Abstract
Phylogenetic analysis of avian and other vertebrate fatty acid binding proteins (FABPs) supported the hypothesis that several gene duplications within this family occurred prior to the most recent common ancestor (MRCA) of tetrapods and bony fishes. The chicken genome encodes two liver-expressed FABPs: (1) L-FABP or FABP1; and (2) Lb-FABP. We propose that the latter be designated FABP10, because in our phylogenetic analysis it clustered with zebrafish FABP10. Bioinformatic analysis of across-tissue gene expression patterns in the chicken showed some congruence with phylogenetic relationships. On the basis of expression, chicken FABP genes seemed to form two major groups: (1) a cluster of genes many of which showed predominant expression in the digestive system (FABP1, FABP2, FABP6, FABP10, RBP1, and CRABP1); and (2) a cluster of genes most of which had pre-dominant expression in tissues other than those of the digestive system, including muscle and the central nervous system (FABP3, FABP4, FABP5, FABP7, and PMP2). Since these clusters corresponded to major clusters in the phylogenetic tree as well, it seems a plausible hypothesis that the earliest duplication in the vertebrate FABP family led to the divergence of a gut-specialized gene from a gene expressed mainly in nervous and muscular systems. Data on gene expression in livers of two lines of chickens selected for high growth and low growth showed differences between FABP1 and FABP10 expressions in the liver, supporting the hypothesis of functional divergence between the two chicken liver-expressed FABPs related to food intake.
Keywords: Fatty acid binding protein, Gene expression, Liver proteins, Multi-gene family
1. Introduction
The fatty acid-binding proteins (FABPs) of vertebrates, which bind long-chain fatty acids, are encoded by members of a multi-gene family, often named on the basis of the tissue in which each member was first identified in mammals (Chmurzyńska, 2006; Schaap et al., 2002; Storch and Corsico, 2008). Although showing substantial differentiation in amino acid sequences, known FABP structures have similar tertiary structures, involving a β-barrel within which the ligand-binding cavity is located (Marcelino et al., 2006). In mammals, one member of this family (FABP1 or L-FABP) accounts for 2–5% of total cytosolic protein in liver cells; the same molecule is also expressed in the small intestine, and is believed to be involved in intestinal assimilation of fatty acids (Alpers et al., 2000). Other FABPs have been named for their expression in such tissues as intestine (FABP2 or I-FABP), adipocytes (FABP4 or A-FABP), and brain (FABP7 or B-FABP). In addition to proteins that bind fatty acids, cytosolic proteins that bind retinol and retinoic acid clearly belong to the same family on the basis of sequence homology (Schaap et al., 2002).
Although certain FABPs have been studied in zebrafish and chicken (Karanth et al., 2009; Murai et al., 2009), much remains to be learned regarding the expression and function of the members of this family in non-mammalian vertebrates. In the chicken Gallus gallus, two liver FABPs have been reported, designated L-FABP and Lb-FABP by Murai et al. (2009). Lb-FABP is expressed only in the liver, whereas L-FABP is expressed in liver and intestine (Murai et al., 2009) In experiments with Japanese quail, the expression of the two genes varied with the light cycle and with food deprivation (Murai et al., 2009). In the presence of food, Lb-FABP expression increased markedly at the start of the light cycle and then declined, a pattern not seen in food deprivation (Murai et al., 2009). It is not clear how these two liver FABPs are related to FABPs of other vertebrates, and little is known about functional differentiation of other avian FABPs.
Few evolutionary studies have addressed the question of how avian FABPs are related to those of mammals, amphibians, and bony fishes. The most comprehensive phylogenetic analysis of FABPs published to date (Schaap et al., 2002) included only three avian sequences, all from chicken. More recently, complete or nearly complete genome sequences have become available for two birds, the chicken and the zebrafinch Taeniopygia guttata; the clawed frogs of the genus Xenopus; the zebrafish Danio rerio; and numerous mammals. Exploiting these sequences, we here present a comprehensive phylogeny of vertebrate FABPs in order to reconstruct the relationships of avian members of this family to those of other vertebrate classes. Schaap et al. (2002) estimated gene duplication times in the FABP family by assuming a molecular clock. In the present study, because of the availability of sequences from numerous genomes, we are able to reconstruct gene duplication times relative to the divergence of major vertebrate clades, without the assumption of a molecular clock. In addition, we use gene expression data to examine functional divergence of FABP members in the chicken. By relating expression patterns to the phylogenetic tree, we reconstruct the patterns of evolutionary differentiation of avian FABPs.
2. Materials and methods
2.1. Sequence analyses
FABP family member sequences were identified by BLASTP homology search and downloaded from the Genbank database. The phylogenetic analyses presented below were based on 87 FABP sequences representing bony fishes, amphibians, birds, and mammals. In preliminary analyses, many additional mammalian sequences were included. However, for ease of presentation, the range of mammalian diversity was represented by sequences from human Homo sapiens, mouse Mus musculus, opossum Monodelphis domestica, and platypus Ornithorhynchus anatinus. Birds were represented by the genome sequences of chicken and zebrafinch, as well as individual sequences from the pheasant Phasianus colchis, duck Anas platyrhynchos, and goose Anser anser. Xenopus tropicalis represented amphibians, while the zebrafish represented bony fishes.
Sequences were aligned by the CLUSTAL X program (Thompson et al., 1997) at the amino acid sequence level. All sites at which the alignment postulated a gap were excluded from phylogenetic analyses; the resulting data set included 118 aligned amino acid sites. A Bayesian phylogenetic tree was reconstructed using the program MrBayes (Ronquist and Huelsenbeck, 2003) with the JTT + gamma substitution model that takes into account rate heterogeneity across sites (Jones et al. 1992; Rodriguez et al., 1990). 1,000,000 generations were run for four chains, with trees sampled every 100 generations. Consensus tree and respective Bayesian posterior probabilities were inferred from the last 5000 sampled trees. Phylogenetic trees were also reconstructed by the neighbor-joining (NJ) method (Saitou and Nei, 1987), based on the JTT + gamma amino acid distance using MEGA 5 (Tamura et al., 2011). The interior branch test with bootstrap estimation of the standard error of branch lengths, was used to assess the reliability of branching patterns in the NJ tree (Nei and Kumar, 2000).
2.2. Gene expression data
From the GEO database, we downloaded accession GSE12974, which provides microarray expression data on 41,534 array features in 20 tissues from pooled samples of adult healthy chickens. The following tissues were used: bursa of Fabricius, cerebellum, cerebral cortex, eye, femur with bone marrow, gallbladder, gizzard, heart, intestine, kidney, liver, lung, muscle, ovary, oviduct, skin, spleen, stomach, testis, and thymus. The data were obtained from two-color experiments with two different tissues hybridized to each array, and each tissue arrayed in replicate with dye swaps. The data analyzed were arcsine-transformed normalized intensity values. We applied hierarchical cluster analysis to data on expression of 11 FABP family members across the 20 tissues, using Ward's (1963) linkage method. We used a jack-knife approach to assess the reliability of clustering patterns in the resulting dendrogram. This approach involved systematically leaving out each one of the 20 tissues and performing the cluster analysis based on the remaining 19 tissues. The percentage of the 20 jack-knife samples that included a given cluster provided a measure of the strength of support in the data for that cluster.
To obtain further evidence regarding FABP expression in the liver, we analyzed microarray data on gene expression in livers of two lines of chickens selected for high growth and low growth (GEO accession GSE7254). Four biological replicates were used for each genotype at six different ages (1, 3, 5, 7, 9 and 11 weeks post-hatching). Expression data (Loess-normalized ratios) for FABP family members were analyzed by two-way ANOVA using line (high growth or low growth) and week post-hatching as factors.
3. Results
3.1. Phylogenetic analyses
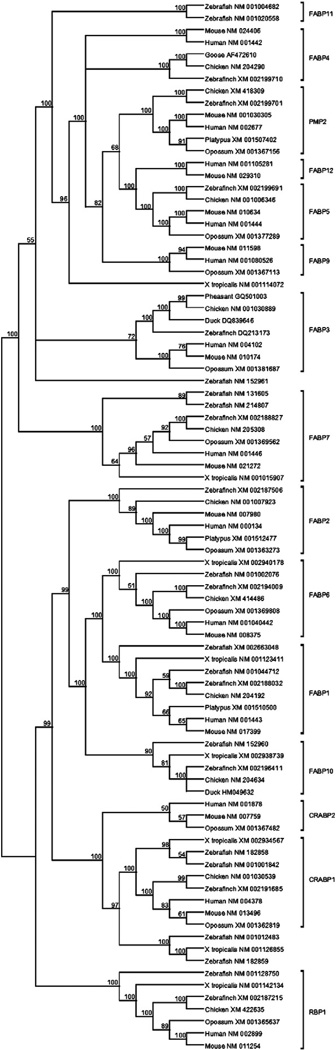
A Bayesian phylogenetic tree of 87 FABP amino acid sequences of vertebrates showed several clusters of genes including apparent orthologs from birds and mammals, often along with sequences from amphibians and bony fishes as well (Fig. 1). The NJ analysis yielded a similar overall topology (Supplementary Fig. S1). Therefore, in most cases, it was easy to identify the apparent mammalian ortholog of the avian sequences, and in those cases we refer to the avian sequence by the name of the apparent mammalian ortholog (Table 1). In each of FABP1, FABP2, FABP5, FABP6, FABP7, PMP2, RBP1, and CRABP1, the apparent avian and mammalian orthologs belonged to a cluster supported by 100% posterior probability (Fig. 1). In the case of FABP3, the apparent avian and mammalian orthologs belonged to the same cluster, but support for the cluster was only 72% (Fig. 1). By contrast, whether mammalian and avian FABP4 should be considered orthologs was unresolved (Fig. 1).
Fig. 1.
Bayesian phylogenetic tree of vertebrate FABPs. Numbers on branches are posterior probabilities.
Table 1.
Chicken FABP family members.
| Symbol | Protein | Refseq mRNA | Chromosome | High expression scoresa |
|---|---|---|---|---|
| FABP1 | Fatty acid-binding protein 1, liver (L-FABP) | NM_204192 | 4 | Liver, intestine |
| FABP2 | Fatty acid-binding protein 2, intestinal (I-FABP) | NM_001007923 | 4 | Intestine |
| FABP3 | Fatty acid-binding protein 3, muscle and heart | NM_001030889 | 23 | Heart, stomach, gizzard, femur, skin, cerebral cortex, kidney, lung, eye, ovary, oviduct, bursa |
| FABP4 | Fatty acid-binding protein 4, adipocyte (A-FABP) | NM_204290 | 2 | Cerebral cortex, eye, kidney |
| FABP5 | Fatty acid-binding protein 5, psoriasis-associated (epidermal; E-FABP) | NM_001006346 | 2 | Gizzard, cerebral cortex, skin, heart, eye, muscle, stomach, cerebellum |
| FABP6 | Fatty acid-binding protein 6, ileal | XM_414486 | 13 | Intestine |
| FABP7 | Fatty acid-binding protein 7, brain (B-FABP) | NM_205308 | 3 | Cerebral cortex, kidney, eye, spleen |
| PMP2 | Peripheral myelin protein 2 | XM_418309 | 2 | Cerebral cortex, cerebellum |
| FABP10 | Fatty acid-binding protein, liver basic (Lb-FABP) | NM_204634 | 23 | Liver |
| RBP1 | Retinol-binding protein 1, cellular | XM_422635 | 9 | Eye |
| CRABP1 | Cellular retinoic acid-binding protein 1 | NM_001030539 | 10 | Eye |
>1 S.D. above mean level in that tissue for 41,534 array features in GEO accession GSE12974.
The chicken liver-expressed gene whose protein product has been called Lb-FABP (Murai et al., 2009) clustered with a zebrafish gene (NM_152960) that has been designated FABP10 (Fig. 1); and this cluster was supported by 90% posterior probability. Therefore, we use the latter name for this chicken gene (Table 1). The FABP10 cluster was included genes from birds, Xenopus, and zebrafish, but no mammalian sequences (Fig. 1). Conversely, certain mammalian genes (FABP9, FABP12, and CRABP2) lacked any apparent avian orthologs (Fig. 1).
There were two major clusters in the phylogenetic tree, each supported by a highly significant (100% posterior probability) internal branch: (1) the cluster containing FABP1, FABP2, FABP6, FABP10, CRABP1, CRABP2, and RBP1; and (2) the cluster including all remaining members of the family except FABP2 (Fig. 1). These two clusters were also separated by a significant internal branch in the NJ tree (Supplementary Fig. S1). Since both clusters included sequences from zebrafish, the phylogenetic analysis supported the hypothesis that these two major subfamilies of the FABP family arose by gene duplication prior to most recent common ancestor (MRCA) of tetrapods and bony fishes. Each of FABP1, FABP2, FABP6, FABP7, FABP10, RBP1, and CRABP1 formed a cluster supported by 100% posterior probability and including sequences from zebrafish and one or more of the tetrapods (Fig. 1). Therefore the phylogeny supported the hypothesis that each of these genes arose by gene duplication before the MRCA of tetrapods and bony fishes.
FABP1 and FABP6 clustered together, with 100% posterior probability; and FABP10 fell outside this cluster (Fig. 1). Therefore, although the cluster including avian FABP10 and related amphibian and zebrafish sequences was supported by only 90% posterior probability, the fact that the FABP10 cluster fell outside the FABP1 and FABP6 clusters (Fig. 1) provided strong support for the hypothesis that FABP10 originated before the MRCA of tetrapods and bony fishes. Likewise, mammalian CRABP2 fell outside of a cluster including tetrapod and zebrafish CRABP1, and this position was supported by a significant (97%) posterior probability (Fig. 1). Therefore, even though no zebrafish, amphibian, or avian homolog of CRABP2 was found, the phylogeny supported the hypothesis that CRABP2 originated prior to the MRCA of bony fishes and tetrapods.
Zebrafish FABP11 and a sequence from X. tropicalis (NM_001114072) fell outside a cluster including avian and mammalian FABP4, FABP5, FABP9, FABP12, and PMP2; and this relationship was supported by a significant (100% posterior probability) internal branch (Fig. 1). Therefore, the phylogenetic tree supported the hypothesis that the genes encoding the latter five proteins arose by gene duplication after the MRCA of amniotes and amphibians. The relationships within this amniote-specific cluster were not well resolved (Fig. 1). However, a branch with significant (100%) posterior probability supported the clustering of avian and mammalian FABP5 sequences (Fig. 1). Likewise a branch with significant (100%) posterior probability supported the clustering avian and mammalian PMP2 sequences (Fig. 1). Therefore, the phylogeny supported the hypothesis that both FABP5 and PMP2 arose prior to the MRCA of birds and mammals. Moreover, since mammalian FABP9 and FABP12 fell outside the latter clusters, the phylogenetic tree supported the hypothesis that FABP9 and FABP12 arose prior to the MRCA of birds and mammals.
3.2. Expression across adult tissues
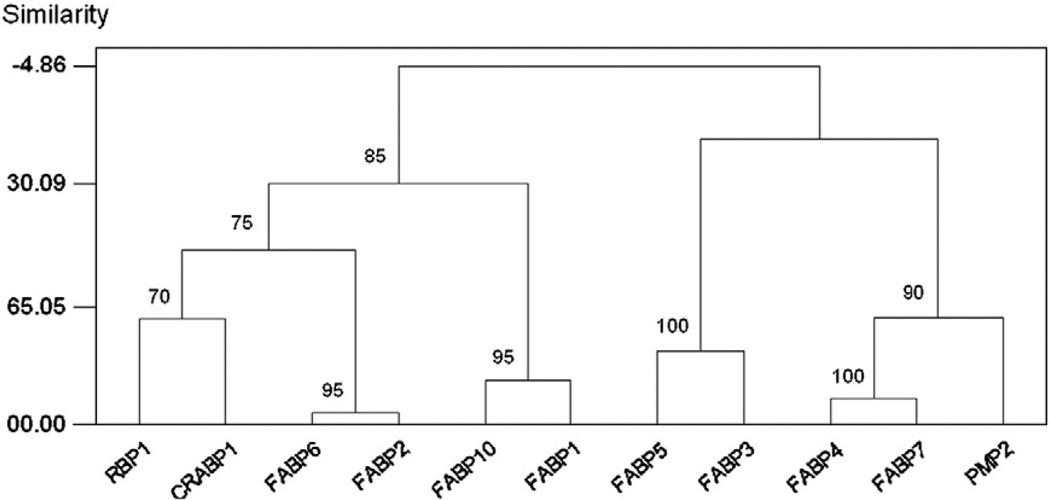
Hierarchical clustering of expression data across 20 adult tissues showed that FABP2 and FABP6, both characterized by high intestinal expression levels (Table 1), were the most similar pair of genes in their expression pattern; and the clustering of these two genes received 95% jack-knife support (Fig. 2). The next most similar pair, with 100% jack-knife support, were FABP4 and FABP7 (Fig. 2), both with high levels of expression in cerebral cortex, eye, and kidney (Table 1). In addition, FABP1 and FABP10, both being highly expressed in the liver (Table 1), clustered with 95% jack-knife support (Fig. 2).
Fig. 2.
Hierarchical clustering of expression data of chicken FABPs across 20 adult tissues. Numbers on the branches represent the percentage of jack-knife samples supporting the branch.
The clustering patterns based on gene expression (Fig. 2) showed both certain broad similarities with the phylogenetic tree (Fig. 1) and a number of differences of detail. In the gene expression data, there were two major clusters, which received 85% jack-knife support: (1) FABP1, FABP2, FABP6, FABP10, CRABP1, and RBP1; and (2) FABP3, FABP4, FABP5, FABP7, and PMP2 (Fig. 2). These corresponded to the two major clusters in the phylogenetic tree (Fig. 1). The former cluster included the following genes with high levels of expression in the liver and/or intestine: FABP1, FABP2, FABP6, and FABP10 (Table 1). Although RBP1 showed highest expression in the eye (Table 1), the tissue with the second highest expression score for RBP1 was the intestine. Although not having high levels of expression in any digestive system tissue, CRABP1 was evidently drawn into this cluster by the clustering algorithm largely because it shared with RBP1 high expression levels in the eye (Table 1).
3.3. Liver expression in high and low growth lines
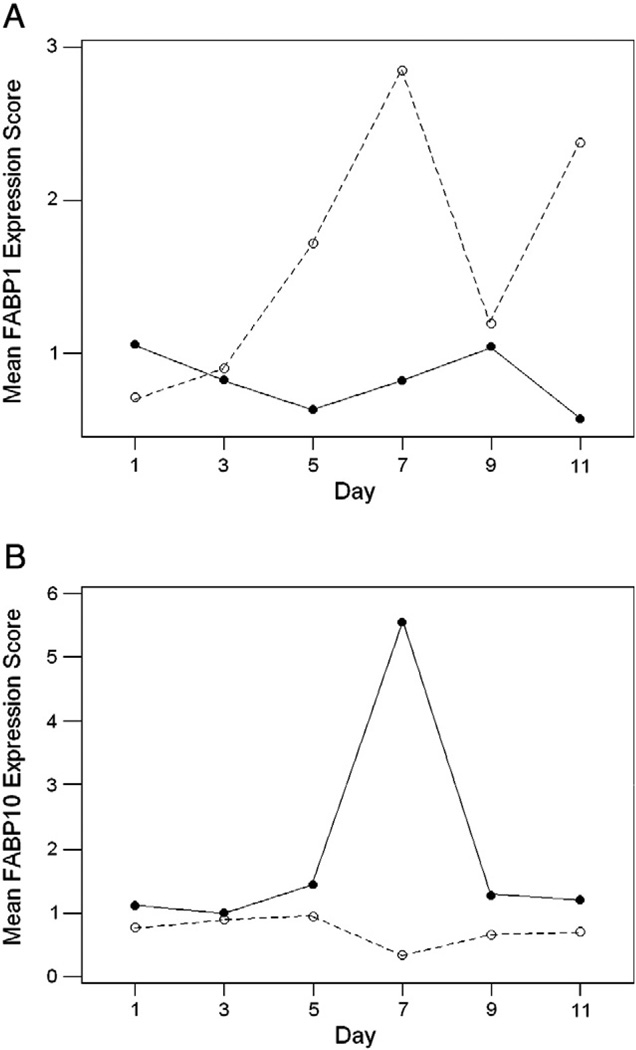
Gene expression data in liver tissue from days 1–11 post-hatching in high growth (HG) and low growth (LG) lines of chicken was analyzed by a factorial ANOVA. There was a significant line-by-day interaction in FABP1 expression (F5,36=8.40; P<0.001; Bonferroni-corrected for multiple testing), indicating a difference between the two lines with respect to the pattern of FABP1 expression over time (Fig. 3A). This difference appeared to be due mainly to a substantial peak in FABP1 expression in days 5–7 in the LG line (Fig. 3A). Likewise there was a significant line-by-day interaction in FABP10 expression (F5,36=11.09; P<0.001; Bonferroni-corrected for multiple testing; Fig. 3B). The latter interaction was due mainly to a substantial increase in FABP10 expression at day 7 in the HG line (Fig. 3B). Data were also available for liver expression of FABP3 and FABP4; neither of the latter two genes showed a significant line-by-day interaction (F5,36=0.44; ns.; and; F5,36=1.88; ns.; respectively).
Fig. 3.
Mean expression scores of (A) FABP1 and (B) FABP2 days 1–11 post-hatching in chicken lines selected for high growth (solid lines) and low growth (dotted lines). There were significant line-by-day interactions in both FABP1 expression (F5,36=8.40; P<0.001) and FABP10 expression (F5,36=11.09; P<0.001).
4. Discussion
Phylogenetic analysis of avian and other vertebrate fatty acid binding proteins supported the hypothesis that several gene duplications within this family occurred prior to the most recent common ancestor (MRCA) of tetrapods and bony fishes. Of the 11 members of this family found in the chicken genome, the phylogenetic analysis provided strong support for the origin of seven of them (FABP1, FABP2, FABP6, FABP7, FABP10, RBP1, and CRABP1) prior to the MRCA of tetrapods and bony fishes. On the other hand, the phylogenetic analysis supported the hypothesis that three of the chicken genes (FABP4, FABP5, and PMP2) originated by gene duplication within the amniotes after the MRCA of amniotes and amphibians. FABP3 was the only chicken member of the family whose time of origin remained unresolved in the phylogeny.
Whether avian FABP4 is orthologous to mammalian FABP4 was unresolved, and the mammalian FABP9, FABP12, and CRABP2 showed no evidence of avian orthologs. Even though CRABP2 genes were found only in mammals, the phylogenetic tree supported the hypothesis that CRABP2 originated prior to the MRCA of tetrapods and bony fishes. The phylogenetic tree supported the hypothesis that FABP9 and FABP12 originated after the MRCA of amniotes and amphibians, but before the MRCA of birds and mammals. Conversely, although FABP10 originated prior to the MRCA of tetrapods and bony fishes, there was no evidence of a mammalian ortholog. Thus, the evolutionary history of the FABP family in birds and mammals has been characterized by episodes of lineage-specific gene loss. There were more cases of loss of orthologs in birds (FABP9, FABP12, and CRABP2) than in mammals (FABP10), a pattern consistent with the greater tendency toward loss of ancestral orthologs in birds than in mammals (Hughes and Friedman, 2008).
Across-tissue gene expression patterns in the chicken showed some congruence with phylogenetic relationships. On the basis of expression, chicken FABP genes seemed to form two major groups (Fig. 2): (1) a group of genes with predominant expression in the digestive system (FABP1, FABP2, FABP6, FABP10, RBP1, and CRABP1; and (2) a group of genes predominantly expressed in tissues other than those of the digestive system, including muscle and the central nervous system (FABP3, FABP4, FABP5, FABP7, and PMP2). Since these clusters corresponded to major clusters in the phylogenetic tree as well (Fig. 1), it seems a plausible hypothesis that the earliest duplication in the vertebrate FABP family led to the divergence of a gut-specialized gene from a gene expressed mainly in nervous and muscular systems. Further duplication of the gut-specialized gene has given rise to distinct functions in the digestive system in the case of FABP1, FABP2, FABP6, and FABP10, while RBP1 and CRABP1 have been co-opted for expression in the eye.
Since the gene duplication events that gave rise to distinct vertebrate FABPs occurred in the distant past, it was not possible to reconstruct the evolutionary forces acting at the time of gene duplication. For example, synonymous nucleotide sites were saturated with changes in comparisons between different chicken FABP genes; thus, testing for the effects of natural selection by comparing synonymous and nonsynonymous substitutions (Hughes and Nei, 1988) was not possible. Our phylogenetic analysis suggested that original gene duplication gave rise to separate gut-specific and neuromuscular FABP genes, which were in turn subsequently duplicated to give rise to more specialized genes. Such a pattern of recurrent increases in specialization after gene duplication is consistent with evolutionary models whereby duplicate genes come to share the functions of a less specialized ancestor (Hughes, 1994; Lynch and Force, 2000).
Analysis of expression data from lines of chicken selected for high and low growth showed significant differences in the liver expression patterns of FABP1 and FABP10, with a peak in FABP1 expression in days 5–7 post-hatching in the low-growth line and a peak of FABP10 expression at day 7 post-hatching in the high-growth line. These different patterns are consistent with evidence from Japanese quail that FABP1 and FABP10 are functionally differentiated (Murai et al., 2009). In adults of the latter species, the expression of FABP10 was found to be induced by feeding to a greater extent than was that of FABP1 (Murai et al., 2009). The higher expression of FABP10 early in post-hatching growth in a high-growth line of chickens is further consistent with the hypothesis that this protein plays a specific role in the liver in response to food intake.
Supplementary Material
Acknowledgments
This research was supported by grants GM43940 to A.L.H. and GM86782-01A1 to H.P. from the National Institutes of Health.
Abbreviations
- FABP
fatty acid binding protein
- JTT
Jones–Taylor–Thornton model
- MRCA
most recent common ancestor
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.gene.2011.09.016.
References
- Alpers DH, Bass NM, Engle MJ, DeSchryver-Keeskemeti K. Intestinal fatty acid-binding protein may favor differential apical fatty acid binding in the intestine. Biochim. Biophys. Acta. 2000;1448:352–362. doi: 10.1016/s1388-1981(99)00200-0. [DOI] [PubMed] [Google Scholar]
- Chmurzyńska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J. Appl. Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B Biol. Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Genome size reduction in the chicken has involved massive loss of ancestral protein-coding genes. Mol. Biol. Evol. 2008;25:2681–2688. doi: 10.1093/molbev/msn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at MHC class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Karanth S, Lall SP, Denovan-Wright EM, Wright JM. Differential transcriptional modulation of duplicated fatty acid-binding protein genes by dietary fatty acids in zebrafish (Danio rerio): evidence for subfunctionalization or neofunctionalization of duplicated genes. BMC Evol. Biol. 2009;9:219. doi: 10.1186/1471-2148-9-219. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino AM, Smock RG, Gierasch LM. Evolutionary coupling of structural and functional sequence information in the intracellular lipid-binding protein family. Proteins. 2006;63:373–384. doi: 10.1002/prot.20860. [DOI] [PubMed] [Google Scholar]
- Murai A, Furuse M, Kitaguchi K, Kusumoto K, Nakanishi Y, Kobayashi M, Horio F. Characterization of critical factors influencing gene expression of two types of fatty acid-binding proteins (L-FABP and Lb-FABP) in the liver of birds. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 2009;154:216–223. doi: 10.1016/j.cbpa.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Rodriguez F, Oliver JF, Marí A, Medina JR. The general stochastic model of nucleotide substitution. J. Theor. Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schaap FG, van der Vusse GJ, Glatz JF. Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol. Cell. Biochem. 2002;239:69–77. [PubMed] [Google Scholar]
- Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu. Rev. Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Diggins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.