Abstract
The Saccharomyces cerevisiae Set1/COMPASS was the first histone H3 lysine 4 (H3K4) methylase identified over ten years ago. Since then, it has been demonstrated that Set1/COMPASS and its enzymatic product, H3K4 methylation, is highly conserved across the evolutionary tree. Although there is only one COMPASS in yeast, human cells bear at least six COMPASS family members each capable of methylating H3K4 with non-redundant functions. In yeast, the monoubiquitination of histone H2B by Rad6/Bre1 is required for proper H3K4 and H3K79 trimethylations. This histone crosstalk and its machinery are also highly conserved from yeast to human. In this review, the process of histone H2B monoubiquitination-dependent and independent histone H3K4 methylation as a mark of active transcription, enhancer signatures, and developmentally poised genes will be discussed. The misregulation of histone H2B monoubiquitination and H3K4 methylation results in the pathogenesis of human diseases including cancer. Recent findings in this regard will also be examined.
Keywords: Histone methylation, chromatin, MLL, Set1, COMPASS, Rad6, Bre1, histone monoubiquitination, chromosomal translocations and leukemia
1 - Introduction
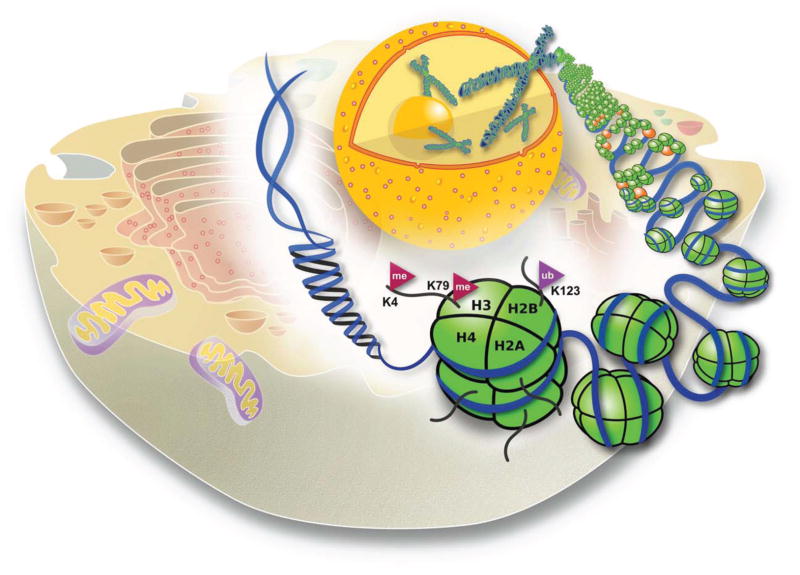
The very long linear sequences of nucleotides within the DNA possesse the genetic blueprint required for the creation of all organisms, with the exception of RNA viruses. The several meters of DNA constituting our genome must remain functional and accessible to the transcriptional machinery, yet be protected and packaged within a very minute space within the nucleus of our cells. To achieve such dynamic packaging of this genomic information, the DNA is found in complex with an equal mass of histone proteins to form nucleosomes, the basic unit of chromatin (Figure 1).
Figure 1. Chromatin, histone modifications and gene expression.
The very long eukaryotic DNA is compacted within the cell nucleus through its interactions with histones, forming the nucleosomes. Structural studies of nucleosomes demonstrated that the histone N-terminal tails protrude outward beyond the gyres of the DNA. Many of the amino acid residues within the histone tails can be posttranslationally modified providing a landing pad for a diverse array of transcription factors, chromatin remodelers and DNA- interacting proteins to regulate gene expression. In this figure, sites of posttranslational modifications on histone tails for H3K4 methylation and H2B monoubiquitination by the COMPASS family and the Rad6/Bre1 complex, respectively, are shown.
The eukaryotic genome is compacted at several levels. The first level requires the wrapping of DNA around the outside of an octamer of two of each histones: H2A, H2B, H3 and H4, or some variant of these canonical histones to form nucleosomes (1–3). Using electron microscopy, the array of nucleosomes on DNA was observed as a series of “beads-on-a-string” or the 11nm model. This “beads-on-a-string” structure of nucleosomes can further interact through short-range inter-nucleosomal exchanges to compact the nucleosomal array to a structure known as the 30nm fiber. To form the fully packaged metaphase chromosomes, linker histone proteins, such as histone H1, further compact the 30nm fiber into stable chromatin fibers, which can then be further packaged into metaphase chromosomes (Figure 1).
Upon packaging, the DNA within the nucleus is found in both lightly and tightly packed regions of chromatin. Based on a series of cytogenetic observations by light microscopy, Emil Heitz reported the presence of darkly stained chromatin in the nucleus, which he referred to as “heterochromatin.” Heitz found that the heterochromatin remained condensed throughout the cell cycle (4). This observation was in contrast to the behavior of the euchromatic regions (the lightly packed regions), which are subjected to cycles of condensation and decondensation at different stages of the cell cycle. Initial studies by Heitz suggested that heterochromatin is devoid of genes, however, functional studies in Drosophila established that genes such as rolled and light reside within the heterochromatin (5, 6).
In the early 1930s, H.J. Muller described the phenomenon of position-effect variegation (PEV) of gene expression which describes that the expression of genes in Drosophila that is brought near heterochromatin is silenced (7). PEV, which was originally observed in insects, has since been demonstrated to exist in other organisms including plants and mammals and is a general mechanism for the regulation of gene expression. The variegation in gene expression is thought to be regulated by a ‘spreading effect’ of factors from the adjacent heterochromatin. This explanation for the molecular mechanism of PEV presumes that the juxtaposed gene locus is condensed to form a transcriptionally inactive state through the movement of factors or posttranslational modifications of factors from heterochromatic regions to regions within its vicinity. In the late 1970s, studies by Grigliatti and colleagues in Drosophila demonstrated that the variegation in gene expression in Drosophila is a function of histone gene multiplicity or expression levels, suggesting for the first time that chromatin and its basic components, the “histones,” may have a fundamental role in the process of regulating gene expression (8, 9).
Nucleosomes and histones also represent an analogous inhibitory effect to transcription in other cell types in addition to Drosophila. For example, attenuation of histone levels, and thereby nucleosome levels, by genetic means in yeast cells resulted in the activation of genes that were otherwise inactive (10, 11). Not only were nucleosomes found to be inhibitory to transcription, their presence as being wrapped around DNA can control the interaction of the DNA binding factors. Although the DNA sequences recognized by the DNA binding proteins are found throughout the genomes of organisms, the actual in vivo interaction of the DNA binding proteins with chromatin is highly specific and is only found on precise regions. For example, the Repressor-Activator Protein 1 (Rap1) is found to be associated with certain limited sites on chromatin that have regions with the potential to act as promoters, although its primary DNA sequence consensus binding site is found throughout the genome (12). Similar to these in vivo studies, fundamental in vitro biochemical studies demonstrated that nucleosomes are inhibitory to the initiation of transcription by RNA polymerase II (Pol II) (13–15). Biochemical studies also demonstrated that even if Pol II is initiated, the presence of nucleosomes, or just histone tetramers, provide a blockage to productive transcription-elongating Pol II (16, 17). The above findings in consensus with other findings in the field suggested that not only does chromatin regulate the initiation and elongation steps of gene expression in vivo, but also that chromatin is pivotal in modulating the interaction of DNA binding proteins with its consensus binding sites on DNA. Indeed, using DNaseI hypersensitivity mapping, Weintraub and Groudine provided the seminal concept that nucleosomes can block the access of the transcription factors to DNA, and the regulatory regions thereby controlling gene expression, initiation of DNA replication, recombination and other processes requiring DNA access (18).
To define the mechanism of heterochromatin formation and the molecular machinery involved in the regulation of gene expression from heterochromatin and other developmental loci such as the homeotic gene-containing regions, genetic studies were performed in Drosophila and other organisms. From these studies, the trithorax (trx) group and the Polycomb (Pc) group of genes were identified to play opposing roles in homeotic gene expression (19–21). The suppressor and enhancer screens of PEV in Drosophila identified Suppressors and Enhancers of position-effect variegation [Su(var)] and [E(var)], respectively. Many of the genes found from the above screens encode for proteins bearing a 130- to 140-amino-acid motif called the SET domain (12–13). This domain takes its name from the Drosophila proteins Su(var)3–9, Enhancer of zeste (E(z)), and trx (22, 23). Many of the SET domain-containing proteins have now been demonstrated to possess histone or lysine methyltransferase (H/or KMTase) activity.
Detailed structural studies of nucleosomes demonstrated that the N-terminal tails of each of the histones protrude outward beyond the gyres of the DNA (24) (Figure 1). Many amino acid residues within the histone tails can be posttranslationally modified; and the modifications of histone tails could provide a landing pad for a diverse array of transcription factors, chromatin remodelers and DNA-interacting proteins to regulate gene expression. The covalent modifications of histones to date include acetylation, phosphorylation, ADP-ribosylation, biotinylation, ubiquitination, and methylation (25–29). Although, the list of SET domain-containing proteins with identified KMTase activity towards histones is growing, so far, this list only includes eight classes of enzymes, KMT1-KMT8 (30).
The KMT2 class and its first member, Set1, in yeast Saccharomyces cerevisiae were isolated within the Set1/COMPASS functioning as a histone H3K4 methylases. Given the limitation in space, I will only focus my attention in this review on the full description of the KMT2 class and its histone substrate. I will discuss our current knowledge on how histone H3K4 methylation by the KMT2 class is regulated via histone H2B monoubiquitination implemented by the Rad6/Bre1 complex, and how these marks function as a sign of active transcription, enhancer signatures, and are present and mark the developmentally poised genes in mammalian cells.
2 - Molecular and biochemical properties of Set1/COMPASS
2.1 - Biochemical purification of Set1/COMPASS as the first histone H3K4 methylase
Early genetic studies in Drosophila identified two families of proteins, the trithorax group (trxG) and the Polycomb group (PcG), to play a central role in the regulation of gene expression throughout development (31). The clustered homeotic (Hox) genes in the Bithorax and Antennapedia gene complexes in Drosophila are some of the known targets of the trxG and PcG proteins. The Drosophila trx gene was discovered based on the identification of several mutations in this gene that resulted in the partial transformation of the halteres into wings (32). The halteres are small knobbed-like structures found behind the wings in Drosophila functioning as a gyroscope informing the insect about position of the body throughout flight. Mutations in trxG and PcG genes resembled mutations in the Hox genes, leading to the suggestion that trxG proteins function as positive effectors of gene expression while the PcG proteins function in the repression of gene expression (33, 34).
Most of the trxG and PcG proteins are conserved in mammals functioning within similar pathways to those of their Drosophila counterparts (35, 36). The first mammalian homolog of Drosophila trx, the Mixed Lineage Leukemia (MLL) gene, was cloned based on the identification of its random translocations found in patients suffering from hematological malignancies, including acute myeloid and lymphoid leukemia, AML and ALL, respectively (37–40). Despite the critical role of the MLL gene product in the pathogenesis of hematological malignancies, very little was known about the molecular and biochemical properties of MLL until the genetic and biochemical studies of its yeast homolog, Set1.
To begin to define the biochemical function(s) of MLL in the hope of a better understanding of its role in leukemic pathogenesis, initial studies from our laboratory in the late 1990s set out to purify MLL-containing complexes. Our approach was to fractionate nuclear extracts and follow MLL by immunoblotting. Cloning and sequencing of MLL predicted a 400 kDa polypeptide on Western, but it was much later that it was determined that MLL is proteolytically cleaved in vivo, resulting in N-terminal (~150kDa) and C-terminal (~200 kDa) fragments that can associate non-covalently in cells (41). While we were unable to detect full-length MLL at its expected electrophoretic mobility (~400kDa), and therefore could not identify MLL complexes from mammalian cells, we had identified Set1 of yeast Saccharomyces cerevisiae as a MLL homolog (42). We used conventional chromatographic methods to isolate a yeast Set1-containing complex from 300 liters of yeast nuclear extract (42). This complex was named COMPASS for Complex of Proteins Associated with Set1, and it was demonstrated to be a histone H3 lysine 4 (H3K4) methylase. Set1/COMPASS was the first H3K4 methylase identified and is capable of catalyzing the mono-, di- and trimethylation of H3K4 (42–45).
2.2 - Kinetic properties and contribution of Set1/COMPASS subunits in H3K4 mono-, di- and trimethylation
Set1 alone is not active as a KMTase, as Set1 within COMPASS is the active form of the enzyme. Indeed, any alteration to the C-terminal domain of Set1 containing its SET domain, such as insertion of tags including TAP, HA or Flag, results in the full loss of complex formation and the activity of the enzyme (43, 44). Set1/COMPASS consists of eight subunits including Set1, Cps60, Cps50, Cps40, Cps35, Cps30, Cps25, and Cps15 (Figure 2; Table 1), with each of the subunits having a specific function in the assembly and regulation of the pattern of H3K4 mono-, di and trimethylation by the enzyme (Table 1). Several subunits of Set1/COMPASS including Set1, Cps50 and Cps30 are essential for complex formation/stability; and full KMTase activity of the complex as yeast cells lacking these subunits are defective in H3K4 mono-, di- and trimethylation. The Cps35 subunit of COMPASS (also known as SWD2) is the only essential subunit of the complex in yeast and it is shared with other complexes including the cleavage and polyadenylation factor (CPF) functioning in transcription termination on snoRNA genes (46–48). The essentiality of Cps35 for growth is due to its requirement within CFP and not in COMPASS, as the lethality due to the lack of Cps35 is suppressed by the overexpression of the C-terminal fragment of Sen1, a superfamily I helicase functioning in snoRNA termination (47). The Cps35 null strains in a Sen1-overexpression-background demonstrate a significant reduction in H3K4 di- and trimethylation levels, with no detectable change in H3K4 monomethylation. Therefore, the Cps35 subunit of Set1/COMPASS is required for proper H3K4 di- and trimethylation, but not monomethylation. The Cps40 subunit of the complex is required for proper H3K4 trimethylation, as in its absence, yeast cells lack over 80% of H3K4 trimethylation with very little effect on H3K4 mono- and dimethylation (45). The Cps25 and Cps60 subunits of the complex appear to be required for proper H3K4 di- and trimethylation, but not monomethylation (45, 49, 50).
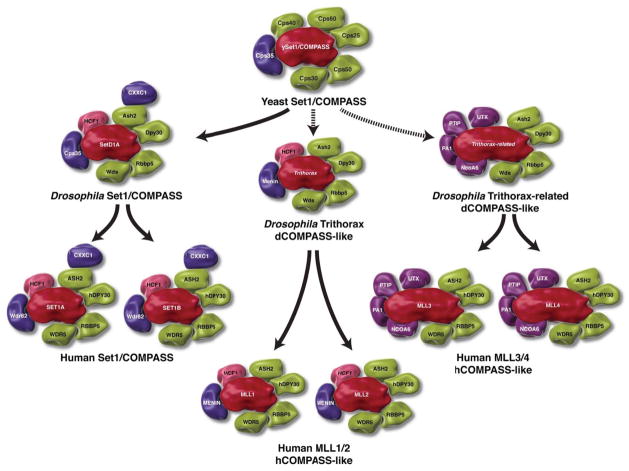
Figure 2. The subunit composition of the COMPASS family from yeast to human.
The yeast Set1/COMPASS is the founding member of the family of COMPASS H3K4 methyltransferases. There is only one Set1/COMPASS in yeast capable of methylating histone H3 on its fourth lysine (H3K4). From yeast to Drosophila, COMPASS is divided into three family members. The Set1/COMPASS of Drosophila is the direct descendent of the yeast Set1 complex (shown by solid arrow). There are two COMPASS-like complexes in Drosophila (shown by dotted arrow); one Trithorax (trx)-containing complex and the other Trithorax-related-containing complex (trr). The SET domain-containing enzymes from yeast to Drosophila are shown in RED, and the conserved subunits between yeast and Drosophila are shown in GREEN. The complex specifics are shown in BLUE and PURPLE. From Drosophila to human cells, the COMPASS family is divided into six members. The mammalian Set1A and B are direct homologs of yeast and Drosophila Set1 and are found in Set1A-B/COMPASS (shown by solid arrow). The subunit composition and function of Trx of Drosophila is divided between MLL1 and MLL2 in the mammalian cells. Both MLL1 and MLL2 are found in COMPASS-like complexes. The subunit composition and function of Trr of Drosophila is divided between MLL3 and MLL4 in the mammalian cells, both of which are also found in COMPASS-like complexes. All six mammalian COMPASS family members are capable of methylating H3K4. The known common subunits shared between yeast, Drosophila and the human complexes are shown in GREEN. Cps35 in yeast and Drosophila and its homolog of mammalian Wdr82 shown in BLUE, are found only in Set1A-B/COMPASS. Menin shown in BLUE is found in complex with Trx and the MLL1-2/COMPASS-like complexes. The shared subunits among the trr and the MLL3-4 complexes, Utx, PTIP, PA1 and NCOA6 are shown in PURPLE.
Table 1.
Subunit composition and biological activities of COMPASS family from yeast to human.
| ySet1/COMPASS | Drosophila COMPASS/COMPASS-like Complexes | Mammalian COMPASS/COMPASS-like Complexes | Biological Activities |
|---|---|---|---|
| Set1 | Set1; Trx; Trr | Set1A/B; MLL1-4 | The catalytic subunits |
|
| |||
| Cps60 (Bre2) | Ash2 | Ash2 | Required for H3K4me3 |
|
| |||
| Cps50 (Swd1) | dRbbp5 | dRbbp5 | Required for assembly |
|
| |||
| Cps40 (Spp1) | dCxxC1 | CxxC1 | Components of Set1 complexes |
|
| |||
| Cps35 (Swd2) | dWdr82 | Wdr82 | |
|
| |||
| Cps30 (Swd3) | Wds | Wdr5 | Required for assembly |
|
| |||
| Cps25 (Sdc1) | Dpy30 | Dpy30 | Required for H3K4me2/3 |
|
| |||
| HCF1 | HCF1 | Components of Set1 complexes | |
|
| |||
|
|
|
Trx and MLL1/2 specific | |
|
| |||
| PTIP; PA1; NCOA6; UTX | PTIP; PA1; NCOA6; UTX | Trr and MLL3/4 specific | |
2.3 - The pattern of H3K4 methylation by Set1/COMPASS and its dependence on the rate of transcription elongation by RNA Pol II
Biochemical screens in yeast identified a role for the RNA Pol II C-terminal domain (CTD) kinases Ctk1, Ctk2, and Ctk3 (the Ctk complex) in the regulation of H3K4 monomethylation by Set1/COMPASS (51, 52). These studies demonstrated that the loss of the Ctk complex kinase activity in yeast cells results in reduced histone H3K4 monomethylation levels followed by a global increase in the histone H3K4 trimethylation levels (51). Given the fact that Set1/COMPASS associates with transcribing Pol II through its interactions with the Paf1 complex (53, 54) and that there is an established role for the Ctk complex in transcriptional elongation control through CTD phosphorylation, a possible role for the rate of transcription elongation by Pol II in dictating the pattern of H3K4 mono-, di- and trimethylation by Set1/COMPASS was tested (51). These in vivo studies demonstrated that as Set1/COMPASS travels with transcribing Pol II during the promoter escape, where Pol II transcribes at a slower rate, therefore, the chromatin within such regions are mostly trimethylated as Set1/COMPASS has a longer window of opportunity to methylate its substrate, the nucleosomes. However, in regions where Pol II transcribes at a faster rate, during productive elongation, Set1/COMPASS spends less time with its substrate, and therefore, chromatin within such areas are monomethylated (51). In support of such observations, in vitro enzymological studies demonstrated that the onset of monomethylation on an unmethylated histone H3 by Set1/COMPASS is virtually immediate, while the onset of trimethylation occurs upon extended time of association between the histone H3 tail and Set1/COMPASS, further supporting the notion that the pattern of H3K4 trimethylation by the enzyme requires an extended interaction with its substrate (51).
3 - Identification of the metazoans Set1/Trx/MLL in COMPASS-like complexes functioning as H3K4 methylases
3.1 - Biochemical purification of MLL and Set1/COMPASS-like complexes from Drosophila to human
The COMPASS family of H3K4 methylases is highly conserved from yeast to plant and to human (42, 43, 55–58). Three independent genes in Drosophila: dSet1, trithorax (trx) and trithorax-related (Trr), are homologs to yeast Set1 (Mohan et al., 2011) (Figure 2; Table 1). The dSet1 is the direct descendent of yeast Set1, while trx and Trr are more distantly related to yeast Set1 (Figure 2). Although it was initially reported that trx is found in a heterotrimeric complex with CBP and SBF1 (59), our studies have demonstrated that dSet1, trx and Trr are all three found in COMPASS-like complexes capable of methylating histone H3K4 (Mohan et al., 2011). Each enzyme localizes to hundreds of sites on polytene chromosomes, but the deletion or RNAi-mediated knockdown of either one of these genes results in lethality in Drosophila, indicating that the H3K4 methylase function of each complex is non-redundant. Loss of dSet1 results in global losses of H3K4 di- and trimethylation (60, 61), indicating that trx and Trr may have more specialized functions. dSet1, trx and Trr each interact with a set of core COMPASS subunits, but there are also subunits unique to each complex (61). Importantly, each of these unique subunits has an ortholog in the mammalian version of its COMPASS-like complex; mammals, however, have two homologous complexes for each of the Drosophila complexes.
In human cells, Set1A, Set1B, MLL1, MLL2, MLL3 and MLL4 are homologs of yeast Set1 (62, 63) (Figure 2; Table 1). Following the identification of Set1/COMPASS in yeast as the first H3K4 methylase (42–45), the affinity purification of the menin tumor suppressor protein, which is the product of the MEN1 gene, resulted in the isolation of MLL2 and many of the human homologs of the yeast COMPASS subunits within the same complex (56). This biochemical study demonstrated that not only is MLL2 associated within a COMPASS-like complex, but it can also function as a histone H3K4 methylase (56). Subsequent studies demonstrated that the MLL1-4 are all found in COMPASS-like complexes capable of mono-, di- and trimethylating H3K4 (64–71) (Figure 2; Table 1). The Set1A and Set1B proteins in human cells were also identified in COMPASS-like compositions functioning as an H3K4 methylase (66, 72, 73). These studies established the fact that Set1/COMPASS, found initially in yeast, is highly conserved from yeast to human, both in composition and function. Detailed biochemical studies have demonstrated that each COMPASS-like complex in metazoans carries shared subunits that are similar to their yeast counterpart. Additionally, the metazoan complexes also possess complex-specific subunits bringing about target specificity and functioning as interaction modules for each complex (Figure 2; Table 1). Details of the composition of the human COMPASS-like complexes are described below.
3.2 - Composition of metazoan COMPASS-like complexes
The shared subunits between yeast COMPASS and the metazoan Set1/MLL/COMPASS-like complexes include ASH2 (related to Cps60), RbBP5 (related to Cps50), Wdr5 (related to Cps30) and Dpy30 (related to Cps25) (Figure 2; Table 1) (62, 74). Metazoan Set1/COMPASS possesses CxxC and Wdr82, which are related to yeast Cps40 and Cps35 respectively, however, these proteins are not detected stoichiometrically within the MLL1-4 complexes (62, 66, 73, 75, 76). The menin tumor suppressor protein, whose gene, MEN1, is mutated in familial multiple endocrine neoplasia type 1, together with the lens epithelium derived growth factor (LEDGF) are found to be associated with both the MLL1 and MLL2 complexes. However, these proteins are absent in the MLL3 and MLL4 complexes (56, 66). The histone H3K27 demethylase UTX, the Pax Transactivation domain-Interacting Protein (PTIP), nuclear receptor coactivator (NCOA6), and PTIP-associated 1 (PA1) are specifically found within the MLL3-4/COMPASS-like complexes (64–66, 69, 71, 77–79).
3.3 - Functional diversity of the metazoans’ COMPASS-like complexes
Following the identification of MLL1 within a COMPASS-like complex functioning as an H3K4 trimethylase (56), the vast majority of H3K4 methylation studies were focused on MLL1’s activity. However, biochemical and cell biological studies in mammalian cells demonstrated that the attenuation of Wdr82 (a Set1A/B specific subunit) levels in cells results in the reduction of the Set1A/B protein levels, but not the core components or MLL1 (66, 80). This loss in the Wdr82 levels is followed by a global loss in H3K4 trimethylation levels with very little alteration in the H3K4 di- and trimethylation levels (66). This study demonstrated that: 1) mammalian Wdr82, the Set1A/B specific factor, is required for the majority of proper H3K4 trimethylation by Set1, and not for the MLL COMPASS-like complexes; 2) Set1A/B complexes in mammalian cells are the major H3K4 methylases; and 3) MLL1-4 do not function globally as the major regulators of H3K4 trimethylation, but rather must have specific functions. Recently, the generality of the role of Set1/COMPASS as a major histone H3K4 methylase in other organisms was confirmed (60, 61).
Studies in Caenorhabditis elegans confirmed that many of the components of the COMPASS family are highly conserved in C. elegans as well (81). Similar to studies in mammalian cells (58), the loss of the Set1 homolog in C. elegans resulted in a major decrease in bulk H3K4 trimethylation, however, removal of the only MLL1-4 homolog in C. elegans, Set-16, did not have a global effect on the pattern of H3K4 methylation (81). Recent studies have demonstrated a specific role for the components of Set1/COMPASS in C. elegans, specifically in the regulation of the expression of the gustatory neurons ASEL and ASER, a bilaterally symmetric neuron pair that is functionally lateralized and occur stereotypically (82). This study demonstrated that Set1/COMPASS has a role in the specification of a left-right asymmetry in this sensory neuron pair. Several shared components of the COMPASS family in C. elegans, including Ash2, Wdr5 and Rbbp5 are required for ASE laterality defects (82). Furthermore, Wdr82, which is specific to Set1/COMPASS and not the MLL1-4/COMPASS-like complexes, is also required for ASE laterality (82), suggesting a specific role for Set1/COMPASS in this process. Since C. elegans lacks the Trx/MLL1-2 complexes (please see below) (83), this function of Set1/COMPASS could be shared between trx and Set1 in C. elegans.
3.4 - Mammalian MLL1/MLL2, the Drosophila homologs of trithorax involved in the positive regulation of homeotic gene expression
Early studies by Korsmeyer and colleagues demonstrated that MLL1 is required for proper segment identity in mammals, and that it positively regulates Hox gene expression (84). The targeted deletion of both copies of MLL1 in mouse embryonic stem (ES) cells by homologous recombination resulted in embryonic lethality (84, 85). MLL1 heterozygous (+/−) mice demonstrated retarded growth, hematopoietic abnormalities, and similar to the loss of the trx gene in Drosophila, demonstrated bidirectional homeotic transformations (84). Reduction of MLL1 levels in MLL1 heterozygous (+/−) animals also demonstrated abnormalities in the anterior boundaries of Hoxa7 and Hoxc9 expression, whereas their posterior expression was lost in MLL −/− embryos. Subsequent cellular studies confirmed a role for MLL1 as a regulator of Hox gene expression and linked MLL’s specific role in this process to its role in leukemic pathogenesis (86–89).
Given the importance of the histone methyltransferase activity of MLL1 in leukemogenesis, and its existence in just one of the six COMPASS-like complexes, the global pattern of H3K4 methylation was tested in mouse embryonic fibroblasts (MEFs) wildtype and null type for MLL1 (77). Surprisingly, these studies demonstrated that MLL1 is required for the H3K4 trimethylation of only less than 5% of the promoters carrying this modification (77). The Gene Ontology (GO) analysis of the MLL1 H3K4 methylation targets demonstrated that many of these genes include developmental regulators such as Hox genes, however, not all Hox genes require MLL1 for their proper H3K4 methylation (77). Also, the loss of MLL1 resulted in decreased levels of Pol II recruitment, decreased expression and a concomitant loss of H3K4 methylation at these genes. Analysis of the entire Hox cluster established that MLL1 is only required for the methylation of a subset of Hox genes. However, the deletion of Menin, which is a component of both the MLL1 and MLL2 complexes (Figure 2), resulted in the abolishment of the majority of H3K4 methylation at the entire Hox loci. This finding established that MLL1 and MLL2 are the functional homologs of the Drosophila trx gene, as the loss of the MLL3/MLL4 and/or the Set1 complexes demonstrated little to no effect on the H3K4 methylation or expression of this homeotic cluster (77). Similar findings in Drosophila also suggest that Set1/COMPASS is the major H3K4 methylase and that trx and Trr/COMPASS-like complexes have limited and specific genomic targets (61).
3.5 - Mammalian MLL3/MLL4: the homologues of the Drosophila trithorax-related (Trr)
The Drosophila trithorax-related (trr) gene was identified based on the homology of its SET-domain with trx, and cloned using a degenerate PCR strategy (90, 91). Similar to trx, mutations or deletions of the trr gene result in embryonic lethality indicating that the function of these two related genes are not redundant. Indeed, the loss of the Trr function does not result in homeotic transformations as seen with the loss-of-function of trx, and no dominant interaction has been reported between trr1 and the hypomorphic alleles of either Polycomb (Pc) or trx, revealing that Trr is not a major regulator of the expression of homeotic genes (74, 90).
The major steroid hormone pathway receptor in Drosophila, the ecdysone receptor (EcR), co-localizes extensively with Trr on polytene chromosomes (92). Furthermore, Trr and EcR can be co-immunoprecipitated in an ecdysone-dependent manner (92). Following the demonstration that the SET-domain of Trr can also function as a histone H3K4 methylase (92), it was demonstrated that association of Trr and EcR on chromatin in an ecdysone-dependent manner is followed by the trimethylation of histone H3K4 of EcR target promoters. More importantly, in trr alleles lacking the SET-domain of Trr, the H3K4 trimethylation levels on EcR target genes are significantly abridged suggesting that the catalytic activity of Trr is required for ecdysone-dependent gene activation through H3K4 methylation in vivo (92).
Cloning of one of the human homologs of Drosophila Trr, the MLL4/ALR gene (93), was followed by the purification of its complex from human cells in a COMPASS-like composition capable of interacting with the estrogen receptor (94). The ER alpha directly binds to MLL4’s two LXXLL motifs near its C terminus in a ligand-dependent manner (94). Similar to EcR in Drosophila, the MLL4 complex is recruited to the promoters of the ER alpha target genes following estrogen stimulation, and the expression of the ER alpha target genes is abridged upon inhibition of MLL4 expression. Subsequently, other complexes containing MLL3/MLL4 were also identified, and demonstrated that MLL3/4 to exist in COMPASS-like complexes (64–66, 69, 71, 78) (Figure 2). Overall, studies on the MLL3/MLL4 complexes have demonstrated that these COMPASS family members play an important role as co-activators of nuclear transport signaling, adipogenesis and immunoglobulin class switching (71, 94–98). The prominent role of the MLL3/4 and Trr complexes in nuclear receptor coactivation indicates specialization among different classes of the MLL1-4 proteins. Indeed, C. elegans, which has a reduced homeotic gene cluster (99) lacks a MLL1/2/Trx-like gene, but does have Set1 and MLL3/4/Trr genes. Interestingly, C. elegans has an expanded number of nuclear receptors encoded in their genome, perhaps explaining why this organism has MLL3/4 (Set-16), but not MLL1/2 genes (83, 100).
3.6 - A histone H3K27 demethylase as a component of the Trr/MLL3/MLL4 complexes
As the histone H3K4 methylase function of trx in Drosophila and MLL1-2 in mammalian cells is considered to be a positive regulator of homeotic gene expression, the histone H3K27 methylation by the Polycomb group of proteins (PcG) is considered to be a negative regulator of this process. The Jumonji C domain-containing histone demethylase, UTX, functions as a histone H3K27 demethylase (78, 79, 101–104). Surprisingly, UTX was identified to co-purify with the MLL3/MLL4 complexes (64, 65, 69, 78) and to function during retinoic acid signaling events (78). Similar complexes containing Trr and UTX have also been observed in Drosophila (61) and have been demonstrated to function in cell growth control, where it interacts with the Notch and Rbf pathways to suppress tumorigenesis (104). A similar phenomenon was observed in C. elegans and in human cells (105). It is not clear at this time why the Trr/MLL3/MLL4-containing complexes carry H3K27 demethylase activity within their complexes and why the Set1A/B and Trx/MLL1/MLL2 complexes lack this activity. H3K27 methylation machinery and a MLL3-like protein can be found even in unicellular organisms such as ciliated protozoa, indicating an ancient role for antagonism between H3K4 and H3K27 methylation that goes beyond regulating Hox gene patterning (83). Recent genome-wide studies of PcG proteins indicate that these proteins play a much larger role than to repress Hox genes (106–108). Perhaps the MLL3/4 H3K4 methyltransferases together with H3K27 demethylase activity regulate a distinct form of antagonizing PcG function that differs from the Trx-Hox paradigm. MLL3 and MLL4, as well as UTX, are frequently mutated genes in human cancers (109–112). Therefore, determining the way that these two classes of enzymes interact with each other to regulate gene expression is an important area of future research.
3.7- Metazoan COMPASS subunit Dpy-30 in dosage compensation
Dosage compensation is a process by which the expression of X-linked genes is equalized between males and females. In mammals, this is achieved by silencing one of the two X chromosomes in females. In Drosophila, genes on the single X chromosome in males are transcribed at twice the rate to achieve the same transcriptional output from two X chromosomes in females. In C. elegans, the two X chromosomes in hermaphrodites are transcribed at half the level of the X chromosome in XO males (113). In each lineage, along with the evolution of an autosome into a sex chromosome, there was also the co-option of global chromatin-modifying machinery for the purpose of dosage compensation. In mammals, the machinery includes proteins involved with heterochromatin formation that are co-opted for silencing one of the X chromosomes (forming the cytologically visible Barr body) (114). In Drosophila, the MSL complex implements H4K16 acetylation on the male X chromosome to achieve dosage compensation. In both Drosophila and mammals, a related H4K16 acetylation complex modifies histones in both males and females on all chromosomes for proper gene expression genome-wide (115). In C. elegans, a condensin complex (called the DCC for dosage compensation complex) shares subunits with the chromosome compaction machinery necessary for the faithful segregation of the mitotic and meiotic chromosomes (113, 116–119). While mutations in many of the DCC genes cause hermaphrodite-specific lethality, rare survivors have a shortened or dumpy morphology. Another gene whose mutation gives a dumpy phenotype is dpy-30 (120, 121). However dpy-30 is also essential for the development of XO males, indicating an additional, more general function. C. elegans DPY-30 is homologous to Cps25 found in yeast COMPASS and to Dpy30 found in Drosophila and the mammalian COMPASS family. Co-immunoprecipitation experiments and genome-wide profiling studies demonstrate that C. elegans DPY-30 associates not only with COMPASS and COMPASS-like complexes, where it is essential for the H3K4me3 mark, but also functions in the DCC, independently of other COMPASS subunits (122). As a member of the DCC, DPY-30 plays an essential role in the loading of DCC components onto X chromosomes of hermaphrodites. Thus, the same subunit that is required for H3K4me3, a mark of active transcription from yeast to mammals, is also a component of a complex that reduces transcription through chromatin condensation. The existence of DPY-30 in the context of the DCC has the potential to provide clues for the function of DPY-30 and its homologs of the COMPASS family, due to unique genetic tools developed over the last 25 years in studies of C. elegans dosage compensation.
4- Development of a biochemical screen (GPS) in yeast for the identification of the molecular pathway of histone H3K4 methylation
4.1 - Identification of histone H2B monoubiquitination by Rad6 as a signal for H3K4 trimethylation by Set1/COMPASS
A proteomic screen called Global Proteomic analysis in S. cerevisiae (GPS) was devised to identify factors and molecular pathway(s) required for proper histone H3K4 methylation by Set1/COMPASS (123). This method takes advantage of the availability of the collection of the yeast non-essential deletion mutants (124, 125) and antibodies generated toward H3K4 mono-, di-, and trimethlyated H3K4 to identify factors required for implementation of this histone modification in cells (123). In GPS, extracts from strains deleted for each of the non-essential yeast genes from the collection are generated and analyzed by SDS/PAGE followed by Western blot analysis. This method allows the identification of the protein coding genes in the yeast genome required for Set1/COMPASS function.
The first hit from this biochemical screen was the demonstration of the first trans-tail histone crosstalk (126). This study demonstrated that histone H2B monoubiquitination by the E2-conjugating enzyme, Rad6, is required for proper H3K4 methylation by Set1/COMPASS (126). At the same time, another independent study also reported the role of H2B monoubiquitination by Rad6 in H3K4 methylation (127). These studies confirmed that H2B monoubiquitination by Rad6 is required for H3K4 di- and trimethylation by Set1/COMPASS and that methylation of H3K4 is downstream of the H2B monoubiquitination function of Rad6 (45, 126, 127). The histone H2B monoubiquitination crosstalk is also required for proper histone H3K79 trimethylation by the non-set domain-containing enzyme, Dot1 (128–130). Indeed, global genomic studies have demonstrated that promoters within the yeast genome bearing H2B monoubiquitination possess H3K79 trimethylation and that promoters lacking H2B monoubiquitination only bear H3K79 mono- and dimethylation, further solidifying a direct link for this trans-histone tail crosstalk throughout the yeast genome (131).
4.2 - Bre1, the E3-conjugating enzyme, is required for the association of Rad6 with chromatin and H2B monoubiquitination
Following the identification of the role of histone H2B monoubiquitination by Rad6 in regulating H3K4 methylation, investigators were searching to identify the E3-conjugating enzyme required for Rad6 function in chromatin modification and transcription. Our GPS screen identified the C3HC4 ring finger-containing protein, Bre1, as a factor required for proper H3K4 di- and trimethylation by COMPASS or H3K79 methylation by Dot1 (130). Deletion or generation of a single point mutation in the ring finger of Bre1 rendered this E3 ligase inactive in H2B monoubiquitination and resulted in the loss of H3K4 and H3K79 methylations in strains bearing such mutations (130). In support of this observation, biochemical purification of Rad6 demonstrated that this enzyme exists in complexes with several different E3 ligases, including Bre1 (130). This study demonstrated the existence of physical interactions between Rad6 and Bre1 (130). In strains lacking Bre1, Rad6 is not able to interact with chromatin, and therefore, the levels of H2B monoubiquitination and H3K4 and H3K79 trimethylation are lost, suggesting that Bre1 is required for the recruitment of Rad6 to chromatin on genes (53, 130). Bre1 and its interacting factor, Lge1, were also identified to be required for proper H2B monoubiquitination and H3K4 methylation (132). This study further suggested a function for Bre1/Lge1-dependent H2B monoubiquitination in the control of cell size in yeast (132).
Studies in yeast demonstrated that a subunit of Set1/COMPASS, Cps35 (also known as Swd2), associates with the enzyme and chromatin in an H2B monoubiquitination-dependent manner and that the interaction between Cps35 and monoubiquitinated nucleosomes appears to be indirect as Cps35 is not capable of interacting with free ubiquitin (76, 133). Given the fact that Cps35 is required for proper H3K4 di- and trimethylation (47, 48, 76), such studies suggest that Cps35 can function in regulating histone H2B monoubiquitination/H3K4 methylation crosstalk. Similarly, other studies have also found a role for Cps35/Swd2 as a factor within Set1/COMPASS regulating monoubiquitination/H3K4 methylation crosstalk (134). In support of these observations, the Drosophila and human homologue of Cps35, a protein known as Wdr82, is also required for Set1/COMPASS function in H3K4 trimethylation (58).
4.3 - The conserved role of Bre1 in Set1/COMPASS function from yeast to human
Both Rad6 and its E3-conjugating enzyme, Bre1, are highly conserved in structure and function from yeast to human cells, demonstrating the power of genetic and biochemical screens in yeast in identifying the molecular machinery required for proper H3K4 methylation (3, 83, 135–139). In Drosophila, Bre1 was identified through genetic screens as a factor that is required for the proper expression of Notch target genes during development (139). Similar to yeast cells (130), depletion of Bre1 levels in Drosophila results in a reduction of H2B monoubiquitination levels followed by a reduction in histone H3K4 and H3K79 methylation levels (61, 130, 138). In addition to a role in the regulation of the Notch signaling pathway, Bre1 and H2B monoubiquitination also function in Wnt signaling and the maintenance of multiple types of adult stem cells (138, 140, 141).
Although initial studies in mammalian cells suggested that the RING domain-containing E3 ubiquitin ligase, Mdm2, which ubiquitinates the p53 tumor suppressor protein, can also monoubiquitinate histone H2B both in vivo and in vitro (142), it has been firmly established that similar to yeast, human Bre1 is the E3 ligase functioning with human Rad6 to regulate the global pattern of H2B monoubiquitination (135–137). These studies demonstrated that the machinery required for the proper regulation of H2B monoubiquitination is highly conserved from yeast to human (3). Indeed, when using recombinant nucleosomes containing a wildtype or monoubiquitinated H2B version, the human Set1/COMPASS containing Wdr82 (mammalian homolog of yeast Cps35) used the H2B monoubiquitinated substrate with higher kinetics and efficacy than the non-monoubiquitinated version, indicating the conservation of function of H2B monoubiquitination in H3K4 trimethylation by Set1/COMPASS from yeast to human (137).
4.4 - Histone H2B monoubiquitination by Rad6/Bre1 in transcription elongation control
Although H2B monoubiquitination is required for proper histone H3K4 and H3K79 trimethylation from yeast to human, this modification and its machinery have recently been linked to other pathways. The global analysis of the pattern of H2B monoubiquitination in yeast demonstrated that this mark is found on other regions of chromatin lacking some or all of the histone marks mentioned above (131). For example, histone H3K4 trimethylation is only associated with the promoters of active genes, however, histone H2B monoubiquitination is found covering the entire body of an actively transcribed gene (131). Histone H3K79 trimethylation appears to be associated with genes that are on average larger in length versus H3K79 dimethylation, which is found on the promoters and associates with genes that are on average shorter in length (131). These findings suggested that dimethylation of histone H3K79 is associated with cell cycle regulation and G1/S transition, however, H3K79 trimethylation does not appear to play a role in this process. Subsequent studies demostrated that histone H3K79 trimethylation is required for the Wnt signaling in metazoans and gene expression in yeast (138, 143).
In vitro studies using reconstituted transcription systems demonstrated that histone H2B monoubiquitination works cooperatively with the transcription elongation factor FACT to regulate the elongation properties of RNA Pol II (144). In support of this observation, studies in the fission yeast, Schizosaccharomyces pombe, established that the loss of H2B monoubiquitination is associated with defects in nuclear structure, cell growth and septation. Chromatin immunoprecipitation experiments analyzing the localization properties of RNA Pol II in S. pombe cells lacking H2B monoubiquitination confirmed that the loss of H2B monoubiquitination results in an altered pattern of Pol II distribution and histone occupancy in the wake of Pol II transcription in gene coding regions (145). These in vivo findings further suggest that H2B monoubiquitination could regulate the rate and the process of transcriptional elongation control (145). Whether H2B monoubiquitination and/or Rad6/Bre1 can directly regulate the elongation properties of Pol II, or whether H2B monoubiquitination of nucleosomes alters transcription elongation due to changing the positioning and/or occupancy of nucleosomes, in front of and/or in the wake of transcribing polymerase, remains to be resolved.
4.5 - Histone H2B monoubiquitination by Rad6/Bre1 in DNA double-strand break repair by homologous recombination
Histone H2B monoubiquitination by Rad6/Bre1 has also been proposed to be required for DNA double-strand break repair by homologous recombination (146, 147) (146). The protein kinase ATM is central to the DNA damage response (DDR) as it phosphorylates many of the factors found within the DDR network. In human cells, double-stranded DNA breaks induce histone H2B monoubiquitination catalyzed by Rad6/Bre1, and this process is regulated by the ATM-dependent phosphorylation of the Rnf20 and Rnf40 subunits of the human Bre1 complex to form a heterodimer (146). Histone H2B monoubiquitination was demonstrated to be involved in the two major double-stranded repair pathways: nonhomologous end-joining and homologous recombination repair (146).
In another independent study, the RNF20 subunit of human Bre1 was shown to be recruited to double-stranded DNA break sites independently of H2AX. This study showed that the histone H2B monoubiquitination-dependent methylation of H3K4 at double-stranded break sites is involved in the recruitment SNF2h functioning in homologous recombination repair and the enhanced sensitivity to radiation damage (147). However, these studies did not identify which COMPASS family member is involved in this process. Given the requirement of H2B monoubiquitination for Set1A-B/COMPASS function in human cells, Set1/COMPASS could be the appropriate methyltransferase over the MLL1-4/COMPASS family.
Although the exact molecular mechanism for Rad6/Bre1 and histone H2B monoubiquitination in DNA damage repair is still in its infancy, these studies point to the diverse implementation, regulation and biological outcome for this highly conserved process. Given the fact that genetic and biochemical studies in yeast have been very fruitful in this regard, studies concentrating on defining the role of the Rad6/Bre1 complex in DNA double-stranded break repair and in homologous recombination in yeast could be highly informative.
4.6 - The RNA polymerase II elongation factor, the Paf1 complex, regulating Set1/COMPASS function
Biochemical affinity purifications, with yeast RNA Pol II as the bait, identified the Polymerase Associated Factor 1 (PAF1) as a Pol II-interacting protein (148). Deletion of Paf1 in yeast resulted in several general transcriptional phenotypes including a defect in the induction of the galactose-regulated genes, suggesting a general role for Paf1 in transcriptional control (148). Another factor, Rtf1, was genetically identified to function as a RNA Pol II elongation factor in yeast (149). Further biochemical studies demonstrated that both Paf1 and Rtf1 and a few other factors including Cdc73, Ctr9 and Leo1 interact within a same complex, named the Paf1 complex, controlling transcriptional initiation and elongation in yeast (150–153).
Very little was known about the exact molecular function of the Paf1 complex in transcription until the subunits of this complex appeared as hits in the GPS biochemical screen when exploring for factors required for proper H3K4 and H3K79 methylation (54). Several components of the Paf1 complex including Paf1, Rtf1, Cdc73 and Ctr9 were identified as factors whose deletion resulted in a reduction in H3K4 and H3K79 trimethylation levels (54). The Paf1 complex regulates H3K4 methylation at several levels. The components of the Paf1 complex are required for proper H2B monoubiquitination by Rad6/Bre1, since in the absence of the Paf1 complex, both Rad6 and Bre1 are recruited to chromatin. However, Rad6/Bre1 are not functional in H2B monoubiquitiation until Paf1 is recruited to the same site (53). Not only does the Paf1 complex function by regulating H2B monoubiquitination through Rad6/Bre1, it also plays a role as a landing pad for Set1/COMPASS on RNA Pol II promoting H3K4 methylation on chromatin within the transcribed regions of the genes (53, 54). These early studies proposed that the Paf1 complex plays a role as a “platform” for the association of the histone-modifying machinery with the transcribing RNA Pol II (53, 54, 154). Indeed, subsequent studies identified other factors such as Set2, in addition to COMPASS, that required the Paf1 complex for their association and methylase function (155).
Similar to the subunits of Set1/COMPASS or Rad6/Bre1, which were initially identified in yeast to play a role in histone H3 and H2B modifications and shown to be functionally conserved in mammalian cells (42, 43, 53, 126, 127, 130, 135, 137, 144), the Paf1 complex is also highly conserved in structure and function from yeast to mammalian cells (144, 156–159). Even the “platform” function of the Paf1 complex, as a site of recruitment for Set1/COMPASS (53, 154), is conserved in mammalian cells. The Paf1 complex in human cells can associate and recruit the MLL1/COMPASS-like complex to transcribing RNA Pol II (160, 161). However, given the fact that the Paf1 complex associates with all transcriptionally active sites in mammalian cells, this finding does not describe how MLL, which is only required for H3K4 methylation on a small subset of genes (77), is specifically recruited by the Paf1 complex to such loci.
5 - Histone H3K4 methylases in development and disease pathogenesis
5.1 - “Licensed to Elongate” a model for leukemic pathogenesis through MLL1 translocations
Among the four MLLs, the MLL1 gene is the subject of frequent chromosomal translocations associated with acute myeloid and lymphoid leukemia (162–164). Chromosomal translocations involving the MLL1 gene in childhood malignancies account for ~10% of all detectable translocation events, and many of the patients bearing such chromosomal translocations are at high risk of relapse and require aggressive treatment (162, 164). Upon translocation, the C-terminal half of MLL is replaced by the translocating chromosomes and the entire SET domain of MLL is lost as a result of such translocations. Although the chromosomal copy of MLL within the translocations loses its SET domain, the SET domain of MLL from the other chromosomal copy of MLL appears to be required for leukemic pathogenesis (165). A role for MLL1 translocations as a cause of leukemic pathogenesis is well established, as knock-in or immortalization models of cancer have demonstrated that MLL1 translocations result in the pathogenesis of hematological malignancies with varying latencies of disease based on the translocation partners of MLL (162, 166–169). Although MLL’s contribution to leukemic pathogenesis was known for a long time, the exact molecular mechanism and contribution of so many of the seemingly unrelated MLL partners in leukemic pathogenesis was not clear until recently.
There are a large number of MLL translocation partners with very little sequence similarities (170). As diverse as the number of MLL partners in leukemia, are the numbers of models proposed describing how MLL1 translocations in so many unrelated genes result in the pathogenesis of leukemia (171–174). The ELL protein, which is one of the major translocation partners of MLL, was the first MLL-associated partner in leukemia for which a molecular and biochemical function was determined (175, 176). Over fifteen years ago, ELL was biochemically purified and demonstrated to function as a RNA Pol II elongation factor (175). ELL increases the catalytic rate of transcription elongation by RNA Pol II by suppressing transient pausing by the enzyme (175). Given the fact that biochemically, ELL functions as a Pol II elongation factor, and that ELL is one of the many MLL partners found in leukemia, it was proposed over fifteen years ago that the misregulation of the elongation stage of transcription could be central to leukemic pathogenesis through MLL translocations (175–177). Biochemical and genetic studies with human ELL 1–3 and the sole copy of ELL in Drosophila established that ELL can function as a RNA Pol II elongation factor both in vitro and in vivo (175, 178–183). However, it was not clear how MLL translocations, to so many unrelated genes, resulted in leukemia.
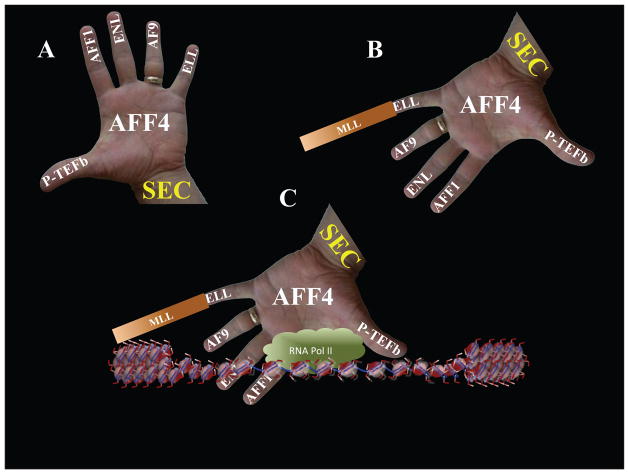
Following the biochemical purifications of many of the MLL-chimeras including MLL-ELL, MLL-ENL, MLL-AF9 and MLL-AFF1, it was demonstrated that AFF4, itself a partner of MLL in leukemia, is found as a common factor among the complexes purified from these translocations (184). Further biochemical purification of AFF4 resulted in the demonstration that many of the MLL translocation partners are found associated with the elongation factors ELL and P-TEFb within a complex called the Super Elongation Complex (SEC) (184). It is the translocation of MLL into SEC that is involved in the misrecruitment of SEC to MLL target genes, perturbing transcription elongation checkpoints at these loci, and resulting in leukemic pathogenesis (Figure 3) (83, 162, 184, 185). Not only is SEC involved in the regulation of transcription of the MLL target genes in leukemia, this elongation complex plays a central role in regulating transcription elongation control and gene expression during development, in response to environmental stimuli, and in HIV Tat-dependent transactivation (186–188).
Figure 3. MLL translocation into Super Elongation Complex in leukemic pathogenesis.
The MLL gene is found in a variety of chromosomal translocations associated with childhood leukemia. The ELL protein was the first translocation partner of MLL for which a biological function was demonstrated. Biochemically, ELL was identified as a RNA polymerase II elongation factor and it was proposed then that the elongation stage of transcription could be central in the pathogenesis of leukemia through MLL translocations. We now know that many of the MLL translocation partners such as ENL, AF9, AFF1, AFF4 are found in a super elongation complex (SEC) with ELL and the RNA polymerase II elongation factor P-TEFb. It has been proposed that the translocation of MLL into SEC results in the mistargeting of SEC to MLL-regulated genes and the misregulating of their expression without appropriate transcriptional elongation checkpoints.
5.2 – COMPASS family in the regulation of lifespan and aging
In addition to marking actively transcribed regions and the regulation of developmentally controlled genes, histone H3K4 methylation machineries have recently been linked with lifespan in a germline-dependent manner in C. elegans (189). Using a directed RNAi screen in fertile worms, the ASH2L subunit of the COMPASS complexes of C. elegans was demonstrated to play a role in the regulation of lifespan. Based on homology searching, it has been proposed that C. elegans possesses two of the three classes of the COMPASS family of H3K4 methylases (83). One is Set-2 of C. elegans, which is very similar to the Drosophila Set1 and human Set1A/B genes. The other is C. elegans Set-16, which bears resemblance to Drosophila Trr and human MLL3/4. The loss of Trx/MLL1-2 in C. elegans can be explained by the fact that this organism does not possess a large number of homeotic genes for development.
Along this line, reductions in the levels of other members of Set1/COMPASS in C. elegans, including Wdr5 and Set-2 (Set1 homolog), resulted in extension of lifespan, suggesting that an excess of H3K4 methylation is detrimental for longevity (189). In support of this observation, loss of the histone H3K4 demethylase RBR-2 in C. elegans was demonstrated to be also required for normal lifespan. Given the fact that histone H3K4 trimethylation by Set1/COMPASS requires proper H2B monoubiquitination by Rad6/Bre1 from yeast to human (3, 76, 190), it will of great interest to determine whether this pathway is also required for longevity in C. elegans, and whether di- and/or trimethylation is required for this process. This pathway can be further dissected by identification of the conserved role of Wdr82, which is specifically required for H3K4 trimethylation by Set1/COMPASS. The determination of the role of these factors in the specific regulation of di- and trimethylation in the regulation of longevity will also be of great interest.
5.3 - Histone H3K4 methylation by the Trr/COMPASS family in nuclear receptor transactivation, and cancer pathogenesis
The Trr family of H3K4 methylases in Drosophila and their human homologs, the MLL3/4 complexes, have been shown to function in diverse processes. Initially for Drosophila Trr and later for mammalian MLL3/4, it was demonstrated that they function in the nuclear hormone transactivation pathway. The ecdysone receptor (EcR), which is the major steroid hormone pathway receptor in Drosophila, was demonstrated to co-localize extensively with Trr in an ecdysone-dependent manner (92). Furthermore, Trr and the EcR can be isolated within a common complex in an ecdysone-dependent manner (92). Following these initial observations in Drosophila, it was demonstrated that the human homolog of Trr, the MLL3/4 family of the H3K4 methylases, also functions within the nuclear hormone pathway (94). This class of H3K4 methylases was demonstrated to work in the crosstalk between the ATPase-dependent chromatin remodeling complexes and efficient nuclear receptor transactivation.
In other studies, the MLL3/4 family members have been linked to cancer through interactions with p53 and the DNA damage pathway (191). Recently, frequent mutations in MLL4 were associated with non-Hodgkin’s lymphoma (110). Follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL) are the most common non-Hodgkin’s lymphomas. DNA sequence analysis from samples from patients suffering from FL and DLBCL demonstrated that about 32% of DLBCL and 89% of FL cases had somatic mutations in the MLL4 gene (110). Given the widespread role of MLL1 in leukemic pathogenesis and recent findings about MLL3/4 in cancer, further investigation of the roles of these families of methyltransferases will better our understanding of the Trr/MLL3-4 family of histone H3K4 methyltransferases.
6 - Concluding Remarks
It has been almost ten years since the original report on the biochemical purification of Set1/COMPASS as the yeast homolog of Drosophila trx and mammalian MLL (42) and its biochemical characterization as the first histone H3K4 methylase (42–44, 192). As described in this review, there has been a watershed of information regarding the COMPASS family of H3K4 methylases and the biological role of this histone modification from yeast to humans. We now know that Set1/COMPASS is highly conserved from yeast to human both enzymatically and structurally. Histone H3K4 trimethylation in yeast was shown to mark the promoters of the actively transcribed regions (53, 54, 193) and this mark is highly conserved from yeast to human (194). Although there is only one Set1/COMPASS in yeast, there are three COMPASS-like complexes in Drosophila, which are the Set1, trx and Trr/COMPASS-like complexes (Figure 2; Table 1). The activities of these three family members of the H3K4 methylases of Drosophila are expanded in mammalian cells. The activity of Set1/COMPASS is shared by the Set1A-B/COMPASS complexes. The trx and Trr activities are shared by the MLL1-2 and MLL3-4 COMPASS-like complexes, respectively (Figure 2). The H3K4 methylase activities of Set1, trx, and Trr of Drosophila and Set1A-B, MLL1-2 and MLL3-4 of mammals, are highly specific and non-redundant as the deletion of any of these methylases results in lethality, indicating that these enzymes cannot compensate for one another (84, 195–197).
Although we have learned an extensive amount of information about the process of implementation and the removal of the different patterns of histone H3K4 methylation during the past ten years, there is still much to be learned about the biology and reasons behind the diversity of the COMPASS family in metazoan cells. For example, Set1/COMPASS of yeast can implement the mono-, di- and trimethylation of H3K4, and several subunits of COMPASS are required to regulate this process (Table 1). However, in mammalian cells the different patterns of H3K4 methylations have been attributed to different biological functions. For example, as with yeast, H3K4 trimethylation has become a universal mark of actively transcribed regions in metazoans. And yet, other patterns of H3K4 methylation by the metazoan COMPASS family have been associated with other possible biological processes. Histone H3K4 monomethylation is becoming a landmark for the identification of active enhancers (198, 199). It is not clear which COMPASS family member is involved in the implementation of H3K4 monomethylation on the enhancers and what the role of this mark is in enhancer organization and function within the genomes.
Another example of diversity and variation in the H3K4 methylation patterns, is its co-existence with histone H3K27 trimethylation, which is referred to as bivalency (200). Promoters for developmentally regulated genes in mammalian stem cells are marked by both H3K4 trimethylation and H3K27 methylation. Such bivalently marked regions are thought to be required for proper regulation and the stem cells’ commitment to one fate or another. However, it is not clear which COMPASS family member is involved in the creation of bivalently marked promoters on developmentally controlled genes or how such regions function during development (201). Furthermore, other organisms such as Xenopus tropicalis, which share similar trithorax and polycomb group proteins, do not appear to use bivalently marked promoters throughout development (201, 202). Are bivalently marked promoters a mammalian specific phenomenon? Future research on COMPASS family members should include the molecular characterization of the mechanism of the recruitment of the COMPASS family to different loci within the genome. Due to the lack of reliable antibodies towards Set1A-B, MLL1-2 and MLL3-4, it is not clear at this time how different COMPASS family members are recruited to different loci. Early studies demonstrated that in the absence of MLL1-2, the Hox cluster in mouse embryonic fibroblasts lose their H3K4 methylation and concomitantly cease to express these loci (77). This finding suggested that unlike Set1/COMPASS, which implements H3K4 methylation subsequent to transcriptional activation, H3K4 methylation implemented by the MLL1-2/COMPASS family is instructive to transcription. How is it that the MLL1-2 complexes, which are found within almost identical complexes, are recruited to different loci within the Hox cluster? How is it that Set1A-B and MLL3-4 activities are excluded from these loci? Future studies addressing the above questions should employ molecular genetics, and biochemical and cell biological approaches on the COMPASS family in diverse model systems. The results from these studies will greatly add to our knowledge of the molecular complexity and functional diversity of this highly conserved ancient machinery.
Acknowledgments
I am indebted to many of my colleagues for their conversations and suggestions while writing this chapter. I am grateful my colleague Edwin Smith for his critical reading of this review and valuable comments and input. Also I thank Laura Shilatifard for editorial assistance and Julia Schulze for help with the illustrations. I apologize to my many colleagues whose work I was not able to cite in this review due to space limitations. Studies in my laboratory on the COMPASS family of histone H3K4 methylases and childhood leukemia are supported by NIH grants R01CA150265, R01GM069905 and R01CA89455, the Alex’s Lemonade Stand Foundation for Childhood Cancer and the Stowers Institute for Medical Research.
References
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–71. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heitz E. Das Heterochromatin der Moose. Jahrb Wiss Botanik. 1928;69:762–818. [Google Scholar]
- 5.Hilliker AJ, Holm DG. Genetic analysis of the proximal region of chromosome 2 of Drosophila melanogaster. I. Detachment products of compound autosomes. Genetics. 1975;81:705–21. doi: 10.1093/genetics/81.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasuhara JC, DeCrease CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10958–63. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller HJ, Altenburg E. The Frequency of Translocations Produced by X-Rays in Drosophila. Genetics. 1930;15:283–311. doi: 10.1093/genetics/15.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore GD, Procunier JD, Cross DP, Grigliatti TA. Histone gene deficiencies and position--effect variegation in Drosophila. Nature. 1979;282:312–4. doi: 10.1038/282312a0. [DOI] [PubMed] [Google Scholar]
- 9.Moore GD, Sinclair DA, Grigliatti TA. Histone Gene Multiplicity and Position Effect Variegation in DROSOPHILA MELANOGASTER. Genetics. 1983;105:327–44. doi: 10.1093/genetics/105.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–45. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 11.Han M, Kim UJ, Kayne P, Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–8. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–34. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 13.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–10. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 14.Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 15.Workman JL, Roeder RG. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–22. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 16.Chang CH, Luse DS. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J Biol Chem. 1997;272:23427–34. doi: 10.1074/jbc.272.37.23427. [DOI] [PubMed] [Google Scholar]
- 17.Ujvari A, Hsieh FK, Luse SW, Studitsky VM, Luse DS. Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J Biol Chem. 2008;283:32236–43. doi: 10.1074/jbc.M806636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–56. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 19.Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–6. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 20.Mahmoudi T, Verrijzer CP. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene. 2001;20:3055–66. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- 21.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 22.Stassen MJ, Bailey D, Nelson S, Chinwalla V, Harte PJ. The Drosophila trithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech Dev. 1995;52:209–23. doi: 10.1016/0925-4773(95)00402-m. [DOI] [PubMed] [Google Scholar]
- 23.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–31. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 25.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–16. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 26.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–79. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 27.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 29.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Molecular cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 31.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 32.Ingham PW, Whittle R. Trithorax: a new homeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Molecular and General Genetics. 1980;179:607–14. [Google Scholar]
- 33.Ingham PW. Genetic control of the spatial pattern of selector gene expression in Drosophila. Cold Spring Harbor symposia on quantitative biology. 1985;50:201–8. doi: 10.1101/sqb.1985.050.01.026. [DOI] [PubMed] [Google Scholar]
- 34.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 35.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–8. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 38.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 39.Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R, III, Patel Y, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci U S A. 1991;88:10735–9. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nature reviews Cancer. 2010;10:721–8. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–94. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, et al. COMPASS, a histone H3 (Lysine 4) methyl transferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 44.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–48. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–56. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Nedea E, He X, Kim M, Pootoolal J, Zhong G, et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–10. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 47.Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, et al. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–87. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 48.Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol. 2004;24:2932–43. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlichter A, Cairns BR. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. Embo J. 2005;24:1222–31. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–12. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 51.Wood A, Shukla A, Schneider J, Lee JS, Stanton JD, et al. Ctk complex-mediated regulation of histone methylation by COMPASS. Mol Cell Biol. 2007;27:709–20. doi: 10.1128/MCB.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao T, Shibata Y, Rao B, Laribee RN, O’Rourke R, et al. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol Cell Biol. 2007;27:721–31. doi: 10.1128/MCB.01628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–42. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 54.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–9. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 55.Jiang D, Kong NC, Gu X, Li Z, He Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011;7:e1001330. doi: 10.1371/journal.pgen.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–97. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 57.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 58.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, et al. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–4. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 60.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. The EMBO journal. 2011 doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, et al. COMPASS-like complexes in Drosophila. Mol Cell Biol. 2011 doi: 10.1128/MCB.06092-11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–8. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho YW, Hong T, Hong S, Guo H, Yu H, et al. PTIP associates with MLL3-and MLL4-containing histone H3 lysine 4 methyltransferase complex. The Journal of biological chemistry. 2007;282:20395–406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Molecular and cellular biology. 2007;27:1889–903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Molecular and cellular biology. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes & development. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Molecular and cellular biology. 2004;24:5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Developmental cell. 2007;13:580–92. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 71.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Molecular and cellular biology. 2003;23:140–9. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. The Journal of biological chemistry. 2007;282:13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 73.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–31. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 74.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–9. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–18. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, et al. Histone Crosstalk between H2B Monoubiquitination and H3 Methylation Mediated by COMPASS. Cell. 2007;131:1084–96. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 77.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–85. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee MG, Villa R, Trojer P, Norman J, Yan KP, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 79.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 80.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Molecular and cellular biology. 2008;28:609–18. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li T, Kelly WG. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 2011;7:e1001349. doi: 10.1371/journal.pgen.1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002109. doi: 10.1371/journal.pgen.1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–72. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 85.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA. 1998;95:10632–6. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes & development. 2003;17:2298–307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Molecular and cellular biology. 2004;24:617–28. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 89.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6629–34. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sedkov Y, Benes JJ, Berger JR, Riker KM, Tillib S, et al. Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mech Dev. 1999;82:171–9. doi: 10.1016/s0925-4773(98)00246-9. [DOI] [PubMed] [Google Scholar]
- 91.Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, et al. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, et al. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426:78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, et al. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997;15:549–60. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- 94.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281:15714–20. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 95.Lee J, Saha PK, Yang QH, Lee S, Park JY, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19229–34. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–23. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim DH, Lee J, Lee B, Lee JW. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyl transferase activity. Molecular endocrinology. 2009;23:1556–62. doi: 10.1210/me.2009-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Molecular endocrinology. 2009;23:610–9. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aboobaker AA, Blaxter ML. Hox Gene Loss during Dynamic Evolution of the Nematode Cluster. Current biology: CB. 2003;13:37–40. doi: 10.1016/s0960-9822(02)01399-4. [DOI] [PubMed] [Google Scholar]
- 100.Robinson-Rechavi M, Maina CV, Gissendanner CR, Laudet V, Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. Journal of molecular evolution. 2005;60:577–86. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- 101.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–94. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 102.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. The EMBO journal. 2010;29:1401–11. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, et al. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Molecular and cellular biology. 2008;28:1041–6. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol. 2010;30:2485–97. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, et al. The histone demethylase UTX enables RB-dependent cell fate control. Genes & development. 2010;24:327–32. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature genetics. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 107.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS genetics. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome research. 2011;21:216–26. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ashktorab H, Schaffer AA, Daremipouran M, Smoot DT, Lee E, Brim H. Distinct genetic alterations in colorectal cancer. PloS one. 2010;5:e8879. doi: 10.1371/journal.pone.0008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011 doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed]
- 111.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lieb JD, Capowski EE, Meneely P, Meyer BJ. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science. 1996;274:1732–6. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- 114.Escamilla-Del-Arenal M, da Rocha ST, Heard E. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Human genetics. 2011;130:307–27. doi: 10.1007/s00439-011-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laverty C, Lucci J, Akhtar A. The MSL complex: X chromosome and beyond. Current opinion in genetics & development. 2010;20:171–8. doi: 10.1016/j.gde.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 116.Chuang PT, Albertson DG, Meyer BJ. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–74. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 117.Chuang PT, Lieb JD, Meyer BJ. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–9. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- 118.Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–77. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 119.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsu DR, Chuang PT, Meyer BJ. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development. 1995;121:3323–34. doi: 10.1242/dev.121.10.3323. [DOI] [PubMed] [Google Scholar]
- 121.Hsu DR, Meyer BJ. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 1994;137:999–1018. doi: 10.1093/genetics/137.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes & development. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schneider J, Dover J, Johnston M, Shilatifard A. Global proteomic analysis of S. cerevisiae (GPS) to identify proteins required for histone modifications. Methods Enzymol. 2004;377:227–34. doi: 10.1016/S0076-6879(03)77013-X. [DOI] [PubMed] [Google Scholar]
- 124.Giaever G, Chu AM, Ni L, Connelly C, Riles L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 125.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, et al. Life with 6000 genes. Science. 1996;274:546, 63–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 126.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–71. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 127.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 128.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002;277:34655–7. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 129.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 130.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–74. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 131.Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, et al. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–41. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–6. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 133.Zheng S, Wyrick JJ, Reese JC. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol. 2010;30:3635–45. doi: 10.1128/MCB.00324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vitaliano-Prunier A, Menant A, Hobeika M, Geli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–71. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 135.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–11. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 136.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–70. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 137.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–89. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–86. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 140.Smith E, Shilatifard A. Developmental biology. Histone cross-talk in stem cells. Science. 2009;323:221–2. doi: 10.1126/science.1168660. [DOI] [PubMed] [Google Scholar]
- 141.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–51. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–9. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 143.Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, et al. Dot1 and Histone H3K79 Methylation in Natural Telomeric and HM Silencing. Mol Cell. 2011;42:118–26. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pavri R, Zhu B, Li G, Trojer P, Mandal S, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–17. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 145.Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–47. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 41:529–42. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–28. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 148.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–76. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–47. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–80. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 152.Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–67. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, et al. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. Embo J. 2002;21:1764–74. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem. 2003;278:26303–6. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 155.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–18. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–73. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang P, Bowl MR, Bender S, Peng J, Farber L, et al. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol Cell Biol. 2008;28:2930–40. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, et al. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103:11970–4. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–21. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–63. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–8. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 163.Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 164.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–9. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 165.Thiel AT, Blessington P, Zou T, Feather D, Wu X, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–59. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Corral J, Lavenir I, Impey H, Warren AJ, Forster A, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–61. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 167.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–74. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 168.DiMartino JF, Miller T, Ayton PM, Landewe T, Hess JL, et al. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood. 2000;96:3887–93. [PubMed] [Google Scholar]
- 169.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J. 1997;16:4226–37. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem. 2005;95:429–36. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- 171.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–54. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 173.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 174.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 175.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–6. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 176.Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–46. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 177.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 178.Shilatifard A, Duan DR, Haque D, Florence C, Schubach WH, et al. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc Natl Acad Sci U S A. 1997;94:3639–43. doi: 10.1073/pnas.94.8.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor. ELL3 J Biol Chem. 2000;275:32052–6. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- 180.Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. Embo J. 2001;20:6104–14. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gerber MA, Shilatifard A, Eissenberg JC. Mutational analysis of an RNA polymerase II elongation factor in Drosophila melanogaster. Mol Cell Biol. 2005;25:7803–11. doi: 10.1128/MCB.25.17.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A. 2008;105:8575–9. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–37. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.He N, Liu M, Hsu J, Xue Y, Chou S, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–38. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–51. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–98. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–7. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–5. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee J, Kim DH, Lee S, Yang QH, Lee DK, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci U S A. 2009;106:8513–8. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 193.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 194.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 195.Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 8 doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 329:917–23. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 199.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–9. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 201.Herz HM, Nakanishi S, Shilatifard A. The curious case of bivalent marks. Dev Cell. 2009;17:301–3. doi: 10.1016/j.devcel.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 202.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, et al. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–34. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]