Abstract
Primary IGH translocations involving seven recurrent partner loci and oncogenes are present in about 40% of multiple myeloma tumors. Secondary IGH rearrangements, which occur in a smaller fraction of tumors, usually are complex structures, including insertions or translocations that can involve three chromosomes, and often with involvement of MYC. The main approach to detect IGH rearrangements is interphase – but sometimes metaphase – FISH strategies that use a telomeric variable region probe and a centromeric constant region/Eα enhancer or 3′ flanking probe to detect a separation of these two probes, or a fusion of these probes with probes located at nonrandom partner sites in the genome. We analyzed 18 myeloma cell lines for detection discrepancies among Vysis, Cytocell, and in-house IGH probe sets that hybridize with differing sequences in the IGH locus. There were no detection discrepancies for the three telomeric IGH probes, or for unrearranged IGH loci or primary IGH translocations using the centromeric IGH probes. However, the majority of complex IGH rearrangements had detection discrepancies among the three centromeric IGH probes.
INTRODUCTION
Multiple myeloma (MM), a monoclonal tumor of post-germinal center B cells, is the second most common hematological malignancy (Siegel et al., 2013). It is generated sporadically at an average rate of 1% per year from premalignant monoclonal gammopathy of undetermined significance (MGUS) (Kyle et al., 2010). In about 40% of MGUS and MM, it is thought that a chromosome rearrangement involving the IGH locus at 14q32 is an initiating event (Fonseca et al., 2004; Kuehl and Bergsagel, 2012). These primary IGH translocations usually are reciprocal translocations that involve seven recurrent partner loci/oncogenes: CCND1, 11q13; CCND2, 12p13; CCND3, 6p21; MAF, 16q23; MAFB, 20q12; MAFA, 8q24.3; and MMSET/FGFR3, 4p16. In most cases, it appears that one of the 3′ IGH enhancers (Eα1 or Eα2) on der(14) dysregulates an oncogene that can be located up to 1 Mb from the enhancer, but less often the Eμ enhancer can dysregulate an oncogene, e.g., MMSET on der(4)t(4;14) (Bergsagel and Kuehl, 2001; Gabrea et al., 2006). Secondary IGH rearrangements, usually associated with dysregulation of an oncogene mediated by the Eα1 and/or Eα2 enhancers, often have a number of characteristics that distinguish them from primary translocations: 1) they rarely involve one of the seven primary partner chromosomal loci but often involve MYC (8q24.1); 2) they usually are complex non-reciprocal rearrangements, including insertions or translocations that can involve three chromosomes, and sometimes with associated duplications or inversions; 3) the IGH breakpoints rarely are associated with disruption of sequences that are subject to B-cell specific DNA modification processes (Gabrea et al., 2006).
There are two main approaches that have been used to detect IGH rearrangements by FISH of metaphase chromosomes or interphase nuclei: break apart FISH, i.e., separation of telomeric and centromeric probes within or flanking the IGH locus; or fusion FISH, i.e., one or both of these same probes separately juxtaposed with probes from a partner chromosome locus (Ahmann et al., 1998; Avet-Loiseau et al., 2001; Avet-Loiseau et al., 2002; Smadja et al., 2003; Fonseca et al., 2004; Ross et al., 2005; Chiecchio et al., 2006; Lopez-Corral et al., 2011). Two IGH break-apart probes are available commercially, and several labs have developed their own probes for research purposes. Importantly, these various probes are located at different positions within or flanking the IGH locus. The goal of this study was to determine the efficiency with which different probe sets can detect primary and secondary IGH rearrangements in MM.
MATERIALS AND METHODS
Multiple Myeloma Cell Lines (MMCL)
Eighteen independent MMCL were used for this study. This panel of MMCL was selected based on previous findings of IGH rearrangements (Gabrea et al., 2008) or unpublished results. Two MMCL had no IGH rearrangements (L363 and XG-6), two MMCL had primary IGH translocations (H929 and KHM-11), and 14 MMCL had secondary and/or complex IGH rearrangements – and in some cases primary IGH translocations (DP6-DJ, U266, JK-6L, KP-6, MM1-144, VP-6, JJN3, KMS11, LP-1, AMO-1, EJM, OPM-1, MOLP-8, and KMS12-BM).
Plasma cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and Pen Strep (Gibco). For cell lines DP6-DJ and VP-6 the medium was supplemented with IL-6 (Peprotech, Rocky Hill, NJ; final concentration 2 ng/ml). Metaphase spreads were prepared according to standard cytogenetic procedures, with the use of a temperature and humidity controlled chamber (Thermotron, Venturedyne Ltd., Pewaukee, WI) set at 21° C and 40–50% humidity.
Probe Preparation and Hybridization
The directly labeled Vysis LSI IGH (Abbott Molecular, Abbott Park, IL) and Cytocell IGH Break Apart (Rainbow Scientific Inc., Windsor, CT) commercial probes were used according to the manufacturer’s protocol. In-house probes were labeled by nick translation with either biotin-16-dUTP or digoxigenin-11-dUTP (Gabrea et al., 2008). Labeled probes (250 ng of each) were co-precipitated with 50-fold excess of Cot-1 DNA (Invitrogen) and resuspended in 10 μl of hybridization solution (50% formamide plus 10% dextran sulfate in 1x SSC). After denaturation (80°C for 10 min) and preannealing (37°C for 60 min), the hybridization mixture was placed on slides and incubated at 37°C overnight.
FISH
FISH was carried out after the slides were aged at ambient temperature for 3–5 days. When necessary, slides were pretreated with pepsin (Sigma, St. Louis, MO), 20 μl for 3 min at 37° C followed by post-fixation in formaldehyde. The slides were denatured in 70% formamide/2x SSC at 80° C for 2 min and air dried before application of the probe mixture. For hybridization, slides were placed into a humidified chamber and incubated overnight at 37°C. Slides hybridized with in-house probes were washed, incubated with avidin-FITC/rhodamine-antidigoxigenin dual detection reagent (Vector Labs, Burlingame, CA). All slides hybridized with commercial probes were also counterstained with DAPI Slow Fade Gold antifade reagent (Invitrogen, Eugene, OR).
Microscopic analysis was performed using a Leica DMXRA fluorescence microscope equipped with filters for DAPI, FITC, TRITC, and a custom-made triple band pass filter for DAPI, FITC, and TRITC (Chroma Technology, Bellows Falls, VT). Image acquisition was accomplished using a cooled charge coupled device (CCD) camera (Sensys, Photometrics, Tucson, AZ) attached to the microscope and LEICA QFISH software. At least 5, but usually 10–20 metaphases, were analyzed for each probe set in each cell line. For each metaphase evaluated, the number of paired and discrete CH and VH signals and the total number of CH and VH signals in the metaphase were enumerated. The predominant IGH signal configuration was determined for each cell line and each IGH probe hybridization and described as follows, xF yCH zVH, where x is the number of fusion/paired (F) signals, y is the number of discrete CH signals, and z is the number of discrete VH signals.
Additional SKY and FISH data available for derivative chromosomes were used for analysis (Gabrea et al., 2008; Shou et al., 2000 and unpublished).
Mate Pair Libraries
The Illumina Mate Pair Library Preparation kit was used for library construction following the manufacturer’s instructions and two samples were run on one lane of an Illumina HiSeq2000 with 50 bp reads. The sequences were aligned to hg19 using BWA and a BAM file containing only the clustered discordant reads. Breakpoints in the MYC locus with more than five discordant reads were identified in this small BAM file by visual inspection using IGV program. Identical breakpoints that were recurrent in 22 MMCL and 17 non-MM tumor cell lines were deemed to represent germline structural variations and were excluded from analysis.
RESULTS
Hybridization Using Three Different IGH Probe Sets
Two commercially available probe sets, Vysis LSI IGH and Cytocell IGH Break Apart probes, and one in-house VH/CH probe set (Gabrea et al., 2008; Shou et al., 2000) were used for FISH analyses. Table 1 and Figure 1 provide the genomic locations of the three IGH probe sets and the three enhancer elements: Eμ, Eα1, and Eα2. The Vysis 3′ Flanking probe does not detect Eα1 and Eα2 enhancer sequences. By contrast, the Cytocell IGH C and in-house CH probes detect both enhancer regions. Constant/enhancer region or 3′ flanking probes are referred to as CH, and variable region or 5′ flanking probes as VH in Figures, Tables, and text. In each cell line, metaphase FISH results obtained with the three different probe sets were assessed for the presence of detection discrepancies among the probe sets. There were no VH discrepancies among the three probe sets, but in some cases a CH signal was detected by the Cytocell and in-house probes but not by the Vysis probes.
Table 1.
Locationsa of FISH Probes and IGH Enhancers on Chromosome 14
| FISH probe or enhancer | Start | End | Size (kb) |
|---|---|---|---|
| CH-BAC | 106,016,640 | 106,069,726 | 53.1 |
| Cytocell IGH C | 105,999,340 | 106,123,465 | 124.1 |
| Vysis LSI 3′ Flanking | 105,665,462 | 105,947,774 | 282.3 |
| VH cosmid | 107,213,664 | 107,247,079 | 33.4 |
| Cytocell IGH V | 106,667,215 | 107,284,022 | 616.8 |
| Vysis LSI IGHV | 106,441,408 | 107,268,415 | 827.0 |
| Eα2 | 106,032,614 | 106,048,676 | 16.1 |
| Eα1 | 106,152,458 | 106,167,601 | 15.1 |
| Eμ | 106,328,218 | 106,328,537 | 0.3 |
chromosome 14 locations from GRCH37/hg19 assembly
Figure 1.
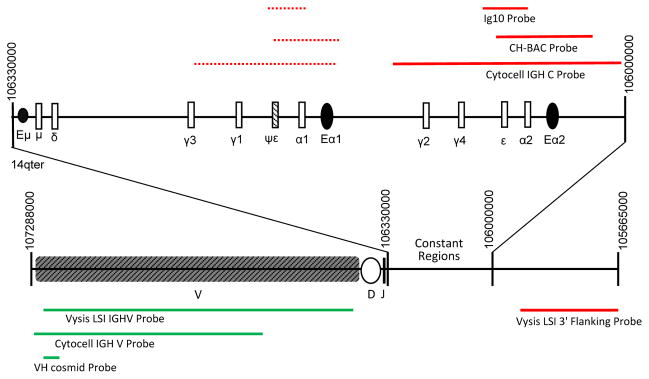
The locations of break apart probes in the germline IGH locus on chromosome 14. Open boxes indicate constant regions, and black ovals indicate IGH enhancer regions. Constant region/3′probes are shown in red and variable region/5′ probes are shown in green. Dashed lines represent areas of cross-hybridization. The approximate position of the Ig10 cosmid, a 3′ probe used in many other studies, but not in this study, is based on its initial description (Flanagan and Rabbits, 1982).
Detection of Normal CH/VH Fusion Signals and Primary Translocations
In MMCL without any IGH rearrangements (L363 and XG-6) or with primary IGH translocations only [t(4;14)(p16;q32) in H929 and KHM-11], no discrepancies between the IGH break-apart probes sets were observed (data not shown). Moreover, in the group of 14 MMCL that have secondary and/or complex IGH rearrangements, fusion signals on unrearranged IGH loci, and split of CH and VH signals for primary IGH translocations were detected with all three probe sets (Table 2). Previously, we showed that U266 has an atypical primary IGH rearrangement generated by insertion of Eα1 enhancer sequences near Cyclin D1 during IGH switch recombination (Gabrea, et al. 1999); this insertion was detected with the in-house CH and Cytocell IGH C probes but not with the Vysis LSI 3′ flanking probe.
Table 2.
Metaphase FISH Analyses of 14 Myeloma Cell Lines with Three Pairs of IGH Probesa
| Cell lineb | Normal fusion signalc | Primary translocations | Secondary or complex IgH rearrangements | |||
|---|---|---|---|---|---|---|
| #CH,VH signalsd | Rearrangements | #CH,VH signalsd | Rearrangements | Rearrangement typee | ||
| DP6-DJ | 2F | 0 | 0 | 1CH (0CH) | ins CH at 8q24 on der(8)(CH+,VH−,CMYC+,wcp14−) | I |
| U266 | 2F | 0 | 0 | 1CH (0CH) | ins CH at 11q13 on der(11)t(4;11)(?;q14)(CH+,VH−,CCND1+) | I |
| VP-6 | 1F | 1CH;1VH | t(14;16)(q32;q23)(CH+,VH−,CMAF+;CH−,VH+) | 2CH (0CH) | ins CH at 8q24 on der(8)(CH+,VH−,CMYC+,wcp14−) x2 | I |
| KMS12-BM | 2F | 1CH;1VH 1VH |
t(11;14)(CH−,VH+;CH+,VH−,CCND1+)f der(?11)t(?11;14)(CH−,VH+) |
2CH 2CH 1CH 1CH |
ins CH at 8q24 on
der(8)(CH+,VH−,CMYC+, wcp14−)
x2 der(?)t(?;8)(CH+,VH−,CMYC+,wcp14−) x2 ins CH on der(?) (CH+,VH−,wcp14−) (long chromosome) ins CH on der(?)(CH+,VH−,wcp14−)(small chromosome) |
I [I] I [I] |
| JJN3 | 0 | 1CH;1VH 1VH |
t(14;16)(q32;q23)(CH+,VH−,CMAF+;CH−,VH+) der(16)t(14;16)(q32;q23)(CH−,VH+) |
2CH
(0CH) 2CH (1CHg) |
ins CH at 8q24 on
der(8)(CH+,VH−,CMYC+,wcp14−)
x2 der(14)t(8;14;16)(CMYC+,CH+,wcp8+;wcp14+,CHg+,MAF+) |
I [I+PT] |
| MM1-144 | 0 | 1CH;1VH | t(14;16)(q32;q23)(CH+,VH−,CMAF+;CH−,VH+) | 1CH (0CH) | ins CH at BP on der(3)t(3;8)(?;q24)(CH+,CMYC+) | CI |
| KMS11 | 0 | 1CH;1VH 1CH;1VH |
t(4;14)(p16;q32)(CH−,VH+,MMSET+;CH+,VH−,FGFR3+) t(14;16)(q32;q23)(CH+,VH−,CMAF+;CH−,VH+) |
2CH (0CH) | ins CH at BPs on der(?;8;8;?)(wcp14−,CH+,CMYC+,wcp8+, CMYC+,CH+,wcp14−) | CI |
| EJM | 1F | 1CH 2VH |
der(14)t(7;14;20)(CH+,VH−,MAFB+) der(20)t(14;20)(q32;q12)(CH−,VH+) x2 |
1CH | ins CH at BP on der(18)t(7;18)(CH+,VH−,wcp14−) | CI |
| MOLP-8 | 2F | 2CH;2VH | t(11;14)(VH+;CH+,VH−,CCND1+)f x2 | 2CH 2CH |
ins CH on
der(4)(wcp14−,CH+,VH−,wcp8−,CMYC+)
x2 ins CH on der(17)t(11;17)(q13;?p13)(wcp14−,CH+,VH−,wcp11+) x2 |
CI CI |
| AMO-1 | 1F | 0 | 0 |
2CH
(0CH) 2VH 1CH 1CH 2VH |
ins CH at BP on der(8)t(8;12)
(CH+,VH−,CMYC+,CCND2+)
x2 der(12)(CH−,VH+) x2 der(14)t(8;14)(q24;q32)(CH+,VH−,CMYC+) der(14)t(8;14)inv(14)(CH+,VH−,CMYC+) der(?)(CH−,VH+)x2 |
CI T [T] |
| OPM-1 | 0 | 2CH;2VH | der(14)t(4;14)(p16;q32)(CH−,VH+;CH+,VH−)f x2 | 2CH | der(8)t(1;14;8)(CH+,VH−,CMYC+) x2 | T |
| LP-1 | 0 | 2CH | der(14)t(4;14)(p16;q32)(CH+,VH−,FGFR3+) x2 |
3F
(0CH3VH) 2CH (0CH) 4F (2F2VH) |
der(8)t(3;8)(q24;p23)(CMYC+,wcp14−,CH+,VH+)
x3 ins CH on der(?)(wcp14−,CH+,VH−) x2 der(1)t(1;8)(wcp14−,CH+,VH+;wcp8+,CMYC+,wcp14−,CH+,VH+) x2 |
VT1A I [VT1A]+VT2 |
| KP-6 | 2F | 0 | 0 | 1F | der(17)(CH+,VH+,wcp14−) | VT2 |
| JK-6L | 1F | 0 | 0 | 1F1CH (1CH1VH) | t(8;14)(q24;q32)(CH+,VH+;CH+,CMYC+) | VT1B |
Cell lines in bold have one or more detection discrepancies among the three probe sets.
Normal fusion signals (F) are defined as paired CH/VH signals seen on a normal chromosome 14 or on a derivative chromosome
#CH, VH signals is the total number of paired (F) and discrete CH or VH signals observed for different IGH rearrangements. Detection discrepancies (only with the Vysis LSI 3′ Flanking probe) are shown in parentheses, with the bold CH signals identified with the in-house and Cytocell probes but not by the Vysis probe.
Type of rearrangement: simple CH insertion (I); complex insertion (CI); non-variant translocation not involving a primary partner (T); primary translocation (PT); variant translocations (VT1a, VT1b, VT2). Brackets indicate rearrangements already accounted for. See text for additional details.
Derivative which contains VH signal underwent further rearrangement.
CH signal originated from primary translocation.
Discrepancies Among Probe Sets in Detection of Secondary or Complex IGH Rearrangements
Nine of the 14 (64%) cell lines with complex or secondary rearrangements show discrepancies for one or more CH signals (Table 2). The total number of independent complex rearrangements was 18, and discrepancies in detection of CH signals were observed for 10 of them (56%), always due to the detection of CH signals by the Cytocell and in-house probes but not the Vysis probe.
Detection of CH Insertions
Discrepancies were found for eight insertions present in eight different cell lines (DP6-DJ, U266, VP-6, JJN3, MM1-144, KMS11, AMO-1, and LP-1), whereas no discrepancies were found for five insertions present in three cell lines (KMS12-BM, EJM, and MOLP-8) (Table 2). Examples of CH insertions that were detected by the Cytocell and in-house probes but not by the Vysis probe are shown in Figure 2A and 2B. Two karyotypic types of CH insertions have been identified in MM cell lines and patient tumor samples: simple and complex insertions (Gabrea et al., 2008). Simple CH insertions are present in an otherwise apparently normal region of another chromosome: usually chromosome 8 near the MYC locus (in cell lines DP6-DJ, VP6, KMS12-BM, and JJN3), but sometimes chromosome 11 near CCND1 (in cell line U266) or other chromosomes (KMS12-BM and LP-1). Complex insertions involve CH loci inserted into or near the breakpoint junction between different chromosomes, e.g., between chromosomes 3 and 8 on der(3)t(3;8) in the MM.1 cell line (Table 2). Discrepant detection of insertions was seen for both kinds of CH insertions: five of seven simple CH insertions and three of six complex insertions were not detected by the Vysis probe (Table 2).
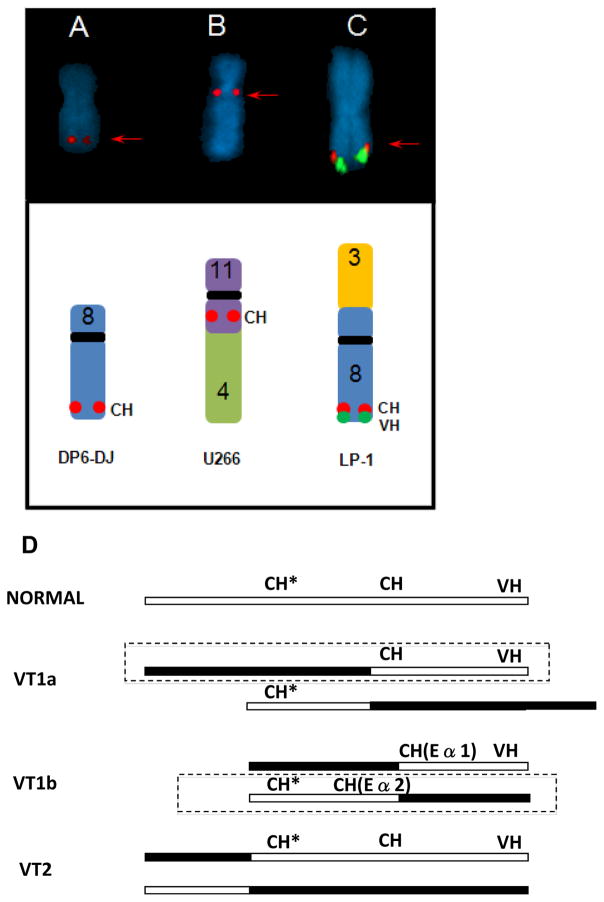
Figure 2.
Examples of complex IGH rearrangements. (A–C) Red arrows indicate signals that are detected by the Cytocell/in-house probes, but not detected with the Vysis probe. (A) der(8) with CH insertion. (B) der(11)t(4;11)(?;q14) with CH inserted at 11q13. (C) der(3)t(3;8) VT1a variant IGH translocation (CH and VH signals near chromosome arm 8q telomere) (D) anatomy of VT1a, VT1b, and VT2 variant IGH translocations where the open box depicts chromosome 14 and closed box another chromosome, CH includes Eα1 or Eα2 enhancer sequences detected by in-house or Cytocell probes, CH* represents sequences detected by 3′ Vysis Probe, and VH represents sequences detected by all 3 VH probes. The boxed derivative chromosome is expected to have a target gene dysregulated by Eα1 or Eα2. Note that the CH probe hybridizes to Eα1 and Eα2 sequences, consistent with the presence of CH signals on both derivative chromosomes in the VT1b variant IGH translocation.
Detection of Secondary IGH Translocations
Six secondary IGH translocations were present in five cell lines (AMO-1, OPM-1, LP-1, KP-6, and JK-6L). Two types of secondary IGH translocations have been defined previously: non-variant and variant (Gabrea et al., 2008). Non-variant secondary IGH translocations have a separation of CH and VH sequences due to breakpoints between them. Two of the six secondary translocations studied were non-variant translocations (indicated by T in Table 2). For both MMCL (AMO-1 and OPM-1), MYC was located near CH at the chr14:chr8 junction. Both non-variant translocations were detected by all three probe sets.
Variant IGH translocations are identified by CH and VH co-localization at the telomere of another chromosome. Depending on the site of the breakpoint relative to the CH locus, we further divided variant IGH translocations into two types. Variant translocations of the first type (VT1) had breakpoints within or centromeric to Eα1 or within or minimally centromeric of Eα2, whereas variant translocations of the second type (VT2) had breakpoints on chromosome 14 substantially centromeric to the Ea2 enhancer (Fig. 2D; Discussion).
A variant translocation (VT1a) in cell line LP-1 was detected as a fusion of MYC, CH, and VH signals with Cytocell and in-house probe sets, and fusion of MYC and VH signals but no CH signal with the Vysis probe set (Table 2, Fig. 2C). A variant translocation (VT1b) in cell line JK-6L was detected by the Cytocell and in-house probe sets as a fusion of MYC and CH signals on der(14) but a fusion of CH and VH signals on der(8); the Vysis probe also detected a MYC/CH fusion signal on der(14) but only a VH signal on der(8) (Table 2). The VT1a and VT1b translocations in LP1 and JK6L, respectively, have different functional implications (see Fig. 2D; Discussion).
Two variant translocations of the second type (VT2) were detected similarly by all three sets of probes as a fusion of CH and VH signals on der(1)t(1;8) in cell line LP-1 and on der(17) in cell line KP-6 (Table 2).
DISCUSSION
In previous studies, we performed metaphase FISH assays with our in-house CH probe (Fig. 1, Table 1) to identify apparent insertions of 3′ IGH enhancers (Eα1 and Eα2) into simple or complex loci in 16 of 47 MMCL and 8 of 48 advanced primary tumors (Gabrea et al., 2008). Virtually all of these insertions were undetectable by chromosome 14 painting probes, indicating that the inserted sequences were <5 Mb. The cloned breakpoints of the IGH insertion near the CCND1 gene in the U266 MMCL indicated that the Vysis LSI 3′ Flanking Probe, which is located more than 250 kb centromeric to the Eα2 enhancer (Fig. 1, Table 2), would not detect this insertion (Gabrea et al., 1999). Since most analyses to detect IGH rearrangements in MM tumors have used the Vysis probes, we decided to do metaphase FISH analyses on a panel of 18 MMCL (Table 2) to compare our in-house probes with the commercial Vysis and Cytocell probe sets (Fig. 1, Table 1). As summarized in the Results and Table 2, we found no detection discrepancies among the three VH probe sets. We also found no discrepancies among the three CH probe sets in detecting VH/CH fusion signals on unrearranged IGH loci or in detecting separate VH and CH signals for primary IGH translocations. Since it has been well documented that primary IGH translocations almost always have breakpoints in the IGH switch regions, or less often telomeric to the Eμ enhancer (Bergsagel and Kuehl 2001; Gabrea et al., 2006; Walker et al., 2013), it is not surprising that all three probe sets would give concordant results.
By contrast, the majority of simple (5 of 7) or complex (3 of 6) insertions involving CH sequences are detected by the in-house CH probe or the Cytocell IGH C probe but not by the Vysis LSI 3′ Flanking probe. Consistent with the FISH results for insertions detected with the in-house CH probe, it is assumed that most insertions include the Eα1 and/or Eα2 enhancer and that it would not be surprising if some insertions did not include sequences detected by the Vysis probe. However, with the exception of the primary IGH insertion in U266, the precise IGH sequence content of insertions had not been previously determined in MM. From mate pair sequencing data we have determined the sequence content of four IGH insertions that are consistent with the FISH results in three MMCL. In JJN3, the inserted IGH sequences (chr14: 106.024–106.069 Mb) include the Eα2 enhancer but not sequences detected by the Vysis probe. Similarly, in AMO-1, an insertion of IGH sequences (chr14:106.115–106.330 Mb) includes the Eα1 enhancer but not sequences detected by the Vysis probe. By contrast, in MOLP-8, both IGH insertions include sequences that are detected by all three CH probes: chr14:105.751–106.174 Mb on der(4) and chr14:105.661–106.304 Mb on der(17).
Ross et al. (2005) considered the possibility that the Vysis probe used in their interphase FISH experiment might have missed small insertions, and therefore compared the Vysis LSI 3′ Flanking Probe with the cosIg10 probe on 56 MM tumors but found no discrepancy for these two probe sets. By contrast, we found that 8 of 48 advanced MM tumors had IGH insertions (Gabrea et al., 2008), so that we would predict that the IGH insertion in 4 or 5 of these advanced MM tumors would not have been detected with the Vysis probe (i.e., assuming that the fraction of IGH insertions missed by the Vysis probe is the same for MMCL and MM tumors). There are several ways to explain the apparent discrepancy between our results for advanced MM tumors and the results obtained by Ross et al. (2005). First, although the precise sequences included in the cosIg10 probe are not known, the original description of this cosmid (Flanagan and Rabbitts, 1982) suggests that it does not include sequences that would hybridize with Eα1 or Eα2 (Fig. 1), and therefore might not detect some insertions. Second, the prevalence of IGH insertions probably increases with the stage of the disease (16 of 47 in MMCL and 8 of 48 in advanced MM tumors which mostly had a high proliferation index and for which substantial quantities of metaphase chromosomes were available for analysis) (Gabrea et al., 2008). The MM samples analyzed by interphase FISH by Ross et al. (2005) mostly represented earlier disease stages, and therefore may have had a low prevalence of IGH insertions.
Secondary translocations were detected by metaphase FISH without discrepancies by all three probe sets in cases of non-variant IGH translocations (T) and variant IGH translocations with a breakpoint centromeric to the sequences detected by the 3′ Vysis probe (VT2). Mate pair sequences for KP6 show that the chromosome 14 breakpoint is at 104.601 Mb, i.e., about 1.5 Mb centromeric to the IGH locus, and for LP1 the chromosome breakpoint is at 103.492 Mb, which is about 2.5 Mb centromeric to the IGH locus.
The two kinds of type 1 variant IGH translocations, both of which have detection discrepancies among the 3 probe sets, represent a more complex situation. The VT1a variant IGH translocation is exemplified by LP-1, which has a fusion of MYC, CH, and VH signals with the in-house and Cytocell probes but only a MYC/VH fusion signal with the Vysis probe, indicating that the chromosome 14 breakpoint is centromeric to the Eα1 and/or Eα2 enhancers but telomeric to sequences detected by the 3′ Vysis flanking probe (Fig. 2C). As a result interphase FISH studies using the break apart strategy would not detect the VT1a translocation with the in-house or Cytocell probe sets, but would detect this translocation as a discrete VH signal using the Vysis probe. Primary IGH translocations rarely, if ever, have the VT1a variant IGH translocation pattern.
The VT1b variant IGH translocation is exemplified by JK-6L which has a t(8;14) translocation that is similar to a non-variant t(8;14) translocation, i.e., a discrete CH signal on der(14) with all three probe sets, except that der(8) has a VH/CH fusion signal with the in-house and Cytocell probes but only a VH signal with the Vysis probe (Table 2 and Fig. 2D). Presumably Eα2 targets MYC on der(14), whereas Eα1 does not target an oncogene on der(8). In this case, interphase FISH analyses using the break apart strategy would detect a discrete CH signal on der(14) with all three probe sets, whereas only the Vysis probe set would detect the discrete VH signal on der(8). The VT1b variant IGH translocation pattern sometimes is found with primary IGH translocations, but occurs in only a small minority of MM tumors.
In summary, there were no detection discrepancies for the three variable region probes, or for unrearranged IGH loci or primary IGH translocations using the three constant region/3′IGH probes. However, the majority of complex IGH rearrangements had detection discrepancies among the three constant region/3′IGH probes, with the Vysis probe frequently not identifying sequences identified by the Cytocell or in-house probes.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Health, National Cancer Institute, Center for Cancer Research
References
- Ahmann GJ, Jalal SM, Juneau AL, Christensen ER, Hanson CA, Dewald GW, Greipp PR. A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet. 1998;101:7–11. doi: 10.1016/s0165-4608(97)00058-7. [DOI] [PubMed] [Google Scholar]
- Avet-Loiseau H, Daviet A, Brigaudeau C, Callet-Bauchu E, Terre C, Lafage-Pochitaloff M, Desangles F, Ramond S, Talmant P, Bataille R. Cytogenetic, interphase, and multicolor fluorescence in situ hybridization analyses in primary plasma cell leukemia: a study of 40 patients at diagnosis, on behalf of the Intergroupe Francophone du Myelome and the Groupe Francais de Cytogenetique Hematologique. Blood. 2001;97:822–825. doi: 10.1182/blood.v97.3.822. [DOI] [PubMed] [Google Scholar]
- Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL, Minvielle S, Bataille R. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99:2185–2191. doi: 10.1182/blood.v99.6.2185. [DOI] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- Chiecchio L, Protheroe RK, Ibrahim AH, Cheung KL, Rudduck C, Dagrada GP, Cabanas ED, Parker T, NIGHtingale M, Wechalekar A, Orchard KH, Harrison CJ, Cross NC, Morgan GJ, Ross FM. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia. 2006;20:1610–1617. doi: 10.1038/sj.leu.2404304. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Rabbitts TH. Arrangement of human immunoglobulin heavy chain constant region genes implies evolutionary duplication of a segment containing gamma, epsilon and alpha genes. Nature. 1982;300:709–713. doi: 10.1038/300709a0. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, Davies FE, Drach J, Greipp PR, Kirsch IR, Kuehl WM, Hernandez JM, Minvielle S, Pilarski LM, Shaughnessy JD, Jr, Stewart AK, Avet-Loiseau H. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- Gabrea A, Bergsagel PL, Chesi M, Shou Y, Kuehl WM. Insertion of excised IGH switch sequences causes overexpression of cyclin D1 in a myeloma tumor cell. Molecular Cell. 1999;3:119–123. doi: 10.1016/s1097-2765(00)80180-x. [DOI] [PubMed] [Google Scholar]
- Gabrea A, Leif Bergsagel P, Michael Kuehl W. Distinguishing primary and secondary translocations in multiple myeloma. DNA Repair (Amst) 2006;5:1225–1233. doi: 10.1016/j.dnarep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Gabrea A, Martelli ML, Qi Y, Roschke A, Barlogie B, JDS, Sawyer JR, Kuehl WM. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes, Chromosomes & Cancer. 2008;47:573–590. doi: 10.1002/gcc.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest. 2012;122:3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle RA, Durie BG, Rajkumar SV, Landgren O, Blade J, Merlini G, Kroger N, Einsele H, Vesole DH, Dimopoulos M, San Miguel J, Avet-Loiseau H, Hajek R, Chen WM, Anderson KC, Ludwig H, Sonneveld P, Pavlovsky S, Palumbo A, Richardson PG, Barlogie B, Greipp P, Vescio R, Turesson I, Westin J, Boccadoro M. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia. 2010;24:1121–1127. doi: 10.1038/leu.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Corral L, Gutierrez NC, Vidriales MB, Mateos MV, Rasillo A, Garcia-Sanz R, Paiva B, San Miguel JF. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res. 2011;17:1692–1700. doi: 10.1158/1078-0432.CCR-10-1066. [DOI] [PubMed] [Google Scholar]
- Ross FM, Ibrahim AH, Vilain-Holmes A, Winfield MO, Chiecchio L, Protheroe RK, Strike P, Gunasekera JL, Jones A, Harrison CJ, Morgan GJ, Cross NC. Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia. 2005;19:1634–1642. doi: 10.1038/sj.leu.2403857. [DOI] [PubMed] [Google Scholar]
- Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Smadja NV, Leroux D, Soulier J, Dumont S, Arnould C, Taviaux S, Taillemite JL, Bastard C. Further cytogenetic characterization of multiple myeloma confirms that 14q32 translocations are a very rare event in hyperdiploid cases. Genes Chromosomes Cancer. 2003;38:234–239. doi: 10.1002/gcc.10275. [DOI] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, Ross FM, Davies FE, Gonzalez D, Morgan GJ. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood. 2013;121:3413–3419. doi: 10.1182/blood-2012-12-471888. [DOI] [PubMed] [Google Scholar]