Abstract
The accurate and comprehensive identification of functional regulatory sequences in mammalian genomes remains a major challenge. Here we describe Site-specific Integration FACS-sequencing (SIF-seq), an unbiased, medium-throughput functional assay for the discovery of distant-acting enhancers. Pluripotent cell reporter assays, targeted single-copy genomic integration, and flow cytometry are coupled with high-throughput DNA sequencing to enable parallel screening of large numbers of DNA sequences. We demonstrate the utility of this method by functionally interrogating >500 kb of mouse and human sequence for enhancer activity and identifying embryonic stem (ES) cell enhancers at pluripotency loci including NANOG. We also demonstrate the effectiveness of the approach in differentiated cell populations through the identification of cardiac enhancers from cardiomyocytes and neuronal enhancers from neural progenitors. SIF-seq is a powerful and flexible method for the de novo functional identification of mammalian enhancers in a potentially wide variety of cell types.
Introduction
Enhancers are noncoding DNA elements that generally act from a distance to activate transcription of a target gene(s) 1. Enhancer activity is frequently tissue- or cell type-specific, with many enhancers active only in one or a few tissues or cell types 2,3. These distance and tissue-specific features of enhancers have complicated their identification and characterization. Despite these technical challenges, there has been great interest in identifying enhancers because they play important roles in development and disease. Enhancer sequence or copy number variants are associated with a variety of human diseases 4,5. Furthermore, a large fraction of disease-associated regions identified through genome-wide association studies (GWAS) fall entirely in noncoding regions of the genome 6,7, and putative enhancers are enriched for disease-associated single nucleotide polymorphisms (SNPs) 8. In mice, individual deletions of enhancers have been shown to considerably alter development 9-13. However, the lack of comprehensive, functionally validated enhancer datasets for most tissues and cell types has prohibited the systematic exploration of their roles in human biology and disease.
Currently, most putative enhancers are identified via chromatin-based assays, such as ChIP-seq or DNase-seq 3,6,8. Such assays predict enhancer elements indirectly based on their association with specific transcription factors, transcriptional coactivators, chromatin structure, or epigenomic marks. One limitation of these approaches is that they are associated with false-positive and negative errors, and putative enhancers predicted this way must be further validated with functional reporter assays 14,15. Because of this limitation and the cell type-specificity of enhancers, there is a pressing need for higher-throughput functional enhancer assays that can be used in a wide variety of cell types and developmental contexts.
To enable unbiased, higher-throughput mammalian enhancer identification in biologically relevant cell types, we developed Site-specific Integration FACS-sequencing (SIF-seq). This method can be used for de novo discovery of mammalian enhancers across large genomic intervals and for medium-throughput validation of putative enhancers predicted by chromatin-based methods. Unlike previous medium- and high-throughput enhancer assays for mammals 16-18, SIF-seq includes the integration of putative enhancers into a single genomic locus 19. Therefore, the activity of enhancers is assessed in a reproducible chromosomal context rather than on a transiently expressed plasmid. Furthermore, by making use of embryonic stem (ES) cells and in vitro differentiation, SIF-seq can be used to assess enhancer activity in a wide variety of disease-relevant cell types.
To demonstrate the utility of this method, we used it to randomly interrogate, at a resolution of ~1 kb, genomic intervals and identify enhancers. We successfully used SIF-seq for the de novo functional identification of ES cell enhancers near genes involved in pluripotency or early embryogenesis (mouse and human Nanog/NANOG, mouse Sall1) and for the identification of cardiomyocyte enhancers near genes that regulate heart development (human MYH6 and MYH7). Furthermore, we demonstrate that SIF-seq can be used to assess the activity of putative enhancers in neural progenitor cells.
Results
SIF-seq Accurately Identifies Mouse ES Cell Enhancers
We first sought to use SIF-seq (Fig. 1) for de novo identification of mouse embryonic stem cell enhancers. We constructed two enhancer test libraries by shearing two Bacterial Artificial Chromosomes (BACs) containing loci of interest into ~1–1.6 kb fragments (Table 1, Supplementary Fig. 1). BAC1 (RP23-225H20) covered ~231 kb of mouse genomic sequence, including the Sall1 gene. In mouse ES cells, this region has a high density of sites that are marked with H3K27ac or p300 (Supplementary Fig. 2) 3, both strong predictors of enhancer activity 14,15. BAC2 (RP24-73P7) contained ~233 kb of mouse sequence encoding several genes, including the pluripotency gene Nanog (Fig. 2b, Supplementary Fig. 3a). The sheared BAC fragments were cloned into a genomic targeting plasmid next to a Venus Yellow Fluorescent Protein (YFP) gene 20 that is under the control of a minimal promoter. The resulting plasmids were then delivered to Hprt-deficient male mouse ES cells, where they were integrated by homologous recombination into the Hprt locus on the X chromosome 19, and drug selection was used to remove any cells that were not correctly targeted. This resulted in ES cell libraries where every cell had exactly one potential enhancer sequence coupled to a reporter gene integrated in single copy at the Hprt locus, a site that has been previously shown to be a suitable neutral region to study the activity of tissue-specific regulatory elements 21.
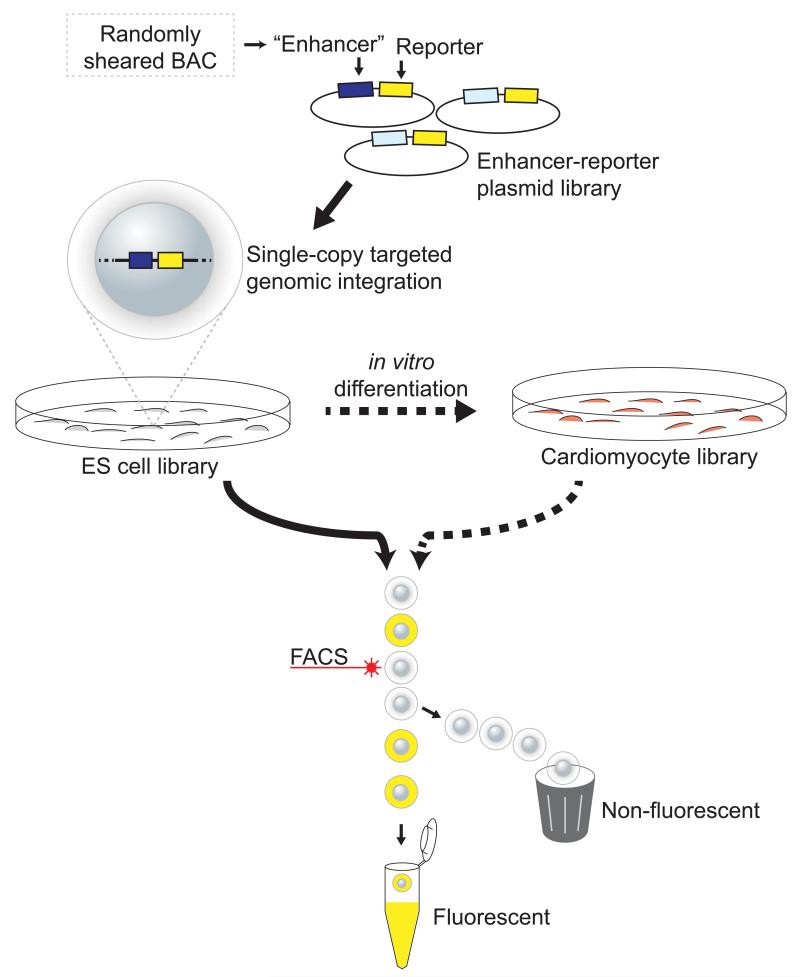
Figure 1.
Overview of Site-specific Integration FACS-seq (SIF-seq). DNA test fragments containing putative enhancers are linked to a fluorescent reporter gene, and these reporter constructs are integrated into a single, reproducible site in the genomes of mouse embryonic stem (ES) cells. This targeted genome integration, combined with drug selection, results in a library of mouse ES cells such that each cell contains exactly one candidate fragment-reporter construct, and each construct is integrated into the same genomic position in each cell. Reporter-expressing cells, which are enriched for active enhancers, are isolated by flow cytometry. Presumptive enhancer sequences are then amplified from these fluorescing cells and sequenced. A high density of sequence reads indicates the presence of an enhancer. For enhancer discovery in a wider variety of cell types, ES cell libraries can be in vitro differentiated prior to sorting.
Table 1.
Summary of Experiments
| Library Name | BAC Name | Size of BAC (bp) | Cell Type |
|---|---|---|---|
| mSall1 | RP23-225H20 | 230,977 | ESC |
| mNanog | RP24-73P7 | 233,215 | ESC |
| hNANOG | RP11-103J24 | 163,875 | ESC |
| hMYH6 and hMYH7 | RP11-929J10 | 223,497 | ESC |
| Cardiomyocyte | |||
| Ultraconserved | N/A | N/A | Neural progenitor |
BAC: bacterial artificial chromosome, bp: base pairs, ESC: embryonic stem cells
Figure 2.
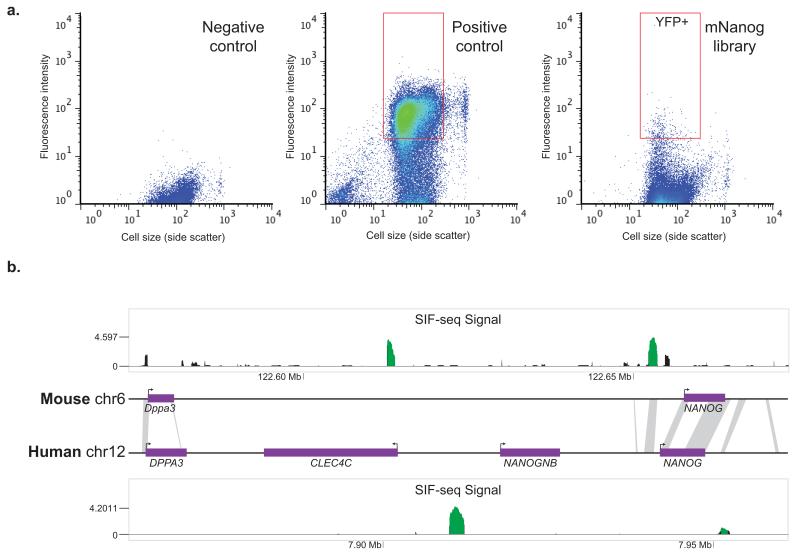
SIF-seq identifies mouse and human ES cell enhancers. a) Representative flow sorting graphs showing YFP reporter gene signal for different cell populations, with each point on the graphs representing a unique cell. Mouse ES cells containing a YFP reporter gene under the control of a DNA sequence with no enhancer activity show no reporter gene expression (Negative control), in contrast to cells containing a strong ES cell enhancer (Positive control). The ES cell library containing random ~1 kb DNA fragments from the mouse Nanog region includes a small population of cells with robust reporter expression (red box), which were isolated and their enhancers identified. b) ES cell enhancers (green peaks) from both the human and mouse NANOG locus were identified using SIF-seq. Grey bars connecting the mouse and human genomic loci indicate the few sites of sequence homology between the two genomes according to the UCSC Genome Browser Net Alignments (genome.ucsc.edu). SIF-seq signal represents log2(normalized fluorescent read depth / normalized input read depth). For simplicity, only SIF-seq signal >0 is shown.
To identify the active ES cell enhancers present in the tested regions, we used fluorescent-activated cell sorting (FACS) to isolate cells with robust reporter expression (Fig. 2a). Populations transfected with two DNA fragments that had no or strong enhancer activity were used as negative and positive controls, respectively, to calibrate the sorting process. Cells from the negative control showed universally low levels of reporter expression, in contrast to the positive control, in which the majority of cells showed very strong YFP expression. Each ES cell library from randomly sheared BACs contained a small population of cells with robust reporter expression and a large population with negligible reporter expression, which is expected considering that any given genomic locus is likely to harbor only a few enhancers active in any given cell type. The YFP-expressing cells, expected to contain an enhancer activating reporter gene expression, were collected by FACS, and the enhancer sequences in these fluorescing cells were amplified by PCR using universal primers that recognize the sequences flanking the enhancer site. Enhancer amplicons were then sequenced using next-generation sequencing technology, and the reads were mapped to the BAC reference sequence. To determine which sequences were tested in each library and to control for biases in the library construction, the candidate sequence positions from an unsorted sample of the library, analogous to a ChIP-seq input sample, were amplified and sequenced in the same manner. Functionally active enhancers were defined as those sequences that showed a statistically significant enrichment in the fluorescing cell population relative to the input control (see Online Methods for a detailed explanation of how statistical significance was determined).
For both BACs, we successfully constructed ES cell libraries containing a diverse collection of DNA fragments that in total randomly covered ~85% of each BAC region (Supplementary Table 1, input samples in Supplementary Figs. 2 and 3a). Both libraries showed strong enrichment of a small number of putative enhancer sequences in the reporter-expressing cell types (Fig. 2b, Supplementary Figs. 2 and 3a). Testing of the same BAC region using different versions of the Hprt targeting plasmid supported the reproducibility of the assay (see Supplementary Note). In the assayed regions, we re-identified a previously described ES cell enhancer ~5 kb upstream of Nanog 22, as well as a new putative enhancer between Nanog and Dppa3, more than 40 kb away from the Nanog transcription start site. We also identified a putative enhancer ~25 kb upstream of Slc2a3 and three putative enhancers downstream of Sall1. Recently published chromatin interaction data support the role of these sites as enhancers by demonstrating a physical interaction between the majority of these putative enhancers and at least one promoter, including those for the genes Nanog, Sall1, Slc2a3, and Gdf3 (Supplementary Fig. 4) 23. These results suggest that SIF-seq is capable of correctly identifying ES cell enhancers present in complex libraries of DNA sequences.
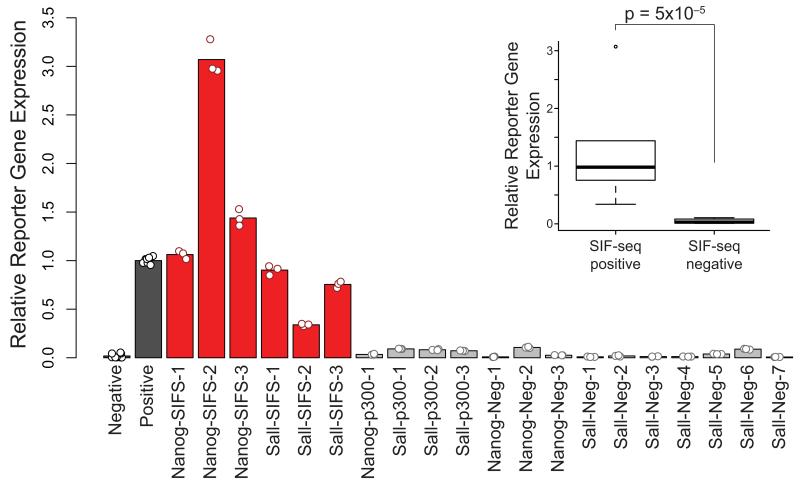
To confirm the accuracy of our enhancer discovery, we individually examined the enhancer activity of the six candidate enhancer sites that were identified by SIF-seq and 14 sites with no predicted activity, including four loci that were negative by SIF-seq but showed strong p300 and/or H3K27ac interaction in mouse ES cell ChIP-seq experiments 3 and ten randomly chosen sites. Each site was cloned, linked to a reporter with a minimal promoter, and integrated into the Hprt locus of mouse ES cells. Reporter gene expression was measured by quantitative reverse transcription PCR (RT-PCR). All six SIF-seq-predicted enhancers showed robust enhancer activity in the validation assay (Fig. 3). This is in contrast to the sequences predicted to be negative by SIF-seq, including the four sites with p300 and/or H3K27ac interaction, all of which had negligible reporter expression. Collectively, the enhancers predicted by SIF-seq drove significantly higher reporter expression than those that were predicted negative (Fig. 3, p = 5×10−5, one-tailed t-test). The high validation rate in these complementary assays demonstrates the accuracy of de novo predictions of enhancers by SIF-seq.
Figure 3.
Validating mouse ES cell enhancers identified by SIF-seq. In order to confirm that SIF-seq accurately identifies enhancers, we selected 6 putative mouse ES cell enhancers predicted by SIF-seq (red bars) and individually tested them for enhancer activity by quantitative RT-PCR. All showed robust reporter expression comparable to the positive control. This is in contrast to sites that did not display enhancer activity by SIF-seq (grey bars), including four sites that had significant p300 and/or H3K27ac interaction. For each sample, reporter gene expression was normalized to the expression of actin and was then normalized to the positive control, which was set at 1. Error bars indicate the standard deviation for technical quantitative RT-PCR replicates (N = 9 for negative control, N = 3 for remaining samples). Positive and negative controls (black bars) are identical to those shown in Figure 2a. Inset: Box plot summarizing RT-PCR results. Putative enhancers identified by SIF-seq drive significantly higher reporter gene expression than SIF-seq negative regions (p = 5×10−5 by one-tailed t-test, whiskers indicate data ranges exclusive of those values represented by individual points).
Identification of Poorly Conserved Human ESC Enhancers
We chose to use mouse ES cells to test the activity of putative enhancers in the context of a native chromosome environment because they are more amenable to targeted genomic alteration than many other mammalian cell types. To explore the potential utility of this approach for mapping and characterizing human noncoding regulatory sequences, we next tested whether SIF-seq using mouse ES cells could be used to identify ES cell enhancers present in the human genome. We built a DNA fragment library from a randomly sheared BAC (RP11-103J24) containing ~160 kb of human sequence encompassing the NANOG gene. Outside of the immediate NANOG locus, the UCSC Genome Browser Net Alignment 24 for this region shows minimal sequence conservation between human and mouse (Fig. 2b). This lack of homology largely prevents the combined use of mouse-derived data and human-mouse sequence orthology to identify distally active enhancers at the human locus. Using SIF-seq, we randomly interrogated enhancer activity in ~1 kb fragments across the BAC region and identified two ES cell enhancers, one just downstream of NANOG and one between NANOGNB and CLEC4C (Fig. 2b, Supplementary Fig. 3b). To confirm that these human sequences are bona fide ES cell enhancers, we validated their activity by individually testing them in mouse ES cells. Both putative enhancers robustly activated reporter gene expression (Supplementary Fig. 5). Furthermore, both sites showed ENCODE chromatin profiles consistent with strong human ES cell enhancer activity (Supplementary Fig. 3b) 25. Of the two human enhancers identified at this locus, neither was identified as an enhancer at the mouse Nanog locus, as they both had little to no sequence conservation to mouse. These data demonstrate the ability of human ES cell enhancers to activate reporter gene expression in mouse ES cells even when the enhancer sequences are not conserved in the mouse genome, thereby highlighting the utility of SIF-seq for the accurate identification of both mouse and human enhancer sequences.
SIF-seq Enhancer Identification in Additional Cell Types
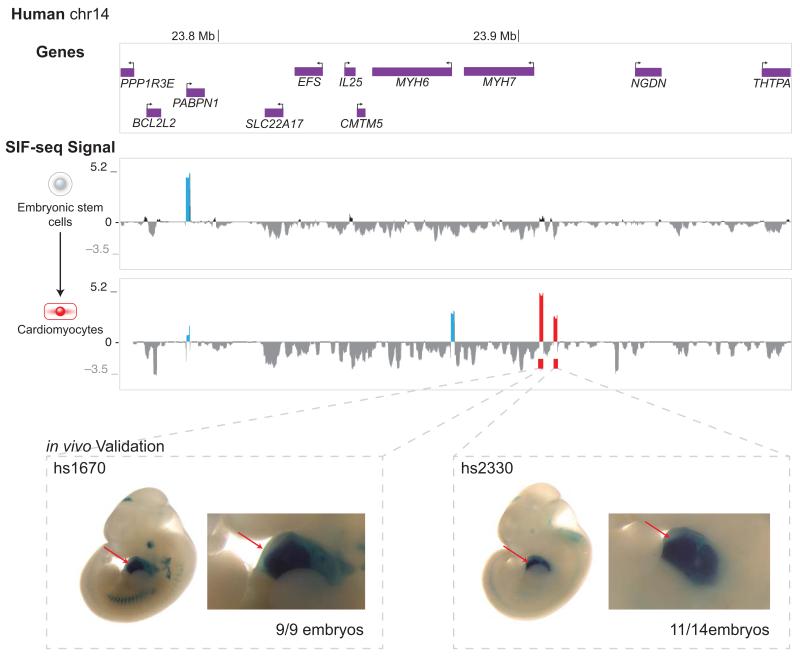
Previously available higher-throughput mammalian enhancer assays rely largely on transient transfection of cells or tissues 16-18, which can increase throughput and sampling depth but severely limits their use to easily transfectable cell types and precludes the discovery of enhancers active in many biologically or disease-relevant cell types. Therefore, we next explored if SIF-seq can be used to identify enhancers in additional, disease-relevant cell types by using in vitro differentiation of ES cells. We constructed libraries by randomly shearing a BAC (RP11-929J10) containing human genes important for heart function, MYH6 and MYH7, and integrated these into the Hprt locus in mouse ES cells as before. SIF-seq was carried out at the initial ES cell stage and upon differentiation to cardiomyocytes. For the MYH6 and MYH7 region, we randomly interrogated enhancer activity in ~1 kb DNA fragments across the BAC at the pluripotent ES cell stage and identified one sequence that was enriched in the reporter expressing cells: the promoter of the ubiquitously-expressed PABPN1 gene (Fig. 4). In ES cells differentiated into cardiomyocytes, four sites were enriched in the reporter expressing cells: the PABPN1 promoter, the MYH6 promoter, and two putative heart enhancers upstream of MYH7. One of these enhancers (hs1670) was previously identified in a larger-scale enhancer screen 13 and drives strong, reproducible reporter gene expression throughout the heart in transgenic mouse reporter assays at embryonic day 11.5 (E11.5) 26 (Fig. 4). We also validated the second putative enhancer, hs2330, using a transgenic mouse assay and found that 11 of 14 embryos had reproducible enhancer activity throughout the heart at E11.5 (Fig. 4). These results provide evidence that mammalian enhancers active in vivo can be identified by performing SIF-seq on ES cells that have been differentiated to mature cell types in vitro.
Figure 4.
SIF-seq accurately identifies cardiac enhancers. The region around human MYH6 and MYH7 was tested with SIF-seq for the presence of ES cell and cardiomyocyte enhancers. In ES cells, only the promoter of the ubiquitously expressed PABPN1 gene (blue peak in SIF-seq ES cell signal plot) was enriched in the reporter-expressing cells. Upon differentiation to cardiomyocytes, two putative enhancers (red peaks) near MYH7, along with the PABPN1 and MYH6 promoters (blue peaks), were enriched in the reporter-expressing cells. Both putative cardiomyocyte enhancers (hs1670 and hs2330) identified through SIF-seq show strong, reproducible activity in the heart (arrow) of transgenic embryonic day 11.5 (E11.5) mice. Mouse embryos have an average crown-rump length of 6mm at E11.5.
Finally, to demonstrate the feasibility of using SIF-seq in neuronal cell types and to show that SIF-seq can be used to validate large numbers of putative enhancers identified by other complementary methods, we pooled together 192 noncoding sequences from throughout the human genome that were identified via comparative genomics approaches as having extreme evolutionary sequence conservation (“ultraconservation”) in vertebrates. These sites had been previously tested for enhancer activity in transgenic E11.5 mice 27,28. The pooled sequences were integrated into the Hprt locus of ES cells as before, and the cells were then differentiated to Nestin-positive neural progenitors. Of the 192 sites, 153 (80%) were successfully tested in the neural progenitor library, and eight were found to be significantly overrepresented in the reporter-expressing cells (Supplementary Fig. 6a). Six of the eight (75%) overrepresented sequences had reproducible enhancer activity in the central nervous system in mice at E11.5 (Supplementary Fig. 6c) 27,28. Despite the in vitro neural progenitor differentiation likely representing an earlier stage in development than E11.5, this is a substantial enrichment of central nervous system enhancers (75% versus 29% of total loci, p<0.05, one-tailed Fisher’s exact test) (Supplementary Fig. 6d). This, together with the results in cardiomyocytes above, demonstrates that SIF-seq can be used to functionally identify enhancers that are active in vivo in a variety of cell types.
Discussion
To address the need for higher-throughput functional assays that assess enhancer activity in a genomic context and that can be used in a wider variety of disease-relevant cell types, we have developed SIF-seq. This method first systematically introduces candidate enhancers into a single reproducible reporter locus in the mouse genome in mouse ES cells and then uses fluorescence-activated cell sorting (FACS) and highly-parallel sequencing to identify those sequences that robustly activate reporter gene expression. We successfully employed the method to test the transcriptional activity of a large series of 1–1.5 kb DNA fragments that in total cover over 500 kb of sequence from the human and mouse genomes. By exploiting ES cell differentiation protocols, we accurately mapped tissue-specific enhancers active in ES cells, cardiomyocytes, and neural progenitor cells. This demonstrates that SIF-seq can be used to identify enhancers in a range of biologically or disease-relevant cell types, limited only by currently available stem cell differentiation methods. Using SIF-seq, we found that the ES cell enhancers present at the NANOG locus differed substantially between mouse and human. These experiments clearly demonstrated that human ES cell enhancers that are not present in the mouse genome can still be identified using reporter assays in mouse ES cells. Although we did not explicitly test the activity of species-specific enhancers, such as those derived from certain classes of repetitive elements 29, these results strongly suggest that SIF-seq can be used to identify enhancers from other mammalian genomes where desired cell types are difficult or impossible to obtain.
By performing unbiased enhancer discovery across several genomic loci, we compared SIF-seq and ChIP-seq-based methods for enhancer discovery. In mouse embryonic stem cells, where we tested the most sequence and had access to the most comparable ChIP-seq data 3, we found that all SIF-seq identified enhancers had robust p300 and H3K27ac interactions. However, many sites that had p300 and/or H3K27ac interactions were not identified as enhancers by SIF-seq. In independent validation assays, all six tested SIF-seq-identified mouse ES cell enhancers had activity, in contrast to all four of the tested SIF-seq-negative sites with p300 and/or H3K27ac interactions. These strong validation results indicate that SIF-seq may predict enhancers more accurately than certain chromatin based methods.
The need for higher-throughput assays to directly interrogate enhancer activity has led to the recent development of multiplex methods to functionally assess genetic regulatory elements 16-18,30-33. However, with the exception of one method to test enhancers in Drosophila embryos 32, all of these methods rely on transient delivery of enhancer-reporter plasmids, limiting their use to a small number of easily transfected cell types. Furthermore, many enhancers have been shown to have negligible activity when tested in transient assays but robust activity when integrated into the genome 34-36. This suggests that transient delivery of enhancer-reporter constructs may not recapitulate the native chromatin environment found in chromosomes, which may be necessary for proper gene regulation. SIF-seq improves substantially on these previous methods by assessing mammalian enhancer activity in a genomic context and in a potentially much wider variety of cell types.
Currently, the number of putative enhancers assessed in a single SIF-seq experiment is limited by the efficiency of site-specific genome integration of the reporter construct in mouse ES cells. For the experiments described, genome integration into the Hprt locus occurred in approximately 1 in 105 mouse ES cells, corresponding to approximately 1,500 individual integration events per library. New genome editing technologies, particularly Cas9 37-39, may further improve this integration efficiency and thereby increase the throughput of this approach. The use of Cas9 or other editing technologies could also potentially allow for the use of SIF-seq in already differentiated cell lines that are not readily amenable to targeted genome integration.
Because enhancers play important roles in development and the mounting evidence for their significant contributions to human disease, the identification of enhancer elements in different cell types and under different biological conditions is currently of high priority in biomedical research. The use of SIF-seq will help to surmount the considerable limitations currently curbing the ability to functionally identify or validate large numbers of putative enhancers directly in many disease-relevant cell types. For example, the expanded use of this method has the potential to decrease the need for transgenic mice in testing enhancers active in specific cell types. In addition to enhancer identification and validation, this method should also be easily adapted to study the effects allelic variants have on enhancer activity, a technique that will become increasingly important as whole genome sequencing is progressively adopted in human disease studies. The further use and development of SIF-seq will allow for the more comprehensive study of the roles enhancers play in human health and disease.
Online Methods
Targeting Vectors
Descriptions of the Hprt targeting vectors used are given in Supplementary Note 1, and the vectors have been made publically available through Addgene (plasmids #51291 and #51292).
Constructing Plasmid Libraries
Bacterial Artificial Chromosomes (BACs) were ordered from the BACPAC Resource Center at Children’s Hospital Oakland Research Institute. BAC DNA was sheared with a Bioruptor XL (Diagenode) or a Sonifier II 450 (Branson), and ~1–1.6 kb long DNA fragments were size selected using agarose gel electrophoresis. We note that although we limited our libraries to this size range, DNA fragments can be sheared to a variety of smaller or larger size ranges to identify larger or smaller enhancers, depending upon the specific application. Size-selected DNA was purified using the QIAquick Gel Extraction Kit (Qiagen). Purified DNA was end-repaired using the End-It DNA End-Repair Kit (epicenter) and purified using the QIAquick PCR Purification Kit (Qiagen) according to manufacturer instructions. A-tailing was carried out in a 50 μL reaction containing 1× NEBuffer 2 (New England Biolabs), 15 U Klenow Fragment (3′➜5′ exo-) (New England Biolabs), and 0.2 mM dATP (Roche) and incubated at 37 °C for 30 minutes. The DNA was again purified using the QIAquick PCR Purification Kit (Qiagen) prior to adaptor ligation.
Cloning adaptors were made using the following HPLC-purified oligos: Adaptor-attB1 and Adaptor-attB2 (Supplementary Table 2). Adaptor oligos were mixed in equimolar amounts and prepared by denaturing at 95 °C on a heat block and annealing by allowing the heat block to slowly return to room temperature.
Cloning adaptors were ligated to the library of DNA fragments using the NEBNext Quick Ligation Module (New England Biolabs) according to manufacturer instructions with a 2 μM final concentration of cloning adaptors. The DNA library was purified from the unligated adaptors using either 35 μL of AMPure XP beads (Beckman Coulter) per 50 μL ligation reaction according to manufacturer instructions (hNANOG and mNanog libraries) or agarose gel electrophoresis size selection (all remaining libraries). The DNA concentration of the fragment library was measured using the Qubit dsDNA HS Assay and a Qubit Fluorometer (Life Technologies). Correctly adapted DNA fragments were enriched by PCR amplification in a 50 μL PfuUltra II Fusion HS DNA Polymerase (Agilent) reaction using 5-10 ng of library DNA and the following primers: attB1F and attB2R (Supplementary Table 2). Cycling conditions were as follows: initial 95 °C denaturation for 2 minutes, 20 cycles of amplification (denaturation at 95 °C for 20 seconds, primer annealing at 65 °C for 20 seconds, and extension at 72 °C for 15 seconds), and final extension at 72 °C for 3 minutes.
After amplification, 1–1.6 kb library fragments were again size selected on a 1% agarose gel and QIAquick Gel Extraction Kit purified as described above except that the library was eluted in a final volume of 25 μL. The DNA libraries were cloned into either pSKB1-GW-hsp68-Venus (mSall1 and Ultraconserved libraries) or pSKB1-Venus-H19 (all remaining libraries) using a single tube, two-step Gateway cloning reaction (Life Technologies) as follows: up to 100 ng of the PCR amplicon library was incubated with 200 ng pDONR221 plasmid and 3 μL BP Clonase II Enzyme Mix in a 15 μL total reaction volume. After a room temperature incubation for ~20 hours, 10 μL of this reaction was mixed with 2 μL of 150 ng/μL pSKB1-GW-hsp68-Venus or pSKB1-Venus-H19 and 3 μL LR Clonase II Enzyme Mix and incubated at room temperature for ~20 hours.
Plasmid libraries were transformed by electroporating 1 μL of each LR Clonase II reaction into 50 μL of One Shot TOP10 Electrocomp E. coli (Life Technologies). After a 30 minute recovery incubation in rich SOC medium, the transformed cells were transferred to 500 mL LB medium containing 200 μg/mL ampicillin and grown at 37 °C for ~12.5 hours. Plasmid DNA was isolated using the Plasmid Maxi Kit (Qiagen) with manufacturer recommended protocol modifications for large, low-copy plasmids. Plasmid DNA was linearized with PmeI (NEB) and then purified by phenol-chloroform extraction, washed with 70% ethanol, and resuspended in buffer containing 10 mM Tris-HCl and 0.1 mM EDTA with a pH of 7.5.
Making Embryonic Stem Cell Libraries
All experiments used the E14Tg2a.4 40 male mouse ES cell line, which has a 36 kb X chromosome deletion that removes the first two exons of the Hprt gene. Cells were grown under feeder free conditions on gelatin coated plates and fed standard ES cell medium: Knockout DMEM (Life Technologies) containing 15% fetal bovine serum (HyClone), 2 mM L-glutamine (Life Technologies), 0.1 mM nonessential amino acids (Life Technologies), 0.05 mM 2-mercaptoethanol (Sigma), 1,000 U/mL ESGRO® LIF (Millipore), and penicillin-streptomycin. Cells were fed daily. Approximately 20 ug of linearized plasmid DNA was transfected into 1–1.5×107 ES cells in 0.8 mL HEPES buffered saline (Sigma-Aldrich) using a Gene Pulser Xcell™ (Bio-Rad) set to 250 V and 500 μF. Ten such transfections were performed for each library. Correctly targeted cells were selected by the addition of Hypoxanthine-aminopterin-thymidine (HAT) Supplement (Life Technologies) to the ES cell medium for 3–10 days, beginning 24 hours after transfection. Following HAT selection, cells were fed ES cell medium containing 1× HT supplement (Life Technologies) for two days. Cells from the same library were pooled together and expanded on fresh plates prior to sorting.
Sorting
Prior to sorting, cells were washed with phosphate buffered saline (PBS) and harvested with trypsin. Cells were pelleted by centrifugation, the trypsin was removed, and the cells were washed with PBS. Cells were resuspended in 1% w/v saline by repeated pipetting and passed through a 0.4 μm strainer to ensure single cell suspension. Cells were sorted on an Influx cell sorter (BD Biosciences) using Spigot Version 6.1.10 software (BD Biosciences). Flow cytometry metrics were analyzed using FlowJo Version 7.6.3 (TreeStar).
In vitro Differentiation
Mouse ES cell differentiation to cardiac Troponin T-expressing cardiomyocytes was carried out as previously described 41,42. Differentiation to neural progenitors was carried out as previously described 43,44 without the use of cyclopamine, and cells were harvested on Day 14. Differentiated cells were fixed in 4% paraformaldehyde for 15 minutes and washed with PBS prior to sorting.
PCR Amplifying Inserts and Sequencing
DNA was isolated from both YFP-expressing and unsorted control populations of cells using the QIAamp DNA Mini Kit (Qiagen). The enhancer position sites were amplified from the genomic DNA in 50 μL Platinum® Taq DNA Polymerase High Fidelity reactions (Life Technologies) containing the attB1F and attB2R primers (Supplementary Table 2) and up to 100 ng of genomic DNA. Cycling conditions were as follows: initial 94 °C denaturation for 2 minutes followed by 30 cycles of amplification (denaturation at 94 °C for 30 seconds, primer annealing at 55 °C for 30 seconds, and extension at 68 °C for 90 seconds). Five PCR reactions were performed for each sample and pooled prior to subsequent purification with AMPure XP beads. PCR amplicons were sequenced using a PacBio RS (Pacific Biosciences).
Data Analysis and Peak Calling
Sequence reads were aligned to reference sequences using the RS_Resequencing workflow within the Pacific Biosciences SMRT Portal. Mapped read coverage from the sorted and unsorted libraries were scaled by the total number of sequenced bases, and coverage values were increased by a small amount to reduce signal volatility driven by very low coverage. Corrected coverage estimates from both libraries were used to generate log2 ratios for the sorted/unsorted coverage at each base position across the tested region. A sliding-window algorithm was used to identify subregions where the sorted coverage was significantly (p<0.05) enriched versus the unsorted coverage, which represent functionally validated enhancers via this screen. Enriched subregions were required to be at least 800 bp long and to be at least 1.5-fold enriched for sorted versus unsorted coverage. P-values for enriched regions were generated by comparing the highest enrichment value in the enriched subregion to the distribution of enrichment values from the remainder of the full tested region. Computer source code will be provided upon request.
P300 and H3K27ac ChIP-seq was performed previously 3. For H3K27ac, signal and peak calls were obtained directly from the UCSC ENCODE web portal (genome.ucsc.edu). For p300, the resulting fastq data files from ChIP and input library sequencing were downloaded from the UCSC ENCODE web portal, reads were aligned to the mouse genome (mm9) using the BWA aligner (call: bwa aln -t 6 -l 25 mm9 sample.fastq.gz), and peaks were called using MACS (call: macs14 -t chip.bam --control=input.bam -name=chip_output --format=BAM --gsize=mm --tsize=50 --bw=300 --mfold=10,30 --nolambda --nomodel --shiftsize=150 -p 0.00001).
Enhancer Controls and Validation
Enhancer control loci and sites chosen for validation were amplified using primers listed in Supplementary Table 2. Sites validated in ES cells were cloned into an Hprt targeting vector using Gateway cloning and individually targeted to the Hprt locus of E14 cells as above. After HAT selection, cells were transferred and expanded on fresh plates before they were harvested.
For ES cell enhancer validation, whole RNA was isolated from each cell line using the RNAqueous Kit (Life Technologies). RNA was treated with RNase-free DNase (Promega) and reverse transcribed using SuperScript III (Life Technologies) with random hexamer priming. YFP reporter expression was measured on a LightCycler 480 (Roche) using 20 μL manufacturer-recommended LightCycler 480 Probes Master reactions that included 1) primers YFP_F and YFP_R (Supplementary Table 2) to amplify YFP, 2) Universal ProbeLibrary Probe #67 (Roche), and 3) the Mouse ACTB Gene Assay (Roche), to measure actin expression, as a control. Quantitative RT-PCR results were assessed by the 2−ΔΔCt method 45, using actin expression to normalize YFP expression.
Cardiac enhancers were validated in transgenic mouse assays as previously described 26,27. All animal work was reviewed and approved by the LBNL Animal Welfare and Research Committee.
Supplementary Material
Acknowledgments
We would like to thank S. Bronson (Pennsylvania State University) for the pSKB1 plasmid and A. Miyawaki (RIKEN) for the Venus-YFP gene. We would also like to thank R. Malmstrom, K. Singh, and Z. Zhao for technical help. A.V. and L.A.P. were supported by US National Institute of Health (US NIH) grants U01DE020060NIH, R01HG003988, and U54HG006997. D.E.D. was supported by US NIH grant 5T32HL098057 (to Children’s Hospital Oakland Research Institute). B.G.B. was supported by the US NIH Bench to Bassinet Program (U01HL098179). A.K. and B.G. were supported by the UK National Centre for the Replacement, Refinement and Reduction of Animals in Research, the UK Biotechnology and Biological Sciences Research Council, and core support grants by the Wellcome Trust to the CIMR and Wellcome Trust–MRC Cambridge Stem Cell Institute. Research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under US Department of Energy Contract DE-AC02-05CH11231, University of California.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
Accession codes: All sequence files are available through SRA: SRP034877.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinjan DA, Lettice LA. Long-Range Gene Control and Genetic Disease. Adv. Genet. 2008;61:339–388. doi: 10.1016/S0065-2660(07)00013-2. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Cowper-Sal Lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 2012;22:1437–1446. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 10.Yanagisawa H, Clouthier DE, Richardson JA, Charité J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- 11.Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danielian PS, Echelard Y, Vassileva G, McMahon AP. A 5.5-kb enhancer is both necessary and sufficient for regulation of Wnt-1 transcription in vivo. Dev Biol. 1997;192:300–309. doi: 10.1006/dbio.1997.8762. [DOI] [PubMed] [Google Scholar]
- 13.Attanasio C, et al. Fine Tuning of Craniofacial Morphology by Distant-Acting Enhancers. Science. 2013;342 doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotney J, et al. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature Biotechnology. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nature Biotechnology. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bronson SK, et al. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, et al. Vascular bed-specific regulation of the von Willebrand factor promoter in the heart and skeletal muscle. Blood. 2011;117:342–351. doi: 10.1182/blood-2010-06-287987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki A, et al. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kothary R, et al. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 27.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 28.Visel A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward MC, et al. Latent regulatory potential of human-specific repetitive elements. Mol Cell. 2013;49:262–272. doi: 10.1016/j.molcel.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 31.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nature Biotechnology. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gisselbrecht SS, et al. Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos. Nat Methods. 2013;10:774–780. doi: 10.1038/nmeth.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam J, Dong P, Tarpine R, Istrail S, Davidson EH. Functional cis-regulatory genomics for systems biology. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3930–3935. doi: 10.1073/pnas.1000147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuan DY, Solomon WB, London IM, Lee DP. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human ‘beta-like globin’ genes. Proc Natl Acad Sci USA. 1989;86:2554–2558. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser P, Hurst J, Collis P, Grosveld F. DNaseI hypersensitive sites 1, 2 and 3 of the human beta-globin dominant control region direct position-independent expression. Nucleic Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hug BA, Moon AM, Ley TJ. Structure and function of the murine beta-globin locus control region 5′ HS-3. Nucleic Acids Res. 1992;20:5771–5778. doi: 10.1093/nar/20.21.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skarnes WC. Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Meth. Enzymol. 2000;328:592–615. doi: 10.1016/s0076-6879(00)28420-6. [DOI] [PubMed] [Google Scholar]
- 41.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaspard N, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 44.Gaspard N, et al. Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc. 2009;4:1454–1463. doi: 10.1038/nprot.2009.157. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.