ZBTB20 promotes long-lived plasma cell survival after primary activation under alum but not TLR2- and TLR4-activating adjuvant conditions.
Abstract
The duration of antibody production by long-lived plasma cells varies with the type of immunization, but the basis for these differences is unknown. We demonstrate that plasma cells formed in response to the same immunogen engage distinct survival programs depending on the adjuvant. After alum-adjuvanted immunization, antigen-specific bone marrow plasma cells deficient in the transcription factor ZBTB20 failed to accumulate over time, leading to a progressive loss of antibody production relative to wild-type controls. Fetal liver reconstitution experiments demonstrated that the requirement for ZBTB20 was B cell intrinsic. No defects were observed in germinal center numbers, affinity maturation, or plasma cell formation or proliferation in ZBTB20-deficient chimeras. However, ZBTB20-deficient plasma cells expressed reduced levels of MCL1 relative to wild-type controls, and transgenic expression of BCL2 increased serum antibody titers. These data indicate a role for ZBTB20 in promoting survival in plasma cells. Strikingly, adjuvants that activate TLR2 and TLR4 restored long-term antibody production in ZBTB20-deficient chimeras through the induction of compensatory survival programs in plasma cells. Thus, distinct lifespans are imprinted in plasma cells as they are formed, depending on the primary activation conditions. The durability of vaccines may accordingly be improved through the selection of appropriate adjuvants.
Plasma cells are terminally differentiated B lymphocytes that secrete large quantities of antibodies. During the initial stages of a T cell–dependent antibody response, plasma cells are found in the extrafollicular regions of secondary lymphoid organs (Fagraeus, 1948). These extrafollicular plasma cells are responsible for the initial surge in antibody levels after immunization or infection, but are thought to survive for only several days before undergoing apoptosis (Jacob et al., 1991; Smith et al., 1994; Sze et al., 2000). A second wave of plasma cells that express high-affinity antibodies is generated from the germinal center reaction (Han et al., 1995; Smith et al., 1997; Phan et al., 2006). Affinity-matured plasma cells egress from secondary lymphoid organs to seed the BM, where they can persist for many years (Slifka et al., 1995, 1998; Manz et al., 1997; Hargreaves et al., 2001; Pabst et al., 2005; Kabashima et al., 2006). These long-lived plasma cells are solely responsible for maintaining antigen-specific serum antibodies long after clearance of infection or vaccination (Manz et al., 1998; Slifka et al., 1998; Cambridge et al., 2003; Ahuja et al., 2008; DiLillo et al., 2008).
The ontogeny of long-lived plasma cells indicates that signals received within the germinal center reaction confer longevity. Potential mechanisms for determining longevity include the induced expression of chemokine receptors, such as CXCR4 and S1PR1, which allow plasma cells to egress to the BM and access survival cytokines (Benner et al., 1981; Hargreaves et al., 2001; Hauser et al., 2002; Kabashima et al., 2006). One of the survival cytokines, APRIL, binds to its receptor BCMA and activates plasma cell–intrinsic antiapoptotic factors such as MCL1 (Moreaux et al., 2004; O’Connor et al., 2004; Belnoue et al., 2008; Peperzak et al., 2013). XBP1 and ATG5 are also essential for plasma cell survival because of their roles in regulating ER stress (Reimold et al., 2001; Hu et al., 2009; Pengo et al., 2013). Factors that establish and maintain plasma cell identity, such as BLIMP1, are also required for long-term antibody responses (Shapiro-Shelef et al., 2005).
Clearly, however, additional pathways that fine-tune the survival of plasma cells remain to be discovered. The duration of antibody production and plasma cell lifespan varies widely with the specific vaccine or infection, yet the basis for these differences remains unknown (Amanna et al., 2007; Amanna and Slifka, 2010). Multiple recent clinical studies have shown that protection against malaria and Pertussis wanes rapidly after vaccination, leading to high rates of infection and mortality in previously immunized children (Misegades et al., 2012; Olotu et al., 2013). Thus, an understanding of the particular features of vaccines and host responses that confer durable antibody production is of utmost importance.
In previous work, we found that ZBTB20, a member of the Broad complex, tramtrack, bric-a-brac-poxvirus, and zinc finger (BTB-POZ) family of transcriptional repressors, was highly expressed in plasma, germinal center, and memory B cells (Bhattacharya et al., 2007). Members of this family of transcription factors contain an N-terminal BTB-POZ domain that mediates homodimerization and recruitment of nuclear co-repressors, as well as a variable number of zinc finger domains at the C terminus, which mediate DNA binding (Melnick et al., 2002). Prior studies have shown that ZBTB20 regulates pancreatic β cell function, neuronal development in the hippocampus, and transcription of α-fetoprotein (Xie et al., 2008, 2010; Sutherland et al., 2009; Nielsen et al., 2010; Zhang et al., 2012). However, the physiological importance of elevated ZBTB20 expression in activated B cells remained unknown. Here, we demonstrate that ZBTB20 is required for long-term antibody production and plasma cell persistence specifically after alum-adjuvanted immunization. In contrast, maintenance of antibody production after immunization with TLR ligand–containing adjuvants is ZBTB20 independent. We conclude that long-lived plasma cells generated in response to the same antigen can access distinct survival programs, depending on the initial activation conditions experienced by B cells in germinal centers.
RESULTS
ZBTB20 is highly expressed in activated B cells but is not required for B cell development
Our previous microarray experiments demonstrated elevated expression of ZBTB20, a member of the BTB-POZ family of transcriptional repressors, after B cell activation (Bhattacharya et al., 2007). To study the functional role of ZBTB20 in B cells, we obtained a gene-trapped mouse embryonic stem cell line carrying an insertion within the Zbtb20 locus. The insertion consists of a splice acceptor, a β-galactosidase–neomycin resistance fusion cassette, and a polyadenylation signal. As this promoterless gene trap is integrated in the intronic region between exons 3 and 4 of Zbtb20 and upstream of the two known translation start sites (Mitchelmore et al., 2002), the insertion is predicted to both report and attenuate ZBTB20 expression.
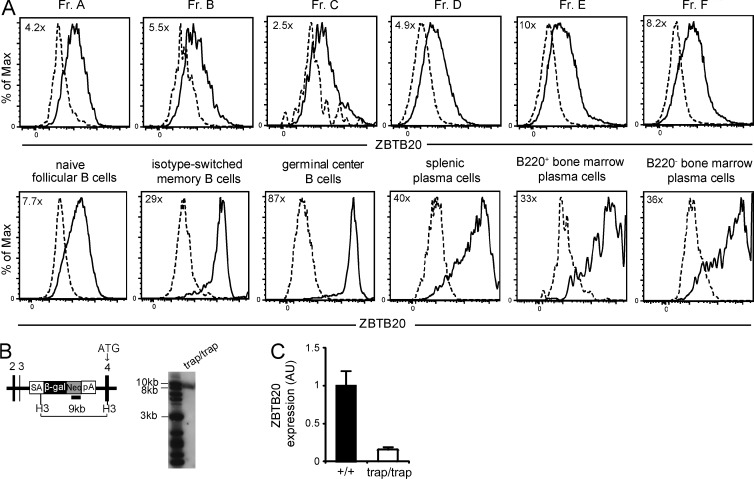
We first generated heterozygous Zbtb20+/trap mice using this gene-trapped embryonic stem cell line. The β-galactosidase activity of the gene-trap can be used as a ZBTB20 transcriptional reporter and measured through cleavage of the substrate fluorescein di–β-galactopyranoside (Nolan et al., 1988). Thus, we first assessed the expression of ZBTB20 in heterozygous B cell progenitors, naive follicular B cells, polyclonal splenic isotype-switched memory B cells, splenic and BM plasma cells, and germinal center B cells from unimmunized mice to confirm the microarray results (Fig. S1). Flow cytometry analysis of Zbtb20+/trap cells showed that ZBTB20 expression was substantially elevated in all activated and memory B cell populations relative to B cell progenitors and the naive B cell compartment (Fig. 1 A). The frequency of CD138hi BM plasma cells was low, consistent with previous studies (Manz et al., 1997; Slifka et al., 1998). To confirm the accuracy of our gating strategy and to exclude the possibility of nonplasma cell contamination within this population, B220− CD138hi cells were double-sorted into α-Igκ– and α-Igλ–coated ELISPOT wells, and IgM- and IgG-containing spots were quantified. Approximately 50% of sorted cells gave rise to detectable spots (not depicted); this is almost certainly an underestimate of the true plasma cell frequency, as viability and postsort purity were unlikely to have been 100%. Using gene-trap–specific Southern blotting, we confirmed the presence of a single insertion only within the Zbtb20 locus (Fig. 1 B).
Figure 1.
ZBTB20 is highly expressed by activated B cells. (A) Flow cytometry analysis of ZBTB20 expression through β-galactosidase expression in Zbtb20+/trap B cell progenitors by the Hardy classification (Fr. A–F), naive B cells, splenic plasma cells, germinal center B cells, isotype-switched memory B cells, and B220+ and B220− BM plasma cells. Gating strategies are shown in Fig. S1. Cells from heterozygous Zbtb20+/trap (solid lines) and control Zbtb20+/+ (dashed lines) mice were both stained with the β-galactosidase substrate fluorescein di-β-galactopyranoside. Values in the top left corner of plots represent fold changes of the mean fluorescence intensities between Zbtb20+/trap and Zbtb20+/+ cells. Data are representative of four independent experiments with one mouse per group per experiment. (B) Map of Zbtb20 locus (left) and Southern blot analysis (right) demonstrating the correct location and copy number of the gene trap insertion. Black numbered rectangles denote exons 2–4 of Zbtb20. The small solid rectangle indicates the location of the Neo-specific probe. ATG, translation start site; β-gal, β-galactosidase; H3, HindIII; Neo, neomycin phosphotransferase; pA, polyadenylation site; SA, splice acceptor. (C) qRT-PCR analysis of ZBTB20 transcript levels in peripheral blood B cells from Zbtb20+/+ (n = 9) and Zbtb20trap/trap (n = 8) fetal liver chimeras. Transcript levels were measured in technical triplicates, and Zbtb20 expression was normalized to the mean Actb expression of each sample. Mean values ± SD are shown in arbitrary units (AU) relative to wild-type B cell levels. Data are representative of two independent experiments.
Consistent with previous studies using independent ZBTB20-deficient mouse strains, homozygous Zbtb20trap/trap mice did not survive to adulthood (Rosenthal et al., 2012; Sutherland et al., 2009). We thus performed all subsequent experiments using fetal liver reconstitutions of 800 cGy–irradiated adult wild-type mice. Peripheral blood B cells derived from homozygous Zbtb20trap/trap fetal liver donors expressed approximately sixfold lower levels of ZBTB20 transcripts than did B cells from Zbtb20+/+ chimeras, confirming the efficacy of the gene trap mutation (Fig. 1 C). We also assessed hematopoietic development through competitive reconstitution assays in which CD45.2 Zbtb20trap/trap and Zbtb20+/+ fetal livers were cotransplanted with wild-type CD45.1 BM. No significant differences were observed between Zbtb20trap/trap and Zbtb20+/+ chimeras in their contribution to progenitors or mature immune cells (Fig. S2 and not depicted). Thus, ZBTB20 is not required for hematopoietic development.
ZBTB20 regulates long-term antibody production through a B cell–intrinsic mechanism
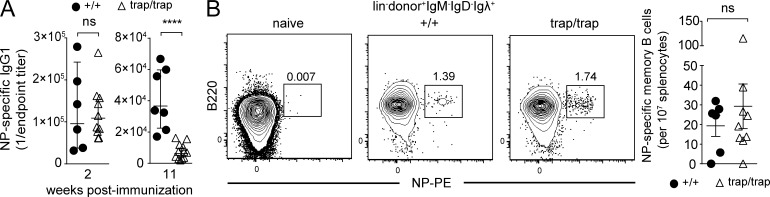
Given that ZBTB20 is dramatically induced in activated B cells, we next examined its role in antibody responses. For this purpose, we immunized fetal liver chimeras with alum-adjuvanted 4-hydroxy-3-nitrophenyl-acetyl (NP) conjugated to chicken gamma globulin (CGG), a T-dependent hapten protein conjugate antigen. Although NP-specific serum antibody titers in Zbtb20trap/trap chimeras were similar to controls at 2 wk after immunization, they were significantly reduced (approximately sixfold) relative to antibody titers in Zbtb20+/+ chimeras at 11 wk after immunization (Fig. 2 A). In contrast, the numbers of NP-specific memory B cells in Zbtb20trap/trap chimeras were similar to controls (Fig. 2 B).
Figure 2.
ZBTB20 is required for the long-term production of antigen-specific antibodies. Fetal liver chimeras were generated by transplanting Zbtb20+/+ or Zbtb20trap/trap fetal liver cells between 14.5 and 17.5 d postcoitus into 8–12-wk-old lethally irradiated wild-type mice. Mice were immunized 8–16 wk after reconstitution. (A) ELISA measurements of serum NP-specific IgG1 titers from Zbtb20+/+ or Zbtb20trap/trap fetal liver chimeras at 2 and 11 wk after alum-adjuvanted NP-CGG immunization (error bars depict geometric means ± 95% confidence interval). Each data point represents a geometric mean of technical duplicates. Data are representative of three experiments, each with two to eight mice per genotype. Statistical significance was determined with the Mann–Whitney test. ns, not significant (P > 0.05); ****, P < 0.0001. (B) Frequencies of donor splenic lineage− (CD3/CD4/CD8/CD11c/Gr-1/Ter119) NP-specific memory B cells from Zbtb20+/+ or Zbtb20trap/trap fetal liver chimeras were determined by flow cytometry at 12–16 wk after alum-adjuvanted NP-CGG immunization. Mean values ± SEM are shown, and examples of gating strategies are shown to the left. Data are representative of two experiments, each with four to nine mice per genotype. Statistical significance was determined with an unpaired Student’s two-tailed t test. ns, not significant (P > 0.05).
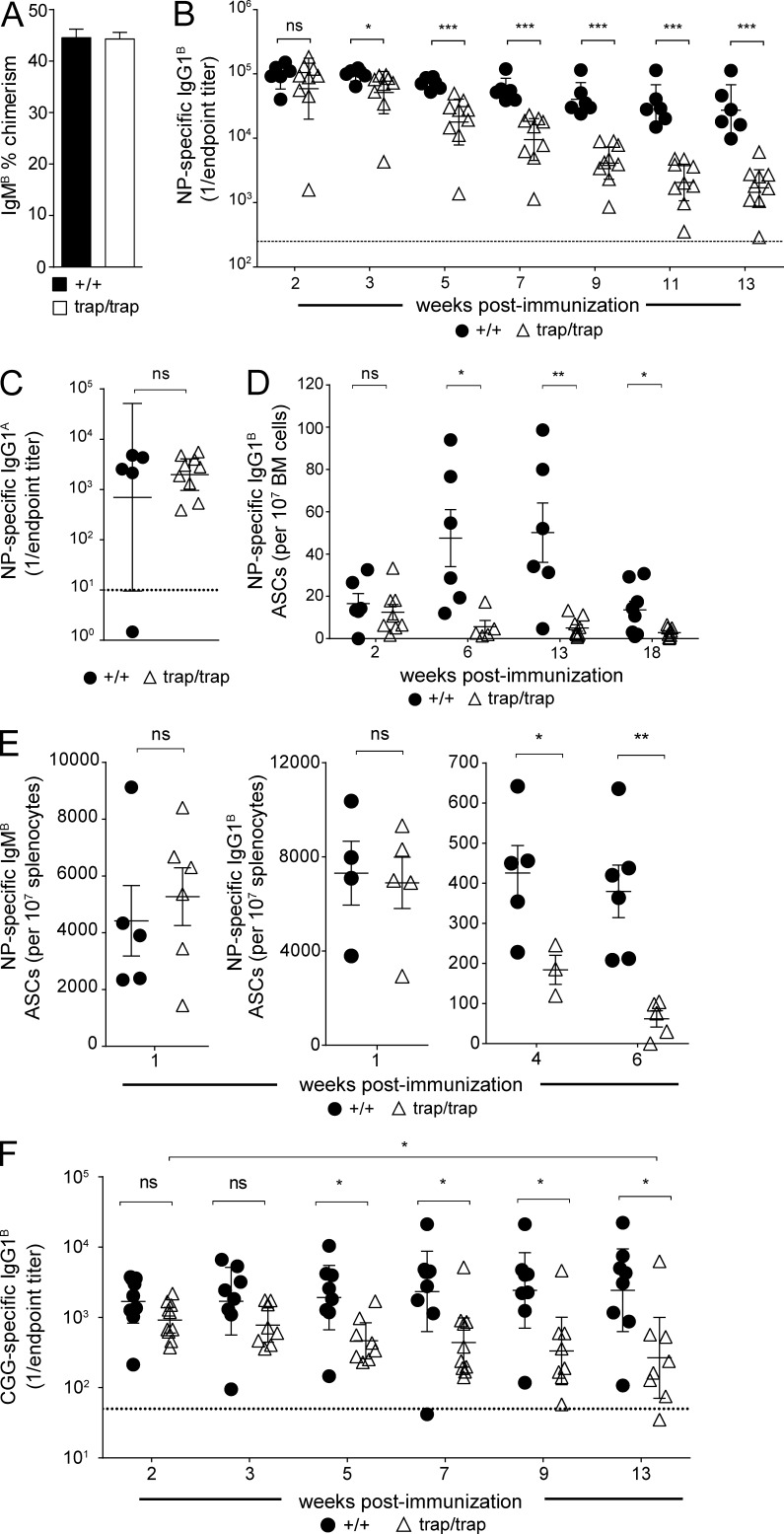
To determine whether the requirement for ZBTB20 in long-term NP-specific antibody production is B cell intrinsic, we performed mixed chimera experiments. Mice were reconstituted with equal numbers of IgHA wild-type fetal liver cells and either IgHB Zbtb20trap/trap or littermate control Zbtb20+/+ fetal liver cells. Allotype-specific secondary antibodies were then used to distinguish donor B cells and antibodies. Donor CD45.2 IgHA and IgHB fetal liver cells contributed equally to the B cell compartment, whereas residual CD45.1 recipient B cells were undetectable (Fig. 3 A). Mixed chimeras were immunized with NP-CGG as above, and analysis of NP-specific serum IgG1B responses demonstrated that antibody titers were normal at 2 wk after immunization (Fig. 3 B). However, similar to the data shown in Fig. 2 A, Zbtb20trap/trap-derived IgG1B antibody titers were progressively lost over time, reaching levels that were 16-fold lower at 13 wk after immunization when compared with control IgG1B titers from Zbtb20+/+ chimeras (Fig. 3 B). Analysis of control NP-specific serum IgG1A titers revealed no differences between Zbtb20+/+ and Zbtb20trap/trap mixed chimeras (Fig. 3 C). These data demonstrate a B cell–intrinsic requirement for ZBTB20 in long-term antibody responses.
Figure 3.
Requirement for ZBTB20 in maintenance of antibody responses is B cell intrinsic. Mixed fetal liver chimeras were generated by transplanting either Zbtb20+/+ or Zbtb20trap/trap IghB fetal liver cells with congenic IghA wild-type (B6.Cg-IghA Thy1AGpi1A) fetal liver cells at a 1:1 ratio into 8–12-wk-old lethally irradiated wild-type mice. Mice were immunized 8–16 wk after reconstitution. Ig allotype–specific secondary antibodies (IgMA vs. IgMB; IgG1A vs. IgG1B) were then used to distinguish donor B cells and antibodies. (A) Peripheral blood B cell IgMB cell chimerism in Zbtb20+/+ (n = 9) or Zbtb20trap/trap (n = 8) mixed fetal liver chimeras was determined by flow cytometry at 7 wk after reconstitution. Mean values ± SEM are shown. Data are representative of three independent experiments. (B and C) ELISA measurements of IgG1B NP-specific serum titers at multiple time points between 2 and 13 wk (B) and IgG1A control wild-type NP-specific serum titers at week 13 (C) from Zbtb20+/+ (n = 6) or Zbtb20trap/trap (n = 9) mixed chimeras after immunization (each point represents one mouse, and error bars depict geometric means ± 95% confidence interval). Data are representative of two independent experiments. Statistical significance was determined by the Mann–Whitney test. ns, not significant (P > 0.05); *, 0.01 < P < 0.05; ***, P < 0.001. (D and E) ELISPOT assays of NP-specific IgHB ASCs in the BM at 2, 6, 13, and 18 wk (D) and in the spleen at 1, 4, and 6 wk (E) from Zbtb20+/+ or Zbtb20trap/trap mixed chimeras after alum-adjuvanted NP-CGG immunization. Mean values ± SEM are shown. Each data point represents a mean of technical triplicates. Each time point has four to nine mice per genotype. Data are cumulative of two independent experiments. Statistical significance was determined with an unpaired Student’s two-tailed t test. ns, not significant (P > 0.05); *, 0.01 < P < 0.05; **, P < 0.01. (F) ELISA measurements of serum CGG-specific IgG1B titers at multiple time points between 2 and 13 wk from Zbtb20+/+ or Zbtb20trap/trap mixed chimeras after alum-adjuvanted NP-CGG immunization (each data point represents one mouse, and error bars depict geometric means ± 95% confidence interval). Data are representative of two experiments, each with 8–10 mice per genotype. Statistical significance was determined by the Mann–Whitney test. ns, not significant (P > 0.05); *, 0.01 < P < 0.05.
As the t1/2 of IgG1 serum antibodies is ∼1 wk (Vieira and Rajewsky, 1988), antibody titers in ZBTB20-deficient chimeras declined at nearly the maximum possible rate 3 wk after immunization (Fig. 3 B). These data suggest an absence of new antibody production at later phases of the response, potentially as the result of reduced numbers of plasma cells. To directly quantify the number of antigen-specific BM plasma cells, we performed ELISPOT analysis at 2, 6, 13, and 18 wk after immunization. NP-specific Zbtb20trap/trap antibody-secreting cell (ASC) numbers were similar to controls at 2 wk after immunization (Fig. 3 D). However, by 6 and 13 wk after immunization, ZBTB20-deficient NP-specific BM plasma cell numbers had failed to expand. As a result, Zbtb20+/+ NP-specific plasma cells outnumbered their Zbtb20trap/trap counterparts by 10-fold (Fig. 3 D). By 18 wk after immunization, most ZBTB20-deficient chimeras lacked detectable numbers of NP-specific BM plasma cells (Fig. 3 D). Similarly, no defects in splenic NP-specific plasma cell numbers were observed at 1 wk after immunization in ZBTB20-deficient chimeras, which are mostly comprised of short-lived plasma cells (Fig. 3 E). However, significant deficiencies in splenic plasma cells were observed in Zbtb20trap/trap chimeras at 4 and 6 wk after immunization relative to wild-type chimeras (Fig. 3 E). These data indicate a B cell–intrinsic requirement for ZBTB20 to accumulate and maintain long-lived plasma cells for enduring antibody responses.
To determine whether ZBTB20 is required for long-term antibody responses to protein antigens, we quantified CGG-specific titers over time. We again observed large defects in serum antibody production at late time points in ZBTB20-deficient chimeras (Fig. 3 F). These defects manifested as a failure to increase CGG-specific antibodies over time, perhaps because monomeric CGG as an ELISA antigen only allows for the detection of high-affinity antibodies, which peak late in the response. Regardless, these data are consistent with the requirement for ZBTB20 to accumulate affinity-matured long-lived plasma cells, irrespective of the antigen.
Zbtb20 deficiency is overcome by inclusion of TLR ligand–containing adjuvants
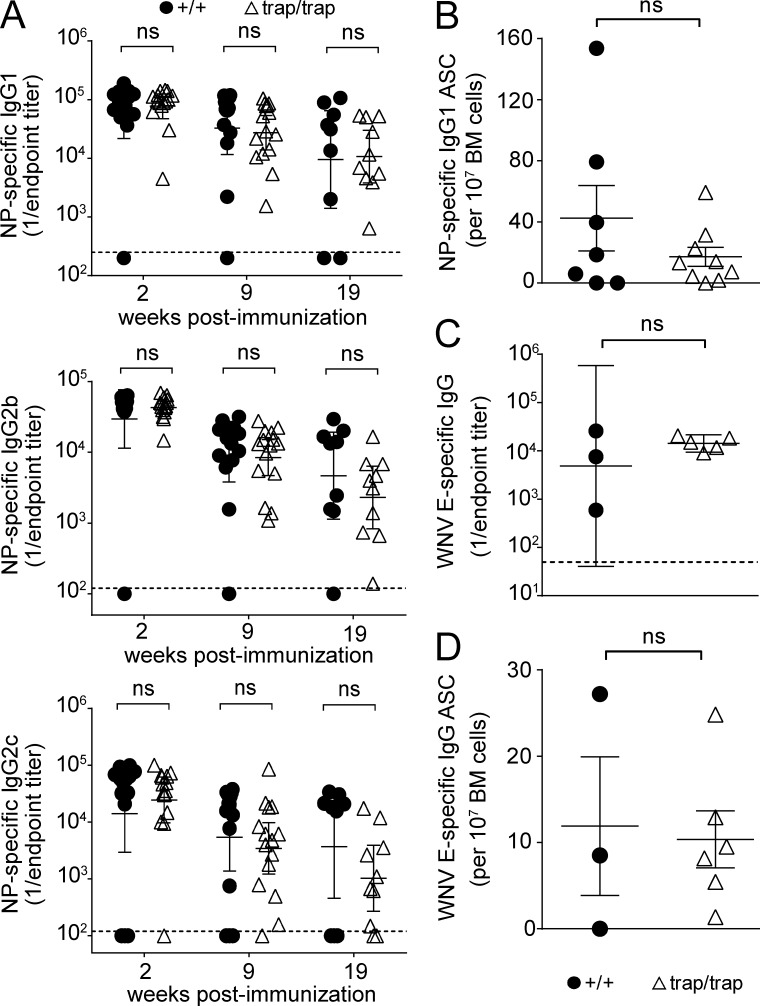
Although the data above indicate that ZBTB20 is required for long-lived antibody responses irrespective of the immunizing antigen, other features of the immunogen may also affect the duration of immunity. Specifically, the use of pathogen-derived adjuvants, in place of aluminum salts, correlates with particularly durable antibody responses in clinical vaccines (Amanna et al., 2007; Marrack et al., 2009). Thus, we tested whether such pathogen-derived adjuvants engage distinct ZBTB20-independent pathways to maintain antibody production. Fetal liver chimeras were immunized with the same dose of NP-CGG as above but adjuvanted with monophosphoryl lipid A, a TLR4 ligand (Martin et al., 2003), and trehalose dicorynomycolate, a TLR2 ligand derived from Mycobacterium tuberculosis (Bowdish et al., 2009). This formulation is very similar to Ribi adjuvant, in which the bulk of long-term antibody production is TLR dependent (Gavin et al., 2006). We examined IgG1, IgG2b, and IgG2c NP-specific titers, as B cells of different isotypes can use distinct survival programs (Dogan et al., 2009; Pape et al., 2011; Wang et al., 2012). For every isotype, Zbtb20trap/trap-derived serum NP-specific antibody titers were similar to controls at all time points examined (Fig. 4 A). Moreover, antigen-specific BM plasma cells persisted for at least 20 wk after immunization in both control and Zbtb20trap/trap chimeras (Fig. 4 B). To test the generality of these findings, we immunized mice with a vaccine against West Nile virus (WNV), which activates TLR3 signaling (Wang et al., 2004; Daffis et al., 2008; Xia et al., 2013). WNV-specific antibody titers at 21 wk and plasma cell numbers at 26 wk after vaccination were similar between Zbtb20+/+ and Zbtb20trap/trap chimeras (Fig. 4, C and D). These results suggest that TLR-based adjuvants use a ZBTB20-independent pathway for long-term antibody production, although we cannot fully exclude the possibility that other components of the formulations may also have adjuvanting effects.
Figure 4.
Immunization with TLR ligand–containing adjuvants bypasses the requirement for ZBTB20. (A) ELISA measurements of serum NP-specific IgG1, IgG2c, and IgG2b titers from Zbtb20+/+ or Zbtb20trap/trap fetal liver chimeras at 2, 9, and 19 wk after immunization with TLR ligand–adjuvanted NP-CGG (error bars depict geometric means ± 95% confidence interval). Data are cumulative from two experiments with 9–14 Zbtb20+/+ and 10–15 Zbtb20trap/trap chimeric mice per time point. Statistical significance was determined by the Mann–Whitney test. ns, not significant (P > 0.05). (B) ELISPOT analysis of NP-specific IgG1 BM ASCs from Zbtb20+/+ (n = 7) or Zbtb20trap/trap (n = 9) fetal liver chimeras at 20 wk after immunization (mean values ± SEM). Statistical significance was determined with an unpaired Student’s two-tailed t test. ns, not significant (P > 0.05). Data are cumulative from two independent experiments. (C) ELISA measurements of serum WNV-specific IgG titers from Zbtb20+/+ (n = 3) or Zbtb20trap/trap (n = 5) fetal liver chimeras at 21 wk after vaccination (error bars depict geometric means ± 95% confidence interval). Statistical significance was determined by the Mann–Whitney test. ns, not significant (P > 0.05). (D) ELISPOT assays of WNV-specific ASCs. WNV E protein–specific IgG-secreting cells in the BM from Zbtb20+/+ (n = 3) or Zbtb20trap/trap (n = 6) chimeric mice were quantified at 26 wk after vaccination. Mean values ± SEM are shown. Statistical significance was determined with an unpaired Student’s two-tailed t test. (C and D) Data are representative of two experiments, each with three to six mice per genotype.
ZBTB20 is dispensable for germinal center function and plasma cell generation
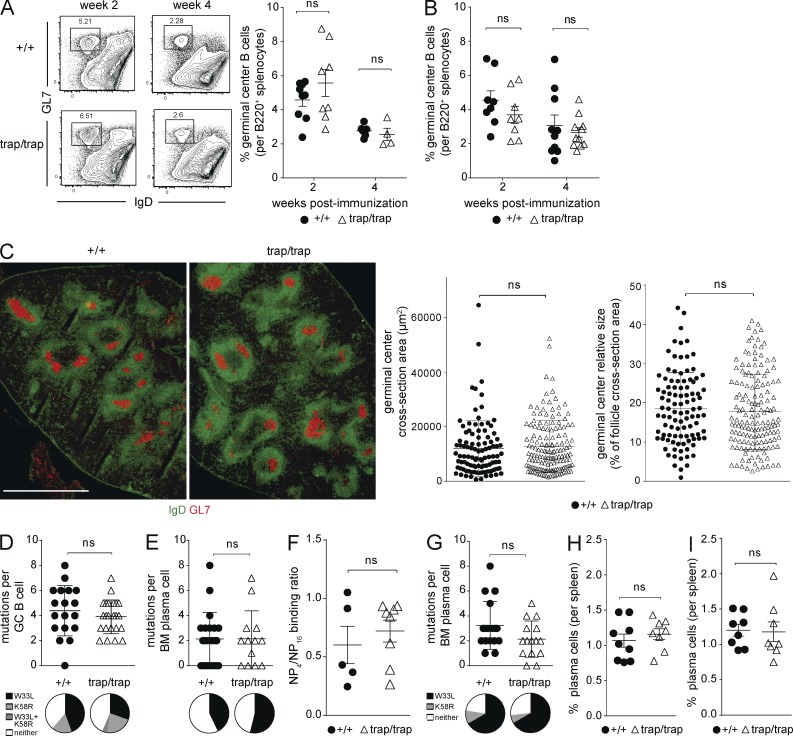
Mutations or conditions that compromise durable antibody production often are linked to defects in the germinal center reaction and subsequent formation of long-lived plasma cells. Of note, TLR ligand adjuvants can induce particularly robust germinal center reactions (Meyer-Bahlburg et al., 2007; Hwang et al., 2009; Kasturi et al., 2011). We thus hypothesized that ZBTB20 deficiency causes defects in germinal center reactions and plasma cell formation and that these defects are overcome by TLR ligand adjuvants. To determine whether premature waning of antibody responses in ZBTB20-deficient chimeras is linked to germinal center defects, we quantified the numbers of germinal centers and their function after alum- or TLR ligand–adjuvanted NP-CGG immunization. The frequencies of germinal center B cells were similar between ZBTB20-deficient and control chimeras at 2 and 4 wk after immunization with alum adjuvants and with TLR ligand adjuvants (Fig. 5, A and B, respectively). Moreover, no significant differences in germinal center sizes were observed histologically when comparing ZBTB20-deficient and control chimeras (Fig. 5 C). Thus, ZBTB20-deficient germinal centers are grossly normal.
Figure 5.
ZBTB20 does not regulate germinal center reactions or plasma cell generation. (A and B) Flow cytometric quantification of Zbtb20+/+ and Zbtb20trap/trap germinal center B cells at 2 and 4 wk after immunization with alum (A)- or TLR ligand–adjuvanted (B) NP-CGG. Example flow cytometry plots and gating strategies are shown to the left in A. Mean values ± SEM are shown along with individual data points. Data are cumulative from two experiments, with 4–9 (A) or 8–11 (B) mice per genotype per time point. Statistical significance was determined with an unpaired Student’s two-tailed t test. ns, not significant (P > 0.05). (C) Microscopic quantification of germinal center areas in splenic sections. Zbtb20+/+ and Zbtb20trap/trap germinal center areas were quantified from frozen splenic sections, obtained 2 wk after alum-adjuvanted NP-CGG immunization and subsequently stained with fluorescently labeled antibodies against GL7 and IgD. Example immunofluorescent microscopy images are shown to the left (bar, 1 mm). Both the germinal center cross section areas (middle) and relative sizes (right) are shown (mean values ± SD). Statistical significance was determined with an unpaired Student’s two-tailed t test. Data are cumulative from two independent experiments. (D and E) Sequencing analysis of heavy chain genes from individual NP-specific Zbtb20+/+ and Zbtb20trap/trap germinal center B cells (D) and BM plasma cells (E) isolated at 2 wk after NP-CGG immunization adjuvanted with alum. Sequences were examined for total (top; error bars depict mean values ± SD) and canonical affinity-enhancing (bottom) somatic mutations. Statistical significance was determined with a Mann–Whitney test. Each data point represents sequence from an individual cell. Data are cumulative from two independent experiments, each with two mice per genotype. (F) ELISA measurements of relative affinity of NP-specific serum antibodies in Zbtb20+/+ and Zbtb20trap/trap fetal liver chimeras. Data were obtained by quantifying the ratio of high-affinity (NP4 binding) to total (NP16 binding) NP-specific antibodies at 11 wk after alum-adjuvanted NP-CGG immunization. Mean values ± SEM are shown along with individual data points. Data are representative of three experiments, each with two to eight mice per genotype. Statistical significance was determined with an unpaired Student’s two-tailed t test. (G) Sequencing analysis of heavy chain genes from individual NP-specific Zbtb20+/+ and Zbtb20trap/trap BM plasma cells isolated at 2 wk after NP-CGG immunization adjuvanted with TLR ligands. Statistical significance was determined with a Mann–Whitney test. Each data point represents sequence from an individual cell. Error bars depict mean values ± SD. Data are cumulative from two independent experiments, each with two mice per genotype. (H and I) Flow cytometric analysis of splenic plasma cells at 2 wk after alum (H)- or TLR ligand–adjuvanted (I) immunization with NP-CGG. Mean values ± SEM are shown. Statistical significance was determined with an unpaired Student’s two-tailed t test. Data are cumulative from two experiments with eight to nine mice per genotype for each adjuvant.
Like ZBTB20, the transcription factor AIOLOS is required for long-term antibody responses. Aiolos−/− animals form and maintain germinal centers, and the extent of somatic hypermutation is normal (Cortés and Georgopoulos, 2004). However, selection and survival of germinal center B cells carrying high-affinity antigen receptors is blocked in Aiolos−/− mice. Because only the high-affinity clones within germinal centers differentiate into long-lived plasma cells, Aiolos−/− mice form only short-lived plasma cells (Cortés and Georgopoulos, 2004; Phan et al., 2006). To test whether ZBTB20 deficiency causes a similar failure in affinity maturation, we analyzed canonical affinity-enhancing W33L and/or K58R mutations in immunoglobulin heavy chain genes from NP-specific cells (Cumano and Rajewsky, 1986; Furukawa et al., 1999). The extent of somatic hypermutation and the frequency of cells carrying affinity-enhancing mutations were similar between ZBTB20-deficient and control germinal center B cells (Fig. 5 D). We next examined whether ZBTB20 is required for the generation of affinity-matured BM plasma cells. For this purpose, we sequenced immunoglobulin heavy chain genes from NP-specific BM plasma cells at 2 wk after alum-adjuvanted immunization. As shown in Fig. 5 E, no significant differences were observed between mutant and wild-type plasma cells after alum-adjuvanted immunization. Moreover, the relative affinities of polyclonal NP-specific serum antibodies at 11 wk after immunization were similar between ZBTB20-deficient and control chimeras (Fig. 5 F). We conclude that affinity maturation and the initial generation of affinity-matured BM plasma cells is unaffected by ZBTB20 deficiency. This phenotype is thus distinct from Aiolos−/− and Cr2−/− mice, in which long-term antibody defects are linked to changes in affinity maturation (Chen et al., 2000; Cortés and Georgopoulos, 2004).
TLR ligand–based immunogens can induce large T cell– and germinal center–independent antibody responses that can persist for many weeks (Bortnick et al., 2012; Bortnick and Allman, 2013). Thus, it is possible that ZBTB20 expression is important for regulating post–germinal center plasma cells but not T-independent long-lived plasma cells formed by TLR-based immunizations. Arguing against this possibility, NP-specific BM plasma cells in both control and ZBTB20-deficient chimeras were extensively mutated and affinity matured after TLR ligand–adjuvanted immunization (Fig. 5 G). Moreover, extrafollicular splenic plasma cell numbers were similar after alum- and TLR ligand–adjuvanted NP-CGG immunization (Fig. 5, H and I, respectively). These results suggest that inclusion of TLR ligands in the adjuvant, rather than increasing the number of T-independent plasma cells, yields plasma cells derived from the germinal center that do not depend on ZBTB20 for their accumulation or maintenance.
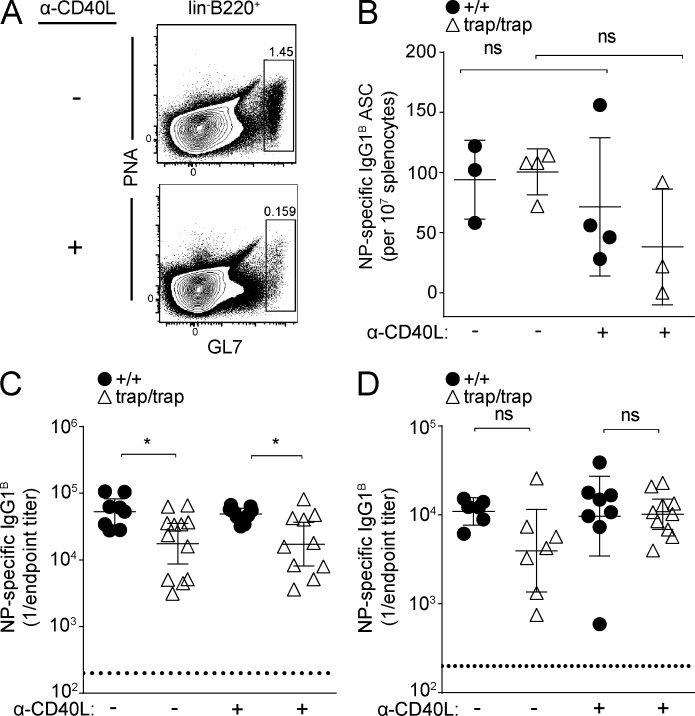
Many long-lived plasma cells are formed late in the germinal center reaction, emerging 2 wk or more after immunization (Han et al., 1995). Some synthetic TLR ligand–containing adjuvants can induce remarkably persistent germinal center reactions, whereas alum adjuvants cannot (Dogan et al., 2009; Kasturi et al., 2011). These persistent germinal center reactions could potentially overcome ZBTB20 deficiency by continuously generating and replenishing BM plasma cells. Thus, to further distinguish defects in plasma cell formation from maintenance, we administered a blocking antibody against CD40L at 2 wk after immunization of ZBTB20-deficient and control chimeras. Injection of this antibody led to the dissolution of germinal center reactions and prevented the formation of new dependent plasma cells (Fig. 6, A and B; Han et al., 1995). Thus, NP-specific antibodies and plasma cells in these α-CD40L–treated animals were largely limited to those already formed at 2 wk after immunization. After alum-adjuvanted immunization, serum antibody deficiencies at 7 wk after immunization were still observed in ZBTB20-deficient chimeras relative to controls that were treated with the CD40L-blocking antibody (Fig. 6 C). In contrast, irrespective of whether mice were treated with the blocking antibody, antigen-specific titers between ZBTB20-deficient and control chimeras remained similar after TLR ligand-adjuvanted immunization (Fig. 6 D). These experiments confirm that BM plasma cells are formed in normal numbers in ZBTB20-deficient chimeras irrespective of the adjuvant used. Instead, these data suggest that intrinsic proliferative or survival differences between alum- and TLR ligand–induced BM plasma cells underlie the selective dependence on ZBTB20.
Figure 6.
ZBTB20 deficiency causes plasma cell–intrinsic defects. (A) Flow cytometric quantification of germinal center B cells after administration of blocking α-CD40L antibody. Wild-type mice were immunized with alum-adjuvanted NP-CGG and then left untreated or injected with 200 µg α-CD40L at days 13, 14, and 15. Frequencies of splenic lineage− (CD4/8/Gr-1/Ter119) germinal center B cells were analyzed as shown at day 16 after immunization. Data are representative of two experiments with two mice per treatment group. (B) ELISPOT analysis of splenic plasma cells at 4 wk after immunization with TLR ligand–adjuvanted NP-CGG. Mice were left untreated or administered α-CD40L at days 13, 14, and 15 as in A. Statistical significance was determined with an unpaired Student’s two-tailed t test. Error bars depict mean values ± SD. Each data point represents one mouse. Data are representative of two experiments, each with three to four mice per group. (C and D) ELISA measurements of serum antibody titers (error bars depict geometric means ± 95% confidence interval) from Zbtb20+/+ and Zbtb20trap/trap fetal liver chimeras immunized with alum (C)- or TLR ligand–adjuvanted (D) NP-CGG and then treated with α-CD40L or left untreated as in A. NP-specific serum antibody titers were quantified at 7 wk after immunization. Data are cumulative from two experiments with 8–12 (C) or 6–10 (D) mice per genotype. Statistical significance was determined with a Mann–Whitney test. ns, not significant (P > 0.05); *, 0.01 < P < 0.05.
ZBTB20 regulates survival of plasma cells
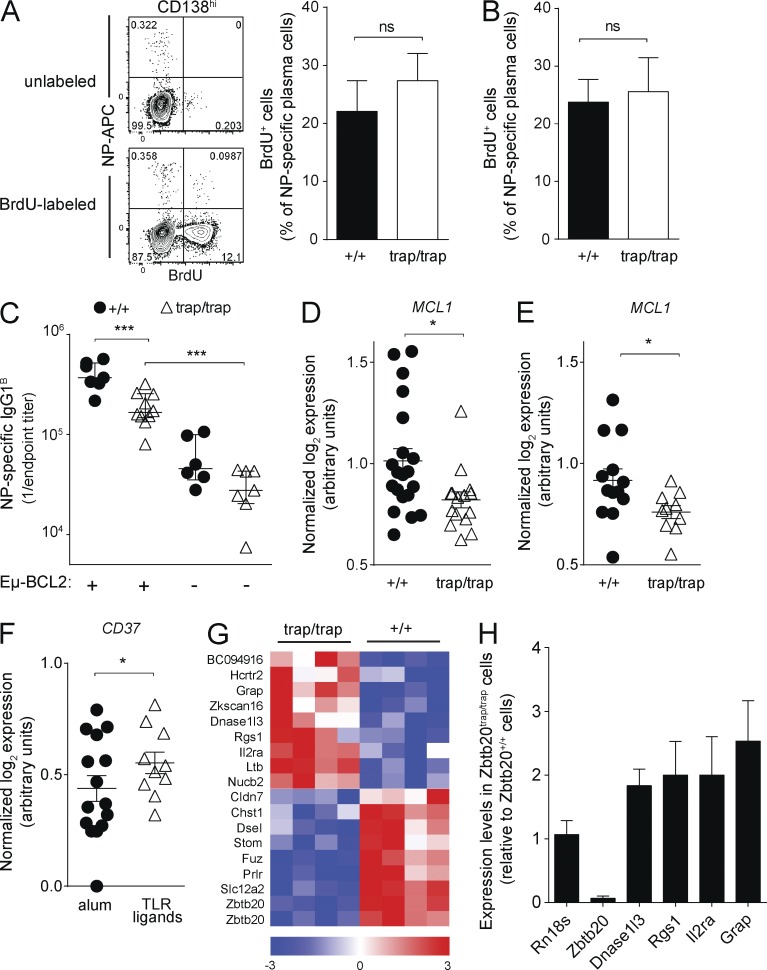
After their migration to the BM, plasma cell precursors proliferate for several weeks and then can survive for years (Benner et al., 1981; Han et al., 1995; Manz et al., 1997; Smith et al., 1997; Slifka et al., 1998). The phenotypes of ZBTB20-deficient plasma cells are consistent with defects in either proliferation or survival. To specifically quantify plasma cell proliferation, ZBTB20-deficient and wild-type chimeras were treated with blocking α-CD40L at 3 wk after alum- or TLR ligand–adjuvanted immunization to minimize new plasma cell formation. These animals were then fed BrdU in the drinking water for 4 d, and the level of incorporation was measured in NP-specific BM plasma cells. Irrespective of whether alum (Fig. 7 A) or TLR ligand (Fig. 7 B) adjuvants were used, ZBTB20-deficient and control NP-specific BM plasma cells incorporated BrdU at similar rates, demonstrating no differences in proliferation. These data argue that survival deficits in plasma cells lead to the selective dependence on ZBTB20 after alum-adjuvanted immunization.
Figure 7.
ZBTB20 regulates survival programs in plasma cells. (A and B) Flow cytometric analysis of BrdU incorporation in NP-specific BM plasma cells. Zbtb20+/+ and Zbtb20trap/trap animals were immunized with alum (A)- or TLR ligand–adjuvanted (B) NP-CGG. Immunized mice were then treated with three daily doses of blocking α-CD40L antibody at 3 wk after immunization and then fed BrdU in the drinking water for 4 d. Gating strategies and data from an immunized control mouse not fed BrdU (top) and an immunized BrdU-fed control mouse (bottom) are shown to the left in A. Data are from either three to four (A) or three to six (B) mice per genotype. Data are cumulative from two independent experiments. Mean values ± SEM are shown. Statistical significance was determined with an unpaired Student’s two-tailed t test. ns, not significant. (C) ELISA measurements of serum NP-specific IgG1 titers from Zbtb20+/+, Zbtb20trap/trap, Zbtb20+/+Eμ-Bcl2, or Zbtb20trap/trapEμ-Bcl2 fetal liver chimeras at 6 wk after alum-adjuvanted NP-CGG immunization (error bars depict geometric means ± 95% confidence interval). Data are cumulative from three independent experiments with 6–10 samples per genotype. Statistical significance was determined with the Mann–Whitney test. ***, P < 0.001. (D and E) Single-cell analysis of MCL1 expression. NP-specific Zbtb20+/+ and Zbtb20trap/trap BM cells were isolated 2 wk after alum-adjuvanted (D; Zbtb20+/+: n = 20; Zbtb20trap/trap: n = 15) or TLR ligand–adjuvanted (E; Zbtb20+/+: n = 13; Zbtb20trap/trap: n = 10) immunization, and transcript levels were assessed. (F) Single-cell analysis of CD37 expression. ZBTB20-deficient, NP-specific BM plasma cells were isolated 2 wk after immunization with alum (n = 15)- or TLR ligand–adjuvanted (n = 10) NP-CGG, and transcript levels were assessed and normalized to expression of the housekeeping gene Rpl13a. For D–F, mean values ± SEM are shown along with individual data points, each representing the expression level in a single cell. Data are cumulative from two independent experiments. Statistical significance was determined with a Mann–Whitney test. *, 0.01 < P < 0.05. (G) Microarray analysis of Zbtb20+/+ and Zbtb20trap/trap polyclonal BM plasma cells. Transcripts with mean expression values at least twofold higher or lower in Zbtb20trap/trap cells are shown in the heat map. Four biological replicates were arrayed for each genotype of BM plasma cells. Red represents high relative expression levels, and blue represents low relative expression levels. (H) qRT-PCR analysis of candidate ZBTB20-regulated transcripts. RNA from polyclonal CD45.2+ Zbtb20+/+ or Zbtb20trap/trap BM plasma cells was isolated for analysis. Transcript levels of each gene were measured in technical triplicates and were normalized to the mean Hprt expression of each sample. Three biologically distinct samples per genotype were measured. Mean values ± SEM are shown.
To functionally test whether ZBTB20-deficient plasma cells are defective in their long-term survival, we first generated Zbtb20+/+ and Zbtb20trap/trap mice carrying an Eμ-Bcl2 transgene, and fetal liver chimeras were established. Mice carrying this particular transgene express BCL2 at high levels only in B cells, and previous studies have shown that BCL2 expression can attenuate plasma cell apoptosis (Strasser et al., 1991; Smith et al., 1994). These Zbtb20trap/trap BCL2-expressing fetal liver chimeras were then immunized with alum-adjuvanted NP-CGG. Unlike their Zbtb20trap/trap counterparts, NP-specific serum antibodies were maintained by Zbtb20trap/trap Eμ-Bcl2 chimeras at higher levels than in Zbtb20+/+ chimeras (Fig. 7 C). The ability of BCL2 expression to elevate serum antibody titers suggests defects in survival of ZBTB20-deficient plasma cells and further argues against defects in formation. Nevertheless, serum antibody titers in Zbtb20trap/trap Eμ-Bcl2 chimeras were reduced relative to Zbtb20+/+ Eμ-Bcl2 chimeras (Fig. 7 C). Similar trends were seen in BM plasma cells through ELISPOT analyses at 12 wk after immunization (not depicted). These data suggest that BCL2 expression leads to a partial rescue and/or that ZBTB20 regulates other parallel survival pathways in plasma cells. There are several candidate parallel cell death pathways that cannot be inhibited by BCL2 expression. For example, death receptor signals can bypass mitochondria and BCL2 to cause cell death (Scaffidi et al., 1998). In addition, BCL2 does not bind the proapoptotic mitochondrial protein NOXA; rather, NOXA uniquely binds antiapoptotic MCL1 and A1 (Chen et al., 2005; Kuwana et al., 2005; Willis et al., 2005; Certo et al., 2006; Kim et al., 2006). Of note, the balance of NOXA and MCL1 expression has been shown to be critically important for plasma and myeloma cell survival (Le Gouill et al., 2004; Qin et al., 2005; Wensveen et al., 2012; Peperzak et al., 2013).
The defects in Zbtb20trap/trap long-lived plasma cell accumulation manifest over the course of several weeks. Thus, it is unlikely that a dramatic deficiency in survival pathway expression, which would likely lead to rapid death, explains the observed phenotypes. We therefore used a single-cell quantitative RT-PCR (qRT-PCR) assay to provide sufficient statistical power to identify candidate ZBTB20-dependent survival pathways other than BCL2 that may be only subtly dysregulated. Individual wild-type and ZBTB20-deficient NP-specific BM plasma cells were sorted at 2 wk after immunization, and transcript levels of several genes involved in apoptosis and in long-term antibody responses were evaluated. No differences were observed in the expression of several factors thought to be important in plasma cell survival, such as IL6ST, CD28, BAK, XBP1, PRDM1, IRF4, CD44, ITGA4, ITGB1, ITGB2, CXCR4, and TNFRSF13C (not depicted). However, expression of MCL1, which is critical for long-lived plasma cell survival (Peperzak et al., 2013), was statistically significantly reduced in NP-specific, ZBTB20-deficient plasma cells after alum-adjuvanted immunization (Fig. 7 D). Transcription factors regulate programs of genes, and it is unlikely that any single downstream gene can fully explain the ZBTB20 phenotype; nevertheless, the reduction in MCL1 expression may contribute to the defects in Zbtb20trap/trap plasma cells. Consistent with this interpretation, Peperzak et al. (2013) have shown that plasma cells express higher levels of MCL1 than other B cells and are uniquely sensitive to perturbations in MCL1 levels.
One possible explanation for the adjuvant-specific requirement for ZBTB20 is that it regulates survival genes only when B cells are activated under certain conditions, e.g., in NP-specific plasma cells generated in response to alum-adjuvanted antigen but not when using TLR ligand adjuvants. Arguing against this possibility, however, MCL1 expression was also reduced in ZBTB20-deficient plasma cells formed by TLR ligand–adjuvanted immunization (Fig. 7 E). A second possible explanation is that ZBTB20 regulates the same targets in all plasma cells, but that TLR ligands engage compensatory survival pathways. As an example of such a pathway, TLR ligands preferentially induce CD37 in ZBTB20-deficient plasma cells over those formed by alum-adjuvanted immunization (Fig. 7 F). A recent study has shown the importance of this gene in maintaining long-lived plasma cells (van Spriel et al., 2012).
Because of the extreme paucity of antigen-specific BM plasma cells (typically <100 cells can be isolated per Zbtb20trap/trap chimera), it is technically challenging to perform unbiased microarray experiments to identify additional ZBTB20 targets. Thus, under the assumption that ZBTB20 regulates relevant targets in all BM plasma cells, irrespective of their mode of formation, we performed microarray analyses on polyclonal Zbtb20+/+ and Zbtb20trap/trap populations. Several genes were clearly dysregulated by ZBTB20 deficiency (Fig. 7, G and H). Although the collective dysregulation of multiple genes likely underlies the ZBTB20-deficient phenotype, we noted in particular that RGS1 expression was elevated in ZBTB20-deficient plasma cells. Previous studies have shown that the proper expression of RGS1 is essential for plasma cell localization and responsiveness to prosurvival chemokines (Bowman et al., 1998; Moratz et al., 2000, 2004; Shi et al., 2002). These data suggest that ZBTB20 regulates survival pathways in all long-lived plasma cells but that specific modes of primary activation induce compensatory pathways that can overcome ZBTB20 deficiency.
DISCUSSION
Durable antibody production is a critical feature of successful vaccines. As such, a deeper understanding of the molecular basis of plasma cell survival could guide the rational improvement of vaccines, yet relatively little is known on this subject. Here we have shown that long-term survival of plasma cells after immunization with antigen and alum, the most prevalent adjuvant used in vaccines (Marrack et al., 2009), is ZBTB20 dependent. In contrast, long-lived plasma cells produced in response to TLR-based adjuvants are ZBTB20 independent. To our knowledge, this is the first direct evidence that plasma cells engage distinct survival programs, which, in turn, are highly dependent on the adjuvant used during immunization.
Most adjuvants are thought to act only indirectly on B cells, and not at all on plasma cells once they have been generated (Richard et al., 2008). Therefore, it was surprising for us to find that ZBTB20 is an alum-specific, B cell–intrinsic factor required for long-term antibody responses. Alum initially activates macrophages and monocytic inflammatory DC precursors (Jordan et al., 2004; Eisenbarth et al., 2008; Franchi and Núñez, 2008; Kool et al., 2008a,b; Li et al., 2008; McKee et al., 2009). These APCs prime CD4+ T lymphocytes, which then migrate into B cell follicles and germinal centers to assist in antibody responses (Mannhalter et al., 1985; Ansel et al., 1999; Breitfeld et al., 2000; Schaerli et al., 2000; Crotty et al., 2003; Allen et al., 2007; Yu et al., 2009; Victora et al., 2010). We suspect that ZBTB20 regulates the capacity of B cells to receive help from Tfh cells in germinal centers after priming by alum-activated myeloid cells, in some way augmenting the plasma cell survival program to generate a persistent antibody response. The implication of this proposed mechanism is that BM plasma cell lifespan is imprinted during ontogeny within the germinal center reaction (Tarlinton, 2006; Amanna and Slifka, 2010).
It is unclear why the ability to mount durable antibody responses after challenge with aluminum salts would be under a strong selection pressure during evolution. However, alum adjuvants trigger responses that are in many ways similar to those elicited by clinically important helminth infections, such as schistosomiasis. Like alum, Schistosoma mansoni infections induce a heavily Th2-biased response and activate the NLRP3 inflammasome but do not activate TLR signaling (Kane et al., 2008; Ritter et al., 2010). Future studies will determine whether ZBTB20 is required for durable antibody responses to these types of infections. It also remains to be seen whether other transcription factors are required for durable antibody responses to infections that do activate TLRs, yet it is worth noting that expression of ZBTB32, another BTB-POZ family transcription factor, is induced in plasma cells specifically upon LPS stimulation, but not after alum-adjuvanted immunization (Bhattacharya et al., 2007; Yoon et al., 2012). Several studies have demonstrated the ability of T-independent antigens to induce long-lived plasma cell formation (Taillardet et al., 2009; Racine et al., 2011; Bortnick et al., 2012; Foote et al., 2012). Because our current study focused on the antibody response toward T-dependent antigens, additional work will be necessary to determine whether ZBTB20 or other transcription factors are critical for long-term plasma cell survival in the T-independent response. Our data raise the intriguing possibility that distinct transcription factors are used by B cells to regulate durable antibody immunity in response to different types of infections.
In this regard, other transcription factors have been linked to the generation of durable antibody responses, yet ZBTB20 is the first reported to specifically control plasma cell lifespan. Unlike ZBTB20, the transcription factor AIOLOS is required for the formation of long-lived plasma cells, and this requirement is likely secondary to its role in upstream germinal center reactions (Cortés and Georgopoulos, 2004). Other transcription factors, such as BLIMP1, IRF4, and XBP1, are required for differentiation and secretory programs in all ASCs, including short-lived plasma cells (Reimold et al., 2001; Shapiro-Shelef et al., 2003, 2005; Klein et al., 2006; Sciammas et al., 2006). Thus, through functional analyses of genes upstream and downstream of ZBTB20, novel pathways that specifically regulate the lifespan of plasma cells and the duration of immunity may be revealed in future studies.
Although vaccine development and optimization have historically been performed empirically, modern approaches have the potential to provide molecular guides to rationally improve vaccines. Recent studies on both human influenza and yellow fever vaccines have demonstrated the feasibility and clinical value in such molecular approaches (Querec et al., 2009; Nakaya et al., 2011). The induction of lifelong protection is a particularly important metric for a vaccine’s success, yet little is known about the immunogenic or molecular determinants that drive an ultra-durable antibody response. The discovery of ZBTB20 as an adjuvant-specific transcriptional regulator of plasma cell survival provides a genetic foothold to identify pathways that differentially regulate durable versus transient vaccine responses. Importantly, our findings also justify further research on the use of pathogen-derived adjuvants to elicit maximally durable antibody responses.
MATERIALS AND METHODS
Mice.
All animal procedures were approved by the Animal Studies Committee at Washington University in St. Louis. C57BL/6N and B6.SJL mice were purchased from the National Cancer Institute, and B6.Cg-IghA Thy1AGpi1A mice were purchased from the Jackson Laboratory. Zbtb20+/trap mice were generated using a gene-trapped C57BL6/N mouse embryonic stem cell line (IST12271A5; Texas A&M Institute for Genomic Medicine) by the Transgenic Knockout Microinjection Core facility at Washington University in St. Louis. Mice were first crossed to C57BL6/N (National Cancer Institute) for one generation and then maintained through heterozygous intercrosses. In Fig. 1 A, wild-type controls were B6.SJL mice. For all other experiments, littermate controls were used for fetal liver reconstitutions. Genotyping of Zbtb20 alleles was performed using primers specific for the wild-type allele, 5′-CTCAGCCTCTGTGGCACATCACAGCCACTT-3′ (forward) and 5′-GTTCCTTATCTGCCCTACCTTATCTCATG-3′ (reverse); and a reverse primer specific for the gene-tapped allele, 5′-CTTGCAAAATGGCGTTACTTAAGC-3′.
Fetal liver reconstitutions.
Fetal livers were collected from Zbtb20+/+ or Zbtb20trap/trap embryos between 14.5 and 17.5 d postcoitus, homogenized into single-cell suspensions, and transplanted by retroorbital injection into 8–12-wk-old 800 cGy–irradiated B6.SJL-Ptprca Pepcb or B6.Cg-IghA Thy1AGpi1A mice. Mice were immunized 8–16 wk after reconstitution.
Immunizations.
Mice were immunized intraperitoneally with a single dose of 100 µg NP-CGG (hapten protein ratio: 15:22; Bioresearch Technologies) either in 5% precipitated aluminum potassium sulfate (Thermo Fisher Scientific) in phosphate buffered saline or in an equal volume (100 µl) of Sigma Adjuvant System (containing monophosphoryl lipid A and trehalose dicorynomycolate; Sigma-Aldrich). For WNV, mice were vaccinated intraperitoneally with two doses of 100 µl West Nile–Innovator vaccine (Pfizer Animal Health) on two consecutive days.
Antibodies.
The following purified monoclonal antibodies were purified from hybridoma supernatants by Bio X Cell: 2C11 (anti-CD3), GK1.5 (anti-CD4), 53-6.7 (anti-CD8), 1D3 (anti-CD19), 6B2 (anti-B220), 8C5 (anti–Gr-1), M1/70 (anti–Mac-1), TER119 (anti-Ter119), A20.1.7 (anti-CD45.1), AL1-4A2 (anti-CD45.2), E13-161-7 (anti–Sca-1), and A7R34 (anti–IL-7 receptor α). Purified antibodies were conjugated with biotin, Pacific Blue, Alexa Fluor 488, Alexa Fluor 647, or Alexa Fluor 680 (Life Technologies) according to the manufacturer’s instructions. The following antibodies were purchased from eBioscience: A2F10 (anti-Flk2), B3B4 (anti-CD23) conjugated with PE, II/41 (anti-IgM) conjugated with PerCP–eFluor 710, PK136 (anti-NK1.1) conjugated with PerCP-Cy5.5, 2B8 (anti-CD117), RA3-6B2 (anti-B220), M1/70 (anti-CD11b), RB6-8C5 (anti-Gr-1) conjugated with PE-Cy7, 11-26 (anti-IgD) conjugated with FITC, eBio129c (anti-PDCA-1), AA4.1 (anti-CD93) conjugated with APC, N418 (anti-CD11c), RA3-6B2 (anti-B220) conjugated with APC–eFluor 780, 11-26 (anti-IgD) conjugated with eFluor 450, and GL-7 (anti–GL-7) conjugated with biotin. The following antibodies were purchased from BioLegend: 93 (anti-CD16/32) conjugated with PerCP/Cy5.5, 16-10A1 (anti-CD80) conjugated with PE-Cy5, 145-2C11 (anti-CD3) conjugated with Alexa Fluor 488, 7E9 (anti-CD21/35) conjugated with Pacific Blue, and RMG1-1 (anti-IgG1) conjugated with APC. The following antibodies were purchased from BD: 281-2 (anti-CD138), S7 (anti-CD43), 53-6.7 (anti-CD8a), AF6-78 (anti-IgMB) conjugated with PE, DS-1 (anti-IgMA) conjugated with FITC, 140706 (anti-CCR6) conjugated with Alexa Fluor 647, 6C3 (anti-BP1) and 281-2 (anti-CD138) conjugated with biotin, and unconjugated R26-46 (anti-Igλ). Streptavidin conjugated with Qdot 605 and goat anti–rat IgG (H+L) conjugated with Alexa Fluor 633 were purchased from Life Technologies.
Cell isolation and analysis.
Splenic plasma cells were isolated from spleens at 7 d after immunization, and germinal center B cells were isolated from spleens at 14 or 28 d after immunization; NP-specific splenic memory B cells were isolated from spleens at 12–16 wk after immunization; donor-derived polyclonal BM plasmas were isolated from BM of fetal liver chimeras 8 mo after reconstitution. For splenic IgG1+ plasma cell isolation, depletion of non-B cells was performed by labeling with monoclonal antibodies specific to CD3, CD4, CD8, Gr-1, and TER119 and plating onto Petri dishes coated with goat anti–rat IgG (SouthernBiotech). Plates were incubated at 4°C for 30 min, and nonadherent cells were collected. For BM CD138+ plasma cell enrichment, cells were labeled with biotinylated CD138 and enriched with streptavidin microbeads on LS columns (Miltenyi Biotec). For splenic NP-specific memory B cell enrichment, splenocytes were labeled with unconjugated anti-Igλ and enriched with anti–rat IgG MicroBeads (Miltenyi Biotec) on LS columns. Igλ-enriched B cells were then stained with NP40-PE (Biosearch Technologies). All B cell subsets were then purified according to the schemes shown in Fig. 1 on a FACSAria II (BD). β-Galactosidase activity was measured using FluoReporter lacZ Flow Cytometry kit (Life Technologies) according to the manufacturer’s instruction.
Serological analysis.
ELISA plates were coated overnight with 5 µg/ml NP16-BSA or NP4-BSA (Biosearch Technologies) or with WNV E protein (gift of M. Diamond, Washington University in St. Louis, St. Louis, MO), and serial dilutions of sera were plated onto coated plates. Technical duplication was performed for every serum sample. Wells were stained with biotinylated anti-IgG1A (10.9), anti-IgG1B (B68-2), anti-IgG1 (nonallotype specific; A85-1), anti-IgG2c (5.7), or anti-IgG2b (R12-3), purchased from BD. Wells were then stained with streptavidin-conjugated horseradish peroxidase (BD), and tetramethylbenzidine (Dako) was used to detect peroxidase reactivity, 2N H2SO4 was used to quench the reaction, and optical densities were quantified at 450 nm. The end-point titer of each sample was determined using Prism software (GraphPad Software) from a one-phase exponential decay curve defined as the dilution that generates an OD450 value of the background plus 3 SD.
ELISPOT assays.
For the detection of antigen-specific ASCs, MultiScreen filter plates (EMD Millipore) were coated with 50 µg/ml NP16-BSA or with 40 µg/ml of recombinant WNV E protein in PBS overnight. Splenocytes at 2 wk after immunization were seeded at 4.0 × 105 cells/well; BM cells of all time points and splenocytes after 2 wk were seeded at 5–10 × 106 cells/well. Cells were cultured in 100 µl RPMI/5% FBS. Technical triplication was performed for every sample. Plates were incubated overnight at 37°C. Wells were stained with biotinylated anti-IgG1A, anti-IgG1B, or anti–mouse IgG (γ-chain specific; Sigma-Aldrich) and then with streptavidin-conjugated horseradish peroxidase (BD). Spots were developed using 3-amino-9-ethyl-carbazole (Sigma-Aldrich), and the reaction was quenched by rinsing wells with water. Spots were counted using an ImmunoSpot S6 Analyzer (CTL Laboratories).
RNA extraction, cDNA synthesis, and qRT-PCR.
RNA extraction was performed either with TRIzol (Life Technologies) or with RNeasy Mini kit (QIAGEN). First strand cDNA synthesis was performed with SuperScript III Reverse transcription kit using oligo (dT) primers or random hexamers (Life Technologies) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Green PCR master mix (Applied Biosystems) on a Prism 7000 Sequence Detection System (Applied Biosystems). Primer sequences are as follows: Zbtb20, 5′-CAGCCAAACAGAACTACGTCA-3′ (forward) and 5′-GCGTCACCATGTGCTTGATA-3′ (reverse); β-actin, 5′-CCTGAACCCTAAGGCCAAC-3′ (forward) and 5′-ACAGCCTGGATGGCTACG-3′ (reverse); Rn18S, 5′-AAGACGGACCAGAGCGAAAG-3′ (forward) and 5′-GCCAGTCGGCATCGTTTATG-3′ (reverse); Hprt, 5′-TCTTTGCTGACCTGCTGGATT-3′ (forward) and 5′-TTATGTCCCCCGTTGACTGAT-3′ (reverse); Dnase1l3, 5′-TGGTTTGGTTCCATTCCCCC-3′ (forward) and 5′-CACTGGCTTCTCACATCCGT-3′ (reverse); Rgs1, 5′-AATGCAGTGGTCTCAGTCTCT-3′ (forward) and 5′-CCTCACAAGCCAACCAGAAT-3′ (reverse); Il2ra, 5′-TGAAGTGTGGGAAAACGGGG-3′ (forward) and 5′-GCAGGAAGTCTCACTCTCGG-3′ (reverse); and Grap, 5′-TACAGCTTCCAGGCTACCGA-3′ (forward) and 5′-CTTGGGAACAAAACCCTCCG-3′ (reverse).
Immunofluorescence.
Unfixed spleens were frozen in O.C.T. compound, and 10-µm-thick sections were cut with a Microm HM 550 cryostat (Thermo Fisher Scientific). Sections were fixed in acetone at −20°C for 5 min and stained with anti–GL7-PE and anti–IgD-FITC (BD). Fluorescent images were scanned with a NanoZoomer 2.0-HT equipped with a Fluorescence Imaging Module (Hamamatsu Photonics). Germinal center areas were quantified using NDP.view software (Hamamatsu Photonics). Distinct GL7+ germinal center and surrounding IgD+ follicle perimeters were manually defined, and areas were calculated. Three Zbtb20+/+ and Zbtb20trap/trap chimeras each were analyzed. Six sections of ∼10-mm2 area, spaced at least 120 µm apart from each other in the z-dimension, were analyzed for each animal. For presentation, contrast adjustment was applied equally to the representative images of both genotypes shown in Fig. 4 B using Photoshop CS3 (Adobe).
α-CD40L treatment, BrdU labeling, and analysis.
Three doses of 200 µg α-CD40L antibody (MR1; Bio X Cell) were intravenously administered on the indicated days. BrdU (Sigma-Aldrich) was given to mice in drinking water at 2 mg/ml for 4 d after MR1 treatment. On day 26 after immunization, CD138+ BM plasma cells were enriched, stained for surface expression of B220 and CD138, and then processed and stained for incorporated BrdU with the FITC BrdU Flow kit (BD) according to the manufacturer’s instructions. NP-APC used in intracellular staining was made by conjugating allophycocyanin (Sigma-Aldrich) with 4-hydroxy-3-nitrophenylacetyl-O-succinimide ester (Biosearch Technologies) as previously described (McHeyzer-Williams and McHeyzer-Williams, 2004).
Single-cell BCR sequencing.
The procedure of sequencing the VH region of the dominant V186.2-containing NP-specific IgG1 BCRs at single-cell levels has been previously described (McHeyzer-Williams et al., 1991). In brief, single NP-APC–binding IgG1+ germinal center B cells or BM plasma cells were sorted directly into a Superscript III cDNA synthesis reaction containing random hexamers (Life Technologies). cDNA products were then used as templates to amplify the VH region of V186.2-containing IgG1 transcripts through nested PCR using HotStar Taq Polymerase (QIAGEN). The PCR products were then gel-purified and sequenced using the inner primers. Mutations on amplified BCR sequences were then identified by comparison with germline sequences through the NCBI IgBLAST server. The primers of the first round of the nested PCR are 5′-GGATGACTCATCCCAGGGTCACCATGGAGT-3′ (forward) and 5′-GCTGTATCATGCTCTTCTTG-3′ (reverse), and the primers of the second round are 5′-CCAGGGGCCAGTGGATAGAC-3′ (forward) and 5′-GGTGTCCACTCCCAGGTCCA-3′ (reverse).
Microarrays and analysis.
For all samples, cells were sorted directly into RLT buffer (QIAGEN). RNA was then purified with the RNeasy Mini kit (QIAGEN), and the amount and integrity were quantified on a Bioanalyzer (Agilent Technologies). A minimum RNA integrity value of 3.8 was required to proceed with sample processing. cDNA amplification was performed using Ovation Pico WTA System V2 (NuGEN). 5 µg of amplified cDNA per sample was used for fragmentation and labeling using Encore Biotin Module (NuGEN) according to the manufacturer’s instructions. Labeled cDNA was hybridized to GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix) by the Genome Technology Access Center core facility at Washington University in St. Louis. Microarrays were required to meet certain quality assurance standards to be included in subsequent analysis. Perfect-match probe signals were required to be two- to fourfold higher than background probe signals for each array. Relative log expression signal distributions were required to be narrow and equal for all arrays, with median values equaling 0, thus indicating consistent signal intensity among groups. Area under the curve for positive versus negative controls was >0.88 for all arrays, demonstrating robust signal-to-noise ratios. Bacterial spike-in controls validated equal and efficient hybridization for all arrays. Quality control analysis was performed on raw array data using Expression Console software (Affymetrix). Analysis of microarray data was performed using Arraystar software (DNASTAR). Unsupervised hierarchical clustering analysis was performed on the genomic analysis server GenePattern (Broad Institute). Pearson correlation was chosen for distance measure, and pairwise complete linkage was chosen as the clustering method for sample or gene clustering.
Single-cell qRT-PCR.
All reagents for real-time qRT-PCR were acquired based on a two-step single-cell gene expression analysis protocol developed by Fluidigm. In brief, single NP-APC–binding BM plasma cells from mixed wild-type IgHa: Zbtb20trap/trap IgHb chimeras were directly sorted into an RT reaction assembly containing the SuperScript VILO cDNA Synthesis kit (Invitrogen), SUPERase-In RNase inhibitor (Ambion), T4 Gene 32 protein (New England BioLabs), and 10% NP-40 (Thermo Scientific). After the RT step, a specific target amplification step was performed to enrich the cDNAs of interest with TaqMan PreAmp Master Mix (Applied Biosystems) and gene-specific primers. Unincorporated primers were subsequently removed by Exonuclease I treatment (New England Biolabs, Inc.). Real-time quantitative PCR was performed on a 96.96 Dynamic Array integrated fluidic circuit (Fluidigm) with single-cell cDNA samples, gene-specific primers, and SsoFast EvaGreen supermix with low ROX (Bio-Rad Laboratories) in a BioMark HD reader (Fluidigm) by the Genome Technology Access Center core facility at Washington University in St. Louis. Sequences of gene-specific primers for both specific target amplification and real-time PCR are as follows: MCL1, 5′-AGGACGAAACGGGACTGG-3′ (forward) and 5′-AAAGCCAGCAGCACATTTCT-3′ (reverse); CD37, 5′-TTGCCTCAGCCTCATCAAGTA-3′ (forward) and 5′-GCAGTGGCACGAAGGACAAA-3′ (reverse); Rpl13a, 5′-CCATTGTGGCCAAGCAGGTA-3′ (forward) and 5′-TCGGGAGGGGTTGGTATTCA-3′ (reverse); and NeoR, 5′-GCGTTGGCTACCCGTGATA-3′ (forward) and 5′-GGAGCGGCGATACCGTAAA-3′ (reverse). Raw log2 expression values were extracted using Singular software (Fluidigm) and then normalized by dividing by the log2 RPL13A expression value from the corresponding well. Wells without detectable RPL13A expression were excluded from further analysis. The genotype of each cell was determined by the presence (Zbtb20trap/trap) or absence (wild type) of neomycin expression.
Statistics.
Means, geometric means, SEM, 95% confidence interval, unpaired Student’s two-tailed t tests, Mann–Whitney tests, and one-phase exponential decay curve-fitting for end-point dilution estimation were calculated with Prism software.
Online supplemental material.
Fig. S1 shows the gating strategy for cells shown in Fig. 1. Fig. S2 shows the gating strategy demonstrating normal B cell and other immune cell development in Zbtb20trap/trap chimeras. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131821/DC1.
Supplementary Material
Acknowledgments
We thank the Transgenic, Microinjection, and Knockout core facility, which was supported in part by National Institutes of Health (NIH) grant P30AR48335, for blastocyst injections of ES cells. We thank the Genome Technology Access Center at Washington University in St. Louis School of Medicine for microarray experiments. The Center is partially supported by National Cancer Institute Cancer Center support grant P30CA91842 to the Siteman Cancer Center and by the Institute for Clinical and Translational Science/Clinical and Translational Science Award grant UL1RR024992 from the National Center for Research Resources (NCRR). We thank G. London for assistance with the NanoZoomer. The NanoZoomer instrument and core facility was supported by the Hope Center Alafi Neuroimaging Lab and Neuroscience Blueprint Interdisciplinary Center Core award to Washington University in St. Louis (P30 NS057105).
Y. Wang was supported by a predoctoral fellowship from the Cancer Biology Pathway program and the Siteman Cancer Center. D. Bhattacharya is a New York Stem Cell Foundation–Robertson Investigator. Funding for this project was provided by the Children’s Discovery Institute of Washington University in St. Louis and St. Louis Children’s Hospital (to D. Bhattacharya). This research was also supported by the New York Stem Cell Foundation, NIH grant R01AI099108, and a Research Scholar grant from the American Cancer Society (125091-RSG-13-252-01-LIB; to D. Bhattacharya). This publication is solely the responsibility of the authors and does not necessarily represent the official view of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, NCRR, or NIH.
The authors declare no competing financial interests.
Author contributions: Y. Wang and D. Bhattacharya performed all experiments, analyzed all data, and wrote the paper.
Footnotes
Abbreviations used:
- ASC
- antibody-secreting cell
- BTB-POZ
- Broad complex, tramtrack, bric-a-brac-poxvirus, and zinc finger
- CGG
- chicken gamma globulin
- NP
- 4-hydroxy-3-nitrophenyl-acetyl
- qRT-PCR
- quantitative RT-PCR
- WNV
- West Nile virus
References
- Ahuja A., Anderson S.M., Khalil A., Shlomchik M.J. 2008. Maintenance of the plasma cell pool is independent of memory B cells. Proc. Natl. Acad. Sci. USA. 105:4802–4807 10.1073/pnas.0800555105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C.D., Okada T., Tang H.L., Cyster J.G. 2007. Imaging of germinal center selection events during affinity maturation. Science. 315:528–531 10.1126/science.1136736 [DOI] [PubMed] [Google Scholar]
- Amanna I.J., Slifka M.K. 2010. Mechanisms that determine plasma cell lifespan and the duration of humoral immunity. Immunol. Rev. 236:125–138 10.1111/j.1600-065X.2010.00912.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanna I.J., Carlson N.E., Slifka M.K. 2007. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357:1903–1915 10.1056/NEJMoa066092 [DOI] [PubMed] [Google Scholar]
- Ansel K.M., McHeyzer-Williams L.J., Ngo V.N., McHeyzer-Williams M.G., Cyster J.G. 1999. In vivo–activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 190:1123–1134 10.1084/jem.190.8.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnoue E., Pihlgren M., McGaha T.L., Tougne C., Rochat A.F., Bossen C., Schneider P., Huard B., Lambert P.H., Siegrist C.A. 2008. APRIL is critical for plasmablast survival in the bone marrow and poorly expressed by early-life bone marrow stromal cells. Blood. 111:2755–2764 10.1182/blood-2007-09-110858 [DOI] [PubMed] [Google Scholar]
- Benner R., Hijmans W., Haaijman J.J. 1981. The bone marrow: the major source of serum immunoglobulins, but still a neglected site of antibody formation. Clin. Exp. Immunol. 46:1–8 [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Cheah M.T., Franco C.B., Hosen N., Pin C.L., Sha W.C., Weissman I.L. 2007. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179:6808–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortnick A., Allman D. 2013. What is and what should always have been: long-lived plasma cells induced by T cell-independent antigens. J. Immunol. 190:5913–5918 10.4049/jimmunol.1300161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortnick A., Chernova I., Quinn W.J., III, Mugnier M., Cancro M.P., Allman D. 2012. Long-lived bone marrow plasma cells are induced early in response to T cell-independent or T cell-dependent antigens. J. Immunol. 188:5389–5396 10.4049/jimmunol.1102808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish D.M., Sakamoto K., Kim M.J., Kroos M., Mukhopadhyay S., Leifer C.A., Tryggvason K., Gordon S., Russell D.G. 2009. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog. 5:e1000474 10.1371/journal.ppat.1000474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman E.P., Campbell J.J., Druey K.M., Scheschonka A., Kehrl J.H., Butcher E.C. 1998. Regulation of chemotactic and proadhesive responses to chemoattractant receptors by RGS (regulator of G-protein signaling) family members. J. Biol. Chem. 273:28040–28048 10.1074/jbc.273.43.28040 [DOI] [PubMed] [Google Scholar]
- Breitfeld D., Ohl L., Kremmer E., Ellwart J., Sallusto F., Lipp M., Förster R. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192:1545–1552 10.1084/jem.192.11.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambridge G., Leandro M.J., Edwards J.C., Ehrenstein M.R., Salden M., Bodman-Smith M., Webster A.D. 2003. Serologic changes following B lymphocyte depletion therapy for rheumatoid arthritis. Arthritis Rheum. 48:2146–2154 10.1002/art.11181 [DOI] [PubMed] [Google Scholar]
- Certo M., Del Gaizo Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S.A., Letai A. 2006. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 9:351–365 10.1016/j.ccr.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Chen L., Willis S.N., Wei A., Smith B.J., Fletcher J.I., Hinds M.G., Colman P.M., Day C.L., Adams J.M., Huang D.C. 2005. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 17:393–403 10.1016/j.molcel.2004.12.030 [DOI] [PubMed] [Google Scholar]
- Chen Z., Koralov S.B., Gendelman M., Carroll M.C., Kelsoe G. 2000. Humoral immune responses in Cr2−/− mice: enhanced affinity maturation but impaired antibody persistence. J. Immunol. 164:4522–4532 [DOI] [PubMed] [Google Scholar]
- Cortés M., Georgopoulos K. 2004. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J. Exp. Med. 199:209–219 10.1084/jem.20031571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S., Kersh E.N., Cannons J., Schwartzberg P.L., Ahmed R. 2003. SAP is required for generating long-term humoral immunity. Nature. 421:282–287 10.1038/nature01318 [DOI] [PubMed] [Google Scholar]
- Cumano A., Rajewsky K. 1986. Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP. EMBO J. 5:2459–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Samuel M.A., Suthar M.S., Gale M., Jr, Diamond M.S. 2008. Toll-like receptor 3 has a protective role against West Nile virus infection. J. Virol. 82:10349–10358 10.1128/JVI.00935-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Hamaguchi Y., Ueda Y., Yang K., Uchida J., Haas K.M., Kelsoe G., Tedder T.F. 2008. Maintenance of long-lived plasma cells and serological memory despite mature and memory B cell depletion during CD20 immunotherapy in mice. J. Immunol. 180:361–371 [DOI] [PubMed] [Google Scholar]
- Dogan I., Bertocci B., Vilmont V., Delbos F., Mégret J., Storck S., Reynaud C.A., Weill J.C. 2009. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 10:1292–1299 10.1038/ni.1814 [DOI] [PubMed] [Google Scholar]
- Eisenbarth S.C., Colegio O.R., O’Connor W., Sutterwala F.S., Flavell R.A. 2008. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 453:1122–1126 10.1038/nature06939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagraeus A. 1948. The plasma cellular reaction and its relation to the formation of antibodies in vitro. J. Immunol. 58:1–13 [PubMed] [Google Scholar]
- Foote J.B., Mahmoud T.I., Vale A.M., Kearney J.F. 2012. Long-term maintenance of polysaccharide-specific antibodies by IgM-secreting cells. J. Immunol. 188:57–67 10.4049/jimmunol.1100783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Núñez G. 2008. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1β secretion but dispensable for adjuvant activity. Eur. J. Immunol. 38:2085–2089 10.1002/eji.200838549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Akasako-Furukawa A., Shirai H., Nakamura H., Azuma T. 1999. Junctional amino acids determine the maturation pathway of an antibody. Immunity. 11:329–338 10.1016/S1074-7613(00)80108-9 [DOI] [PubMed] [Google Scholar]
- Gavin A.L., Hoebe K., Duong B., Ota T., Martin C., Beutler B., Nemazee D. 2006. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 314:1936–1938 10.1126/science.1135299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Hathcock K., Zheng B., Kepler T.B., Hodes R., Kelsoe G. 1995. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J. Immunol. 155:556–567 [PubMed] [Google Scholar]
- Hargreaves D.C., Hyman P.L., Lu T.T., Ngo V.N., Bidgol A., Suzuki G., Zou Y.R., Littman D.R., Cyster J.G. 2001. A coordinated change in chemokine responsiveness guides plasma cell movements. J. Exp. Med. 194:45–56 10.1084/jem.194.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A.E., Debes G.F., Arce S., Cassese G., Hamann A., Radbruch A., Manz R.A. 2002. Chemotactic responsiveness toward ligands for CXCR3 and CXCR4 is regulated on plasma blasts during the time course of a memory immune response. J. Immunol. 169:1277–1282 [DOI] [PubMed] [Google Scholar]
- Hu C.C., Dougan S.K., McGehee A.M., Love J.C., Ploegh H.L. 2009. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 28:1624–1636 10.1038/emboj.2009.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.Y., Park C., Harrison K., Kehrl J.H. 2009. TLR4 signaling augments B lymphocyte migration and overcomes the restriction that limits access to germinal center dark zones. J. Exp. Med. 206:2641–2657 10.1084/jem.20091982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Kassir R., Kelsoe G. 1991. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J. Exp. Med. 173:1165–1175 10.1084/jem.173.5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M.B., Mills D.M., Kappler J., Marrack P., Cambier J.C. 2004. Promotion of B cell immune responses via an alum-induced myeloid cell population. Science. 304:1808–1810 10.1126/science.1089926 [DOI] [PubMed] [Google Scholar]
- Kabashima K., Haynes N.M., Xu Y., Nutt S.L., Allende M.L., Proia R.L., Cyster J.G. 2006. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. J. Exp. Med. 203:2683–2690 10.1084/jem.20061289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane C.M., Jung E., Pearce E.J. 2008. Schistosoma mansoni egg antigen-mediated modulation of Toll-like receptor (TLR)-induced activation occurs independently of TLR2, TLR4, and MyD88. Infect. Immun. 76:5754–5759 10.1128/IAI.00497-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi S.P., Skountzou I., Albrecht R.A., Koutsonanos D., Hua T., Nakaya H.I., Ravindran R., Stewart S., Alam M., Kwissa M., et al. 2011. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 470:543–547 10.1038/nature09737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Rafiuddin-Shah M., Tu H.C., Jeffers J.R., Zambetti G.P., Hsieh J.J., Cheng E.H. 2006. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8:1348–1358 10.1038/ncb1499 [DOI] [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Kool M., Pétrilli V., De Smedt T., Rolaz A., Hammad H., van Nimwegen M., Bergen I.M., Castillo R., Lambrecht B.N., Tschopp J. 2008a. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J. Immunol. 181:3755–3759 [DOI] [PubMed] [Google Scholar]
- Kool M., Soullié T., van Nimwegen M., Willart M.A., Muskens F., Jung S., Hoogsteden H.C., Hammad H., Lambrecht B.N. 2008b. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J. Exp. Med. 205:869–882 10.1084/jem.20071087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T., Bouchier-Hayes L., Chipuk J.E., Bonzon C., Sullivan B.A., Green D.R., Newmeyer D.D. 2005. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 17:525–535 10.1016/j.molcel.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Le Gouill S., Podar K., Amiot M., Hideshima T., Chauhan D., Ishitsuka K., Kumar S., Raje N., Richardson P.G., Harousseau J.L., Anderson K.C. 2004. VEGF induces Mcl-1 up-regulation and protects multiple myeloma cells against apoptosis. Blood. 104:2886–2892 10.1182/blood-2004-05-1760 [DOI] [PubMed] [Google Scholar]
- Li H., Willingham S.B., Ting J.P., Re F. 2008. Cutting edge: inflammasome activation by alum and alum’s adjuvant effect are mediated by NLRP3. J. Immunol. 181:17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhalter J.W., Neychev H.O., Zlabinger G.J., Ahmad R., Eibl M.M. 1985. Modulation of the human immune response by the non-toxic and non-pyrogenic adjuvant aluminium hydroxide: effect on antigen uptake and antigen presentation. Clin. Exp. Immunol. 61:143–151 [PMC free article] [PubMed] [Google Scholar]
- Manz R.A., Thiel A., Radbruch A. 1997. Lifetime of plasma cells in the bone marrow. Nature. 388:133–134 10.1038/40540 [DOI] [PubMed] [Google Scholar]
- Manz R.A., Löhning M., Cassese G., Thiel A., Radbruch A. 1998. Survival of long-lived plasma cells is independent of antigen. Int. Immunol. 10:1703–1711 10.1093/intimm/10.11.1703 [DOI] [PubMed] [Google Scholar]
- Marrack P., McKee A.S., Munks M.W. 2009. Towards an understanding of the adjuvant action of aluminium. Nat. Rev. Immunol. 9:287–293 10.1038/nri2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Michalek S.M., Katz J. 2003. Role of innate immune factors in the adjuvant activity of monophosphoryl lipid A. Infect. Immun. 71:2498–2507 10.1128/IAI.71.5.2498-2507.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams L.J., McHeyzer-Williams M.G. 2004. Analysis of antigen-specific B-cell memory directly ex vivo. Methods Mol. Biol. 271:173–188 [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams M.G., Nossal G.J., Lalor P.A. 1991. Molecular characterization of single memory B cells. Nature. 350:502–505 10.1038/350502a0 [DOI] [PubMed] [Google Scholar]
- McKee A.S., Munks M.W., MacLeod M.K., Fleenor C.J., Van Rooijen N., Kappler J.W., Marrack P. 2009. Alum induces innate immune responses through macrophage and mast cell sensors, but these sensors are not required for alum to act as an adjuvant for specific immunity. J. Immunol. 183:4403–4414 10.4049/jimmunol.0900164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick A., Carlile G., Ahmad K.F., Kiang C.L., Corcoran C., Bardwell V., Prive G.G., Licht J.D. 2002. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22:1804–1818 10.1128/MCB.22.6.1804-1818.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Bahlburg A., Khim S., Rawlings D.J. 2007. B cell intrinsic TLR signals amplify but are not required for humoral immunity. J. Exp. Med. 204:3095–3101 10.1084/jem.20071250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misegades L.K., Winter K., Harriman K., Talarico J., Messonnier N.E., Clark T.A., Martin S.W. 2012. Association of childhood pertussis with receipt of 5 doses of pertussis vaccine by time since last vaccine dose, California, 2010. JAMA. 308:2126–2132 10.1001/jama.2012.14939 [DOI] [PubMed] [Google Scholar]
- Mitchelmore C., Kjaerulff K.M., Pedersen H.C., Nielsen J.V., Rasmussen T.E., Fisker M.F., Finsen B., Pedersen K.M., Jensen N.A. 2002. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J. Biol. Chem. 277:7598–7609 10.1074/jbc.M110023200 [DOI] [PubMed] [Google Scholar]
- Moratz C., Kang V.H., Druey K.M., Shi C.S., Scheschonka A., Murphy P.M., Kozasa T., Kehrl J.H. 2000. Regulator of G protein signaling 1 (RGS1) markedly impairs Giα signaling responses of B lymphocytes. J. Immunol. 164:1829–1838 [DOI] [PubMed] [Google Scholar]
- Moratz C., Hayman J.R., Gu H., Kehrl J.H. 2004. Abnormal B-cell responses to chemokines, disturbed plasma cell localization, and distorted immune tissue architecture in Rgs1−/− mice. Mol. Cell. Biol. 24:5767–5775 10.1128/MCB.24.13.5767-5775.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreaux J., Legouffe E., Jourdan E., Quittet P., Rème T., Lugagne C., Moine P., Rossi J.F., Klein B., Tarte K. 2004. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 103:3148–3157 10.1182/blood-2003-06-1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya H.I., Wrammert J., Lee E.K., Racioppi L., Marie-Kunze S., Haining W.N., Means A.R., Kasturi S.P., Khan N., Li G.M., et al. 2011. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 12:786–795 10.1038/ni.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.V., Blom J.B., Noraberg J., Jensen N.A. 2010. Zbtb20-induced CA1 pyramidal neuron development and area enlargement in the cerebral midline cortex of mice. Cereb. Cortex. 20:1904–1914 10.1093/cercor/bhp261 [DOI] [PubMed] [Google Scholar]
- Nolan G.P., Fiering S., Nicolas J.F., Herzenberg L.A. 1988. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc. Natl. Acad. Sci. USA. 85:2603–2607 10.1073/pnas.85.8.2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor B.P., Raman V.S., Erickson L.D., Cook W.J., Weaver L.K., Ahonen C., Lin L.L., Mantchev G.T., Bram R.J., Noelle R.J. 2004. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 199:91–98 10.1084/jem.20031330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olotu A., Fegan G., Wambua J., Nyangweso G., Awuondo K.O., Leach A., Lievens M., Leboulleux D., Njuguna P., Peshu N., et al. 2013. Four-year efficacy of RTS,S/AS01E and its interaction with malaria exposure. N. Engl. J. Med. 368:1111–1120 10.1056/NEJMoa1207564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O., Peters T., Czeloth N., Bernhardt G., Scharffetter-Kochanek K., Förster R. 2005. Cutting edge: egress of newly generated plasma cells from peripheral lymph nodes depends on beta 2 integrin. J. Immunol. 174:7492–7495 [DOI] [PubMed] [Google Scholar]
- Pape K.A., Taylor J.J., Maul R.W., Gearhart P.J., Jenkins M.K. 2011. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 331:1203–1207 10.1126/science.1201730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengo N., Scolari M., Oliva L., Milan E., Mainoldi F., Raimondi A., Fagioli C., Merlini A., Mariani E., Pasqualetto E., et al. 2013. Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 14:298–305 10.1038/ni.2524 [DOI] [PubMed] [Google Scholar]
- Peperzak V., Vikström I., Walker J., Glaser S.P., LePage M., Coquery C.M., Erickson L.D., Fairfax K., Mackay F., Strasser A., et al. 2013. Mcl-1 is essential for the survival of plasma cells. Nat. Immunol. 14:290–297 10.1038/ni.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Paus D., Chan T.D., Turner M.L., Nutt S.L., Basten A., Brink R. 2006. High affinity germinal center B cells are actively selected into the plasma cell compartment. J. Exp. Med. 203:2419–2424 10.1084/jem.20061254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J.Z., Ziffra J., Stennett L., Bodner B., Bonish B.K., Chaturvedi V., Bennett F., Pollock P.M., Trent J.M., Hendrix M.J., et al. 2005. Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res. 65:6282–6293 10.1158/0008-5472.CAN-05-0676 [DOI] [PubMed] [Google Scholar]
- Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10:116–125 10.1038/ni.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine R., McLaughlin M., Jones D.D., Wittmer S.T., MacNamara K.C., Woodland D.L., Winslow G.M. 2011. IgM production by bone marrow plasmablasts contributes to long-term protection against intracellular bacterial infection. J. Immunol. 186:1011–1021 10.4049/jimmunol.1002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., Glimcher L.H. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 412:300–307 10.1038/35085509 [DOI] [PubMed] [Google Scholar]
- Richard K., Pierce S.K., Song W. 2008. The agonists of TLR4 and 9 are sufficient to activate memory B cells to differentiate into plasma cells in vitro but not in vivo. J. Immunol. 181:1746–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter M., Gross O., Kays S., Ruland J., Nimmerjahn F., Saijo S., Tschopp J., Layland L.E., Prazeres da Costa C. 2010. Schistosoma mansoni triggers Dectin-2, which activates the Nlrp3 inflammasome and alters adaptive immune responses. Proc. Natl. Acad. Sci. USA. 107:20459–20464 10.1073/pnas.1010337107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal E.H., Tonchev A.B., Stoykova A., Chowdhury K. 2012. Regulation of archicortical arealization by the transcription factor Zbtb20. Hippocampus. 22:2144–2156 10.1002/hipo.22035 [DOI] [PubMed] [Google Scholar]
- Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K.J., Debatin K.M., Krammer P.H., Peter M.E. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675–1687 10.1093/emboj/17.6.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P., Willimann K., Lang A.B., Lipp M., Loetscher P., Moser B. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192:1553–1562 10.1084/jem.192.11.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., Singh H. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., Calame K. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.I., Savitsky D., Liao J., Calame K. 2005. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J. Exp. Med. 202:1471–1476 10.1084/jem.20051611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G.X., Harrison K., Wilson G.L., Moratz C., Kehrl J.H. 2002. RGS13 regulates germinal center B lymphocytes responsiveness to CXC chemokine ligand (CXCL)12 and CXCL13. J. Immunol. 169:2507–2515 [DOI] [PubMed] [Google Scholar]
- Slifka M.K., Matloubian M., Ahmed R. 1995. Bone marrow is a major site of long-term antibody production after acute viral infection. J. Virol. 69:1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka M.K., Antia R., Whitmire J.K., Ahmed R. 1998. Humoral immunity due to long-lived plasma cells. Immunity. 8:363–372 10.1016/S1074-7613(00)80541-5 [DOI] [PubMed] [Google Scholar]
- Smith K.G., Weiss U., Rajewsky K., Nossal G.J., Tarlinton D.M. 1994. Bcl-2 increases memory B cell recruitment but does not perturb selection in germinal centers. Immunity. 1:803–813 10.1016/S1074-7613(94)80022-7 [DOI] [PubMed] [Google Scholar]
- Smith K.G., Light A., Nossal G.J., Tarlinton D.M. 1997. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 16:2996–3006 10.1093/emboj/16.11.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Cory S. 1991. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 67:889–899 10.1016/0092-8674(91)90362-3 [DOI] [PubMed] [Google Scholar]
- Sutherland A.P., Zhang H., Zhang Y., Michaud M., Xie Z., Patti M.E., Grusby M.J., Zhang W.J. 2009. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol. Cell. Biol. 29:2804–2815 10.1128/MCB.01667-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze D.M., Toellner K.M., García de Vinuesa C., Taylor D.R., MacLennan I.C. 2000. Intrinsic constraint on plasmablast growth and extrinsic limits of plasma cell survival. J. Exp. Med. 192:813–822 10.1084/jem.192.6.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillardet M., Haffar G., Mondière P., Asensio M.J., Gheit H., Burdin N., Defrance T., Genestier L. 2009. The thymus-independent immunity conferred by a pneumococcal polysaccharide is mediated by long-lived plasma cells. Blood. 114:4432–4440 10.1182/blood-2009-01-200014 [DOI] [PubMed] [Google Scholar]
- Tarlinton D. 2006. B-cell memory: are subsets necessary? Nat. Rev. Immunol. 6:785–790 10.1038/nri1938 [DOI] [PubMed] [Google Scholar]
- van Spriel A.B., de Keijzer S., van der Schaaf A., Gartlan K.H., Sofi M., Light A., Linssen P.C., Boezeman J.B., Zuidscherwoude M., Reinieren-Beeren I., et al. 2012. The tetraspanin CD37 orchestrates the α4β1 integrin–Akt signaling axis and supports long-lived plasma cell survival. Sci. Signal. 5:ra82 10.1126/scisignal.2003113 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., Nussenzweig M.C. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P., Rajewsky K. 1988. The half-lives of serum immunoglobulins in adult mice. Eur. J. Immunol. 18:313–316 10.1002/eji.1830180221 [DOI] [PubMed] [Google Scholar]
- Wang N.S., McHeyzer-Williams L.J., Okitsu S.L., Burris T.P., Reiner S.L., McHeyzer-Williams M.G. 2012. Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nat. Immunol. 13:604–611 10.1038/ni.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Town T., Alexopoulou L., Anderson J.F., Fikrig E., Flavell R.A. 2004. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 10:1366–1373 10.1038/nm1140 [DOI] [PubMed] [Google Scholar]
- Wensveen F.M., Derks I.A., van Gisbergen K.P., de Bruin A.M., Meijers J.C., Yigittop H., Nolte M.A., Eldering E., van Lier R.A. 2012. BH3-only protein Noxa regulates apoptosis in activated B cells and controls high-affinity antibody formation. Blood. 119:1440–1449 10.1182/blood-2011-09-378877 [DOI] [PubMed] [Google Scholar]
- Willis S.N., Chen L., Dewson G., Wei A., Naik E., Fletcher J.I., Adams J.M., Huang D.C. 2005. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19:1294–1305 10.1101/gad.1304105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Winkelmann E.R., Gorder S.R., Mason P.W., Milligan G.N. 2013. TLR3- and MyD88-dependent signaling differentially influences the development of West Nile virus-specific B cell responses in mice following immunization with RepliVAX WN, a single-cycle flavivirus vaccine candidate. J. Virol. 87:12090–12101 10.1128/JVI.01469-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Zhang H., Tsai W., Zhang Y., Du Y., Zhong J., Szpirer C., Zhu M., Cao X., Barton M.C., et al. 2008. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc. Natl. Acad. Sci. USA. 105:10859–10864 10.1073/pnas.0800647105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Ma X., Ji W., Zhou G., Lu Y., Xiang Z., Wang Y.X., Zhang L., Hu Y., Ding Y.Q., Zhang W.J. 2010. Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Proc. Natl. Acad. Sci. USA. 107:6510–6515 10.1073/pnas.0912315107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.S., Scharer C.D., Majumder P., Davis C.W., Butler R., Zinzow-Kramer W., Skountzou I., Koutsonanos D.G., Ahmed R., Boss J.M. 2012. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J. Immunol. 189:2393–2403 10.4049/jimmunol.1103371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xie Z., Zhou L., Li L., Zhang H., Zhou G., Ma X., Herrera P.L., Liu Z., Grusby M.J., Zhang W.J. 2012. The zinc finger protein ZBTB20 regulates transcription of fructose-1,6-bisphosphatase 1 and β cell function in mice. Gastroenterology. 142:1571–1580: e6 10.1053/j.gastro.2012.02.043 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.