Abstract
Multiple myeloma (MM) is characterized by the clonal expansion of malignant plasma cells (multiple myeloma cells, MMC), primarily in the bone marrow (BM). Osteolytic bone lesions are detected in 80% of patients, due to increased osteoclastic bone resorption and reduced osteoblastic bone formation. MMC are found closely associated to sites of increased bone resorption. Osteoclasts strongly support MMC survival and vice versa in vitro. To further elucidate the mechanisms involved in osteoclast/MMC interaction, we have identified 552 genes overexpressed in osteoclasts compared to other BM cell subpopulations. Osteoclasts express specifically genes coding for four CCR2-targeting chemokines, and genes coding for MMC growth factors (IGF-1, APRIL). An anti-CCR2 MoAb blocked osteoclast chemoattractant activity for MMC and CCR2-chemokines are also MMC growth factors, promoting MAPK activation in MMC. An anti-IGF-1 receptor MoAb completely blocked the osteoclast-induced survival of MMC suppressing both osteoclast and MMC survival. Specific APRIL or IL-6 inhibitors partially blocked osteoclast-induced MMC survival. These in-vitro data may explain why newly-diagnosed patients whose MMC express high levels of CCR2 present numerous bone lesions.
Taken together, this study displays additional mechanisms involved in osteoclast/MMC interaction and suggests using CCR2 and/or IGF-1 targeting strategies to block this interaction and prevent drug resistance.
Keywords: Aged; Bone Resorption; metabolism; pathology; Cell Communication; genetics; physiology; Cell Movement; genetics; Cells, Cultured; Chemotactic Factors; genetics; metabolism; Disease Progression; Gene Expression Profiling; Humans; Microarray Analysis; Middle Aged; Multiple Myeloma; genetics; metabolism; pathology; Neoplasm Metastasis; Osteoclasts; metabolism; physiology; Receptors, CCR2; genetics; metabolism; physiology
Introduction
Multiple myeloma (MM) is a plasma cell neoplasm characterized by the accumulation of malignant plasma cells primarily in the bone marrow (BM). A majority of patients with MM develop osteolytic bone disease characterized by bone pain, pathologic fractures, and hypercalcemia, due to the disruption of the coupling of osteoclastic bone resorption and osteoblastic bone formation[1]. An increase in bone turnover rate was reported to precede progression from Monoclonal Gammopathy of Undetermined Significance (MGUS) to overt MM[1]. In patients with intramedullary MM, multiple myeloma cells (MMC) develop in close interaction with the BM microenvironment, mainly BM stromal cells[2], endothelial cells[3] and osteoclasts[4,5]. In particular, MMC promote osteoclast formation directly [4,6] or indirectly[7,8] and osteoclasts support MMC survival, producing APRIL or IL-6 particularly[9,10]. Several factors are involved in myeloma bone disease[11], mainly the receptor activator of nuclear transcription factor-B (RANKL)[8], macrophage inflammatory protein-1-α (MIP-1α/CCL3)[12–14], tumor necrosis factorα (TNFα)[15], interleukin-1α (IL-1α)[15], and interleukin-6 (IL-6)[15]. A shift in RANKL vs. osteoprotegerin expression in patients with MM favors osteoclast generation and activation[16] and blocking RANKL decreases tumor burden and bone destruction in patients with MM [4]. MIP-1α/CCL3 is produced by MMC, stromal cells, monocytes and osteoclasts[13,17]. Both osteoclasts and MMC express CCR1, a receptor for MIP-1α/CCL3 receptor, which promotes osteoclast formation and activation and inhibition of MIP-1α/CCL3 decreases markedly both tumor burden and bone destruction in a murine model of MM[14,18].
Given the importance of the interaction of MMC and osteoclasts to promote both MMC growth and osteoclast formation, this study aims to further characterize the cell communication mechanisms between these 2 cell types. We show here that osteoclasts specifically express a set of chemokines targeting CCR2 receptors that are overexpressed by MMC compared to normal plasma cells. At the same time, newly-diagnosed MM patients with a high CCR2 gene expression on MMC exhibit a higher number of bone lesions than patients with a low CCR2 expression on MMC. The chemoattractant activity of osteoclasts on MMC is inhibited by an anti-CCR2 MoAb. These CCR2-chemokines are also myeloma growth factors (MGFs), promoting MAPK activation in MMC. We also show that osteoclasts support MMC by producing IGF-1, APRIL and IL-6. This study underscores the important role of osteoclasts in recruiting MMC and promoting their survival and emphasizes the interest of CCR2 targeting therapies.
Materials and methods
XG- human myeloma cell lines (HMCLs) were obtained as described[19]. SKMM, L363, OPM2, LP1 and RPMI8226 HMCLs were purchased from ATTC (LGC Promochem, France). Multiple Myeloma cells (MMC) were obtained in agreement to the French and German ethical laws. MMC were purified from the BM of 206 patients with newly-diagnosed MM (median age, 59 years) after written informed consent was given. The study has been approved by the ethic boards of Heidelberg University and Montpellier University hospitals. These 206 patients were treated with high dose therapy (HDC) and autologous stem cell transplantation (ASCT) and were termed in the following Heidelberg-Montpellier (HM) series[20]. We also used Affymetrix data of a cohort of 345 purified MMCs from previously untreated patients from the Arkansas Cancer Research Center (ACRC, Little Rock, AR). The patients were treated with total therapy 2[21] and termed in the following ACRC-TT2 series. These data are publicly available via the online Gene Expression Omnibus (Gene Expression Profile of Multiple Myeloma, accession number GSE2658. http://www.ncbi.nlm.nih.gov/geo/. Accessed June 1, 2006). Normal BM plasma cells (BMPCs) and whole BM cells (WBMCs) were obtained from healthy donors after informed consent was given. WBMCs were collected after lysis of red blood cells with NH4Cl. After Ficoll-density gradient centrifugation, plasma cells were purified using anti-CD138 MACS microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). BM environment cells from 7 newly diagnosed patients were obtained after depletion of MMC with anti-CD138 MACS microbeads (Miltenyi Biotech). For 5 newly diagnosed patients, BM T cells, monocytes and polymorphonuclear neutrophils (PMN) were purified. BM cells were labeled with a phycoerythrin (PE)-conjugated anti-CD3 MoAb, Allophycocyanin (APC)-conjugated anti-CD14 MoAb, and a fluorescein isothiocyanate (FITC)-conjugated anti-CD15 MoAb (all from Becton Dickinson, San Jose, CA). CD3+, CD14+ and CD15+ cells were sorted with a FACSAria cell sorter (Becton Dickinson). Memory B cells, polyclonal plasmablasts (PPCs) and BM stromal cell lines (BMSCs) were generated as described previously[9]. The study was approved by the ethics boards of the Medical Faculty of the University of Heidelberg and the University of Montpellier.
Osteoclasts
Osteoclasts were generated as previously described[9]. In brief, peripheral blood mononuclear cells were obtained from 7 patients with MM after informed consent. Cells were cultured at 2.5 × 106 cells/ml in αMEM-10% FCS. After 12 hours of culture, non-adherent cells were eliminated and adherent cells were cultured in αMEM-10% FCS, RANKL (50 ng/ml, PeproTech, EC Ldt, London, UK), M-CSF (25 ng/ml, Peprotech), and 10 nM dexamethasone for 14 days. Before use, osteoclasts were phenotyped by RT-PCR (TRAP and cathepsin K expression), by cytometry (integrin αvβ3 expression) and bone resorbing activity (OsteoLyse assay kit, Cambrex, Emerainville, France).
Flow cytometry analysis
CCR1 and CCR2 expression on HMCLs was evaluated by incubating 5 × 105 cells with PE-conjugated anti-CCR1 or anti-CCR2 MoAbs (Becton Dickinson, Mountain View, CA) in phosphate-buffered saline (PBS) containing 30% human AB serum at 4°C for 30 minutes. For primary samples, cells were double stained with PE-conjugated anti-CCR1 or anti-CCR2 and FITC-conjugated anti-CD138 (Beckman- Coulter) MoAbs. Flow cytometry analysis was carried out on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
In vitro cell migration assay
For Transwell migration assay, 24-well plates with transwell inserts (6.5 mm diameter, 5 μm pore size; Costar Corning Elscolab) and RPMI 1640 medium (Life Technologies) supplemented with 0.5% BSA (Sigma-Aldrich) were used. The inserts were coated with 100 μl human fibronectin solution (Invitrogen) at a concentration of 10 μg/ml in distilled water and incubated for 1h at 37°C and 5% CO2. The solution was removed and the inserts were dried for 2 hrs at 37°C. The lower transwell chamber containing osteoclasts was filled with 600 μl of αMEM-10% FCS. 105 MMC were added to the upper chamber. Cells were then allowed to migrate for 90 min at 37°C in a humid atmosphere (5% CO2). Finally, cells were collected from upper and lower wells and the number of MMC that transmigrated into lower wells was evaluated with a FACS flow cytometer (Becton Dickinson, Mountain View, CA) after incubation with an anti-CD138 monoclonal antibody (MoAb) PE-conjugated at 4°C for 30 min. The frequencies of migrating cells (number of cells that migrated to the lower chamber divided by cell number in the upper + lower chamber) are indicated. For migration inhibition, neutralizing anti-CCR1 and CCR2 MoAbs (R&D Systems, Abington, United Kingdom) or a MAPK inhibitor (PD98059) (Cell Signaling) were used.
Growth assay for myeloma cells
HMCLs were IL-6- and serum-starved for 2 hours and cultured for 4 days in 96-well flat-bottom microtiter plates in serum-free culture medium without cytokine (control), with rIL-6 (2 ng/mL) or with graded CCL2, CCL7, CCL8, CCL13, or CCL23 concentrations. The growth of myeloma cells was evaluated by quantifying intracellular ATP with a Cell Titer Glo Luminescent Assay (Promega, Madison, WI) with a Centro LB 960 luminometer (Berthold Technologies, Bad Wildbad, Germany).
Elisa
The concentrations of BAFF, APRIL, IL-10, IL-6, and IGF-1 were assayed with ELISAs purchased from Bender MedSystems (Burlingame, CA) for BAFF and APRIL[22], from Diaclone (Besançon, France) for IL-6, IL-10 and from R&D Systems for IGF-1 in accordance with the manufacturer’s instructions.
Study of apoptosis
IL-6-dependent HMCLs were starved of IL-6 for 3 hours and cultured in 24-well flat-bottomed microtiter plates at 105 cells/well in RPMI 1640-10% FCS in the presence or not of osteoclasts (2.5 × 104 cells/well), with or without B-E8 anti-IL-6 MoAb (10 μg/ml), TACI-Fc (20 μg/ml), anti-IL-10 MoAb (10 μg/ml), anti-CCR2 MoAb (10 μg/ml) or IGF-1 receptor MoAb (4 μg/ml) (CALBIOCHEM, San Diego, CA). Recombinant IL-6 (2 ng/mL) was added in one culture group as positive control. After 4 days of culture, cells were washed twice in PBS and apoptosis was assayed with FITC-conjugated annexin V labeling (Boehringer). Fluorescence was analyzed on a FACScan flow cytometer (Becton Dickinson).
Preparation of complementary RNA (cRNA) and microarray hybridization
RNA extraction was performed using the RNeasy kit (QIAGEN, Hilden, Germany), the SV-total RNA extraction kit (Promega, Mannheim, Germany) and Trizol (Invitrogen, Karlsruhe, Germany). Labeled cRNA was generated using the small sample labeling protocol vII (Affymetrix, Santa Clara, CA), and hybridized to U133 2.0 plus arrays according to the manufacturer’s instructions. Fluorescence intensities were quantified and analyzed using the GECOS software (Affymetrix).
All microarray data presented in this paper have been deposited in the ArrayExpress public database, under accession numbers E-MEXP-2360 for BMPC[23] and E-TABM-937 for B cells, PPC, MMC and environment population samples.
Real-time RT-PCR
Total RNA was converted to cDNA using the Superscript II reverse transcriptase (Invitrogen, Cergy Pontoise, France). The assays-on-demand primers and probes and the TaqMan Universal Master Mix were used according to the manufacturer’s instructions (Applied Biosystems, Courtaboeuf, France). The measurement of gene expression was performed using the ABI Prism 7000 Sequence Detection System and analyzed using the ABI PRISM 7000 SDS Software. For each set of primers, serial dilutions of a standard cDNA were amplified to create a standard curve, and values of unknown samples were estimated relative to this standard curve in order to assess the PCR efficiency. Ct values were obtained for GAPDH and the respective genes of interest during log phase of the cycle. Gene of interest levels were normalized to GAPDH for each sample (δCt = Ct gene of interest − Ct GAPDH) and compared with the values obtained for a known positive control using the following formula 100/2δδCt where δδCt = δCt unknown − δCt positive control.
Western blot analysis
HMCLs were starved overnight in RPMI 1640-1% bovine serum albumin (BSA) without IL-6, and then incubated with serum-free culture medium, recombinant IL-6 (30 ng/ml), recombinant CCL7 or CCL8 (2 μg/ml) for 10 and 30 minutes. Cells were then processed for western blot analysis as detailed elsewhere[19]. The primary antibodies (phospho-specific antibodies anti-ERK1/2, anti-STAT3 and anti-AKT; New England Biolabs, Beverly, MA, USA) were diluted 1% BSA TBS-T (1:1000 dilution). The primary antibodies were visualized with goat anti-rabbit (Sigma) or goat anti-mouse (Bio-Rad, Hercules, CA) peroxidase-conjugated antibodies using an enhanced chemiluminescence detection system. As a control for protein loading, we used anti-STAT3 (1:2000) (Transduction Laboratories, Lexington, KY), anti-ERK1/2 (1:2000) (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-AKT (New England Biolabs) antibodies.
Statistical analysis
Gene expression data were normalized with the MAS5 algorithm and analyzed with our bioinformatics platform - RAGE http://rage.montp.inserm.fr/[24] and Amazonia, http://amazonia.montp.inserm.fr/) -, the SAM (Significance Analysis of Microarrays) software (as previously described)[19] and the Maxstat package used in R software(http://cran.r-project.org/). Statistical comparisons were done with Mann-Whitney, Chi-square, or unpaired or paired Student’s t-tests.
Results
Identification of cell communication signals involved in MMC/osteoclast interactions
To identify osteoclast-associated factors that could promote MMC survival, we investigated genes overexpressed in osteoclasts compared to either BM CD14 monocytes, BM CD15 polymorphonuclear cells, BM CD34 cells, BM stromal cells, or BM B cells using supervised SAM analysis (ratio ≥ 2, FDR ≤ 1%, 1000 permutations). Genes overexpressed in osteoclasts compared to normal BM plasma cells or primary MMC were also determined. Crossing these gene lists yielded 552 genes/expressed sequence tags (ESTs) significantly overexpressed in osteoclasts compared to the 7 BM cell populations (Supplemental Table S1). These genes were significantly (P < .05) enriched in genes encoding for 2 major pathways - “cellular function and maintenance” and “cell-to cell signaling and interaction” – as well as 10 other pathways (supplementary Figure S1).
Osteoclasts express genes coding for CCR2-chemokines specifically and high CCR2 gene expression in myeloma cells is associated with increased bone lesions
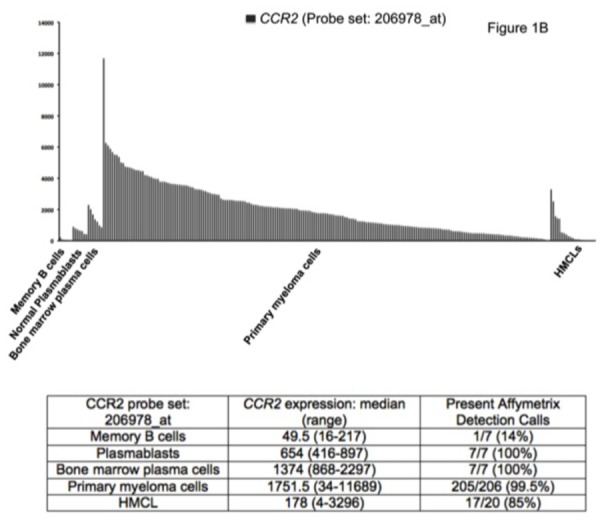
Seven of the 552 osteoclast genes encoded for chemokines (Figure 1A and Table S2). Four of these chemokines (CCL2, CCL7, CCL8 and CCL13) target the CCR2 receptor expressed by myeloma cells [25,26]. One gene encodes for CCL23 chemokine, which is a weak activator of CCR1[27], a chemokine receptor also expressed by myeloma cells[13,14,28]. The 2 other chemokine genes encode for CXCL5 whose receptor – CXCR2 - is not expressed by MMC (see http://amazonia.transcriptome.eu/), and CCL10 whose receptor is unknown. Besides these osteoclast-specific genes, osteoclasts expressed additional chemokine genes also expressed by other components of the bone marrow environment, in particular the MIP1-α/CCL3 gene expressed by monocytes, bone marrow stromal cells and MMC (Supplementary Figure S2) as previously described[29]. Given the very specific expression of genes coding for CCR2-chemokines by osteoclasts and that myeloma cells stimulate bone resorption[30], we questioned whether CCR2 gene expression of MMC from newly diagnosed patients with MM could be linked with increased bone lesions. The CCR2 gene expression on normal and malignant plasma cells is depicted in Figure 1B. The reliability of the Affymetrix probe set (206978_at) to assay for CCR2 gene expression was validated characterizing protein expression by flow cytometry in HMCLs. A strong correlation between CCR2 gene and protein expression was found (r = .90, P < .001, Supplementary Figure S3). Furthermore, CCR2 protein could not be detected in HMCLs with an absent Affymetrix call (Supplementary Figure S3). CCR2 expression was not expressed in normal memory B cells in agreement with its reported suppression by Pax5 B cell transcription factor[31]. CCR2 was expressed by normal plasmablasts, and bone marrow plasma cells and was significantly increased in MMC compared to normal BMPC (P ≤ .05, Figure 1B). We then investigated whether CCR2 expression in MMC could be linked with bone lesions. Using the Maxstat function of R package, a maximum difference in the proportion of patients with major bone structural damage or >3 osteolyses was obtained splitting patients into 2 groups with a CCR2 cutoff = 3735. Major bone structural damage or >3 osteolyses were observed in 55% of the patients with CCR2 signal in MMC > 3735 versus 36% of the patients with CCR2 signal ≤ 3735 (P < .05, Table 1). This CCR2 signal cutoff is 2.7 fold the median value of CCR2 signal in normal plasma cells. Of note, since osteoclasts specifically express the gene coding for CCL23 that activates CCR1 weakly, we also studied whether CCR1 expression in MMC could be linked with bone lesion extent. Contrarily to CCR2, no cutoff of CCR1 gene expression in MMC could identify a group of newly-diagnosed patients with increased bone lesions. An explanation is that various cells in the bone marrow environment could produce other CCR1-targeting chemokines. This is the case for MIP1-α/CCL3 whose gene is expressed by osteoclasts but also by monocytes, stromal cells, and MMC as mentioned above. This data suggests these chemokines could be important to recruit MMC to the bone marrow but not specifically close to osteoclasts.
Figure 1. Chemokine signature of osteoclasts.
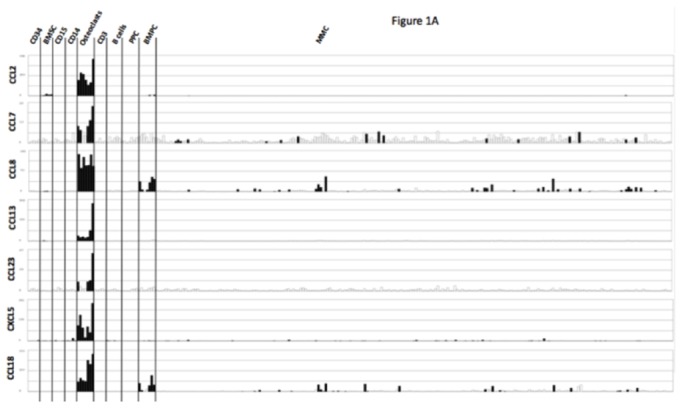
A. Affymetrix CCL2, CCL7, CCL8, CCL13, CCL23, CXCL5, and CCL18 gene expression in BM CD34 cells (n = 5), BM stromal cells (n = 5), purified BM CD15 (n = 5), CD14 (n = 5) and CD3 cells (n = 5), osteoclasts (n = 7), normal memory B cells (n = 6), normal polyclonal plasmablasts (n = 7), normal BM plasma cells (BMPC) (n = 7), and purified myeloma cells from consecutive patients with multiple myeloma (MM) (n = 206).
B. CCR2 expression in memory B cells, normal plasmablasts and normal bone marrow plasma cells from 7 healthy donors, 206 with MM and 20 HMCL.
The Affymetrix signal of the CCR2 probe set 206978_at is indicated on the y axis. CCR2 expression in each sample is indicated by the height of the bar. Samples are ordered from the highest to lowest expression of CCR2 gene from left to right on the x axis. The attached table shows the median value and range of Affymetrix signal and the percentage of samples with a present call.
Table 1. Clinical data of the CCR2high and CCR2low patients.
206 newly-diagnosed patients with MM were separated into 2 subgroups according to CCR2 gene expression in myeloma cells in order to optimize the difference in patients’ proportion with major structural damage or > osteolyses using the Maxstat package. The 2 subgroups represent patients with high (i.e. CCR2 signal > 3735, CCR2high) or low (i.e. CCR2 signal ≤ 3735, CCR2low) CCR2 expression. Data are the percentages of patients within each subgroup with the indicated clinical or biological parameter.
| Categories | CCR2high (n=29) | CCR2low (n=177) | |
|---|---|---|---|
| % of patients in each subgroup | |||
| Bone lesions: Major structural damage or >3 osteolyses | 55%* | 36%* | P = .03 |
| Age ≥ 65 yr | 21% | 19% | NS |
| Kappa light chain | 64% | 64% | NS |
| Lambda light chain | 36% | 34% | NS |
| Non-secreting | 0% | 2% | NS |
| IgA subtype | 21% | 25% | NS |
| B2M ≤ 3.5 mg/liter | 72% | 62% | NS |
| B2M > 5.5 mg/liter | 7% | 17% | NS |
| Lactate dehydrogenase ≥ 240 IU/liter | 18% | 24% | NS |
| Albumin < 35 g/liter | 21% | 35% | NS |
| Hemoglobin < 10 g/dl | 10%* | 32%* | P = .01 |
| C-reactive protein ≥ 5 mg/liter | 38% | 36% | NS |
| CCR2high | CCR2low | |||||
|---|---|---|---|---|---|---|
| Staging | I | II | III | I | II | III |
| Salmon and Durie | 10% | 3% | 87% | 11% | 17% | 72% |
| NS | ||||||
When the percentages were statistically significantly different with a Chisquare test (P ≤ .05), the data are shown in italic.
Role of CCR2-chemokines in the osteoclast chemoattractant activity for myeloma cells
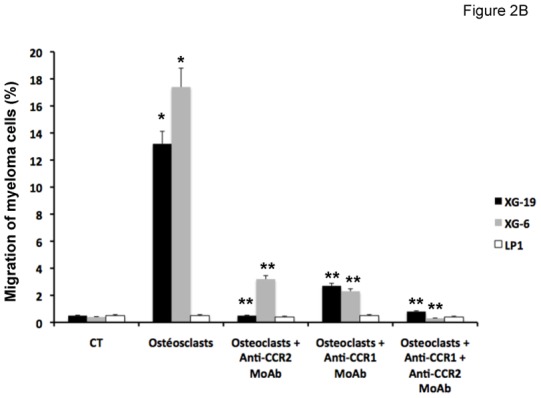
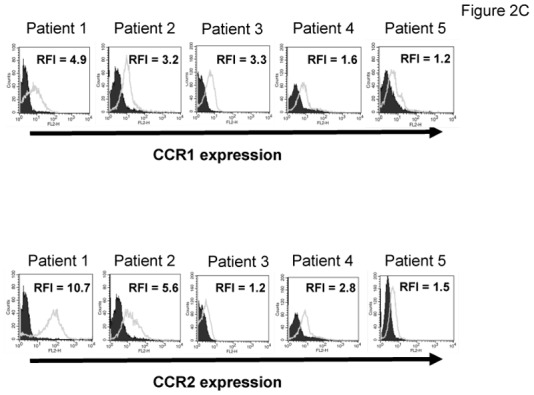
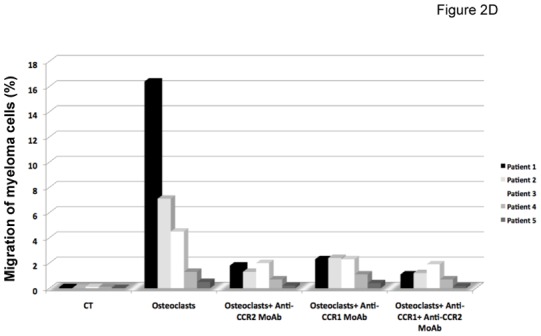
The chemoattractant activity of osteoclasts for MMC was evaluated using the XG-6, XG-19, and LP1 HMCLs. XG-19 and XG-6 myeloma cells express CCR2 gene and protein, unlike LP1 myeloma cells (Figure 2). Osteoclasts could efficiently attract XG-19 and XG-6 cells, unlike LP1 cells in a transwell assay. This chemoattractant activity was inhibited by a neutralizing MoAb to CCR2 by 95% and 81% for the XG-19 and XG-6 cells respectively (P ≤ .05, Figure 2B). As osteoclasts express MIP1-α/CCL3 chemokine gene together with monocytes or bone marrow stromal cells and specifically CCL23 gene, we also investigated a role of CCR1-targeting chemokines in the osteoclast chemoattractant activity. XG-19 and XG-6 cells expressed CCR1 and a MoAb to CCR1 inhibited by 80% and 87% the osteoclast-induced migration of XG-19 and XG-6 cells respectively (P ≤ .05, Figure 2B). Addition of both anti-CCR1 and anti-CCR2 MoAbs further inhibited the osteoclast-mediated migration of myeloma cells (Figure 2B). Similar data were obtained with primary myeloma cells of 5 patients (Figure 2C&D). Primary myeloma cells variably expressed CCR2 or CCR1 assayed by flow cytometry depending on patients’ sample. Osteoclasts could attract more than 4% primary myeloma cells for 3 out of the 5 patients, the highest chemoattraction being observed with primary myeloma cells with the highest CCR2 and CCR1 expression (Figure 2C&D). The osteoclast chemotactic activity was inhibited by a neutralizing anti-CCR2 MoAb (P < .05, Figure 2D). It was also inhibited by a neutralizing anti-CCR1 MoAb (P < .05, Figure 2D)
Figure 2. Gene and protein expression of CCR1 and CCR2 chemokine receptors.
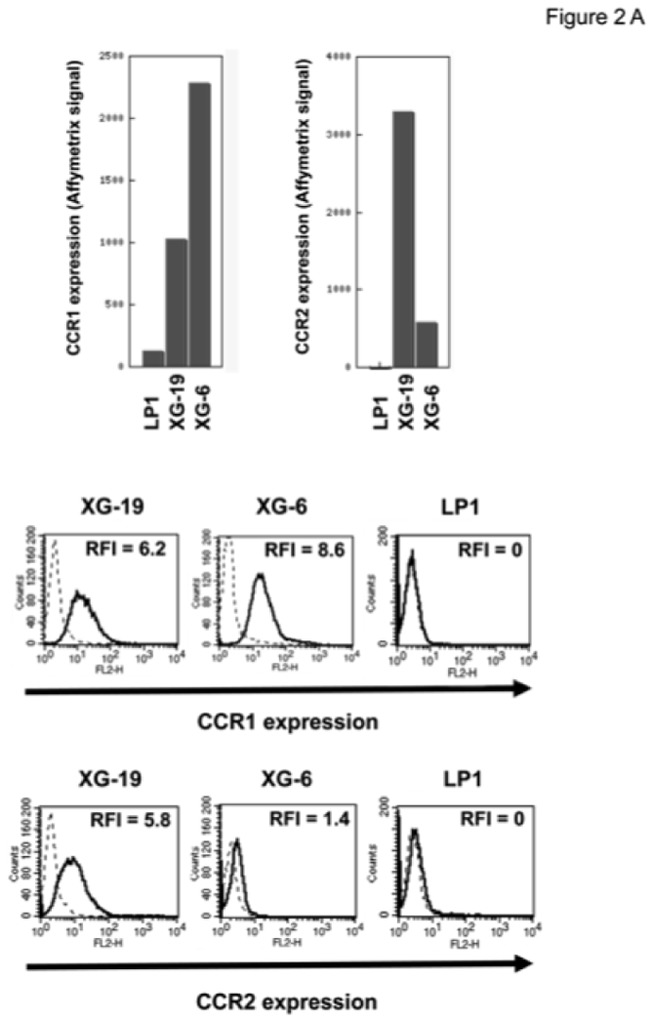
A. Data are Affymetrix signals for CCR1 (probe set 205098_at) and CCR2 (probe set 206978_at) in LP1, XG-19 and XG-6 HMCLs. Membrane expression of CCR1 or CCR2 was evaluated by flow cytometry using PE-conjugated anti-CCR1 or anti-CCR2 MoAbs or isotype-related PE-conjugated control MoAbs. The numbers in the panels are the RFI of the PE-conjugated anti-CCR1 or anti-CCR2 MoAbs compared to the PE-conjugated control MoAbs.
B. The chemoattractant activity of osteoclasts to myeloma cells was assayed using XG-19, XG-6 and LP1 HMCLs. Data are the fraction of MMC in the upper chamber of the transwell that could migrate to the lower chamber. Results are the mean values ± SD of 3 experiments. * indicates a significant difference of MMC migration in MMC/osteoclast cocultures compared to control using a paired Student’s t test (P ≤ .05). ** indicates a significant difference of MMC migration in MMC/osteoclast cocultures with anti-CCR1 and/or anti-CCR2 MoAb compared to MMC/osteoclast cocultures using a paired Student’s t test (P ≤ .05).
C. The expression of CCR1 and CCR2 by CD138+ primary myeloma cells of patients was evaluated by flow cytometry using FITC-conjugated anti-CD138 MoAb and PE-conjugated anti-CCR1 or anti-CCR2 MoAbs labelling. Isotype matched FITC-conjugated or PE-conjugated MoAbs recognizing no human antigens were used as control MoAb. The numbers in the panels are the RFI of the PE-conjugated anti-CCR1 or anti-CCR2 MoAbs compared to the PE-conjugated control MoAbs.
D. The chemoattractant activity of osteoclasts to myeloma cells was assayed using primary myeloma cells. Data are the fraction of primary MMC in the upper chamber of the transwell that could migrate to the lower chamber.
CCR2-chemokines activate MAP kinase pathway in myeloma cells and promote myeloma cell growth and a MAP kinase inhibitor abrogates the chemoattractant activity of osteoclasts to myeloma cells
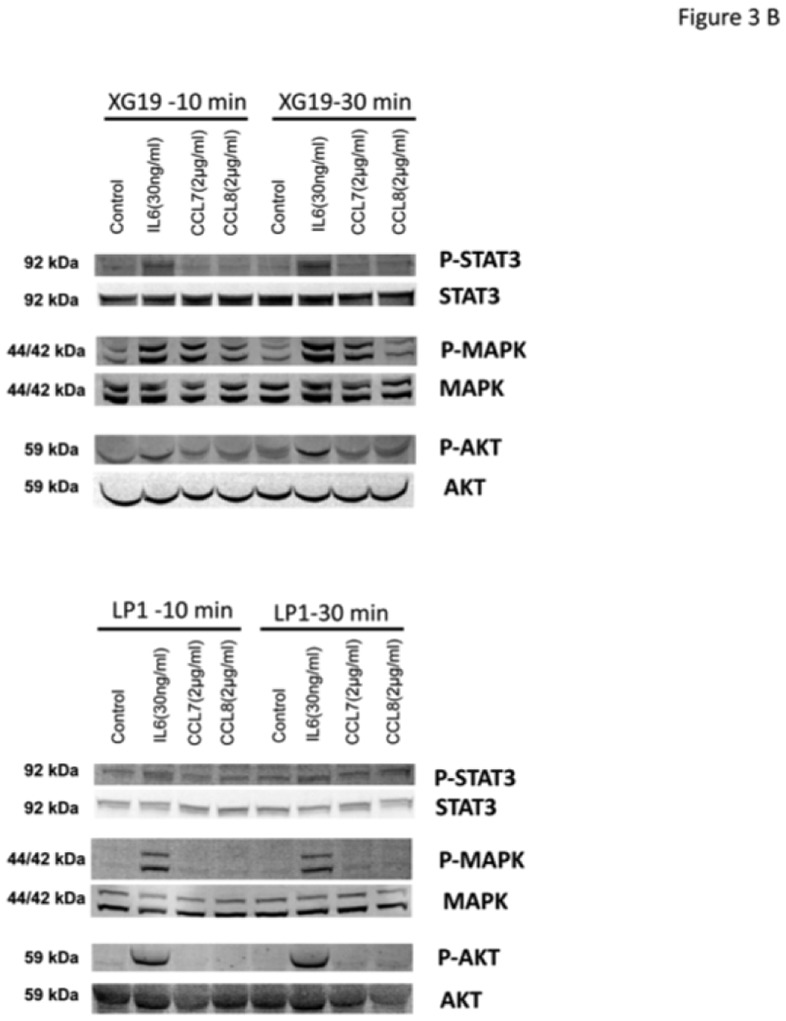
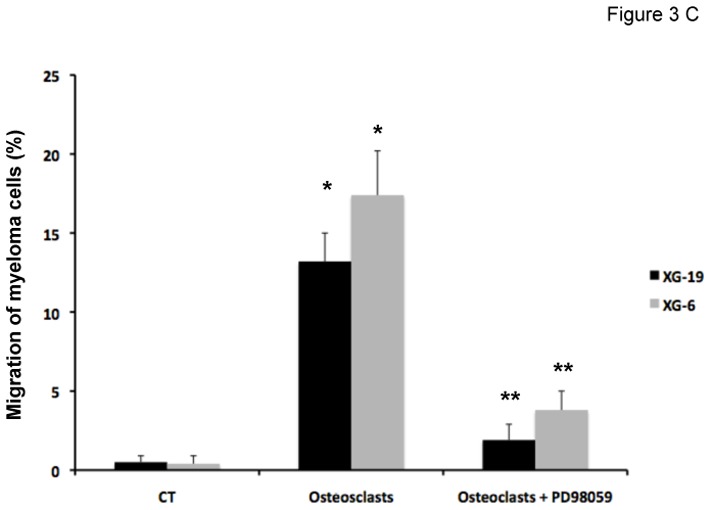
The recombinant CCL7, CCL8, and CCL13 CCR2-chemokines significantly increased the growth of CCR2+ XG-19 cells and only CCL8 that of XG-6 cells (P ≤ .05, Figure 3A). These chemokines did not stimulate the growth of CCR2− LP1 cells (Figure 3A). CCL7 and CCL8 chemokines, which had the highest effects on myeloma cell growth, were assayed for signal transduction. The two chemokines induced MAPK phosphorylation in XG-19 cells. They did not induce STAT3 or AKT phosphorylations whereas IL-6 induced the phosphorylation of STAT3, MAPK and AKT, in agreement with previous data[32]. CCL7 or CCL8 did not induce MAPK phosphorylation in CCR1−CCR2− LP1 cells (Figure 3B). As the osteoclast chemoattractant activity for MMC was inhibited by anti-CCR2 MoAbs and as CCR2-chemokines activated MAPK pathway in MMC, we looked for the effect of a MAP kinase inhibitor on osteoclast chemoattractant activity for MMC. The PD98059 MAP kinase inhibitor dramatically inhibited the migration of myeloma cells towards osteoclasts (Figure 3C).
Figure 3. CCL7, CCL8 and CCL13 support the growth of CCR2+ HMCLs.
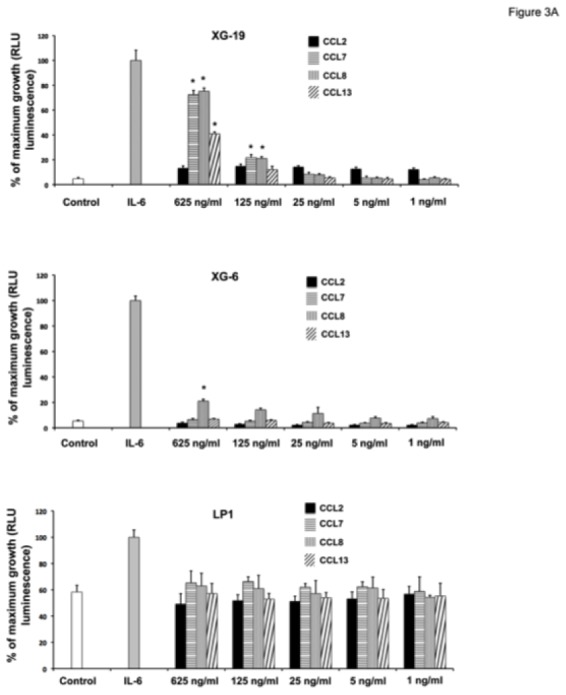
A. XG-19, LP1 and XG-6 were IL-6 starved for 3 hours and cultured either with no cytokine, or in the presence of IL-6 (3 ng/mL) or in the presence of increasing concentrations of CCL2, CCL7, CCL8 or CCL13. Results are the mean ± SD values of the RLU fluorescence determined on sextuplet culture wells. Results are those of one experiment representative of 5. *Mean value is significantly different from that obtained without adding cytokine using a Student t test (P ≤ .05).
B. XG-19 or LP1 cells were starved overnight and cultured without cytokine, or with either IL-6 (30 ng/mL), CCL7 (2 μg/mL), or CCL8 (2 μg/mL) for 10 and 30 minutes at 37°C. Cell lysates were probed by Western blotting with antibodies against phospho-STAT3 (pSTAT3), phospho-ERK1/2 (pMAPK), and phospho-AKT (pAKT). Blots were reprobed with antibodies to STAT3, MAPK, and AKT proteins to quantitate protein loading. Western blots are of one experiment representative of 3.
C. The chemoattractant activity of osteoclasts to XG-19 or XG-6 myeloma cells was assayed with or without a MAPK inhibitor (PD98059). Data are the fraction of primary MMC in the upper chamber of the transwell that could migrate to the lower chamber and are mean values of three experiments. * indicates a significant increase in MMC migration in MMC/osteoclast cocultures compared to MMC alone using a paired Student’s t test (P ≤ .05). ** indicates a significant decrease of MMC migration in MMC/osteoclast cocultures with PD98059 (10 μM) compared to MMC/osteoclast cocultures using a paired Student’s t test (P ≤ .05).
Growth factors produced by osteoclasts
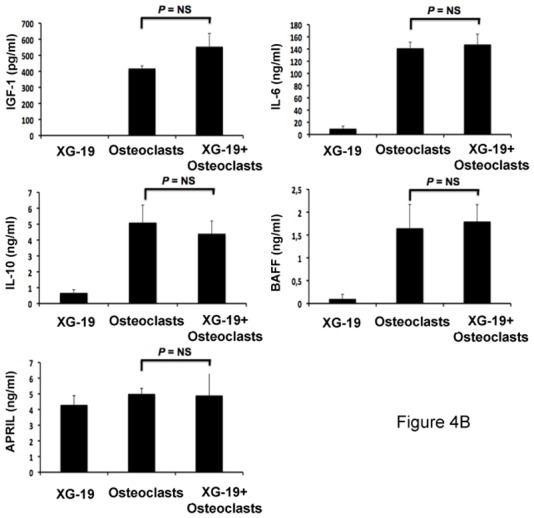
Osteoclasts overexpress genes encoding for previously reported MGFs, including IGF-1[33], IL-10[34] and APRIL[32] (Figures 4A). IL-6 and BAFF genes are also highly expressed by osteoclasts but were not picked in the osteoclast gene list because they are also highly expressed by BM stromal cells (IL-6), monocytes, or polymorphonuclear cells (BAFF)(Figure 4A). Osteoclasts produced IGF-1 or APRIL proteins. 418 ± 16 pg/ml of IGF-1 and 5 ± 0.3 ng/ml of APRIL were detected in 3-day culture supernatants of osteoclasts (Figure 4B), concentrations that are active on myeloma cells[32,33]. Osteoclasts also produced IL-6 (141 ± 13 pg/mL) and BAFF (1.6 ± 0.5 ng/mL) and produced a low amount of IL-10 (5.1 ± 1.1 pg/mL) compared to the known biologically active concentrations [35]. A coincubation of osteoclasts with XG-19 MMC did not significantly affect the production of IGF-1, IL-6, APRIL, BAFF or IL-10 (Figure 4B).
Figure 4. Gene expression of several myeloma cell growth factors.
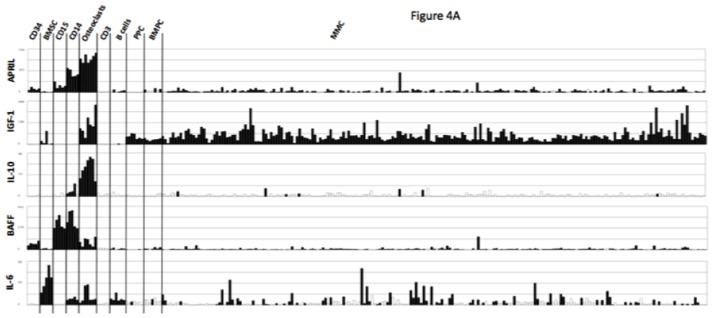
A. Affymetrix APRIL, IGF-1, IL-10, BAFF and IL-6 gene expression in BM CD34 cells (n = 5), BM stromal cells (n = 5), purified BM CD15 (n = 5), CD14 (n = 5) and CD3 cells (n = 5), osteoclasts (n = 7), normal memory B cells (n = 6), normal polyclonal plasmablasts (n = 7), normal BM plasma cells (BMPC) (n = 7), and purified myeloma cells from patients with multiple myeloma (MM) (n = 131).
B. The concentrations of BAFF, APRIL, IL-10, IL-6, or IGF-1 were assayed with an ELISA in 3-day culture supernatant of XG-1 and XG19 cells, osteoclasts and XG-1/osteoclasts and XG-19/osteoclasts cocultures. Results are the mean value of three independent experiments.
To investigate the role of these various growth factors in the survival activity of MMC induced by osteoclasts, we used the XG-19 and XG-20 HMCLs that expressed IL-6R, IL-10R, IGF1R, TACI and BCMA BAFF/APRIL receptors. Upon IL-6 deprivation, XG-19 and XG-20 cells rapidly apoptosed and a coculture with osteoclasts protected them from apoptosis (P = .01). FACS data from one representative experiment are shown in Figure 5A and the mean values ± SD of five experiments in Table 2. The survival promoting activity of osteoclasts was partially inhibited blocking IL-6 using the B-E8 anti-IL-6 MoAb (P = .05 for XG-19 and P = .01 for XG-20) or BAFF/APRIL using TACI-Fc fusion protein (P = .01 for XG-19 and P = .02 for XG-20). Inhibiting IGF-1 using an anti-IGF-1R MoAb completely blocked MMC survival supported by osteoclasts in serum-free culture medium (P = .003 for XG19 and P = .0005 for XG20) (Figure 5A). The anti-IGF-1R MoAb further increased the apoptosis rate observed without osteoclasts, likely by inhibiting the autocrine IGF-1/IGF-1R loop occurring in some HMCLs[33]. A neutralizing anti-IL-10 MoAb had no significant effect. The anti-CCR2 MoAb did not either inhibit the MMC growth activity of osteoclasts, but this anti-CCR2 MoAb partially blocked the growth activity of recombinant CCL7 or CCL8 (Supplementary Figure S4), whereas it blocked efficiently their chemoattractant activity (Figure 2B).
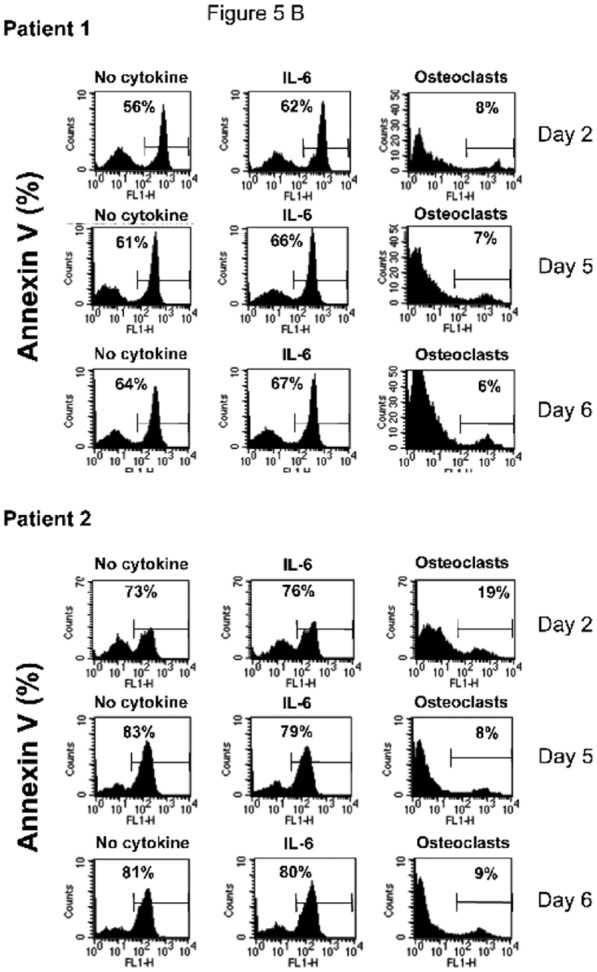
Figure 5. Osteoclasts promote the survival of cytokine-dependent myeloma cell lines.
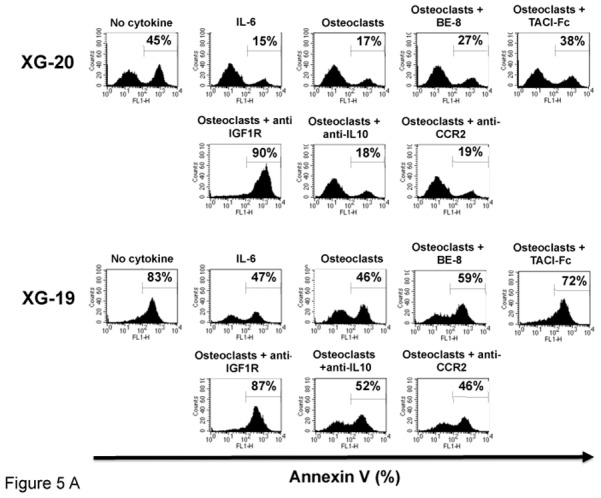
A. XG-19 and XG-20 myeloma cells were cultured at 105 cells/ml without cytokine, or with IL-6 (2 ng/ml), or with 104 osteoclasts with or without anti-IL-6 (BE8) MoAb (10 μg/ml), TACI-Fc (20 μg/ml), anti-IGF-1R MoAb (4 μg/ml), anti-IL-10 MoAb (10 μg/ml) or anti-CCR2 MoAb (10 μg/ml). Cells were recovered after 3 days of culture and apoptotic cells were detected by annexin V staining. Results are those of one experiment representative of five.
B. Purified MMC of patients were cultured at 105 cells/ml without cytokine or in the presence of osteoclasts (1 osteoclast for 4 MMC) or IL-6 (2 ng/ml). Cells were recovered after 2, 5 and 6 days of culture and apoptotic cells were detected by annexin V staining.
Table 2.
XG-19 and XG-20 HMCLs were cultured at 105 cells/ml without cytokine or with IL-6 (2 ng/ml), or with 104 osteoclasts with or without anti-IL-6 (BE8) MoAb (10 mg/ml), TACI-Fc (20 mg/ml), anti-IGF-1R MoAb (4 mg/ml), anti-IL-10 MoAb (10 mg/ml) or anti-CCR2 MoAb (10 mg/ml). Cells were recovered after 3 days of culture and apoptotic cells were detected by annexin V staining. Results are the means ± SD of five experiments.
| XG-20 | XG-19 | |||
|---|---|---|---|---|
| % Annexin V | P value (N = 5) | % Annexin V | P value (N = 5) | |
| No cytokine | 48 ± 9 | 90 ± 3 | ||
| IL-6 | 21 ± 7 | .008 (compared to No cytokine) | 56 ± 4 | .0003 (compared to No cytokine) |
| Osteoclasts | 16 ± 3 | .01 (compared to No cytokine) | 41 ± 9 | .01 (compared to No cytokine) |
| Osteoclasts + B-E8 | 25 ± 2 | .01 (compared to Osteoclasts) | 51 ± 8 | .05 (compared to Osteoclasts) |
| Osteoclasts + TACI-Fc | 29 ± 5 | .02 (compared to Osteoclasts) | 67 ± 8 | .01 (compared to Osteoclasts) |
| Osteoclasts + anti-IGF1R | 84 ± 8 | .0005 (compared to Osteoclasts) | 76 ± 9 | .003 (compared to Osteoclasts) |
| Osteoclasts + anti-IL-10 | 18 ± 2 | NS (compared to Osteoclasts) | 42 ± 4 | NS (compared to Osteoclasts) |
| Osteoclasts + anti-CCR2 | 19 ± 2 | NS (compared to Osteoclasts) | 43 ± 3 | NS (compared to Osteoclasts) |
NS: not significantly different
Osteoclasts promote the survival of primary MMC
Purified primary MMC rapidly apoptosed in vitro within 2 days[36] and adding recombinant IL-6 did not prevent apoptosis. A coculture with osteoclasts completely prevented primary MMC from apoptosis on day 2 and this protective effect lasted until day 6 (Figure 5 B). Given the rarity of primary MMC, it was not possible to add the inhibitors to the various growth factors produced by osteoclasts to identify those that are critical.
Discussion
The aim of this study was to identify some major cell communication signals involved in recruitment of MMC close to osteoclasts and subsequent support of MMC survival[4,37].
We first identified 552 genes that are significantly overexpressed in osteoclasts compared to various cell components of the BM microenvironment and to MMC. Comparing our results with previous gene array analyses of osteoclasts, 22 common genes coding for secreted or extracellular matrix proteins were identified in Pederson’s study[38] and in our current study, in particular CCL7, CCL8, and CXCL5 chemokine genes and RANK, IGF-1 and IL-10 genes (Table S2). This stringent method was initially chosen to limit the size of the osteoclast gene list, being aware that some genes that are highly expressed both in osteoclasts and in another cell population could be eliminated. Our data show this method is of interest to find the signals that may attract MMC specifically to osteoclasts and promote MMC survival.
There are about 50 chemokines[39], some of them being already found to be produced in the bone marrow milieu of patients with MM: mainly CXCL12/SDF-1 (targeting CXCR4 and produced by stromal cells, endothelial cells and myeloma cells) and MIP1α/CCL3 (targeting CCR1 and CCR5 and produced by osteoclasts, bone marrow stromal cells, osteoblasts, monocytes, and myeloma cells [29]). These chemokines are important to recruit circulating MMC into the BM as illustrated for CXCL12/SDF-1[40]. They could be also critical to trigger the fate of a myeloma cell in the presence of gradients of multiple chemokines produced by various environment cells. The current data provide some answer regarding the chemokines targeting CCR1 or CCR2, both receptors being expressed by myeloma cells[28]. These CCR1- or CCR2-chemokines are produced by osteoclasts and we show that CCR1 and CCR2 chemokines contribute to osteoclast chemoattractant activity for myeloma cells in vitro, in agreement with previous data for CCR1-chemokines[29]. In addition, we show that the expression of CCR2-chemokines is very specific to osteoclasts, whereas it was previously shown that various cells including osteoclasts produce the MIP-1α/CCL3 CCR1-chemokine[13,14]. This suggests that a myeloma cell that expresses CCR1 could be recruited close to the various cells producing MIP-1α/CCL3, i.e. osteoclasts, stromal cells, monocytes, or myeloma cells. But if the myeloma cell expresses CCR2, it could be attracted by osteoclasts specifically. Given that osteoclast can recruit and promote survival of CCR2+ myeloma cells, and in turn, myeloma cells promote osteoclast recruitment and activation, it is hardly surprising that the bone lesion number in patients with newly-diagnosed MM was associated with high CCR2 expression in myeloma cells, unlike CCR1 expression. This hypothesis should be further tested injecting CCR2+ or CCR2− myeloma cell lines in immunodeficient mice. If the specificity for human CCR2-targeting chemokine expression is kept by murine osteoclasts, one should expect that CCR2+ myeloma cell lines could create more bone lesions than CCR2− ones and that anti-CCR2 antibodies could prevent bone lesions and even tumor growth. Alternatively, this could be tested in the murine 5T2 multiple myeloma model since these murine myeloma cells express CCR2[41].
Chemokine receptors are G-protein-coupled receptors which can activate various signaling pathways depending on chemokine receptor and cell type: MAPK, PI-3/AKT, Src, JNK, Rho[42,43]. The CCR2-chemokines induced MAPK phosphorylation in myeloma cells, and did not activate Akt or Stat3 pathways, in agreement with the inhibition of osteoclast chemoattractant activity for myeloma cells by PD98059 MAPK inhibitor. High concentrations of recombinant CCR2-chemokines (≥ 125 ng/ml) can trigger the survival of CCR2+ MMC unlike CCR2− MMC.
Osteoclasts also highly expressed genes encoding for some major MGFs and produced high levels of these MGFs: IGF-1, APRIL and IL-6 and produced the corresponding proteins. A high expression of IL-10 gene was found, but no protein production. In agreement with growth factor production, the survival of MMC induced by osteoclast was partly inhibited by IL-6 and APRIL inhibitors, indicating their contribution to osteoclast-induced MMC survival. An interesting finding is the complete inhibition of osteoclast-induced survival of MMC by the anti-IGF-1R MoAb. This may have by two reasons. First of all, IGF-1 is a critical factor for the generation and survival of osteoclasts[44] and the neutralizing anti-IGF-1R decreased osteoclast survival in our culture system (results not shown). Secondly, IGF-1 is a major MGF, both as a paracrine and autocrine growth factor. In particular, the MGF activities of IL-6, HGF, or EGF-family members are inhibited in part by IGF-1R inhibitors, being dependent of an autocrine IGF-1 loop produced by MMCs[33].
Bone lesions in MM are due to an abnormal bone remodeling with increased osteoclast and decreased osteoblast activities[37,45]. In healthy individuals, bone remodeling occurs in closed bone remodeling compartments (BRCs) containing osteoblasts and osteoclasts[46]. These BRCs are protected from bone marrow cells by a canopy of flat cells with osteoblastic markers[46–48]. Red blood cells are found in the BRCs and radiolabelled ferritin is detected in BRCs 5 minutes after injection[47,48]. This indicated that BRCs are linked to the vasculature through capillaries that are directly connected to the BRC canopy[48]. This connection of closed BRCs with the vasculature should insure the recruitment of osteoblasts or osteoclast progenitors, while protecting BRC cells from other bone marrow cells. In patients with MM, the canopy of BRCs is frequently disrupted in association with the extent of bone lesions[48]. This canopy disruption makes a direct contact possible between osteoclasts and MMC.
The current data suggest that osteoclasts can directly recruit MMC by producing CCR2-chemokines, promote MMC survival, growth and drug resistance by producing various growth factors. Vice versa, MMC will further promote osteoclast progenitor recruitment and differentiation producing MIP1-α/CCL3, MIP-1β, and CXCL12 chemokines, IGF-1, and increasing RANKL production by stromal cells[4,37,45,49,50]. Thus osteoclasts and MMC can recruit each other and mutually promote their survival through various mechanisms. It will be interesting to investigate whether this strong cooperation between MMC and osteoclasts is a reflection of a cooperation occurring between osteoclasts and normal plasma cells. Indeed, CCR2 is a plasma cell marker, which is not expressed by B cells. The reason is that CCR2 gene expression is repressed by the Pax5 B cell transcription factor[31]. This negative control by Pax5 is removed when B cells differentiate into plasma cells, due to the repression of Pax5 gene expression by the Blimp1 plasma cell transcription factor[23,31]. After generation in the lymph node and exit into the circulation, plasma cells have to find a niche either in the bone marrow or in mucosa. CXCR4 is one of the critical homing molecules for normal plasma cells to the bone marrow by SDF-1 producing stromal cells[28]. CCR2 could be another critical homing molecule of normal plasma cells towards osteoclasts producing CCR2-chemokines, providing a physiological explanation of its specific induction in plasma cell differentiation. The concentration of CCR2-chemokines as well as plasma cell survival factors produced by osteoclasts – IL-6, APRIL – should be high in the closed BRCs whose canopy is directly linked to a capillary, making the recruitment and survival of these normal plasma cells possible. Using an in-vitro model of normal plasma cell generation[23], we have found that osteoclasts can promote the survival of normal plasma cells (unpublished observations).
In conclusion, we have identified an additional mechanism involved in the interaction of osteoclasts and MMC. Osteoclasts are the main cells in the bone marrow environment that produce various CCR2-chemokines enabling malignant plasma cells attraction. Osteoclasts also produce the major growth factors for MMC – IGF-1, IL-6, APRIL. Targeting the osteoclast/MMC interaction through CCR2 and/or IGF-1 appears to be a promising therapeutic approach in myeloma.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ligue Nationale Contre le Cancer (équipe labellisée 2009), Paris, France, from INCA (n∘RPT09001FFA) and from MSCNET European strep (N∘E06005FF), and in part by grants from the Hopp-Foundation (Germany); the University of Heidelberg (Heidelberg, Germany); the National Center for Tumor Diseases (Heidelberg, Germany); the Tumorzentrum Heidelberg/Mannheim, Germany; the Deutsche Krebshilfe (Bonn, Germany); The Deutsche Forschungsgemeinschaft (DGF) Transregio TRR79 (Bonn, G ermany); Novartis Pharma GmbH, Nürnberg, Germany. Microarray experiments were performed at the Institute of Research in Biotherapy (http://irb.chu-montpellier.fr/en/laboratories_microarray.html) in the Montpellier University Hospital
Footnotes
Author contributions:
MJ performed research and participated in the writing of the paper.
KA completed western-blot experiments.
HD, and GH collected bone marrow samples and clinical data and participated in the writing of the paper.
RT participated in the bioinformatics studies and participated in the writing of the paper.
MP and RG provided with technical assistance.
KB participated in the design of the research and the writing of the paper.
References
- 1.Bataille R, Chappard D, Marcelli C, et al. Recruitment of new osteoblasts and osteoclasts is the earliest critical event in the pathogenesis of human multiple myeloma. J Clin Invest. 1991;88:62–66. doi: 10.1172/JCI115305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corre J, Mahtouk K, Attal M, et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia. 2007;21:1079–88. doi: 10.1038/sj.leu.2404621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ria R, Todoerti K, Berardi S, et al. Gene expression profiling of bone marrow endothelial cells in patients with multiple myeloma. Clin Cancer Res. 2009;15:5369–5378. doi: 10.1158/1078-0432.CCR-09-0040. [DOI] [PubMed] [Google Scholar]
- 4.Yaccoby S, Wezeman MJ, Henderson A, et al. Cancer and the microenvironment: myeloma-osteoclast interactions as a model. Cancer Res. 2004;64:2016–2023. doi: 10.1158/0008-5472.can-03-1131. [DOI] [PubMed] [Google Scholar]
- 5.Andersen TL, Soe K, Sondergaard TE, Plesner T, Delaisse JM. Myeloma cell-induced disruption of bone remodelling compartments leads to osteolytic lesions and generation of osteoclast-myeloma hybrid cells. Br J Haematol. 2010;148:551–61. doi: 10.1111/j.1365-2141.2009.07980.x. [DOI] [PubMed] [Google Scholar]
- 6.Han JH, Choi SJ, Kurihara N, Koide M, Oba Y, Roodman GD. Macrophage inflammatory protein-1alpha is an osteoclastogenic factor in myeloma that is independent of receptor activator of nuclear factor kappaB ligand. Blood. 2001;97:3349–3353. doi: 10.1182/blood.v97.11.3349. [DOI] [PubMed] [Google Scholar]
- 7.Giuliani N, Colla S, Sala R, et al. Human myeloma cells stimulate the receptor activator of nuclear factor-kappa B ligand (RANKL) in T lymphocytes: a potential role in multiple myeloma bone disease. Blood. 2002;100:4615–4621. doi: 10.1182/blood-2002-04-1121. [DOI] [PubMed] [Google Scholar]
- 8.Pearse RN, Sordillo EM, Yaccoby S, et al. Multiple myeloma disrupts the TRANCE/osteoprotegerin cytokine axis to trigger bone destruction and promote tumor progression. Proc Natl Acad Sci U S A. 2001;98:11581–11586. doi: 10.1073/pnas.201394498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreaux J, Cremer FW, Reme T, et al. The level of TACI gene expression in myeloma cells is associated with a signature of microenvironment dependence versus a plasmablastic signature. Blood. 2005;106:1021–1030. doi: 10.1182/blood-2004-11-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piva R, Penolazzi L, Borgatti M, et al. Apoptosis of human primary osteoclasts treated with molecules targeting nuclear factor-kappaB. Ann N Y Acad Sci. 2009;1171:448–456. doi: 10.1111/j.1749-6632.2009.04906.x. [DOI] [PubMed] [Google Scholar]
- 11.Giuliani N, Colla S, Morandi F, et al. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood. 2005;106:2472–2483. doi: 10.1182/blood-2004-12-4986. [DOI] [PubMed] [Google Scholar]
- 12.Oyajobi BO, Mundy GR. Receptor activator of NF-kappaB ligand, macrophage inflammatory protein-1alpha, and the proteasome: novel therapeutic targets in myeloma. Cancer. 2003;97:813–817. doi: 10.1002/cncr.11133. [DOI] [PubMed] [Google Scholar]
- 13.Oba Y, Lee JW, Ehrlich LA, et al. MIP-1alpha utilizes both CCR1 and CCR5 to induce osteoclast formation and increase adhesion of myeloma cells to marrow stromal cells. Exp Hematol. 2005;33:272–278. doi: 10.1016/j.exphem.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Vallet S, Raje N, Ishitsuka K, et al. MLN3897, a novel CCR1 inhibitor, impairs osteoclastogenesis and inhibits the interaction of multiple myeloma cells and osteoclasts. Blood. 2007;110:3744–3752. doi: 10.1182/blood-2007-05-093294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callander NS, Roodman GD. Myeloma bone disease. Semin Hematol. 2001;38:276–285. doi: 10.1016/s0037-1963(01)90020-4. [DOI] [PubMed] [Google Scholar]
- 16.Terpos E, Szydlo R, Apperley JF, et al. Soluble receptor activator of nuclear factor kappaB ligand-osteoprotegerin ratio predicts survival in multiple myeloma: proposal for a novel prognostic index. Blood. 2003;102:1064–1069. doi: 10.1182/blood-2003-02-0380. [DOI] [PubMed] [Google Scholar]
- 17.Abe M, Hiura K, Wilde J, et al. Role for macrophage inflammatory protein (MIP)-1alpha and MIP-1beta in the development of osteolytic lesions in multiple myeloma. Blood. 2002;100:2195–2202. [PubMed] [Google Scholar]
- 18.Choi SJ, Oba Y, Gazitt Y, et al. Antisense inhibition of macrophage inflammatory protein 1-alpha blocks bone destruction in a model of myeloma bone disease. J Clin Invest. 2001;108:1833–1841. doi: 10.1172/JCI13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreaux J, Hose D, Jourdan M, et al. TACI expression is associated with a mature bone marrow plasma cell signature and C-MAF overexpression in human myeloma cell lines. Haematologica. 2007;92:803–811. doi: 10.3324/haematol.10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldschmidt H, Sonneveld P, Cremer FW, et al. Joint HOVON-50/GMMG-HD3 randomized trial on the effect of thalidomide as part of a high-dose therapy regimen and as maintenance treatment for newly diagnosed myeloma patients. Ann Hematol. 2003;82:654–659. doi: 10.1007/s00277-003-0685-2. [DOI] [PubMed] [Google Scholar]
- 21.Barlogie B, Tricot G, Rasmussen E, et al. Total therapy 2 without thalidomide in comparison with total therapy 1: role of intensified induction and posttransplantation consolidation therapies. Blood. 2006;107:2633–2638. doi: 10.1182/blood-2005-10-4084. [DOI] [PubMed] [Google Scholar]
- 22.Rossi JF, Moreaux J, Hose D, et al. Atacicept in relapsed/refractory multiple myeloma or active Waldenstrom’s macroglobulinemia: a phase I study. Br J Cancer. 2009;101:1051–1058. doi: 10.1038/sj.bjc.6605241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jourdan M, Caraux A, De Vos J, et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009;114:5173–5181. doi: 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reme T, Hose D, De Vos J, et al. A new method for class prediction based on signed-rank algorithms applied to Affymetrix microarray experiments. BMC Bioinformatics. 2008;9:16. doi: 10.1186/1471-2105-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vande Broek I, Asosingh K, Vanderkerken K, Straetmans N, Van Camp B, Van Riet I. Chemokine receptor CCR2 is expressed by human multiple myeloma cells and mediates migration to bone marrow stromal cell-produced monocyte chemotactic proteins MCP-1, -2 and -3. Br J Cancer. 2003;88:855–862. doi: 10.1038/sj.bjc.6600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal R, Ghobrial IM, Roodman GD. Chemokines in multiple myeloma. Exp Hematol. 2006;34:1289–1295. doi: 10.1016/j.exphem.2006.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berahovich RD, Miao Z, Wang Y, Premack B, Howard MC, Schall TJ. Proteolytic activation of alternative CCR1 ligands in inflammation. J Immunol. 2005;174:7341–7351. doi: 10.4049/jimmunol.174.11.7341. [DOI] [PubMed] [Google Scholar]
- 28.De Vos J, Hose D, Reme T, et al. Microarray-based understanding of normal and malignant plasma cells. Immunol Rev. 2006;210:86–104. doi: 10.1111/j.0105-2896.2006.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards CM, Zhuang J, Mundy GR. The pathogenesis of the bone disease of multiple myeloma. Bone. 2008;42:1007–1013. doi: 10.1016/j.bone.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roodman GD. New potential targets for treating myeloma bone disease. Clin Cancer Res. 2006;12:6270s–6273s. doi: 10.1158/1078-0432.CCR-06-0845. [DOI] [PubMed] [Google Scholar]
- 31.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 32.Moreaux J, Legouffe E, Jourdan E, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–3157. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprynski AC, Hose D, Caillot L, et al. The role of IGF-1 as a major growth factor for myeloma cell lines and the prognostic relevance of the expression of its receptor. Blood. 2009;113:4614–4626. doi: 10.1182/blood-2008-07-170464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu ZJ, Costes V, Lu ZY, et al. Interleukin-10 is a growth factor for human myeloma cells by induction of an oncostatin M autocrine loop. Blood. 1996;88:3972–3986. [PubMed] [Google Scholar]
- 35.Lu ZY, Zhang XG, Rodriguez C, et al. Interleukin-10 is a proliferation factor but not a differentiation factor for human myeloma cells. Blood. 1995;85:2521–2527. [PubMed] [Google Scholar]
- 36.Gu ZJ, Vos JD, Rebouissou C, et al. Agonist anti-gp130 transducer monoclonal antibodies are human myeloma cell survival and growth factors. Leukemia. 2000;14:188–197. doi: 10.1038/sj.leu.2401632. [DOI] [PubMed] [Google Scholar]
- 37.Sezer O. Myeloma bone disease: recent advances in biology, diagnosis, and treatment. Oncologist. 2009;14:276–283. doi: 10.1634/theoncologist.2009-0003. [DOI] [PubMed] [Google Scholar]
- 38.Pederson L, Ruan M, Westendorf JJ, Khosla S, Oursler MJ. Regulation of bone formation by osteoclasts involves Wnt/BMP signaling and the chemokine sphingosine-1-phosphate. Proc Natl Acad Sci U S A. 2008;105:20764–20769. doi: 10.1073/pnas.0805133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 40.Alsayed Y, Ngo H, Runnels J, et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109:2708–2717. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanderkerken K, Vande Broek I, Eizirik DL, et al. Monocyte chemoattractant protein-1 (MCP-1), secreted by bone marrow endothelial cells, induces chemoattraction of 5T multiple myeloma cells. Clin Exp Metastasis. 2002;19:87–90. doi: 10.1023/a:1013891205989. [DOI] [PubMed] [Google Scholar]
- 42.Thelen M, Stein JV. How chemokines invite leukocytes to dance. Nat Immunol. 2008;9:953–959. doi: 10.1038/ni.f.207. [DOI] [PubMed] [Google Scholar]
- 43.Rubin JB. Chemokine signaling in cancer: one hump or two? Semin Cancer Biol. 2009;19:116–122. doi: 10.1016/j.semcancer.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Nishida S, Elalieh HZ, Long RK, Halloran BP, Bikle DD. Role of IGF-I signaling in regulating osteoclastogenesis. J Bone Miner Res. 2006;21:1350–1358. doi: 10.1359/jbmr.060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2009;23:10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hauge EM, Qvesel D, Eriksen EF, Mosekilde L, Melsen F. Cancellous bone remodeling occurs in specialized compartments lined by cells expressing osteoblastic markers. J Bone Miner Res. 2001;16:1575–1582. doi: 10.1359/jbmr.2001.16.9.1575. [DOI] [PubMed] [Google Scholar]
- 47.Eriksen EF, Eghbali-Fatourechi GZ, Khosla S. Remodeling and vascular spaces in bone. J Bone Miner Res. 2007;22:1–6. doi: 10.1359/jbmr.060910. [DOI] [PubMed] [Google Scholar]
- 48.Andersen TL, Sondergaard TE, Skorzynska KE, et al. A physical mechanism for coupling bone resorption and formation in adult human bone. Am J Pathol. 2009;174:239–247. doi: 10.2353/ajpath.2009.080627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka Y, Abe M, Hiasa M, et al. Myeloma cell-osteoclast interaction enhances angiogenesis together with bone resorption: a role for vascular endothelial cell growth factor and osteopontin. Clin Cancer Res. 2007;13:816–823. doi: 10.1158/1078-0432.CCR-06-2258. [DOI] [PubMed] [Google Scholar]
- 50.Yaccoby S, Pennisi A, Li X, et al. Atacicept (TACI-Ig) inhibits growth of TACI(high) primary myeloma cells in SCID-hu mice and in coculture with osteoclasts. Leukemia. 2008;22:406–413. doi: 10.1038/sj.leu.2405048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


