Abstract
Efforts to develop more effective therapies for acute leukemia may benefit from high-throughput screening systems that reflect the complex physiology of the disease, including leukemia stem cells (LSCs) and supportive interactions with the bone-marrow microenvironment. The therapeutic targeting of LSCs is challenging because LSCs are highly similar to normal hematopoietic stem and progenitor cells (HSPCs) and are protected by stromal cells in vivo. We screened 14,718 compounds in a leukemia-stroma co-culture system for inhibition of cobblestone formation, a cellular behavior associated with stem-cell function. Among those that inhibited malignant cells but spared HSPCs was the cholesterol-lowering drug lovastatin. Lovastatin showed anti-LSC activity in vitro and in an in vivo bone marrow transplantation model. Mechanistic studies demonstrated that the effect was on-target, via inhibition of HMGCoA reductase. These results illustrate the power of merging physiologically-relevant models with high-throughput screening.
INTRODUCTION
The hematopoietic niche is a complex mixture of heterogeneous cell types that supports normal hematopoiesis and is hypothesized to play a chemoprotective role in the treatment of acute myeloid leukemia (AML), possibly contributing to the failure of standard of care chemotherapy to cure at least half of adult patients with this disease1-3. Within AML is a population of cells with the capacity for self-renewal, disease initiation, and disease propagation termed leukemia stem cells (LSCs)4. These cells are less sensitive to mainstay AML chemotherapies such as daunorubicin and cytarabine5,6 and are particularly responsive to a number of supportive stromal factors, including interleukin-6 (IL-6), stromal cell-derived factor-1 (SDF-1), interleukin-8 (IL-8), and angiopoietin-13,7, further blunting the cytotoxic effects of chemotherapy.
Strategies to target LSC dependencies within the context of the bone-marrow microenvironment are therefore attractive, however, two major obstacles have made such therapeutic targeting challenging in practice. First, many of the liabilities identified to date in leukemia cells also exist in normal hematopoietic stem and progenitor cells (HSPCs) due to the biological similarity of these populations1,8. Illustrating this, the dose-limiting toxicity for standard of care AML treatments, including cytarabine and daunorubicin, is toxicity to normal HSPCs9,10. As such, discovering therapeutics that target LSCs but spare HSPCs is difficult. Second, to date there has not been a way to model complex phenotypes of primary leukemia cells within the bone-marrow niche in a manner compatible with high-throughput small-molecule screening. Such screening requires that cells be grown in microtiter plates with a reproducible, automated readout. This is particularly problematic in the case of LSCs and HSPCs, whose stem-associated properties are recognized ex vivo via the formation of ”cobblestone areas” (the burrowing of primitive cells beneath a layer of stromal fibroblasts, forming phase dark areas of Cobblestone Area-Forming Cells (CAFCs) organized in a tight association), generally requiring a highly trained eye to detect microscopically by phase contrast11-13.
We reasoned that a high-throughput screening system capable of supporting primary cells in the context of a simulated bone-marrow niche might enable the discovery of leukemia-selective compounds not otherwise identified using standard cell line-based viability screens. We report here the development of such a system involving the co-culture of primary LSC-enriched cells with bone-marrow stromal cells, coupled to an automated machine-learning algorithm capable of recognizing the CAFC phenotype. A small-molecule screen identified novel compounds that inhibited leukemic CAFCs while sparing normal HSPCs, as well as compounds previously established as LSC-selective. A subset of the compounds identified were not readily apparent by traditional cell line screening, illustrating the limitations of conventional methods. These experiments demonstrate the feasibility of physiologically-relevant small-molecule screening within a niche-like microenvironment. Moreover, the panel of compounds identified may represent starting points for new types of AML therapies.
RESULTS
Sustaining Primary Leukemia in a Niche-like Environment
To generate primary leukemia cells for high-throughput study, we used a well-characterized mouse model of human AML driven by the oncogene MLL-AF93,8,14. The LSC-enriched fraction of cells (hereafter referred to as “LSCe cells”) was isolated from the bone marrow of leukemic animals by fluorescence-activated cell sorting (FACS) (Supplementary Results, Supplementary Figs. 1a, b and Methods). Importantly, as LSCe cells are difficult to maintain over extended periods in vitro when cultured in isolation8, we developed a co-culture system to support these cells ex vivo and to enable cobblestone area formation. Historically, ex vivo maintenance of normal HSCs has required co-culture with supportive stroma, and stem-cell activity has been most faithfully quantified ex vivo by cobblestone area formation in the Cobblestone Area-Forming Cell (CAFC) assay or by colonies arising from cobblestone areas in the Long-Term Culture-Initiating Cell assay12,13. Primary leukemia cells have similarly been examined11; however, these assays have not been attempted at high-throughput scale. Toward that goal, we plated dsRed+ LSCe cells in 384-well format onto two types of supportive GFP+ bone marrow-derived stromal cells in order to identify reproducible effects: primary bone marrow mesenchymal stromal cells derived from actin-GFP mice or GFP-expressing bone marrow stroma-derived OP9 cells (see Methods).
LSCe cells co-cultured with either stroma grew robustly in the absence of cytokine supplementation, forming distinct cellular aggregates beneath the stroma indicative of cobblestone area formation (Fig. 1a). Moreover, cell culture media that had been preconditioned by OP9 stromal cells for 3 days augmented cobblestone area formation beneath this type of stromal monolayer, suggesting that secreted factors contribute to the CAFC phenotype. We also found that the enrichment for stem-cell activity using c-Kithi produced higher frequencies of CAFCs, as predicted, relative to c-Kitlo cells seeded at the same density (Supplementary Fig. 1c and Methods). To affirm the in vivo leukemia-initiating potential of the LSCe cells in our model, we co-cultured 25, 100, or 500 LSCe cells with bone marrow stromal cells and transplanted the contents into lethally irradiated mice 6 days later. All recipient animals developed leukemia within 30 days, indicating the intrinsic leukemogenicity of the LSCe cells and its persistence in the high-throughput co-culture system (Supplementary Fig. 1d).
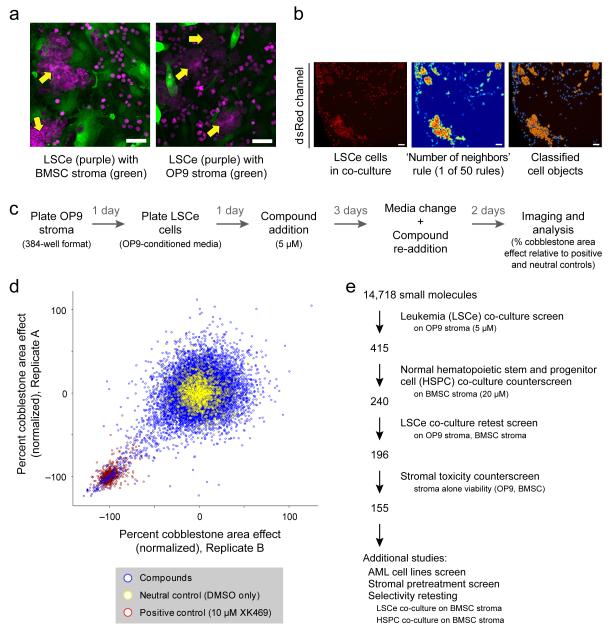
Figure 1. A High-Throughput System for Probing Primary, Stem-enriched Leukemia Cells Within a Stromal Niche.
(a) LSCe cells generate cobblestone area-forming cell (CAFC (arrows)) and non-CAFC morphologies when plated with bone marrow stroma (primary BMSCs or OP9 cells), at the 6-day assay endpoint. Qualitative images are shown as a visual introduction to the devised assay. The red channel has been pseudocolored to accommodate colorblindness. Scale bars represent 50 μM. (b) A representative example of the “Number of neighbors” rule, one of 50 computational rules that together identify CAFCs. A raw image of LSCe cells in co-culture (left, dsRed channel) is converted to a heatmap (middle) reflecting the number of adjacent cells (red = many, blue = few), identifying cell “objects “(right; see Methods) that are part of a cobblestone area (orange = CAFC, blue = non-CAFC). Qualitative images are shown as a conceptual introduction to the computational method. Scale bars represent 50 μM. (c) Schematic of the small-molecule co-culture screen protocol. (d) The co-culture screen data for 14,718 compounds (blue) added at ~5 μM, in duplicate. Values were normalized to the means of both neutral (DMSO carrier, yellow, set at 0% effect) and positive (10 μM XK469, a topoisomerase IIβ inhibitor, red, set at −100% effect) controls. (e) Schematic summarizing the filtering steps used to define high priority compounds. The number of compounds prioritized at each step is shown.
To enable high-throughput studies, we also developed an image analysis pipeline by incorporating CellProfiler software15 to enumerate the CAFC phenotype in the dsRed channel based on the most discriminating morphological features of the CAFCs. The machine-learning algorithm16 used several hundred manually-selected example images of CAFC and non-CAFC cell images to generate a set of 50 morphological ‘rules’ (for example, shape, intensity, texture, and cell neighbor relationships) that defined CAFCs (Fig. 1b, Supplementary Table 1, Methods, and Supplementary Fig. 1e). Tests on an independent set of images indicated an accuracy sufficient for screening, with a sensitivity of 86% and a specificity of 87% compared to manual review by experts (see Methods).
Small-Molecule Screening for Leukemia Selectivity
We screened in duplicate 14,718 small-molecule compounds including ~1,920 bioactive molecules, 1,600 natural products, and 2,880 compounds generated via diversity oriented synthesis (DOS)17 (Fig. 1c, Supplementary Table 2, and Methods), and determined that compounds inhibiting CAFCs by at least 85% were detected with 98% confidence (Supplementary Fig. 1f). Using this approach, we identified 415 compounds that inhibited leukemic CAFCs in co-culture (Fig. 1d and Methods).
We next excluded 145 of the 415 hits that caused gross stromal cell toxicity and/or killed normal HSPCs, based on data from a screen of the same compounds at ~20 μM on HSPCs (Linlo Sca-1+ c-Kit+ CD48lo, from β-actin-dsRed mice) co-cultured with primary bone marrow stromal cells (Stewart, A.L. et al., in preparation; see Methods).
240 of the remaining compounds were retested in 8-point dose response (from ~150 nM to 20 μM) on LSCe cells in co-culture with OP9 stromal cells and in co-culture with bone marrow stromal cells. 196 compounds exhibited a dose response with an EC50 ≤ 5 μM in the presence of one of the stromal types, and 139 compounds displayed activity in the presence of both types (see Methods). To exclude the possibility that CAFC inhibition was a result of direct stromal toxicity, we also tested the compounds in 8-point dose on OP9 or bone marrow stromal cell monolayers in the absence of LSCe cells using a CellTiter-Glo (Promega) viability readout. 36 compounds exhibited significant stromal toxicity at concentrations below 20 μM (Supplementary Fig. 1g) and were excluded from further study. Together, these filters identified155 prioritized compounds that inhibited LSCe cells relative to normal HSPCs and lacked significant toxicity to two bone-marrow stromal cell types (see schematic Fig. 1e and Supplementary Table 3).
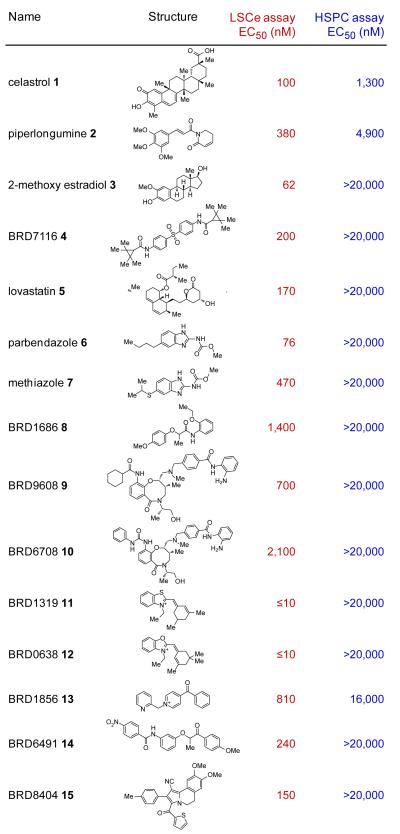
We more rigorously assessed the selectivity of the prioritized hits by examining the compounds at 8 concentrations against both normal HSPCs and LSCe cells, each co-cultured on primary bone marrow stromal cells. A subset, prioritized on the basis of potency and selectivity, was subsequently re-examined (see Methods). Several of these compounds displayed greater than 100-fold selectivity against leukemia cells compared to normal HSPCs (Fig. 2). Importantly, compounds known to have preferential activity against LSCs, including celastrol, a triterpenoid antioxidant compound18, and parthenolide, a sesquiterpene lactone19, were among the 155 validated hits from the screen (Fig. 2 and Supplementary Table 3). Moreover, parthenolide is known to have activity against leukemia stem cells in vivo20. We also identified 2-methoxyestradiol, a microtubule inhibitor known to lack myelosuppressive side effects due to its inability to bind the tubulin protein expressed in HSPCs21 (Fig. 2). These results indicate that the LSCe co-culture assay yields physiologically-relevant insights and starting points for therapeutic discovery. Among the novel compounds we identified were two benzimidazole carbamates, parbendazole and methiazole, used as antihelminthic agents in cattle feed (Fig. 2 and Supplementary Fig. 2a).
Figure 2. Prioritized Screening Hits Display Selective Toxicity to LSCe Cells Relative to HSPCs in Co-culture.
The names and structures of 15 prioritized small-molecule screening hits are shown, with EC50 values (from dose-response curves across 8 concentrations) for both LSCe cells (as percent cobblestone area effect; 2 replicates per concentration) and HSPCs (as total cells per well; 6 replicates per concentration) co-cultured under identical conditions on primary BMSC stromal cells. Celastrol, piperlongumine, and 2-methoxy estradiol were previously identified, validating the screening approach.
To determine if the co-culture assay could reveal compounds not readily identified in standard screens of AML cell line viability, we tested the 155 compounds on six human AML cell lines (U937, THP-1, NOMO-1, SKM-1, NB4, and OCI-AML3), two of which (NOMO-1, THP-1) harbor the MLL-AF9 oncogene present in the primary leukemia cells used in our initial screen. WE tested the compounds at 8 concentrations in the absence of co-culturing using standard conditions and a viability readout (CellTiter-Glo). Whereas a subset of compounds, such as parbendazole and methiazole, also displayed potent activity in the AML cell lines (EC50 < 0.625 μM for each of 6 cell lines; Supplementary Fig. 2b and Supplementary Table 3), other compounds showed greater than 10-fold higher potency on primary LSCe cells in co-culture compared to their mean potency across the AML cell lines (Supplementary Table 3). We also identified compounds with greater activity against the cell lines than the co-cultured LSCe cells, indicating that primary LSCe cells are not merely non-specifically more sensitive than established cell lines (Supplementary Table 3). As described below, we next selected two compounds for more extensive follow-up because they were exemplary of the novel biology exposed by the screen: BRD7116 as an example of cell-non-autonomous anti-leukemia activity, and lovastatin as an example of compounds that would have been overlooked with conventional cell line-based screening.
BRD7116 Displays Cell-non-autonomous Anti-leukemia Activity
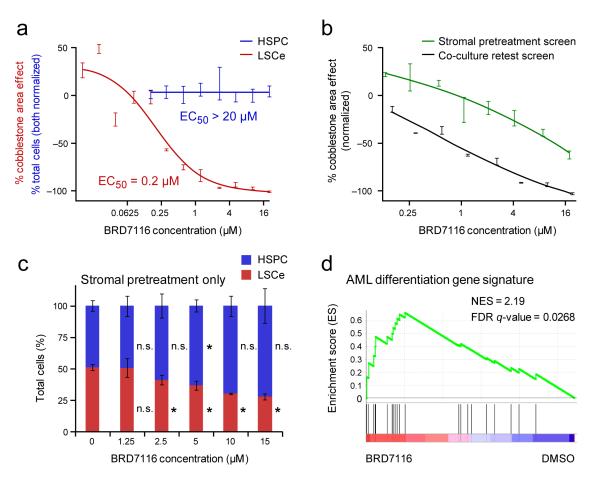
To identify compounds that inhibit LSCe cells via activity on the stromal cells (a property that would not be discoverable using standard cell line screening) we pretreated OP9 stroma at 8 concentrations for 3 days, and washed the stroma prior to the addition of LSCe cells, and then imaged the co-cultures 6 days later (Supplementary Fig. 3a). We hypothesized that inhibition of cobblestone area formation under these conditions would be a result of activity of the compound on the stromal cells, as the LSCe cells were not directly exposed. All 155 hits were tested in this way. Troglitazone, a peroxisome proliferator-activated receptor-γ (PPAR-γ) agonist used for the treatment of diabetes22, inhibited leukemic CAFCs in the stromal pretreatment screen yielding a dose-response curve similar to that observed in co-culture (Supplementary Fig. 3b). This inhibition was accompanied by a dose-dependent adipocytic change in the stroma (Supplementary Fig. 3c), consistent with the established ability of PPAR-γ agonists to regulate adipogenesis23. OP9 stromal cells can readily differentiate into adipocytes24, and adipocytes are known to antagonize hematopoietic cell self-renewal25. Consistent with a cell-non-autonomous mechanism, none of the AML cell lines tested was sensitive to troglitazone at concentrations as high as 35 μM (Supplementary Fig. 3d).
A novel compound, BRD7116, a bis-aryl sulfone, also showed evidence of stroma-mediated anti-LSCe activity. BRD7116 exhibited an EC50 of 200 nM for LSCe cells in co-culture (Fig. 3a), whereas it displayed limited activity against normal HSPCs (Fig. 3a) and AML cell lines (~50% inhibition at 20 μM) (Supplementary Fig. 3e). BRD7116 also showed activity against patient-derived, primary human leukemia cells (Supplementary Fig. 3f). Furthermore, pretreatment of the stroma partially recapitulated the leukemic CAFC inhibition observed in co-culture (Fig. 3b) without evidence of altered stromal cell viability (CellTiter-Glo). In contrast to troglitazone, this niche effect was not accompanied by an obvious change in stromal morphology.
Figure 3. Novel Small Molecule BRD7116 Selectively Targets LSCe Cells by Cell-Autonomous and Cell-Non-Autonomous Mechanisms.
(a) Dose-response effects of bis-aryl sulfone hit BRD7116 on LSCe cells (red, in duplicate) and normal HSPCs (blue, 6 replicates per dose) co-cultured on primary BMSC stroma, shown overlaid. BRD7116 was applied to the co-cultures in both cases. (b) The effects of treating only the stromal monolayer prior to co-culture are compared to the effects of treating both the stroma and LSCe cells together in co-culture. The data shown is from the stromal pretreatment secondary screen in which OP9 stromal cells were treated with BRD7116 for three days prior to LSCe cell plating (green). The LSCe co-culture retest data with OP9s (black) is also shown. Both screens were performed in duplicate. (c) The effects of treating only the stromal monolayer with BRD7116 for three days prior to the plating of admixed LSCe cells and HSPCs on primary BMSC stroma (as mean +/− SEM of quadruplicate replicates). These two hematopoietic populations are quantified as percent of total hematopoietic cells under each stromal pretreatment condition. * p < 0.05; n.s. = not significant, for effects relative to DMSO (0 μM BRD7116) controls. (d) Compared to DMSO control, gene expression changes present in LSCe cells after 6 hours of direct treatment with 5 μM BRD7116 are significantly enriched for an AML differentiation gene signature, as defined by all-trans retinoic acid (ATRA) treatment of human AML cells, by Gene Set Enrichment Analysis (GSEA).
We next directly compared the relative inhibition of LSCe cells and HSPCs under internally controlled conditions using a 3-component co-culture system. We added dsRed+ LSCe cells and GFP+ HSPCs (from ubiquitin C-GFP mice) simultaneously to uncolored bone marrow stromal cells pretreated for 3 days with BRD7116. In contrast to DMSO pretreatment controls, BRD7116 pretreatment of the stroma preferentially inhibited the LSCe cells compared to HSPCs (Fig. 3c, with stromal viability quantifications shown in Supplementary Fig. 3g).
Having observed a cell-non-autonomous effect, we next explored whether BRD7116 had any cell-autonomous effects on LSCe cells. We exposed LSCe cells for 6 hours to 5 μM BRD7116 in suspension, then performed gene-expression profiling. Using Gene Set Enrichment Analysis (GSEA)26,27, we found that treatment of LSCe cells with BRD7116 induced transcriptional changes consistent with myeloid differentiation, as defined by all-trans retinoic acid (ATRA) treatment of ATRA-sensitive human AML cells28 (Fig. 3d). The mechanism by which BRD7116 has this differentiation-inducing effect remains to be determined.
Lovastatin Kills LSCs by HMG-CoA Reductase Inhibition
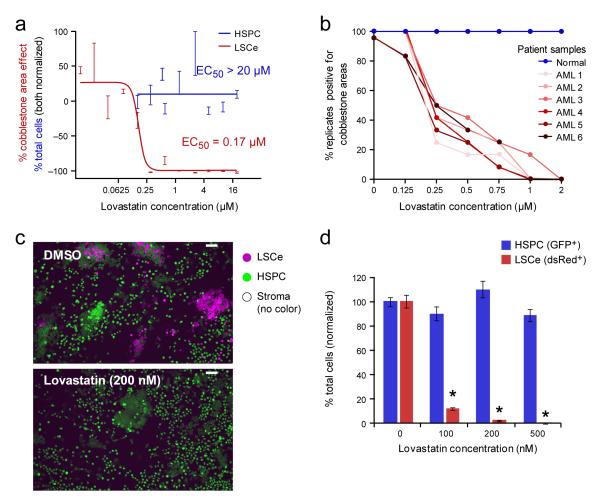
Lovastatin, an FDA-approved drug in wide clinical use for hypercholesterolemia, also emerged from the co-culture screen, inhibiting LSCe cobblestone area formation with an EC50 < 200 nM while showing limited activity against human and murine AML cell lines (EC50 > 2,000 nM) and no apparent toxicity to normal HSPCs at concentrations as high as 20 μM (Fig. 4a and Supplementary Figs. 4a, b, c). Of note, the inhibitory effects of lovastatin were cell-autonomous, as lovastatin did not score in the stromal pretreatment assay described above. In addition, LSCe cells generated using an alternative oncogene, MOZ-TIF229, were also sensitive to lovastatin in co-culture (EC50 < 250 nM) (Supplementary Fig. 4d), indicating that the effect was not MLL-AF9-specific.
Figure 4. Lovastatin Selectively Inhibits Murine and Human Leukemia Cells in Co-culture.
(a) Lovastatin displays leukemia-selective activity (red dose-response curve, in duplicate) compared to HSPCs (blue, 6 replicates per dose) in co-culture with BMSCs. (b) The effects of lovastatin on CAFC activity of primary human CD34+ cells isolated from either normal or leukemic patient samples. Co-cultures were treated for 6 days, and then rinsed. The fraction of replicate co-cultures containing cobblestone areas at the 5-week assay endpoint (2 weeks for FLT3-ITD sample) is shown (n ≥ 6). The clinical characteristics of the AML samples are as follows, including whether each AML case is primary (P) or directly related to previous cytotoxic therapy, as therapy-related (TR) leukemia is associated with a worse prognosis than de novo disease. AML 1: FLT3-ITD+ (P), AML 2: NPM1 mutation (P), AML 3: CBFB-MYH11 fusion (P), AML 4: trisomy 11, del(17p) (TR), AML5: CEPBα double mutation (P), AML 6: MLL-AF9 fusion (TR). (c) Representative images of admixed LSCe cells and HSPCs co-cultured with uncolored primary BMSCs at 5 days of exposure to lovastatin. The red channel has been pseudocolored to accommodate colorblindness. Scale bars represent 50 μM. (d) Quantification of the effect (mean +/− SEM of quadruplicate replicates) is also shown and is representative of two independent experiments. * p < 0.001 for LSCe cells relative to DMSO (0 μM lovastatin) controls.
We next extended the lovastatin studies to primary human leukemia cells, using longer-duration CAFC assays. CD34+ cells purified from umbilical cord blood or leukemic CD34+ cells purified from the bone marrow of leukemic patients11,12 were co-cultured with stromal cells and treated for 6 days with lovastatin or DMSO, rinsed, and cobblestone area formation was assessed at 5 weeks (2 weeks for a leukemia harboring the FLT3-ITD oncogene30). Lovastatin inhibited cobblestone area formation for all 6 patient-derived AML samples with potencies comparable to that seen in the murine co-culture system, whereas no inhibition of normal HSPC cobblestone area formation was observed under these same conditions (Fig. 4b and Supplementary Table 4).
To directly compare the relative inhibition of LSCe cells and HSPCs under internally controlled conditions, we plated heterotypic cultures consisting of dsRed+ LSCe cells, GFP+ HSPCs and uncolored primary BMSCs. 24 hours later we treated the co-cultures with 200 nM lovastatin or DMSO and assessed them 5 days later. In contrast to the DMSO controls, lovastatin selectively inhibited the dsRed+ leukemia cells while sparing the GFP+ HSPCs, which continued to display normal, CAFC morphologies (Fig. 4c, d).
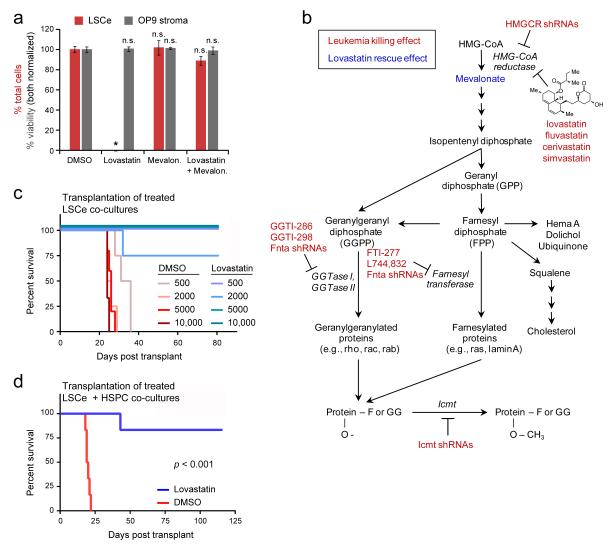
Statins act to lower cholesterol levels by inhibiting HMG-CoA reductase (HMGCR), the enzyme that catalyzes the rate-limiting step of cholesterol biosynthesis31. To evaluate the possibility that lovastatin was acting by an alternative mechanism, we tested the ability of mevalonate, the metabolite immediately downstream of HMGCR, to rescue the observed lovastatin effects. Consistent with an HMGCR on-target mechanism, mevalonate fully rescued the anti-leukemia effects of lovastatin in co-culture (Fig. 5a), and five additional statins (simvastatin, fluvastatin, cerivastatin, rosuvastatin and atorvastatin) selectively inhibited LSCe cells relative to normal HSPCs (Supplementary Table 5).
Figure 5. Sensitivity of LSCe Cells to HMGCR Inhibition.
(a) The addition of 2 mM mevalonate to co-cultures of LSCe cells with OP9 stroma rescued the anti-leukemia effect of 1 μM lovastatin, with no stromal toxicity observed. Data are mean +/− SEM of triplicate replicates, and are representative of two independent experiments. P-values shown are relative to DMSO control. * p < 0.001, n.s. = not significant. (b) Schematic summarizing genetic and pharmacologic findings for the mevalonate pathway in LSCe cells in vitro (co-culture experiments) and in vivo (RNAi pooled screen). (c) Limiting dilution analysis of varying numbers of LSCe cells pretreated in co-culture with BMSCs for 24 hours with DMSO control or 5 μM lovastatin prior to syngeneic transplantation into mice (n ≥ 3 as shown). (d) Ex vivo treatment for 48 hours with 5 μM lovastatin impaired the ability of LSCe cells co-cultured with BMSCs to induce leukemia upon transplantation into mice relative to DMSO treatment (n = 6). See Supplementary Figure 5 for effects on admixed normal HSPCs.
To further establish that the lovastatin effect was mediated by HMGCR inhibition, we next performed an in vivo pooled RNAi screen32,33 targeting Hmgcr and downstream components of the mevalonate pathway (Supplementary Fig. 5a). LSCe cells were transduced with a lentiviral shRNA pool consisting of shRNAs targeting 8 genes (with a minimum of 4 different shRNAs per gene) and 7 control shRNAs (Supplementary Fig.5b and Methods). After 24 hours, we harvested half of the cells and transplanted the other half into sub-lethally irradiated mice. We harvested the bone marrow of the recipient mice two weeks later at leukemia onset, and performed massively parallel sequencing of genomic DNA to determine the representation of each shRNA in the leukemia cells after two weeks in vivo compared to the representation in the cells prior to transplantation. Consistent with our in vitro findings with statins, we observed that multiple shRNAs targeting Hmgcr were strongly depleted (≥ 20-fold relative to control hairpins) at two weeks, consistent with a role for HMGCR in propagating leukemia in vivo (Fig. 5b, Supplementary Fig. 5c, Supplementary Table 6, and Methods).
In addition, shRNAs targeting Fnta, the common alpha subunit of both geranylgeranyl transferase I (GGTase I) and farnesyl transferase (FT), and shRNAs targeting isoprenylcysteine carboxyl methyltransferase (Icmt) were strongly depleted in vivo (Fig. 5b). GGTase I and FT transfer prenylation moieties generated in the mevalonate biosynthetic pathway to CAAX proteins, and Icmt processes these moieties, stabilizing the moieties on target proteins34. Accordingly, small-molecule farnesyl transferase inhibitors (FTIs) and geranylgeranyl transferase inhibitors (GGTIs) likewise inhibited LSCe cells in the co-culture system to various degrees (Fig. 5b and Supplementary Figs. 5d, e).
Because lovastatin was developed to inhibit the synthesis of cholesterol, its pharmacologic properties have been optimized to achieve high concentrations in the liver, but not elsewhere31. In particular, bone-marrow exposure following oral lovastatin administration to humans and mice is predicted to be low35. To address the effects of lovastatin on LSCe cell leukemogenicity in vivo, we performed syngeneic bone-marrow transplantation studies following 24-hour ex vivo exposure of LSCe cell co-cultures to lovastatin or DMSO control. Limiting dilution analysis revealed that all mice receiving 500 to 10,000 co-cultured LSCe cells in the DMSO group developed leukemia by 5 weeks, whereas only one animal in the lovastatin group developed leukemia by 11.5 weeks (Fig. 5c).
We next extended the transplantation experiments to HSPCs and LSCe cells co-cultured together using a 3-component system. We exposed heterotypic co-cultures containing dsRed+ LSCe cells, uncolored CD45.1+ HSPCs, and GFP+ BMSCs to lovastatin or DMSO for 48 hours then transplanted en masse with untreated wild-type helper splenocytes (CD45.1+ CD45.2+) into lethally irradiated CD45.2+ recipient animals. We observed that mice transplanted with lovastatin-treated mixed cultures survived substantially longer than the DMSO control mice (Fig. 5d), all of which developed leukemia by 3 weeks. Furthermore, CD45 chimerism analysis revealed that long-term hematopoietic engraftment at 16 weeks of the admixed normal HSPCs was comparable to that of DMSO-treated HSPCs co-cultured without LSCe cells (Figs. 5f, g), indicating that lovastatin did not impair normal HSPC function. Taken together, these experiments support the possibility that HMGCR inhibition can selectively target leukemia stem cells in the bone-marrow niche while preserving normal hematopoiesis.
DISCUSSION
The earliest attempts to screen for anti-cancer small molecules were conducted in living laboratory animals. While such approaches are highly physiologic, they are not amenable to high-throughput screening and can only interrogate compounds with favorable pharmacologic properties. More recently, there has been a dramatic shift toward ultra-high-throughput biochemical screens that lack biological complexity but allow for the low-cost screening of massive numbers of compounds. Though this advanced type of screening has identified new AML therapies such as FLT3 inhibitors36, serendipitous discoveries more than a half century ago remain the standard of care for treatment of AML (e.g., daunorubicin and cytarabine)37.
Experimental models that more faithfully reflect the biological complexity of primary disease may facilitate the discovery of next-generation AML therapeutics. Prior efforts have begun to work toward that goal2,38-40, but to date have not brought primary stem-enriched mammalian cancer cells, stromal cell support, and stem-associated readouts to bear on high-throughput small-molecule screening. The approach herein reflects an initial effort to integrate these principles, and may provide physiologic mimicry while enabling large-scale testing of therapeutic candidates. We note the importance of the follow-up studies in naturally-arising, primary human leukemia cells as means to ensure that the findings were generalizable, as the screening system could in principle identify compounds specific to MLL-AF9 or to the ectopic overexpression system. Accordingly, the screening system can be extended to other primary murine and human leukemia models and can also be scaled to larger collections of compounds. To facilitate similar efforts in other laboratories, we have made the cobblestone area quantification algorithms and image analysis software freely available at http://www.cellprofiler.org/published_pipelines.shtml.
The most interesting class of compounds to emerge from the screen was those that selectively killed malignant cells in a manner not readily observable by conventional cell line screening. The existence of such compounds raises the possibility that the biology modeled here is fundamentally distinct from that modeled by cell lines adapted to long-term in vitro culture. Among such compounds are those that killed LSCe cells via an effect on the stroma cells, such as BRD7116. The mechanism by which BRD7116 impairs the stroma’s ability to support leukemia cells remains an important question for future study.
Lovastatin, an FDA-approved drug, was found to preferentially inhibit LSCs compared to normal HSPCs irrespective of leukemia genotype. Genetic, biochemical, and pharmacologic evidence reported here support the notion that lovastatin kills leukemia cells via inhibition of its known target, HMG-CoA reductase (HMGCR). Importantly, the in vivo selection against Hmgcr shRNAs in leukemia within the native bone-marrow niche supports the physiologic relevance of the in vitro co-culture system, linking the mechanistic target identified for lovastatin sensitivity in vitro to leukemia propagation in vivo.
Together, the preclinical observations described herein form a compelling case for statins as potential anti-AML therapeutics. While the potential for statins to yield anti-cancer effects has been proposed41-43, studies of their effects on primary LSCs within the context of a supportive microenvironment have not been reported. Importantly, statins as a drug class have been optimized for activity within the liver, consistent with their primary use as cholesterol-lowering agents. The clinical testing of the hypothesis that HMGCR inhibition will result in anti-leukemia activity in patients will require a drug that achieves concentrations in the bone marrow at or above concentrations observed herein to inhibit murine and human primary AML cells in co-culture (e.g., above the EC50 of ~200 nM for lovastatin). While bone marrow drug concentrations have not been reported, the highest plasma concentration achieved (Cmax) for lovastatin at standard dosing (40mg daily) is only ~50 nM, with a half-life of 2.9 hours35, and would thus not be expected to be efficacious. A phase 1 clinical trial of pravastatin in AML found preliminary evidence of an anti-leukemia effect, prompting an ongoing phase 2 trial44. Of note, pravastatin did not score in our LSCe co-culture assay or in a recently reported model of FLT3-mutated AML45, consistent with its well-documented low uptake by non-liver cells31.
Thus, the anti-leukemia potential of mevalonate pathway inhibition has yet to be fully explored, warranting additional clinical studies. Careful measures of bone marrow pharmacokinetics and ideally pharmacodynamic measures of HMGCR inhibition should therefore be considered. We do note that a recent epidemiological study found that statin use begun prior to a cancer diagnosis is associated with reduced cancer-related mortality46, but whether this result indicates an effect on tumor cells (versus other beneficial effects on the host) is unknown.
A key future research goal should be to identify the effector molecules whose modification via the HMGCR pathway explains the anti-leukemia effects of statins. One candidate would be farnesylation or geranylation of RAS proteins. Despite promising initial studies with farnesyltransferase inhibitors, it was later found that KRAS and NRAS could remain active via geranylgeranyl transferase-mediated prenylation in the presence of farnesyltransferase inhibitors47,48, likely explaining their lack of clinical efficacy49. Subsequent efforts to simultaneously inhibit both proteins, while highly active against pancreatic cancer cells, proved unacceptably toxic in animals50. Importantly, it is possible that the anti-leukemia effect of statins is explained by a mechanism independent of, or complementary to, RAS prenylation. A systematic assessment of mevalonate-dependent effects will likely provide key insight into leukemia cell sensitivities.
The statin result reported here is just one example of multiple compounds found to have preferential leukemia-killing properties compared to normal HSPCs. These discoveries reflect the power of bringing more physiologically-relevant, complex biology to small-molecule screening for AML. We therefore suggest that the approach of reconstructing tissue-like interactions of primary, heterologous cells should be extended more broadly in the search for novel cancer therapeutics.
ONLINE METHODS
Mouse Maintenance and Transplantation
All mouse experiments were in accordance with an IUCAC-approved animal protocol. Recipient mice were either sublethally (1 × 5.5 Gy [550 rads]) or lethally irradiated (2 × 5.5 Gy [550 rads]) prior to tail vein transplantation. Transplanted cells were rinsed and resuspended in 500 μl Hank’s balanced salt solution (HBSS) and loaded into 27½ gauge syringes. Leukemia onset was assessed daily.
Primary Murine LSC-enriched (LSCe) Leukemia Cells and Normal Hematopoietic Stem and Progenitor Cells (HSPCs)
To generate LSCe cells, the MLL-AF9 fusion (kindly provided by Dr. S. A. Armstrong) was retrovirally transduced as described1 into primary granulocytemonocyte progenitors (GMPs; Linlo, Sca-1, c-Kit+, FcYRIIhi, CD34hi) isolated from β-actin-dsRed transgenic mice (6051, Jackson Laboratory) containing a Red Fluorescent Protein gene (DsRed.MST) under the control of the chicken β-actin promoter coupled with the cytomegalovirus (CMV) immediate early enhancer, accompanied by polyadenylation sites. The cells were then transplanted into wild-type C57BL/6 mice and the resulting primary leukemias were serially transplanted through secondary, tertiary, and quaternary recipient mice, yielding large numbers of LSCe cells with a predictable disease latency. Cell staining was performed using murine antibodies for CD3, CD4, CD8, B220, CD19, IL-7R, and Ter119 (biotin-tagged), streptavidin-APCcy7, c-Kit (APC83), Sca-1 (biotin-tagged or APCcy7), CD34 (FITC), and FcγRII (PEcy7), and Hoechst 33258 viability reagent. For assay development, LSCe cells were isolated using a Hoechst− dsRed+ Linlo Sca-1− c-Kittop 5% FcγRIIhi CD34hi gating strategy with the c-Kit gate expanded to c-Kittop 10-20% during primary screening.as described1. As all of the dsRed+ leukemia cells in the quaternary transplants were Linlo and Sca-1− and nearly all of the dsRed+ c-Kithi cells fell into the FcγRIIhi CD34hi gate, a simplified Hoechst− dsRed+ c-Kittop 5% gating strategy was utilized for further experiments, with the c-Kit gate expanded to c-Kittop 10-20% during primary screening. This same gating strategy was used for the comparison of c-Kithi and c-Kitlo cells in co-culture, with the c-Kit gate set as c-Kittop 25% and c-Kitbottom 25%, respectively..
Normal HSPCs (DsRed+ Lin−Sca1+c-Kit+CD48−) were isolated from β-actin-dsRed mouse bone marrow (fully backcrossed to the C57BL/6J background) using biotin-conjugated anti-mouse lineage antibodies CD4, CD8, CD3, B220, Gr-1, Mac-1, Ter-119 and CD48 and Dynabead magnetic separation (Invitrogen) per the manufacturer’s protocol. The resulting Lin/CD48-depleted cells were stained with streptavidin APC-Cy7, c-Kit-APC, Sca-1-FITC, and CD48-Pacific Blue antibodies. A Lin−Sca1+c-Kit+ (LSK) gating was used for secondary studies utilizing HSPCs from CD45.1 or GFP+ mice. FACS plots were created using FlowJo software (Tree Star, Inc).
High-Throughput Co-Culture Assay Methods
See Supplementary Note 1.
Human Cobblestone Area-Forming Cell (CAFC) Assays
Cells were obtained from bone marrow and/or peripheral blood of AML patients with informed consent obtained under the procedures of the Memorial Sloan-Kettering Institutional Review Board during initial presentation of disease (“primary”) or at relapse (“therapy-related”).Normal cord blood was obtained from the New York Blood Center. Mutations and cytogenetic abnormalities were determined via fluorescence in situ hybridization (FISH), karyotyping, and DNA sequencing (FLT3, NPM1, CEPBα, KIT). Samples were centrifuged over Ficoll-Paque PLUS (GE Heathcare) step gradients (2000 x g for 30 minutes), yielding mononuclear cells, then CD34+ cells were isolated using immunomagnetic beads (MACS Cell Separation, Miltenyi Biotec), except for the NPM1 mutation sample which was found to be CD34− by flow cytometry. MS-5 mouse bone marrow-derived stromal cells (kindly provided by Dr. Itoh, Dept. of Biology, Niigata University, Japan) were plated in 96-well format (20,000 per well in α-MEM containing 10% FCS) and kept at at 37°C/ 5% CO2. One day later, CD34-enriched cells (normal or leukemic) were added in 100μL of fresh co-culturing media at a density determined to generate one cobblestone per well after 5 weeks in neutral control wells. The following day, 50μL media containing drug in DMSO was added to each well for a final DMSO concentration of 0.2%. Six days later, the wells were rinsed and fresh media was added. The co-cultures were then maintained with one subsequent half media change at 3 weeks and assessed for cobblestones at 5 weeks (2 weeks for FLT3-ITD sample2). A cobblestone was defined as an instance of at least 6 tightly packed cells beneath the MS-5 stromal monolayer3. The co-culture media formulation consisted of α-Eagle minimum essential medium, 12.5% fetal calf serum (S11550, Atlanta Biologicals), 12.5% horse serum (Gemini Bio Products), 200 mM glutamine, 1 mM monothioglycerol (Sigma Cell Culture), 1 μM hydrocortisone (Sigma), and 20ng/ml human recombinant IL-3 (Amgen).
OP9 and Primary Murine Bone Marrow Mesenchymal Stromal Cells (BMSCs)
OP9 stromal cells (ATCC) were transduced with (GFP)-labeled by retrovirus and cultured in α-MEM without nucleosides (500mL, 36453, Stem Cell Technologies) with sodium bicarbonate (14.6mL, 25080-094, Gibco), L-glutamine (5mL, 071000, StemCell Technologies), β-mercaptoethanol (2.5mL, ES-007-E, Chemicon International), FBS (100mL), and penicillin/streptomycin (Pen-Strep). OP9 cells were maintained at low to medium density at 37°C/ 5% CO2 and periodically replaced with new stocks to ensure optimal LSCe support. ‘OP9 conditioned medium’, was isolated from OP9 cells grown for three days.
Primary, CD105+ bone marrow stromal cells were isolated from actin-GFP or ubiquitin C-GFP (000329 or 4353, Jackson Lab) or wild-type C57BL/6 mice. Crushed bones were washed in 0.5% fetal bovine serum (HyClone) in PBS without magnesium and calcium (Gibco), filtered through 70-μm, subject to red blood cell lysis with ACK buffer (Lonza) for 3-4 minutes, then resuspended in ‘BMSC medium’ (400mL α-MEM (StemCell Technologies), 100mL FBS (HyClone) and 5mL Pen-Strep). Cells were seeded into 150-cm2 tissue culture flasks (25 ml media volume each, 3 flasks per mouse), and placed in a 33°C/ 5% CO2 incubator for 2-3 days, then a media change was performed. 8-14 days later, the cultures were rinsed, split, by brief trypsinization (0.25%), pooled, filtered through a 70 μm filter, replated at 3-4 million cells per 150-cm2 tissue culture flask, and grown another 3-4 days until nearly confluent. The cells were then trypsinized, filtered, and resuspended in 0.5% FBS in PBS with biotin-conjugated anti-mouse CD105 antibody (eBioscience) for 15-30 minutes and isolatde using Dynabead M-280 streptavidin-linked magnetic beads (Invitrogen)for 15-30 minutes at 4°C with agitation and rinsed once . The CD105+cell fraction was replated at 1-2 million cells per 150-cm2 tissue-culture flask and incubated at 33°C/ 5% CO2 for 2-3 days before being used for short-term experimentation.
96-well Assay Setup for Secondary Studies
For 96-well format secondary studies, GFP+ OP9 cells (13,500 per well) were plated onto gelatin coated 96-well plates (3904, Corning). 24 hours later, the media was removed and 2,000 sorted LSCe cells were added in 100μL of OP9 co-culture cocktail media, 50μL of media containing compound(s) was added approximately 24 hours later to 0.2% final DMSO working concentration, then a full media change and compound re-addition was performed 3 days later. For 96-well BMSC co-cultures, 16000 BMSCs were plated onto fibronectin-coated 96-well plates in 100uL BMSC medium. Three days later, 2000 leukemia cells were plated in 50μL BMSC medium added to the existing media per well. The following day, compounds were added in 20μL medium and incubated for 5 days. For both types of stroma, the plates were imaged at 4x or 10x magnification at the completion of 5 days of total compound exposure (6 days of LSCe co-culture). Total viable leukemia cells were counted using the Cell Scoring Application Module in MetaXpress software (Molecular Devices).
Stromal Toxicity Screen
OP9 or BMSC stromal cells were plated in white bottom, 384-well plates (6,750 or 2,000 cells respectively; 3570, Corning). After 24 hours compounds were added (8 concentrations), the plates were incubated for 72 hours at 37°C/ 5% CO2, and viability was quantified using a luminescence measure of cellular ATP (CellTiter-Glo, Promega). 36 compounds were removed from the 196 tested due to stromal toxicity at concentrations below 20 μM (mean signal ≤ 3 standard deviations below DMSO control with BMSCs, 5 standard deviations with OP9s).
Selectivity Retesting of LSCe Co-culture and HSPC Co-culture Effects
The remaining 155 prioritized compounds were tested (8-12 concentrations) in duplicate on LSCe cells in 384 well format (300 per well) and on HSPCs (400 cells per well) with BMSCs at 33°C/ 5% CO2. One subset was retested on LSCe cells with BMSCs using freshly-sourced compound and another subset (shown in Figure 2) was retested on HSPCs with BMSCs at 8 concentrations. Compounds prioritized during these steps were selected based on the degree of selectivity relative to HSPCs, potency of LSCe cell effects, and novelty. BRD2761 (5-chloro-7-((4-ethylphenyl)(pyridin-2-ylamino)methyl)quinolin-8-ol) at 22 μM was used as a positive control for HSPC toxicity.
Human AML Cell Line Viability Screens
Human AML cell lines (ATCC) were cultured under standard conditions in medium specified by ATCC. 3000 cells in 30 μL were plated into 384-well plates. 16 hours later, 100 nl of compounds in DMSO were added, the plates were incubated at 37°C/ 5% CO2 for 72 hours, and then analyzed using CellTiter-Glo. A two-point normalization was performed per screening plate using the mean of DMSO wells (set at 0% effect) and the mean of the cytotoxic positive control wells (0.4 μM staurosporine, set at −100% effect). The independent cell line viability screen was performed as described (Basu, Bodycombe, and Cheah et al., submitted). Cells were plated at an optimal density determined during assay development in 384-well format and incubated overnight at 37°C/5% CO2. Compounds were pin-transferred into duplicate assay plates and incubated for 72 hours. Cellular viability was assessed using CellTiter-Glo.
Stromal Pretreatment Screen
OP9 cells were plated into monolayers in 384-well plate format as in the original co-culture screen. 24 hours later, 100 nl of compounds in pure DMSO at various concentrations were added, the plates were incubated at 37°C/ 5% CO2 for three days, the wells were washed twice with PBS to a 600-fold dilution, then LSCe cells were plated. Media was changed 3 days later, and the plates were imaged and analyzed 2 days after that. Of note, the pretreatment assay may underestimate the cell-non-autonomous effect of the compound on LSCe cells. Media conditioned by untreated OP9 cells was added to support the leukemia cells at the time of LSCe cell plating, as required for efficient cobblestone area formation, potentially restoring stromally secreted factors affected by pretreatment.
For the pretreatment studies using primary BMSC stroma, GFP+ HSPCs (from UbiquitinC-GFP mice) and LSCe cells (dsRed+) were plated together (1200 HSPCs, 1200 LSCe cells, each in 25μl media) in 384-well format onto BMSC stromal monolayers (isolated from wild-type C57BL/6 mice) that were previously treated with BRD7116 for the three days prior at 33°C/ 5% CO2. The treated monolayers were rinsed twice with PBS before the plating of the hematopoietic cells. 5 days later, the wells were imaged in the dsRed and GFP channels at 10x magnification and total cell areas (per well) in each channel were quantified using MetaXpress software.
Compounds
See Supplementary Table 2 for compounds screened. Compounds were diluted to a final working concentration of 0.2% DMSO. Sources of compounds are described in Supplementary Table 3. Lovastatin was purchased from Analyticon Discovery (lovastatic acid, NP-001236; primary, retest, and secondary screens) and Sequoia Research Products (lovastatin, SRP015851; secondary experiments). The top 155 compounds (see Supplementary Table 3) were picked for high-throughput screening from Broad small-molecule libraries and were (insert symbol: greater than or equal to) 75% pure. For secondary studies, we determined the purity of BRD7116 and lovastatin to be greater than 99% pure, with lovastatin existing as an 80/20 mixture of the closed prodrug and free acid forms. Tandem Liquid Chromatography/Mass Spectrometry (LC/MS) was performed on a Waters 2795 separations module and 3100 mass detector. Compound purity was determined by monitoring UV absorbance at 210 nm followed by detection of the expected molecular ion.
Gene Expression Analysis
RNA was extracted (RNeasy kit 74104, Qiagen) from 250,000 LSCe cells treated with either 5 μM BRD7116 or DMSO control (n = 5, 2, respectively) for 6 hours in suspension in 150 μL of IMDM, 10% FBS, and 1% Pen-Strep and prepared (Illumina® TotalPrep™-96 RNA Amplification Kit, Applied Biosystems, PN #4393543) for hybridization to the Illumina’s MouseRef-8 v2.0 Expression BeadChip per the manufacturer’s protocol. Illumina’s BeadArray Reader was used to measure the fluorescence intensity at each addressed bead location according to manufacturer’s instruction. Raw data were normalized by cubic spline method implemented in Illumina Normalizer module of GenePattern analysis tool kit (www.broadinstitute.org/genepattern), and converted into human gene symbols based on the orthologous gene mapping table provided by Jackson laboratory (www.jax.org). For each compound, a ranked list of genes was created by comparing the treated samples to DMSO control samples. The genes were ordered using the signal-to-noise statistic (the difference of means in each group scaled by the sum of standard deviations computed over 3 treatment replicates). The ranked lists were analyzed using Gene Set Enrichment Analysis36 (GSEA) at http://www.broadinstitute.org/gsea/index.jsp.
Other Statistics
Statistical significance was assessed using the two-tailed t-Test (two-sample assuming unequal variances) in Excel (Microsoft). P-values were as stated in legends, where “not significant” indicates a p-value greater than 0.09. Kaplan-Meier survival analysis was performed using the log-rank (Mantel-Cox) test in Prism 5 (GraphPad). All other statistical analysis except where noted was performed with R (www.r-project.org) or Excel. All averages were calculated as means unless otherwise noted, with error bars denoting standard error of the mean (SEM).
Murine Myeloid Cell Line Studies
The murine myeloid cell lines (all ATCC) were maintained as specified by ATCC at 37°C/ 5% CO2. Each line was plated under standard conditions in 96-well format (15,000 cells per well in 150μL media). 18 hours later, 20μL of media was added containing lovastatin or DMSO to a final DMSO concentration of 0.2%. 72 hours later, cellular viability was assessed using CellTiter-Glo reagent. The assay was also read at 5 days with no apparent shift in EC50 observed.
MOZ-TIF2 LSCe Cell Co-culture Studies
Primary MOZ-TIF2 LSCe cells were generated and isolated as described39 as for MLL-AF9 LSCe cells but with one additional round of transplantation. Viable MOZ-TIF2 cells within BMSC (GFP+) co-cultures were quantified after trypsinization using flow cytometry and counting beads (5000 per well; C36950, Life Technologies). All other assay parameters were as described above for MLL-AF9 LSCe cells with BMSCs in 96-well format.
In vitro Studies of Admixed HSPCs and LSCe cells in Co-culture
BMSC monolayers (from C57BL/6 mice) were plated into 384-well plates in 30μl as described, then 800 LSCe cells (dsRed+) and 1600 normal HSPCs (GFP+; C57BL/6 UbiquitinC-GFP mice) were added 3 days later (in 10μL media each). 24 hours later, lovastatin (5 μM) or DMSO was added, the plates were incubated for five days after which the wells were imaged in the dsRed and GFP channels at 10x magnification and total cell areas (per well) in each channel were quantified using MetaXpress software.
Mevalonate Rescue of Lovastatin Treatment
96-well co-culture assays of LSCe cells with OP9 stroma were performed as described, with treatment with 2 mM mevalanolactone (M4667, Sigma-Aldrich) and/or 1 μM lovastatin. Both reagents were added simultaneously and the DMSO carrier concentration was kept constant across all wells.
In Vivo Pooled RNAi screen
Detailed viral packaging protocols for generating arrayed virus for use in pooled shRNA studies can be found at http://www.broadinstitute.org/rnai/public/resources/protocols. In replicates of 5, LSCe cells suspended in 400 μl (5 million/ml) in IMDM, 10% FBS, 10 ng/ml mIL-3 (Peprotech), 10 ng/ml mIL-6 (Peprotech), 20 ng/ml BMSCF (Peprotech), and 5 μg/ml polybrene (Sigma Aldrich) were infected with 400 μl of pooled virus in 12-well plates (spinfection at 2,500 rpm, 37°C for 90 minutes). Two hours later, 800μl of fresh IMDM, 10% FBS, 10 ng/ml mIL-3, 10 ng/ml mIL-6, 20 ng/ml BMSCF was added to each well. After 24 hours, each well was split and half of the cells were frozen for processing and the other half was transplanted into sublethally irradiated mice. Two weeks later, the recipient mice were sacrificed and bone marrow was harvested for DNA sequencing (QIAamp Blood Mini Kit, Qiagen).
The shRNA region was PCR amplified from the purified genomic DNA by combining 5 μL primary PCR primer mix, 4 μL dNTP mix, 1x Ex Taq buffer, 0.75 μL of Ex TaqDNA polymerase (TaKaRa), and 6 μg genomic DNA in a total reaction volume of 50 μL. Thermal cycler PCR conditions consisted of heating samples to 95°C for 5 min; 15 cycles of 94 °C for 30 sec, 65 °C for 30 sec, and 72 °C for 20 sec; and 72 °C for 5 min. PCR reactions were them pooled by sample. A secondary PCR step was performed containing 5 μM of common barcoded 3′ primer, 8 μL dNTP mix, 1x Ex Taq buffer, 1.5 μL Ex TaqDNA polymerase, and 30 μL of the primary PCR mix for a total volume of 90 μL. 10 μL of independent 5′ barcoded primers were then added into each reaction, after which the 100 μL total volume was divided into two 50 μL final reactions. Conditions for secondary PCR were: 95 °C for 5 min; 15 cycles of 94 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 20 sec; and 72 °C for 5 min. Individual 50 μL reactions were then re-pooled. Reactions were then run on a 2% agarose gel and intensity-normalized. Equal amounts of samples were then combined and gel-purified using a 2% agarose gel. Massively parallel sequencing was then performed using a custom sequencing primer under standard Illumina conditions. The raw read counts for each shRNA were normalized to the total reads, and the calculated fold change of normalized reads between the two time points was divided by the mean fold change of all the control shRNAs over the same time points. A gene scored as a hit if ≥ 2 unique shRNAs specific to that gene had a greater than 20-fold change relative to control shRNAs. The 7 control shRNAs used in the pooled in vivo RNAi screen:
| TRC Clone ID | shRNA Sequence |
|---|---|
| TRCN0000072203 | 5′-CCGG-CGCGTGATGAACTTCGAGGAC-CTCGAG- GTCCTCGAAGTTCATCACGCG-TTTTTG-3′ |
| TRCN0000072229 | 5′-CCGG-GCGATCGTAATCACCCGAGTG-CTCGAG- CACTCGGGTGATTACGATCGC-TTTTTG-3′ |
| TRCN0000072236 | 5′-CCGG-CCAACGTGACCTATCCCATTA-CTCGAG- TAATGGGATAGGTCACGTTGG-TTTTTG-3′ |
| TRCN0000072245 | 5′-CCGG-TCACAGAATCGTCGTATGCAG-CTCGAG- CTGCATACGACGATTCTGTGA-TTTTTG-3′ |
| TRCN0000072253 | 5′-CCGG-ACACTCGGATATTTGATATGT-CTCGAG- ACATATCAAATATCCGAGTGT-TTTTTG-3′ |
| TRCN0000072256 | 5′-CCGG-ACGCTGAGTACTTCGAAATGT-CTCGAG- ACATTTCGAAGTACTCAGCGT-TTTTTG-3′ |
| TRCN0000072258 | 5′-CCGG-TGTCCGGTTATGTAAACAATC-CTCGAG- GATTGTTTACATAACCGGACA-TTTTTG-3′ |
The gene hits identified in the pooled in vivo RNAi screen:
| Gene | CloneID | Fold- Depletion |
shRNA Sequence |
|---|---|---|---|
| Hmgcr | TRCN0000173340 | 161 | 5′-CCCGG-GCAAGTGATTACCCTGAGTTT- CTCGAG-AAACTCAGGGTAATCACTTGC TTTTTTG-3′ |
| TRCN0000173464 | 91 | 5′-CCCGG-GCGAGTGCATTAGCAAAGTTT- CTCGAG-AAACTTTGCTAATGCACTCGC- TTTTTTG-3′ |
|
| TRCN0000173543 | 21 | 5′-CCCGG-GTAACCCAAAGGGTCAAGATA- CTCGAG-TATCTTGACCCTTTGGGTTAC- TTTTTTG-3′ |
|
| Fnta | TRCN0000192669 | 611 | 5′-CCCGG-GCTATCACTGTAACCAACTAA- CTCGAG-TTAGTTGGTTACAGTGATAGC- TTTTTTG-3′ |
| TRCN0000202095 | 115 | 5′-GCCGG-CAGAGCAGAATGGGCTGATAT- CTCGAG-ATATCAGCCCATTCTGCTCTG- TTTTTTG-3′ |
|
| Icmt | TRCN0000287908 | 53 | 5′-CCGG-GAGAACATCTTCTGGCCAGAA- CTCGAG-TTCTGGCCAGAAGATGTTCTC- TTTTTG-3′ |
| TRCN0000295336 | 29 | 5′-CCGG-GATGGTGGTCTTCGGAGAATG- CTCGAG-CATTCTCCGAAGACCACCATC TTTTTG-3′ |
shRNA Knockdown Efficacy
Ba/F3 cells maintained in RPMI (11875135, Invitrogen) with 10% FBS, 10 ng/mL murine IL3 (Peprotech), 1x Pen-Strep (15140-122, Gibco), and 5 μg/mL polybrene (Sigma) were spinfected. Three controls shRNAs (LUC-58, RFP-03, LacZ-29) were used in addition to the Hmgcr shRNAs. 24 hours later, puromycin (2 μg/mL, Sigma) was added. The cells were harvested three days later. RNA was isolated with aTurbocapture assay (Qiagen) and processed to cDNA using a Sensiscript kit (Qiagen). All quantitative PCRs were performed using the Taqman assay (Applied Biosystems) on a 384-well qPCR machine (ABIPrism) with knockdown determined using ΔΔCT analysis and the Gapdh reference gene.
Chemical Inhibition of Geranylgeranyl and Farnesyl transferases
Co-culture assays of LSCe cells with OP9 stroma were performed in 96-well format with FTI-277 (10μM, F9803, Sigma-Aldrich), FTI L744,832 (5μM, BML-G242, Enzo Life Sciences), GGTI-286 (10μM, 345878, Calbiochem), GGTI-298 (10μM, 345883, Calbiochem), all at 0.2% DMSO.
Syngeneic Transplantation Experiments
Co-cultures of LSCe cells with BMSCs were plated in 96-well format with 500, 100, or 25 LSCe cells per well. 6 days later, each well was rinsed with PBS, trypsinized, and transplanted en masse into a sublethally irradiated recipient mouse. For the limiting dilution experiments with and without lovastatin treatment, co-cultures of LSCe cells with BMSC (GFP+) stroma were plated with 500 to 10,000 LSCe cells per well, and treated with 5 μM lovastatin or DMSO 24 hour later. After 5 days each well was rinsed with PBS, trypsinized, and transplanted into a sublethally irradiated recipient mouse.
For the triple co-cultures, BMSC monolayers (GFP+) were plated into 96-well plates, then 10,000 freshly isolated LSCe cells (dsRed+) and 10,000 normal HSPCs were added. Lovastatin (5 μM) or DMSO was added 1 day later. 48 hours later, the wells were trypsinized and transplanted along with 2 million helper splenocytes from 6 month old CD45.1+CD45.2+ mice into lethally irradiated, CD45.2+recipient mice . Mice were monitored daily for the presence of leukemia and those that succumbed to leukemia prior to 16 weeks were euthanized. The multi-lineage long-term engraftment of the co-cultured HSPCs (CD45.1−) was quantified by FACS analysis of the peripheral blood of mice alive after 16 weeks and compared to mice that received HSPCs co-cultured with DMSO in the absence of LSCe cells. CD45.1+ B, T, and myeloid cells were also quantified using FACS. Antibodies include B220 (clone Ra3-6b2, BD), CD3 (17A2, eBioscience), and Gr-1/CD11b (RB6-8C5, BD; M1/70, Biolegend), CD45.1 (A20, BD), and CD45.2 (552950, BD).
The Gene Expression Omnibus accession number for the gene expression data reported in this paper is GSE____.
Supplementary Material
ACKNOWLEGEMENTS
We wish to thank L. VerPlank, V. Raksakulthai, A. Liberzon, M. Palmer, G. Cowley, J. Burbank, P. Aspesi, Jr., M. Bliss-Moreau, D. Barker, B. Wagner, A. Krivtsov, S. A. Armstrong, R. Karmacharya, J. Perez, R. Onofrio, D. Thomas, R. Busanelli, D. Auclair, K. Masson, J. Du, C. Moore, M. Kharas, Y. Hoshida, D. Bachovchin, A. Mullally, J. Grabarek, A. Fraser, A. Basu, J. Cheah, N. Bodycombe, C. Mulrooney, S. Johnston, G. Walzer, D. Wilpitz, A. Bracha, A. Fabian, C. Hon, J. McGrath, C. Hartland, M. Hickey, T. R. Jones, M. Bray, K. Sokolnicki, R. Okabe, M. Paktinat, M. McConkey, L. Gaffney, L. Solomon, and K. Rose, Broad Compound Management, the Golub, Gilliland, and Ebert laboratories for scientific discussions, technical expertise, and/or reagents. This work was funded by grants from the Starr Cancer Consortium, the National Institutes of Heath (U54CA112962, U01HL1004402, T32 HL007623, N01-CO-12400, R01 GM089652, RL1-CA133834, 20XS139, RL1HG004671, RL1CA133834, RL1GM084437, UL1RR024924), and the National Science Foundation (DBI 1148823). The content of this publication is solely the responsibility of the authors and does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. T.R.G. and S.L.S. are Investigators at the Howard Hughes Medical Institute.
Footnotes
ADDITIONAL INFORMATION Supplementary Information includes Supplementary Results, which includes five figures, six tables, and one Supplementary Note.
AUTHOR CONTRIBUTIONS A.E.C., A.F.S., A.L.S., A.M.S., B.L.E., D.G.G., D.T.S., K.A.H., M.A.S.M., P.G.M., S.L.S., S.M. and T.R.G. designed the project strategy and interpreted results. A.E.C., A.L.S., A.M.S., D.G.G., D.J.L., D.T.S., J.A.B., J.E.H., J.M.N., K.A.H., N.J.T., P.G.M., S.L.S., S.M., S.Sh, T.H., T.K. and Z.T. developed the assay. A.L.S., D.J.L., J.A.B., J.M.N., K.A.H., M.Du, N.J.T., P.A.C., P.G.M., S.C., S.Sh and T.H. conducted small-molecule screening. A.L.S., A.R.K., B.M., C.H., D.J.L., D.P.S., F.A., J.A.B., J.H.S., J.M.N., K.A.H., L.P.C., M.Da, M.Du, M.J., N.J.T., P.G.M., R.N., R.P., S.C., S.Sh, T.H. and V.D. conducted follow-up studies. A.M.S., B.L.E., K.A.H., P.G.M. and T.R.G. drafted the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests. D.G.G. is a full-time employee of Merck & Company, Inc.
REFERENCES
- 1.Konopleva M, Tabe Y, Zeng Z, Andreeff M. Therapeutic targeting of microenvironmental interactions in leukemia: mechanisms and approaches. Drug Resist Updat. 2009;12:103–13. doi: 10.1016/j.drup.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMillin DW, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16:483–9. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–68. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 5.Guzman ML, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98:2301–7. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 6.Costello RT, et al. Human acute myeloid leukemia CD34+/CD38− progenitor cells have decreased sensitivity to chemotherapy and Fas-induced apoptosis, reduced immunogenicity, and impaired dendritic cell transformation capacities. Cancer Res. 2000;60:4403–11. [PubMed] [Google Scholar]
- 7.Despeaux M, et al. Critical features of FAK-expressing AML bone marrow microenvironment through leukemia stem cell hijacking of mesenchymal stromal cells. Leukemia. 2011;25:1789–93. doi: 10.1038/leu.2011.145. [DOI] [PubMed] [Google Scholar]
- 8.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 9.Plosker GL, Faulds D. Epirubicin. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic use in cancer chemotherapy. Drugs. 1993;45:788–856. doi: 10.2165/00003495-199345050-00011. [DOI] [PubMed] [Google Scholar]
- 10.Stentoft J. The toxicity of cytarabine. Drug Saf. 1990;5:7–27. doi: 10.2165/00002018-199005010-00003. [DOI] [PubMed] [Google Scholar]
- 11.Terpstra W, et al. Long-term leukemia-initiating capacity of a CD34-subpopulation of acute myeloid leukemia. Blood. 1996;87:2187–94. [PubMed] [Google Scholar]
- 12.Breems DA, Blokland EA, Neben S, Ploemacher RE. Frequency analysis of human primitive haematopoietic stem cell subsets using a cobblestone area forming cell assay. Leukemia. 1994;8:1095–104. [PubMed] [Google Scholar]
- 13.Ploemacher RE, van der Sluijs JP, van Beurden CA, Baert MR, Chan PL. Use of limiting-dilution type long-term marrow cultures in frequency analysis of marrow-repopulating and spleen colony-forming hematopoietic stem cells in the mouse. Blood. 1991;78:2527–33. [PubMed] [Google Scholar]
- 14.Corral J, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–61. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 15.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones TR, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A. 2009;106:1826–31. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–9. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 18.Hassane DC, et al. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 2008;111:5654–62. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman ML, et al. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 2005;105:4163–9. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman ML, et al. An orally bioavailable parthenolide analog selectively eradicates acute myelogenous leukemia stem and progenitor cells. Blood. 2007;110:4427–35. doi: 10.1182/blood-2007-05-090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escuin D, et al. The hematopoietic-specific beta1-tubulin is naturally resistant to 2-methoxyestradiol and protects patients from drug-induced myelosuppression. Cell Cycle. 2009;8:3914–24. doi: 10.4161/cc.8.23.10105. [DOI] [PubMed] [Google Scholar]
- 22.Memon RA, et al. Up-regulation of peroxisome proliferator-activated receptors (PPAR-alpha) and PPAR-gamma messenger ribonucleic acid expression in the liver in murine obesity: troglitazone induces expression of PPAR-gamma-responsive adipose tissue-specific genes in the liver of obese diabetic mice. Endocrinology. 2000;141:4021–31. doi: 10.1210/endo.141.11.7771. [DOI] [PubMed] [Google Scholar]
- 23.Gimble JM, et al. Peroxisome proliferator-activated receptor-gamma activation by thiazolidinediones induces adipogenesis in bone marrow stromal cells. Mol Pharmacol. 1996;50:1087–94. [PubMed] [Google Scholar]
- 24.Wolins NE, et al. OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J Lipid Res. 2006;47:450–60. doi: 10.1194/jlr.D500037-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Naveiras O, et al. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–63. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 28.Park DJ, Vuong PT, de Vos S, Douer D, Koeffler HP. Comparative analysis of genes regulated by PML/RAR alpha and PLZF/RAR alpha in response to retinoic acid using oligonucleotide arrays. Blood. 2003;102:3727–36. doi: 10.1182/blood-2003-02-0412. [DOI] [PubMed] [Google Scholar]
- 29.Huntly BJ, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–96. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Moore MA, Dorn DC, Schuringa JJ, Chung KY, Morrone G. Constitutive activation of Flt3 and STAT5A enhances self-renewal and alters differentiation of hematopoietic stem cells. Exp Hematol. 2007;35:105–16. doi: 10.1016/j.exphem.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Gazzerro P, et al. Pharmacological actions of statins: a critical appraisal in the management of cancer. Pharmacol Rev. 2012;64:102–46. doi: 10.1124/pr.111.004994. [DOI] [PubMed] [Google Scholar]
- 32.Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41:1133–7. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinopoulos PA, Papavassiliou AG. Multilevel modulation of the mevalonate and protein-prenylation circuitries as a novel strategy for anticancer therapy. Trends Pharmacol Sci. 2007;28:6–13. doi: 10.1016/j.tips.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Bellosta S, Paoletti R, Corsini A. Safety of statins: focus on clinical pharmacokinetics and drug interactions. Circulation. 2004;109:III50–7. doi: 10.1161/01.CIR.0000131519.15067.1f. [DOI] [PubMed] [Google Scholar]
- 36.Langdon WY. FLT3 signaling and the development of inhibitors that target FLT3 kinase activity. Crit Rev Oncog. 2012;17:199–209. doi: 10.1615/critrevoncog.v17.i2.50. [DOI] [PubMed] [Google Scholar]
- 37.Weiss RB. The anthracyclines: will we ever find a better doxorubicin? Semin Oncol. 1992;19:670–86. [PubMed] [Google Scholar]
- 38.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh JR, et al. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5:236–43. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachlos E, et al. Identification of drugs including a dopamine receptor antagonist that selectively target cancer stem cells. Cell. 2012;149:1284–97. doi: 10.1016/j.cell.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 41.Dimitroulakos J, et al. Increased sensitivity of acute myeloid leukemias to lovastatin-induced apoptosis: A potential therapeutic approach. Blood. 1999;93:1308–18. [PubMed] [Google Scholar]
- 42.Li HY, Appelbaum FR, Willman CL, Zager RA, Banker DE. Cholesterol-modulating agents kill acute myeloid leukemia cells and sensitize them to therapeutics by blocking adaptive cholesterol responses. Blood. 2003;101:3628–34. doi: 10.1182/blood-2002-07-2283. [DOI] [PubMed] [Google Scholar]
- 43.Osmak M. Statins and cancer: Current and future prospects. Cancer Lett. 2012;324:1–12. doi: 10.1016/j.canlet.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Kornblau SM, et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high-dose Ara-C: a phase 1 study. Blood. 2007;109:2999–3006. doi: 10.1182/blood-2006-08-044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams AB, et al. Fluvastatin inhibits FLT3 glycosylation in human and murine cells and prolongs survival of mice with FLT3/ITD leukemia. Blood. 2012;120:3069–79. doi: 10.1182/blood-2012-01-403493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen SF, Nordestgaard BG, Bojesen SE. Statin use and reduced cancer-related mortality. N Engl J Med. 2012;367:1792–802. doi: 10.1056/NEJMoa1201735. [DOI] [PubMed] [Google Scholar]
- 47.Whyte DB, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–64. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 48.Rowell CA, Kowalczyk JJ, Lewis MD, Garcia AM. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem. 1997;272:14093–7. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- 49.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–91. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lobell RB, et al. Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 2001;61:8758–68. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.