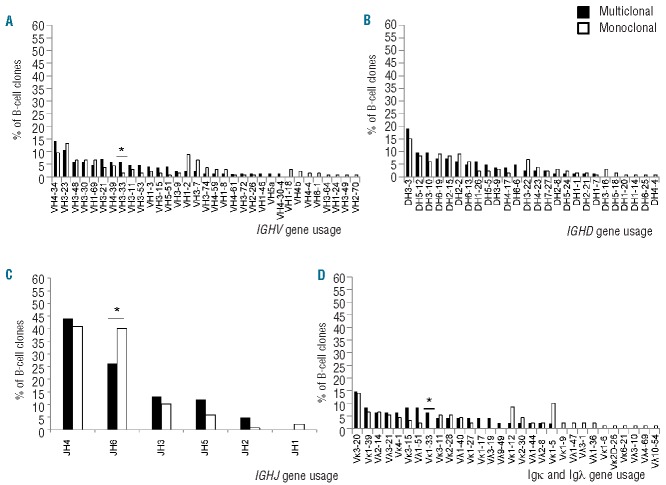
The molecular profile of the BCR of CLL-like MBLlo, MBLhi and CLL B-cell clones and of B-cell clones other than CLL from multiclonal versus monoclonal cases was very similar (Table 3 and Online Supplementary Table S5). No statistically significant differences in multiclonal versus monoclonal VH gene usage were found for most groups. Despite this general behavior, CLL-like MBLhi B-cell clones from multiclonal cases less frequently showed usage of the DH1, DH4 and DH7 gene families than B-cell clones from monoclonal cases; in addition, JH6 genes were also less frequently used by CLL B-cell clones from multiclonal versus monoclonal cases (Table 3). Overall, 33 functional IGHV gene rearrangements were identified from which 12 (V4-34, V3-23, V3-48, V3-30, V1-69, V3-21, V4-39, V3-33, V3-11, V3-53, V1-2, V3-7) were highly represented among the B-cell clones (≥5% of all B-cell clones corresponding to ≥4 and ≥5 B-cell clones sharing the same IGHV gene in multiclonal and monoclonal cases, respectively) (Figure 1A). Interestingly, 11 of these IGHV genes were found at similar frequencies within the clones of multiclonal versus monoclonal cases, while the V3-33 gene was typically associated with multiclonal cases (6% versus 1%, P=0.03). Regarding IGHD genes, no significant differences were observed between B-cell clones from multiclonal and monoclonal cases, the D3-3, D5-12, D3-10, D6-19, D2-15, and D2-2 genes being the most frequently used and shared by both groups of B-cell clones (Figure 1B). Among IGHJ genes, significant differences were only observed for the JH6 gene, which was more frequently used in monoclonal cases (40% versus 26%; P=0.03) (Figure 1C).
Table 3.
Molecular characteristics of the B-cell receptor (BCR) of chronic lymphocytic leukemia (CLL)-like monoclonal B-cell lymphocytosis (MBL)lo, MBLhi and CLL B-cell clones from monoclonal versus multiclonal cases.
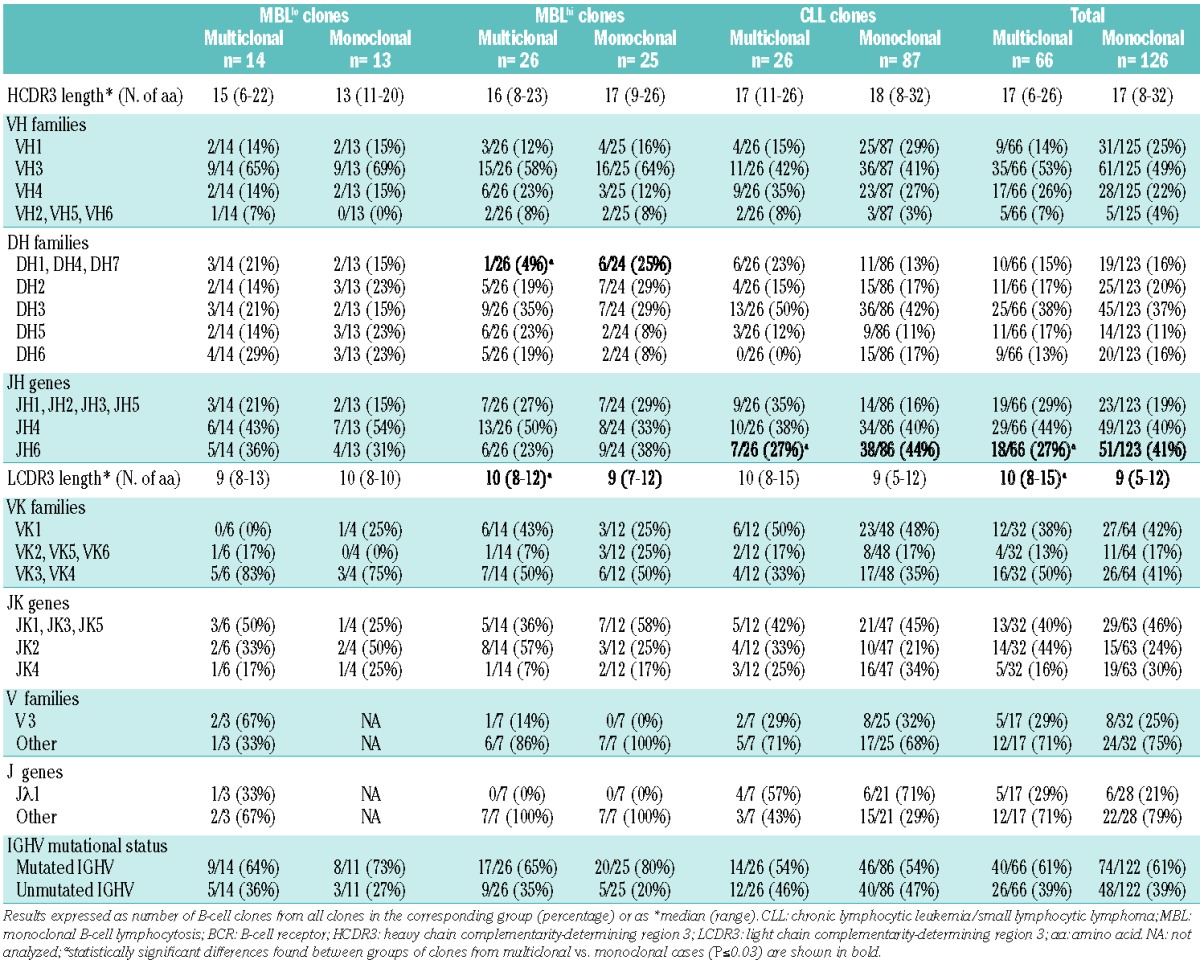
Figure 1.
Frequency of IGHV (A), IGHD (B), IGHJ (C) and both IGKV and IGLV (D) genes in multiclonal versus monoclonal CLL and non-CLL-like B-cell clones. Diagrams show the relative frequency of each IG gene in multiclonal compared to monoclonal B-cell clones (black and white bars, respectively). *Statistically significant differences were found between the multiclonal vs. monoclonal subgroups (P<0.05).
Except for slightly longer LCDR3 sequences of the IGKV and IGLV genes found among B-cell clones from multiclonal versus monoclonal cases, especially among CLL-like MBLhi clones (Table 3), no other significant differences were found in the molecular characteristics of the immunoglobulin light chain genes, either among CLL-like or non-CLL like B-cell clones from multiclonal versus monoclonal cases (Table 3 and Online Supplementary Table S5). Regarding IGKV and IGLV genes, only the VK1-33 gene was associated with multiclonal cases (6% versus 0%; P=0.02) (Figure 1D)
Molecular features of phylogenetically related B-cell receptors of B-cell clones from multiclonal cases
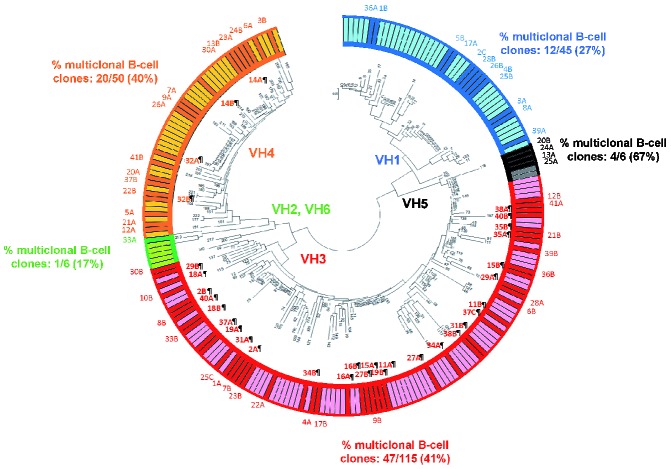
Thirty-two of the 85 B-cell clones from individual multiclonal cases were phylogenetically closely related and had exactly the same IGHV family (IGHV3 in 28 B-cell clones and IGHV4 in 4 B-cell clones) (Figure 2). Of note, this subgroup of B-cell clones frequently showed IGHV3 gene usage (28/85, 33%) and they displayed shorter HCDR3 sequences than other (multiclonal and monoclonal) B-cell clones: 13 (6–25) versus 17(9–26) and 16 (8–32) amino acids (P=0.001 and P=0.004, respectively). In addition, they also showed a higher frequency of del(13q14.3) compared to B-cell clones from multiclonal cases expressing phylogenetically unrelated IGHV families (41% versus 17%, respectively; P=0.05). Moreover, a slightly higher frequency of multiclonal cases whose coexisting clones were cytogenetically altered was found among phylogenetically closely related clones versus phylogenetically unrelated clones from multiclonal cases (53% versus 34%, respectively; P=0.06). Interestingly, a trend towards an increased percentage of IGHV-mutated B-cell clones among phylogenetically related B-cell clones from multiclonal cases compared to other B-cell clones from multiclonal cases was also found (70% versus 54%, respectively; P=0.1). Interestingly, most of the coexisting phylogenetically related clones had a CLL-like phenotype (10/16 cases, identified in Online Supplementary Table S3 by the ¶ symbol), while in 4/16 multiclonal cases, one CLL-like B- cell clone coexisted with one non-CLL B-cell clone, (2 marginal zone lymphomas, 1 lymphoma of mucosa-associated lymphoid tissue and 1 hairy cell leukemia clones from cases 29, 32, 37 and 38, also identified in Online Supplementary Table S3 with the ¶ symbol). In a minority of cases (2/16), the two coexisting phylogenetically related clones were both non-CLL-like, their phenotype being consistent with follicular lymphoma and lymphoma of mucosa-associated lymphoid tissue (case 16¶ and case 34¶, respectively, in Online Supplementary Table S3).
Figure 2.
Sequence distance cladogram of IGHV gene usage in CLL-like and non-CLL-like B-cell clones from multiclonal (dark colored bars in the outside circle) and monoclonal (light colored bars in the outside circle) cases. Five major branches were found in the sequence distance cladogram (i.e VH1, VH5, VH3, VH2-VH6, VH4). B-cell clones from individual multiclonal cases are represented by numbers; from them, those phylogenetically closely related B-cell clones, which share the same IGHV family, are specifically identified by bold numbers in the inner part of the circle and the symbol ¶. Of note, B-cell clones from multiclonal cases 14¶, 16¶ and 35¶ belong to closely located sub-branches of the cladogram, with their IGHV sequences having amino acid identity of 79%, 76% and 69%, respectively. In turn, B-cell clones from the multiclonal case 32¶ belong to the VH4 major branch with IGHV sequences whose amino acid identity is 69%. Finally, the other B-cell clones from multiclonal cases – cases 2¶, 11¶, 15¶, 18¶, 19¶, 27¶, 29¶, 31¶, 34¶, 37¶, 38¶ and 40¶– belong to the VH3 major branch, having IGHV sequences with amino acid identity > 60% (68%, 73%, 73.4%, 61%, 79%, 70%, 63%, 77%, 69.9%, 70%, 72% and 68.4%, respectively).
Homology of the HCDR3 region between B-cell clones coexisting in multiclonal cases versus non-coexisting (monoclonal) B-cell clones
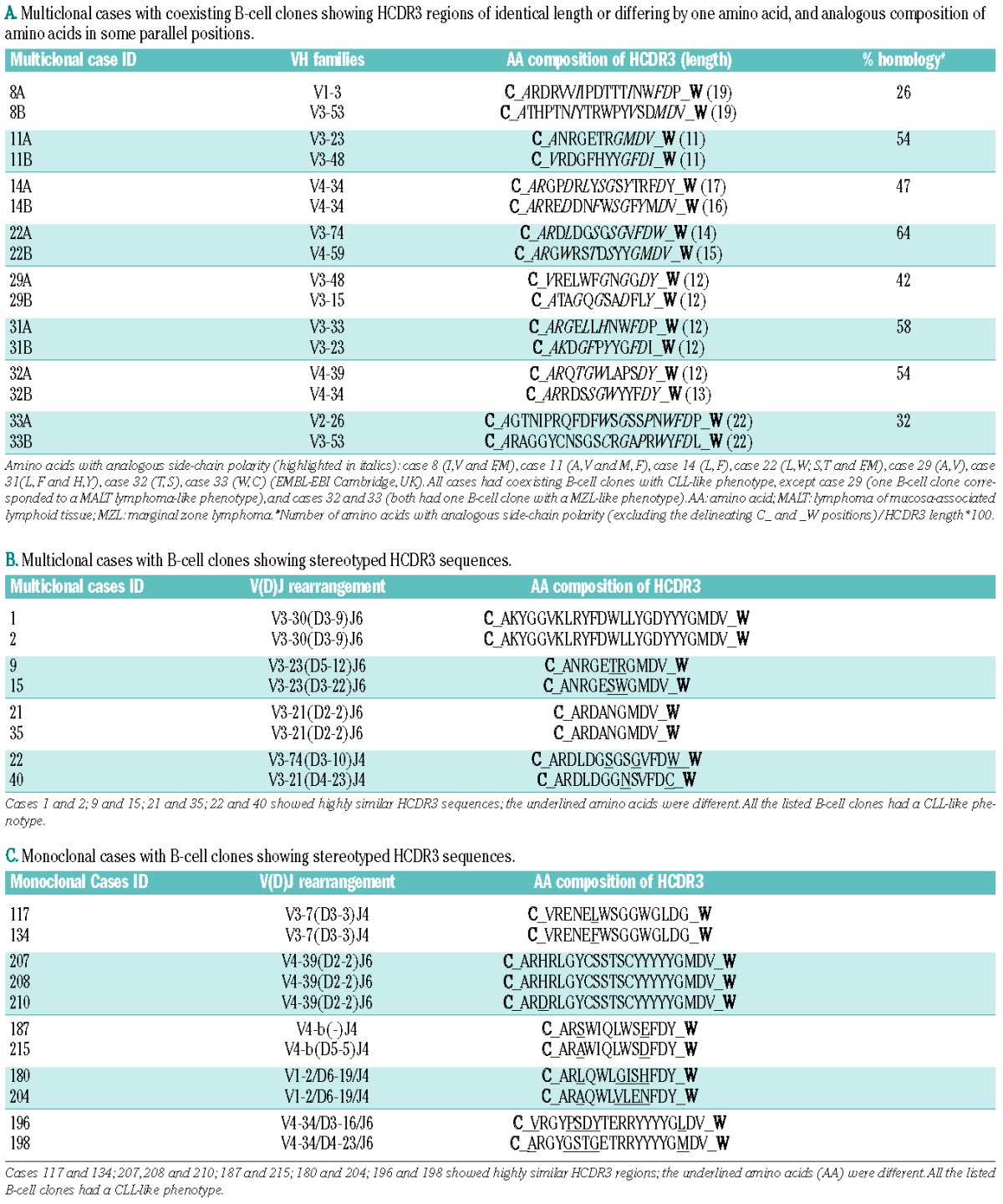
The HCDR3 amino acid sequence from coexisting B-cell clones had the same length or differed by just one amino acid in 8/41 multiclonal cases analyzed (19%) (Table 4A). The homology of all these case-paired HCDR3 regions was calculated as the number of identical amino acids or amino acids with an analogous side-chain polarity [(excluding the anchor second-CYS104 (C_) and the J-TRP 118 (_W) amino acid positions that delineate the HCDR3 region)] divided by the corresponding HCDR3 length (Table 4A). It is worth noting that the amino acid composition of HCDR3 sequences of the same length (±1 amino acid) that belonged to the same or evolutionary, highly-related VH families (e.g. VH3-48, VH3-21, VH3-11)33 (n=57) from monoclonal cases (Online Supplementary Table S6) showed a tendency towards a lower homology than that of multiclonal cases: median of 37% (range: 11% to 71%) versus 50% (range: 26% to 64%), respectively; (P=0.1). Since stereotyped sequences are widely represented in CLL,34 we further analyzed the frequency of stereotyped HCDR3 sequences in multiclonal cases (Table 4B) versus monoclonal cases (Table 4C). Interestingly, the number of multiclonal cases showing the same or highly similar stereotyped HCDR3 sequences was significantly higher than that of monoclonal cases: 8/41 (19%) versus 11/143 (8%), respectively (P=0.001). Furthermore, the amino acid composition of HCDR3 sequences from monoclonal cases with stereotyped HCDR3 sequences showed clearly less identical and/or conserved positions than those found among multiclonal cases (underlined amino acids in Tables 4B and 4C).
Table 4.
Multiclonal cases with coexisting B-cell clones sharing HCDR3 features (A). Multiclonal (B) and monoclonal (C) cases with B-cell clones showing stereotyped HCDR3 amino acid sequences. A. Multiclonal cases with coexisting B-cell clones showing HCDR3 regions of identical length or differing by one amino acid, and analogous composition of amino acids in some parallel positions.
Discussion
Multiclonal expansions of phenotypically aberrant B-cell clones (MBLlo) have been reported as frequently present in the general population35; of note, multiclonal expansions of immunophenotypically normal B cells can also be found in non-malignant diseases, such as autoimmune disorders and inflammatory responses against several infectious agents (e.g. Helicobacter pylori, hepatitis C virus).36,37 Whether clonal expansions of aberrant B cells found in otherwise healthy individuals (MBLlo) reflect a prominent reactive process against potent antigenic stimuli with unknown clinical relevance, or whether they represent an early (multi)clonal manifestation of a BCR-dependent neoplastic event, still remains to be established. In this regard, it should be noted that between 30% and 40% of such cases show cytogenetic changes shared by clinical MBL and CLL, e.g. del(13q). Of note, among other large structural chromosomal alterations, clonal mosaicism involving del(13q14) has also been recently found in peripheral blood cell populations from otherwise healthy individuals, particularly among subjects with more advanced age (around 2–3% in the elderly), but its potential relationship with MBL and CLL remains unknown.38,39 Compared to the typical (monoclonal) MBL and B-CLPD, coexisting B-cell clones from multiclonal MBL and B-CLPD may potentially have a greater probability of interacting with common immunological determinants. However, there is still little information about the potential existence of shared BCR features in cases showing two or more coexisting B-cell clones versus monoclonal cases.
In the present study, we analyzed for the first time the molecular and cytogenetic features of a large group (n=85) of coexisting, but unrelated, B-cell clones from a series of 41 multiclonal MBL and B-CLPD cases, in comparison to 143 monoclonal cases. Overall, the former clones more frequently showed cytogenetic and hematologic features which are typical of the earliest MBL stages and/or initial phases of CLL.18,36,37 Accordingly, B-cell clones from multiclonal cases more frequently corresponded to MBL cases, whereas B-cell clones from monoclonal cases were more frequently found to correspond to overt CLL. Of note, these findings do not contradict the apparent discrepancy between such associations and our previous observation among CLL patients of a worse clinical outcome for multiclonal cases carrying non-CLL clones,11 as this latter study was restricted to cases of overt CLL. In addition, multiclonal cases were also associated with lower clonal B-cell counts in peripheral blood, a lower number of cytogenetically altered clones, particularly of those carrying del(13q), and a decreased frequency of clones with two or more alterations. Of note, clonal expansions of non-CLL like B-cell clones were also more frequently observed in multiclonal than in monoclonal cases, such expansions corresponding mainly to indolent lymphomas (e.g. marginal zone lymphoma) which have been associated with chronic immune responses.40,41
Collectively, these results support the notion that the presence of multiple B-cell clones in the same individual more closely reflects the earlier stages of the disease. If this holds true and chronic antigen stimulation is involved in the onset of MBL and B-CLPD, as has recently been suggested for MBL based on epidemiological studies,42 it could be hypothesized that B-cell clones coexisting in multiclonal cases would show more closely related BCR features than B-cell clones from monoclonal cases. In this regard, our results point out the existence of a slightly higher level of HCDR3 homology among B-cell clones from multiclonal versus monoclonal cases. In fact, in around one fifth of all multiclonal cases, the coexisting B-cell clones showed a high homology in their HCDR3 amino acid sequences; this also held true when we compared the homology of the HCDR3 sequences of these multiclonal cases against those of monoclonal cases whose B-cell receptors were restricted to the same and/or ontogenetically related IGHV families. In addition, the frequency of stereotyped HCDR3 was also higher in multiclonal versus monoclonal cases. Such more closely related BCR features would be found independently of whether common antigens or superantigens are specifically involved, although the former would potentially lead to a higher HCDR3 homology, whereas superantigens could contribute to a greater frequency of usage of specific IGHV, IGHD and/or IGHJ genes.13,43
In the present study, we found a similar frequency of IGHV gene usage between coexisting multiclonal and non-coexisting monoclonal B-cell clones in association with a lower frequency of DH1, DH4 and DH7 as well as JH6 families in multiclonal versus monoclonal B-cell clones. Overall, these results suggest that no single antigen or superantigen is involved in common in MBL and B-CLPD. This is further supported by the relatively low percentage of alignment (≈50%) of the IGHV amino acid sequences observed among the different clonal B-cell populations analyzed, since such potential antigens -including superantigens - would require interaction with highly conserved sites at the IGHV/HCDR3 regions of the BCR.44 Interestingly however, the higher representation of DH1, DH4, DH7 and JH6 IGH gene segments in monoclonal versus multiclonal B-cell clones, together with the slightly higher levels of HCDR3 homology observed among coexisting (multiclonal) versus non-coexisting (monoclonal) B-cell clones from MBL, CLL and other B-CLPD cases, would indicate that still non-random selection of specific HCDR3, DH and JH segments could exist in the MBL and CLL repertoire of both multiclonal and monoclonal cases, which could reflect antigen-driven selection and expansion of specific B-cell clones, at the MBL and/or CLL stages.45
In this regard, based on the phylogenetic proximity of their BCR, we could further identify, within the B-cell clones from multiclonal cases, a considerably represented subgroup of B-cell clones showing preferential usage of IGHV3 genes and shorter HCDR3 sequences carrying a significantly higher number of IGHV mutations versus the unrelated clones. These results add support to the involvement of a common antigen, at least in this specific subset of cases.46 Interestingly, these “phylogenetically-related” B-cell clones coexisting in multiclonal cases showed a significantly higher frequency of del(13q) than B-cell clones expressing other IGHV genes. These observations further suggest that the BCR features of this subset of coexisting multiclonal B-cell clones could also contribute to determining the probability and/or type of cytogenetic progression occurring at the earliest stages of the disease, as previously suggested by our group18 and others.47,48 Further long-term, longitudinal studies are required to confirm this hypothesis, since multiple productive IGHV gene rearrangements may also underlie clonal drift leading to selection for more aggressive clones whose proportions would change over time.49
In summary, based on the molecular features of the BCR and the cytogenetic profile of B-cell clones from the multiclonal versus monoclonal cases of MBL, CLL and other B-CLPD analyzed here, it may be concluded that multiclonality is typically associated with early stages of B-CLPD; at the same time it appears to reflect more closely an antigen-driven nature of MBL and B-CLPD, with potential involvement of multiple and diverse antigenic determinants.
Acknowledgments
The authors would like to thank Maria Luz Sánchez and Paloma Bárcena for expert assistance in FACS-sorting experiments, Ana Rasillo, María Laura Gutiérrez and Ana Balanzategui for expert assistance in cytogenetic/molecular studies, and María Jara, Belén Espinosa and Cristina Jimenez for technical assistance. The authors also thank Alfonso Romero and Paulino Fernandez-Navarro for their assistance in the coordination with the Primary Health Care Group of Salamanca, as well as all members of the Primary Health Care Group of Salamanca for the Study of MBL, who were directly responsible for collection of samples from the cohort of individuals with CLL-like low count monoclonal B-cell lymphocytosis included in this study.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
AH was supported by a grant from the Fundação Para a Ciência e Tecnologia of Portugal (SFRH/BD/31609/2006); ARC was partly supported by a grant from Fundación Científica de la Asociación Española contra el Cáncer (AECC-2008) and by a grant from Red Temática de Investigación Cooperativa en Cáncer del Instituto de Salud Carlos III - FEDER (RD12/0036/0048). The research was supported by the following grants: Red Temática de Investigación Cooperativa en Cáncer (RTICC) del Instituto de Salud Carlos III - FONDOS FEDER (RD06/0020/0035 and RD12/0036/0048); FIS PI06/0824-FEDER, PS09/02430-FEDER and FIS PI12/00905-FEDER, from the Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, Madrid, Spain; GRS206/A/08 from the Gerencia Regional de Salud de Castilla y León and Ayuda al Grupo GR37 de Excelencia de Castilla y León, Consejería de Educación; SAN/1778/2009, Consejería de Sanidad, Junta de Castilla y León, Valladolid, Spain and FS/1-2010 Fundación Memoria D. Samuel Solórzano, Universidad de Salamanca, Salamanca, Spain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Catovsky D. Chronic lymphoproliferative disorders. Curr Opin Oncol. 1995; 7(1):3–11 [PubMed] [Google Scholar]
- 2.Matutes E, Owusu-Ankomah K, Morilla R, Garcia Marco J, Houlihan A, Que TH, et al. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia. 1994;8(10):1640–5 [PubMed] [Google Scholar]
- 3.Montserrat E. Chronic lymphoproliferative disorders. Curr Opin Oncol. 1997;9(1):34–41 [DOI] [PubMed] [Google Scholar]
- 4.Orfao A, Almeida J, Sanchez ML, San Miguel JF. Immunophenotypic diagnosis of leukemic B-cell chronic lymphoproliferative disorders other than chronic lymphocytic leukemia. Chronic Lymphocytic Leukemia, p173 Series: Contemporary Hematology Humana Press; 2004 [Google Scholar]
- 5.Armes JE, Angus P, Southey MC, Battaglia SE, Ross BC, Jones RM, et al. Lymphoproliferative disease of donor origin arising in patients after orthotopic liver transplantation. Cancer 1994;74(9):2436–41 [DOI] [PubMed] [Google Scholar]
- 6.Cleary ML, Sklar J. Lymphoproliferative disorders in cardiac transplant recipients are multiclonal lymphomas. Lancet 1984;2(8401):489–93 [DOI] [PubMed] [Google Scholar]
- 7.Schmitt-Graff A, Hummel M, Anagnostopoulos I, Stoltenburg G, Stein H. [Primary brain lymphoma in acquired immunodeficiency syndrome. Immunophenotype and molecular pathologic characterization in stereotactic biopsy, autopsy and cerebrospinal fluid cytology]. Pathologe. 1995;16(1):75–80 [DOI] [PubMed] [Google Scholar]
- 8.Lefebvre C, Fabre B, Vettier C, Rabin L, Florin A, Wang J, et al. Composite splenic marginal zone lymphoma and mantle cell lymphoma arising from 2 independent B-cell clones. Hum Pathol. 2007;38(4):660–7 [DOI] [PubMed] [Google Scholar]
- 9.Sanchez ML, Almeida J, Lopez A, Sayagues JM, Rasillo A, Sarasquete EA, et al. Heterogeneity of neoplastic cells in B-cell chronic lymphoproliferative disorders: biclonality versus intraclonal evolution of a single tumor cell clone. Haematologica. 2006;91(3):331–9 [PubMed] [Google Scholar]
- 10.Woda BA, Knowles DM., 2nd Nodular lymphocytic lymphoma eventuating into diffuse histiocytic lymphoma: immunoperoxidase demonstration of monoclonality. Cancer. 1979;43(1):303–7 [DOI] [PubMed] [Google Scholar]
- 11.Sanchez ML, Almeida J, Gonzalez D, Gonzalez M, Garcia-Marcos MA, Balanzategui A, et al. Incidence and clinicobiologic characteristics of leukemic B-cell chronic lymphoproliferative disorders with more than one B-cell clone. Blood. 2003;102(8):2994–3002 [DOI] [PubMed] [Google Scholar]
- 12.Lanasa MC, Allgood SD, Volkheimer AD, Gockerman JP, Whitesides JF, Goodman BK, et al. Single-cell analysis reveals oligoclonality among ‘low-count’ monoclonal B-cell lymphocytosis. Leukemia. 2010;24(1):133–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiorazzi N, Ferrarini M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 2011;117(6):1781–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadzidimitriou A, Agathangelidis A, Darzentas N, Murray F, Delfau-Larue MH, Pedersen LB, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood. 2011;118(11):3088–95 [DOI] [PubMed] [Google Scholar]
- 15.Kostareli E, Sutton LA, Hadzidimitriou A, Darzentas N, Kouvatsi A, Tsaftaris A, et al. Intraclonal diversification of immunoglobulin light chains in a subset of chronic lymphocytic leukemia alludes to antigen-driven clonal evolution. Leukemia. 2010;24(7):1317–24 [DOI] [PubMed] [Google Scholar]
- 16.Sutton LA, Kostareli E, Hadzidimitriou A, Darzentas N, Tsaftaris A, Anagnostopoulos A, et al. Extensive intraclonal diversification in a subgroup of chronic lymphocytic leukemia patients with stereotyped IGHV4-34 receptors: implications for ongoing interactions with antigen. Blood. 2009;114(20):4460–8 [DOI] [PubMed] [Google Scholar]
- 17.Coelho V, Krysov S, Steele A, Sánchez Hidalgo M, Johnson PW, Chana PS, et al. Identification in CLL of circulating intraclonal subgroups with varying B-cell receptor expression and function. Blood. 2013;122(15):2664–72 [DOI] [PubMed] [Google Scholar]
- 18.Henriques A, Rodriguez-Caballero A, Nieto WG, Langerak AW, Criado I, Lecrevisse Q, et al. Combined patterns of IGHV repertoire and cytogenetic/molecular alterations in monoclonal B lymphocytosis versus chronic lymphocytic leukemia. PLoS One. 2013;8(7):e67751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenze D, Berg E, Volkmer-Engert R, Weiser AA, Greiner A, Knorr-Wittmann C, et al. Influence of antigen on the development of MALT lymphoma. Blood. 2006;107(3):1141–8 [DOI] [PubMed] [Google Scholar]
- 20.Bahler DW, Miklos JA, Swerdlow SH. Ongoing Ig gene hypermutation in salivary gland mucosa-associated lymphoid tissue-type lymphomas. Blood. 1997;89(9):3335–44 [PubMed] [Google Scholar]
- 21.Cabras AD, Candidus S, Fend F, Kremer M, Schulz S, Bordi C, et al. Biclonality of gastric lymphomas. Lab Invest. 2001;81(7):961–7 [DOI] [PubMed] [Google Scholar]
- 22.Konoplev S, Lin P, Qiu X, Medeiros LJ, Yin CC. Clonal relationship of extranodal marginal zone lymphomas of mucosa-associated lymphoid tissue involving different sites. Am J Clin Pathol. 2010;134(1):112–8 [DOI] [PubMed] [Google Scholar]
- 23.Sabattini E, Bacci F, Sagramoso C, Pileri SA. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica. 2010;102(3):83–7 [PubMed] [Google Scholar]
- 24.Swerdlow S.H. WHO classification of tumours of haematopoietic and lymphoid tissues, p439, 4th edn Lyon, France: International Agency for Research on Cancer, 2008 [Google Scholar]
- 25.Nieto WG, Teodosio C, Lopez A, Rodriguez-Caballero A, Romero A, Barcena P, et al. Non-CLL-like monoclonal B-cell lymphocytosis in the general population: prevalence and phenotypic/genetic characteristics. Cytometry B Clin Cytom. 2010;78(Suppl 1):S24–34 [DOI] [PubMed] [Google Scholar]
- 26.Kalina T, Flores-Montero J, van der Velden VH, Martin-Ayuso M, Bottcher S, Ritgen M, et al. EuroFlow standardization of flow cytometer instrument settings and immunophenotyping protocols. Leukemia. 2012;26(9):1986–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26(9):1908–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF, et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009;114(1):33–7 [DOI] [PubMed] [Google Scholar]
- 29.Sanchez ML, Almeida J, Vidriales B, Lopez-Berges MC, Garcia-Marcos MA, Moro MJ, et al. Incidence of phenotypic aberrations in a series of 467 patients with B chronic lymphoproliferative disorders: basis for the design of specific four-color stainings to be used for minimal residual disease investigation. Leukemia. 2002;16(8):1460–9 [DOI] [PubMed] [Google Scholar]
- 30.Quijano S, Lopez A, Rasillo A, Sayagues JM, Barrena S, Sanchez ML, et al. Impact of trisomy 12, del(13q), del(17p), and del(11q) on the immunophenotype, DNA ploidy status, and proliferative rate of leukemic B-cells in chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2008;74(3):139–49 [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez D, Gonzalez M, Alonso ME, Lopez-Perez R, Balanzategui A, Chillon MC, et al. Incomplete DJH rearrangements as a novel tumor target for minimal residual disease quantitation in multiple myeloma using real-time PCR. Leukemia. 2003;17(6):1051–7 [DOI] [PubMed] [Google Scholar]
- 32.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003; 17(12):2257–317 [DOI] [PubMed] [Google Scholar]
- 33.Darzentas N, Hadzidimitriou A, Murray F, Hatzi K, Josefsson P, Laoutaris N, et al. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia. 2010;24(1):125–32 [DOI] [PubMed] [Google Scholar]
- 34.Tsakou E, Agathagelidis A, Boudjoghra M, Raff T, Dagklis A, Chatzouli A, et al. Partial versus productive immunoglobulin heavy locus rearrangements in chronic lymphocytic leukemia: implication for B-cell receptor stereotypy. Mol Med. 2012;18:138–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almeida J, Nieto WG, Teodosio C, Pedreira CE, López A, Fernández-Navarro P, et al. CLL-like B-lymphocytes are systematically present at very low numbers in peripheral blood of healthy adults. Leukemia. 2011;25(4):718–22 [DOI] [PubMed] [Google Scholar]
- 36.Dolcetti R, Boiocchi M. Cellular and molecular bases of B-cell clonal expansions. Clin Exp Rheumatol. 1996;14(Suppl 14):S3–13 [PubMed] [Google Scholar]
- 37.Racanelli V, Sansonno D, Piccoli C, D’Amore FP, Tucci FA, Dammacco F. Molecular characterization of B cell clonal expansions in the liver of chronically hepatitis C virus-infected patients. J Immunol. 2001;167(1):21–9 [DOI] [PubMed] [Google Scholar]
- 38.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012; 44(6):642–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arcaini L, Merli M, Volpetti S, Rattotti S, Gotti M, Zaja F. (2012) Indolent B-cell lymphomas associated with HCV infection: clinical and virological features and role of antiviral therapy. Clin Dev Immunol. 2012; 2012:638185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaacson PG. Mucosa-associated lymphoid tissue lymphoma. Semin Hematol. 1999;36(2):139–47 [PubMed] [Google Scholar]
- 42.Casabonne D, Almeida J, Nieto WG, Romero A, Fernández-Navarro P, Rodriguez-Caballero A, et al. Common infectious agents and monoclonal B-cell lymphocytosis: a cross-sectional epidemiological study among healthy adults. PLoS One. 2012;7(12):e52808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bikos V, Darzentas N, Hadzidimitriou A, Davis Z, Hockley S, Traverse-Glehen A, et al. Over 30% of patients with splenic marginal zone lymphoma express the same immunoglobulin heavy variable gene: ontogenetic implications. Leukemia. 2012;26(7):1638–46 [DOI] [PubMed] [Google Scholar]
- 44.Silverman GJ. B cell superantigens: possible roles in immunodeficiency and autoimmunity. Semin Immunol. 1998;10(1):43–55 [DOI] [PubMed] [Google Scholar]
- 45.Volpe JM, Kepler TB. Large-scale analysis of human heavy chain V(D)J recombination patterns. Immunome Res. 2008;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosner K, Winter DB, Tarone RE, Skovgaard GL, Bohr VA, Gearhart PJ. Third complementarity-determining region of mutated VH immunoglobulin genes contains shorter V, D, J, P, and N components than non-mutated genes. Immunology. 2001;103(2):179–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vardi A, Dagklis A, Scarfò L, Jelinek D, Newton D, Bennett F, et al. Immunogenetics shows that not all MBL are equal: the larger the clone, the more similar to CLL. Blood. 2013;121(22):4521–8 [DOI] [PubMed] [Google Scholar]
- 48.Stamatopoulos K. CLL: promiscuity leads to risks. Blood. 2009;114(17):3508–9 [DOI] [PubMed] [Google Scholar]
- 49.Plevova K, Skuhrova Francova H, Burckova K, Brychtova Y, Doubek M, Pavlova S, et al. Multiple productive immunoglobulin heavy chain gene rearrangements in chronic lymphocytic leukemia are mostly derived from independent clones. Haematologica. 2014; 99(2):329–38 [DOI] [PMC free article] [PubMed] [Google Scholar]